Abstract
Histone deacetylases (HDACs) are believed to regulate gene transcription by catalyzing deacetylation reactions. HDAC3 depletion in mouse liver upregulates lipogenic genes and results in severe hepatosteatosis. Here we show that pharmacologic HDAC inhibition in primary hepatocytes causes histone hyperacetylation but does not upregulate expression of HDAC3 target genes. Meanwhile, deacetylase-dead HDAC3 mutants can rescue hepatosteatosis and repress lipogenic genes expression in HDAC3-depleted mouse liver, demonstrating that histone acetylation is insufficient to activate gene transcription. Mutations abolishing interactions with the nuclear receptor corepressor (NCOR or SMRT) render HDAC3 nonfunctional in vivo. Additionally, liver-specific knockout of NCOR, but not SMRT, causes metabolic and transcriptomal alterations resembling those of mice without hepatic HDAC3, demonstrating that interaction with NCOR is essential for deacetylase-independent function of HDAC3. These findings highlight non-enzymatic roles of a major HDAC in transcriptional regulation in vivo and warrant reconsideration of the mechanism of action of HDAC inhibitors.
INTRODUCTION
Small molecules that inhibit histone deacetylases (HDACs) show promise in treating many diseases, including cancer, neurodegenerative, cardiovascular, and metabolic diseases (Gryder et al., 2012; Kazantsev and Thompson, 2008). A variety of HDAC inhibitors (HDIs) are under clinical investigation with two already approved for treating lymphoma (Gryder et al., 2012). Yet, the mechanism of action for HDIs is not clear and very controversial (Wanczyk et al., 2011). For example, upregulation of p21 (CIP1/WAF1) gene expression have been widely observed in cancer cells upon treatment of various HDIs, and is held as a prevalent explanation for how HDIs cause cell cycle arrest (Ocker and Schneider-Stock, 2007). However, knockdown of p21 or its upstream regulator p53 fails to rescue cell cycle progression defects in fibroblast cells depleted of HDAC1 and HDAC2 (Wilting et al., 2010). Such lack of knowledge on the genuine pharmacological targets of HDIs poses the major challenge for their development as drugs (Kazantsev and Thompson, 2008).
Numerous genetic mouse models have established that HDACs play pivotal roles in a plethora of biological processes including embryonic development, cardiovascular health and energy metabolism (Finkel et al., 2009; Haberland et al., 2009). HDACs fall into several classes based on their catalytic mechanism and sequence homology (Yang and Seto, 2008). Class I, II, and IV HDACs depend on the zinc (Zn) metal for their enzymatic activities, whereas class III sirtuins require NAD (nicotine adenine dinucleotide) as a co-factor (Sauve et al., 2006). Class I HDACs form multiple-protein nuclear complexes, with HDAC 1 and 2 found in the NuRD (nucleaosome remodeling and deacetylating), Sin3, and CoREST (co-repressor for element-1-silencing transcription factor) complexes (Yang and Seto, 2008). HDAC3, another class I HDAC, exists in a distinct complex that contains either NCOR (nuclear receptor corepressor) or its homolog SMRT (silencing mediator of retinoic and thyroid receptors) (Goodson et al., 2005; Perissi et al., 2010).
HDAC3 not only forms a complex with NCOR/SMRT but also requires interaction with the DAD (deacetylase activating domain) of NCOR/SMRT for its enzyme activity (Guenther et al., 2001). The recently published structure of HDAC3 co-crystallized with a short DAD peptide reveals an inositol tetraphosphate molecule Ins(1,4,5,6)P4 (IP4) embedded at the interface between HDAC3 and DAD, which likely serves as a ‘intermolecular glue’ to stabilize the interaction (Watson et al., 2012). Binding to IP4 and DAD triggers a conformational change in HDAC3 that makes the catalytic channel accessible to the substrate (Arrar et al., 2013; Watson et al., 2012). Consistent with this structural model, combined mutations on residues that interact with IP4, including Y478A in NCOR and Y470A in SMRT, completely abolish deacetylase activities of HDAC3 in mice (You et al., 2013). Interestingly, knock-in mice bearing these mutations in the DADs of both NCOR and SMRT (NS-DADm) live to adulthood despite undetectable deacetylase activity in the embryo, whereas global deletion of HDAC3 is embryonic lethal (Bhaskara et al., 2008; You et al., 2013). This suggests a deacetylase-independent function of HDAC3 for survival. However, it is not known whether such function is restricted to embryonic development, whether it is directly related to transcriptional regulation, or what the underlying mechanism is.
We have previously shown that nuclear receptor Rev-erbs recruit HDAC3 to the genome in liver and that acute liver-specific knockout of HDAC3 by injecting HDAC3f/f mice with AAV (adeno-associated virus) expressing Cre recombinase causes histone hyperacetylation at genome-wide HDAC3 binding sites, upregulates lipogenic genes near HDAC3 binding sites, and leads to remarkable hepatosteatosis (Feng et al., 2011; Sun et al., 2011). The lipid metabolic phenotype in these mice can be completely rescued by re-expression of HDAC3 at its endogenous levels in the liver using an AAV vector, which creates an excellent in vivo phenotype-rescue system for functional analysis of structure-based HDAC3 mutations (Sun et al., 2012). Here we integrate this system with epigenomic approaches and novel genetic mouse models to provide new mechanistic insights into HDAC3 biology.
RESULTS
HDI-dependent histone hyperacetylation does not upregulate gene expression as seen in HDAC3-depletion
Genetic deletion of HDAC3 in adult mouse livers either through AAV in a liver-specific manner or by an inducible Mx1-Cre transgenic line in a whole-body manner results in prominent hepatosteatosis and severe liver hypertrophy (Knutson et al., 2008; Sun et al., 2012). These findings not only demonstrate the importance of HDAC3 in maintaining normal adult liver function, but also raise the concern of hepatotoxicity for pan-HDIs. However, hepatosteatosis is not a prevalent side effect of most pan-HDIs in patients or animals (Chateauvieux et al., 2010; Subramanian et al., 2010; Zhang et al., 2012). To evaluate the outcome of continuous HDAC inhibition, we compared HDIs with ex vivo HDAC3 knockout in primary hepatocytes for altered gene expression.
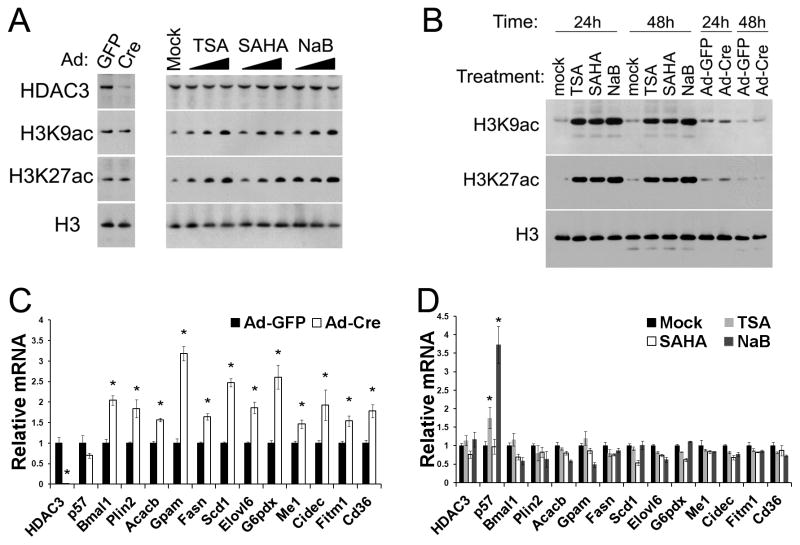
Primary hepatocytes isolated from HDAC3f/f mice were infected with adenovirus (Ad) expressing either GFP or Cre. Total cell lysates (Figure 1A) or histone extracts (Figure 1B) were harvested at different time after HDAC3 depletion and were analyzed by western blot. Global histone acetylation on histone 3 lysine 9 (H3K9ac) and lysine 27 (H3K27ac) was not changed despite efficient depletion of HDAC3 proteins. This is not surprising since the HDAC3 cistrome only constitutes a very small fraction of the total genome (Feng et al., 2011), and is consistent with the lack of global histone acetylation changes following knockout or knockdown of a specific HDAC (Bradner et al., 2010; Montgomery et al., 2008; Oehme et al., 2009). Many HDAC3 target genes were upregulated, including those involved in circadian rhythm and lipid synthesis relevant to HDAC3 in vivo physiology (Sun et al. 2012), demonstrating the validity of this ex vivo system for characterizing hepatic HDAC3 function (Figure 1C). In comparison, treating hepatocytes with different pan-HDIs including Trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), or sodium butyrate (NaB) caused an expected dose-dependent and persistent increase in global histone acetylation on both H3K9 and H3K27 (Figures 1A and 1B). Histone acetylation at HDAC3 binding sites near several HDAC3 target genes were also increased by pan-HDIs to a similar or higher degree compared to HDAC3 depletion (Figures S1A and S1B). However, the expression of HDAC3 target genes was generally not increased by these pan-HDIs, suggesting that histone hyperacetylation per se is not sufficient to activate gene transcription (Figure 1D).
