Abstract
Accurate quantification of positron emission tomography (PET) is important for diagnosis and assessment of cancer treatment. The low spatial resolution of PET imaging induces partial volume effect to PET images that biases quantification. A PET partial volume correction method is proposed using high-resolution, anatomical information from magnetic resonance images (MRI). The corrected PET is pursued by removing the convolution of PET point spread function (PSF) and by preserving edges present in PET and the aligned MR images. The correction is implemented in a Bayesian's deconvolution framework that is minimized by a conjugate gradient method. The method is evaluated on simulated phantom and brain PET images. The results show that the method effectively restores 102 ± 7% of the true PET activity with a size of greater than the full-width at half maximum of the point spread function. We also applied the method to synthesized brain PET data. The method does not require prior information about tracer activity within tissue regions. It can offer a partial volume correction method for various PET applications and can be particularly useful for combined PET/MRI studies.
Keywords: partial volume effects, deconvolution, PET/MRI
INTRODUCTION
Positron emission tomography (PET) can produces quantitatively accurate measurement of tissue radioactivity concentration. A variety of studies have shown PET's capability to differentiate between benign and malignant tumors, determine tumor stages, and assess tissue response to therapy. However, despite continual improvement of the physical characteristics of PET, it is stilled limited by low spatial resolution compared with high-resolution magnetic resonance imaging (MRI) or computed tomography (CT). The limited spatial resolution induces partial volume effect to PET which biases the quantitative measurement of regional radioactivity concentration. Experiments on ECAT (Siemens/CTI, Knoxville, TN) have shown the ratio of peak measurement to true radioactivity (recovery coefficient (RC)) is 0.13 for a 6.4 mm inner diameter hot spheroid with object-to-background ratio of 12.6 in a water-filled elliptical cylinder [1]. Such bias results in incorrect estimation of true local radioactivity concentration and could eliminate quantitative change in tissue metabolism or physiology as a function of time [2].
In order to generate PVC corrected PET images, a variety of methods have been proposed and generally can be classified into two categories: model fitting methods and anatomical image-guided methods. Model fitting based methods simultaneously recover the size and the activity of a small spheroid tumor by minimizing the distance between PET images and convolution of the spheres with a point spread function [3]. The method assumes there are only two regions with uniform activities: background and a spheroid-shape tumor. The unknown are the spheroid location and diameter, radioactivity of the tumor and background, 3D Gaussian modeling of the system's PSF. These fitting methods can be considered as a type of iterative deconvolution method using greatly simplified prior information about the object geometry and activity distribution. These simplifications make it difficult to apply the method to images having objects with various shapes. Although the methods do not require anatomic information, initial knowledge about the object location is necessary for an optimization approach to correctly converge. The other strategy is to use high resolution anatomical images as an atlas of radioactivity distribution to guide the correction. PET and aligned, high-resolution, anatomical images can be acquired by the emergence of combined PET/CT and PET/MRI machines as well as the development of image registration methods. With aligned anatomical images, these methods [4,5,6] segment the anatomical images into a number of non-overlapping compartments and assume each compartment has distinct and uniform activity distribution in PET. So there are only N unknown variables corresponding to compartments in the anatomical image matrix. Convolution of this matrix with point spread function should consistent with the acquired PET images. Therefore, partial volume correction can be solved as a matrix inversion or least square fitting problem. These methods depend on the accuracy of classification of anatomical images and probably miss the subtle uptake change within a compartment because of the limited number of classes for segmentation.
In this paper, we develop an MRI-guided partial volume correction method but eliminate segmentation of MR images. We hypothesize that the corrected PET image is not only the deconvolution version of observed PET but also has maximal edge correspondence to the aligned MRI. As the aligned MRI provides high contrast and high resolution structure information to PET, the proposed method restores the true PET by deconvolution but maintains the edge information from registered MRI. Our goal is to not only enable accurate activity measurement in interesting regions, but also yield a corrected image at the pixel level. The new method is validated using both synthesized and experimental PET data.
METHOD
2.1. PET partial volume effects
There are two distinct phenomena resulting in partial volume effect to PET [7]. First is the relatively low spatial resolution of PET so that a voxel could consist of different structures and undermine subresolution heterogeneity. This tissue fraction effect can be attenuated by an imaging system with higher special resolution. However, the imaging physics, the detector design and reconstruction process impose a theoretical resolution limitation to PET. The second is a point spread function (PSF) that blurs PET images. The response of a scanner to a point source of radioactivity shows a bell shape instead of a perfect impulse function. This point spreading effect causes activity spillover between regions so that the imaged activity is not only from the source at the voxel but also from its neighboring structures. Mathematically, the effect is modeled as a convolution of a point spread function. It can be corrected in principle by a deconvolution procedure if the PSF of the scanner has been characterized. Deconvolution is an ill-pose problem and often leads to noise amplification and severe ringing artifacts. Therefore, prior information about imaging objects and noise are usually required to regularize the process.
