Abstract
African-born Australians are a recognised “priority population” in Australia's Sixth National HIV/AIDS Strategy. We compared exposure location and route for African-born people living with HIV (PLHIV) in Victoria, Australia, with HIV-1 pol subtype from drug resistance assays and geographical origin suggested by phylogenetic analysis of env gene. Twenty adult HIV positive African-born Victorian residents were recruited via treating doctors. HIV exposure details were obtained from interviews and case notes. Viral RNA was extracted from participant stored plasma or whole blood. The env V3 region was sequenced and compared to globally representative reference HIV-1 sequences in the Los Alamos National Library HIV Database. Twelve participants reported exposure via heterosexual sex and two via iatrogenic blood exposures; four were men having sex with men (MSM); two were exposed via unknown routes. Eight participants reported exposure in their countries of birth, seven in Australia, three in other countries and two in unknown locations. Genotype results (pol) were available for ten participants. HIV env amplification was successful in eighteen cases. HIV-1 subtype was identified in all participants: eight both pol and env; ten env alone and two pol alone. Twelve were subtype C, four subtype B, three subtype A and one subtype CRF02_AG. Reported exposure location was consistent with the phylogenetic clustering of env sequences. African Australians are members of multiple transnational social and sexual networks influencing their exposure to HIV. Phylogenetic analysis may complement traditional surveillance to discern patterns of HIV exposure, providing focus for HIV prevention programs in mobile populations.
Introduction
African-born Australians have been recognised as a “priority population” in Australia's Sixth National HIV/AIDS Strategy, published in 2010 [1]; they are diagnosed late with HIV compared to Australian-born people living with HIV (PLHIV) and are over-represented among new diagnoses of HIV [2]–[4].
National surveillance data indicates that heterosexual sex is the most common exposure route reported among African-born Australians diagnosed with HIV, as it is in populations on the African continent [3], [5], although the proportion of African-born people diagnosed with AIDS in Australia includes a higher proportion of MSM than is reported amongst African migrants in European AIDS surveillance figures [6], [7] and there is growing recognition of the importance of sex between men as an epidemiological factor in low-income countries, including Sub-Saharan Africa [8], [9]. European studies suggest most African-born PLHIV are infected in their countries of origin, prior to migration [10] however Australia is more geographically distant from Africa and has a more regulated migration process that results in the de facto exclusion of most prospective immigrants with HIV [11]. All adults and some children applying for permanent residency are screened for HIV, as are some temporary entrants (those working in health care and students from Sub-Saharan Africa intending to stay for at least one year); those diagnosed with HIV are denied a visa except the minority who are granted a waiver of the health requirement [11], [12]. The experience of migration-related HIV screening has led some African Australians to view HIV as a problem left behind when migrating to Australia: a view that is at odds with the surveillance data described above [13]–[15]. When planning HIV prevention and testing strategies it is therefore important to understand when African-born Australians are exposed to HIV with respect to their initial migration, and whether exposure occurs within Australia or abroad.
Some information about HIV exposure in African Australian communities can be obtained through routine passive surveillance procedures, but passive surveillance data is based on information obtained during routine clinical consultation—the accuracy and reliability of which information may be compromised by numerous patient, clinician and structural factors influencing the clinical interaction, during initial and subsequent visits [16]–[18]. More detailed information about individual and population level factors influencing exposure requires active data collection. Interviews, surveys and field observation provide valuable epidemiological data, but molecular techniques such as phylogenetic analysis may provide complementary information to clarify the location and route of HIV acquisition and its timing with respect to migration.
HIV-1 subtype C accounts for more than 50% of all infections globally [19], and dominates HIV epidemics in Sub-Saharan Africa, India, Papua New Guinea, and parts of South America. Two circulating recombinant forms (CRFs), CRF01_AE and CRF02_AG, are each responsible for 5% of HIV cases. In Sub-Saharan Africa the distribution of HIV subtypes is diverse: subtype A is found in western, eastern and central Africa; subtype C in western and southern Africa; subtype G in the western and central regions; and subtype D in the central and eastern regions. CRF02_AG is concentrated in the western region. Other subtypes such as F, H, J and K are mainly found in central Africa [20], [21]. Subtype B is the subtype most commonly reported from Western Europe, North America and Australia, regardless of reported risk factor, although a recent paper describing non-B subtypes in Australia linked both CRF01_AE and subtype C with MSM transmission and reported subtype C to be the most common subtype among females [22].
The ability to describe the genetic similarities and differences between HIV strains has contributed to our understanding of the epidemiology of HIV, when phylogenetic data have been compared with epidemiological data obtained from history or observation of HIV exposure and transmission events. Sources of demographic and epidemiological data have usually included questionnaires completed by treating physicians [23], [24], surveillance notification data [22], [25]–[27], routine clinical case notes [10], [28] or cohort study data [29], [30]. Less commonly, researchers have conducted direct interviews or administered questionnaires to PLHIV to obtain information about travel and exposure [23], [31], [32]. Full genome sequencing has provided the most detailed phylogenetic discrimination, enabling identification of recombinant strains and discriminating between closely related strains [33]; this has been most valuable in outbreak investigations and forensic settings, where it has been necessary to discriminate between closely-related strains isolated from individuals from whom detailed information was obtained about travel, social contacts, sexual and other risk behaviours [32], [34]. However, many phylogenetic studies have used genetic sequencing of pol gene conveniently obtained from antiretroviral resistance genotyping undertaken in routine clinical practice to detect mutations in reverse transcriptase (RT) or protease (PR); sequences from recently-infected or drug-naive patients have been preferred, to avoid genetic variation arising from drug selection pressure [24], [27]–[30], [35], [36]. Other researchers have used gag or env sequences, for which genetic variation is less subject to drug-related selection pressure in PLHIV already on treatment [10], [25], [26]. Limited sequencing of the highly variable region of env has provided useful epidemiological information in a range of social and geographical settings [10], [23], [25], [26], [28], [32], [37]–[39].
This study aimed to describe the location and route of HIV infection for African-born residents of Victoria and to compare the self-reported history of HIV exposure with the likely geographical origin of the HIV suggested by phylogenetic analysis. We hope that the results of this study will help inform the development and implementation of strategies to detect prevalent HIV infections amongst African immigrants exposed prior to migration and to prevent incident infections amongst those not already exposed, either through exposure within Australia or during subsequent travel abroad.
