Abstract
Cells undergo dynamic remodeling of the cytoskeleton during adhesion and migration on various extracellular matrix (ECM) substrates in response to physiological and pathological cues. The major mediators of such cellular responses are the heterodimeric adhesion receptors, the integrins. Extracellular or intracellular signals emanating from different signaling cascades cause inside-out signaling of integrins via talin, a cystokeletal protein that links integrins to the actin cytoskeleton. Various integrin subfamilies communicate with each other and growth factor receptors under diverse cellular contexts to facilitate or inhibit various integrin-mediated functions. Since talin is an essential mediator of integrin activation, much of the integrin crosstalk would therefore be influenced by talin. However, despite the existence of an extensive body of knowledge on the role of talin in integrin activation and as a stabilizer of ECM-actin linkage, information on its role in regulating inter-integrin communication is limited. This review will focus on the structure of talin, its regulation of integrin activation and discuss its potential role in integrin crosstalk.
Introduction
The communication of extracellular matrix (ECM) with intracellular cytoskeleton is crucial for regulating cell adhesion, cell shape change and cell migration. Such communication depends heavily on integrins, a large family of noncovalent heterodimeric (α/β) adhesion receptors [65], [68]. Integrins function by engaging ECM ligands through their large extracellular domains and actin-binding proteins through their short cytoplasmic tails (CT), thereby linking ECM with the cytoskeleton (Fig1). Talin [19], [24], [63], [82], the focus of the article, together with filamin [95] and α-actinin [77], [127], [160], [139], are known to be the key players in this linkage, which can bind directly and simultaneously to both actin and integrin CTs. Numerous other intracellular proteins also connect integrins and the actin cytoskeleton but indirectly via shared binding partners. These extensive integrin-actin networks of protein-protein interactions coalesce to form discrete structures, focal adhesions, podosomes or analogous structures, that constitute dynamic hubs of adhesive and signaling activities [85], [146], [177].
Fig1. Bidirectional signaling across integrins.
Agonist stimulation, signaling via G protein coupled receptors (GPCRs) and/or growth factor receptors can lead to “inside-out” signaling of integrins. Extracellular domains of resting integrins open up to bind extracellular matrix ligands (L). This leads to “outside-in” signaling of integrins resulting in cytoskeletal remodeling and downstream signaling cascades.
The effects of talin on integrin function are broad. It transduces signals across integrins in both the inside-out and outside-in directions and it also influences the organization of the actin network and the composition of focal adhesions [1], [28], [45], [75], [76], [110], [147], [178]. Much of the recent studies on talin have emphasized its unique role in the inside-out signaling of integrins; i.e., their transformation from their basal or “resting” state to a more “active” state in which they can engage their cognate ECM ligands more efficiently (integrin activation). Less emphasized but clearly documented is the influence of talin on integrin outside-in signaling (Fig 1). Much less is known concerning the role of talin in the crosstalk between integrins of the same or different integrin subfamilies or with other signaling pathways. This review will summarize recent advance on talin structure and its control of integrin function and will touch upon its role in integrin crosstalk.
Talin expression, structure and subcellular distribution
Talin was discovered three decades ago as a protein highly enriched at cell adhesion sites [20]. The Tln gene and its orthologs can be traced from vertebrates back to protists [149]. Of the two mammalian isoforms of talin, Tln1 is ubiquitously expressed, being most abundant in the heart and scarce in the brain. Tln2 is enriched in the heart and brain with lower levels detected in the skeletal muscle, liver and lung [115]. Although the Tln1 and Tln2 isoforms in mammals share 74% identity, they do not fully compensate for each other. For example, in skeletal muscle or heart specific knockout models and during epithelial embryogenesis, when Tln1 is inactivated, Tln2 levels do not increase and existing levels do not compensate for Tln1 [29], [101], [108]. Moreover, Tln1 levels remain unelevated and do not compensate functionally for Tln2 in Tln2 deficient mice [29]. Gastrulation defects result in early embryonic lethality of Tln1 global knockout mice while Tln2 knockout mice are viable and fertile although early and severe myopathy in skeletal muscles is observed in the Tln2 knockout which is not prominent in the skeletal muscle specific Tln1 knockout [17], [29]. Platelet or endothelial specific Tln1 deficiency results in severe phenotypes in mouse models [114], [57], [122], [131], although some compensation could be demonstrated by exogenous Tln2 in endothelial cells and upregulated Tln2 expression was observed in undifferentiated embryonic stem cells [83], [179]. The vital role of talin in integrin activation and cell adhesion has been established using multiple model organisms [17], [30], [31], [115] as well as tissue-specific inactivation of the Tln1 gene [108], [114], [122].
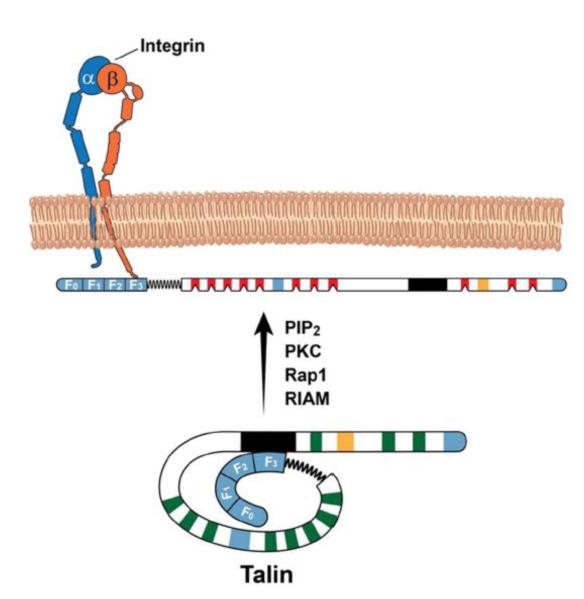
The human talin (Q9Y490, UniProtKB) monomer is a 270 kDa protein composed of 2541 amino acids and consists of an N-terminal head of 433 amino acids (talin-H) followed by a much larger rod domain (talin-R) (Fig 2). Talin-H contains four subdomains, F0, F1, F2 and F3 with F1-F3 being homologous to a typical FERM domain. The F0 and F1 subdomains exhibit ubiquitin-like folds [53] and form a novel linear arrangement with F1-F3 subdomains as opposed to the cloverleaf arrangement typical of most FERM domain proteins [40]. The F1 subdomain also has a 30 amino acid insert that was not visualized in the X-ray structure of talin-H domain [42]. The F3 subdomain is critical to many functions of talin as it binds to many proteins, including integrins [25], phosphatidylinositol-4-phosphate 5-kinase gamma 90 (PIPkinaseγ90) [35], [98] and layilin [15]. Talin F2-F3 has also been implicated in binding to focal adhesion kinase, FAK [26], [90] although the biophysical evidence for such binding is lacking. Using nuclear magnetic resonance (NMR) spectrometry, we failed to detect any significant interaction of 15N-labeled talin F2-F3 with 8-fold excess of a FAK peptide comprising of residues 1011-1042 (Yang et al, unpublished results), a region that was indicated to be the FAK binding site for talin [90]. The F1 and F2 domains anchor to the membrane, which likely position the F3 in an orientation facilitating β integrin engagement [4], [40]. Talin-R consists of a series of helical bundles which contain multiple protein binding sites for vinculin [48], RIAM [93], [55], [91], integrin[163], a THATCH domain and a dimerization sequence at the C-terminus [61]. Notably, talin has been found to contain three actin binding sites: one at talin-H and two at talin-R [32] but the structural details of how these sites mediate actin crosslinking and regulate actin remodeling remain unknown. Table 1 provides a list of talin binding partners and the specific regions within talin to which they bind.
Fig 2. Structure of talin.
Talin can be subdivided into head (talin-H) and rod (talin-R) regions. Talin-H is comprised of F0, F1, F2 and F3 domains while the talin-R has ~11 vinculin- and 3 actin-binding sites (in blue). The vinculin binding sites are dormant (in green) and are likely mechanoactivated (in red). PIP2, PKC, Rap1 and or RIAM can relieve the autoinhibitory effect of talin-R (1654-2344aa; black region) on F3 domain, promoting talin binding to β CT. The secondary integrin binding site is in orange.
The overall organization of talin, monomer versus dimer, may be different in specific tissues and/or species. Studies of talin isolated from chicken gizzard revealed that it is monomeric at low protein concentrations at physiological ionic strength [113], [172]. However, studies showed that talin isolated from human platelets exists predominantly as an antiparallel homodimer [50]. Low ionic strength induces a more globular shape to the chicken talin monomer while, at physiological or greater ionic strengths, it assumes an extended and filamentous conformation [113], [172]. Structural studies have revealed that the dimerization domain (2496-2529 residues) is important for the function of its C-terminal actin binding site [45].