Figure 1. HDI-dependent histone hyperacetylation does not upregulate gene expression as seen in HDAC3-depletion.
(A) Primary hepatocytes from HDAC3f/f mice were treated with adenovirus (Ad) expressing GFP or Cre, or increasing concentrations of HDAC inhibitors (HDIs) including Trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), or sodium butyrate (NaB). Cells were directly lysed in SDS sampling buffer after 48 h followed by immunoblot analysis. (B) Primary hepatocytes were treated with adenovirus or the highest concentrations of HDIs. Histones were acid extracted after the indicated time and analyzed by immunoblot. (C) Quantitative RT-PCR (RT-qPCR) analysis of primary hepatocytes after 48 h since HDAC3 depletion. * P < 0.05 compared with GFP. (D) RT-qPCR analysis of primary hepatocytes treated for 48 h with the highest concentrations of HDIs. p57, cyclin-dependent kinase inhibitor 1C; Bmal1, aryl hydrocarbon receptor nuclear translocator-like; Plin2, perilipin 2; Acacb, acetyl-CoA carboxylase beta; Gpam, glycerol-3-phosphate acyltransferase, mitochondrial; Fasn, fatty acid synthase; Scd1, stearoyl-Coenzyme A desaturase 1; Elovl6, fatty acid elongase 6; G6pdx, glucose-6-phosphate dehydrogenase X-linked; Me1, malic enzyme 1; Cidec, fat specific protein 27; Fitm1, fat storage-inducing transmembrane protein 1; Cd36, fatty acid translocase. * P < 0.05 compared with the mock treatment group. All error bars, s.e.m. from triplicate plates. See also Figure S1.
These results are consistent with previous findings that gene expression changes elicited by pan-HDIs are moderate and do not necessarily resemble those caused by HDAC depletion (Lopez-Atalaya et al., 2013; Mullican et al., 2011). In addition, genetic depletion of histone acetyltransferases (HATs) in mouse fibroblasts drastically abolishes histone acetylation, but only causes mild changes in gene expression (Kasper et al., 2010). These findings raise the possibility that histone acetylation may only correlates with, but does not necessarily cause, active gene transcription. In keeping with this notion, some catalytically-inactive mutants of HATs are able to rescue growth defects caused by HAT knockout in yeast (Sterner et al., 2002). While it is understandable that many HATs may have enzyme-independent functions, given their large size (typically >200 kDa) suitable for scaffolding roles and multiple-domain architecture responsible for interacting many proteins, HDACs are smaller proteins (typically <70 kDa) and it would be surprising if the deacetylase enzymatic activities do not fully account for the phenotype caused by HDAC depletion. Therefore, to complement the HDI-based pharmacological approach, we next genetically dissected HDAC3-mediated transcriptional repression by structure-function analysis in vivo.
Mutations Y298F (YF) and K25A (KA) abolish HDAC3 enzymatic activity by distinct mechanisms
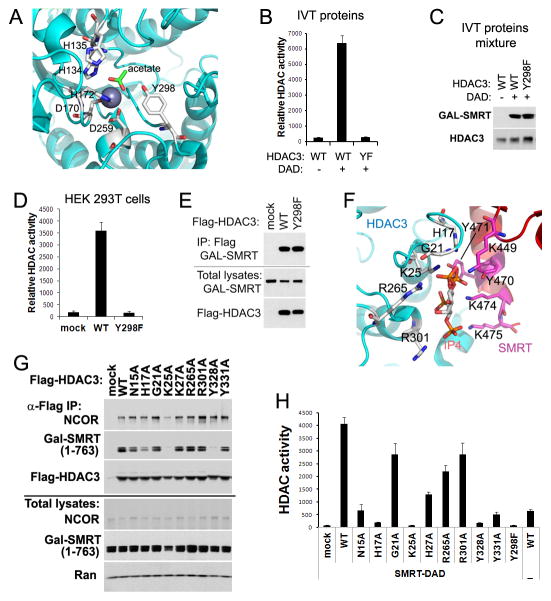
Crystal structures of HDACs revealed that the highly conserved Tyr residue (Y298 in HDAC3) is located within the active site and is catalytically essential in stabilizing the tetrahedral intermediate and polarizing the substrate carbonyl for nucleophilic attack in coordination with Zn ion (Figures 2A and S2) (Lombardi et al., 2011; Watson et al., 2012). Mutation of Y298F (YF) rendered the in vitro-translated (IVT) HDAC3 proteins completely inactive in the presence of a truncated SMRT protein (amino acid 1–763) containing DAD, as measured by a fluorescence-based HDAC assay using peptide substrate (Figures 2B and 2C). To further address whether YF lost deacetylase activity within cells, Flag-tagged HDAC3 was co-expressed along with DAD in HEK 293T cells. An HDAC assay of anti-Flag immunoprecipitates showed that YF does not have detectable deacetylase activity (Figure 2D), consistent with a previous report that Y298H substitution in HDAC3 completely eliminates deacetylase activity against radioactively labeled histones (Lahm et al., 2007). The same Y>F substitution in HDAC8 was also inactivating and was used to crystallize the substrate-bound HDAC8, because the enzyme failed to finish the catalytic transition and trapped its substrate in the catalytic pocket (Vannini et al., 2007). As expected, the interaction between HDAC3 and DAD was not affected by YF (Figure 2E).
Figure 2. Mutations Y298F (YF) and K25A (KA) abolish HDAC3 enzymatic activity by distinct mechanisms.
(A) Key residues within the HDAC3 catalytic site. (B) In vitro-translated (IVT) HDAC3 wild-type (WT) or YF mutant were mixed with IVT GAL-tagged SMRT-DAD (amino acid 1–763), followed by HDAC enzyme assay. Values are fluorescence signal intensities. (C) Immunoblot analysis of the above IVT proteins mixtures. (D) Flag-tagged HDAC3 and GAL-tagged SMRT (1–763) were co-expressed in HEK 293T cells. Cell lysates were immunoprecipitated with Flag antibodies followed by HDAC assay. (E) Immunoblot analysis of the above immunoprecipitates (IP). (F) Potential residues involved in binding of HDAC3 with SMRT-DAD and inositol phosphates Ins(1,4,5,6)P4 (IP4). (G) Flag-HDAC3 mutants and SMRT (1–763) were co-expressed in HEK 293T cells. Cell lysates were immunoprecipitated with Flag antibodies followed by Immunoblot analysis. (H) HDAC assay of the above immunoprecipitates. All error bars, s.e.m. from triplicate assays. See also Figure S2.
Another approach to eliminate HDAC3 deacetylase activity is to mutate key residues required for its interaction with DAD. The crystal structure suggests several residues that could directly contact DAD or the IP4 molecule (Figure 2F) (Watson et al., 2012). HDAC3 harboring an Ala substitution on each of these residues was co-expressed with DAD in HEK 293T cells. K25A (KA) disrupted deacetylase activity and interaction with DAD (Figures 2G and 2H), consistent with the structure that K25 is nested in the center of the HDAC3-IP4 interface and likely forms hydrogen bonds with multiple oxygen atoms of IP4. The essential role of K25 in DAD-binding supports the role of IP4 as ‘intermolecular glue’ (Watson et al., 2012). Loss of deacetylase activity in K25A mutant is consistent with the undetectable HDAC3 enzyme activity in NS-DADm mice (You et al., 2013) and support the interdependence between DAD and IP4 in activating HDAC3 enzymatic activity (Arrar et al., 2013).
