SUMMARY
The E. coli replisome stalls transiently when it encounters a lesion in the leading-strand template, skipping over the damage by reinitiating replication at a new primer synthesized downstream by the primase. We report here that template unwinding and lagging-strand synthesis continue downstream of the lesion at a reduced rate after replisome stalling, that one replisome is capable of skipping multiple lesions, and that the rate limiting steps of replication restart involve the synthesis and activation of the new primer downstream. We also find little support for the concept that polymerase uncoupling, where extensive lagging-strand synthesis proceeds downstream in the absence of leading-strand synthesis, involves physical separation of the leading-strand polymerase from the replisome. Instead, our data indicate that extensive uncoupled replication likely results from a failure of the leading-strand polymerase still associated with the DNA helicase and the lagging-strand polymerase that are proceeding downstream to reinitiate synthesis.
INTRODUCTION
The DNA replication machinery (replisome) faces considerable challenges during the process of chromosome duplication. Protein road blocks, in the form of frozen protein complexes, or stalled RNA polymerases, are known to stall and/or derail the replication machinery at high frequency (Gupta et al., 2013; Merrikh et al., 2011). The replisome must also negotiate a multitude of structurally unrelated DNA lesions that are generated during normal cell growth. Consequently the replisome has evolved many strategies — tailored to the specific type of barrier — to ensure that chromosome duplication is rapidly and accurately completed.
The Escherichia coli replisome is a large multi-subunit molecular machine that possesses all of the enzymatic activities required to catalyze rapid and processive DNA replication (reviewed in (McHenry, 2011)). The chromosome is unwound by the hexameric DnaB helicase translocating 5′ → 3′ on the lagging-strand template (LeBowitz and McMacken, 1986). The τ subunit of the DnaX clamp loader complex plays a central role in anchoring replisome components by binding both the leading- and lagging-strand DNA polymerases and DnaB (Gao and McHenry, 2001; Kim et al., 1996a, b). On undamaged DNA, the leading strand is synthesized continuously, whereas the lagging strand is synthesized discontinuously in 1-2 kb long Okazaki fragments that are initiated by repeated priming by the primase, DnaG, and subsequent β-clamp assembly catalyzed by the DnaX complex.
Provided that DnaB translocation is not impeded, lagging-strand lesions are efficiently bypassed (McInerney and O'Donnell, 2004; Nelson and Benkovic, 2010); the lagging-strand polymerase simply cycles to a new RNA primer downstream of the lesion, leaving behind a single-strand (ss)DNA gap that can be filled subsequently by either RecA-dependent recombination pathways or specialized translesion DNA polymerases (Kuzminov, 1999). The fate of the replisome following a collision with a leading-strand lesion is less clear. Following UV irradiation of E. coli cells both proficient and deficient in nucleotide excision repair, DNA replication was inhibited (Rupp and Howard-Flanders, 1968; Setlow et al., 1963; Swenson and Setlow, 1966). Consistent with these observations, Rudolph et al., (2007) showed that movement of all pre-existing replication forks was delayed following UV exposure. These results demonstrate that UV irradiation significantly inhibits the progress of E. coli replication forks in vivo and, assuming that lagging-strand lesions are efficiently bypassed, are consistent with a model whereby leading-strand lesions block replication fork progression.
In contrast, the leading-strand reinitiation model postulates that leading-strand synthesis can be reinitiated downstream of template damage by a priming event on the leading-strand template (Rupp, 1996). This model was born out of the observations that replication does not stop completely following UV-irradiation, and that the nascent DNA subsequently synthesized is considerably shorter than unirradiated controls and contains single-stranded gaps (Iyer and Rupp, 1971; Rupp and Howard-Flanders, 1968). The first mechanistic evidence that leading-strand priming could occur outside of the origin of replication came from experiments conducted on model replication fork substrates lacking a nascent leading strand. Origin-independent replisome loading via the PriC-pathway (reviewed in (Heller and Marians, 2006b)) resulted in the formation of a functional replisome that catalyzed coupled leading- and lagging-strand synthesis via a de novo leading-strand priming event (Heller and Marians, 2006a). Recent experiments conducted in vitro on a DNA template containing a single, site-specific leading-strand cyclobutane pyrimidine dimer (CPD), revealed that the replisome stalls transiently at, or close to the site of damage. However, despite the absence of any of the known proteins required for origin-independent replisome assembly (Heller and Marians, 2006b), replication was not permanently arrested and, following a lag, coupled synthesis was seen to resume downstream of the lesion (Yeeles and Marians, 2011) with the net effect being that the replisome jumped over the leading-strand lesion.
Multiple reactions must be coordinated at the replication fork to enable coupled replication (i.e., coordinated leading- and lagging-strand synthesis catalyzed by one replisome) to be reinitiated. (1) Following collision with the damage, sufficient ssDNA must be exposed on the leading-strand template to enable primer synthesis to occur. (2) Recruitment of DnaG, presumably via an interaction with DnaB, and subsequent primer synthesis. (3) Assembly of a new β-clamp around the primer-template. And (4) recruitment of a leading-strand polymerase and the resumption of leading- and lagging-strand synthesis. In this study we have sought to investigate how and when these processes occur and the relative contributions that they make to the overall kinetics of the leading-strand lesion skipping reaction that we reported previously (Yeeles and Marians, 2011). Our data reveal that the replisome stalls, or slows significantly, a short distance downstream of the lesion, with lagging-strand synthesis frequently continuing beyond the site of damage. The data also suggest that the rate limiting steps for leading-strand reinitiation involve primer synthesis and the subsequent assembly of the new β clamp, and that once replication is reinitiated, normal replication rates are resumed and the replisome is capable of responding to a second lesion in the leading-strand template. We also report the unexpected finding that a DnaX complex lacking the domains required to interact with DnaB can enhance the efficiency of leading-strand replication restart, a result that is consistent with β-clamp assembly being a key rate-limiting step in the leading-strand reinitiation pathway. Finally, we demonstrate that during polymerase “uncoupling,” when extensive lagging-strand synthesis occurs downstream of the lesion in the absence of leading-strand synthesis, the leading-strand polymerase appears to remain associated with the lagging-strand polymerase and the replication fork helicase.