In resting state cells, talin distribution is random and diffuse[9] and is autoinhibited from binding to integrins [49], [52], [156] and membrane [5], [156]. Talin-R plays a major role to structurally restrain the talin-H binding to the plasma membrane and integrin [156]. Upon agonist stimulation, talin is rapidly localized to the plasma membrane [10]. Protein kinase C alpha (PKCα), Rap1A, Rap1 effector RIAM, and phosphatadylinositol 4, 5 biphosphate (PIP2) have been shown to play important roles in the talin membrane localization and activation of its integrin binding function [58], [96], [156], [144]. The crystal structure of autoinhibited talin (pdb 4F7G) provides a clear atomic view of the talin-R and F3 interface [136] that is distinct from that proposed in an earlier model [52]. This crystal structure, along with the NMR binding studies suggests a steric and electrostatic mechanism to relieve the autoinhibition of talin. Specifically, the structure reveals that talin F2-F3 in complex with talin-R prevents the membrane association of talin F2-F3. In the autoinhibited talin complex, the negative charges on talin-R are repelled by negative phosphatidylserine on the inner leaflet of the plasma membrane. Higher concentration of negatively charged PIP2 enriched at sites in the membrane is likely to create an attractive force for positive charges on talinF2-F3. Interaction of talin F2-F3 with membrane misaligns the talinR-F3 interface, overcoming the autoinhibition and allowing integrin tail to dock onto talin F3, thus promoting the active conformation of talin via a pull-push mechanism [156]. The most recent model of talin depicts it to be a compact doughnut- shaped dimer with talin-R forming the ring and talin-H occupying the ‘hole’ in the center. This model is based on a combination of electron microscopy, small angle X-ray scattering, NMR and the known X-ray structures of talin fragments [54]. This model implies that for integrin and vinculin to bind talin, large movements of talin domains may be necessary [54]. Actin based mechanical forces are thought to play a role in exposing the vinculin binding sites on talin, but the spatio-temporal and molecular details of how this is accomplished remain to be resolved. The precise mechanisms of how PKC, Rap1A, and RIAM recruit and potentially activate talin are also not clearly understood. Recent studies have shown that multiple RIAM molecules can bind to the sites in talin-R where vinculin also binds, suggesting a turnover mechanism by which RIAM initially recruits talin to the integrin site and is later replaced by vinculin during focal adhesion assembly [55]. Calpain, a calcium regulated protease, can cleave the Q433-Q434 peptide bond in talin [138], thereby liberating talin-H to bind integrin β3CT with a much higher affinity than intact talin [175] and leading to integrin activation. Calpain cleavage of talin at Q433-Q434 as well as K2493-K2494 in talin-R has also been shown to regulate focal adhesion turnover [7], [42]. However, talin activation and consequent integrin activation can occur in the absence of calpain cleavage as ionophore-mediated αLβ2 [38] and RIAM induced β3 integrin activation [58] do not require proteolysis of intact talin by calpain. Thus, there may be several pathways for exposing the integrin binding site in intact talin, and the role of calpain may be geared more to the regulation of focal adhesion turnover.
Talin-integrin interaction
Among the 40 integrin CT binding proteins, talin is unique for its ability to bind and activate integrins. Another family of β integrin CT binding proteins, Kindlins, is also essential for integrin activation [134] and is discussed further below. Interactions of talin-H with the CT of β1, β2, β3, β5 and β7 integrins have been reported [21], [25], [130], [132], [145], [151] but the reported affinity varies widely. Solution NMR was used to estimate the dissociation constants (Kd) of talin F3-β CT to be in the μM range with the highest affinity for β1D, followed by β7, β3 and β1A CT [2], [3], [4]. As a point of reference, the Kd of talin F3 for β3 CT was estimated to be 273μM by NMR. Surface plasmon resonance (SPR) estimated a Kd of talin F2- F3 for the β3 [24] and β2 CT to be 91nM and 12.5 nM [125], respectively. A Kd of talin F2-F3 for β1 was estimated to be 67nM by pulldown assays [16]. The variable affinities probably arise from different technologies, different talin fragments, differences in talin and β CT sequences [3], and sample preparations. Notably, talin binding affinity to β3 CT increases dramatically in the presence of phospholipids [72], [117], suggesting that membrane plays a crucial role in elucidating the physiologically relevant talin-integrin binding. On the other hand, it does appear that the affinity of talin for integrin β CT is of relatively low affinity, which may provide a biological control mechanism so that integrin activation states can be inter-converted readily during cyclic cell adhesion and migration processes. The primary integrins mediating leukocyte (β2 and β7) and platelet (αIIbβ3) functions require rapid transitions between resting and activated states while the ubiquitous β1 integrins are present in a partially active state [59], [68]. Thus, the higher affinity of talin-H for the β2, β3 and β7 integrins may be needed to ensure rapid and efficient integrin activation. The two talin isoforms also exhibit different affinities for the same β CT [2-4]. The Tln2 isoform binds β3 and β1A (non-muscle splice variant) less robustly than Tln1. The tighter association of Tln2 with β1D (muscle specific β1 integrin) may be necessary to withstand high mechanical stress of muscle contraction exerted on myotendinous junctions [3]. Talin has also been reported to bind to the α subunit of the platelet integrin αIIbβ3, but the functional significance of this interaction has not been elucidated [47], [82], [136].
The major integrin binding site of talin is on talin-F3. The F3 domain recognizes two regions in integrin β CT: one on the NPxY/F motif, a conserved sequence in the midsection of most β CTs [21], [43], and a second in the membrane proximal region [165], [168], [171], as represented in Fig 3. The membrane proximal interaction of talin interferes with the integrin β CT interaction with α CT thereby causing the unclasping of the CT complex and initiating integrin activation as initially shown for integrin αIIbβ3 [168]. This mechanism has been also demonstrated with other integrin α/β heterodimers as well [4], [11], [81], [167]. This unclasping process extends into the membrane, where interaction between the α/β transmembrane helices is also disrupted and ultimately induces conformational changes in the extracellular domain of the integrin [79], [89], [176], [180]. Talin binding to the membrane and integrin β3 CT has recently been shown to alter the transmembrane domain topology of the interacting α and β chains which may provide an additional mechanism for transmission of conformational change across the membrane and into the extracellular domain [80]. However, the precise topology of the transmembrane domains of integrin α and β subunits remain controversial [86]. Readers are referred to reviews for further details of the biophysical nature of integrin CT interactions (e.g.[170]).
Fig 3. Regions on different β integrin CTs to which talin has been reported to bind.
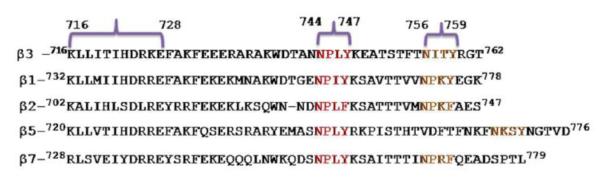
Talin binds to the conserved membrane proximal region and the first NPxY/F motif (shown in red) on β CTs, as shown by the regions in brackets. However, the talin binding region may well extend to residues beyond these highlighted regions on the β CT. The second NPxY/F motif is shown in orange.
In addition to the integrin binding site within talin-H, which is known as IBS-1, there is also a second integrin binding site (IBS-2) on talin-R. IBS-2 has not been implicated in regulating integrin activation. An IBS-2 containing talin-R fragment (1974-2293 amino acids) was found to have a binding affinity for β3 and β1 CT similar to that of talin-H [47]. However, this IBS-2-β3 and β1 integrin binding is impeded by residues C-terminal to IBS-2, making the overall affinity of the rod domain weaker than talin-H [47]. An affinity of 34.5nM of talin-R for β3 CT was reported [141], which is higher than published values talin-H [175]. This may suggest that there are two inhibitory mechanisms at play that regulate integrin-talin binding: (i) one involving talin-R(1654-2344 residues) and talin FERM domain that masks the β CT binding site on talin H [49] and (ii) C terminal rod (2300-2541 residues) and IBS-2 (1974-2293 residues) that lowers the strength of the IBS-2-β CT binding. The role of talin-H is to bind and activate integrins, and talin-R may be important in recruitment of adhesion molecules to focal adhesion [169]. It has also been proposed that IBS-1 activates integrins while IBS-2 stabilizes the integrintalin link, freeing talin-H to then interact with other binding partners such as PIPKIγ90 [41].
Regulation of talin and its interaction with integrins
Autoinhibition provides one mechanism to control the dynamics of talin-mediated integrin activation [49], [156]. Competition with negative regulators filamin [78], [123], ICAP-1 [102], β CT phosphorylation [2], [128] and lipid binding [49], [51], [72] provide additional mechanisms that regulate talin-mediated integrin activation and cytoskeletal-integrin linkage. Talin competes with filamin, a negative regulator of integrin activation [22], [33], [70], [123] or ICAP-1 for overlapping binding sites in integrin β CT. Integrin β CT that have higher affinity for these negative regulators bind talin less well [22], [78], [102]. Phosphorylation of tyrosine residues in the NPxY/F motif on β CT diminishes talin binding and favors binding of other partners, such as Dok1, which binds preferentially to the phosphorylated β CT [2]. Release of inhibitory restraint of talin-R on the talin PTB domain by PIP2 exposes sites involved in integrin activation [49]. This exposure facilitates talin F2-F3 binding to lipids within the plasma membrane thereby orientating talin for more favorable binding to β3CT [72], [117].
Early studies established that talin can be phosphorylated by PKC on serine (S) and threonine residues [99], [100], [8], [8], and subsequent studies indicated that talin was also a substrate for calyculin [121] and CdK5 [64]. Mass spectrometry revealed about 30 phosphorylation sites in talin [137], but only limited data support a functional significance to such post-translational modification. Phosphorylation of S425 within talin-H by Cdk5 prevents binding to and consequently ubiquitination by Smurf1, thereby regulating the intracellular turnover of talin [64]. However, the physiological relevance of phosphorylation of talin in terms of effects on integrin-related activities is less clear. Thrombin stimulated platelets leading to enhanced PKC mediated talin-H phosphorylation is associated with talin redistribution but was independent of its proteolytic cleavage and integrin interaction [10]. In chicken embryo fibroblasts, although stress fiber and focal contact organization was unperturbed by talin phosphorylation, precursors of focal contacts had reduced actin and talin-rich protrusions [8]. It has also been suggested that PKC-mediated talin phosphorylation may be involved in focal adhesion disassembly [164].