The KA mutation also reduced the binding of HDAC3 to endogenous full-length NCOR (Figure 2G). The residual binding is probably mediated by the second HDAC3-interacting domain in the middle region of NCOR/SMRT independent of the N-terminal DAD, which has been shown to interact with HDAC3 without enabling enzyme activities (Guenther et al., 2001; Li et al., 2000; Wen et al., 2000).
Deacetylase-dead HDAC3 mutants rescue derangement of both gene transcription and lipid metabolism in HDAC3-depleted liver
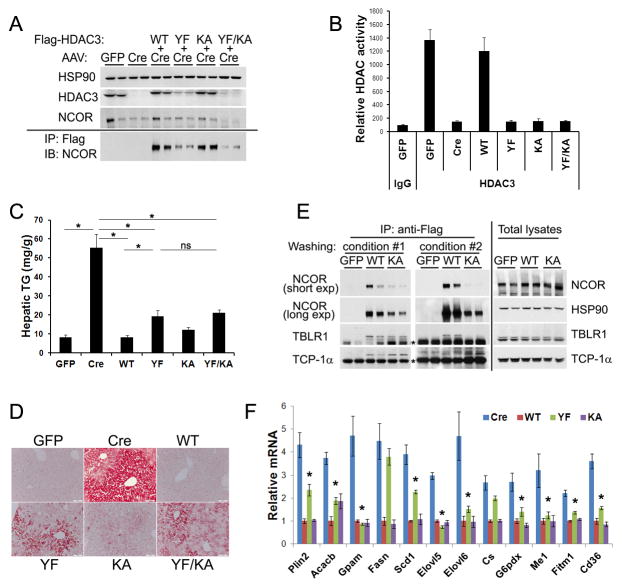
We further characterized YF, KA, and combined YF/KA mutations in an in vivo phenotype-rescue model. Intravenous injection of HDAC3f/f mice with AAV expressing Cre under a hepatocyte-specific thyroxine-binding globulin promoter (AAV-Tbg-Cre) depletes hepatic HDAC3, upregulates lipogenic genes, and results in hepatosteatosis without obvious inflammatory responses during the time frame examined (Sun et al., 2012). Here we engineered Flag-tagged HDAC3 into the same AAV-Tbg vector and co-injected it along with AAV-Tbg-Cre. When expressed at endogenous protein levels, the wild-type (WT) exogenous HDAC3 completely rescued fatty liver phenotype (Figures 3A–D) and repressed most genes that are upregulated upon HDAC3 depletion, as measured by microarray analysis (Figure S3A). This condition serves as the positive control for the subsequent mutation analysis.
Figure 3. Deacetylase-dead HDAC3 mutants rescue derangement in both gene transcription and lipid metabolism in HDAC3-depleted liver.
(A) HDAC3f/f mice were injected with AAV-Tbg-GFP or AAV-Tbg-Cre along with AAV vectors expressing Flag-tagged HDAC3 mutants. Total liver lysates (upper panel), or anti-Flag immunoprecipitates (IP) after washing with buffer containing 1% NP-40 (lower panel), were analyzed by immunoblot (IB). (B) Liver protein lysates were immunoprecipitated with either HDAC3 antibodies or normal IgG, followed by HDAC assay, n = 4. (C) Hepatic triglyceride (TG) measurement, n = 4. * P < 0.05. ns, not significant. (D) Oil red O (ORO) staining of livers. (E) Immunoprecipitation from the liver lysates were washed with more stringent buffer containing either 1% NP-40 plus 1% sodium deoxycholate (condition #1) or 1% NP-40 plus 1% sodium deoxycholate plus 0.1% SDS (condition #2), followed by immunoblot analysis with different exposure time (exp). * indicates non-specific signals. (F) RT-qPCR analysis of livers from mice described above, n = 4. * P < 0.05 between Cre and YF. All error bars, s.e.m. See also Figures S3 and S4.
Although the same dosages of AAV-HDAC3 were administered for all mutants, HDAC3 proteins containing YF mutation, either alone or in combination with other mutations, were expressed at much lower levels (Figure 3A). The exact reason for such sub-physiological expression is unclear and may be related to changes in protein stability, given that the mRNA levels of these mutants were the same (Figure S3B). HDAC3 proteins were immunoprecipitated with anti-HDAC3 antibodies from liver lysates and subjected to an HDAC assay, which showed that they were indeed deacetylase-dead in vivo (Figure 3B). Surprisingly, all three deacetylase-dead mutants rescued the fatty liver to a large degree, as demonstrated by quantification of hepatic triglycerides (TG) (Figure 3C) and Oil Red O (ORO) staining of neutral lipids in liver sections (Figure 3D). The rescued phenotype in the YF and YF/KA conditions was particularly striking, given their lower protein levels.
The KA mutant remained bound to full-length NCOR despite disrupted binding to DAD under the standard washing conditions in the presence of 1% NP-40 (Figure 3A), suggesting that the second interacting domain in NCOR is sufficient for recruiting HDAC3 in vivo (Guenther et al., 2001; Li et al., 2000; Wen et al., 2000). Such interaction was readily diminished, but not completely abolished, by washing the Flag immunoprecipitates with buffers containing higher detergent concentrations, suggesting that the interaction is stable but of lower affinity than the HDAC3-DAD interaction (Figure 3E). Binding of HDAC3 to TBLR1, another component of the NCOR/SMRT complex, followed the same pattern as HDAC3-NCOR interaction, consistent with the notion that it is mediated through NCOR or SMRT (Figure 3E) (Yoon et al., 2003; Zhang et al., 2002). Interestingly, as HDAC3 was expelled from the NCOR/SMRT complex by harsher washing conditions, we noted increased abundance of HDAC3 within the chaperone TCP-1 ring complex (TRiC) (Figure 3E), suggesting that the TRiC serves as the reservoir for free HDAC3. This is in agreement with previous findings that several key components of the TRiC bind to HDAC3 and exist in a complex distinct from the NCOR/SMRT complex (Guenther et al., 2002; Joshi et al., 2013; Li et al., 2006).
Consistent with the rescue of the metabolic phenotype, YF and KA mutants repressed most lipogenic genes that are upregulated upon HDAC3 depletion (Figure 3F). The extent of lipogenic gene repression correlated well with the residual hepatosteatosis phenotype, with YF repressing most genes to a large degree and KA repressing almost all genes to the same degree as WT. The difference between YF and KA is likely due to the lower protein levels of YF. Efforts to boost YF protein levels by injecting 10-fold higher dosage of the AAV-Tbg-HDAC3 (YF) still resulted in significantly lower protein levels and similar profiles of gene expression as well as hepatic lipid content (Figures S4A–D). Taken together, the deacetylase-dead KA mutant almost completely rescued the metabolic derangement and gene transcriptional alteration in HDAC3-depleted liver, whereas the deacetylase-dead YF rescued these deficits to a large degree despite its lower protein level. These data suggest that the in vivo function of HDAC3 in liver is largely independent of its deacetylase activities.
It should be noted that not all HDAC3 target genes were repressed to the same degree by catalytically inactive mutants (Figures 3F and S3C). Also, there was still residual hepatic steatosis in the K25A rescued liver, albeit to a very limited degree (Figures 3C and 3D). These findings suggest that the catalytic activity is required for some aspects of HDAC3 function, and may be even more important in another tissue or a different physiological condition.