RESULTS
DnaB and Lagging-strand Synthesis Progress Forward Slowly After the Replisome Encounters a Leading-strand Template Lesion
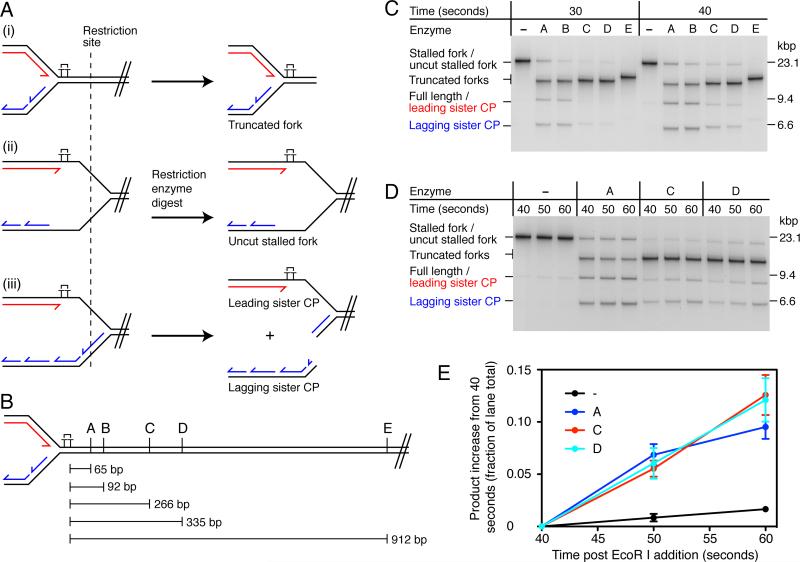
To investigate the complete structure of a stalled fork, we utilized the unidirectional replication system that we developed previously (Yeeles and Marians, 2011) to generate a population of stalled replication forks for subsequent analysis. Origin-dependent replication is initiated on oriC-containing plasmid templates (Figure S1A) in the absence of a topoisomerase to form early-replication intermediates (ERIs). Replisomes are then released synchronously from the ERIs by cleavage with the restriction enzyme EcoRI. The counter-clockwise-moving fork is blocked by the Tus:TerB complex, enabling progression of the clockwise-moving fork to be monitored. Reaction products are digested post-replicatively with PvuI to remove the origin and counter-clockwise-moving fork, leaving an essentially linear template. In this system, stalled replication forks are resolved to generate fully-replicated products (restarted leading strands and lagging strands) (Yeeles and Marians, 2011). To ensure that stalled forks were the predominant starting reaction species, reactions on the CPD-C template (Figure S2) were arrested at early time points (30 and 40 sec). Restriction enzyme mapping was used to investigate fork architecture. Three potential structures that could be generated were considered (Figure 1A). (i) The replisome could arrest in close proximity to the CPD, without extensive unwinding of the template downstream from the damage. (ii) DnaB-catalyzed template unwinding could continue beyond the lesion in the absence of both leading- and lagging-strand synthesis, generating regions of ssDNA on both template strands. (iii) Uncoupled replication beyond the CPD - where lagging-strand synthesis continues in the absence of leading-strand synthesis – would generate a region of ssDNA specifically on the leading-strand template. Each fork structure will give a distinctive cleavage pattern when digested post-replicatively with enzymes that cut a short distance downstream from the CPD (Figure 1A) and analysis with multiple enzymes that cleave at different positions will provide information about the location where the replisome stalls (Figure 1B).
Figure 1. Lagging-strand Synthesis and Template Unwinding Proceed Slowly After the Replisome Stalls at a Leading-strand Lesion.
(A) Illustration of replication fork structures that could be generated following fork stalling. (i) The replisome arrests at the site of damage and little template unwinding occurs downstream. (ii) Template unwinding continues downstream of the damage in the absence of leading- and lagging-strand synthesis. (iii) Template unwinding and lagging-strand synthesis continue downstream of the damage in the absence of leading-strand synthesis. Different fork architectures can be identified by restriction mapping with enzymes that are located downstream of the lesion. A single cleavage event on the lagging-strand sister will generate leading- and lagging-strand sister cleavage products, denoted leading sister CP and lagging sister CP.
(B) Schematic illustrating the positions relative to the CPD of the restriction sites used for fork mapping. Enzyme A, KpnI; B, PstI; C, BsaI; D, AhdI and E, ApaLI.
(C) Restriction mapping of stalled replication forks. Standard replication reactions were conducted using the CPD-C template (Figures 2A and S2) for the indicated times. Following quenching, stalled forks were digested with the indicated restriction enzymes and analyzed by native gel electrophoresis. Digested products were identified by two-dimensional electrophoresis (Figure S1B). Note that the leading sister CP migrates to a position indistinguishable from that of full-length products (fully replicated lagging strands and restarted leading strands).
(D) Pulse-chase reaction followed by restriction mapping. The experiment was conducted as in (C), except that a 25-fold molar excess of unlabelled dGTP (chase) was added 30-sec post EcoRI addition.
(E) Quantification of the data shown in panel (D). Data has been normalized by subtracting the initial value (40 sec) from the remaining time points. Error bars represent the standard error of the mean (SEM) from three independent experiments.
Restriction digests generated up to three novel cleavage products (Figure 1C, e.g., enzyme A), the patterns of which were influenced by both the restriction enzyme site location and the replication reaction time length. To identify these products, stalled forks were digested with enzyme A and analyzed by two-dimensional gel electrophoresis (Figure S1B). The product that migrated in the location of the original stalled fork contained both the leading-strand stall fragment and Okazaki fragments, demonstrating that it was a stalled replication fork resistant to enzyme A digestion (Figure 1A (ii)). The band migrating directly below the original fork, but above the position of full-length duplex DNA, also contained both leading- and lagging-strand products, identifying it as a truncated fork, where the downstream unreplicated region of the template had been removed (Figure 1A (i)). The 9.5 kbp band (leading sister CP) was composed exclusively of the nascent leading strand, whereas the faster migrating species of approximately 6.5 kbp (lagging sister CP) consisted mainly of Okazaki fragments. These species were likely generated by a single cleavage event on the lagging strand at the stalled fork (Figure 1A (iii)), liberating the lagging-strand sister, while the leading-strand sister remained associated with the downstream region of the template and therefore migrated in a position similar to that of full-length duplex DNA. We saw little evidence for the interconversion of replication fork structures by branch migration during the course of restriction digests, perhaps because of the replisome remaining resident on the templates. Consistent with this idea, removal of replication proteins prior to restriction digests resulted in the conversion of uncut forks to truncated forks (data not shown).
For reactions arrested 30-sec post EcoRI cleavage, only enzymes A and B produced significant amounts of leading- and lagging-sister cleavage products, whereas enzymes C, D and E generated truncated forks almost exclusively (Figure 1C, 30 sec). Thus the majority of replication forks either stalled, or significantly reduced in speed, such that they had not yet reached the enzyme C site 266 bp downstream from the CPD. The data also demonstrate that in the majority of cases where the replisome reached a restriction site, uncoupled replication (Figure 1A, Fork iii) was more prevalent than template unwinding in the absence of synthesis, as leading- and lagging-sister cleavage products were more prominent than uncut forks (Fork iI) for enzymes A and B. Comparison of the digest patterns at 30 and 40 sec revealed an increase in leading- and lagging-sister cleavage products at 40 sec for enzymes A, B, C, and D. Only enzyme E, which maps 912 bp downstream from the CPD, failed to generate such products when forks were arrested 40-sec post EcoRI cleavage.
