Abstract
Eukaryotic cells compartmentalize their biochemical processes within organelles, which have specific functions that must be maintained for overall cellular health. As the site of aerobic energy mobilization and essential biosynthetic activities, mitochondria are critical for cell survival and proliferation. Here, we describe mechanisms to control the quality and quantity of mitochondria within cells with an emphasis on findings from the budding yeast, Saccharomyces cerevisiae. We also describe how mitochondrial quality and quantity control systems that operate during cell division affect lifespan and cell cycle progression.
Keywords: mitochondria, inheritance, lifespan, cell division, yeast
Organelle inheritance during asymmetric cell division
Cell polarization is achieved by the asymmetric distribution of cellular constituents along a cellular axis. This process creates subcellular domains such as the leading edge of motile cells, apical and basolateral aspects of epithelial cells, and neurological and immunological synapses. Cell polarization is also critical for asymmetric cell division, a process that underlies diversity during development. Emerging studies have revealed mechanisms for controlling both the amount and functional state of mitochondria in distinct subcellular domains in polarized cells, which in turn affects cell fitness and function.
Model systems provide a foundation for understanding mitochondrial quality and quantity control during asymmetric cell division. At the onset of cell division in the budding yeast Saccharomyces cerevisiae, a bud site is selected on the surface of the mother cell. The cytoskeleton is then polarized towards that site, which leads to delivery of cellular constituents to the bud for bud formation and growth (Fig. 1). Organelle movements in mammalian cells depend on both microtubules and actin filaments. However, most organelles move along actin cables, bundles of actin filaments that align along the mother-bud axis, in yeast [1].
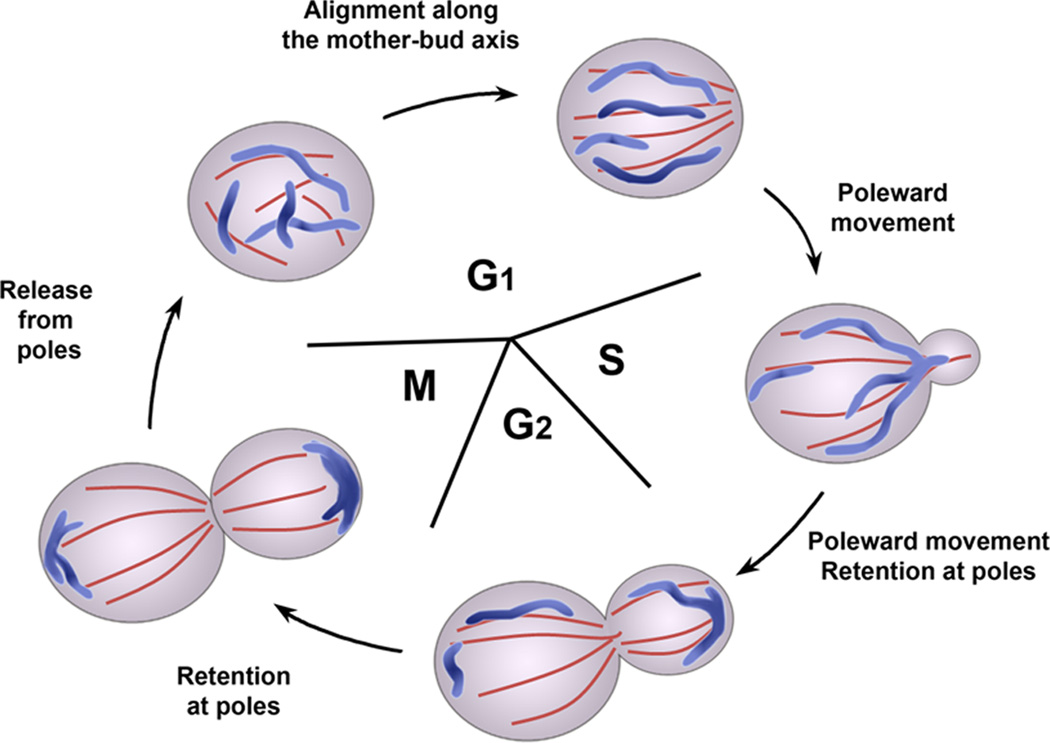
Figure 1. The mitochondrial inheritance cycle in budding yeast.
Since mitochondria are essential organelles that must be produced from pre-existing mitochondria, there are mechanisms to ensure that daughter cells receive mitochondria. In budding yeast, segregation of mitochondria between mother and daughter cells occurs by cytoskeleton-dependent movements of the organelle that resemble those of chromosome movement: mitochondria undergo poleward movement toward the bud tip and the distal tip of the mother cell, followed by anchorage at the poles. These movements result in segregation of the organelle during cell division.
In the case of mitochondrial inheritance, the organelle aligns along polarized actin cables during G1 phase [2]. During bud growth in S and G2 phases and through the end of the cell division cycle, mitochondria undergo actin cable-dependent poleward movements, either toward the bud (anterograde movement) or away from the bud (retrograde movement) [3]. In addition, mitochondria accumulate and are immobilized at the mother cell tip (the pole opposite to the site of bud emergence), and the bud tip [4–7]. Finally, after cytokinesis, mitochondria are released from the poles and redistributed throughout the cytoplasm [2] (Figure 1).
Here, we discuss mitochondrial motility and immobilization during inheritance in yeast, and discuss how these processes lead to quantity control (ensuring that a daughter cell inherits mitochondria) and quality control (ensuring that the inherited mitochondria are functional). In particular, we focus on how checkpoints inhibit cell cycle progression in response to defects in mitochondrial inheritance, and how this machinery promotes preferential inheritance of fitter mitochondria in daughter cells, in turn affecting lifespan.
Mitochondrial motility during inheritance in budding yeast
A central player in mitochondrial function is a protein complex originally referred to as the mitochore that consists of Mdm10p, Mdm12p, and Mmm1p [8]. Mitochore subunits were originally identified as proteins required for mitochondrial morphology and inheritance [9]. Early studies also revealed a role for the mitochore in linking mitochondria to the actin cytoskeleton for movements leading to inheritance [8, 10]. Later, Mdm34p was identified as a member of the complex and additional roles were discovered, including linking mitochondria to ER and mediating assembly of beta barrel proteins in the mitochondrial outer membrane (MOM) [11–13]. This complex is also referred to as ERMES, for ER-mitochondria encounter structure [11].