Deacetylase-dead HDAC3 rescues HDAC3-dependent transcriptional repression despite failing to repress genome-wide histone acetylation
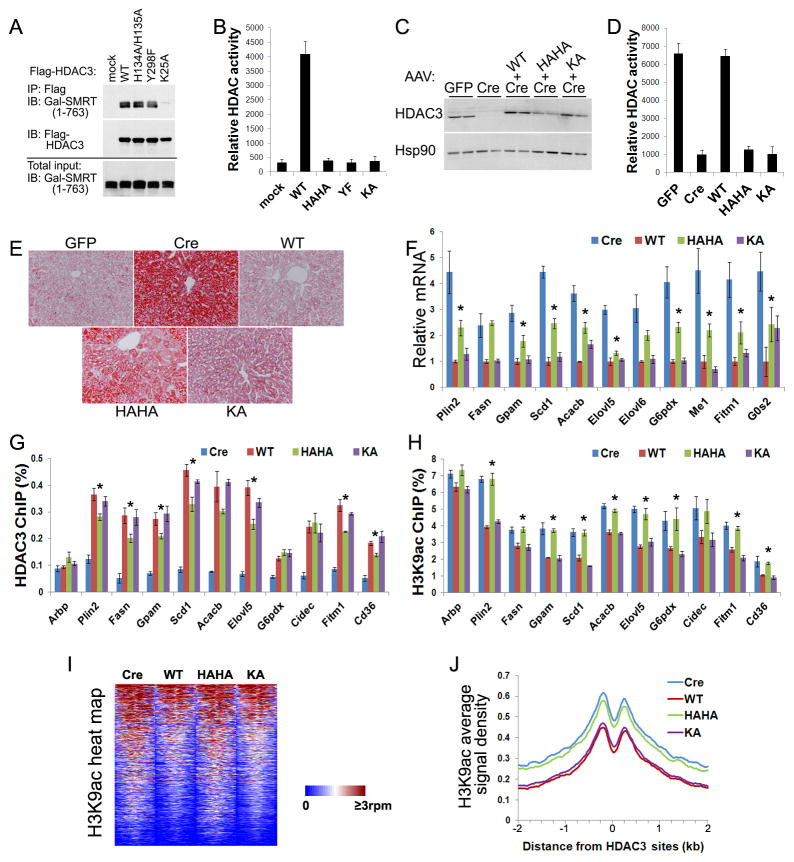
To overcome the sub-physiological expression of the YF mutant, we sought to construct another mutant of the catalytic site. Two highly-conserved tandem His residues (H134 and H135 in HDAC3) are located close to the Zn ion and catalytically important (Figures 2A and S2). They serve as a general base and a general electrostatic catalyst donating a proton to the epsilon nitrogen atom on the substrate lysine, which leads to collapse of the tetrahedral intermediate (Lombardi et al., 2011). Ala substitution of these two His (HAHA) rendered HDAC3 completely inactive in HEK 293T cells without affecting interaction with the SMRT (1–763) containing DAD (Figures 4A and 4B). Similar to YF, HAHA was also expressed at sub-physiological levels when introduced into liver by AAV (Figure 4C). The lack of deacetylase activity in HAHA was confirmed by an HDAC assay using HDAC3 proteins immunoprecipitated from the liver lysates (Figure 4D). Like YF, HAHA rescued fatty liver and repressed lipogenic genes to a large degree (Figures 4E, 4F, S5A, and S5B).
Figure 4. Deacetylase-dead HDAC3 rescues HDAC3-dependent transcriptional repression despite failing to repress genome-wide histone acetylation.
(A) GAL-tagged SMRT (1–763) and Flag-HDAC3 were co-expressed in HEK 293T cells. Cell lysates were immunoprecipitated with Flag antibodies followed by immunoblot analysis. (B) HDAC assay of the above immunoprecipitates in triplicates. (C) HDAC3f/f mice were injected with AAV-Tbg-GFP or AAV-Tbg-Cre along with AAV vectors expressing Flag-tagged HDAC3 mutants. Total liver lysates were analyzed by immunoblot analysis. (D) Liver lysates were immunoprecipitated with anti-HDAC3 antibodies and assayed for enzyme activities, n = 4. (E) ORO staining of livers. (F) RT-qPCR analysis of livers, n = = 4. * P < 0.05 between Cre and HAHA. (G) Livers were subjected to chromatin immunoprecipitation (ChIP) with HDAC3 antibodies followed by qPCR analysis using primers for HDAC3 sites near the indicated genes, n = 4. * P < 0.05 between WT and HAHA. (H) ChIP-qPCR analysis with antibodies against acetylated lysine 9 of histone 3 (H3K9ac), n = 4. * P < 0.05 between WT and HAHA. (I) Heat map of H3K9ac ChIP-seq signals from −1 kb to +1 kb surrounding the center of HDAC3 sites within 50 kb of genes that are upregulated upon HDAC3 depletion. rpm, reads per million. (J) Average H3K9ac signals, represented by reads per bp in 10 million total reads, from −2 kb to +2 kb surrounding the center of the same HDAC3 sites as shown above. G0S2, G0/G1 switch 2; Arbp, ribosomal protein, large, P0, also known as 36B4. All error bars, s.e.m. See also Figure S5.
We next addressed how these different mutants affect chromatin recruitment of HDAC3 and histone acetylation. ChIP-qPCR analysis was performed using primers specific for the previously-determined HDAC3 sites near target lipogenic genes (Feng et al., 2011). Chromatin occupancy of HAHA showed modest but significant reduction in most HDAC3 sites, with the degree of changes likely an effect of poor protein expression (Figure 4G). KA has normal chromatin occupancy, supporting the notion that the second interacting domain in NCOR/SMRT is sufficient for recruiting HDAC3. Therefore, loss of IP4 binding with DAD in NS-DADm mice may induce a conformational change in NCOR/SMRT that affects their interaction with HDAC3 through the second domain, resulting in reduced HDAC3 recruitment and mild steatosis in NS-DADm livers (You et al. 2013). H3K9 acetylation levels at the examined sites were high in the presence of HAHA to a similar degree as HDAC3 knockout, as expected from the loss of deacetylase activity (Figure 4H). Interestingly, histone acetylation levels were low in the presence of KA to a similar degree as WT, even though KA does not have ability to actively deacetylate histones (Figure 4H).
To generate nonbiased acetylation profiles, we subjected DNA from the H3K9ac ChIP to sequencing (ChIP-seq). Consistent with the ChIP-qPCR results, genome-wide H3K9ac levels at HDAC3 binding sites near its target genes were high in the presence of HAHA to a similar degree as in HDAC3 knockout, and were low in the presence of KA to a similar degree as in WT (Figures 4I and 4J). The fact that HAHA rescued fatty liver and repressed HDAC3 target genes to a large degree in spite of its histone hyperacetylation profile demonstrates that histone acetylation is not sufficient to activate gene transcription. This raises the question whether histone acetylation is really the cause for gene transcription or merely a bystander event associated with increased chromatin accessibility near actively transcribed genes. Several lines of evidence favor the second argument. (1) Acetylation on different Lys residues of histones display an “all-or-none” pattern lacking obvious combinatorial complexity that is required for function as a “code” (Rando, 2012); (2) mutation analysis in yeast shows that Arg substitutions of histone Lys residues generate overall moderate phenotypes in gene transcription, even though they would also disrupt other modifications such as methylation, ubiquitination, or sumolyation on the same lysine residues (Bedford and Brindle, 2012); (3) ablation of histone acetylation is not accompanied by equally reduced levels of gene expression upon HAT knockout (Kasper et al., 2010); (4) some HAT enzyme-dead mutants remain functional (Sterner et al., 2002); (5) in vitro nucleosome reconstitution analysis shows that histone acetylation has only subtle effects on chromatin remodeling (Neumann et al., 2009); (6) gene expression changes elicited by HDIs are moderate and do not necessarily resemble those caused by HDACs depletion (Figure 1) (Mullican et al., 2011). Finally, the notion that histone acetylation is a bystander outcome of active gene transcription readily reconciles the apparent paradox of histone hypoacetylation in the presence of deacetylase-dead HDAC3 KA mutant. KA represses gene transcription in a deacetylase-independent manner, and the histone hypoacetylation is the outcome of such transcription repression. Histone hyperacetylation in the presence of HAHA is likely a combined effect of the abolished deacetylase activity and the low protein level. Taken together, genetic and pharmacological manipulation of HDAC3 demonstrate that HDAC3 target genes can remain repressed despite histone hyperacetylation, suggesting that deacetylase-independent function of HDAC3 mediates gene repression and that histone hyperacetylation is not sufficient to activate gene transcription.
