Abstract
Manganese (Mn) is an essential trace element for plants. Recently, the genes responsible for uptake of Mn in plants were identified in Arabidopsis and rice. However, the mechanism of Mn distribution in plants has not been clarified. In the present study we identified a natural resistance-associated macrophage protein (NRAMP) family gene in rice, OsNRAMP3, involved in Mn distribution. OsNRAMP3 encodes a plasma membrane-localized protein and was specifically expressed in vascular bundles, especially in phloem cells. Yeast complementation assay showed that OsNRAMP3 is a functional Mn-influx transporter. When OsNRAMP3 was absent, rice plants showed high sensitivity to Mn deficiency. Serious necrosis appeared on young leaves and root tips of the OsNRAMP3 knockout line cultivated under low Mn conditions, and high Mn supplies could rescue this phenotype. However, the necrotic young leaves of the knockout line possessed similar levels of Mn to the wild type, suggesting that the necrotic appearance was caused by disturbed distribution of Mn but not a general Mn shortage. Additionally, compared with wild type, leaf Mn content in osnramp3 plants was mostly in older leaves. We conclude that OsNRAMP3 is a vascular bundle-localized Mn-influx transporter involved in Mn distribution and contributes to remobilization of Mn from old to young leaves.
Introduction
Manganese (Mn) is an essential metal nutrient in most organisms. In plants, Mn plays an important role in photosystem II and is a required cofactor for a variety of enzymes [1]. Mn deficiency can reduce plant growth and increase susceptibility to low temperature and pathogen infection [2]. Despite its importance, the amount of Mn required by a plant is relatively low; however, the capacity for Mn uptake always exceeds this requirement and excess Mn can be particularly toxic to plant growth [3]. In general, the uptake and detoxification of Mn is well balanced in plants. Many gene families have been identified as involved in Mn uptake or detoxification of excess Mn.
Much of our understanding on Mn uptake in plants comes from the complementation tests on yeast mutant strain Δsmf1, which is deficient in Mn uptake [4]. SMF1, a gene from natural resistance-associated macrophage protein (NRAMP) family in yeast (Saccharomyces cerevisiae), is responsive to the high-affinity Mn2+ accumulation into yeast cell. In plants, NRAMP family genes also contribute to Mn2+ transportation or translocation. AtNRAMP1 and OsNRAMP5 encode plasma-located proteins that are major high-affinity Mn transporters in Arabidopsis and rice (Oryza sativa L.), respectively [5], [6]. AtNRAMP3 and AtNRAMP4, two other NRAMP members in Arabidopsis, are targeted to vacuolar membrane and operate in the retrieval of Mn2+ from vacuoles in leaf mesophyll cells under Mn deficiency [7]. Shown to be functional orthologs of AtNRAMP3 and AtNRAMP4, TcNRAMP3 and TcNRAMP4 from the metal hyperaccumulator Thlaspi caerulescens are both tonoplast proteins and can complement the Δsmf1 yeast mutant phenotype [8]. When expressed in yeast strains, LeNRAMP1 and LeNRAMP3 from tomato (Lycopersicon esculentum) are targeted to vesicle and tonoplast, respectively, and can retrieve the phenotype of yeast mutant Δsmf1 [9].
Mn always shares the same transporters with iron (Fe) in plants [3], [5], [10]. ZIP [zinc-regulated transporter/iron-regulated transporter (ZRT/IRT)-related proteins] genes and YSL (yellow stripe 1-like) genes are two well-known transporter families of Fe in plants. In recent years, many genes from these two families have been shown capable of transporting Mn. For example, AtIRT1 is an Arabidopsis ZIP transporter with a broad substrate range, including Mn2+ [11]. Besides AtIRT1, six other ZIP genes (AtZIP1, AtZIP2, AtZIP3, AtZIP5, AtZIP6 and AtZIP9) from Arabidopsis were recently shown to be functional in transporting Mn2+ in yeast [12]. Several members of the ZIP family from plant species other than Arabidopsis also showed the ability to transport Mn2+, including LeIRT1 and LeIRT2 from tomato, MtZIP4 and MtZIP7 from Medicago truncatula, and HvIRT1 from barley (Hordeum vulgare) [13]–[15]. In rice, no ZIP genes have been shown to transport Mn2+, but two members from the YSL family (OsYSL2 and OsYSL6) showed activity in Mn2+ transport [16], [17].
Unlike NRAMP and ZIP gene families, cation exchanger (CAX) genes in plants are always involved in tolerance of Mn toxicity. In Arabidopsis, six CAX genes have been identified. AtCAX1 and AtCAX2 were the first CAX genes reported in plants and are known calcium (Ca) transporters [18]. However, expression studies in tobacco (Nicotiana tabacum) and yeast showed that AtCAX2 is also involved in Mn2+ transport [19]. The protein encoded by AtCAX2 is targeted to vacuolar membrane and confers Mn2+ tolerance in Arabidopsis. Furthermore, in a yeast Mn-tolerance screen with an Arabidopsis cDNA library, AtCAX2 was the only gene identified out of the 105 transformants that could suppress the Mn toxicity phenotype [20]. However, other CAX genes in Arabidopsis did not show Mn2+ transport activity. But interestingly, when expressed in yeast with an N-terminally truncated form, AtCAX5 could mediate Mn2+ transport, suggesting that CAX genes in Arabidopsis may control the Mn2+ transport activity by an autoregulatory region at the N-terminus [21]. Five CAX genes have been identified in rice. The OsCAX3 and N-terminal truncated OsCAX1a from rice could confer tolerance to Mn when expressed in yeast [22]. In addition to the CAX gene family, the endomembrane-type Ca-ATPase (ECA) gene family is another one involved in both Ca2+ and Mn2+ transport. In Arabidopsis, AtECA1 and AtECA3 showed Mn transport activity when expressed in yeast [23], [24]. Interestingly, protein encoded by AtECA1 is mainly targeted on endoplasmic reticulum membrane and increases tolerance of plants to Mn toxicity; however, AtECA3 encodes a Golgi-localized protein and is required in Arabidopsis under Mn-deficient conditions [25].