Materials and Methods
Participant recruitment
A case series of African-born adults living with HIV in Victoria was conducted, in which history of reported exposure to HIV was obtained and examined in the context of phylogenetic analysis of HIV strains isolated from participants. Participants were recruited via treating doctors who informed eligible patients about the study and sought permission to provide contact details to the research team. A member of the research team contacted potential participants to describe the study. Written informed consent was obtained from those agreeing to participate.
Ethics statement
The study was designed and conducted in consultation with several African community-based organisations. The human research ethics committees of Melbourne Health and Alfred Health Ethics approved the study protocol. Consent was obtained separately and specifically for the research team to access stored blood samples for the purpose of genetic sequencing of HIV and/or for the collection for this purpose of fresh blood via venepuncture, if suitable stored blood samples were unavailable. Participants were reimbursed AUD $25.00. All information was de-identified prior to analysis.
Participant interviews and review of case notes
Self-reported location of HIV exposure was obtained both directly from interviews, and indirectly from recorded case notes on exposure histories elicited at HIV diagnosis and during subsequent consultations. If country of exposure was not named either in interview or case notes, the location of exposure was recorded as “unknown”. The route of HIV exposure was classified as: male-to-male sex (with or without a history of injecting drug use) (MSM+/−IDU); heterosexual sex; injecting drug use (IDU), and other/unknown. HIV risk factors of source partners were recorded for those participants with a sexual exposure to HIV. Details of circumstances of exposure were obtained from interviews and from case notes. Although year of migration was not specifically asked (due to prevailing political tensions regarding migration of PLHIV) most participants volunteered this information [40], [41]. If participants had undergone a previous HIV genotypic drug resistance assay through one of the two certified Victorian reference laboratories, the genetic information for the pol gene was obtained from the relevant databases. In addition, for participants that consented to either access to stored plasma or providing a fresh blood samples env subtyping was performed.
Details of HIV-related symptoms and diagnoses were obtained through interview as well as case notes. Results of investigations were obtained from case notes. Late diagnosis was defined as either a CD4 count <350 cells/µL or an AIDS-defining illness at HIV diagnosis; this category included cases of advanced HIV disease at diagnosis, who had a CD4 count <200 cells/µL or an AIDS-defining condition at HIV diagnosis [42]. AIDS-defining conditions were those listed in the surveillance definition of AIDS published by the Centers for Diseases Control and Prevention, other than a CD4 count <200 cells/µL in the absence of opportunistic infection [43].
Laboratory procedures
Stored participant plasma samples were obtained from the Victoria Infectious Diseases Reference Laboratories (VIDRL) and the Burnet Institute (Victorian HIV Blood and Tissue Storage Bank). Fresh whole blood samples were collected in 10 mL EDTA tubes during venepuncture for routine clinical blood tests.
Viral RNA was extracted from plasma using the QIAamp Viral RNA Mini Kit (Qiagen, Germany) and from whole blood using the High Pure Viral Nucleic Acid kit (Roche, United Kingdom) as per manufacturers' instructions. For all extracts, a nested reverse transcription PCR (RT-PCR) was used to amplify the V3 region of the HIV env (corresponding to nucleotides 6959–7374 on HXB2 reference genome, accession number K03455), following the methodology previously used by our team for a phylogenetic study of HIV amongst Vietnamese-Australian people injecting drugs [44], [45]. Briefly, the reverse transcriptase and first 35 amplification cycles was performed using the OneStep RT-PCR kit (Qiagen, Germany) and 0.6 µM of outer primers, EnvF1 and EnvR1, which corresponded to nucleotides 6847–6871 and 7365–7389, respectively. 10 µl of template RNA was added to a final reaction volume of 25 µl before cycling commenced. As per manufacturers instructions, cycling parameters included an initial step of 50°C for 30 minutes to allow for reverse transcription, prior to the DNA polymerase activation step of 95°C for 15 minutes. Thirty-five cycles of 94°C for 30 secs, 40°C for 30 secs and 72°C for 2 minutes and a 10 minute final extension at 72°C completed the amplification.
Nested PCR was performed with 0.5 µM inner primers EnvF2 and EnvR2, corresponding to nucleotides 6959–6983 and 7350–7374 respectively. Platinum Pfx polymerase (Invitrogen) was used according to manufacturers instructions with 2.5 µl of the first round PCR product as template in a final volume of 50 µl. Cycling parameters were as above with the omission of the reverse transcription step. The length of the final product was 415bp. PCR product purification, sequencing and sequence assembly was performed as previously described [25].
Sequence analysis
To determine subtype, consensus sequences were aligned in MEGA version 5 software suite with reference strains of each HIV-1 group M isolate and a phylogenetic tree constructed to determine subtype (data not shown) [46]. For further analysis, the consensus sequence for each isolate was subject to the Basic Local Alignment Search Tool (BLAST) program within the National Centre for BioInformatics (NCBI) software suite to determine the most closely related published isolates. The three most similar sequences were downloaded. Once final sequences and subtypes were determined, all downloaded sequences, plus a selected reference sequence of each subtype were aligned with the study sequences using CLUSTAL W in MEGA version 5. This alignment was exported and used to infer a neighbour-joining phylogenetic tree on the basis of pairwise genetic distances calculated using the two-parameter algorithm described by Kimura and Bull [47]–[49]. Statistical support for the tree topology was assessed by bootstrap analysis with 1000 replicates [45]. A bootstrap value of 70% or greater was used to define a phylogeny cluster. Sequences were submitted to the GenBank database under the accession numbers GU211906 to GU211923.
Statistical analysis
Standard descriptive statistical methods were used to describe the demographic and epidemiological characteristics of the participants. Groups were compared using Student's t test or the Wilcoxon rank-sum test for continuous variables and the chi-square test or Fisher's exact test for categorical variables, as appropriate. A p value less than 0.05 was considered statistically significant. The demographic and clinical characteristics of the case series participants, along with some details of HIV exposure, are presented in another manuscript by the authors [50]. Analysis was carried out using the statistical analysis software package Stata 10.