Loss of interaction with NCOR/SMRT renders HDAC3 completely nonfunctional in vivo
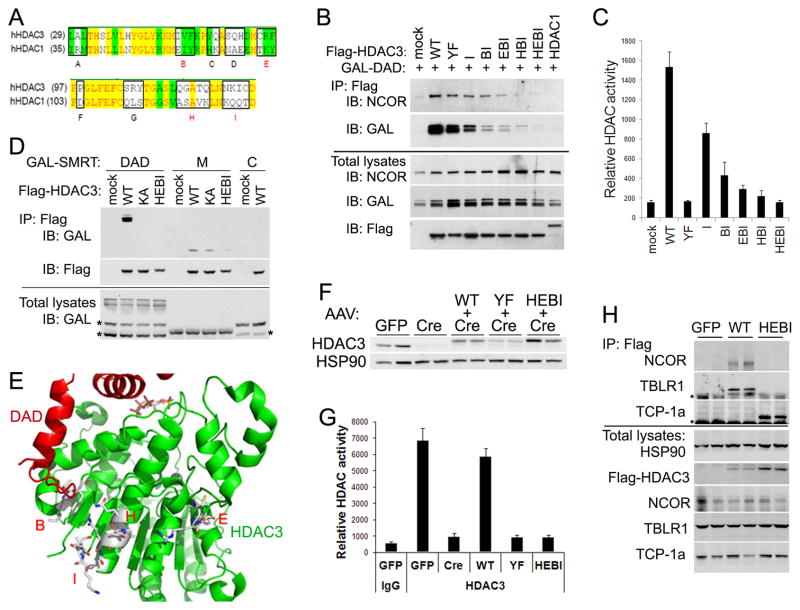
What mediates the deacetylase-independent function of HDAC3? One possibility is that HDAC3 may recruit other epigenome-modifying enzymes such as methyltransferases to the chromatin (Hohl et al., 2013; Stender et al., 2012). However, histone methylation was not changed significantly upon HDAC3 depletion in liver at several HDAC3 sites (Figure S6). Another possibility is that HDAC3 plays a scaffolding role in maintaining the integrity of the corepressor complex through interacting with other proteins. If this is true, abolishing the ability of HDAC3 to interact with the corepressor complex would wipe out the in vivo function of HDAC3. Since none of the tested mutations abolished the physical interaction between HDAC3 and NCOR/SMRT, we sought to identify key residues in HDAC3 for such interaction. Previous truncation analysis of HDAC3 suggests that the key residues for binding NCOR/SMRT are located in the N-terminal region of HDAC3 (Li, 2006). Considering that HDAC1 does not interact with NCOR/SMRT, sequence alignment of HDAC3 and HDAC1 in those regions identified 9 potential essential residue clusters, named “A” through “I” (Figure 5A). Mutation of each cluster in HDAC3 to the corresponding residues in HDAC1 showed that 4 clusters (namely B, E, H, and I) compromised HDAC3 ability to interact with NCOR/SMRT (Li, 2006). Combined mutations in all 4 clusters abolished interaction with not only DAD but also the full-length NCOR in cells (Figure 5B). The combined mutation in the 4 clusters, named “HEBI”, also abolished deacetylase activity, presumably due to loss of interaction with DAD (Figure 5C). To test directly whether HEBI disrupts the interaction with the second domain in the middle region (M) of NCOR/SMRT (Guenther et al., 2001; Li et al., 2000; Wen et al., 2000), truncated SMRT proteins expressed from HEK 293T cells were mixed with HDAC3 and subjected to immunoprecipitation analysis. HEBI disrupted interaction with both DAD and the second domain, while KA only disrupted interaction with DAD (Figure 5D). The HEBI mutations encompass several residues facing outward on the exterior alpha helixes, likely contributing to protein-protein interactions (Figure 5E). Thus the HEBI mutant was annotated as “HDAC3 with Enzyme and Binding activities Inactivated” to distinguish from other mutants aiming to disrupt only the enzyme activity.
Figure 5. A mutant HDAC3 loses interactions with not only DAD but also the full-length NCOR/SMRT.
(A) Sequence alignment of HDAC3 and HDAC1. Identical and similar residues are highlighted in yellow and green respectively. Clusters of residues chosen for mutagenesis are boxed and labeled “A”– “I”. (B) GAL-SMRT (1–763) containing DAD and Flag-tagged HDAC3 mutants were co-expressed in HEK 293T cells. Cell lysates were immunoprecipitated with Flag antibodies and analyzed by immunoblot. (C) HDAC assay of the above immunoprecipitates in triplicates. (D) Protein lysates from HEK 293T cells expressing either a GAL-tagged SMRT truncation or a Flag-tagged HDAC3 mutant were mixed together, immunoprecipitated with Flag antibodies, and analyzed by immunoblot. DAD, the N-terminal 1–763 region of SMRT; M, the middle 1531–1961 region of SMRT containing the second interaction domain with HDAC3; C, the C-terminal 1961–2473 region of SMRT that does not interact with HDAC3 serving as a negative control. * Non-specific signals. (E) HEBI mutations on the crystal structure of HDAC3. (F) HDAC3f/f mice were injected with AAV-Tbg-GFP or AAV-Tbg-Cre along with AAV vectors expressing Flag-tagged HDAC3 mutants. Total liver lysates were analyzed by immunoblot analysis. (G) The liver lysates were immunoprecipitated with either anti-HDAC3 antibodies or normal IgG, followed by HDAC enzyme assay, n = 3–4. (H) The liver lysates were immunoprecipitated with Flag antibodies and washed with buffer containing 1% NP-40, followed by immunoblot analysis. * indicates non-specific signals. All error bars, s.e.m. See also Figure S5.
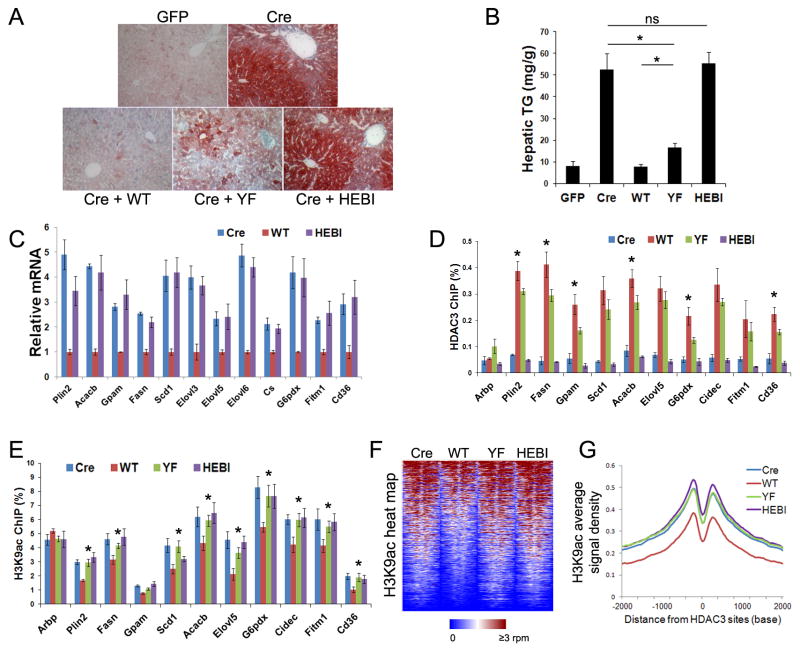
When introduced into the HDAC3-depleted liver by AAV, HEBI was expressed at slightly higher levels than endogenous HDAC3 protein, distinct from the catalytic site mutant YF (Figure 5F). Despite its higher levels, HEBI lacked any detectable deacetylase activity and completely lost interaction with NCOR as well as TBLR1 (Figures 5G and 5H). Interestingly, it had stronger interaction with the TCP-1a, in keeping with the notion that HDAC3 is shunted into TriC when it loses interaction with the corepressor complex (Figure 3E). HEBI completely lost ability to rescue the hepatosteatosis phenotype in HDAC3-depleted livers (Figures 6A and 6B). HEBI was also completely non-functional in terms of repressing expression of HDAC3 target genes (Figure 6C) and occupancy on the chromatin (Figure 6D), suggesting that binding to NCOR/SMRT is essential for genomic recruitment of HDAC3 and subsequent transcriptional repression.
Figure 6. Loss of interaction with NCOR renders hepatic HDAC3 completely nonfunctional in vivo.