We considered two potential explanations for the digest pattern changes observed between 30 and 40 sec. Because stalled-fork production was not saturated at 30 sec (data not shown), replisomes released later from ERIs — between 30 and 40 sec — might have arrested further downstream from the CPD, giving rise to the additional leading- and lagging-sister cleavage products. Alternatively, rather than the replisome coming to a complete halt following collision with the damage, uncoupled replication might have continued downstream from the damage before coupled synthesis was resumed by leading-strand repriming. To distinguish between these two possibilities, pulse-chase experiments were conducted where a 25-fold excess of unlabeled dGTP was added 30 sec post-EcoRI cleavage to prevent further incorporation of [α-32P]dGTP. This enabled the fate of replication forks stalled within the first 30 sec of the reaction to be monitored (Figures 1D and 1E). (The elevated dGTP concentration did not affect either the kinetics of the reaction or the distribution of products, Figure S3.) Aliquots were taken at 40, 50 and 60 sec post-EcoRI addition for restriction enzyme analysis. When stalled forks were digested with enzymes A, C, and D, both the leading- and lagging-sister cleavage products increased in intensity across the time courses (Figures 1D and E), and this increase appeared to be coupled to a decrease in the truncated stalled fork (Figure 1D, enzyme A). Crucially, for all three enzymes the increase in leading- and lagging-sister cleavage products was considerably greater (~5-fold) than the increase in full-length duplex products (fully-resolved replication products consisting of both lagging strands and restarted leading strands) for undigested samples (Figure 1E). As such, the digest pattern changes could not be accounted for by cleavage of fully-resolved replication products that were synthesised during the chase. Rather, we conclude they result from cleavage of forked structures generated by continued uncoupled replication occurring downstream from the CPD. This uncoupled replication appears to occur at a reduced rate compared to that reported for coupled DNA synthesis (500-1000 bp s−1), given that, although the population of replisomes have travelled at least a few hundred base pairs downstream from the CPD, they have not yet reached the end of the template (2.7 kb beyond the CPD, which would be replicated in 5-6 seconds at 500 bp s−1) during the 20-second chase.
Replication Rates Do Not Change Significantly Following the Reinitiation of Leading-strand Synthesis
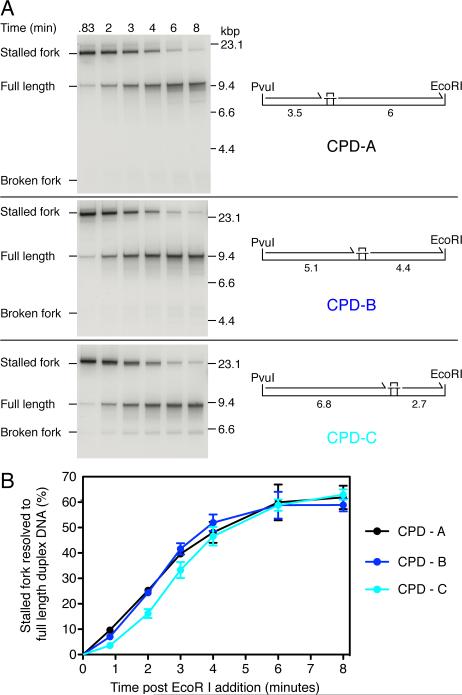
Full-length product formation on a single CPD-containing template was delayed by up to several minutes relative to an undamaged control template (Yeeles and Marians, 2011). The cause of this delay may simply result from replication occurring more slowly downstream from the CPD, delaying the arrival of the replisome at the distal end of the template. We estimate that replication rates would have to slow at least 10-fold to less than 50 bp s−1 to give rise to the observed lag. Alternatively, if normal replication rates are resumed following leading-strand reinitiation, the lag might be the consequence of a rate-limiting step, or series of steps, that occur prior to the reinitiation of leading-strand synthesis, for example, priming on the leading-strand template, or assembly of a new β clamp around the primed leading-strand template. To test these hypotheses, we constructed three single-CPD templates where the position of the CPD was varied relative to the EcoRI and PvuI sites (Figures 2A, S1A, and S2). Should the rate of DNA synthesis following leading-strand reinitation slow significantly enough to dictate the overall reaction kinetics (>10-fold), the kinetics by which full-length products are produced should vary significantly between the three templates. However, if normal replication rates are resumed following leading-strand reinitiation, similar overall kinetics would be expected, given that the maximum predicted difference in replication time for the CPD-distal region at a rate of 500 bp s−1 would be 6 seconds between CPD – A (6 kb) and CPD – C (2.7 kb).
Figure 2. The Kinetics of Full-length DNA Production Are Independent of the CPD Location.
(A) Pulse-chase analysis of replication on three templates where the location of the CPD was varied as indicated. Reactions were conducted under standard replication conditions with [α-32P]dGTP. Forty seconds post-EcoR I addition, a 25-fold excess (1 mM) of unlabelled dGTP was added to prevent further incorporation of labeled nucleotide and aliquots were withdrawn at the indicated times post-EcoR I cleavage. The maximum lengths (kbp) of the putative leading-strand stall and restart products are illustrated.
(B) Quantification of pulse-chase experiments in (A). Error bars represent the SEM from a minimum of three independent experiments.
Restart products were generated on all three templates (Figure S2B, denaturing). Furthermore, uncoupled products – where template unwinding and only lagging-strand synthesis continue to the distal end of the template – were not detected (Figure S2B, native), indicating that in almost all cases where the replisome reached the end of a template, leading-strand synthesis was reinitiated. Thus the location of the CPD does not significantly influence the efficiency of leading-strand reinitiation. To obtain more accurate kinetic information about the rate by which stalled forks are resolved to generate full-length products (lagging strands and restarted leading strands) pulse-chase experiments were conducted (Figure 2). The kinetics of full-length DNA production were very similar for the three templates tested, with product formation saturating approximately 4-5 minutes post EcoRI addition (Figure 2). No correlation was observed between the time taken to fully replicate a template and the length of the CPD distal region, demonstrating that the rate of DNA synthesis does not slow significantly following leading-strand reinitiation downstream from a CPD. The overall kinetics of the reaction appear therefore to be dictated by a rate limiting step(s) that occurs prior to the resumption of coupled DNA replication.