Results
The Victorian HIV surveillance database (Centre for Population Health, Burnet Institute) indicated that 145 African-born individuals had been notified to the Victorian Department of Human Services (DHS) with newly diagnosed HIV infection by 2005 (Guy R, personal communication: Notifications of HIV diagnosis in Victoria: Country of birth in Africa; extract from Victorian HIV Registry. 2005 February 14). Surveillance records included the name and location of referring doctors and initial specialty clinics to which cases were referred, but no details of subsequent consultations or emigration from Victoria. Preliminary enquiries of the HIV specialty clinics, about newly diagnosed patients referred to their services, suggested that approximately 80 African-born patients with HIV were attending these clinics in 2005. Recruitment was undertaken during 2006 and 2007. Treating doctors approached 36 individuals in the course of their routine clinic visits, of whom 31 agreed to be contacted by the research team. Twenty-two individuals consented to take part in the project. Two withdrew from the study, leaving a small sample of 20 participants. Participants were interviewed between 9 May 2006 and 23 July 2007. Case notes were reviewed between 7 September 2006 and 4 March 2009. The time between HIV diagnosis and interview ranged from six months to 16 years, with a median of 63.5 months.
Fourteen participants were male and six were female. Nine were born in Ethiopia; two each born in Mauritius, Zimbabwe, and Kenya; one each born in Eritrea, Zambia, Ghana, Tanzania and South Africa. Twelve individuals were exposed via heterosexual sex, four via male-to-male sex and two via health-care related blood exposure. None were exposed via IDU. For two participants the route of exposure was unknown (Table 1). The median age at HIV diagnosis was 39 years (range 25 to 51). Median CD4 count at HIV diagnosis was 145 cells/µL, with an interquartile range (IQR) of 60 to 320. Median viral load at diagnosis was 4.76 log10 copies/mL (IQR 4.29 to 5.00). One participant was diagnosed during seroconversion. Fourteen were diagnosed late, including ten diagnosed with advanced HIV disease. Ten were asymptomatic at diagnosis; one experienced seroconversion illness; nine were symptomatic, of which five had AIDS-defining illness (Table 2). Eighteen were diagnosed in Australia and two in New Zealand (both before migration to Australia). Five were diagnosed during migration-related HIV screening (two in New Zealand and three in Australia). Eleven had previously tested negative for HIV (five during migration-related screening). Five had most recently tested negative in Australia and six abroad. Seven (50%) of the 14 diagnosed late had previously tested negative, including four (29%) who tested negative in Australia. Nine reported no previous HIV test before diagnosis (Table 1). Fifteen were on antiretroviral therapy at the time of interview. Eighteen (90%) reported their current state of health as “excellent” or “good.” Details of clinical state and treatment have been published elsewhere [50].
Table 1. African-born Victorian residents with HIV: participant characteristics, HIV testing history, exposure and HIV-1 subtype.
| Demographic characteristics | HIV testing history | HIV exposure | HIV subtype[1] | Accession No. | Comments on exposure | ||||||||
| Case No. | Gender | Age at diagnosis (yrs) | Country of birth | Year of arrival | Year of HIV test | Exposure route | Self-reported exposure location | ||||||
| Last negative | First positive | Interview | Case notes | pol | env | ||||||||
| CS01 | M | 51 | South Africa | Unknown | 1987 | 2001 | MSM+/−IDU | Australia/Europe | Unknown | — | B | GU211906 | Negative HIV test in Australia 14 years before diagnosis. Multiple male sexual contacts for many years in Australia, Europe. |
| CS02 | M | 40 | Eritrea | 1995 | 2003 | 2005 | Heterosexual | Kenya/Australia | Unknown | — | C | GU211907 | Negative HIV test in Australia 2 years before diagnosis. No details of source partner. Travel history not available. |
| CS05 | M | 36 | Ethiopia | 1994 | 1994 | 2002 | Heterosexual | Kenya | Unknown | C | C | GU211908 | Last ngeative test in Kenya 8 years before diagnosis. Case notes: heterosexual contact with woman from high-prevalence country. Interview: no sexual source mentioned, only low-risk health care-related blood exposures. |
| CS06 | F | 27 | Kenya | 1997 | 1997 | 1998 | Other | Kenya | Kenya | — | A | GU211909 | Last negative test in Kenya 1 year before diagnosis. Multiple health-care related blood exposures in Kenya. Male partner in Kenya HIV negative. |
| CS07 | M | 40 | Mauritius | Unknown | 2004 | 2004 | MSM+/−IDU | Australia | Australia | B | B | GU211910 | Documented seroconversion after unprotected male-to-male sex in Australia. |
| CS08 | M | 38 | Ethiopia | 1996 | 1996 | 2002 | MSM+/−IDU | Australia | Unknown | B | B | GU211911 | Negative HIV test in UK 6 years prior to diagnosis. Male-to-male sex in Australia. |
| CS09 | F | 32 | Zimbabwe | 1999 | 1998 | 2002 | Other | Zimbabwe | Unknown | — | C | GU211912 | Two health-care related blood exposures in Zimbabwe after last negative test in Zimbabwe 3 years before diagnosis. |
| CS10 | F | 43 | Zambia | 1989 | No prior test | 2000 | Unknown | Australia | Unknown | — | C | GU211913 | One male partner (later found to be HIV+) since 20 years prior to HIV diagnosis. No blood-borne exposure. No previous HIV test. |
| CS11 | M | 48 | Ethiopia | Unknown | No prior test | 2005 | Unknown | Ethiopia | Unknown | — | C | GU211914 | No exposure history available |
| CS12 | M | 34 | Kenya | Unknown | No prior test | 2006 | Heterosexual | Kenya | Kenya | A1 | A | GU211915 | Heterosexual contact in Kenya with female partner from high-prevalence country. |
| CS13 | F | 25 | Ethiopia | 1998 | No prior test | 2001 | Heterosexual | Unknown | Uganda/Ethiopia | C | — | Not sequenced | Heterosexual contact in Uganda/Ethiopia. |
| CS14 | M | 47 | Ethiopia | 1996 | 1995 | 2006 | Heterosexual | Australia | Unknown | — | C | GU211916 | Negative HIV test in Australia 11 years prior to diagnosis. Unknown source partner. |
| CS15 | M | 42 | Tanzania | Unknown | No prior test | 1995 | Heterosexual | Europe | Unknown | — | A | GU211917 | Multiple heterosexual contacts in Europe and Tanzania. |
| CS16 | F | 27 | Ethiopia | Unknown | 1990 | 1997 | Heterosexual | Unknown | Ethiopia | C | — | Not sequenced | Possible heterosexual contact or needle exposure in Ethiopia after negative test in Sudan 7 years before diagnosis |
| CS17 | M | 41 | Ethiopia | 1993 | 1993 | 2002 | Heterosexual | Australia | Unknown | — | C | GU211918 | Source partner known HIV+. Negative HIV test in Sudan 11 years prior to diagnosis. |
| CS18 | M | 30 | Ethiopia | Unknown | No prior test | 1998 | Heterosexual | Ethiopia | Ethiopia | C | C | GU211919 | Heterosexual exposure in Ethiopia. |
| CS19 | M | 26 | Ethiopia | 1990 | No prior test | 1990 | Heterosexual | Ethiopia | Ethiopia | C | C | GU211920 | Possible heterosexual or low-risk blood exposure in Ethiopia. |
| CS20 | F | 31 | Ghana | 2005 | No prior test | 2005 | Heterosexual | Ghana | Ghana | AG | A | GU211921 | Male sexual partners in Ghana. No other exposure history available. |
| CS21 | M | 47 | Zimbabwe | Unknown | 2000 | 2002 | Heterosexual | PNG | PNG | — | C | GU211922 | Heterosexual sex with PNG-born female partner in PNG after last negative test in Australia 2 years before diagnosis. |
| CS22 | M | 40 | Mauritius | Unknown | No prior test | 1992 | MSM+/−IDU | Australia | Unknown | B | B | GU211923 | Multiple male-to-male sexual exposures in Australia. |
[1] pol subtype from antiretroviral susceptibility genotype assay; env subtype from sequencing and phylogenetic analysis of V3 (see Methods).