(A) ORO stain of livers from HDAC3f/f mice injected with the indicated AAV vectors. (B) Hepatic triglyceride (TG) measurement, n = 4. (C) RT-qPCR analysis of livers from mice described above, n = 4. (D) The livers were subjected to ChIP with HDAC3 antibodies followed by qPCR analysis using primers specific for HDAC3 sites near the indicated genes, n = 4. * P < 0.05 between WT and YF. (E) ChIP-qPCR analysis of the livers with H3K9ac antibodies, n = 4. * P < 0.05 between WT and any of the other three groups. (F) Heat map of H3K9ac ChIP-seq signals from −1 kb to +1 kb surrounding the center of HDAC3 sites within 50 kb of genes that are upregulated upon HDAC3 depletion. (G) Average H3K9ac signals, represented by reads per bp in 10 million total reads, from −2 kb to +2 kb surrounding the center of HDAC3 sites. All error bars, s.e.m.
ChIP-qPCR and ChIP-seq profiling revealed that YF behaved in the similar manner as HAHA in all analyses, as expected since both mutants affect the catalytic site of HDAC3 (Figures 6E–G). Histone acetylation is elevated in the presence of HEBI and YF to a similar degree as in HDAC3 knockout livers, suggesting that the in vivo function of HDAC3, albeit independent of deacetylase activities, requires interacting with the NCOR/SMRT complex.
Liver-specific knockout of NCOR causes metabolic and transcriptomal alterations closely resembling those of mice without hepatic HDAC3
If the NCOR/SMRT complex is indeed required for HDAC3 in vivo function, knockout of NCOR and/or SMRT in the liver should recapitulate the phenotype of the HDAC3 knockout. To this end, we have studied mouse lines containing floxed alleles of either NCOR or SMRT (Figure S7A). Administration of AAV-Tbg-Cre in SMRTf/f mice depleted SMRT in liver (Figures 7A and S7B), but did not affect expression of HDAC3 target genes and did not cause hepatosteatosis (Figures 7A and 7B). By contrast, depletion of NCOR in liver markedly upregulated expression of HDAC3 target genes involved in lipogenesis without altering HDAC3 levels (Figures 7C and 7D). There was ectopic accumulation of lipids within NCOR-depleted livers and reciprocal reduction of hepatic glycogen content (Figures 7E and 7F), closely resembling the metabolic changes observed in HDAC3-depleted livers (Knutson et al., 2008; Sun et al., 2012).
Figure 7. Liver-specific knockout of NCOR, but not SMRT, causes metabolic and transcriptomal alterations resembling those of mice without hepatic HDAC3.
(A) RT-qPCR analysis of livers from SMRTf/f mice injected with AAV-Tbg-GFP or AAV-Tbg-Cre, n = 2–3, * P < 0.05 compared to the GFP group. (B) ORO staining of livers from SMRTf/f mice injected with AAV. (C) Immunoblot analysis of livers from NCORf/f mice injected with AAV-Tbg-GFP or AAV-Tbg-Cre. (D) RT-qPCR analysis of livers from NCORf/f mice injected with AAV, n = 4. * P < 0.05 compared to GFP. (E) ORO staining of livers from NCORf/f mice injected with AAV. (F) Hepatic TG and glycogen measurement in NCORf/f; AAV mice, n = 4–5, * P < 0.05 compared to GFP. (G) Heat map of microarray results from NCOR or HDAC3 depleted livers. Genes that were upregulated (294 genes) or downregulated (272 genes) in HDAC3-depleted livers versus their wild-type controls were selected (fold-change > 1.4, q < 0.05), and were sorted by fold-changes in NCOR-depleted livers versus their controls. (H) Gene ontology analysis using top 200 upregulated genes (by fold-change) in either the HDAC3 KO versus WT array or the NCOR KO versus WT array, with p < 0.05. All error bars, s.e.m. See also Figure S7.
Transcriptome profiling revealed that the majority of genes repressed by HDAC3 also tended to be upregulated upon depletion of NCOR, demonstrating the necessity of NCOR in HDAC3-mediated transcription repression (Figure 7G). The overall milder transcriptomal changes in NCOR depleted livers suggest a partial compensation from SMRT. In contrast, among genes downregulated upon HDAC3 depletion, roughly the same percentage were up- or down- regulated upon NCOR depletion, suggesting that those gene expression changes are likely indirect effects of HDAC3 depletion. Genes repressed by either HDAC3 or NCOR were highly enriched in lipid and fatty acid metabolism, consistent with the similar lipid metabolic phenotypes in NCOR and HDAC3 depleted livers (Figure 7H).
Genome-wide occupancy of SMRT in liver did not display oscillation throughout the day (Figure S7C), whereas the hepatic NCOR cistrome shows robust circadian rhythm that is in-phase with HDAC3 (Feng et al., 2011), suggesting that NCOR plays a more important role than SMRT in genomic recruitment of HDAC3 in liver. SMRT may still contribute to physiological recruitment of HDAC3, and indeed a modest increase in NCOR protein levels in SMRT-depleted livers may contribute to the lack of steatosis phenotype (Figure S7B). Nonetheless, the lack of an obvious metabolic phenotype in liver-specific SMRT knockout mice suggests that extrahepatic tissues such as adipose are responsible for the observed metabolic alterations in SMRT heterozygous mice or SMRT knock-in mice bearing mutations in receptor-interacting domains (RIDs) (Fang et al., 2011; Nofsinger et al., 2008; Reilly et al., 2010; Sutanto et al., 2010). The hepatosteatosis phenotype in NCOR liver-specific knockout mice is in contrast to the normal hepatic lipid content in the whole-body knock-in mice with mutated NCOR DAD (N-DADm) and liver-specific knock-in mice bearing NCOR with two RIDs truncated, although both mouse models show a modest increase in lipogenic gene expression (Alenghat et al., 2008; Astapova et al., 2008). These findings suggest that both DAD and the two RIDs contribute to, but are not absolutely required for, NCOR function in vivo. Of note, genomic occupancy of NCOR and SMRT in liver is not affected by HDAC3 depletion (You et al., 2013). Taken together, these results demonstrate that although deacetylase enzymatic activity is dispensable, interaction with NCOR is required for the in vivo function of HDAC3 in liver.
DISCUSSION
Genes for catalytically dead enzymes, bearing mutations at key catalytic residues, are found throughout the genome for almost all enzyme families with conserved sequences across different species (Adrain and Freeman, 2012). Such genomic arrangement not only suggests the prevalent existence of enzyme-independent functions for these pseudoenzymes, but also provides insights into how active enzymes evolve from their dead homologues or even visa versa (Adrain and Freeman, 2012; Leslie, 2013). Here we demonstrate that studying catalytically-inactive mutant enzymes in an in vivo phenotype-rescue setting is an efficient and powerful approach to uncover and characterize enzyme-independent functions.
The importance of the HDACs family has gained increasing recognition over the past decade. Intriguingly, Class IIa HDACs, including HDAC4, -5, -7 and -9, have no enzymatic activity due to a His substitution on the key catalytic Tyr residue (corresponding to Y298 in HDAC3) and therefore are actually pseudoenzymes (Lahm et al., 2007). The deacetylase activity observed in class IIa HDACs purified from cellular contexts is dependent on HDAC3 that is physically associated with them (Fischle et al., 2002). These findings have led to the notion that class IIa HDACs mainly play scaffolding roles in recruiting HDAC3 to their substrates (Mihaylova et al., 2011; Schapira, 2011). The present study takes this scenario one step further by demonstrating that the deacetylase activity is actually dispensable for HDAC3 functions in vivo, suggesting that we should look beyond such scaffolding functions for class IIa HDACs. In line with this concept, several class IIa HDACs are able to exert their cellular functions without scaffolding any deacetylation reactions when overexpressed in vitro in cultured cells (Chatterjee et al., 2011; Ma and D’Mello, 2011; Yang et al., 2011; Zhou et al., 2000).
The concept is by no means limited to class II HDACs. Class I HDAC8 and its deacetylase-dead mutant, can interfere with the ubiquitination machinery to the same degree when overexpressed in cells (Lee et al., 2006). Transgenic overexpression of deacetylase-dead mutants of either HDAC1 or HDAC3 in mouse heart causes cardiomyopathy to the same degree of severity as overexpression of their respective wild-type enzymes, suggesting that deacetylase-independence is generalizable to other class I HDACs, although potential overexpression artifacts cannot be ruled out in this experimental setting (Potthoff, 2007). In addition, HDIs do not block some cellular functions of overexpressed HDAC3 in cultured cells (Gupta et al., 2009). Deacetylase-independent functions have also been suggested for class III HDACs in overexpression cell culture models (Shah et al., 2012; Zhang et al., 2009, 2010). These findings merit further investigation into whether and to what extent the deacetylase enzyme activity may contribute to the biological function of each HDAC in vivo.