The cation diffusion facilitator (CDF) gene family also contributes to Mn tolerance. Four CDF genes (ShMTP1–ShMTP4), the first CDF genes identified in plants, were screened from Stylosanthes hamate for enhancing Mn2+ tolerance by expressing a cDNA library in yeast [26]. Further studies on ShMTP1 suggested that it encoded a tonoplast protein in Arabidopsis and conferred Mn2+ tolerance through internal sequestration. There are 12 members in the CDF family in Arabidopsis, and proteins encoded by four of these genes (AtMTP8–AtMTP11) are closely related to the protein encoded by ShMTP1 [27]. Furthermore, knockout of AtMTP11 in Arabidopsis resulted in hypersensitivity to high Mn2+, suggesting that AtMTP11 can confer plant Mn tolerance [27], [28].
Most knowledge concerning Mn in plants was obtained from studies focused on uptake and tolerance, but little is known about the mechanisms of distribution and translocation of Mn in plants. In this study, we identified another NRAMP gene from rice, OsNRAMP3, involved in Mn distribution. OsNRAMP3 encoded a plasma membrane-localized protein with activity in transporting Mn and was expressed specifically in vascular bundles, especially phloem cells. Knocking out of OsNRAMP3 in rice resulted in high sensitivity to Mn deficiency and disturbed Mn distribution in leaves. These data suggested that OsNRAMP3 played an important role in Mn distribution in rice.
Materials and Methods
Plant Materials
The knockout line of OsNRAMP3 and wild type were based on O. sativa L. ssp. japonica cv. DongJin background. The osnramp3 mutant is a T-DNA insert mutant ordered from the POSTECH RISD database (http://www.postech.ac.kr/life/pfg/risd/) [29]. Promoter analysis and expression profile analyzed by real-time PCR were based on O. sativa L. ssp. japonica cv. Zhonghua 11.
Subcellular Localization of OsNRAMP3
The coding sequence of OsNRAMP3 for subcellular localization was amplified using cv. Zhonghua11 total cDNA as template and primers 5′-gactgaattcatgagcggcccaatgcaa-3′ (forward) and 5′-gactggtaccatcgagatcagaagcagttcgct-3′ (reverse), which were designed based on the sequence of LOC_Os06g46310.1 from the RGAP database (http://rice.plantbiology.msu.edu/). The PCR product was cleaved using EcoR1 and Kpn1 and ligated into pM999–GFP with correct direction. The construct was transformed into Arabidopsis mesophyll protoplasts and observed using a confocal laser scanning microscope (TCS SP2; Leica) after incubation at 22°C for 12–24 h [30].
GUS Staining Assay
To construct the OsNRAMP3-promoter:GUS plasmid, 2.1 kb of genomic sequence located upstream of the OsNRAMP3 initiation codon was amplified by PCR from cv. Zhonghua 11 genomic DNA using primers Pnr5-F (5′-agctgcagatgcgccaaaatactgaat-3′) and Pnr5-R (5′-tcggatcctgcaagaaccctcaagact-3′). The amplified promoter fragment was digested by BamH1 and Pst1 and introduced into vector pDX2181 in the correct direction [31]. The construct was transformed into cv. Zhonghua 11 by Agrobacterium tumefaciens-mediated transformation [32]. The transgenic plant tissues were incubated in X-Gluc strain buffer at 37°C for 4 h [33].
Hydroponic Experiments
Hydroponic experiments were performed according to the standard rice culture solution (1.44 mM NH4NO3, 0.3 mM NaH2PO4, 0.5 mM K2SO4, 1.0 mM CaCl2, 1.6 mM MgSO4, 0.17 mM Na2SiO3, 50 µM Fe-EDTA, 0.06 µM (NH4)6Mo7O24, 15 µM H3BO3, 8 µM MnCl2, 0.12 µM CuSO4, 0.12 µM ZnSO4, 29 µM FeCl3 and 40.5 µM citric acid at pH 5.5) [34]. The solution was renewed every 5 d.
RNA Extraction and Real-time PCR
Total RNA was extracted using TRizol reagent (Invitrogen). Of total RNA, 3 µg was used to synthesize the first-strand cDNAs in 20 µl of reaction mixture using M-MLV reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was performed using the SYBR Premix Ex Taq™ (TaKaRa) with the following gene-specific primers for OsNRAMP3: rqNR3-F (5′-tcagcagcgaactgcttctgatct-3′) and rqNR3-R (5′-atcagctggctaactctttgggct-3′). The rice Ubiquitin 5 gene was used as the internal control with the following primers: qUbq-F (5′-aaccagctgaggcccaaga-3′) and qUbq-R (5′-acgattgatttaaccagtccatga-3′). The real-time PCR reaction was performed on an Applied Biosystems 7500 PCR instrument.
Elemental Analysis
Shoots and roots were harvested separately and roots were washed with distilled water twice before sampling. After drying at 80°C for 3 d, all samples were digested in 65% nitric acid in a MARS6 microwave (CEM) at a temperature gradient of 120–180°C for 45 min, and then diluted in deionized water. The metal contents of the samples were determined by inductively coupled plasma–mass spectrometry (ICP-MS; Agilent 7700 series, USA).
Yeast Complementation Assay
The cDNA fragments containing an entire open reading frame of OsNRAMP3 or AtNRAMP1 were amplified by the following primers: ycNR3-F (5′-gactggtaccatgagcggcccaatgcaac-3′) and ycNR3-R (5′-ctgagaattcctaatcgagatcagaagcagttcg-3′) for OsNRAMP3; and ycAtNR1-F (5′-gactggtaccatggcggctacaggatctggac-3′) and ycAtNR1-R (5′-gactggtacctcagtcaacatcggaggtagat-3′) for AtNRAMP1. The fragments were firstly cloned into pEGM-T Easy vector (Promega) and digested with Kpn1 and EcoR1 for OsNRAMP3 or EcoR1 only for AtNRAMP1, and then correctly introduced into vector pYES2. The resulting plasmids were transformed into yeast strains. The yeast strains used in this study were Δsmf1 (Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; YOL122c::kanMX4) and its wild-type BY4741 (MATa; his3Δ 1; leu2Δ 0; met15Δ 0; ura3Δ 0). Yeast cells were transformed according to standard procedures (Invitrogen). Δsmf1 complementation testing by drop-spotting assays was performed on synthetic defined (SD)-Ura medium, which contained 2% galactose, 0.67% yeast nitrogen base without MnSO4 (Krackeler Scientific Inc), 0.2% appropriate amino acid, 2% agar, 50 mM 2-morpholinoethanesulfonic acid and supplemented without or with 2.5, 5 or 10 µM ethylene glycol bis(2-aminoethyl) tetraacetic acid (EGTA) at pH 6.0.