Integrin avidity modulation by talin
While the α/β CT unclasping mechanism is believed to be primarily a trigger of affinity modulation, talin also induces clustering of integrins leading to avidity modulation. Avidity modulation results from lateral movement of integrins in the plasma membrane to form integrin-rich microdomains or clusters. Integrin clusters exhibit high avidity for ligands, which are themselves multimeric, allowing for multivalent adhesive linkages. Agonist stimulation of platelets or exogenous talin head expression in CHO cells leads to formation of integrin-enriched clusters. Ligand binding to integrins in these clusters may then also promote conformational change to a high affinity state [18]. This scenario reconciles the data from Drosophila integrins where talin seems to be mainly responsible for integrin clustering rather than affinity modulation [17], [60]. In Drosophila, talin is not required to stabilize integrins on the cell surface or recruit them to muscle junctions. Instead, the major role of talin is to connect ECM bound integrins to the cytoskeleton [17], [41], [112], [62], [169]. Formation of microclusters has also been implicated in the function of integrin αLβ2. ICAM-1 binds well to αLβ2 on T-lymphocytes where microclusters can be visualized but not to dendritic cells, which lack such densely packed integrin patches. Talin has been shown to stabilize clusters of αLβ2 in a high affinity state [153]. The post-occupancy signaling from occupied integrins, which, among other events, strengthen their linkages to the actin cytoskeleton, depends upon clustering of integrins even when activated via affinity modulation. This is supported by studies that showed that PIP2 mediated release of talin-H from talin-R is a prerequisite for integrin activation and clustering on immobilized ECM ligands [27], [144]. Receptor clustering was found to precede talin recruitment in Drosophila [162]. In skeletal muscle specific Tln1-/- & Tln2-/- mice, talin was shown to be important for clustering of α7, αv and β1 integrins in myotendinous junctions [29]. In other words, while there are theoretical distinctions between affinity and avidity modulation, these processes overlap in responding cells, and talin’s role in integrin activation may occur via both affinity and avidity modulation, and specific pathways leading to integrin activation may be more prominent in certain tissues, cells or developmental stages [29], [101].
Talin and integrin outside-in signaling
Integrins are adhesion receptors that are capable of bidirectional signaling. Just as intracellular signals can be transduced across the membrane to activate the ligand binding function of integrins, ligand binding and other external environmental cues can also be transmitted via ligated integrins to the β CT and downstream signaling cascades within the cell that ultimately impact growth, survival, differentiation and proliferation of the cell. These events are referred to as “outside-in signaling” (see Fig. 1). Clustered integrins promote phosphorylation and therefore activation of FAK. Activated FAK can bind Src, a non-receptor tyrosine kinase, or other Src family kinases, (SFK) forming complexes which recruit additional proteins to focal adhesion sites. Signaling via the FAK-Src module affects early stages of adhesion [118], and promotes cell spreading [69], [73], but also can destabilize focal adhesions to allow migration of cells [44]. Thus, many outside-in signaling events from occupied integrins depend on or lead to rearrangements in the actin cytoskeleton. For example, platelet dependent clot retraction depends on occupancy of integrin αIIbβ3 and the associated rearrangements of the actin cytoskeleton [129]. In view of the importance of talin in connecting integrins to the cytoskeleton, it would appear that talin should assist in such outside-in responses, and there are data to support this notion [57]. However, the specific details as to how talin is involved in outside-in signaling remain uncertain. In one scenario, talin-H may dissociate from integrin β CT to facilitate integrin outside-in signaling. This mechanism may very well be involved since only talin-R but not talin-H was detected in focal adhesions in Drosophila [162], [161] and Tln1-/- mouse embryonic fibroblasts [112]. This process could be assisted by calpain [42], [7] or PIP2 [144] which separate talin-H from talin-R and is known to play critical role in integrin outside-in signaling [32], [68]. Furthermore, since β CT tyrosine phosphorylation dissociates talin from integrin [2], the notion that there is a rearrangement of talin-mediated integrin-actin connection upon integrin activation and occupancy is supported. Talin-H dissociation would also allow integrin CT to bind other proteins such as kinases and phosphatases known to trigger intracellular signaling pathways, induce cellular responses and cytosleketal rearrangements [68]. However, since most outside-in signaling responses are transduced by clustered integrins, intact talin is needed to bridge integrins and actin at some point during the outside-in signaling response. There is also evidence that not all outside-in responses are talin dependent. In CHO cells, talin recruitment was found to be a post-integrin activation event, promoting cell spreading by inhibiting the integrin β1A-FLNA interactions [123]. GPCR signaling can increase the affinity of α4β1 integrins on monocytes [66], but this increase in affinity appears to be independent of talin, kindlin3 or α-actinin. Rather, these integrin β binding partners, including talin, seem to stabilize the bonds of α4β1 integrin clusters with the cytoskeleton [67]. Thus, it is likely that the role of talin changes throughout the course of outside-in signaling responses.
Integrins are primary mediators of mechanotransduction. In response to the application of force, talin is recruited to integrin clusters. Thus, under strain, talin’s role may not be as an integrin activator, but as a mediator of mechanotransduction signals and strengthener of cytoskeletal connections [140]. This interpretation was supported by studies involving single molecule tracking and super-resolution microscopy in living cells, which suggested that transport of integrins into focal adhesions was talin independent. Integrins diffusing into these focal adhesions were postulated to be subsequently activated by talin recruited from the cytosol. This sequence of events would then lead to fibronectin engagement by the activated integrins in the focal adhesions [143]. Talin may also have functions other than integrin activation. In Tln1-/- embryonic fibroblasts, their adhesive phenotype was promoted only by an intact IBS-2 in talin-R [112]. At focal adhesions, only talin-R and not talin-H was observed [62]. In mammary epithelial cells, talin-R was found to be instrumental in recruiting FAK, vinculin and p21 to regulate cell proliferation [169]. An alternative pathway was demonstrated in which integrin clusters recruited FAK in nascent adhesions without a contribution from talin [90]. In certain tissue- or developmental- specific murine models, integrin activation was unaffected even upon talin knockout [29], [101]. Thus, talin can maintain cytoskeletal integrity [29]) or prevent degradation of β1 integrins besides activating integrins per se [101].
Role of talin in crosstalk involving integrins
The crosstalk between signaling pathways creates complex and interwoven networks of communication, and integrins talk to many different pathways. By virtue of its capacity to control integrin activation and signaling, a role of talin in such integrin-mediated crosstalk can be deduced although direct experimental evidence for talin’s involvement is limited. Integrins αVβ3 and α5β1 and their activation [56], [140], [173] are necessary for formation of a fibronectin-rich ECM, but the role of integrins in crosstalk extends beyond communication with the ECM. Signaling by integrins is intricately coordinated with that of various receptor tyrosine kinases (RTKs) and other growth factor receptors. Distinct integrin-growth factor receptor associations may drive different signaling events in response to specific growth factors [142]. These interactions permit bidirectional crosstalk between integrins and RTKs and other growth factor receptors during cell migration, cancer metastasis, angiogenesis and embryonic development (see reviews by [39], [71], [155] and [157] . Since these interactions reciprocally influence signaling via RTK and integrins and is further influenced by the activation states of integrins, this crosstalk will likely be dependent on talin.
Crosstalk may also be used to describe how integrin engagement influences the function of other integrin subfamily members. Such integrin mediated crosstalk may enhance or inhibit the function of the targeted integrin. Typically, cell surface levels of the targeted integrin are not reduced but rather the control is exerted via a signaling mechanism [135], [150]. As an example of the positive cooperativity between integrins, αvβ1 integrins were found to work in concert with α5β1 integrin to promote spreading of cells on fibronectin [109]. More extensively studied has been the capacity of one integrin to suppress the activity of another integrin. Prerequisites for the suppressive effects of one integrin on another are the ligand-occupied high affinity conformation and an intact β CT [36]. Most emphasis has been given to the communication between β3 and β1 CTs with the β3 CT functioning as the negative regulator [13], [14], [36], [87], [152]; i.e., β3 exerted a trans-dominant effect on β1. These effects have been demonstrated using both adherent, HEK, HUVEC [150], and suspension K562 cells [13], [14] with inhibition of α5β1 mediated migration on and phagocytosis of fibronectin by ligand-occupied αvβ3. In addition to the requisite role of the β CT, a vital role of the β transmembrane domain in this crosstalk has been demonstrated [150].
Chimeric β integrins have been used to probe endogenous integrin function [87], [152] under the premise that the chimeric receptors could compete with the endogenous integrins for cytoplasmic proteins that are needed for integrin mediated functions. Building upon this hypothesis, Calderwood et al. [23] concluded that it was competition for talin that underlies the trans-dominant inhibition of integrin activation. They showed that mutated β CT defective in talin binding was unable to mediate trans-dominant inhibition and this suppression was reversed by overexpression of integrin activating fragments of talin. Thus, the sequestration of talin by the suppressive species is both necessary and sufficient for trans-dominant inhibition of integrin activation. Talin overexpression was shown to relieve the suppression of specific integrins plated on non-canonical ligands [126]. Ligand occupied β1 integrins have been demonstrated to activate PKA [125] that can suppress flow-induced activation of αvβ3 integrins [126]. PKC is involved in agonist stimulated αIIbβ3 integrin activation on platelets [74]. PKC activated Rap1 [97], [111] can interact with RIAM [88] to promote integrin activation by recruiting talin to the plasma membrane [58]. Thus, the RIAM-talin axis could also be involved in inter-integrin communication. Hence, a model of talin mediated integrin crosstalk can be envisaged whereby stimulating signals unfold the talin molecule to expose talin-H, leading to affinity modulation of integrins via binding of talin-F3 to β integrin CT. Upon ligand engagement and integrin clustering, signaling pathways lead to either inhibition of other integrin species, or amplification of activating signals for talin to further augment integrin activation (Fig 4).
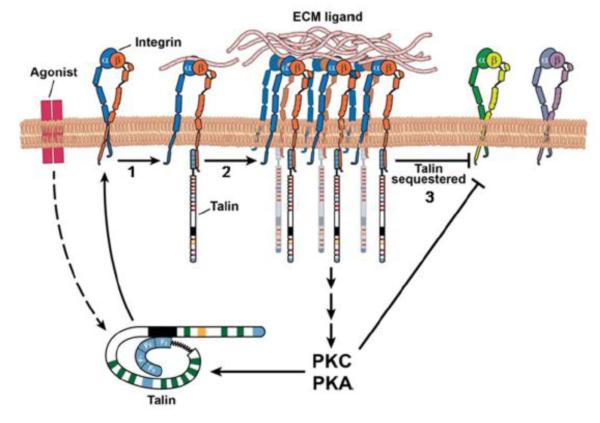
Fig 4. Model of talin mediated integrin crosstalk.
(1) Agonists, extracellular or intracellular signals lead to unraveling of the autoinhibited talin by PIP2, RIAM, Rap1 or PKC. (2) Unmasked talin-F3 binds to and activates β integrin CT. (3) Engagement of multivalent ECM ligands via clustered and activated integrins can lead to sequestration of talin, thereby inhibiting other integrin species. Concomitant signaling via PKC or PKA may also contribute to transdominant inhibition of integrins or perpetuation of integrin activation via talin.