Table 2. African-born Victorian residents with HIV: stage at diagnosis.
| Stage at diagnosis | CD4 count (cells/µL) | Symptoms and (CD4 count) at HIV diagnosis | ||||
| Seroconversion | Asymptomatic | Non-AIDS* | AIDS† | Details of symptoms | ||
| Unknown | CS22‡ | |||||
| CS19‡ | Genital herpes | |||||
| >350 | CS21 (996) | — | ||||
| CS06 (855) | — | |||||
| CS16 (678) | Fever | |||||
| CS07 (647) | Seroconversion illness | |||||
| Late HIV diagnosis § | 350–200 | CS09 (320) | — | |||
| CS15 (250) | — | |||||
| CS10 (247) | Appeared unwell to mother | |||||
| CS20 (233) | — | |||||
| Advanced HIV disease ** | <200 | CS17 (150) | — | |||
| CS02 (14) | Non-Hodgkin lymphoma | |||||
| CS18 (120) | — | |||||
| CS05 (120) | Cerebral toxoplasmosis | |||||
| CS11 (118) | — | |||||
| CS13 (60) | Shingles | |||||
| CS01 (40) | PCP†† | |||||
| CS08 (30) | Cerebral toxoplasmosis | |||||
| CS14 (3) | PCP, CMV,‡‡ oesophageal candidiasis | |||||
| CS12 (2) | Pneumonia | |||||
Non-AIDS defining condition.
Condition fitting definition of acquired immune deficiency syndrome used in national HIV/AIDS surveillance (Kaldor J & McDonald A. JAIDS. 2003;32 Suppl 1).
CD4 count at HIV diagnosis not recorded.
Late HIV diagnosis: CD4 count <350 cells/µL at HIV diagnosis, including “advanced HIV disease”.
Advanced HIV disease: CD4 count <200 cell/µL at diagnosis or AIDS at HIV diagnosis.
Pneumocystis jiroveci pneumonia.
Cytomegalovirus retinitis or disease other than liver, spleen or nodes.
No blood sample was obtainable from CS13. CS16 did not consent to the use of stored or fresh blood for HIV sequencing, but did consent to the use of information from case notes and investigations. Both of these patients had prior evidence of clinical ARV resistance as determined by an HIV drug resistance genotyping assay, from which pol sequence results (subtype C) were obtained, but no env sequence data were available for phylogenetic analysis.
Interview data named location of exposure in 18 cases (in two of these case, two regions were named), with two cases reporting “unknown” location. Case notes recorded named exposure location in nine cases (in one case, two regions were named), with eleven cases recording “unknown” location. Reported location of exposure from interview and case notes was consistent in seven cases. In the remaining 13 cases, exposure location was named in either interviews or case notes alone. In no case did the location of exposure reported from the interview contradict the case notes. The reported location of exposure were: Australia in six cases; country of birth in eight cases; Australia or country of birth in one case; a third location (other than country of birth or Australia) in three cases; and either Australia or a third location in two cases. Overall, 11 (55%) reported exposure in Africa, eight (40%) in Australia and three (15%) in another region (Europe or PNG) (Table 1).
Ten participants had undergone HIV genotyping for drug resistance testing; genotyping laboratories provided pol sequences for all these participants. Fifteen stored plasma samples were accessed and three whole blood samples were collected from participants for env sequence analysis. Extraction and amplification from the samples were attempted and amplification and sequencing was successful for all samples.
Subtype was obtained solely from env in ten cases and from pol alone in two cases. Of the ten pol sequences, five were subtype C, three subtype B, one subtype A and one subtype AG. Of the 18 env sequences, ten were subtype C; four subtype B and four subtype A. For eight participants, both pol and env genetic information were available. HIV-1 subtype was identical in seven of these cases; the exception was CS20, where CRF02_AG was reported in the pol region and subtype A in the env region. This participant was likely to be infected with CRF02_AG, in which the env region is almost entirely comprised of subtype A sequence. Taking both pol and env sequences together, 12 (60%) participants had HIV-1 subtype C, four (20%) subtype B, three (15%) subtype A and one (5%) CRF02_AG (Table 1).
Three participants with subtype A were exposed via heterosexual sex. One (CS12) reported exposure in his country of birth; the second (CS15) had multiple female sexual contacts in Europe and his country of birth. The third participant (CS06) had multiple health-care associated blood exposures in her country of birth. The participant with CRF02_AG (CS20) had multiple heterosexual contacts in her country of birth.