For metal determination, yeast cells transformed with different vectors were grown for 30 h starting from OD600 of 0.01 on SD-Ura liquid medium supplemented with 0.02 µM Mn at pH 6.0. Galactose instead of glucose was used here for induction of the GAL promoter. Cells were harvested by centrifugation and washed three times with deionized water (MilliQ; Millipore). All samples were dried at 80°C for 3 d, and then used to determine metal contents by ICP-MS.
Results
Expression Profiles of OsNRAMP3 in Different Tissues of Rice
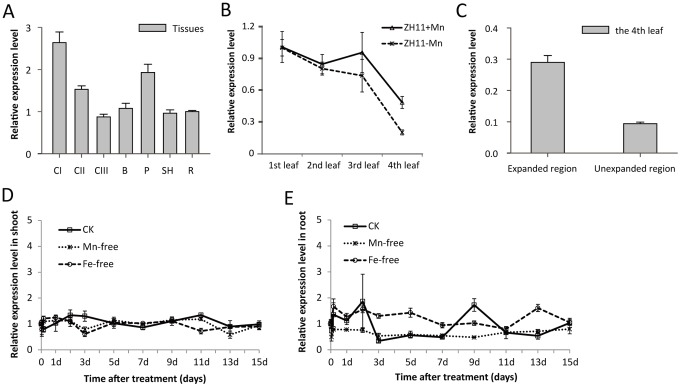
Seven different tissues from O. sativa L. ssp. japonica cv. Zhonghua 11 were used to investigate the expression pattern of OsNRAMP3. We found OsNRAMP3 transcript levels to be similar in most tissues and slightly higher in panicles and the first culm (Figure 1A). We next detected transcript levels of OsNRAMP3 in different leaves (Figure 1B). Four leaves from the same tiller were analyzed here, in which the first leaf was the oldest and the fourth leaf was youngest and partly wrapped in sheath. Interestingly, the expression level of OsNRAMP3 increased with leaf age under both Mn-sufficient and Mn-deficient conditions. In addition, the fourth leaf showed a differential expression pattern of clearly lower OsNRAMP3 expression in unexpanded compared to expanded regions (Figure 1C).
Figure 1. Expression analysis of OsNRAMP3 by real-time RT-PCR.
(A) The relative expression level of OsNRAMP3 in different rice tissues. C1: first culm; CII: second culm; CIII: third culm; B: leaf blade; P: panicle; SH: leaf sheath; R: root. (B) Transcripts of OsNRAMP3 of different leaves under different Mn conditions. The leaves detected here were sampled from the same tiller of rice plants – with the first leaf the oldest and the fourth leaf the youngest and partly wrapped in the leaf sheath. The plants were cultivated hydroponically under normal conditions for two weeks and then shifted to Mn-replete or Mn-free conditions for an additional one week. ZH11: cv. Zhonghua 11. (C) Expression analysis of OsNRAMP3 in different regions of the fourth leaf. (D and E) Kinetics of the response of OsNRAMP3 to Mn or Fe deficiency in shoot (D) and root (E) of rice plants. The plants were cultivated hydroponically under normal conditions for two weeks and then shifted to different conditions for treatment. Data are means ± SD (n = 3).
The NRAMP family genes in Arabidopsis are always induced by Fe or Mn deficiency [5], [35]. Time-course experiments under Mn and Fe deficiency were used to test the response of OsNRAMP3 to Mn and Fe deficiency, respectively – real-time PCR revealed that OsNRAMP3 was little affected by Mn or Fe deficiency (Figure 1D and 1E). In addition, expression did not respond to different Mn supplies (Figure S1).
Identifying the OsNRAMP3 Knockout Line
OsNRAMP3 contains 13 exons and 12 introns (Figure 2A), encoding a protein with 550 amino acids. To investigate the biological function of OsNRAMP3, we obtained a T-DNA insertion line from POSTECH [29]. The T-DNA was inserted into the twelfth exon. Transcripts of OsNRAMP3 could not be detected in the line (Figure 2B), suggesting it was a knockout mutant of OsNRAMP3.
Figure 2. Identification of the knockout line of OsNRAMP3.
(A) The structure of OsNRAMP3. OsNRAMP3 contains 13 exons and 12 introns, and a T-DNA inserted into the 12th exon of osnramp3 mutant plants. (B) The determination of accumulation of OsNRAMP3 in osnramp3 plants by RT-PCR. (C) Phenotypic analysis of knockout line of OsNRAMP3. The plants were cultivated hydroponically under normal conditions for two weeks and then shifted to different Mn supplies for an additional two weeks. Three levels of Mn were applied: 0.08 µM (a), 8 µM (b) and 800 µM (c). The general condition for growth of plants in the three Mn treatments was photographed (d). Careful observations were performed on roots (e) and leaves (f) of wild type and osnramp3 plants at 0.08 µM Mn supply. The red arrow indicates the necrotic area that appeared in osnramp3 roots. In (f), the left leaves are from wild type and the right leaves from osnramp3; 1–4 leaves were from the same tillers, with 1-leaf the oldest and 4-leaf the youngest.
As NRAMP genes always play important roles in transporting Mn, we firstly investigated the response of osnramp3 plants (the knockout line of OsNRAMP3) to different Mn supplies (Figure 2C, panels a–c). Interestingly, when plants were cultivated at low Mn supply (0.08 µM), osnramp3 plants showed obviously reduced growth compared with wild type. Careful observation revealed serious necrosis in young leaves and roots tips of osnramp3 plants, but not in older leaves (Figure 2C, panels d–f).