The Replisome Restarts Replication Downstream From a Second Leading-strand CPD
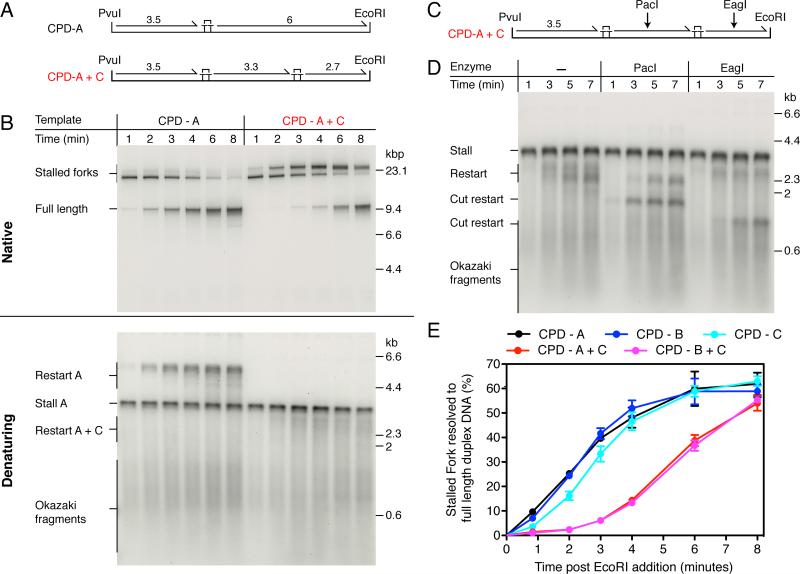
A key predication of the above model is that the presence of a second CPD in the leading-strand template should delay the arrival of a replisome at the distal end of the template, even if leading-strand synthesis is efficiently reinitiated downstream of a second lesion. To test this idea, two DNA templates were synthesized that each contained two site-specific leading-strand template lesions (Figures 3A and S4A). We first compared replication on the single damage CPD-A and the double damage CPD-A+C templates. One minute post EcoRI addition, the replication products were almost indistinguishable, with the products from both templates consisting predominantly of a stalled replication fork (Figure 3B). As the reactions proceeded, some clear differences became apparent. Instead of full-length duplex products being generated two minutes post EcoRI addition, as is the case for the CPD-A template, a novel product migrating more slowly than the original stalled fork was formed in reactions on the CPD-A+C template. This product was generated with kinetics very similar to those of full-length product formation with the CPD-A template and it migrated in an identical position to the stalled fork generated on the single-damage CPD-C template (Figure S2, CPD-C). The data can be explained if replication is restarted downstream from the first CPD, but the replisome then stalls again when it encounters the second CPD located further along the template. Remarkably — having reached a peak at approximately 4 min — the second stalled fork begins to be resolved and full-length duplex DNA products are formed. If leading-strand reinitiation is occurring downstream of both lesions, distinct restart products should be detectable in a denaturing gel. Restart products on the CPD-A template migrated as a smear up to a length of approximately 6 kb (Figures 3B and S2B, denaturing). This smear was entirely absent when a second leading-strand CPD was located downstream of CPD-A, suggesting that close to 100% of the double-damage template contained the second lesion. In contrast to the restart products on the CPD-A template, those generated on the CPD-A+C template migrated as a broad smear spanning a region from approximately 2-3 kb (Figure 3B, denaturing). To differentiate the restart products generated downstream of CPD-A and CPD-C, reactions were conducted with the CPD-A+C template and products were digested with enzymes that mapped to either the region between the two CPDs (PacI) or downstream of the second CPD (EagI) (Figures 3C and D). Digestion with PacI removed the upper portion of the restart smear and conversely EagI cleavage removed the bottom portion of the smear. This result is consistent with the CPD locations, as the distance between the two CPDs — where the PacI site is located — is 600 bp longer than the distance downstream of the second CPD. Cleavage with either enzyme generated uniform products (Figure 3D, Cut restart), likely due to removal of the heterogeneous 5′ ends of the restart products. The kinetics with which the two cut restart products were generated revealed that PacI-sensitive products were produced earlier in the reaction than EagI-sensitive products, while reciprocal kinetics were observed for resistant products, as would be expected if leading-strand reinitiation is occurring sequentially, first downstream of CPD-A, and then downstream of CPD-C.
Figure 3. The Replisome Can Reinitiate Leading-strand Synthesis Downstream from a Second Polymerization-blocking Lesion.
(A) Single- and double-damage replication templates. The maximum lengths (kbp) of the putative leading-strand stall and restart products are illustrated.
(B) Comparison of replication products generated from the single-damage template, CPD-A, and the double-damage template, CPD-A+C, under standard replication conditions.
(C) Illustration showing the position of the PacI and EagI restriction enzyme sites.
(D) Replication was conducted with the CPD-A+C template under standard conditions for the indicated times. To aid visualization of the shorter restart product downstream of CPD-C, [α-32P]dATP was substituted with [α-32P]dGTP, as the leading-strand template in this region is cytosine rich relative to the region between CPD-A and CPD-C. Following quenching, the reaction products were digested with the indicated enzymes prior to alkaline gel electrophoresis.
(E) Quantification of pulse chase experiments (Figures 2B and S5) comparing the rate of full-length product formation for the single- and double-damage templates. Error bars represent the SEM from a minimum of three independent experiments.
To compare the kinetics by which fully-resolved products were formed on the single and double-damage templates, we again utilized the pulse-chase assay (Figures 3E and S5). For both double-damage templates there was a significant lag prior to the formation of full-length duplex products compared to the single-damage templates. Whereas product formation began to plateau with the single-damaged templates after approximately 4 min, a plateau was reached at about 8 min post EcoRI cleavage with the double-damaged templates (Figures 3E). The data illustrate that the overall reaction kinetics are primarily influenced by the number of CPDs in the leading-strand template, rather than their location, and further highlight the significance of a rate limiting step(s) that occurs prior to the resumption of leading-stand synthesis.
Leading-strand Priming Plays a Critical Role in Dictating the Kinetics of Restart
Leading-strand priming is likely to be one of the earliest events that takes place at the stalled replication fork. As DnaG functions distributively during lagging-strand synthesis (Wu et al., 1992), we reasoned that the concentration of DnaG might modulate leading-strand reinitiation in some way. To test this hypothesis, we conducted DnaG titrations using the CPD-A and CPD-C templates (Figure 4). At low DnaG concentrations a significant proportion of the stalled forks persisted 6 min post EcoRI addition for both templates. As the concentration of DnaG increased, the proportion of stalled forks remaining at 6 min decreased, suggesting that the kinetics of stalled-fork resolution could be dependent on DnaG concentration (Figure 4B). To test this idea further, we compared replication of the CPD-A+C template at 200 nM (a sufficiently high concentration of DnaG to ensure efficient leading-strand reinitiation) and 800 nM DnaG (Figures 4C and S6) and found that full-length product formation was appreciably slower at the lower DnaG concentration. Another striking effect of the concentration of DnaG was on the distribution of fully-resolved replication products. At low DnaG concentrations, replication with the CPD-A template produced unit-length products, in addition to a smear that migrated directly below. Uncoupled products were not detected, even at the lowest concentration of DnaG tested. At higher concentrations of DnaG, the smear became less apparent and the full-length band more prominent (Figure 4B). There are two potential, non-mutually exclusive, explanations for such a result. At lower DnaG concentrations, less frequent priming on the lagging strand could result in ssDNA gaps forming between Okazaki fragments that might affect the migration of replicated lagging strands. Or, on the leading strand, the distance between the CPD and downstream priming site might be increased at lower DnaG concentrations, resulting in larger ssDNA gaps between the 3′ end of the stall product and the 5′ end of the restart product. This idea is supported by the distribution of restart products in the denaturing gel, as a higher proportion of longer products were generated at elevated DnaG concentrations (Figures 4B and 5A, denaturing). Comparison of the reaction products on the CPD-A and CPD-C templates revealed some interesting differences. Below 160 nM DnaG, uncoupled products were generated specifically on the CPD-C template (Figures 4A and B). The distance downstream of the CPD is 2-fold longer on the CPD-A template compared to the CPD-C template (Figure S2A), indicating that the propensity for uncoupled products to be generated is dependent on the distance from the CPD to the end of the template. At higher DnaG concentration, priming appeared to occur closer to the CPD and uncoupled products (Figure 4A) were not generated on either template.