In yeast and mammalian cells, mitochondria-ER interactions are also critical for phospholipid biosynthesis. Recent studies support a role for the mitochore/ERMES in mitochondrial-ER interactions and phospholipid biosynthesis at that site. Mmm1p is a glycoprotein that localizes to the ER, while Mdm10p, and Mdm34p are integral MOM proteins. Survival of cells bearing a deletion in any one of these proteins is dependent upon expression of a chimera that artificially tethers mitochondria to ER. Deletion of MDM10, MMM1, MDM12 or MDM34 also results in slow growth and defects in conversion of phosphatidylserine (PS) to phosphatidylcholine (PC) [11]. However, mitochore/ERMES mutants are still able to transport PS from ER to mitochondria [14, 15]. Thus, while there is evidence for a role for the mitochore/ERMES in PC biosynthesis at ER-mitochondrial contacts, its precise function in lipid biosynthesis is complex. Interestingly, expression of an artificial ER-mitochondria tether restores defects in mitochondrial morphology, cell growth and PS to PC conversion in some but not all mitochore-ERMES mutants [11]. These findings indicate that the mitochore/ERMES functions in other processes in addition to linking mitochondria to ER.
Other studies revealed that overexpression of a Rab-like protein Ypt11p (see below) results in an increase in the amount of mitochondria in the bud, but does not restore mitochondrial morphology in mitochore/ERMES mutants. This led to the proposal that the primary function of the mitochore/ERMES is to control mitochondrial morphology and not link mitochondria to the actin cytoskeleton [14]. On the other hand, mitochondria co-localize with actin cables, bind to F-actin in cell-free systems and undergo bidirectional movement along actin cables in living yeast cells. Moreover, deletion of mitochore/ERMES subunits results in loss of mitochondrial motility in vivo and binding of mitochondria to F-actin in vitro [8, 10]. Thus, another function of the mitochore/ERMES may be to link mitochondria to actin cables for movements leading to inheritance.
Movement of mitochondria from the bud to the mother cell is driven by actin cable dynamics. Actin cables, like actin bundles and networks in filopodia or the leading edge of motile cells, undergo retrograde flow: continuous movement from the bud toward the mother cell tip [16]. Mitochondria undergoing retrograde movement are associated with actin cables undergoing retrograde flow. Moreover, mutations that inhibit retrograde actin cable flow also inhibit retrograde mitochondrial movement. These findings support the model that mitochondria bind to actin cables and use the force of retrograde actin cable flow to move from the bud towards the mother cell [3]. To deliver mitochondria from mother cells to buds, anterograde forces must be generated to overcome the opposing retrograde actin cable flow. The two force generators for anterograde cargo movement in yeast are myosin motor proteins [17] and actin polymerization mediated by the Arp2/3 complex [18]. In S. cerevisiae two class V myosins, Myo2p and Myo4p, transport cargoes along actin cables towards the F-actin barbed ends. Myo2p is the anterograde motor for secretory vesicles, vacuoles, peroxisomes, and late Golgi vesicles, including those that recycle ER components from the Golgi to the ER. Myo4p transports the cortical ER (cER) and mRNA into the bud [19]. Arp2/3 complex and actin polymerization drives endosome movement [20].
The mechanism underlying mitochondrial movement during inheritance is controversial. Here, we summarize findings obtained from analysis of mitochondrial movement in living yeast cells and interactions of isolated mitochondria with actin. Mutations in Myo2p, including those in the cargo-binding domain, result in defects in mitochondrial inheritance and reduced frequency of movement of the organelle across the bud neck [21–23]. Consistent with this, Myo2p–dependent actin binding activity is detected in isolated yeast mitochondria and Myo2p is detected on isolated yeast mitochondria by immunoelectron microscopy [21, 22]. Moreover, targeting of Myo2p as an artificial fusion protein to mitochondria promotes mitochondrial inheritance in MYO2 mutants [22]. Thus, mitochondria may utilize Myo2p for transport across the bud neck [22].
Although Myo2p facilitates the transport of mitochondria across the bud neck, its role in the mother cell is questionable. Mutations in MYO2 that eliminate its motor activity, result in defects in mitochondrial distribution, or inhibit association of Myo2p with mitochondria, have no effect on the velocity of mitochondrial movement in mother cells [4, 22]. It is possible that MYO2 affects the frequency and/or persistence of mitochondrial movement in mother cells without affecting velocity. On the other hand, the frequency and velocity of anterograde mitochondrial movement are severely diminished in yeast carrying mutations in the Arp2/3 complex as is mitochondrial inheritance [24]. Consistent with this, Arp2/3 complex protein and activity localize to mitochondria in living yeast and are recovered with isolated yeast mitochondria [24]. In addition, the H372R mutation in actin, which accelerates Arp2/3-dependent actin polymerization, results in mitochondrial morphology defects and loss of mtDNA [25]. Similarly, increasing the rate of Arp2/3-dependent actin polymerization in mating yeast increases mitochondrial motility, while suppressing this polymerization by deletion of the ARC18 subunit, a non-essential subunit of the Arp2/3 complex, has the opposite effect [26]. Studies on Jsn1p indicate that the defect in mitochondrial motility observed in Arp2/3 complex mutants is not a consequence of Arp2/3 complex function in actin organization and function. Jsn1p, a Pumilio family protein, localizes to mitochondria, can bind to Arp2/3 complex, co-immunoprecipitates with mitochondria-associated Arp2/3 complex and is required for localization of the Arp2/3 complex to mitochondria. Thus, Jsn1p is a receptor for the Arp2/3 complex on yeast mitochondria. Deletion of JSN1 results in defects in recruitment of Arp2/3 complex to mitochondria and defects in anterograde mitochondrial movement, but has no major effect on actin cable abundance or polarity or on association of mitochondria with actin cables [3, 27].
Therefore, what are the roles of Myo2p and Arp2/3 complex in mitochondrial inheritance? One possibility is that they may act at specific locations. Anterograde movements of mitochondria in the mother cell may depend on Arp2/3-dependent actin polymerization. In this case, new actin filament branches produced by Arp2/3 complex on the mitochondrial surface are bundled in parallel with the existing actin cables, which guide motility in the anterograde direction along actin cables [3]. Transport across the bud neck may require Myo2p function [23, 28]. Since the bud neck is a bottleneck for movement of all cargos in yeast, transport of mitochondria across this site may require the more robust force-generating capabilities of a myosin motor.