Crosstalk between talin and other integrin binding proteins
Kindlins, a three member family of mammalian proteins, have emerged as important regulators of integrin activation [104]. Talin and kindlins cooperate for optimal integrin activation [12], [103]. Deficiencies/ mutations of kindlins, either in mice [116], [120], [166], or humans [84], [106], [107], [159], can have profound effects on integrin mediated cellular responses. Like talin, kindlins are FERM domain containing proteins. Also like talin, kindlins use their F3 (PTB-like domain) to bind to NPxY/F motifs in integrin β CT. However, talin binds to the membrane proximal of these motifs whereas kindlins engage the membrane distal NPxY/F motif (shown in orange in Fig 3) in β CT. Kindlin-2, talin and β integrin CTs can exist in a ternary complex, showing that talin and kindlin-2 can bind β integrin CT simultaneously and can function as “co-activators” of integrin αIIbβ3 [12]. The mechanism underlying such co-activation is presently unresolved. It has been suggested that talin mediates early stages of integrin activation while kindlin-3 is crucial for induction of the high affinity state of integrins [94]. At this point, it is unclear whether talin binds to the β CT first and facilitates kindlin binding or vice-versa. Based on the displacement model of integrin activation [70], migfilin, which binds to and may be recruited by kindlin to integrin adhesions, could dislodge filamin bound to and make β integrin CT accessible to talin and kindlin binding. Intracellular signaling thus triggered may facilitate further cooperation between talin and kindlin allowing for continuous inter-integrin communication. This will finally result in a robust integrin activation cascade (Fig 5). For more details on kindlins and their roles in integrin activation, readers are referred to recent reviews on this topic [105], [119], [133].
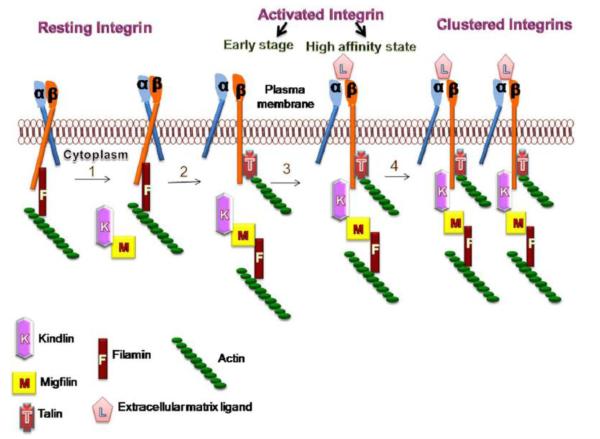
Fig 5. Crosstalk between talin and kindlin in displacement model of integrin activation.
(1) In the first step, filamin bound to resting integrins can be displaced from β CT by migfilin, possibly recruited by kindlins to the vicinity of integrin tails. (2) This makes integrin β CT available for binding by talin at the proximal NPxY/F motif, which unclasps the integrins. 3) Kindlin binding converts the early stage of activated integrin to a high affinity state, allowing ligand binding. 4) Cooperative talin and kindlin binding to β CT may lead to crosstalk between integrins leading to clustered integrins and outside-in-signaling. It is still not clear if migfilin remains associated with kindlin after kindlin-β CT engagement and what are the events that trigger dissociation of migfilin-filamin contacts during this process.
Concluding Remarks
Talin is a key regulator of the communication between the actin cytoskeleton and the ECM. This function depends on the capacity of talin to serve as a binding partner of multiple proteins, including actin and other actin binding proteins and integrins. Integrin engagement by talin controls their capacity to alter their activation state by affinity and/or avidity modulation. This interaction also assists in mediating the outside-in signaling that occurs through ligated integrins. Integrins speak to one another and to a variety of other membrane receptor systems and the downstream signaling that they orchestrate. Talin, by virtue of its direct association with and regulation of integrins, would appear to be integrally involved in this crosstalk. To comprehend the full role of talin in such crosstalk, mutations of specific functions of talin, some of which have been reported [41] and others that still need to be developed, should be characterized for their effects on the function of other cellular receptor systems with an eye on ultimately testing the consequences of such crosstalk in model cell systems and whole organisms.
Highlights.
Talin modulates affinity and avidity of integrins for extracellular matrix ligands
Role of structural components of talin in modulation of integrin activation
Integrins communicate with each other and other growth factor receptors
Signaling via talin-mediated activation can suppress or promote integrin function
Table 1.
Binding partners of talin (talin H is 1-433 aa; talin-R is 482-2541 aa; FERM domain is 86-400aa, comprising of F1, F2 and F3 domains; IBS-Integrin Binding site; ABSActin Binding site; DD-Dimerization Domain; aa-amino acids)
| Interacting protein | Binding region on talin (amino acid residues) |
References |
|---|---|---|
| Actin | ||
| ABS-1 (102-435) | [92] | |
| ABS-2 (951-1327) | [61] | |
| ABS-3 (2300-2541) | [45], [61], [110], [148], [154] | |
| β-integrin | ||
| IBS-1 (206-435) | [24] | |
| IBS-2 (1974-2293) | [47], [112], [141], [163], [174] | |
| Talin (full length) | DD (2496-2529) | [45] |
| Focal adhesion kinase | 225-357 | [26], [90] |
| Layilin | 280-435 | [15], [170] |
| PIPkinaseɣ 90 | 308-400 | [6], [34], [35] |
| PIPkinaseɣ 661 | 150-480 | [98] |
| Phospholipids (acidic) | 385-406 | [37], [51], [124] |
| RIAM | 655-786 (R2) | [55] |
| 787-911 (R3) | ||
| 1461-1580 (R8) | ||
| 1974-2140 (R11) | ||
| Smurf1 | 393-433 | [64] |
| Synemin | 1327-1948 | [158] |
| 1359-1659 | [46] | |
| Talin F3 | 1655-1822 | [49] |
| Vinculin | Helices 4,8,9,11,12,27,33,36 ,46,50, 58 | [48], [55] |
Acknowledgements
Thanks to David R. Schumick (Center for Medical Art and Photography, Cleveland Clinic) for assistance with figures. The authors’ work on talin is supported by NIH grant HL073311.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Anthis NJ, Campbell ID. The tail of integrin activation. Trends Biochem. Sci. 2011;36:191–198. doi: 10.1016/j.tibs.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthis NJ, Haling JR, Oxley CL, Memo M, Wegener KL, Lim CJ, Ginsberg MH, Campbell ID. Beta integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation. J. Biol. Chem. 2009;284:36700–36710. doi: 10.1074/jbc.M109.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthis NJ, Wegener KL, Critchley DR, Campbell ID. Structural diversity in integrin/talin interactions. Structure. 2010;18:1654–1666. doi: 10.1016/j.str.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, Campbell ID. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28:3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banno A, Goult BT, Lee H, Bate N, Critchley DR, Ginsberg MH. Subcellular localization of talin is regulated by inter-domain interactions. J. Biol. Chem. 2012;287:13799–13812. doi: 10.1074/jbc.M112.341214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barsukov IL, Prescot A, Bate N, Patel B, Floyd DN, Bhanji N, Bagshaw CR, Letinic K, Di PG, De CP, Roberts GC, Critchley DR. Phosphatidylinositol phosphate kinase type 1gamma and beta1-integrin cytoplasmic domain bind to the same region in the talin FERM domain. J. Biol. Chem. 2003;278:31202–31209. doi: 10.1074/jbc.M303850200. [DOI] [PubMed] [Google Scholar]
- 7.Bate N, Gingras AR, Bachir A, Horwitz R, Ye F, Patel B, Goult BT, Critchley DR. Talin contains a C-terminal calpain2 cleavage site important in focal adhesion dynamics. PLoS. One. 2012;7:e34461. doi: 10.1371/journal.pone.0034461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckerle MC. The adhesion plaque protein, talin, is phosphorylated in vivo in chicken embryo fibroblasts exposed to a tumor-promoting phorbol ester. Cell Regul. 1990;1:227–236. doi: 10.1091/mbc.1.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertagnolli ME, Beckerie MC. Evidence for the selective association of a subpopulation of GPIIb-IIIa with the actin cytoskeletons of thrombinactivated platelets. J. Cell Biol. 1993;121:1329–1342. doi: 10.1083/jcb.121.6.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertagnolli ME, Locke SJ, Hensler ME, Bray PF, Beckerle MC. Talin distribution and phosphorylation in thrombin-activated platelets. J Cell Sci. 1993;106(Pt 4):1189–1199. doi: 10.1242/jcs.106.4.1189. [DOI] [PubMed] [Google Scholar]
- 11.Bhunia A, Tang XY, Mohanram H, Tan SM, Bhattacharjya S. NMR solution conformations and interactions of integrin alphaLbeta2 cytoplasmic tails. J. Biol. Chem. 2009;284:3873–3884. doi: 10.1074/jbc.M807236200. [DOI] [PubMed] [Google Scholar]
- 12.Bledzka K, Liu J, Xu Z, Perera HD, Yadav SP, Bialkowska K, Qin J, Ma YQ, Plow EF. Spatial coordination of kindlin-2 with talin head domain in interaction with integrin beta cytoplasmic tails. J. Biol. Chem. 2012;287:24585–24594. doi: 10.1074/jbc.M111.336743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blystone SD, Graham IL, Lindberg FP, Brown EJ. Integrin αVβ1 differentially regulates adhesive and phagocytic functions of the fibronectin receptor α5β1. J. Cell Biol. 1994;127:1129–1137. doi: 10.1083/jcb.127.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blystone SD, Lindberg FP, LaFlamme SE, Brown EJ. Integrin β3 cytoplasmic tail is necessary and sufficient for regulation of α5 phagocytosis by αvβ3 and integrin- associated protein. J. Cell Biol. 1995;130:745–754. doi: 10.1083/jcb.130.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borowsky ML, Hynes RO. Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J. Cell Biol. 1998;143:429–442. doi: 10.1083/jcb.143.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouaouina M, Lad Y, Calderwood DA. The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate beta1 and beta3 integrins. J. Biol. Chem. 2008;283:6118–6125. doi: 10.1074/jbc.M709527200. [DOI] [PubMed] [Google Scholar]