Figure 4. Primase Concentration Influences the Rate of Leading-strand Reinitiation.
(A) Illustration of the replication products generated following restarted replication, or complete uncoupled replication downstream from the CPD.
(B) Titration of DnaG using standard reaction conditions for 6 min.
(C) Quantification of full-length replication products (restarted leading strands and lagging strands) from pulse-chase experiments conducted using 200 nM and 800 nM DnaG on the CPD-A+C template. Note that complete uncoupled replication is not observed at either concentration of DnaG (Figure S6). Error bars represent the SEM from three independent experiments.
Figure 5. The DnaX Complex Enhances the Efficiency By Which Leading-strand Synthesis is Reinitiated Downstream of a Lesion.
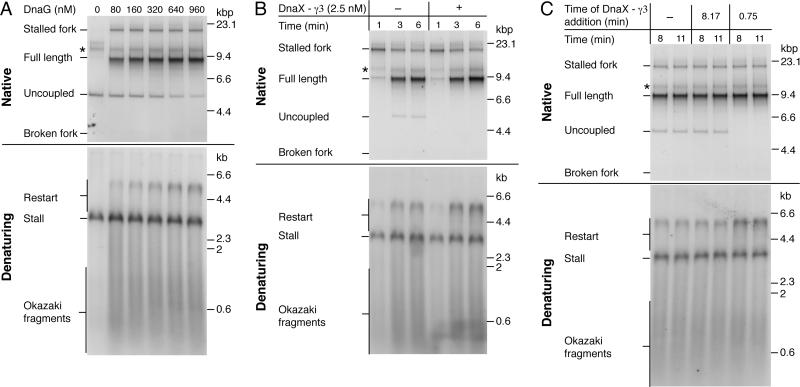
(A) DnaG titration following column isolation of ERIs. Reactions were conducted for 6 min on the CPD-A template.
(B) Column-isolated replication reactions using the CPD-A template and 1 μM DnaG in the presence or absence of DnaX-γ3 complex that was added 45 sec post EcoRI.
(C) Column-isolated replication reactions conducted at 250 nM DnaG with the CPD-A template. DnaX-γ3 complexes (2.5 nM) were added at different time points following EcoRI addition.
* ERIs that are labeled but not extended.
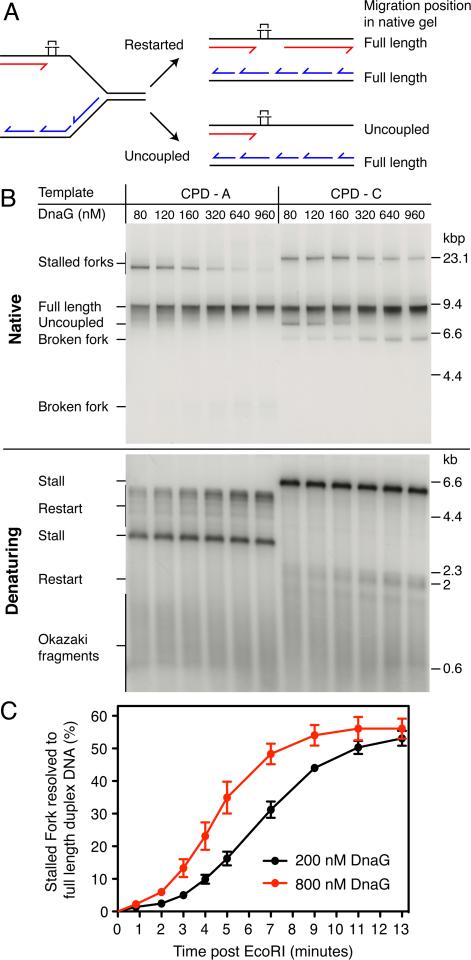
Replisome-independent Assembly of β on the Downstream Primer Enhances the Efficiency of Leading-strand reinitiation
Following primer synthesis, β-clamp assembly is likely to be one of the next events that occurs before leading-strand reinitiation. To gain insight into the factors that affect this reaction, we isolated ERIs and their associated replisomes from non-DNA associated proteins by gel filtration (Yeeles and Marians, 2011). This method serves to remove unbound Pol III* (which contains the clamp-loader DnaX complex), that may be able to influence the assembly of β around newly-primed DNA. Following column isolation the distributively acting proteins — SSB, DnaG, and β — were added back and ERIs were released by EcoRI cleavage. In the absence of DnaG neither lagging-strand synthesis nor leading-strand reinitiation were observed and the entire CPD distal region was unwound to generate uncoupled products (Figure 5A, native) (Yeeles and Marians, 2011). As DnaG concentration increased, uncoupled products decreased and there was a concomitant increase in restart products (Figure 5A, denaturing and figure S7). In contrast to bulk reactions with the CDP-A template, even at the highest concentration of DnaG assayed (960 nM), some replisomes reached the end of the template without reinitiating leading-strand synthesis, suggesting that reinitiation was occurring less efficiently following column isolation.
We speculated that assembly of the β-clamp might have been compromised relative to bulk reactions because of the removal of unbound Pol III* that may have been aiding β-clamp assembly around the new leading-strand primer. If unbound Pol III* was indeed performing such a function, then a DnaX complex unable to interact with either DnaB or DNA polymerase III might be able to substitute. The DnaX complex consists of three copies of the dnaX gene product (τ or γ), χ, ψ, δ and δ′. τ is the full-length product of dnaX, whereas γ is a C-terminally truncated protein that lacks the last 213 amino acids (Flower and McHenry, 1990; Tsuchihashi and Kornberg, 1990). The C-terminal region of τ binds both DnaB and the α-subunit of Pol III core (Dallmann et al., 2000), thereby ensuring that helicase movement is coupled to leading- and lagging-strand synthesis (Kim et al., 1996a). Despite lacking the domains required for these interactions, γ can be constituted into functional clamp-loader complexes. DnaX-γ3 complexes reconstituted in vitro can efficiently assemble β-clamps around primer-template DNA (Downey and McHenry, 2010), although they are unable to form fully-functional replisomes (Kim et al., 1996a). Column-isolated reactions were conducted in the presence or absence of a DnaX-γ3 complex that was added 45-sec post EcoRI addition. In the presence of DnaX-γ3 complex, no uncoupled products were observed in the native gel and there was a clear increase in the intensity of restart products (Figure 5B). This suppression of uncoupled-product formation may have resulted from the DnaX-γ3 complex increasing the rate of β-clamp assembly so that leading-strand synthesis was reinitiated prior to DnaB reaching the end of the template. If this was indeed the case, addition of DnaX-γ3 complex should have no effect on uncoupled products that have already been generated. To test this idea, uncoupled products were synthesized in an 8-min reaction, DnaX-γ3 complex was added 10 sec later, and the reactions incubated for an additional 170 sec (Figure 5C). This essentially post-replicative addition of DnaX-γ3 complex had no appreciable affect on the uncoupled products and there was no visible increase in restart compared to lanes lacking DnaX-γ3 complex. The data supports the hypothesis that the replisome must still be resident on the DNA template in order for DnaX-γ3 complex to enhance the efficiency of restart.