Anchorage of mitochondria at the cell cortex
Localized anchorage of mitochondria promotes inheritance of the organelle by buds and retention of the organelle in mother cells. Since actin cables undergo retrograde flow, anchorage of mitochondria in the bud tip ensures that the organelle is retained in buds. Other studies indicate that mitochondria are also anchored at specific sites in the mother cell. Here, we discuss region-specific anchorage of mitochondria in yeast and a role for mitochondrial-ER interactions and specific proteins in these processes.
Recent studies indicate that mitochondria are anchored in the bud tip by interactions with ER. Yeast cortical ER (cER) is a reticular network of ER that underlies and is anchored to the plasma membrane [7, 29, 30]. Super-resolution light microscopy revealed that mitochondria in the yeast bud tip are associated with cER sheets, while electron microscopy revealed that mitochondria can be deformed into thin tubular extensions from their point of contact with cER in the bud tip, implying tension at the point of contact between the two organelles [7].
Recent studies also support a role for two proteins that bind to the Myo2p cargo-binding domain, Mmr1p and Ypt11p, in bud tip anchorage of mitochondria in yeast. Mmr1p was originally identified as a protein that can bind to mitochondria and Myo2p, localize to the bud tip, and is required for normal mitochondrial distribution [31]. Indeed, yeast with mutations in MMR1 and MYO2 exhibit similar defects in mitochondrial distribution. Mmr1p shows some similarity to Dsl1p, which is part of a complex that tethers COPI vesicles to ER [32]. Moreover, Mmr1p localizes to punctate structures between mitochondria and cER at the bud tip and can be recovered with mitochondria and ER upon subcellular fractionation. Finally, deletion of MMR1 results in defective immobilization of mitochondria in the bud tip, whereas its overexpression causes excessive accumulation of mitochondria at that site [7, 31]. These findings support the model that Mmr1p tethers mitochondria to cER in the bud tip, which results in anchorage and accumulation of the organelle at that site. Since Mmr1p binds to Myo2p, and requires this binding to localize to the bud tip, Myo2p may contribute to mitochondrial distribution by mediating actin-dependent transport of Mmr1p to the bud tip (Fig. 2).
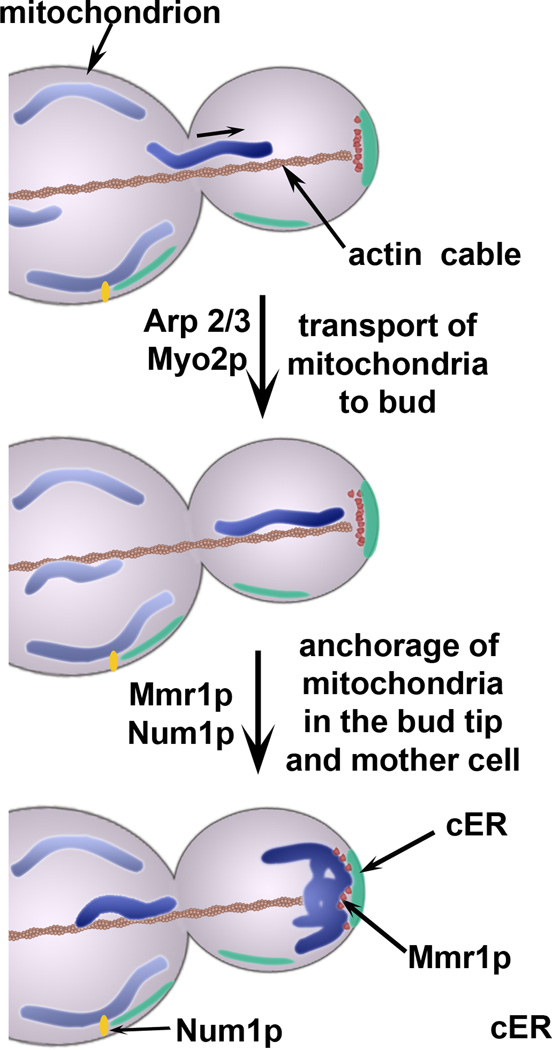
Figure 2. Mitochondrial motility and anchorage in budding yeast.
Mitochondria undergo movement from mother cells to buds using actin cables as tracks and force generation by Arp 2/3 complex and actin polymerization and by Myo2p, a type V myosin. In the yeast bud tip, mitochondria are anchored and accumulate on a cortical ER (cER) sheet. Anchorage of mitochondria to cER in the bud tip is dependent upon Mmr1p, a protein that undergoes Myo2p–dependent localization to the bud, where it is present at the interface between mitochondria and cER in the tip bud. Another mitochondrial anchorage complex consists of foci containing Num1p and Mdm36p. These foci are found at the cell cortex in mother cells and in large buds. Num1p directly interacts with the plasma membrane through its pleckstrin homology (PH) domains, and is also closely apposed to the cER [40].
Ypt11p is a Rab-like protein that can bind to the Myo2p cargo-binding domain and is required for anchorage of mitochondria in the bud tip and for localization of cER in the bud [6, 33, 34]. Specifically, deletion of YPT11 results in defects in accumulation of mitochondria and cER in the bud, while overexpression of YPT11 has the opposite effect [33–35]. While it is clear that Ypt11p is required for accumulation and therefore anchorage of mitochondria in the bud tip, a point of controversy is whether the primary target for Ypt11p is mitochondria or cER. Because Ypt11p is not an abundant protein, its localization is not known. However, artificial targeting of Ypt11p to mitochondria, but not to ER, can suppress the mitochondrial distribution defects observed upon deletion of YPT11 [36]. Thus, Ypt11p may affect mitochondrial distribution through interactions with mitochondria and not ER. On the other hand, Ypt11p localizes to cER in the bud when overexpressed [34]. Moreover, deletion of YPT11 has no effect on the velocity of mitochondrial movement. However, ypt11Δ exhibit defects in accumulation of mitochondria in the bud tip and in cER inheritance [6, 34]. This raises the possibility that Ypt11p affects mitochondrial anchorage in the bud tip through effects on localization of cER in the bud. Indeed, other studies indicate that Ypt11p is a cargo adapter that binds to Ret2p on COPI-containing late Golgi vesicles and links these vesicles to Myo2p for transport to the bud tip [37]. Ret2p also localizes to Golgi-derived ER recycling vesicles, so it is possible that Myo2p and Ypt11p transport these ER retrieval vesicles to the bud tip, and that these in turn contribute to anchorage of mitochondria at that site.