2.2. PET imaging model
Assume that PET image formation is a linear and space-invariant process for the investigated radioactivity levels and that only signal-independent Gaussian noise is present on the images. PET imaging can be modeled as a convolution operation as the following equation
| (1) |
Where r is a 3-element vector ([x, y, z]T) representing a point in a three-dimensional space or a 2-element vector ([x, y]T) in two dimensions. i(r) is the post-reconstructed PET image from a true radioactivity distribution o(r). h(r) is a 3D point spread function. n(r) is Gaussian noise and the operator ⊗ denotes a convolution. The equation can be expressed in the Fourier domain a
| (2) |
k is the conjugate spatial frequency. The Fourier transformation of each variable is denoted by the capitalization. As the convolution becomes a multiplication in the frequency domain, the computation is greatly speeded using a fast Fourier transformation algorithm.
2.3. Bayesian deconvolution framework
The goal of partial volume correction is to restore the true radioactivity o(r) by deconvolution of observed PET image i(r). Direct deconvolution is an ill-posed problem and leads to noise amplification and severe ring artifacts. In order to find a unique and stable solution, prior information about o(r) is required to regularize the minimization. Viewing a PET image as a probabilistic mapping of an object's radioactivity distribution at each pixel, partial volume correction is to maximize a posteriori probability of the true PET activity (o) providing the observed image (i), PSF and a prior information about o. According to the Bayesian’ theorem, the posteriori probability is expressed as
| (3) |
p(o|i) is the posteriori probability of the true PET activity (o) accompanied with an observation (i). p(i) is the probability of observing PET i and is a constant. p(o) is prior information about true PET activity. p(i|o) is the posterior probability density of observing image (i) given true PET activity (o). Based on the assumption of the signal-independent Gaussian noise model, p(i|o) is denoted as
| (4) |
is a gaussian noise variance which could be estimated from background or tissue regions with uniform radioactivity distribution and assumed to be spatial independent.
2.4. Prior probability from MRI anatomical information
PET partial volume correction depends on the prior information of true activity p(o). We propose to use prior information drawn from Markov random fields (MRF) which models intensity interaction between pixels and accounts for local smoothness in image restoration. The prior information is described by Gibbs formulation as
| (5) |
where Uc(o) is Gibbs potential defined on each possible set c of voxels, G is the normalizing factor. Cliques C determine the range of pixel interaction. In proposed method only the interaction between neighboring pixels is considered and the prior information is expressed as
| (6) |
N(r) is neighborhood around pixel r that is 26 voxels in 3D or 8 pixels in 2D. drk is the Euclidian distance between voxel r and k. wrk is a weighting coefficient. The prior information constrains the local smoothness between neighboring pixels, and wrk determines the weight to enforce this regularization, for examples, wrk=1 will leads equal intensity between two neighboring pixel but wrk=0 will release any intensity difference constraint.
We manipulate wrk for an appropriate prior regularization for PET partial volume correction. In PET image, true radioactivity is smooth at two pixels with intensity difference less than a small threshold μ but discontinuous when two pixels having intensity difference higher than a great threshold ν. The prior information is similar to edge preserving deconvolution. However, PET is contaminated by significant partial volume effect, it is difficult to select threshold to recover the edge. MRI images provides high contrast, high resolution anatomical images and aligned MRI provides same structural scene with PET. Therefore, true radioactivity edge should present at the edges of MRI as a region belonging to one tissue class in aligned MRI should have same tracer activity. Whereas there are situations neighboring different tissue classes in MRI have same radioactivity in PET which will be accounted by the local smoothing regularization from the small radioactivity μ. MRI edges are determined by a MRI intensity threshold κ, and an edge is considered to exist if the neighboring MRI pixel intensity is greater than κ. Based on these observations, wrk is defined as
| (7) |
Where Δork = or – ok and Δlrk = MRIr – MRIk . μ and ν are radioactivity thresholds with μ < ν. If |Δork smaller than μ meaning there is no edge between point r and k, wrk ≈ 1 to switch on the constraint for intensity. If |Δork| greater than ν meaning there is an edge between point r and k, wrk ≈ 0 to switch off the intensity constraint. For absolute activity difference between μ and ν, MRI edge information is employed to ensure that there is an edge in o if MRI presents an edge defined as |Δlrk| is greater than the threshold κ (wrk ≈ 0).