Leading-strand Synthesis Remains Coupled to DnaB Unwinding Following DnaX-γ3 Complex-facilitated Leading-strand Reinitiation
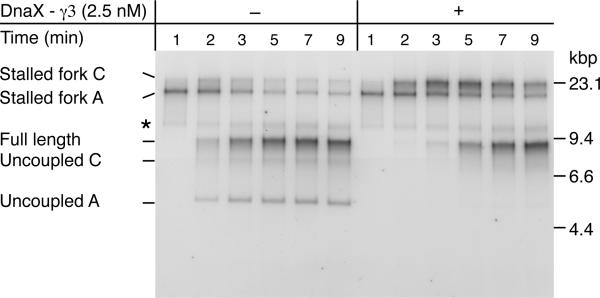
To further investigate how the DnaX-γ3 complex was facilitating leading-strand restart, we compared replication on the double-damage CPD-A+C template in the presence or absence of the complex. A low DnaG concentration (200 nM) was used to reduce the efficiency of leading-strand reinitiation (Figure 5A) and therefore maximize the potential effects of DnaX-γ3 addition. Uncoupled products were generated where leading-strand synthesis was not reinitiated downstream of either lesion (Figure 6, Uncoupled A), and the second stalled fork (Figure 6, Stalled fork C), generated by replisomes that restarted downstream of the first CPD (CPD-A), was less prominent than in bulk reactions. At later time points, a second uncoupled product was formed (Figure 6, Uncoupled C), presumably when replisomes failed to reinitiate leading-strand synthesis downstream of the second CPD (CPD-C) in reactions where leading-strand synthesis was reinitiated downstream of the first CPD. Addition of DnaX-γ3 complex had several effects on the reaction. Uncoupled products were suppressed and more replication forks stalled at the second CPD (Figure 6, compare Stalled fork C −/+ DnaX-γ3 complex). This result indicates that leading-strand synthesis is required for CPD recognition and replisome stalling, which is not unexpected given that DnaB translocates on the opposite strand of the duplex.
Figure 6. Leading-strand Synthesis Remains Coupled to DnaB Template Unwinding Following Replication Restart.
Column-isolated replication reactions conducted with the CPD-A+C template at 200 nM DnaG in the presence or absence of DnaX-γ3 complex that was added 45 sec post EcoRI. * ERIs that are labeled but not extended.
Another striking difference was manifest in the kinetics with which fully-resolved replication products (uncoupled and full length) were formed. Addition of DnaX-γ3 slowed the production of these species. For example, uncoupled A and full-length products were visible within 2-3 minutes in its absence, but full-length products were not readily detected until 5 minutes in its presence. Because of the low DnaG concentration, the majority of replisomes failed to reinitiate leading-strand synthesis downstream of CPD-A in the absence of DnaX-γ3. Essentially, under these conditions, most replisomes appeared not to ‘see’ the second CPD. The reaction proceeded in a manner more similar to a column-isolated reaction on a single-damage template, than it did a bulk reaction on a double-damage template, and this was also reflected in the kinetics (compare figures 5B and 6 - DnaX-γ3 complex and figure 3B). The major effect of added DnaX-γ3 appears to be increased leading-strand reinitiation, which in turn leads to increased stalling at the second CPD. Therefore it seems highly likely that it is this additional replisome-stalling event that delays the production of full-length products relative to complete uncoupled replication beyond CPD-A, which is the pathway that predominates in the absence of DnaX-γ3. Furthermore, as recognition of the second CPD by the leading-strand polymerase delayed the arrival of DnaB at the distal end of template (as inferred by the production of fully-replicated products), the data strongly suggest that forward movement of DnaB remained coupled to leading-strand synthesis following DnaX-γ3 facilitated leading-strand reinitiation.
DISCUSSION
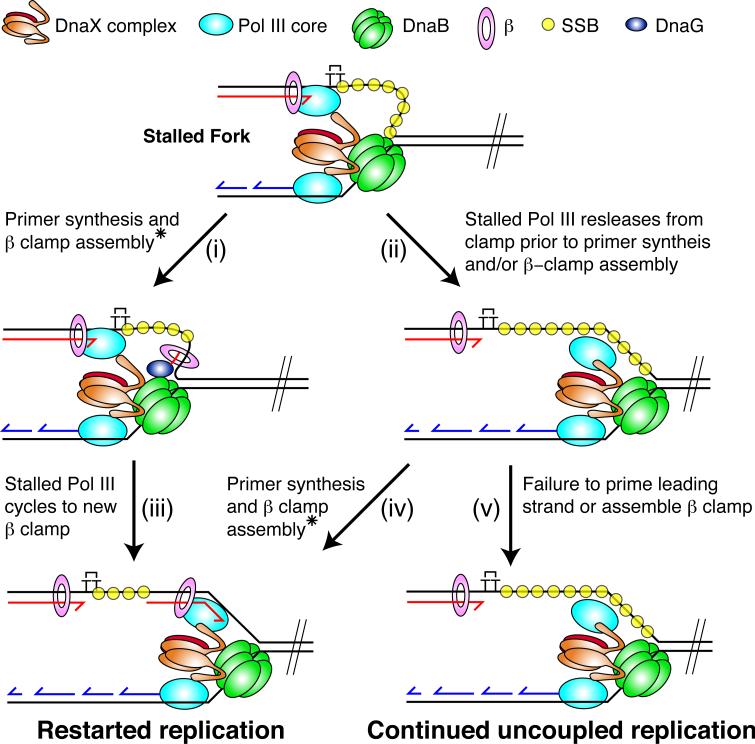
Our results show that following replisome collision with a leading-strand CPD, DnaB and lagging-strand synthesis advance past the site of damage at the majority of replication forks. Continued template unwinding is likely to be crucial for leading-strand reinitiation, as it ensures sufficient ssDNA is generated on the leading-strand template to enable primer synthesis to occur. Previous studies have shown that leading-strand synthesis, coupled to DnaB via the C-terminus of τ, is required for optimal DnaB unwinding rates of up to 1000 bps−1. Replisomes lacking τ move at a markedly reduced rate of ~ 35 bps−1, and the C-terminus of τ alone is not sufficient to stimulate the unwinding rate of DnaB in the absence of replication (Dallmann et al., 2000; Kim et al., 1996a, b). If, following replisome collision with a lesion, DnaB was to simply continue unwinding the template at a reduced rate of 35 bps−1, 1050 bp would on average be unwound within the first 30 sec. Yet pulse-chase experiments showed that 30 sec after addition of the chase the vast majority of templates remained intact 266 bp downstream of the damage, implying that DnaB had yet to advance beyond this point. Movement of DnaB therefore appears to be restricted in the immediate aftermath of the replisome's collision. If the stalled leading-strand polymerase remained associated with both the DNA template and the τ-subunit of the clamp loader, it could potentially form a tether that might be inhibitory to DnaB movement. In this scenario, any continued template unwinding beyond the site of damage would generate a loop of ssDNA on the leading-strand template that would be expected to persist until the polymerase either dissociated from the template or the clamp loader (Figure 7, Stalled fork). Such a tether may be required to prevent DnaB advancing too far ahead of the damage site prior to the reinitiation of leading-strand synthesis.