To balance inheritance between mothers and buds, mitochondria are also tethered in the tip and cortex of the mother cell. The mechanism underlying anchorage of mitochondria in the mother cell tip is not well understood. Recent works indicate that Num1p and Mdm36p link mitochondria to the mother cell cortex [38–40]. Mdm36p is a mitochondrial protein with roles in mitochondrial morphology and division that, until recently, were not well understood [40]. Num1p is a cortical protein that supports the dynein-dependent migration of the nucleus into the bud [41] and maintains normal mitochondrial morphology and distribution [42, 43]. Cortical localization of Num1p is dependent on its C-terminal pleckstrin homology domain, while the N-terminal coiled-coil domain is essential for nuclear and mitochondrial functions [40, 44]. ER may also play a role in mitochondrial tethering in the mother cell cortex, since ER-resident proteins co-purify with Num1p, and ER was found in close proximity with Num1p–containing structures [40].
Tethering of mitochondria at strategic sites may also affect mitochondrial network dynamics. It has been proposed that mitochondrial anchorage by Num1p complex together with cytoskeleton-dependent forces provide tension for Dnm1-dependent mitochondrial fission [45]. Consistent with this, a subset of Dnm1p colocalizes with Num1p [40, 43], and deletion of NUM1 and MDM36 results in mitochondrial fission defects [38, 43]. While only a small decrease in mitochondrial fission activity was found in num1Δ cells [40], a growth defect in cells lacking both mitochondrial network dynamics and the Num1p was detected, and could be rescued by expression of a chimeric mitochondria-cortex tether. As described below, tethering and fusion/fission machineries also exert mitochondrial quality control and contribute to the lifespan of daughter cells.
Mitochondrial quality control
Yeast model two forms of eukaryotic cellular aging. Chronological lifespan (CLS), the survival time of stationary-phase non-dividing yeast cells, is a model for stress resistance in post-mitotic cells. Replicative lifespan (RLS), the number of times that a cell can divide prior to senescence, is a model for aging of division-competent cells. One intuitive concept is that babies are born young, independent of the age of their parents. This process, mother-daughter age asymmetry, also occurs in budding yeast. Mother cells age with each budding cycle; however, daughter cells for the most part are born young, with a full RLS. During aging, mitochondria utilize several quality control mechanisms to ensure that the inherited mitochondria are functional (Box1). Below, we describe a role for mitochondrial quality control in yeast lifespan control and mother-daughter age asymmetry.
Box 1: Mechanisms for mitochondrial quality control.
Studies in mammalian cells and fungi have provided a foundation for understanding mechanisms for mitochondrial quality control and how defects in these processes can lead to neurodegenerative diseases and diabetes. These studies revealed two levels of control: mitochondrial repair mechanisms and mechanisms to identify and eliminate mitochondria that are beyond repair. Several repair mechanisms are active in mitochondrial quality control including mitochondrial fusion, which repairs low-functioning mitochondria by intraorganellar complementation; molecular chaperones, which bind to and stabilize unfolded proteins; and proteases both within and outside the organelle that degrade damaged mitochondrial proteins including the proteasome in the cytosol, and mitochondrial AAA+ proteases and Pim1/Lon [69–71].
Mitophagy and mitochondrial fusion and fission have been implicated in elimination of mitochondria that are beyond repair [72]. In pancreatic beta cells, mitochondria with low membrane potential (Δ ψ) are segregated from those with high Δ ψ . This segregation occurs, in part, because mitochondria with low Δ ψ can undergo fission but cannot undergo fusion. These low-functioning mitochondria are then eliminated by mitophagy [73].
Defects in mitochondrial quality control are well documented in Parkinson’s disease (PD) [72, 74]. Pink1 (Pten-induced kinase) and Parkin (an E3 ubiquitin ligase) are central regulators of mitochondrial homeostasis, especially in the substantia nigra, the area in the brain affected in PD [75, 76]. According to a recent model for mitochondrial quality control in PD, Pink1 is imported and degraded in functioning mitochondria, but not in mitochondria with no Δ ψ. Pink1 that accumulates on the surface of mitochondria with no Δ ψ recruits Parkin to the MOM. Mitochondria-associated Parkin then ubiquitinates mitochondrial proteins, leading to mitophagic elimination of the dysfunctional mitochondria [75, 77]. Recent studies indicate that the mitofusin Mfn2 is a substrate and receptor for Parkin on mitochondria, which may serve to inhibit fusion of poorly functioning mitochondria with other mitochondria in addition to targeting poor functioning mitochondria for mitophagy (Chen and Dorn, Science 2013).
The function of cytoplasmic Parkin has been more elusive. Recently Shin et al showed that Parkin catalyzes ubiquitination, which leads to degradation, of PARIS (PARkin Interacting Substrate) zinc finger protein. PARIS represses the expression of PGC-1, a transcriptional co-activator involved in cellular energy homeostasis and mitochondrial biogenesis. These observations support another mechanism for Parkin in PD: Parkin mutations may compromise mitochondrial quality control by repressing biogenesis of new, fully functioning mitochondria.
Microvesicles produced from mitochondria are the basis of novel mechanisms for mitochondrial quality control. Recent studies indicate that stress-induced mitochondria-derived microvesicles contain oxidized proteins and are a selectively targeted to lysosomes or peroxisomes in yeast. Interestingly, production of these microvesicles is independent of the fission GTPase DRP1 [78]. Mitochondria-derived microvesicles can be a source of rejuvenation. Recent studies documented connexin-dependent transfer of mitochondria-derived microvesicles from bone marrow-derived stromal cells to alveolar epithelial cells during acute lung injury, which results in increased alveolar ATP concentrations and reduced injury [79].
These studies revealed mechanisms for mitochondrial quality control and show how failure of these systems can lead to disease. Indeed, defects in mitochondrial quality control have been linked to neurodegenerative diseases including Parkinson’s disease, spinocerebellar ataxia, spastic paraplegia and peripheral neuropathies, and metabolic diseases including type II diabetes and non-alcoholic and alcoholic steatosis [80, 81].