Nine of the 12 participants with subtype C were exposed via heterosexual sex, one (CS09) via health-care associated blood exposure and two (CS11 and CS10) via unknown routes. Three participants reported heterosexual exposure in Australia: one (CS02) had a negative HIV test in Australia two years prior to diagnosis but did not report a specific source partner (he also had heterosexual contact in Kenya, but attributed his HIV infection to exposure in Australia); another (CS14) reported heterosexual contact in Australia and had last tested negative abroad 11 years prior to diagnosis; the third (CS17) had an HIV-positive source partner and had last tested negative to HIV abroad eleven years prior to diagnosis). Five participants (CS05, CS13, CS16, CS18, CS19) reported heterosexual exposure in Africa (CS16 reported exposure in Ethiopia, possibly via heterosexual contact or health care-associated blood exposure; she did not consent to the use of stored or fresh blood samples for genetic sequencing). One participant (CS21) reported heterosexual contact in Papua New Guinea with a local-born female partner. One participant (CS09) reported health care-associated blood exposures in her country of birth and had never been sexually active. One male participant (CS11) reported exposure in Ethiopia via an unknown route. One female participant (CS10) reported exposure in Australia via an unknown route; however she also reported that her only sexual partner for the twenty years preceding her diagnosis, who had migrated with her from Africa to Australia several years previously, had been diagnosed with HIV after migration; she denied any exposure to blood products and had not previously been tested for HIV; the likelihood appears strong that she had in fact acquired HIV from her long-term male partner, before or after migration.
All four participants with subtype B viruses were exposed via male-to-male sex. One (CS07) was certainly exposed in Australia, having a documented seroconversion after a high-risk sexual contact. Two were likely exposed in Australia, reporting multiple male sexual contacts in Australia; one (CS08) had a negative HIV test abroad six years prior to his diagnosis and the other (CS22) had not previously been tested for HIV. The remaining man (CS01) had multiple male sexual contacts in Europe and Australia and had not been tested for HIV prior to his diagnosis.
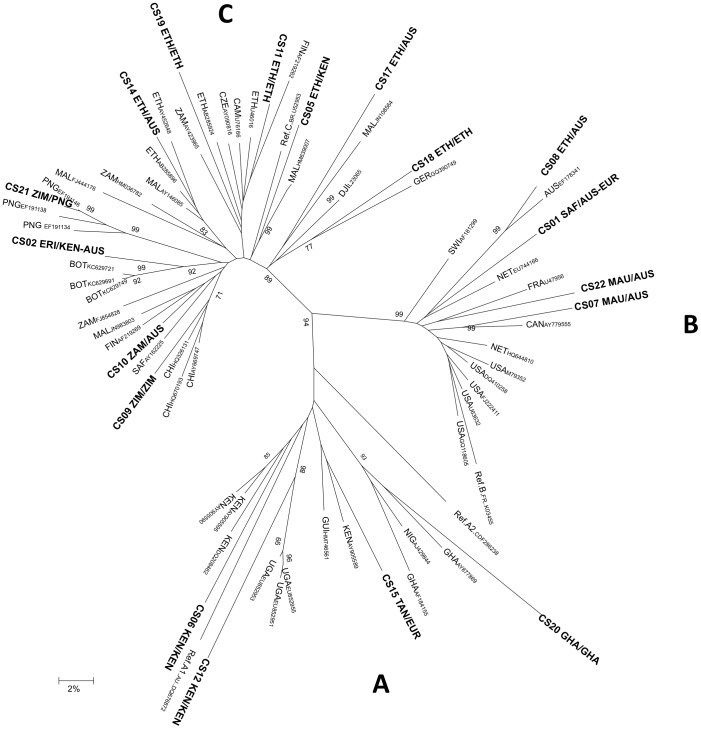
The phylogenetic tree produced from the analysis of env sequence alignments is presented in Figure 1. Of the 18 cases with available env sequences, 15 clustered with reference sequences from the reported country of exposure. In the remaining three cases (CS10, CS14 and CD17), the env isolate clustered with reference sequences from the country of birth. Isolates from the three individuals identified with subtype A were all related to the “East African” cluster. The single CRF02_AG strain from Ghana (CS20) was linked to the “West African” A cluster. The four subtype B isolates grouped with other subtype B isolates from the USA, Australia and Europe; these isolates did not appear to cluster with any more specific geographic region. Nine of the ten cases with subtype C clustered with South-East African sequences, while the remaining sequence was closely associated with Papua New Guinean sequences, consistent with reports of heterosexual sex with a woman from PNG.
Figure 1. African-born Victorian residents with HIV: phylogenetic analysis of V3 env sequences and reference subtype A, B and C sequences.
Neighbour joining tree reflecting the genetic relationship between HIV-1 isolates from African-born Victorians, the most homologous HIV-1 strains and selected reference strains. Study isolates are indicated in bold followed by Country of Birth/Country of reported exposure. Significant bootstrap values are indicated at the relevant node. Sequences were analysed over a 369bp region corresponding to the HIV-1 envelope region and spanning nucleotides 6984–7353 (HXB2 coordinates). The scale bar represents 2% genetic distance. Country codes: AUS – Australia; BOT- Botswana; CAN – Canada; CHI-China; CZE – Czech Republic; DJI – Djibouti; ERI – Eritrea; ETH – Ethiopia; EUR – Europe; FIN- Finland; FRA – France; GER – Germany; GHA – Ghana; GUI – Guinea Bissau; KEN – Kenya; MAL – Malawi; NET – The Netherlands; PNG – Papua New Guinea; SAF – South Africa; SWI – Switzerland; TAN – Tanzania; USA – United States of America; 23 – Uganda; 24 – Zambia; 25 – Zimbabwe.
Discussion
This small study shows how phylogenetic analysis may complement clinical data in ascertaining the location and circumstances of HIV exposure amongst African immigrants in Victoria, Australia. The results demonstrated a preponderance of non-B HIV-1 subtypes and a high degree of sequence diversity. Exposure histories were consistent with phylogenetic analysis. This study contributes to current understanding of the geographical and social complexities influencing the epidemiology of HIV in Victoria's African communities.
Most participants had non-B subtypes of HIV-1, particularly subtype C (the most commonly occurring subtype worldwide) or subtype A (distributed widely across Africa) [19]–[21]. However, little similarity was found between participant sequences within each subtype. These data contrast to findings from our earlier study of Vietnamese-born IDU in Australia, who were more uniformly infected with the Vietnamese subtype, CRF01_AE [25]. The predominance and diversity of non-B HIV-1 strains has been previously noted in African immigrant populations in Europe, Canada and Australia [22], [36], [51], [52], reflecting the huge genetic diversity of HIV within Africa [53].