Figure 7. Model for Stalled Fork Resolution by Leading-strand Reinitiation and Uncoupling.
The model is described in the Discussion.
* Assembly of the new β-clamp around the leading strand primer can be catalyzed either by the DnaB-associated DnaX complex or an exogenous DnaX complex not associated with the replisome (i.e DnaX-γ3).
Resolution of stalled-replication forks yielded either restarted leading-strand, or uncoupled products (Figure 7), and the balance between these two events was influenced by leading-strand priming frequency, the efficiency of β-clamp assembly on the downstream primer, and the length of the CPD distal region. Furthermore, the overall reaction kinetics, i.e., the rate at which fully-replicated products were synthesized, was influenced by the concentration of DnaG. If forward translocation by DnaB is indeed limited by continued association with the stalled leading-strand polymerase, priming and subsequent β-clamp assembly on the loop of ssDNA generated should enable leading-strand reinitiation to occur in close proximity to the damage (Figure 7, i, and iii). This reaction would likely be favored at high DnaG concentrations, when stochastic priming should be more frequent, and may explain why there is a larger population of longer restart products at elevated DnaG concentrations (Figure 4B). It may also explain why the overall reaction kinetics are influenced by DnaG concentration, as more frequent priming could lead to accelerated cycling of the stalled leading-strand polymerase and therefore faster release of the replisome to continue downstream replication. It is tempting to speculate that leading-strand priming in such a loop could also help to facilitate β-clamp assembly by the replisome associated clamp-loader, negating the need for an exogenous complex. However, because some uncoupled products were still observed at high DnaG concentrations following column isolation in the absence of DnaX-γ3 complex, it is likely that exogenous DnaX complex would also be required to participate in this reaction occasionally.
Alternatively, the leading-strand polymerase could dissociate from the original β-clamp prior to priming and clamp assembly, releasing any potential restrictions on DnaB translocation and enabling it to unwind the downstream region of the template (Figure 7, ii). In this scenario the speed at which DnaB unwinds DNA is currently unknown. However, from our data it can be inferred that the uncoupled replisome moves at least as fast as previous estimates for DnaB unwinding in the absence of leading-strand synthesis (35 bps−1), because uncoupled products were generated on the CPD-A template within the first 3 min of column-isolated reactions (Figure 5B), which involves unwinding of an approximately 6 kbp region. Uncoupled products will be generated if priming and/or β-clamp assembly are not accomplished prior to DnaB reaching the end of the template (Figure 7, v). Alternatively, if leading-strand primer synthesis and clamp assembly do occur prior to complete template unwinding, leading-strand reinitiation could still be achieved (Figure 7, iv), and our experimental observations suggest this possibility. In bulk, uncoupled products were generated at low DnaG concentrations on the CPD-C but not CPD-A templates, where the downstream regions of the templates are 2.7 and 6 kbp, respectively. Thus, under these conditions, the propensity to generate uncoupled products is inversely related to the length of the template distal to the CPD. This effect is as predicted if complete uncoupled replication beyond the damage is a function of the time taken for DnaB to unwind the template, compared to the rates of primer synthesis and β-clamp assembly. The result would also be explained if DnaB either lacks the processivity or necessary unwinding rate to fully unwind the downstream region of the CPD-A template during the course of the reactions. However, this is unlikely given that uncoupled products are efficiently generated within 3 min in column-isolated reactions on the CPD-A template. Two previous studies concluded that the E. coli replisome predominantly uncouples following collision with a leading-strand lesion (Higuchi et al., 2003; Pages and Fuchs, 2003). However, these experiments were conducted on templates with damage-distal regions of only up to 1.8 kbp that, based on findings presented herein, may have precluded the detection of leading-strand reinitiation.
The efficiency of leading-strand reinitiation in vivo remains to be determined, as does the extent to which replication becomes uncoupled in its absence. Even if uncoupling distances were to be limited in vivo, perhaps by nucleoid-associated proteins such as RNA Polymerase, it seems likely that significant ssDNA would still be generated. The fate of the original replisome in this situation is currently unknown. Should it dissociate, origin-independent replisome loading by PriC would enable direct restart by repriming (Heller and Marians, 2006a). Replication could also be reinitiated by fork remodeling and damage removal, or by translesion DNA synthesis.
Our study has revealed the unexpected finding that an exogenous DnaX-γ3 complex can participate in the reinitiation of leading-strand synthesis, despite there currently being no available evidence that the complex is able to interact directly with the replisome. We considered two distinct mechanisms by which DnaX-γ3 complex might be facilitating restart. Either the complex could be assembling β-clamps around leading-strand primers that were missed by the replisome-associated DnaX complex, or the DnaX-γ3 complex could be unloading the β-clamp associated with the stalled leading-strand polymerase, which would potentially accelerate polymerase cycling and enhance restart. We strongly favor the first mechanism for several reasons. The β-unloading reaction that is catalyzed by DnaX-γ3 complex is relatively slow, with a rate constant of 0.015 s−1, and a KM of 280 nM (Leu et al., 2000), far in excess of the 2.5 nM used in these experiments. Additionally, association of the leading-strand polymerase with τ during replication has been shown to protect the β-clamp from unloading by exogenous DnaX complex (Kim et al., 1996c). Thus, under these conditions, it seems unlikely that DnaX-γ3 complex would be able to unload β-clamps at a sufficient rate to influence leading-strand reinitation significantly on the timescale of our experiments. In vivo, the DnaX complex participates in multiple reactions in addition to its role at the replication fork, such as nucleotide excision repair, post-replicative translesion synthesis, and β-clamp unloading (Leu et al., 2000). The ability to perform such functions is in part because of the reasonably high expression levels of the individual components. In fact, several studies (Leu et al., 2000; Reyes-Lamothe et al., 2010) have shown that many DnaX-complex subunits are more abundant than Pol III core, and therefore a proportion of the complexes found within the cell are likely not to be associated with the replicative polymerase. Given these high expression levels, and the low concentration of DnaX-γ3 that is required to enhance restart in vitro, it seems probable that such a reaction should be able to function in vivo following replisome collision with leading-strand damage.