Mitochondrial quality control and its effect on lifespan
General mechanisms for mitochondrial quality control in metazoans and yeast are described in Box 1. Here, we describe the role for two mechanisms for mitochondrial quality control during aging in budding yeast, mitochondrial protein repair and fusion/fission. Pim1p/Lon is a conserved ATP-dependent protease in the mitochondrial matrix with functions, including chaperone activity for respiratory complex assembly and mitochondrial turnover of misfolded proteins and aggregates. Deletion of PIM1 decreases RLS in yeast and leads to an oxidizing cytosolic environment, consistent with unrepaired damage to mitochondrial oxygen-handling proteins that are a source of reactive oxygen species (ROS). Pim1p activity is also decreased in aged yeast cells [46] suggesting that protein repair by Pim1p is a mechanism for mitochondrial quality control during aging.
In addition to protein repair, mitochondria must also undergo proper fusion/fission events to ensure mitochondrial quality control. The mitofusins, Fzo1p and Mgm1p, mediate outer and inner membrane fusion, respectively, while Dnm1p drives mitochondrial fission. Inhibition of inner membrane fusion shortens RLS and CLS and sensitizes cells to apoptosis in S. cerevisiae [47]. Furthermore, inhibition of outer membrane fusion shortens the lifespan of the fungal model Neurospora crassa [48] [49]. Conversely, inhibition of mitochondrial fission, by deletion of DNM1, extends lifespan in S. cerevisiae and Podospora anserina [50]. These studies support the model that maintaining mitochondria as a continuous reticulum promotes longer lifespan, potentially by intraorganellar complementation of damaged mitochondrial components.
In silico studies modeling mitochondrial network dynamics revealed conditions under which mitochondrial fission and fusion can be harmful [51]. According to the “mitochondrial infectious damage adaptation” model, as cells age, the abundance of damaged mitochondria increases beyond a level that can be repaired by intraorganellar complementation. Instead, mixing of mitochondria can lead to propagation of mitochondrial damage to other mitochondria. Thus, while mitochondrial fusion and fission can promote mitochondrial function, their function in mitochondrial quality control may be more complex than previously appreciated.
Mitochondrial quality control as a mechanism for mother-daughter age asymmetry
Mother-daughter age asymmetry is a consequence of asymmetric yeast cell division. Aging determinants, including extrachromosomal rDNA circles, protein aggregates containing oxidatively damaged or unfolded proteins, and lower-functioning organelles including vacuoles (similar to lysosomes) are selectively retained in mother cells [52–55]. Conversely, rejuvenation determinants, including higher-functioning vacuoles and detoxification factors for ROS, are preferentially inherited by daughter cells [56, 57].
Several findings indicate that the machinery for mitochondrial inheritance can segregate “less fit” mitochondria from fitter mitochondria during yeast cell division. Fluorescence photobleaching studies indicate that mitochondria in the bud form a single continuous reticulum that is physically distinct from mitochondria in mother cells. Measurement of mitochondrial ROS and redox state indicates that the mitochondrial reticulum anchored in the bud tip has less ROS and is more reducing compared to mother cell mitochondria, and that mother cells contain distinct mitochondria that are variable in fitness, and on average are less fit than mitochondria in the bud [58] (Fig. 3).
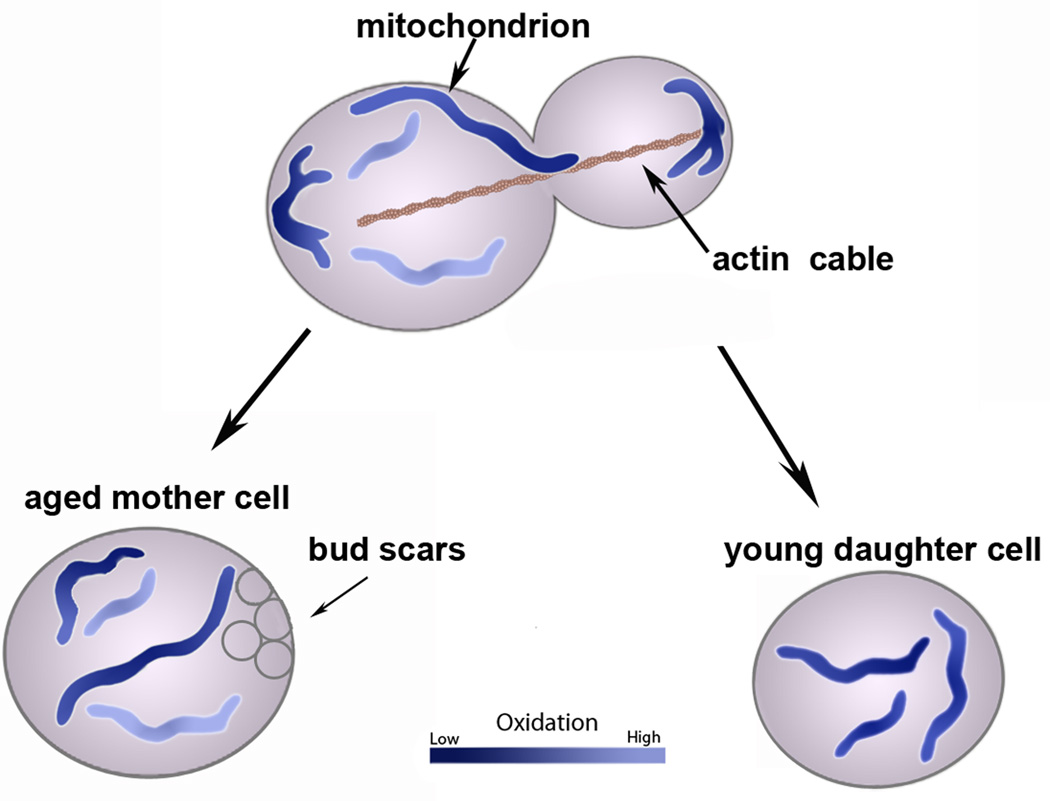
Figure 3. Mitochondrial quality control during inheritance and anchorage in budding yeast.