Our current findings have profound implications for mechanistic characterization of small molecule HDIs. If HDACs do not require deacetylase activity for most of their functions in vivo, they may not be the de facto targets of HDIs. Almost all existing HDIs exert their inhibiting activities by chelating the Zn metal in the active site of HDACs (Gryder et al., 2012). Besides HDACs, there are over 300 Zn-dependent enzymes encompassing all of the six main enzyme families, whose active sites usually share a common tetrahedral [(XYZ)-Zn-OH2] structure in which the Zn ion is coordinated by three amino acid residues with the fourth site occupied by a catalytically-important water molecule or a hydroxide group (Parkin, 2004). It is likely that HDIs interfere with other Zn enzymes or other metalloproteins besides HDACs in vivo, which is genuinely responsible for their pleiotropic therapeutic effects.
This idea is in keeping with several observations. Transcriptomal profiling of HDIs-exposed cells revealed overall minimal changes in gene expression and quite different patterns in response to different pan-HDIs (Halsall et al., 2012; Lopez-Atalaya et al., 2013). In fact, some effects of HDIs can be independent of gene expression changes (Wardell et al., 2009). In many animal and cell culture models, HDI treatment does not phenocopy HDAC knockout or knockdown, and in some cases even generates opposite phenotypes. For example, while HDIs have anti-cancer effects in an almost universal manner against a wide range of tumors, HDAC1 depletion promotes teratoma formation (Lagger et al., 2010), HDAC1 and HDAC2 knockdown facilitates leukemogenesis in pre-leukemic mice (Santoro et al., 2013), and HDAC3 knockout in liver results in hepatocellular carcinoma (Bhaskara et al., 2010). NCOR and SMRT also suppress breast and prostate cancers, consistent with their functions in repressing gene transcription mediated by estrogen and androgen receptors (Keeton and Brown, 2005; Qi et al., 2013). Last but not least, although recent cancer genomic studies powered by advanced DNA sequencing technologies have implicated many transcription factors and epigenomic modifiers in carcinogenesis, few mutations have been found in HDACs that are associated with any types of malignancies, even though some HDIs have been approved for treating cancers and many more show similar promise (Garraway and Lander, 2013; Suvà et al., 2013).
EXPERIMENTAL PROCEDURES
Mice
HDAC3f/f mice were described previously (Mullican et al., 2011). NCORf/f and SMRTf/f mice were obtained from MCI/ICS (Mouse Clinical Institute–Institut Clinique de la Souris, Illkirch, France; http://www.ics-mci.fr/). NCORf/f mice contained floxed exon 11 (Yamamoto et al., 2011). SMRTf/f mice (ICS # K175/DG34/EUMO15) contained floxed exon 4 (Figure S7A). AAV2/8-Tbg-HDAC3 vectors containing mutations were intravenously injected together with AAV2/8-Tbg-Cre in adult mice for rescue experiments, using AAV2/8-Tbg-GFP as a negative control. Details were described in Supplemental Experimental Procedures.
Cell culture and DNA constructs
Primary hepatocytes were isolated from HDAC3f/f mice and treated with adenovirus or HDIs. Details were described in Supplemental Experimental Procedures. Site-directed mutagenesis was performed using Stratagene kit.
Immunoprecipitation, immunoblot, and HDAC assay
Primary hepatocytes were either lyased directly in Laemmli sample buffer or acid extracted. Immunoprecipitation, immunoblot, and antibodies were described in Supplemental Experimental Procedures. HDAC assay was conducted using a fluorescence kit (Active Motif) following manufacture’s instruction.
RT-qPCR, microarray, ChIP-qPCR, ChIP-seq, and computational analysis
These procedures were described previously (Feng et al., 2011) and detailed in the Supplemental Experimental Procedures.
Statistics
To determine significance differences between two groups, student’s two-tail t-test was used for all experiments except the microarray.
Accession numbers
The following data were deposited in Gene Expression Omnibus: microarray in HDAC3f/f; AAV-Cre versus AAV-Cre + AAV-HDAC3-WT at 2-weeks post-injection (GSE 49386) and NCORf/f; AAV-Cre versus AAV-GFP (GSE 49387); H3K9ac ChIP-seq in two rescue experiments (GSE 49365) and SMRT ChIP-seq at 5 pm versus 5 am (GSE 51045).
Supplementary Material
ARTICLE HIGHLIGHTS.
Catalytically inactive HDAC3 mutants are functional in vivo
Histone acetylation is not sufficient to activate gene transcription
Loss of binding with corepressor NCOR renders HDAC3 nonfunctional
Liver-specific knockout of NCOR resembles HDAC3 depletion
Acknowledgments
We thank Dr. David Steger for critical reading of the manuscript, Jarrett Remsberg for images of crystal structure, and Cristina Lanzillotta for technical assistance. We thank the Penn Diabetes Center (DK19525) Functional Genomics Core for sequencing and Viral Vector Core for AAV production. We thank Penn Digestives Disease Center Morphology Core (DK050306) for histology studies and Molecular Profiling Core for microarray analysis. This work was supported by K99DK099443 (to ZS) and R37DK43806 (to MAL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adrain C, Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol. 2012;13:489–498. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bućan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrar M, Turnham R, Pierce L, De Oliveira CAF, McCammon JA. Structural insight into the separate roles of inositol tetraphosphate and deacetylase-activating domain in activation of histone deacetylase 3. Protein Sci. 2013;22:83–92. doi: 10.1002/pro.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astapova I, Lee LJ, Morales C, Tauber S, Bilban M, Hollenberg AN. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc Natl Acad Sci USA. 2008;105:19544–19549. doi: 10.1073/pnas.0804604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford DC, Brindle PK. Is histone acetylation the most important physiological function for CBP and p300? Aging (Albany NY) 2012;4:247–255. doi: 10.18632/aging.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, Hiebert SW. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, Mak R, Tanguturi SK, Mazitschek R, Haggarty SJ, Ross K, Chang CY, Bosco J, West N, Morse E, et al. Chemical genetic strategy identifies histone deacetylase 1 (HDAC1) and HDAC2 as therapeutic targets in sickle cell disease. Proc Natl Acad Sci USA. 2010;107:12617–12622. doi: 10.1073/pnas.1006774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateauvieux S, Morceau F, Dicato M, Diederich M. Molecular and therapeutic potential and toxicity of valproic acid. J Biomed Biotechnol. 2010;2010 doi: 10.1155/2010/479364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee TK, Idelman G, Blanco V, Blomkalns AL, Piegore MG, Jr, Weintraub DS, Kumar S, Rajsheker S, Manka D, Rudich SM, et al. Histone deacetylase 9 is a negative regulator of adipogenic differentiation. J Biol Chem. 2011;286:27836–27847. doi: 10.1074/jbc.M111.262964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Suh JM, Atkins AR, Hong SH, Leblanc M, Nofsinger RR, Yu RT, Downes M, Evans RM. Corepressor SMRT promotes oxidative phosphorylation in adipose tissue and protects against diet-induced obesity and insulin resistance. Proc Natl Acad Sci USA. 2011;108:3412–3417. doi: 10.1073/pnas.1017707108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Goodson M, Jonas BA, Privalsky MA. Corepressors: custom tailoring and alterations while you wait. Nucl Recept Signal. 2005;3:e003. doi: 10.1621/nrs.03003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Sodji QH, Oyelere AK. Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed. Future Med Chem. 2012;4:505–524. doi: 10.4155/fmc.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 2002;16:3130–3135. doi: 10.1101/gad.1037502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Ho PC, Ha SG, Lin YW, Wei LN. HDAC3 as a molecular chaperone for shuttling phosphorylated TR2 to PML: a novel deacetylase activity-independent function of HDAC3. PLoS ONE. 2009;4:e4363. doi: 10.1371/journal.pone.0004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsall J, Gupta V, O’Neill LP, Turner BM, Nightingale KP. Genes are often sheltered from the global histone hyperacetylation induced by HDAC inhibitors. PLoS ONE. 2012;7:e33453. doi: 10.1371/journal.pone.0033453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Wagner M, Reil JC, Müller SA, Tauchnitz M, Zimmer AM, Lehmann LH, Thiel G, Böhm M, Backs J, et al. HDAC4 controls histone methylation in response to elevated cardiac load. J Clin Invest. 