EXPERIMENTAL PROCEDURES
DNA Templates and Replication Proteins
DNA templates containing site-specific cyclobutane pyrimidine dimers were synthesized using M13-JY13 (see Supplementary Information for construction) ssDNA as described previously (Yeeles and Marians, 2011). For sequences of the three CPD-containing primers used see Supplementary Information. For templates containing two site-specific CPDs, two CPD-containing oligonucleotides were simultaneously annealed to the ssDNA and templates were synthesized as described (Yeeles and Marians, 2011). Replication proteins were purified as described previously (Yeeles and Marians, 2011).
Replication Reactions
Complete details can be found in Supplementary Information. Standard (bulk) replication reactions (10-40 μl) were essentially as described in Yeeles and Marians (2011). Reactions were initiated by the addition of replication proteins. After a 2-min incubation at 37 °C, [α-32P]dATP was added and ERIs were released by the addition of EcoRI-HF (NEB). Reactions were stopped at the indicated times by the addition of a stop buffer that contained a 10-fold excess of AMP-PNP over ATP and 2′, 3′-dideoxyribonucleoside 5′-triphosphates over dNTPs. Reactions were then digested with PvuI and analyzed by gel electrophoresis. For pulse-chase reactions, [α-32P]dATP was substituted with [α-32P]dGTP. Label was chased by the addition of a 25-fold excess of unlabeled dGTP. Full-length replication products were quantified using ImageGuage software. The counts for full-length products at each time point were divided by the total lane counts at 50 seconds.
For column-isolated reactions, replisome-associated ERIs were generated in standard replication reactions (30 μl). Following a 2-min incubation at 37 °C, KCl was added to a final concentration of 100 mM and the entire sample was applied to a 3 × 195 mm Sepharose-4B column equilibrated and developed in replication reaction buffer that included three dNTPs at 4 μM and 0.5 μM ATP. Fractions containing ERIs were pooled, SSB, DnaG, and β were added back and replication reactions were performed and analyzed as above.
Replication Fork Mapping Experiments
Fork mapping experiments were performed exactly as described for standard replication reactions. Following incubation at 37 °C, samples were quenched in 3 volumes of a 1.33X stop buffer containing: 50 mM Hepes-KOH pH 8, 75 mM potassium glutamate, 10 mM Mg(OAc)2, 10 mM DTT, 100 μg/ml BSA, and a 20-fold molar excess of ddNTPs and ATP-γ-S. Reaction products were immediately digested for 10 min with PvuI and additional enzymes where indicated. Following quenching with 30 mM EDTA, samples were treated with 0.25% SDS and 0.2 mg/ml proteinase K for 30 min at 37 °C. Products were separated by electrophoresis through 0.8% native gels as described previously.
Supplementary Material
HIGHLIGHTS.
Uncoupled replication occurs downstream of a leading-strand lesion prior to restart
Priming and clamp assembly are rate limiting for leading-strand reinitiation
Rapid replication rates are resumed following leading-strand reinitiation
A single replisomxe can bypass multiple leading-strand lesions
ACKNOWLEDGMENTS
These studies were supported by NIH grant GM34557. We thank Tom Kelly, Carolina Gabbai, and Gideon Coster for dicussion and critical reading of the manuscript and Charles McHenry (University of Colorado, Boulder) for the gift of the DnaXγ3 complex.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Dallmann HG, Kim S, Pritchard AE, Marians KJ, McHenry CS. Characterization of the unique C terminus of the Escherichia coli tau DnaX protein. Monomeric C-tau binds alpha AND DnaB and can partially replace tau in reconstituted replication forks. J. Biol. Chem. 2000;275:15512–15519. doi: 10.1074/jbc.M909257199. [DOI] [PubMed] [Google Scholar]
- Downey CD, McHenry CS. Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader. Mol. Cell. 2010;37:481–491. doi: 10.1016/j.molcel.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower AM, McHenry CS. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc. Nat'l. Acad. Sci. U. S. A. 1990;87:3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, McHenry CS. tau binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of tau, binds the replication fork, helicase, DnaB. J. Biol. Chem. 2001;276:4441–4446. doi: 10.1074/jbc.M009830200. [DOI] [PubMed] [Google Scholar]
- Gupta MK, Guy CP, Yeeles JT, Atkinson J, Bell H, Lloyd RG, Marians KJ, McGlynn P. Protein-DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proc. Nat'l. Acad. Sci. U. S. A. 2013;110:7252–7257. doi: 10.1073/pnas.1303890110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006a;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 2006b;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells. 2003;8:437–449. doi: 10.1046/j.1365-2443.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- Iyer VN, Rupp WD. Usefulness of benzoylated naphthoylated DEAE-cellulose to distinguish and fractionate double-stranded DNA bearing different extents of single-stranded regions. Biochim. Biophys. Acta. 1971;228:117–126. doi: 10.1016/0005-2787(71)90551-x. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996a;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. tau couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J. Biol. Chem. 1996b;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- Kim S, Dallmann HG, McHenry CS, Marians KJ. Tau protects beta in the leading-strand polymerase complex at the replication fork. J. Biol. Chem. 1996c;271:4315–4318. doi: 10.1074/jbc.271.8.4315. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- Leu FP, Hingorani MM, Turner J, O'Donnell M. The delta subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J. Biol. Chem. 2000;275:34609–34618. doi: 10.1074/jbc.M005495200. [DOI] [PubMed] [Google Scholar]
- McHenry CS. DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- McInerney P, O'Donnell M. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 2004;279:21543–21551. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470:554–557. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SW, Benkovic SJ. Response of the bacteriophage T4 replisome to noncoding lesions and regression of a stalled replication fork. J. Mol. Biol. 2010;401:743–756. doi: 10.1016/j.jmb.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages V, Fuchs RP. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science. 2003;300:1300–1303. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Lloyd RG. Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev. 2007;21:668–681. doi: 10.1101/gad.417607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp WD. DNA Repair Mechanisms. In: Neidhart FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magazanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella Cellular and Molecular Biology. ASM), pp; Washington, D.C.: 1996. pp. 2277–2294. [Google Scholar]
- Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- Setlow RB, Swenson PA, Carrier WL. Thymine Dimers and Inhibition of DNA Synthesis by Ultraviolet Irradiation of Cells. Science. 1963;142:1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- Swenson PA, Setlow RB. Effects of ultraviolet radiation on macromolecular synthesis in Escherichia coli. J. Mol. Biol. 1966;15:201–219. doi: 10.1016/s0022-2836(66)80221-8. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc. Nat'l. Acad. Sci. U. S. A. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Zechner EL, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J. Biol. Chem. 1992;267:4030–4044. [PubMed] [Google Scholar]
- Yeeles JT, Marians KJ. The Escherichia coli replisome is inherently DNA damage tolerant. Science. 2011;334:235–238. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.