Healthier mitochondria (dark purple tubules) are preferentially retained at the cell poles. Mitochondria that are anchored and accumulate at these poles have less superoxide and are more reducing than mitochondria elsewhere in the cell. It is currently unclear what mechanisms regulate this quality control; however, it is clear that mitochondrial ROS levels affect lifespan and mother-daughter age asymmetry. Old mother cells accumulate aging factors, including mitochondria with high ROS.
Interestingly, mutations that inhibit anchorage of mitochondria in the bud tip also compromise the segregation of fit from less fit mitochondria, and cause a loss of mother-daughter age asymmetry [58, 59]. Wild-type cells give rise to daughter cells with a range of RLS described by a bell-shaped distribution. In contrast, mmr1Δ and ypt11Δ cells give rise to two populations of daughter cells: those that are short-lived or long-lived compared to wild-type cells. Long-lived mmr1Δ cells typically have healthier mitochondria with less ROS than those of wild-type cells or short-lived mmr1Δ cells and typically give rise to long-lived cells. Conversely, short-lived mmr1Δ cells have mitochondria with more ROS than wild-type or long-lived mmr1Δ cells, and give rise only to short-lived daughter cells. Thus, anchorage of mitochondria in the bud tip is necessary for segregation of fit from less fit mitochondria during yeast cell division, which in turn affects lifespan control. Given the newly appreciated function of NUM1 in retention of mitochondria in the mother cell [40], NUM1 may also affect lifespan by contributing to the retention of less fit mitochondria in mother cells.
Yet to be determined is how the machinery for mitochondrial inheritance exerts mitochondrial quality control. Since mitochondria must actively overcome the opposing force of retrograde actin cable flow to be transported from mother cell to bud, this actin flow may serve as a filter to prevent less fit mitochondria from entering the bud. It is also possible that the anchorage machinery in the bud tip preferentially binds to fitter organelles, while that the anchorage machinery in the mother cell may preferentially bind to less fit organelles. These models are not mutually exclusive. Finally, recent evidence indicates that other organelles influence mitochondrial fitness and overall cell aging in S. cerevisiae [55]. As mother cells age, vacuolar acidity decreases while daughter cells from aging mother cells contain acidic vacuoles. Interestingly, the decrease in vacuolar acidity correlates with loss of mitochondrial membrane potential and normal morphology. Thus, multiple factors, including inheritance and interaction with other organelles, influence mitochondrial quality control, ultimately affecting lifespan.
Mitochondrial quantity control during cell cycle progression
Checkpoints ensure that critical processes at each phase of the cell cycle are correctly completed before progression to the next phase. The best characterized checkpoints consist of 1) a sensor that monitors a specific cell division event, 2) a signal transduction pathway that receives signals from the sensor, and 3) targets or effectors that arrest the cell cycle in response to defects, repair of defects and/or trigger cell death when repair is not possible. Below we describe mechanisms that monitor the presence and quantity of mitochondrial membranes and DNA (mtDNA), and inhibit cell cycle progression in response to defects in mitochondrial quantity control.
Monitoring of mitochondrial content in daughter cells
Emerging studies support the existence of mechanisms for mitochondrial quantity control during inheritance in budding yeast [59]. Quantitative analysis of mitochondrial volume in living yeast revealed that mitochondrial network size increases with increasing cell size in buds and decreases with increasing age in mother cells. Interestingly, regardless of the mother's age or mitochondrial content, all buds attained the same average ratio of mitochondrial volume to daughter cell size.
Another example of cellular mitochondrial quantity control is a checkpoint that monitors mitochondrial content in buds and blocks cell cycle progression at cytokinesis when daughter cells fail to inherit mitochondria [60]. This mitochondrial inheritance checkpoint is controlled by a conserved checkpoint signaling pathway, the mitotic exit network. The mechanism for monitoring mitochondrial content in yeast daughter cells is not well understood. However, it is possible that the mechanism that controls mitochondrial content in buds also serves as a sensor for the mitochondrial inheritance checkpoint.
A cell cycle checkpoint that monitors mtDNA in daughter cells
mtDNA encodes subunits of the electron transport chain and F1Fo-ATPase, and RNAs required for mitochondrial protein synthesis. Mutations of human mtDNA have clinical manifestations in the brain, heart, skeletal muscle, kidney, and endocrine system, and are linked to aging and age-associated neurodegenerative diseases [61, 62]. Moreover, changes in mtDNA copy number occur in a number of primary human cancers and correlate with cancer progression [63, 64].
Recent studies revealed a mtDNA inheritance checkpoint in yeast, which inhibits progression from G1 to S phase of the cell cycle in response to the absence of mtDNA in buds [60]. Interestingly, this G1 to S transition defect is not a consequence of loss of mitochondrial respiration or mtDNA-encoded respiratory chain components. Indeed, yeast that contain mtDNA with no coding information exhibit wild-type G1 to S progression. Thus, the checkpoint machinery monitors mtDNA itself, not genes encoded by mtDNA.
Previous studies revealed that DNA damage checkpoint proteins including Rad53p (mammalian Chk2) regulates mtDNA in yeast and mammalian cells. Deletion of RAD53 alters mtDNA copy number in yeast [65–67]. Inhibition of ATM, an upstream activator of Chk2 and site of mutation in the neurodegenerative disorder ataxia telangiectasia (A–T), results in a reduction in mtDNA copy number in mammalians cells [68]. It was also shown that loss of mtDNA activates the kinase activity of Rad53p, which is required for regulation of cell cycle progression in response to mtDNA loss [60]. These findings indicate that the mtDNA inheritance checkpoint is regulated by a conserved checkpoint signaling pathway. Since proteins in the DNA damage checkpoint also regulate mtDNA content in mammalian cells [68], it is possible that this checkpoint is conserved. Other studies indicate that a decrease in mtDNA copy number produced by deletion of Abf2p, a high mobility group mtDNA binding protein, results in delayed cell cycle progression [66]. Moreover, a delay in cell cycle progression produced by mutation of RAD53 or growth on a non-fermentable carbon source, results in an increase in mtDNA copy number [66]. Thus, it is possible that the rate of cell cycle progression is regulated by the size of the pool of heritable mtDNA, and that complete loss of mtDNA results in a complete block in the cell cycle.