2013;123:1359–1370. doi: 10.1172/JCI61084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper LH, Lerach S, Wang J, Wu S, Jeevan T, Brindle PK. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- Keeton EK, Brown M. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-alpha and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol Endocrinol. 2005;19:1543–1554. doi: 10.1210/me.2004-0395. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagger S, Meunier D, Mikula M, Brunmeir R, Schlederer M, Artaker M, Pusch O, Egger G, Hagelkruys A, Mikulits W, et al. Crucial function of histone deacetylase 1 for differentiation of teratomas in mice and humans. EMBO J. 2010;29:3992–4007. doi: 10.1038/emboj.2010.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfí A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sengupta N, Villagra A, Rezai-Zadeh N, Seto E. Histone deacetylase 8 safeguards the human ever-shorter telomeres 1B (hEST1B) protein from ubiquitin-mediated degradation. Mol Cell Biol. 2006;26:5259–5269. doi: 10.1128/MCB.01971-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie M. Molecular biology “Dead” enzymes show signs of life. Science. 2013;340:25–27. doi: 10.1126/science.340.6128.25. [DOI] [PubMed] [Google Scholar]
- Li Y. PhD Dissertation. University of Pennsylvania; 2006. Insights into histone deacetylase 3 activation and its new function in mitosis. [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006;20:2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi PM, Cole KE, Dowling DP, Christianson DW. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr Opin Struct Biol. 2011;21:735–743. doi: 10.1016/j.sbi.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Ito S, Valor LM, Benito E, Barco A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, D’Mello SR. Neuroprotection by histone deacetylase-7 (HDAC7) occurs by inhibition of c-jun expression through a deacetylase-independent mechanism. J Biol Chem. 2011;286:4819–4828. doi: 10.1074/jbc.M110.146860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullican SE, Gaddis CA, Alenghat T, Nair MG, Giacomin PR, Everett LJ, Feng D, Steger DJ, Schug J, Artis D, et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 2011;25:2480–2488. doi: 10.1101/gad.175950.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, Van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofsinger RR, Li P, Hong SH, Jonker JW, Barish GD, Ying H, Cheng SY, Leblanc M, Xu W, Pei L, et al. SMRT repression of nuclear receptors controls the adipogenic set point and metabolic homeostasis. Proc Natl Acad Sci USA. 2008;105:20021–20026. doi: 10.1073/pnas.0811012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocker M, Schneider-Stock R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int J Biochem Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Oehme I, Deubzer HE, Wegener D, Pickert D, Linke JP, Hero B, Kopp-Schneider A, Westermann F, Ulrich SM, Von Deimling A, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15:91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- Parkin G. Synthetic analogues relevant to the structure and function of zinc enzymes. Chem Rev. 2004;104:699–767. doi: 10.1021/cr0206263. [DOI] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Potthoff M. PhD Dissertation. The University of Texas Southwestern Medical Center; Dallas: 2007. MEF2 and HDAC proteins regualte strated muscle development and remodeling. [Google Scholar]
- Qi J, Tripathi M, Mishra R, Sahgal N, Fazil L, Ettinger S, Placzek WJ, Claps G, Chung LWK, Bowtell D, et al. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell. 2013;23:332–346. doi: 10.1016/j.ccr.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SM, Bhargava P, Liu S, Gangl MR, Gorgun C, Nofsinger RR, Evans RM, Qi L, Hu FB, Lee CH. Nuclear receptor corepressor SMRT regulates mitochondrial oxidative metabolism and mediates aging-related metabolic deterioration. Cell Metab. 2010;12:643–653. doi: 10.1016/j.cmet.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro F, Botrugno OA, Dal Zuffo R, Pallavicini I, Matthews GM, Cluse L, Barozzi I, Senese S, Fornasari L, Moretti S, et al. A dual role for Hdac1: oncosuppressor in tumorigenesis, oncogene in tumor maintenance. Blood. 2013;121:3459–3468. doi: 10.1182/blood-2012-10-461988. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Schapira M. Structural biology of human metal-dependent histone deacetylases. Handb Exp Pharmacol. 2011;206:225–240. doi: 10.1007/978-3-642-21631-2_10. [DOI] [PubMed] [Google Scholar]
- Shah ZH, Ahmed SU, Ford JR, Allison SJ, Knight JRP, Milner J. A deacetylase-deficient SIRT1 variant opposes full-length SIRT1 in regulating tumor suppressor p53 and governs expression of cancer-related genes. Mol Cell Biol. 2012;32:704–716. doi: 10.1128/MCB.06448-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Wang X, Bloom MH, Simon GM, Berger SL. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J Biol Chem. 2002;277:8178–8186. doi: 10.1074/jbc.M108601200. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Bates SE, Wright JJ, Espinoza-Delgado I, Piekarz RL. Clinical Toxicities of Histone Deacetylase Inhibitors. Pharmaceuticals. 2010;3:2751–2767. doi: 10.3390/ph3092751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Feng D, Everett LJ, Bugge A, Lazar MA. Circadian epigenomic remodeling and hepatic lipogenesis: lessons from HDAC3. Cold Spring Harb Symp Quant Biol. 2011;76:49–55. doi: 10.1101/sqb.2011.76.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutanto MM, Ferguson KK, Sakuma H, Ye H, Brady MJ, Cohen RN. The silencing mediator of retinoid and thyroid hormone receptors (SMRT) regulates adipose tissue accumulation and adipocyte insulin sensitivity in vivo. J Biol Chem. 2010;285:18485–18495. doi: 10.1074/jbc.M110.107680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfí A, De Francesco R, Steinkühler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 2007;8:879–884. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanczyk M, Roszczenko K, Marcinkiewicz K, Bojarczuk K, Kowara M, Winiarska M. HDACi--going through the mechanisms. Front Biosci. 2011;16:340–359. doi: 10.2741/3691. [DOI] [PubMed] [Google Scholar]
- Wardell SE, Ilkayeva OR, Wieman HL, Frigo DE, Rathmell JC, Newgard CB, McDonnell DP. Glucose metabolism as a target of histone deacetylase inhibitors. Mol Endocrinol. 2009;23:388–401. doi: 10.1210/me.2008-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Santos GM, Schwabe JWR. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting RH, Yanover E, Heideman MR, Jacobs H, Horner J, Van der Torre J, DePinho RA, Dannenberg JH. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29:2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Williams EG, Mouchiroud L, Cantó C, Fan W, Downes M, Héligon C, Barish GD, Desvergne B, Evans RM, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nature Reviews Molecular Cell Biology. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Tse AKW, Li P, Ma Q, Xiang S, Nicosia SV, Seto E, Zhang X, Bai W. Inhibition of androgen receptor activity by histone deacetylase 4 through receptor SUMOylation. Oncogene. 2011;30:2207–2218. doi: 10.1038/onc.2010.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You SH, Lim HW, Sun Z, Broache M, Won KJ, Lazar MA. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol. 2013;20:182–187. doi: 10.1038/nsmb.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Pan Y, Cao Y, Lai PBS, Liu L, Chen GG, Yun J. Histone deacetylase inhibitors facilitate dihydroartemisinin-induced apoptosis in liver cancer in vitro and in vivo. PLoS ONE. 2012;7:e39870. doi: 10.1371/journal.pone.0039870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Li S, Cruz P, Kone BC. Sirtuin 1 functionally and physically interacts with disruptor of telomeric silencing-1 to regulate alpha-ENaC transcription in collecting duct. J Biol Chem. 2009;284:20917–20926. doi: 10.1074/jbc.M109.020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhang R, Chen HZ, Liu JJ, Jia YY, Zhang ZQ, Yang RF, Zhang Y, Xu J, Wei YS, Liu DP, et al. SIRT1 suppresses activator protein-1 transcriptional activity and cyclooxygenase-2 expression in macrophages. J Biol Chem. 2010;285:7097–7110. doi: 10.1074/jbc.M109.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Richon VM, Rifkind RA, Marks PA. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc Natl Acad Sci USA. 2000;97:1056–1061. doi: 10.1073/pnas.97.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.