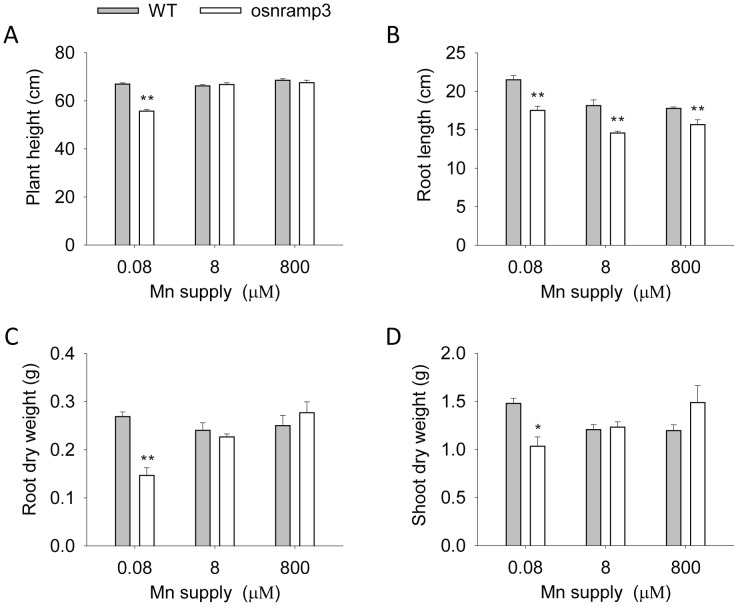
After harvest, the plant height, root length and dry weight of samples were recorded. At low Mn (0.08 µM) condition, the plant heights and root lengths of osnramp3 showed obvious decreases (Figure 3A and 3B); and dry weights of the roots and shoots in osnramp3 were 54 and 70% of wild type, respectively (Figure 3C and 3D), which confirmed the observed phenotype (Figure 2). When supplied with medium (8 µM) or high (800 µM) Mn, root of osnramp3 plants were shorter than that of wild type but there were no differences in plant height or dry weight.
Figure 3. Effects of different Mn supplies on the growth of knockout plants of OsNRAMP3.
The plants were cultivated hydroponically under normal conditions for two weeks and then shifted to three different Mn supplies for an additional two weeks. The plant heights (A) and root lengths (B) were investigated. Then the roots (C) and shoots (D) were harvested separately and their dry weights recorded. Data are means ± SD (n = 15). One and two asterisks indicate values that are significantly different from wild type at P<0.05 and P<0.01, respectively (by t-test).
Metal Determination in Roots and Shoots
Since the knockout line of OsNRAMP3 was sensitive to low Mn, we determined the Mn contents of roots and shoots of osnramp3 and wild-type plants. At a 0.08 µM Mn supply, both roots and shoots of the knockout line accumulated significantly more Mn than the wild type (Figure 4A and 4B). There was no apparent difference in Mn contents between osnramp3 and wild-type plants at higher Mn supplies. Absence of OsNRAMP3 also did not affect magnesium (Mg), Ca, Fe, copper (Cu), zinc (Zn) and cadmium (Cd) contents of roots and shoots of rice plants cultivated under normal conditions (Figure S2).
Figure 4. Determination of Mn concentration in wild type and osnramp3 plants.
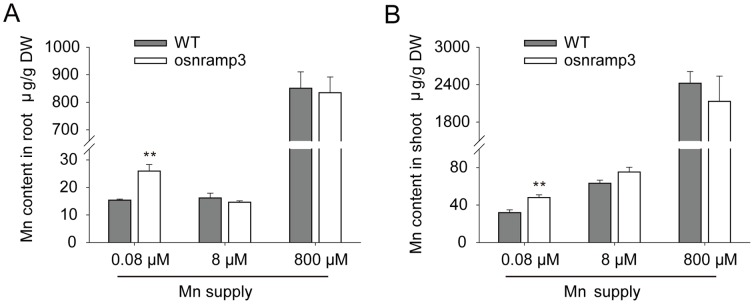
Elemental analysis was performed by ICP-MS on roots (A) and shoots (B) of wild type or osnramp3 plants grown for two weeks at three different Mn supplies after two weeks of normal conditional cultivation by hydroponics. Data are means ± SD (n = 3). One and two asterisks indicate values that are significantly different from wild type at P<0.05 and P<0.01, respectively (by t-test).
Knockout of OsNRAMP3 Resulted in Disturbed Distribution of Mn in Leaves
It seemed contradictory that osnramp3 mutant plants possessed higher Mn contents in roots and shoots but showed reduced growth under low Mn conditions compared with wild-type plants. To determine how this occurred, we next examined the distribution of Mn contents in different leaves for 0.08 or 8 µM Mn supplies. In general, Mn contents increased with leaf age in the treated and control plants. However, at a 0.08 µM Mn supply, osnramp3 plants showed significantly higher Mn contents in the first and second leaves than wild-type plants; but there were no significant differences in third and fourth leaves (Figure 5A). Under 8 µM Mn conditions, higher Mn contents were also observed in first leaves of osnramp3 compared with wild-type plants (Figure 5B); however, this difference was not observed in other leaves. The results here revealed that the absence of OsNRAMP3 in rice caused the Mn accumulation in older leaves, suggesting that OsNRAMP3 was important for Mn distribution in leaves.
Figure 5. Metal analysis of different leaves in wild type and osnramp3 plants.