- 17.Brown NH, Gregory SL, Rickoll WL, Fessler LI, Prout M, White RA, Fristrom JW. Talin is essential for integrin function in Drosophila. Dev. Cell. 2002;3:569–579. doi: 10.1016/s1534-5807(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 18.Bunch TA. Integrin alphaIIbbeta3 activation in Chinese hamster ovary cells and platelets increases clustering rather than affinity. J. Biol. Chem. 2010;285:1841–1849. doi: 10.1074/jbc.M109.057349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burn P, Kupfer A, Singer SJ. Dynamic membrane-cytoskeletal interactions: specific association of integrin and talin arises in vivo after phorbol ester treatment of peripheral blood lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 1988;85:497–501. doi: 10.1073/pnas.85.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burridge K, Connell L. A new protein of adhesion plaques and ruffling membranes. J. Cell Biol. 1983;97:359–367. doi: 10.1083/jcb.97.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calderwood DA, Fujioka Y, de Pereda JM, Garcia-Alvarez B, Nakamoto T, Margolis B, McGlade CJ, Liddington RC, Ginsberg MH. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. USA. 2003;100:2272–2277. doi: 10.1073/pnas.262791999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, Ginsberg MH. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat. Cell Biol. 2001;3:1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 23.Calderwood DA, Tai V, Di PG, De CP, Ginsberg MH. Competition for talin results in trans-dominant inhibition of integrin activation. J. Biol. Chem. 2004;279:28889–28895. doi: 10.1074/jbc.M402161200. [DOI] [PubMed] [Google Scholar]
- 24.Calderwood DA, Yan B, de Pereda JM, Garcia-Alvarez B, Fujioka Y, Liddington RC, Ginsberg MH. The phosphotyrosine binding (PTB)-like domain of talin activates integrins. J. Biol. Chem. 2002;277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 25.Calderwood DA, Zent R, Grant R, Rees DJ, Hynes RO, Ginsberg MH. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 26.Chen HC, Appeddu PA, Parsons JT, Hildebrand JD, Schaller MD, Guan JL. Interaction of focal adhesion kinase with cytoskeletal protein talin. J. Biol. Chem. 1995;270:16995–16999. doi: 10.1074/jbc.270.28.16995. [DOI] [PubMed] [Google Scholar]
- 27.Cluzel C, Saltel F, Lussi J, Paulhe F, Imhof BA, Wehrle-Haller B. The mechanisms and dynamics of (alpha)v(beta)3 integrin clustering in living cells. J. Cell Biol. 2005;171:383–392. doi: 10.1083/jcb.200503017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collier NC, Wang K. Purification and properties of human platelet P235. A high molecular weight protein substrate of endogenous calcium-activated protease(s) J. Biol. Chem. 1982;257:6937–6943. [PubMed] [Google Scholar]
- 29.Conti FJ, Monkley SJ, Wood MR, Critchley DR, Muller U. Talin 1 and 2 are required for myoblast fusion, sarcomere assembly and the maintenance of myotendinous junctions. Development. 2009;136:3597–3606. doi: 10.1242/dev.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cram EJ, Clark SG, Schwarzbauer JE. Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J. Cell Sci. 2003;116:3871–3878. doi: 10.1242/jcs.00705. [DOI] [PubMed] [Google Scholar]
- 31.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 32.Critchley DR, Gingras AR. Talin at a glance. J. Cell Sci. 2008;121:1345–1347. doi: 10.1242/jcs.018085. [DOI] [PubMed] [Google Scholar]
- 33.Das M, Ithychanda SS, Qin J, Plow EF. Migfilin and filamin as regulators of integrin activation in endothelial cells and neutrophils. PLoS. One. 2011;6:e26355. doi: 10.1371/journal.pone.0026355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Pereda JM, Wegener KL, Santelli E, Bate N, Ginsberg MH, Critchley DR, Campbell ID, Liddington RC. Structural basis for phosphatidylinositol phosphate kinase type Igamma binding to talin at focal adhesions. J. Biol. Chem. 2005;280:8381–8386. doi: 10.1074/jbc.M413180200. [DOI] [PubMed] [Google Scholar]
- 35.Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, De CP. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- 36.Diaz-Gonzalez F, Forsyth J, Steiner B, Ginsberg MH. Trans-dominant inhibition of integrin function. Mol. Biol. Cell. 1996;7:1939–1951. doi: 10.1091/mbc.7.12.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dietrich C, Goldmann WH, Sackmann E, Isenberg G. Interaction of NBDtalin with lipid monolayers. A film balance study. FEBS Lett. 1993;324:37–40. doi: 10.1016/0014-5793(93)81527-7. [DOI] [PubMed] [Google Scholar]
- 38.Dreolini L, Takei F. Activation of LFA-1 by ionomycin is independent of calpain-mediated talin cleavage. Biochem. Biophys. Res. Commun. 2007;356:207–212. doi: 10.1016/j.bbrc.2007.02.100. [DOI] [PubMed] [Google Scholar]
- 39.Eliceiri BP. Integrin and growth factor receptor crosstalk. Circ. Res. 2001;89:1104–1110. doi: 10.1161/hh2401.101084. [DOI] [PubMed] [Google Scholar]
- 40.Elliott PR, Goult BT, Kopp PM, Bate N, Grossmann JG, Roberts GC, Critchley DR, Barsukov IL. The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–1299. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellis SJ, Pines M, Fairchild MJ, Tanentzapf G. In vivo functional analysis reveals specific roles for the integrin-binding sites of talin. J. Cell Sci. 2011;124:1844–1856. doi: 10.1242/jcs.083337. [DOI] [PubMed] [Google Scholar]
- 42.Franco SJ, Rodgers MA, Perrin BJ, Han J, Bennin DA, Critchley DR, Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, Ginsberg MH, Liddington RC. Structural determinants of integrin recognition by talin. Mol. Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 44.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 45.Gingras AR, Bate N, Goult BT, Hazelwood L, Canestrelli I, Grossmann JG, Liu H, Putz NS, Roberts GC, Volkmann N, Hanein D, Barsukov IL, Critchley DR. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gingras AR, Bate N, Goult BT, Patel B, Kopp PM, Emsley J, Barsukov IL, Roberts GC, Critchley DR. Central region of talin has a unique fold that binds vinculin and actin. J. Biol. Chem. 2010;285:29577–29587. doi: 10.1074/jbc.M109.095455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gingras AR, Ziegler WH, Bobkov AA, Joyce MG, Fasci D, Himmel M, Rothemund S, Ritter A, Grossmann JG, Patel B, Bate N, Goult BT, Emsley J, Barsukov IL, Roberts GC, Liddington RC, Ginsberg MH, Critchley DR. Structural determinants of integrin binding to the talin rod. J. Biol. Chem. 2009;284:8866–8876. doi: 10.1074/jbc.M805937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gingras AR, Ziegler WH, Frank R, Barsukov IL, Roberts GC, Critchley DR, Emsley J. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J. Biol. Chem. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- 49.Goksoy E, Ma Y-Q, Wang X, Kong X, Perera D, Plow EF, Qin J. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldmann WH, Bremer A, Haner M, Aebi U, Isenberg G. Native talin is a dumbbell-shaped homodimer when it interacts with actin. J. Struct. Biol. 1994;112:3–10. doi: 10.1006/jsbi.1994.1002. [DOI] [PubMed] [Google Scholar]
- 51.Goldmann WH, Niggli V, Kaufmann S, Isenberg G. Probing actin and liposome interaction of talin and talin-vinculin complexes: a kinetic, thermodynamic and lipid labeling study. Biochemistry. 1992;31:7665–7671. doi: 10.1021/bi00148a030. [DOI] [PubMed] [Google Scholar]
- 52.Goult BT, Bate N, Anthis NJ, Wegener KL, Gingras AR, Patel B, Barsukov IL, Campbell ID, Roberts GC, Critchley DR. The structure of an interdomain complex that regulates talin activity. J. Biol. Chem. 2009;284:15097–15106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goult BT, Bouaouina M, Elliott PR, Bate N, Patel B, Gingras AR, Grossmann JG, Roberts GC, Calderwood DA, Critchley DR, Barsukov IL. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. 2010;29:1069–1080. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goult BT, Xu XP, Gingras AR, Swift M, Patel B, Bate N, Kopp PM, Barsukov IL, Critchley DR, Volkmann N, Hanein D. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: Implications for talin activation. J. Struct. Biol. 2013 doi: 10.1016/j.jsb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goult BT, Zacharchenko T, Bate N, Tsang R, Hey F, Gingras AR, Elliott PR, Roberts GC, Ballestrem C, Critchley DR, Barsukov IL. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J. Biol. Chem. 2013 doi: 10.1074/jbc.M112.438119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green JA, Berrier AL, Pankov R, Yamada KM. beta1 integrin cytoplasmic domain residues selectively modulate fibronectin matrix assembly and cell spreading through talin and Akt-1. J. Biol. Chem. 2009;284:8148–8159. doi: 10.1074/jbc.M805934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haling JR, Monkley SJ, Critchley DR, Petrich BG. Talin-dependent integrin activation is required for fibrin clot retraction by platelets. Blood. 2011;117:1719–1722. doi: 10.1182/blood-2010-09-305433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han J, Lim CJ, Watanabe N, Soriani A, Ratnikov B, Calderwood DA, Puzon-McLaughlin W, Lafuente EM, Boussiotis VA, Shattil SJ, Ginsberg MH. Reconstructing and Deconstructing Agonist-Induced Activation of Integrin alphaIIbbeta3. Curr. Biol. 2006;16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 59.Hato T, Yamanouchi J, Tamura T, Yakushijin Y, Sakai I, Yasukawa M. Cooperative role of the membrane-proximal and -distal residues of the integrin beta3 cytoplasmic domain in regulation of talin-mediated alpha IIb beta3 activation. J. Biol. Chem. 2008;283:5662–5668. doi: 10.1074/jbc.M707246200. [DOI] [PubMed] [Google Scholar]