Concluding Remarks
Mitochondrial quality and quantity control contribute to age-associated disorders including neurodegenerative and metabolic diseases. In addition, mtDNA mutations or changes in copy number are implicated in disease and aging [61–64]. Several important aspects of mitochondrial biology have been elucidated in budding yeast, including fission/fusion factors, mitochondrial biogenesis, mitophagy and protein import. However, several important questions remain to be answered (Box 2) regarding mitochondrial quality and quantity control, which will lead to a greater understanding of mitochondria during both cellular homeostasis and age-associated deterioration, and will provide a foundation for understanding quality and quantity control of other organelles. Although yeast produce a bud whereas many mammalian cells undergo symmetric cell division, the mechanisms underlying segregation of mitochondria during yeast cell division may serve as models for understanding asymmetric cell division events during development, oogenesis and stem cell division. They also provide a foundation for understanding the asymmetric localization of mitochondria in polarized cells, such as those forming the neuronal and immunological synapses. Thus, the lessons learned from the budding yeast system will inspire further studies in mammalian cells, and will suggest potential targets for development of therapeutics for diseases associated with defects in mitochondrial function.
Box 2: Outstanding Questions in the Field.
While many mitochondrial quality control mechanisms have been identified, one fundamental question is the relative contribution of each of these pathways to mitochondrial maintenance. Moreover, while it is clear that the machinery for mitochondrial inheritance promotes inheritance of the fittest mitochondria by daughter cells, the mechanisms underlying this process and the criteria that define mitochondrial fitness are not well understood. Emerging studies support a role for ER and vacuoles in mitochondrial inheritance and quality control. However, the mechanisms underlying these processes and a role for organelle crosstalk in the inheritance and quality control of organelles other than mitochondria are yet to be determined.
There are also outstanding questions regarding mitochondrial quantity control during inheritance. Are the mitochondrial and mtDNA inheritance checkpoints conserved in mammalian cells? While it is clear that there are mechanisms to monitor mitochondrial content in developing daughter cells during yeast cell division and to regulate cell cycle progression in response to defects in mitochondrial quantity control, the sensors for mitochondrial content and how these sensors communicate with the cell cycle regulatory machinery and regulate mitochondrial quantity control are not well understood. With respect to the mtDNA inheritance checkpoint, it is not clear how information regarding mtDNA content in daughter cells is transmitted from the mitochondrial matrix, where mtDNA resides, across both mitochondrial membranes and into the nucleus, where Rad53p resides.
Highlights.
-
·
Fitter mitochondria accumulate in buds, affecting mother and daughter cell lifespan
-
·
Mitochondrial repair, dynamics and anchorage control quality of the organelle
-
·
Conserved cell cycle checkpoints respond to defects in mitochondrial inheritance
Acknowledgements
We thank the members of the Pon laboratory for technical assistance and valuable discussions, and Wolfgang Pernice for figure preparation. This work was supported by awards from the HHMI (56006760) to JDV and from the National Institutes of Health (NIH) (GM045735, GM045735S1 and GM096445) and the Ellison Medical Foundation (AG-SS-2465-10) to LP. GM45735S1 was issued from the NIH under the American Recovery and Reinvestment Act of 2009.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon VR, et al. Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. Cell Motil Cytoskeleton. 1997;37:199–210. doi: 10.1002/(SICI)1097-0169(1997)37:3<199::AID-CM2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Fehrenbacher KL, et al. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr Biol. 2004;14:1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Simon VR, et al. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang HC, et al. A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr Biol. 1999;9:1111–1114. doi: 10.1016/s0960-9822(99)80480-1. [DOI] [PubMed] [Google Scholar]
- 6.Boldogh IR, et al. A type V myosin (Myo2p) and a Rab-like G-protein (Ypt11p) are required for retention of newly inherited mitochondria in yeast cells during cell division. Mol Biol Cell. 2004;15:3994–4002. doi: 10.1091/mbc.E04-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swayne TC, et al. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr Biol. 2011;21:1994–1999. doi: 10.1016/j.cub.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boldogh IR, et al. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 10.Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meisinger C, et al. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. Embo J. 2007;26:2229–2239. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meisinger C, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen TT, et al. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 2012;13:880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voss C, et al. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang HC, Pon LA. Actin cable dynamics in budding yeast. Proc Natl Acad Sci U S A. 2002;99:751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman MA, Spudich JA. The myosin superfamily at a glance. J Cell Sci. 2012;125:1627–1632. doi: 10.1242/jcs.094300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammer JA, 3rd, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nature reviews. Molecular cell biology. 2012;13:13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 20.Robertson AS, et al. Functions of actin in endocytosis. Cell Mol Life Sci. 2009;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altmann K, et al. The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J Cell Biol. 2008;181:119–130. doi: 10.1083/jcb.200709099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortsch J, et al. The myosin-related motor protein Myo2 is an essential mediator of bud-directed mitochondrial movement in yeast. J Cell Biol. 2011;194:473–488. doi: 10.1083/jcb.201012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eves PT, et al. Overlap of cargo binding sites on myosin V coordinates the inheritance of diverse cargoes. J Cell Biol. 2012;198:69–85. doi: 10.1083/jcb.201201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boldogh IR, et al. Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc Natl Acad Sci U S A. 2001;98:3162–3167. doi: 10.1073/pnas.051494698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKane M, et al. A mammalian actin substitution in yeast actin (H372R) causes a suppressible mitochondria/vacuole phenotype. J Biol Chem. 2005;280:36494–36501. doi: 10.1074/jbc.M506970200. [DOI] [PubMed] [Google Scholar]
- 26.Senning EN, Marcus AH. Actin polymerization driven mitochondrial transport in mating S. cerevisiae. Proc Natl Acad Sci U S A. 2010;107:721–725. doi: 10.1073/pnas.0908338107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehrenbacher KL, et al. A role for Jsn1p in recruiting the Arp2/3 complex to mitochondria in budding yeast. Mol Biol Cell. 2005;16:5094–5102. doi: 10.1091/mbc.E05-06-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride HM. Mitochondrial-ER tethering: the inheritance of a functional unit. Curr Biol. 2011;21:R949–R951. doi: 10.1016/j.cub.2011.10.043. [DOI] [PubMed] [Google Scholar]