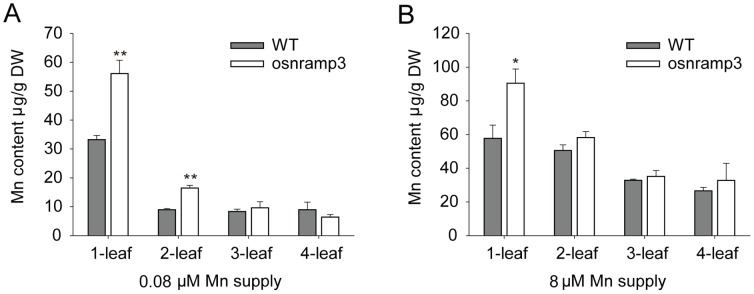
Different leaves were sampled from wild type and osnramp3 plants grown for two weeks at 0.08 (A) or 8 (B) µM Mn supplies after two weeks of normal conditional cultivation by hydroponics and elemental analysis was performed by ICP-MS. The 1–4 leaves were harvested from the same tiller of wild type or osnramp3 plants, and represented leaves from oldest to youngest respectively. Data are means ± SD (n = 3). One and two asterisks indicate values are significantly different from wild type at P<0.05 and P<0.01, respectively (by t-test).
In addition, the contents of Ca, Fe and Cd in older leaves were slightly, but not significantly, higher than that of wild type (Figure S3). There were no differences in Mg, Cu and Zn contents in corresponding leaves between wild type and osnramp3 plants.
Expression of OsNRAMP3 was Specific to Vascular Bundles
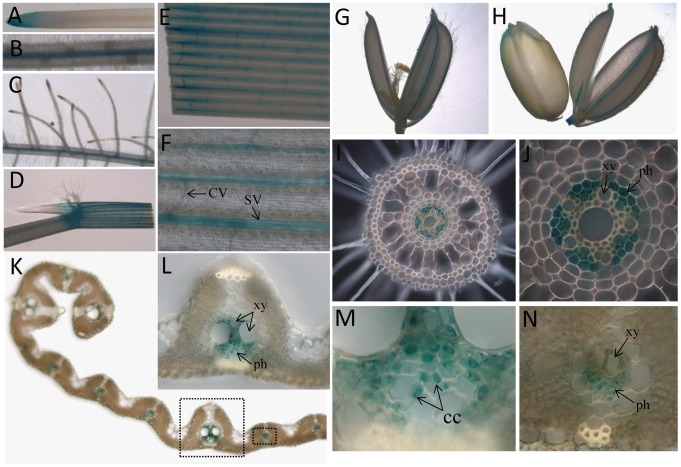
To further investigate the expression pattern of OsNRAMP3 in different tissues, a 2106-bp promoter region of OsNRAMP3 was used to direct beta-glucuronidase (GUS) expression in rice plants. GUS activity was analyzed for different rice tissues. Although there was high GUS activity in various tissues, such as roots (Figure 6A, 6B and 6C), ligules and sheaths of leaves (Figure 6D), leaf blades (Figure 6E and 6F) and grain hulls (Figure 6G), it was specific to vascular bundles of these tissues. In addition, GUS activity was also found in root tips and dorsal vascular bundles of endosperm (Figure 6A and 6H). To determine the exact location of GUS activity, we made transverse sections of roots and leaves after GUS staining. High-magnification observation of roots showed that GUS activity was specific in phloem cells (Figure 6I and 6J). In leaves, GUS activity was mainly detected in phloem cells as also for roots (Figure 6K, 6L, 6M and 6N). An enlarged view of the phloem region revealed particularly strong GUS activity in companion cells (Figure 6M). In addition, some parenchyma cells between phloem and xylem also showed GUS activity in large vascular bundles (Figure 6L).
Figure 6. Histochemical staining of GUS activity in rice plants transformed with the construct OsNRAMP3-promoter:GUS.
(A) Root tip; (B) mature root at 2 cm from tip; (C) lateral root; (D) leaf sheath, ligule and auricle; (E) leaf blade; (F) partly enlarged view of E; (G) hull at heading stage; (H) endosperm and hull at 25 d after heading; (I) transverse section of root, as described in B; (J) high-magnification of I; (K) transverse section of leaf blade, as described in E; (L) detail of a large vascular bundle from K; (M) enlarged view of phloem region of L; and (N) high-magnification of a small vascular bundle from K. All samples except hulls and endosperm were harvested from rice plants grown hydroponically for three weeks under normal conditions. CV: commissural vein; SV: small vascular bundle; LV: large vascular bundle; xy: xylem; ph: phloem; cc: companion cells.
OsNRAMP3 Encodes a Plasma Membrane-localized Protein
To study the subcellular localization of the OsNRAMP3 protein, a vector expressing an OsNRAMP3–GFP fusion under the control of cauliflower mosaic virus (CaMV) 35S promoter was constructed and transformed into Arabidopsis protoplasts. The OsNRAMP3–GFP expression in the protoplasts was examined with confocal microscopy 16 h after transformation. The fluorescence showed that OsNRAMP3 was a plasma membrane-localized protein (Figure 7).
Figure 7. Subcellular localization of OsNRAMP3 protein.
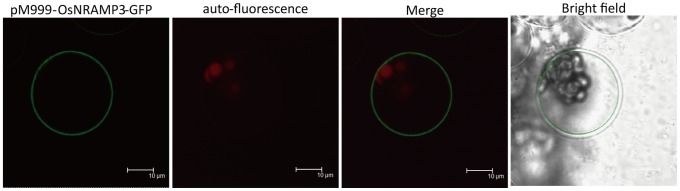
Subcellular localization of OsNRAMP3 protein was determined in Arabidopsis protoplasts. The confocal images were acquired using a confocal laser scanning microscope (TCS SP2; Leica).