The genetic heterogeneity of study sequences and diversity of reported exposures located participants within separate transmission networks. All but one of these networks appeared to be based in Africa, indicating the epidemiological continuity between African populations within Africa and the African diaspora [26], [54]. The exception was the env subtype C sequence of CS21, which appeared to belong to a transmission network based in PNG (the country of his reported exposure) rather than one based in his country of birth. This case illustrated the additional information obtained through phylogenetic analysis, beyond simple serological categorisation by subtype, although serological assays may be useful in preliminary screening of samples [10].
Heterosexual sex was the commonest exposure route for these African immigrant PLHIV, as it has been amongst African immigrants in the UK [10]. However four (20%) reported acquisition through male-to-male sex, highlighting the importance of this route of exposure in African populations within Africa and the diaspora [5], [9]. The association between subtype B and exposure through sex between men is consistent with historical patterns in Australia [51] and with a recent study of HIV infections diagnosed in Victoria 2005–2010; however, non-B subtypes of HIV-1 have been isolated from native-born and foreign-born MSM in studies from Australia and abroad, whilst “African” transmission networks unsurprisingly include individuals not categorised or self-identified as “African” [22], [55]. Subtype B sequences isolated from all four MSM participants resembled env subtype B sequences from Europe, Australia and the US, but were dissimilar to each other. This was consistent with the geographically diverse histories of sexual exposure and with existing evidence of multiple, superimposed, transnational sexual networks mainly (but not exclusively) including MSM [23], [26], [29], [35].
Just over half of the participants reportedly acquired HIV in Africa, but almost a third reported acquisition in Australia. The diverse exposure locations and high proportion (70%) diagnosed late, justify the inclusion of African migrants as a “priority population” in the Australian HIV response [1]. Timely diagnosis (for African settlers arriving in Australia with prevalent HIV infection) must be complemented by prevention, for other African Australians, of HIV exposure after migration. Health promotion about HIV in African communities should address the effect of migration-related HIV screening on subsequent risk perception in Australia [13]–[15].
This study is limited by its small size. However as the number of African-born people diagnosed with HIV in Victoria is not large, this sample size represents a substantial proportion of this population. Additional complexity arises from the high proportion of late diagnoses among African-born cases of HIV living in Victoria, which suggest that testing rates may be too low to provide a solid basis for estimates of HIV prevalence in Victoria's African communities [2]. This study provides no evidence for a locally based cluster of HIV transmission among African Australians living in Victoria, although demonstrating the absence of any such cluster would require a much larger study.
One strength of this study is the collection of geographical and temporal details exposure as well as detailed demographic information. Participation was likely influenced by the degree of engagement of eligible patients with their treating doctors. Ascertainment of details regarding HIV exposure was subject to recall bias and willingness to disclose sensitive information. The extent of participation also limited the samples available for sequencing. Sequencing confined to the V3 region of env limited the discriminatory power of the phylogenetic analysis and the ability to explore recombination [28]. However, genetic analysis in this study was used primarily to investigate the correlation between self-reported location of HIV exposure and genetic characteristics of HIV-1 strains circulating in the reported location of exposure. Limited sequencing of highly variable env region was probably sufficient for this purpose and has been successfully used in this context in previous molecular epidemiological studies [37]–[39]. The heterogeneity of sample env sequences and the absence of reported contacts between cases suggests that little additional epidemiological information would have been obtained by sequencing the whole genome.
Our study shows that international networks of social and sexual contact are likely to affect exposure to HIV for African Australians, within Australia and abroad. Phylogenetic data can complement existing surveillance methods in the development of more focused and effective strategies for HIV prevention and timely diagnosis of HIV infection in this mobile population.
Acknowledgments
We gratefully acknowledge all the participants of this study. The authors would also like to acknowledge the contributions of Chris Birch, William Malouk Daw, Sahra Hussein, Tenenet Taye, Abraha Gebremariam Mamo, Shangale Ali, Mohanad Hakim, Shiraz Hakim, Neveen Hanna, Peter Stanley, Tony Korman, Anne Mijch, Suzanne Garland, Christopher Fairley, Kirsty Buising, Tim Read, Tina Schmidt, Ian Woolley, Kerri Boyd, Christine Bowtell-Harris, Rebecca Guy, Keflemariam Yohannes, Carol El-Hayek, Mirella Ozols, Christalla Hajisava, Paulette Manton, Gina Barri-Rewell, Ann McIntyre, Lisa Natoli, Marion Brown, Jamileh Abu-Duhou, Jim Black, Sandy Gifford, Graham Brown, Martha Morrow, Martha McIntyre, Pascale Allotey, Virginia De Crespigny, Jeanette Venkataya, Nick Christopher, Liz Nicol, Mary Ljubanovic, Jenny Lewis, Tim Spelman, Maelenn Gouillou, Rhiannon Palmer, Lisa Morris and Vicki Edouard.
Funding Statement
The authors are grateful for the financial and material support provided by the Centre for Clinical Research Excellence in Infectious Diseases and the Department of Medicine; the financial support received from the Victorian Government Department of Human Services, the Victor Hurley Research Fund and the Australasian Society for Infectious Diseases; and the material support provided by the Burnet Institute, the Victorian Infectious Diseases Laboratories and the Victorian Infectious Diseases Service. The authors acknowledge the funding for institutional infrastructure provided by the Victorian Department of Innovation and Industry and the Australian National Health and Medical Research Council. Suzanne Crowe holds a Principal Research Fellowship from the National Health and Medical Research Council. Margaret Hellard holds a Senior Research Fellowship from the National Health and Medical Research Council. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program.
References
- 1.Dept Health and Ageing (2010) Sixth National HIV Strategy 2010–2013. Canberra: Commonwealth of Australia.
- 2. Lemoh C, Guy R, Yohannes K, Lewis J, Street A, et al. (2009) Delayed diagnosis of HIV infection in Victoria 1994 to 2006. Sex Health 6: 117–22. [DOI] [PubMed] [Google Scholar]
- 3.The Kirby Institute (2012) HIV, viral hepatitis and sexually transmissible infections in Australia Annual Surveillance report 2012. Sydney: The Kirby Institute for infection and immunity in society, University of New South Wales.