- 29.Voeltz GK, et al. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West M, et al. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh T, et al. Mmr1p is a mitochondrial factor for Myo2p–dependent inheritance of mitochondria in the budding yeast. EMBO J. 2004;23:2520–2530. doi: 10.1038/sj.emboj.7600271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itoh T, et al. Complex formation with Ypt11p, a rab-type small GTPase, is essential to facilitate the function of Myo2p, a class V myosin, in mitochondrial distribution in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:7744–7757. doi: 10.1128/MCB.22.22.7744-7757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buvelot Frei S, et al. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10p and Ypt11p. Mol Cell Biol. 2006;26:7299–7317. doi: 10.1128/MCB.02405-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frederick RL, et al. Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics. 2008;178:825–837. doi: 10.1534/genetics.107.083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewandowska A, et al. Mitochondrial association, protein phosphorylation and degradation regulate the availability of the active Rab GTPase, Ypt11, for mitochondrial inheritance. Mol Biol Cell. 2013 doi: 10.1091/mbc.E12-12-0848. http://www.molbiolcell.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arai S, et al. Ypt11 functions in bud-directed transport of the Golgi by linking Myo2 to the coatomer subunit Ret2. Curr Biol. 2008;18:987–991. doi: 10.1016/j.cub.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 38.Hammermeister M, et al. Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2443–2452. doi: 10.1091/mbc.E10-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klecker T, et al. The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J Cell Sci. 2013 doi: 10.1242/jcs.126045. http://jcs.biologists.org/ [DOI] [PubMed] [Google Scholar]
- 40.Lackner LL, et al. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci U S A. 2013;110:E458–E467. doi: 10.1073/pnas.1215232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto A, Hiraoka Y. Cytoplasmic dynein in fungi: insights from nuclear migration. J Cell Sci. 2003;116:4501–4512. doi: 10.1242/jcs.00835. [DOI] [PubMed] [Google Scholar]
- 42.Dimmer KS, et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerveny KL, et al. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev Cell. 2007;12:363–375. doi: 10.1016/j.devcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 44.Tang X, et al. A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. J Cell Biol. 2012;196:743–756. doi: 10.1083/jcb.201112017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schauss AC, McBride HM. Mitochondrial fission: a non-nuclear role for Num1p. Current biology : CB. 2007;17:R467–R470. doi: 10.1016/j.cub.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Erjavec N, et al. Deletion of the mitochondrial Pim1/Lon protease in yeast results in accelerated aging and impairment of the proteasome. Free Radic Biol Med. 2013;56:9–16. doi: 10.1016/j.freeradbiomed.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 47.Scheckhuber CQ, et al. Unopposed mitochondrial fission leads to severe lifespan shortening. Cell Cycle. 2011;10:3105–3110. doi: 10.4161/cc.10.18.17196. [DOI] [PubMed] [Google Scholar]
- 48.Kurashima K, et al. A uvs-5 Strain Is Deficient for a Mitofusin Gene Homologue, fzo1, Involved in Maintenance of Long Life Span in Neurospora crassa. Eukaryot Cell. 2013;12:233–243. doi: 10.1128/EC.00226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato A, et al. Deletion of a novel F-box protein, MUS-10, in Neurospora crassa leads to altered mitochondrial morphology, instability of mtDNA and senescence. Genetics. 2010;185:1257–1269. doi: 10.1534/genetics.110.117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheckhuber CQ, et al. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol. 2007;9:99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- 51.Figge MT, et al. Deceleration of fusion-fission cycles improves mitochondrial quality control during aging. PLoS Comput Biol. 2012;8:e1002576. doi: 10.1371/journal.pcbi.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 53.Erjavec N, Nyström T. Sir2p–dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:10877–10881. doi: 10.1073/pnas.0701634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klinger H, et al. Quantitation of (a)symmetric inheritance of functional and of oxidatively damaged mitochondrial aconitase in the cell division of old yeast mother cells. Exp Gerontol. 2010;45:533–542. doi: 10.1016/j.exger.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 55.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012 doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shepard KA, et al. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spokoini R, et al. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2:738–747. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 58.McFaline-Figueroa JR, et al. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10:885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rafelski SM, et al. Mitochondrial network size scaling in budding yeast. Science. 2012;338:822–824. doi: 10.1126/science.1225720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crider DG, et al. Rad53 is essential for a mitochondrial DNA inheritance checkpoint regulating G1 to S progression. J Cell Biol. 2012;198:793–798. doi: 10.1083/jcb.201205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park CB, Larsson NG. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu M. Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sci. 2011;89:65–71. doi: 10.1016/j.lfs.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 64.Cook CC, Higuchi M. The awakening of an advanced malignant cancer: an insult to the mitochondrial genome. Biochim Biophys Acta. 2012;1820:652–662. doi: 10.1016/j.bbagen.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor SD, et al. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:3010–3018. doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lebedeva MA, Shadel GS. Cell cycle- and ribonucleotide reductase-driven changes in mtDNA copy number influence mtDNA Inheritance without compromising mitochondrial gene expression. Cell Cycle. 2007;6:2048–2057. doi: 10.4161/cc.6.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–255. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eaton JS, et al. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J Clin Invest. 2007;117:2723–2734. doi: 10.1172/JCI31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luce K, Osiewacz HD. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat Cell Biol. 2009;11:852–858. doi: 10.1038/ncb1893. [DOI] [PubMed] [Google Scholar]
- 70.Weil A, et al. Unmasking a temperature-dependent effect of the P. anserina i-AAA protease on aging and development. Cell cycle. 2011;10:4280–4290. doi: 10.4161/cc.10.24.18560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voos W. Chaperone-protease networks in mitochondrial protein homeostasis. Biochim Biophys Acta. 2013;1833:388–399. doi: 10.1016/j.bbamcr.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 72.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vives-Bauza C, Przedborski S. Mitophagy: the latest problem for Parkinson’s disease. Trends Mol Med. 2011;17:158–165. doi: 10.1016/j.molmed.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 75.Narendra DP, Youle RJ. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid Redox Signal. 2011;14:1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shin JH, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ziviani E, et al. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soubannier V, et al. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 2012;7:e52830. doi: 10.1371/journal.pone.0052830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Islam MN, et al. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nature medicine. 2012;18:759–765. doi: 10.1038/nm.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. Embo J. 2012;31:1336–1349. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mitchell T, et al. Convergent mechanisms for dysregulation of mitochondrial quality control in metabolic disease: implications for mitochondrial therapeutics. Biochem Soc Trans. 2013;41:127–133. doi: 10.1042/BST20120231. [DOI] [PMC free article] [PubMed] [Google Scholar]