OsNRAMP3 is a Functional Mn-influx Transporter
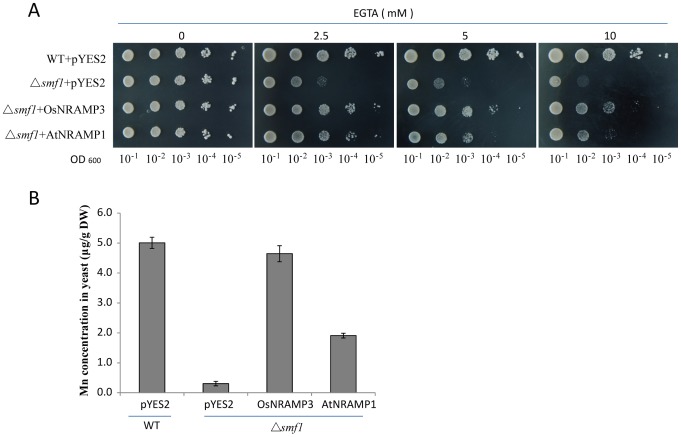
To determine the Mn transport activity of OsNRAMP3, we expressed OsNRAMP3 in yeast mutant strain Δsmf1. Here AtNRAMP1 was used as a positive control, as AtNRAMP1 has been shown to be a functional Mn transporter and can retrieve the growth of Δsmf1 under Mn-limited conditions [5], [36]. The growth of yeast strains Δsmf1 and its wild type transformed with pYES2 (negative control), OsNRAMP3 or AtNRAMP1 were analyzed on Mn-limited medium which controlled by different concentration of ethylene glycol tetraacetic acid (EGTA). The results showed that OsNRAMP3 significantly rescued the growth of Δsmf1 when Mn was limited (Figure 8A), suggesting that OsNRAMP3 possessed high activity for transporting Mn. To further confirm this result, metal concentrations were analyzed in these strains. When OsNRAMP3 was expressed in yeast strain Δsmf1, the Mn contents were almost retrieved to the level of wild type (Figure 8B), strongly indicating that OsNRAMP3 was a Mn-influx transporter.
Figure 8. Complementation assay of OsNRAMP3 on Mn-uptake deficient yeast mutant strain.
(A) Yeast mutant Δsmf1 and its wild type (WT) cells containing pYES2 (vector), OsNRAMP3 or AtNRAMP1 (positive control) grown on synthetic defined (SD)-Ura plates with different EGTA supplies and 2% galactose. (B) Metal determination in Δsmf1 and its wild-type cells containing pYES2, OsNRAMP3 or AtNRAMP1 grown in liquid SD-Ura culture with 0.2 µM Mn supplies and 2% galactose. Data are means ± SD (n = 4).
Discussion
OsNRAMP3 is a Vascular Bundle-located Mn-influx Transporter Involved in Mn Translocation
NRAMP family genes were well known to be related to Mn transport and many of them which encoded plasma-membrane targeted proteins acted as high-affinity transporters to absorb Mn from outside: such as SMF1 from yeast, AtNRAMP1 from Arabidopsis and OsNRAMP5 from rice [4]–[6]. In the present study, we identified OsNRAMP3, another NRAMP gene from rice, with the function of Mn transport. We demonstrated that knockout of OsNRAMP3 resulted in serious reduction in growth of rice plants under low Mn conditions and this reduction could be rescued by higher Mn supply (Figure 2 and 3). Indeed, the contents and translocation of Mn in rice plants were obviously affected by the knockout of OsNRAMP3 (Figure 4 and 5). Consistent with a role of OsNRAMP3 in the Mn translocation are the following findings: (1) OsNRAMP3 encoded a plasma membrane protein, based on the GFP fluorescence from OsNRAMP3–GFP fusion protein transformed into Arabidopsis protoplasts (Figure 7); (2) OsNRAMP3 was an influx Mn transporter, at least in yeast, based on the fact that OsNRAMP3 can rescue the growth and increase Mn contents of yeast when expressed in yeast mutant Δsmf1 (Figure 8); and (3) the expression of OsNRAMP3 was specific to vascular bundles, especially in companion cells of phloem, based on the GUS staining analysis (Figure 6).
Possible Mechanisms of OsNRAMP3 Involvement in Mn Translocation
In general, proteins encoded by NRAMP genes can be divided into two groups: plasma membrane-targeted and intracellular membrane-located proteins. The plasma membrane-targeted proteins from the NRAMP family contribute to uptake of Mn from medium as described above [5], [6]. The intracellular membrane-located proteins always act to retrieve Mn from vacuoles or other organelles: such as AtNRAMP3 and AtNRAMP4 from Arabidopsis [7], TcNRAMP3 and TcNRAMP4 from Thlaspi caerulescens [8], and LeNRAMP1 and LeNRAMP3 from tomato [9]. In this study, OsNRAMP3 showed difference: OsNRAMP3 encoded a plasma membrane protein (Figure 7) but was not involved in Mn uptake. The knockout mutant of OsNRAMP3 was highly sensitive to Mn deficiency (Figure 2C), which were similar to the characteristics of OsNRAMP5 described by Sasaki et al. [6]. However, OsNRAMP3 expression was restricted to vascular bundles rather than the exodermis and endodermis of roots (Figure 6), indicating a different function from OsNRAMP5. In addition, knockout of OsNRAMP3 resulted in Mn contents increasing in roots and shoots of osnramp3 mutant plants (Figure 4), suggesting that OsNRAMP3 was involved in Mn translocation but not Mn uptake.
Young leaves of osnramp3 mutant plants appeared to be withered compared with wild-type plants under low Mn conditions; and the withered appearance could be retrieved by higher Mn supply (Figure 2C), suggesting that the withered leaves of osnramp3 were deficient in the Mn required to maintain growth. However, osnramp3 plants accumulated more Mn in both roots and shoots compared with wild-type plants (Figure 4), which seemed contradictory. Most probably as feedback of signals of lacking Mn in withered leaves, the Mn uptake system of osnramp3 plants was enhanced to absorb more Mn from outside, which caused increased Mn contents in roots and shoots of osnramp3 compared with wild type. Consistent with that, further studies showed that the Mn content of each leaf of osnramp3 plants was not lower than corresponding leaves of wild-type plants (Figure 5A). This suggested that the symptoms of lacking Mn in withered leaves were caused by disturbed distribution of Mn rather than general Mn shortage. GUS analysis in leaves showed that OsNRAMP3 was specifically expressed in phloem cells and some parenchyma cells between phloem and xylem (Figure 6M and 6N), implying that the disturbed distribution of Mn was probably due to sequestrating Mn into vascular bundles of osnramp3 plants. As OsNRAMP3 is an influx Mn transporter, and parenchyma cells in vascular bundles always operate the loading or unloading of ion for xylem [37], [38], the Mn transported via xylem was probably unable to leave the xylem due to the absence of OsNRAMP3 in knockout lines. However, this suggestion requires more supporting evidence.