- 60.Helsten TL, Bunch TA, Kato H, Yamanouchi J, Choi SH, Jannuzi AL, Feral CC, Ginsberg MH, Brower DL, Shattil SJ. Differences in regulation of Drosophila and vertebrate integrin affinity by talin. Mol. Biol. Cell. 2008;19:3589–3598. doi: 10.1091/mbc.E08-01-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hemmings L, Rees DJ, Ohanian V, Bolton SJ, Gilmore AP, Patel B, Priddle H, Trevithick JE, Hynes RO, Critchley DR. Talin contains three actinbinding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci. 1996;109(Pt 11):2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- 62.Himmel M, Ritter A, Rothemund S, Pauling BV, Rottner K, Gingras AR, Ziegler WH. Control of high affinity interactions in the talin C terminus: how talin domains coordinate protein dynamics in cell adhesions. J. Biol. Chem. 2009;284:13832–13842. doi: 10.1074/jbc.M900266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horwitz A, Duggan K, Buck C, Beckerie MC, Burridge K. Interaction of plasma membrane fibronectin receptor with talin -a transmembrane link. Nature. 1986;320:531–533. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- 64.Huang C, Rajfur Z, Yousefi N, Chen Z, Jacobson K, Ginsberg MH. Talin phosphorylation by Cdk5 regulates Smurf1-mediated talin head ubiquitylation and cell migration. Nat. Cell Biol. 2009;11:624–630. doi: 10.1038/ncb1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J. Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyduk SJ, Chan JR, Duffy ST, Chen M, Peterson MD, Waddell TK, Digby GC, Szaszi K, Kapus A, Cybulsky MI. Phospholipase C, calcium, and calmodulin are critical for alpha4beta1 integrin affinity up-regulation and monocyte arrest triggered by chemoattractants. Blood. 2007;109:176–184. doi: 10.1182/blood-2006-01-029199. [DOI] [PubMed] [Google Scholar]
- 67.Hyduk SJ, Rullo J, Cano AP, Xiao H, Chen M, Moser M, Cybulsky MI. Talin-1 and kindlin-3 regulate alpha4beta1 integrin-mediated adhesion stabilization, but not G protein-coupled receptor-induced affinity upregulation. J. Immunol. 2011;187:4360–4368. doi: 10.4049/jimmunol.1003725. [DOI] [PubMed] [Google Scholar]
- 68.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 69.Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- 70.Ithychanda SS, Das M, Ma YQ, Ding K, Wang X, Gupta S, Wu C, Plow EF, Qin J. Migfilin, a molecular switch in regulation of integrin activation. J. Biol. Chem. 2009;284:4713–4722. doi: 10.1074/jbc.M807719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ivaska J, Heino J. Cooperation between integrins and growth factor receptors in signaling and endocytosis. Annu. Rev. Cell Dev. Biol. 2011;27:291–320. doi: 10.1146/annurev-cellbio-092910-154017. [DOI] [PubMed] [Google Scholar]
- 72.Kalli AC, Wegener KL, Goult BT, Anthis NJ, Campbell ID, Sansom MS. The structure of the talin/integrin complex at a lipid bilayer: an NMR and MD simulation study. Structure. 2010;18:1280–1288. doi: 10.1016/j.str.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaplan KB, Swedlow JR, Morgan DO, Varmus HE. c-Src enhances the spreading of src-/- fibroblasts on fibronectin by a kinase-independent mechanism. Genes Dev. 1995;9:1505–1517. doi: 10.1101/gad.9.12.1505. [DOI] [PubMed] [Google Scholar]
- 74.Karniguian A, Grelac F, Levy-Toledano S, Legrand YJ, Rendu F. Collageninduced platelet activation mainly involves the protein kinase C pathway. Biochem. J. 1990;268:325–331. doi: 10.1042/bj2680325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaufmann S, Kas J, Goldmann WH, Sackmann E, Isenberg G. Talin anchors and nucleates actin filaments at lipid membranes. A direct demonstration. FEBS Lett. 1992;314:203–205. doi: 10.1016/0014-5793(92)80975-m. [DOI] [PubMed] [Google Scholar]
- 76.Kaufmann S, Piekenbrock T, Goldmann WH, Barmann M, Isenberg G. Talin binds to actin and promotes filament nucleation. FEBS Lett. 1991;284:187–191. doi: 10.1016/0014-5793(91)80681-r. [DOI] [PubMed] [Google Scholar]
- 77.Kelly DF, Taylor KA. Identification of the beta1-integrin binding site on alpha-actinin by cryoelectron microscopy. J. Struct. Biol. 2005;149:290–302. doi: 10.1016/j.jsb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 78.Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylanne J, Calderwood DA. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 79.Kim C, Lau TL, Ulmer TS, Ginsberg MH. Interactions of platelet integrin {alpha}IIb and {beta}3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood. 2009 doi: 10.1182/blood-2008-10-186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim C, Ye F, Hu X, Ginsberg MH. Talin activates integrins by altering the topology of the beta transmembrane domain. J. Cell Biol. 2012;197:605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 82.Knezevic I, Leisner TM, Lam SCT. Direct binding of the platelet integrin αIIbβ3 (GPIIb- IIIa) to talin - Evidence that interaction is mediated through the cytoplasmic domains of both αIIb and β3. J. Biol. Chem. 1996;271:16416–16421. doi: 10.1074/jbc.271.27.16416. [DOI] [PubMed] [Google Scholar]
- 83.Kopp PM, Bate N, Hansen TM, Brindle NP, Praekelt U, Debrand E, Coleman S, Mazzeo D, Goult BT, Gingras AR, Pritchard CA, Critchley DR, Monkley SJ. Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1. Eur. J. Cell Biol. 2010;89:661–673. doi: 10.1016/j.ejcb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuijpers TW, van d., V, Weterman MA, de BM, Tool AT, van den Berg TK, Moser M, Jakobs ME, Seeger K, Sanal O, Unal S, Cetin M, Roos D, Verhoeven AJ, Baas F. LAD-1/variant syndrome is caused by mutations in FERMT3. Blood. 2009;113:4740–4746. doi: 10.1182/blood-2008-10-182154. [DOI] [PubMed] [Google Scholar]
- 85.Kuo JC, Han X, Hsiao CT, Yates JR, III, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kurtz L, Kao L, Newman D, Kurtz I, Zhu Q. Integrin alphaIIbbeta3 insideout activation: an in situ conformational analysis reveals a new mechanism. J. Biol. Chem. 2012;287:23255–23265. doi: 10.1074/jbc.M112.360966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.LaFlamme SE, Thomas LA, Yamada SS, Yamada KM. Single subunit chimeric integrins as mimics and inhibitors of endogenous integrin functions in receptor localization, cell spreading and migration, and matrix assembly. J. Cell Biol. 1994;126:1287–1298. doi: 10.1083/jcb.126.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1- GTP and mediates Rap1-induced adhesion. Dev. Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 89.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lawson C, Lim ST, Uryu S, Chen XL, Calderwood DA, Schlaepfer DD. FAK promotes recruitment of talin to nascent adhesions to control cell motility. J. Cell Biol. 2012;196:223–232. doi: 10.1083/jcb.201108078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee HS, Anekal P, Lim CJ, Liu CC, Ginsberg MH. Two modes of integrin activation form a binary molecular switch in adhesion maturation. Mol. Biol. Cell. 2013;24:1354–1362. doi: 10.1091/mbc.E12-09-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee HS, Bellin RM, Walker DL, Patel B, Powers P, Liu H, Garcia-Alvarez B, de Pereda JM, Liddington RC, Volkmann N, Hanein D, Critchley DR, Robson RM. Characterization of an actin-binding site within the talin FERM domain. J. Mol. Biol. 2004;343:771–784. doi: 10.1016/j.jmb.2004.08.069. [DOI] [PubMed] [Google Scholar]
- 93.Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 2009;284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lefort CT, Rossaint J, Moser M, Petrich BG, Zarbock A, Monkley SJ, Critchley DR, Ginsberg MH, Fassler R, Ley K. Distinct roles for talin-1 and kindlin-3 in LFA-1 extension and affinity regulation. Blood. 2012;119:4275–4282. doi: 10.1182/blood-2011-08-373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Legate KR, Fassler R. Mechanisms that regulate adaptor binding to betaintegrin cytoplasmic tails. J. Cell Sci. 2009;122:187–198. doi: 10.1242/jcs.041624. [DOI] [PubMed] [Google Scholar]
- 96.Legate KR, Takahashi S, Bonakdar N, Fabry B, Boettiger D, Zent R, Fassler R. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. EMBO J. 2011;30:4539–4553. doi: 10.1038/emboj.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Letschka T, Kollmann V, Pfeifhofer-Obermair C, Lutz-Nicoladoni C, Obermair GJ, Fresser F, Leitges M, Hermann-Kleiter N, Kaminski S, Baier G. PKC-theta selectively controls the adhesion-stimulating molecule Rap1. Blood. 2008;112:4617–4627. doi: 10.1182/blood-2007-11-121111. [DOI] [PubMed] [Google Scholar]
- 98.Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma pphosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- 99.Litchfield DW, Ball EH. Phosphorylation of the cytoskeletal protein talin by protein kinase C. Biochem. Biophys. Res. Commun. 1986;134:1276–1283. doi: 10.1016/0006-291x(86)90388-8. [DOI] [PubMed] [Google Scholar]
- 100.Litchfield DW, Ball EH. Phosphorylation of high molecular weight proteins in platelets treated with 12-O-tetradecanoylphorbol-13-acetate. Biochem. Int. 1990;20:615–621. [PubMed] [Google Scholar]