- 4.Australian Bureau of Statistics (2007) 2006 Census of population and housing. Canberra: Australian Bureau of Statistics. Cat No. 2068.0.
- 5. De Cock K, Jaffe H, Curran J (2012) The evolving epidemiology of HIV/AIDS. AIDS 26: 1205–13. [DOI] [PubMed] [Google Scholar]
- 6. Dore G, Li Y, McDonald A, Kaldor J (2001) Spectrum of AIDS-defining illnesses in Australia, 1992 to 1998: Influence of country/region of birth. J Acquir Immune Defic Syndr 26: 283–90. [DOI] [PubMed] [Google Scholar]
- 7.EuroHIV (2007) HIV/AIDS Surveillance in Europe: end-year report 2006. HIV/AIDS Surveillance in Europe. Saint-Maurice: European Centre for the Epidemiological Monitoring of HIV and AIDS.
- 8. Beyrer C, Baral SD, van Griensven F, Goodreau SM, Chariyalertsak S, et al. (2012) Global epidemiology of HIV infection in men who have sex with men. Lancet 380: 367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith AD, Tapsoba P, Peshu N, Sanders EJ, Jaffe HW (2009) Men who have sex with men and HIV/AIDS in sub-Saharan Africa. Lancet 374: 416–22. [DOI] [PubMed] [Google Scholar]
- 10. Aggarwal I, Smith M, Tatt I, Murad S, Osner N, et al. (2006) Evidence for onward transmission of HIV-1 non-B subtype strains in the United Kingdom. J Acquir Immune Defic Syndr 41: 201–9. [DOI] [PubMed] [Google Scholar]
- 11.Lack S, Geue A, O'Connell E, Kelly P, Slattery D (2007) Administration of the health requirement of the Migration Act 1958. Australian National Audit Office. Audit Report No. 37 2006-7. Canberra: Commonwealth of Australia.
- 12.Dept Immigration and Citizenship (2007) Fact sheet 22: The health requirement. Canberra: Commonwealth of Australia. Available: http://www.immi.gov.au/media/fact-sheets/22health.htm. Accessed 2007 Jan 30.
- 13.Lemoh C, Hellard M, Street A, Biggs BA (2006) Reducing the risk of transmission of HIV/AIDS in African and Arabic-speaking communities in Victoria. Melbourne: The University of Melbourne; McFarlane Burnet Institute for Medical Research and Public Health; Victorian Infectious Diseases Service; Australian Research Centre in Sex, Health and Society.
- 14. Palmer R, Lemoh C, Tham R, Hakim S, Biggs BA (2009) Sudanese women living in Victoria, Australia: health-information-seeking behaviours and the availability, effectiveness and appropriateness of HIV/AIDS information. Diversity Health Care 6: 109–20. [Google Scholar]
- 15. McMichael C, Gifford S (2010) Narratives of sexual health risk and protection amongst young people from refugee backgrounds in Melbourne, Australia. Cult Health Sex 12: 263–77. [DOI] [PubMed] [Google Scholar]
- 16. Sackett DL, Rennie D (1992) The science of the art of the clinical examination. JAMA 267: 2650–2. [PubMed] [Google Scholar]
- 17. Dubois-Arber F, Meystre-Agustoni G, Andre J, De Heller K, Alain P, et al. (2010) Sexual behaviour of men that consulted in medical outpatient clinics in Western Switzerland from 2005–2006: risk levels unknown to doctors? BMC Public Health 10: 528 doi:10.1186/1471-2458-10-528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramsey PG, Curtis R, Paauw DS, Carline JD, Wenrich MD (1998) History-taking and preventive medicine skills among primary care physicians: an assessment using standardized patients. Am J Med 104: 152–8. [DOI] [PubMed] [Google Scholar]
- 19. Hemelaar J, Gouws E, Ghys PD, Osmanov S (2006) Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20: W13–W23. [DOI] [PubMed] [Google Scholar]
- 20. Kandathil AJ, Ramalingam S, Kannangai R, David S, Sridharan G (2005) Molecular epidemiology of HIV. Indian J Med Res 121: 333–44. [PubMed] [Google Scholar]
- 21. McCutchan F (2006) Global epidemiology of HIV. J Med Virol 78: S7–S12. [DOI] [PubMed] [Google Scholar]
- 22. Chibo D, Birch C (2012) Increasing diversity of human immunodeficiency virus type 1 subtypes circulating in Australia. AIDS Res Hum Retroviruses 28: 578–83. [DOI] [PubMed] [Google Scholar]
- 23. de Coul E, Coutinho RA, van der Schoot A, van Doornum GJJ, Lukashov VV, et al. (2001) The impact of immigration on env HIV-1 subtype distribution among heterosexuals in the Netherlands: influx of subtype B and non-B strains. AIDS 15: 2277–86. [DOI] [PubMed] [Google Scholar]
- 24. Monno L, Brindicci G, Lo Caputo S, Punzi G, Scarabaggio T, et al. (2005) HIV-1 subtypes and circulating recombinant forms (CRFs) from HIV-infected patients residing in two regions of central and southern Italy. J Med Virol 75: 483–90. [DOI] [PubMed] [Google Scholar]
- 25. Yirrell DL, Goldberg DJ, Whitelaw J, McSharry C, Raeside F, et al. (1999) Viral subtype and heterosexual acquisition of HIV infections diagnosed in Scotland. Sex Transm Infect 75: 392–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liitsola K, Holmstrom P, Laukkanen T, Brummer-Korvenkontio H, Leinikki, et al (2000) Analysis of HIV-1 genetic subtypes in Finland reveals good correlation between molecular and epidemiological data. Scand J Infect Dis 32: 475–80. [DOI] [PubMed] [Google Scholar]
- 27. Chibo D, Kaye M, Birch C (2012) HIV transmissions during seroconversion contribute significantly to new infections in men who have sex with men in Australia. AIDS Res Hum Retroviruses 28: 460–4. [DOI] [PubMed] [Google Scholar]
- 28. Lospitao E, Alvarez A, Soriano V, Holguin A (2005) HIV-1 subtypes in Spain: a retrospective analysis from 1995 to 2003. HIV Med 6: 313–20. [DOI] [PubMed] [Google Scholar]
- 29. Kouyos RD, von Wyl V, Yerly S, Böni J, Taffé P, et al. (2010) Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J Infect Dis 201: 1488–97. [DOI] [PubMed] [Google Scholar]