Phloem always plays important roles in remobilization of various ions in shoots, such as Zn, Fe, Cu and Mn [39]–[41]. In barley, it was reported that the transpiration stream has little effect on Mn translocation to the youngest leaf, whereas the strong effect on Mn translocation in older leaves [42], implying that major Mn concentration in young leaf is probably transported through the phloem. In the present study, OsNRAMP3 encoded a Mn-influx transporter and was mainly expressed in phloem cells, suggesting that OsNRAMP3 may contribute to remobilization of Mn in shoots. The increased Mn contents in osnramp3 plants mainly occurred in older not younger leaves under low Mn conditions (Figure 5A). Coincidentally, in rice plants, the expression of OsNRAMP3 in leaves slightly increased with leaf aging (Figure 1B). This implied that OsNRAMP3 played an important role in leaf Mn distribution. Together with the phenotype that only young leaves of osnramp3 plants were sensitive to Mn deficiency, the results here strongly suggested that OsNRAMP3 operated the Mn remobilization from old to young leaves via phloem cells.
In rice, OsYSL2 was reported to be a phloem-located metal transporter responsible for long-distance transport of Fe and Mn, especially in seeds [16], [41]. Similarly to OsYSL2, the expression of OsNRAMP3 was also detectable in vascular bundles of endosperm and hulls, suggesting that OsNRAMP3 was required in these tissues. Confirming whether OsNRAMP3 is involved in transporting Mn into seeds requires more detailed study. In roots, the expression of OsNRAMP3 was specific to phloem cells and root tip cells (Figure 6A and 6J). Coincidently, the root tip of knockout plants of OsNRAMP3 became necrotic under low Mn conditions (Fig. 2C, panel e). We investigated the Mn contents in different root regions at different distances from the root tip in rice cv. Zhonghua 11 (data not shown). The root tip possessed the highest Mn contents of the different root regions, suggesting that Mn played an important role in growth of root tips. However, because of serious necrosis in root tips of osnramp3 plants under low Mn conditions, it is difficult to determine the Mn contents in osnramp3 root tips. To determine whether OsNRAMP3 is related to root tip Mn contents and how OsNRAMP3 affects growth of root tips will require further study.
In conclusion, OsNRAMP3 is a vascular bundle-localized influx Mn transporter that is involved in Mn translocation in rice plants. Knockout of OsNRAMP3 disturbed the transport of Mn into young leaves from old leaves and vascular bundle cells, resulting in high sensitivity to low Mn conditions. It is worth noting that when we finished our experiments, we noticed a related but independent work by Yamaji et al. [43], in which OsNRAMP3 was also described as a Mn transporter contributing to Mn distribution in shoots.
Supporting Information
Response of OsNRAMP3 expression to different Mn concentrations. The plants were cultivated hydroponically under normal conditions for two weeks and then shifted to different Mn supplies for an additional two weeks, and then harvested for RNA extraction.
(TIF)
Concentration of various metals in roots and shoots of wild type and osnramp3 plants. The plants were cultivated hydroponically under normal conditions for four weeks. Roots (A) and shoots (B) were harvested separately. Before sampling, roots were washed twice with deionized water. All samples were dried at 80°C for 3 d.
(TIF)
The distribution of various metals in different leaves. Mg (A), Ca (B), Fe (C), Cd (D), Cu (E) and Zn (F) contents were determined in different leaves of wild type and osnramp3 plants cultivated under normal conditions for four weeks. The 1–4 leaves were harvested from the same tiller of wild type or osnramp3 plants, and represented the oldest to youngest leaves, respectively.
(TIF)
Acknowledgments
We thank Professor Jian Xu for kindly providing the pM999–GFP vector.
Funding Statement
This work was supported by grants from the National High Technology Research and Development Program of China (2012AA10A303 and 2012AA10A304), the National Basic Research Program of China (2011CB100304), the National Natural Science Foundation of China (31172017) and transgenic project 2013ZX08001005. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marschner H, Marschner P (2012) Marschner's mineral nutrition of higher plants. London; WalthamMA: Elsevier/Academic Press. xv, 651 p. [Google Scholar]
- 2. Hebbern CA, Pedas P, Schjoerring JK, Knudsen L, Husted S (2005) Genotypic differences in manganese efficiency: field experiments with winter barley (Hordeum vulgare L.). Plant and Soil 272: 233–244. [Google Scholar]
- 3. Pittman JK (2005) Managing the manganese: molecular mechanisms of manganese transport and homeostasis. New Phytol 167: 733–742. [DOI] [PubMed] [Google Scholar]
- 4. Supek F, Supekova L, Nelson H, Nelson N (1996) A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci U S A 93: 5105–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cailliatte R, Schikora A, Briat JF, Mari S, Curie C (2010) High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sasaki A, Yamaji N, Yokosho K, Ma JF (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24: 2155–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lanquar V, Ramos MS, Lelievre F, Barbier-Brygoo H, Krieger-Liszkay A, et al. (2010) Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol 152: 1986–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oomen RJ, Wu J, Lelievre F, Blanchet S, Richaud P, et al. (2009) Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens . New Phytol 181: 637–650. [DOI] [PubMed] [Google Scholar]
- 9. Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P (2003) Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. J Biol Chem 278: 24697–24704. [DOI] [PubMed] [Google Scholar]
- 10. Ishimaru Y, Takahashi R, Bashir K, Shimo H, Senoura T, et al. (2012) Characterizing the role of rice NRAMP5 in Manganese, Iron and Cadmium Transport. Sci Rep 2: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40: 37–44. [DOI] [PubMed] [Google Scholar]
- 12. Milner MJ, Seamon J, Craft E, Kochian LV (2013) Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J Exp Bot 64: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eckhardt U, Mas Marques A, Buckhout TJ (2001) Two iron-regulated cation transporters from tomato complement metal uptake-deficient yeast mutants. Plant Mol Biol 45: 437–448. [DOI] [PubMed] [Google Scholar]