- 101.Liu J, He X, Qi Y, Tian X, Monkley SJ, Critchley DR, Corbett SA, Lowry SF, Graham AM, Li S. Talin1 regulates integrin turnover to promote embryonic epithelial morphogenesis. Mol. Cell Biol. 2011;31:3366–3377. doi: 10.1128/MCB.01403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu W, Draheim KM, Zhang R, Calderwood DA, Boggon TJ. Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation. Mol. Cell. 2013 doi: 10.1016/j.molcel.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma YQ, Qin J, Wu C, Plow EF. Kindlin-2 (Mig-2): a co-activator of beta3 integrins. J. Cell Biol. 2008;181:439–446. doi: 10.1083/jcb.200710196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Malinin NL, Plow EF, Byzova TV. Kindlins in FERM adhesion. Blood. 2010;115:4011–4017. doi: 10.1182/blood-2009-10-239269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Malinin NL, Pluskota E, Byzova TV. Integrin signaling in vascular function. Curr. Opin. Hematol. 2012;19:206–211. doi: 10.1097/MOH.0b013e3283523df0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Malinin NL, Zhang L, Choi J, Ciocea A, Razorenova O, Ma Y-Q, Podrez EA, Tosi M, Lennon DP, Caplin AI, Shurin SB, Plow EF, Byzova TV. A point mutation in kindlin-3 ablates activation of three integrin subfamilies in humans. Nature Med. 2009;15:313–318. doi: 10.1038/nm.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Manevich-Mendelson E, Feigelson SW, Pasvolsky R, Aker M, Grabovsky V, Shulman Z, Kilic SS, Rosenthal-Allieri MA, Ben-Dor S, Mory A, Bernard A, Moser M, Etzioni A, Alon R. Loss of Kindlin-3 in LAD-III eliminates LFA-1 but not VLA-4 adhesiveness developed under shear flow conditions. Blood. 2009;114:2344–2353. doi: 10.1182/blood-2009-04-218636. [DOI] [PubMed] [Google Scholar]
- 108.Manso AM, Li R, Monkley SJ, Cruz NM, Ong S, Lao DH, Koshman YE, Gu Y, Peterson KL, Chen J, Abel ED, Samarel AM, Critchley DR, Ross RS. Talin1 Has Unique Expression versus Talin 2 in the Heart and Modifies the Hypertrophic Response to Pressure Overload. J. Biol. Chem. 2013;288:4252–4264. doi: 10.1074/jbc.M112.427484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marshall JF, Rutherford DC, McCartney AC, Mitjans F, Goodman SL, Hart IR. Alpha v beta 1 is a receptor for vitronectin and fibrinogen, and acts with alpha 5 beta 1 to mediate spreading on fibronectin. J. Cell Sci. 1995;108(Pt 3):1227–1238. doi: 10.1242/jcs.108.3.1227. [DOI] [PubMed] [Google Scholar]
- 110.McCann RO, Craig SW. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Medeiros RB, Dickey DM, Chung H, Quale AC, Nagarajan LR, Billadeau DD, Shimizu Y. Protein kinase D1 and the beta 1 integrin cytoplasmic domain control beta 1 integrin function via regulation of Rap1 activation. Immunity. 2005;23:213–226. doi: 10.1016/j.immuni.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 112.Moes M, Rodius S, Coleman SJ, Monkley SJ, Goormaghtigh E, Tremuth L, Kox C, van der Holst PP, Critchley DR, Kieffer N. The integrin binding site 2 (IBS2) in the talin rod domain is essential for linking integrin beta subunits to the cytoskeleton. J. Biol. Chem. 2007;282:17280–17288. doi: 10.1074/jbc.M611846200. [DOI] [PubMed] [Google Scholar]
- 113.Molony L, McCaslin D, Abernethy J, Paschal B, Burridge K. Properties of talin from chicken gizzard smooth muscle. J. Biol. Chem. 1987;262:7790–7795. [PubMed] [Google Scholar]
- 114.Monkley SJ, Kostourou V, Spence L, Petrich B, Coleman S, Ginsberg MH, Pritchard CA, Critchley DR. Endothelial cell talin1 is essential for embryonic angiogenesis. Dev. Biol. 2011;349:494–502. doi: 10.1016/j.ydbio.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Monkley SJ, Zhou XH, Kinston SJ, Giblett SM, Hemmings L, Priddle H, Brown JE, Pritchard CA, Critchley DR, Fassler R. Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev. Dyn. 2000;219:560–574. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1079>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 116.Montanez E, Ussar S, Schifferer M, Bosl M, Zent R, Moser M, Fassler R. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008;22:1325–1330. doi: 10.1101/gad.469408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moore DT, Nygren P, Jo H, Boesze-Battaglia K, Bennett JS, DeGrado WF. Affinity of talin-1 for the beta3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc. Natl. Acad. Sci. U. S. A. 2012;109:793–798. doi: 10.1073/pnas.1117220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moro L, Dolce L, Cabodi S, Bergatto E, Boeri EE, Smeriglio M, Turco E, Retta SF, Giuffrida MG, Venturino M, Godovac-Zimmermann J, Conti A, Schaefer E, Beguinot L, Tacchetti C, Gaggini P, Silengo L, Tarone G, Defilippi P. Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and p130Cas and leads to phosphorylation of specific EGF receptor tyrosines. J. Biol. Chem. 2002;277:9405–9414. doi: 10.1074/jbc.M109101200. [DOI] [PubMed] [Google Scholar]
- 119.Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 120.Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- 121.Murata K, Sakon M, Kambayashi J, Okuyama M, Hase T, Mori T. Platelet talin is phosphorylated by calyculin A. J. Cell Biochem. 1995;57:120–126. doi: 10.1002/jcb.240570112. [DOI] [PubMed] [Google Scholar]
- 122.Nieswandt B, Moser M, Pleines I, Varga-Szabo D, Monkley S, Critchley D, Fassler R. Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J. Exp. Med. 2007;204:3113–3118. doi: 10.1084/jem.20071827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nieves B, Jones CW, Ward R, Ohta Y, Reverte CG, LaFlamme SE. The NPIY motif in the integrin beta1 tail dictates the requirement for talin-1 in outside-in signaling. J. Cell Sci. 2010;123:1216–1226. doi: 10.1242/jcs.056549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Niggli V, Kaufmann S, Goldmann WH, Weber T, Isenberg G. Identification of functional domains in the cytoskeletal protein talin. Eur. J. Biochem. 1994;224:951–957. doi: 10.1111/j.1432-1033.1994.00951.x. [DOI] [PubMed] [Google Scholar]
- 125.O’Connor KL, Mercurio AM. Protein kinase A regulates Rac and is required for the growth factor-stimulated migration of carcinoma cells. J. Biol. Chem. 2001;276:47895–47900. doi: 10.1074/jbc.M107235200. [DOI] [PubMed] [Google Scholar]
- 126.Orr AW, Ginsberg MH, Shattil SJ, Deckmyn H, Schwartz MA. Matrixspecific suppression of integrin activation in shear stress signaling. Mol. Biol. Cell. 2006;17:4686–4697. doi: 10.1091/mbc.E06-04-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player, Cell Motil. Cytoskeleton. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 128.Oxley CL, Anthis NJ, Lowe ED, Vakonakis I, Campbell ID, Wegener KL. An integrin phosphorylation switch: the effect of beta3 integrin tail phosphorylation on Dok1 and talin binding. J. Biol. Chem. 2008;283:5420–5426. doi: 10.1074/jbc.M709435200. [DOI] [PubMed] [Google Scholar]
- 129.Parise LV. Integrin alpha(IIb)beta(3) signaling in platelet adhesion and aggregation. Curr. Opin. Cell Biol. 1999;11:597–601. doi: 10.1016/s0955-0674(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 130.Patil S, Jedsadayanmata A, Wencel-Drake JD, Wang W, Knezevic I, Lam SC. Identification of a talin-binding site in the integrin beta(3) subunit distinct from the NPLY regulatory motif of post-ligand binding functions. The talin n-terminal head domain interacts with the membrane- proximal region of the beta(3) cytoplasmic tail. J. Biol. Chem. 1999;274:28575–28583. doi: 10.1074/jbc.274.40.28575. [DOI] [PubMed] [Google Scholar]
- 131.Petrich BG, Marchese P, Ruggeri ZM, Spiess S, Weichert RA, Ye F, Tiedt R, Skoda RC, Monkley SJ, Critchley DR, Ginsberg MH. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J. Exp. Med. 2007;204:3103–3111. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pfaff M, Liu S, Erle DJ, Ginsberg MH. Integrin beta cytoplasmic domains differentially bind to cytoskeletal proteins. J. Biol. Chem. 1998;273:6104–6109. doi: 10.1074/jbc.273.11.6104. [DOI] [PubMed] [Google Scholar]
- 133.Q.J. BT, Plow EF. Kindling the Flame of Integrin Activation and Function With Kindlins. 2009;328:323. doi: 10.1097/MOH.0b013e32832ea389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Plow EF, Qin J, Byzova T. Kindling the flame of integrin activation and function with kindlins. Curr. Opin. Hematol. 2009;16:323–328. doi: 10.1097/MOH.0b013e32832ea389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Porter JC, Hogg N. Integrin cross talk: activation of lymphocyte functionassociated antigen-1 on human T cells alters alpha4beta1- and alpha5beta1- mediated function. J. Cell Biol. 1997;138:1437–1447. doi: 10.1083/jcb.138.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Raab M, Daxecker H, Edwards RJ, Treumann A, Murphy D, Moran N. Protein interactions with the platelet integrin alpha(IIb) regulatory motif. Proteomics. 2010;10:2790–2800. doi: 10.1002/pmic.200900621. [DOI] [PubMed] [Google Scholar]
- 137.Ratnikov B, Ptak C, Han J, Shabanowitz J, Hunt DF, Ginsberg MH. Talin phosphorylation sites mapped by mass spectrometry. J. Cell Sci. 2005;118:4921–4923. doi: 10.1242/jcs.02682. [DOI] [PubMed] [Google Scholar]
- 138.Rees DJ, Ades SE, Singer SJ, Hynes RO. Sequence and domain structure of talin. Nature. 1990;347:685–689. doi: 10.1038/347685a0. [DOI] [PubMed] [Google Scholar]