- 30. von Wyl V, Kouyos RD, Yerly S, Boni J, Shah C, et al. (2011) The role of migration and domestic transmission in the spread of HIV-1 non-B subtypes in Switzerland. J Infect Dis 204: 1095–103. [DOI] [PubMed] [Google Scholar]
- 31. de Coul ELMO, Lukashov VV, van Doornum GJJ, Goudsmit J, Coutinho RA (1998) Multiple HIV-1 subtypes present amongst heterosexuals in Amsterdam 1988–1996: no evidence for spread of non-B subtypes. AIDS 12: 1253–5. [DOI] [PubMed] [Google Scholar]
- 32. Hayman A, Moss T, Simmons G, Arnold C, Holmes EC, et al. (2001) Phylogenetic analysis of multiple heterosexual transmission events involving subtype B of HIV type 1. AIDS Res Hum Retroviruses 17: 689–95. [DOI] [PubMed] [Google Scholar]
- 33. Carr J, Laukkanen T, Salminen M, Albert J, Alaeus A, et al. (1999) Characterization of subtye A HIV-1 from Africa by full genome sequencing. AIDS 13: 1819–26. [DOI] [PubMed] [Google Scholar]
- 34. Scaduto DI, Brown JM, Haaland WC, Zwickl DJ, Hillis DM, et al. (2011) Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Natl Acad Sci USA 107: 21242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paraskevis D, Pybus O, Magiorkinis G, Hatzakis A, Wensing AMJ et al. (2009) Tracing the HIV-1 subtype B mobility in Europe: a phylogeographic approach. Retrovirology 6: :49 doi 10.1186/1742-4690-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akouamba B, Viel J, Charest H, Merindol N, Samson J, et al. (2005) HIV-1 genetic diversity in antenatal cohort, Canada. Emerg Infect Dis 11: 1230–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jennes W, Kyongo JK, Vanhommerig E, Camara M, Coppens S, et al. (2012) Molecular epidemiology of HIV-1 transmission in a cohort of HIV-1 concordant heterosexual couples from Dakar, Senegal. PloS One 7 : doi 10.1371/journal.pone.0037402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caridha R, Tran TTH, Gaseitsiwe S, Hung PV, Anh NM, et al. (2012) Short communication phylogenetic characterization of HIV type 1 CRF01_AE V3 Envelope Sequences in Pregnant Women in Northern Vietnam. AIDS ResHum Retroviruses 28: 852–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Graw F, Leitner T, Ribeiro RM (2012) Agent-based and phylogenetic analyses reveal how HIV-1 moves between risk groups: Injecting drug users sustain the heterosexual epidemic in Latvia. Epidemics 4: 104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medew J (2007) Plan to genetically trace reckless HIV infections. The Age April 28, Melbourne: The Age Co. Ltd. Available: http://www.theage.com.au/news/national/plan-to-genetically-trace-reckless-hiv-infections/2007/04/27/1177459980619.html. Accessed 2013 Dec 4.
- 41.Metlikovec J (2007) PM wants HIV migrant ban. Herald Sun April 14, Melbourne: News Digital Media. Database: ProQuest Australia & New Zealand Newstand. ProQuest document ID 360880634. Available: http://search.proquest.com.ezp.lib.unimelb.edu.au/docview/360880634?accountid=12372. Accessed 2013 Dec 4.
- 42. Antinori A, Coenen T, Costagiola D, Dedes N, Ellefson M, et al. (2011) Late presentation of HIV infection: a consensus definition. HIV Med 12: 61–4. [DOI] [PubMed] [Google Scholar]
- 43. Kaldor J, McDonald A (2003) HIV/AIDS surveillance systems in Australia. JAIDS J Acquir Immune Defic Syndr International 32: S18–S23. [DOI] [PubMed] [Google Scholar]
- 44. Ryan C, Elliott J, Middleton T, Mijch A, Street A, et al. (2004) The molecular epidemiology of HIV type 1 among Vietnamese Australian injecting drug users in Melbourne, Australia. AIDS Res Hum Retroviruses 20: 1364–7. [DOI] [PubMed] [Google Scholar]
- 45. Ryan CE, Gare J, Crowe SM, Wilson K, Reeder JC, et al. (2007) The heterosexual HIV type 1 epidemic in Papua New Guinea is dominated by subtype C. AIDS Res Hum Retroviruses. 23: 941–4. [DOI] [PubMed] [Google Scholar]
- 46. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–20. [DOI] [PubMed] [Google Scholar]
- 48. Bull JJ, Huelsenbeck JP, Cunningham CW, Swofford DL, Waddell PJ (1993) Partitioning and combining data in phylogenetic analysis. Syst Biol 42: 384–97. [Google Scholar]
- 49. Hillis DM, Bull JJ (1993) An empirical-test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42: 182–92. [Google Scholar]
- 50. Lemoh CN, Baho S, Grierson J, Hellard M, Street A, et al. (2010) African Australians living with HIV: a case series from Victoria. Sex Health 7: 142–8. [DOI] [PubMed] [Google Scholar]
- 51. Herring BL, Ge YC, Wang B, Ratnamohan M, Zheng F, et al. (2003) Segregation of human immunodeficiency virus type 1 sutypes by risk factor in Australia. J Clin Microbiol 41: 4600–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yebra G, Rivas P, Herrero M, Lopez M, de Mulder M, et al. (2009) Clinical differences and viral diversity between newly HIV type 1-diagnosed African and non-African patients in Spain (2005–2007). AIDS Res Hum Retroviruses 25: 37–44. [DOI] [PubMed] [Google Scholar]
- 53. Papathanasopoulos MA, Hunt GM, Tiemessen CT (2003) Evolution and diversity of HIV-1 in Africa - a review. Virus Genes 26: 151–63. [DOI] [PubMed] [Google Scholar]
- 54. Hughes GJ, Fearnhill E, Dunn D, Lycett SJ, Rambaut A, et al. (2009) Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PloS Pathogens 5: e1000590 doi:10.1371/journal.ppat.1000590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gifford R, Oliveira T, Rambaut A, Pybus O, Dunn D, et al. (2007) Phylogenetic surveillance of viral genetic diversity and the evolving molecular epidemiology of human immunodeficiency virus type 1. J Virol 81: 13050–6. [DOI] [PMC free article] [PubMed] [Google Scholar]