- 14. Lopez-Millan AF, Ellis DR, Grusak MA (2004) Identification and characterization of several new members of the ZIP family of metal ion transporters in Medicago truncatula . Plant Mol Biol 54: 583–596. [DOI] [PubMed] [Google Scholar]
- 15. Pedas P, Ytting CK, Fuglsang AT, Jahn TP, Schjoerring JK, et al. (2008) Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiol 148: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, et al. (2004) OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J 39: 415–424. [DOI] [PubMed] [Google Scholar]
- 17. Sasaki A, Yamaji N, Xia J, Ma JF (2011) OsYSL6 is involved in the detoxification of excess manganese in rice. Plant Physiol 157: 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hirschi KD, Zhen RG, Cunningham KW, Rea PA, Fink GR (1996) CAX1, an H+/Ca2+ antiporter from Arabidopsis . Proc Natl Acad Sci U S A 93: 8782–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ (2000) Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol 124: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schaaf G, Catoni E, Fitz M, Schwacke R, Schneider A, et al. (2002) A putative role for the vacuolar calcium/manganese proton antiporter AtCAX2 in heavy metal detoxification. Plant Biology 4: 612–618. [Google Scholar]
- 21. Edmond C, Shigaki T, Ewert S, Nelson MD, Connorton JM, et al. (2009) Comparative analysis of CAX2-like cation transporters indicates functional and regulatory diversity. Biochemical Journal 418: 145–154. [DOI] [PubMed] [Google Scholar]
- 22. Kamiya T, Akahori T, Maeshima M (2005) Expression profile of the genes for rice cation/H+ exchanger family and functional analysis in yeast. Plant and Cell Physiology 46: 1735–1740. [DOI] [PubMed] [Google Scholar]
- 23. Wu ZY, Liang F, Hong BM, Young JC, Sussman MR, et al. (2002) An endoplasmic reticulum-bound Ca2+/Mn2+ pump, ECA1, supports plant growth and confers tolerance to Mn2+ stress. Plant Physiol 130: 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mills RF, Doherty ML, Lopez-Marques RL, Weimar T, Dupree P, et al. (2008) ECA3, a Golgi-localized P2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis . Plant Physiol 146: 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li XY, Chanroj S, Wu ZY, Romanowsky SM, Harper JF, et al. (2008) A distinct endosomal Ca2+/Mn2+ pump affects root growth through the secretory process. Plant Physiol 147: 1675–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Delhaize E, Kataoka T, Hebb DM, White RG, Ryan PR (2003) Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15: 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, et al. (2007) A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J 51: 198–210. [DOI] [PubMed] [Google Scholar]
- 28. Peiter E, Montanini B, Gobert A, Pedas P, Husted S, et al. (2007) A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc Natl Acad Sci U S A 104: 8532–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22: 561–570. [DOI] [PubMed] [Google Scholar]
- 30. Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- 31. Ye RJ, Zhou F, Lin YJ (2012) Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L ssp.). Plant Cell Reports 31: 1159–1172. [DOI] [PubMed] [Google Scholar]
- 32. Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282. [DOI] [PubMed] [Google Scholar]
- 33. Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo Journal 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory Manual for Physiological Studies of Rice, 3rd ed. International Rice Research Institute, Manila.
- 35. Thomine S, Lelievre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H (2003) AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant Journal 34: 685–695. [DOI] [PubMed] [Google Scholar]
- 36. Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci U S A 97: 4991–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu LL, Tian SK, Zhang J, Yang X, Labavitch JM, et al. (2013) Efficient xylem transport and phloem remobilization of Zn in the hyperaccumulator plant species Sedum alfredii . New Phytologist 198: 721–731. [DOI] [PubMed] [Google Scholar]
- 38. Yamaguchi N, Ishikawa S, Abe T, Baba K, Arao T, et al. (2012) Role of the node in controlling traffic of cadmium, zinc, and manganese in rice. J Exp Bot 63: 2729–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nishiyama R, Kato M, Nagata S, Yanagisawa S, Yoneyama T (2012) Identification of Zn-Nicotianamine and Fe-2'-Deoxymugineic Acid in the Phloem Sap from Rice Plants (Oryza sativa L.). Plant and Cell Physiology 53: 381–390. [DOI] [PubMed] [Google Scholar]
- 40. Zheng LQ, Yamaji N, Yokosho K, Ma JF (2012) YSL16 Is a Phloem-Localized Transporter of the Copper-Nicotianamine Complex That Is Responsible for Copper Distribution in Rice. Plant Cell 24: 3767–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ishimaru Y, Masuda H, Bashir K, Inoue H, Tsukamoto T, et al. (2010) Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J 62: 379–390. [DOI] [PubMed] [Google Scholar]
- 42. Tsukamoto T, Nakanishi H, Kiyomiya S, Watanabe S, Matsuhashi S, et al. (2006) Mn-52 translocation in barley monitored using a positron-emitting tracer imaging system. Soil Science and Plant Nutrition 52: 717–725. [Google Scholar]
- 43. Yamaji N, Sasaki A, Xia JX, Yokosho K, Ma JF (2013) A node-based switch for preferential distribution of manganese in rice. Nat Commun 4: 2442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Response of OsNRAMP3 expression to different Mn concentrations. The plants were cultivated hydroponically under normal conditions for two weeks and then shifted to different Mn supplies for an additional two weeks, and then harvested for RNA extraction.
(TIF)
Concentration of various metals in roots and shoots of wild type and osnramp3 plants. The plants were cultivated hydroponically under normal conditions for four weeks. Roots (A) and shoots (B) were harvested separately. Before sampling, roots were washed twice with deionized water. All samples were dried at 80°C for 3 d.
(TIF)
The distribution of various metals in different leaves. Mg (A), Ca (B), Fe (C), Cd (D), Cu (E) and Zn (F) contents were determined in different leaves of wild type and osnramp3 plants cultivated under normal conditions for four weeks. The 1–4 leaves were harvested from the same tiller of wild type or osnramp3 plants, and represented the oldest to youngest leaves, respectively.
(TIF)