- 139.Roca-Cusachs P, Del RA, Puklin-Faucher E, Gauthier NC, Biais N, Sheetz MP. Integrin-dependent force transmission to the extracellular matrix by alpha-actinin triggers adhesion maturation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1361–E1370. doi: 10.1073/pnas.1220723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roca-Cusachs P, Gauthier NC, Del RA, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rodius S, Chaloin O, Moes M, Schaffner-Reckinger E, Landrieu I, Lippens G, Lin M, Zhang J, Kieffer N. The talin rod IBS2 alpha-helix interacts with the beta3 integrin cytoplasmic tail membrane-proximal helix by establishing charge complementary salt bridges. J. Biol. Chem. 2008;283:24212–24223. doi: 10.1074/jbc.M709704200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ross RS. Molecular and mechanical synergy: cross-talk between integrins and growth factor receptors. Cardiovasc. Res. 2004;63:381–390. doi: 10.1016/j.cardiores.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 143.Rossier O, Octeau V, Sibarita JB, Leduc C, Tessier B, Nair D, Gatterdam V, Destaing O, Albiges-Rizo C, Tampe R, Cognet L, Choquet D, Lounis B, Giannone G. Integrins beta1 and beta3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat. Cell Biol. 2012;14:1057–1067. doi: 10.1038/ncb2588. [DOI] [PubMed] [Google Scholar]
- 144.Saltel F, Mortier E, Hytonen VP, Jacquier MC, Zimmermann P, Vogel V, Liu W, Wehrle-Haller B. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J. Cell Biol. 2009;187:715–731. doi: 10.1083/jcb.200908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sampath R, Gallagher PJ, Pavalko FM. Cytoskeletal interactions with the leukocyte integrin beta2 cytoplasmic tail. Activation-dependent regulation of associations with talin and alpha-actinin. J. Biol. Chem. 1998;273:33588–33594. doi: 10.1074/jbc.273.50.33588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schiller HB, Friedel CC, Boulegue C, Fassler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12:259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schmidt JM, Zhang J, Lee HS, Stromer MH, Robson RM. Interaction of talin with actin: sensitive modulation of filament crosslinking activity. Arch. Biochem. Biophys. 1999;366:139–150. doi: 10.1006/abbi.1999.1204. [DOI] [PubMed] [Google Scholar]
- 148.Senetar MA, Foster SJ, McCann RO. Intrasteric inhibition mediates the interaction of the I/LWEQ module proteins Talin1, Talin2, Hip1, and Hip12 with actin. Biochemistry. 2004;43:15418–15428. doi: 10.1021/bi0487239. [DOI] [PubMed] [Google Scholar]
- 149.Senetar MA, McCann RO. Gene duplication and functional divergence during evolution of the cytoskeletal linker protein talin. Gene. 2005;362:141–152. doi: 10.1016/j.gene.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 150.Simon KO, Nutt EM, Abraham DG, Rodan GA, Duong LT. The alphavbeta3 integrin regulates alpha5beta1-mediated cell migration toward fibronectin. J. Biol. Chem. 1997;272:29380–29389. doi: 10.1074/jbc.272.46.29380. [DOI] [PubMed] [Google Scholar]
- 151.Singh S, D’mello V, van Bergen en HP, Birge RB. A NPxY-independent beta5 integrin activation signal regulates phagocytosis of apoptotic cells. Biochem. Biophys. Res. Commun. 2007;364:540–548. doi: 10.1016/j.bbrc.2007.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Smilenov L, Briesewitz R, Marcantonio EE. Integrin beta 1 cytoplasmic domain dominant negative effects revealed by lysophosphatidic acid treatment. Mol. Biol. Cell. 1994;5:1215–1223. doi: 10.1091/mbc.5.11.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Smith A, Carrasco YR, Stanley P, Kieffer N, Batista FD, Hogg N. A talindependent LFA-1 focal zone is formed by rapidly migrating T lymphocytes. J. Cell Biol. 2005;170:141–151. doi: 10.1083/jcb.200412032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Smith SJ, McCann RO. A C-terminal dimerization motif is required for focal adhesion targeting of Talin1 and the interaction of the Talin1 I/LWEQ module with F-actin. Biochemistry. 2007;46:10886–10898. doi: 10.1021/bi700637a. [DOI] [PubMed] [Google Scholar]
- 155.Somanath PR, Malinin NL, Byzova TV. Cooperation between integrin alphavbeta3 and VEGFR2 in angiogenesis. Angiogenesis. 2009;12:177–185. doi: 10.1007/s10456-009-9141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Song X, Yang J, Hirbawi J, Ye S, Perera HD, Goksoy E, Dwivedi P, Plow EF, Zhang R, Qin J. A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell Res. 2012;22:1533–1545. doi: 10.1038/cr.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Soung YH, Clifford JL, Chung J. Crosstalk between integrin and receptor tyrosine kinase signaling in breast carcinoma progression. BMB. Rep. 2010;43:311–318. doi: 10.5483/bmbrep.2010.43.5.311. [DOI] [PubMed] [Google Scholar]
- 158.Sun N, Critchley DR, Paulin D, Li Z, Robson RM. Identification of a repeated domain within mammalian alpha-synemin that interacts directly with talin. Exp. Cell Res. 2008;314:1839–1849. doi: 10.1016/j.yexcr.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 159.Svensson L, Howarth K, McDowall A, Patzak I, Evans R, Ussar S, Moser M, Metin A, Fried M, Tomlinson I, Hogg N. Leukocyte adhesion deficiency- III is caused by mutations in KINDLIN3 affecting integrin activation. Nat. Med. 2009;15:306–312. doi: 10.1038/nm.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tadokoro S, Nakazawa T, Kamae T, Kiyomizu K, Kashiwagi H, Honda S, Kanakura Y, Tomiyama Y. A potential role for alpha-actinin in inside-out alphaIIbbeta3 signaling. Blood. 2011;117:250–258. doi: 10.1182/blood-2009-10-246751. [DOI] [PubMed] [Google Scholar]
- 161.Tanentzapf G, Brown NH. An interaction between integrin and the talin FERM domain mediates integrin activation but not linkage to the cytoskeleton. Nat. Cell Biol. 2006;8:601–606. doi: 10.1038/ncb1411. [DOI] [PubMed] [Google Scholar]
- 162.Tanentzapf G, Martin-Bermudo MD, Hicks MS, Brown NH. Multiple factors contribute to integrin-talin interactions in vivo. J. Cell Sci. 2006;119:1632–1644. doi: 10.1242/jcs.02859. [DOI] [PubMed] [Google Scholar]
- 163.Tremuth L, Kreis S, Melchior C, Hoebeke J, Ronde P, Plancon S, Takeda K, Kieffer N. A fluorescence cell biology approach to map the second integrin-binding site of talin to a 130-amino acid sequence within the rod domain. J. Biol. Chem. 2004;279:22258–22266. doi: 10.1074/jbc.M400947200. [DOI] [PubMed] [Google Scholar]
- 164.Turner CE, Pavalko FM, Burridge K. The role of phosphorylation and limited proteolytic cleavage of talin and vinculin in the disruption of focal adhesion integrity. J. Biol. Chem. 1989;264:11938–11944. [PubMed] [Google Scholar]
- 165.Ulmer TS, Calderwood DA, Ginsberg MH, Campbell ID. Domain-specific interactions of talin with the membrane-proximal region of the integrin beta3 subunit. Biochemistry. 2003;42:8307–8312. doi: 10.1021/bi034384s. [DOI] [PubMed] [Google Scholar]
- 166.Ussar S, Moser M, Widmaier M, Rognoni E, Harrer C, Genzel-Boroviczeny O, Fassler R. Loss of Kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS. Genet. 2008;4:e1000289. doi: 10.1371/journal.pgen.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vinogradova O, Vaynberg J, Kong X, Haas TA, Plow EF, Qin J. Membrane-mediated structural transitions at the cytoplasmic face during integrin activation. Proc. Natl. Acad. Sci. USA. 2004;101:4094–4099. doi: 10.1073/pnas.0400742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas TA, Plow EF, Qin J. A structural mechanism of integrin αIIbβ3 “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 169.Wang P, Ballestrem C, Streuli CH. The C terminus of talin links integrins to cell cycle progression. J. Cell Biol. 2011;195:499–513. doi: 10.1083/jcb.201104128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wegener KL, Campbell ID. Transmembrane and cytoplasmic domains in integrin activation and protein-protein interactions (review) Mol. Membr. Biol. 2008;25:376–387. doi: 10.1080/09687680802269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 172.Winkler J, Lunsdorf H, Jockusch BM. Energy-filtered electron microscopy reveals that talin is a highly flexible protein composed of a series of globular domains. Eur. J. Biochem. 1997;243:430–436. doi: 10.1111/j.1432-1033.1997.0430a.x. [DOI] [PubMed] [Google Scholar]
- 173.Wu CY, Keivens VM, O’Toole TE, McDonald JA, Ginsberg MH. Integrin activation and cytoskeletal interaction are essential for the assembly of a fibronectin matrix. Cell. 1995;83:715–724. doi: 10.1016/0092-8674(95)90184-1. [DOI] [PubMed] [Google Scholar]
- 174.Xing B, Jedsadayanmata A, Lam SC. Localization of an integrin binding site to the C terminus of talin. J Biol. Chem. 2001;276:44373–44378. doi: 10.1074/jbc.M108587200. [DOI] [PubMed] [Google Scholar]
- 175.Yan B, Calderwood DA, Yaspan B, Ginsberg MH. Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J. Biol. Chem. 2001;276:28164–28170. doi: 10.1074/jbc.M104161200. [DOI] [PubMed] [Google Scholar]
- 176.Yang J, Ma YQ, Page RC, Misra S, Plow EF, Qin J. Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17729–17734. doi: 10.1073/pnas.0909589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat. Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang J, Robson RM, Schmidt JM, Stromer MH. Talin can crosslink actin filaments into both networks and bundles. Biochem. Biophys. Res. Commun. 1996;218:530–537. doi: 10.1006/bbrc.1996.0095. [DOI] [PubMed] [Google Scholar]
- 179.Zhang X, Jiang G, Cai Y, Monkley SJ, Critchley DR, Sheetz MP. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 2008;10:1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhu J, Luo BH, Barth P, Schonbrun J, Baker D, Springer TA. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin alphaIIbbeta3. Mol. Cell. 2009;34:234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]