2.5. Partial volume effect correction with MRI guided information
Partial volume effect Corrected PET is estimated by maximizing a posteriori probability as below
| (8) |
With the definition of p(i|o) and p(o), partial volume effect correction is minimize the following cost function
| (9) |
λ is a parameter to balance the convolution and prior MRF constraint, which is chosen heuristically in this paper but will be simultaneously estimated based on image noise distribution.
Corrected PET o is sought by minimization of cost function J with a conjugate gradient method. Minimization with CG method requires analytical derivatives with respect to the true activity in each pixel, which is given as
| (10) |
Where • denotes a correlation computation. Because PSF (h) is a symmetric gaussian function, the correlation is equivalent to convolution operation. The convolution is computation in Fourier domain mentioned in equation (2) to achieve a better computation speed.
A new conjugate gradient method with guaranteed descent is applied for minimization[8]. Its conjugate gradient scheme guarantees descent by satisfying a new descent condition and avoids jamming happened in Powell method. A line search satisfying the standard wolf condition is used and makes the minimization globally convergent. The method is demonstrated to achieve better performance than common conjugate gradient methods. The conjugate gradient minimization is restarted to steepest descent direction if the CG cannot significantly improve the minimization or reach set iteration number (N=50). PET image is used as the initial value for the conjugate gradient minimization that is ended with no further improvement between two restarts.
2.6. PET Point Spread Function
It is widely accepted that PET PSF can be approximated as an anisotropic 3 dimensional Gaussian function that is written as
| (11) |
σ = σ[σx, σy, σz] is the standard deviation in each direction. The point spread function can be measured by fitting the Gaussian function with PET imaging of a point radioactivity source. Full width at half maximum (FWHM) of different PET machines have been reported and the standard deviation can be related to FWHM as . Although MRI is high resolution and high contrast images, they could also present low level noise and partial volume effect at objects’ edges to complicate the partial volume correction in PET. Therefore, Preprocessing of MRI is required before running the cost function minimization. An edge-preserving diffusion filtering of MRI is applied to attenuate MRI image noise. The diffusion filtering smoothes region with a gradient lower than a threshold but preserves edges with gradients higher than the threshold. We use a threshold that is half of the MRI threshold used in equation (7).
RESULTS
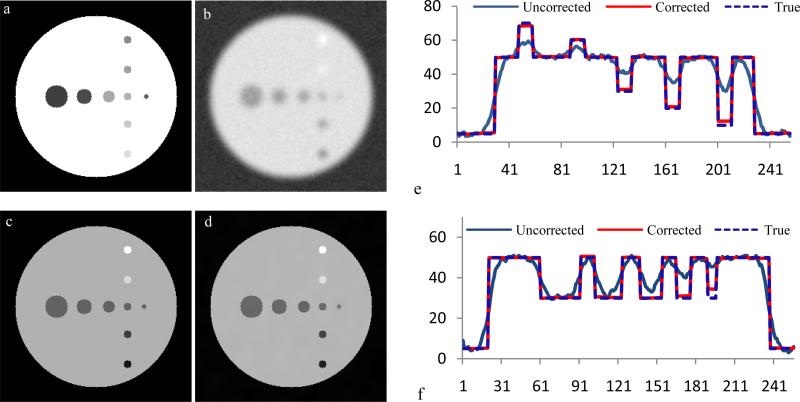
The method is first evaluated on a simulated data in Fig.1. Fig.1a is a simulated MRI for describing a scene that consists of different size of circles in a row whose diameters are 30, 20, 15, 10 and 6 pixels and in a column with diameters of 10 pixels. Different MRI intensity is given for each diameter to label it as different tissue type. A true radioactivity image is simulated in Fig.1c and gives the circles in a row an intensity of 30 for examining PSF blurring effect, and the circles in the column are given intensity of 70, 60, 30, 20 and 10 to test spilling effects. The true activity is added with 10% Gaussian white noise and then convolved with Gaussian function with FWHM=10 pixels to simulate PET imaging. The image is half resolution downsampled and added 10% Gaussian noise to get the final PET image (Fig .1b). In this way, the partial volume effect from low spatial resolution and point spread function blurring is simulated for the PET image and correlated and uncorrelated noise are also added into the image. The PET image is upsampled to the MRI resolution (Fig. 1b) as the first step for partial volume effects correction. The partial volume correction algorithm is applied to achieve the final correction (Fig.1d). The signal along the horizontal line is plotted in Fig.1f to show the activity recovery (O) from the blurring signal (i) compared with the true activity. Fig .1e shows the activity profile along the vertical line.
Fig. 1.
Partial volume correction of simulated data. (a) simulated MRI, (b) simulated PET, (c) the true radioactivity for simulation, (d) corrected PET result, (e) the lines of profile of a vertical line along the objects, and (f) the lines of profile of a horizontal line along the circles. The x-axis of (e) and (f) is the pixel position along the line.
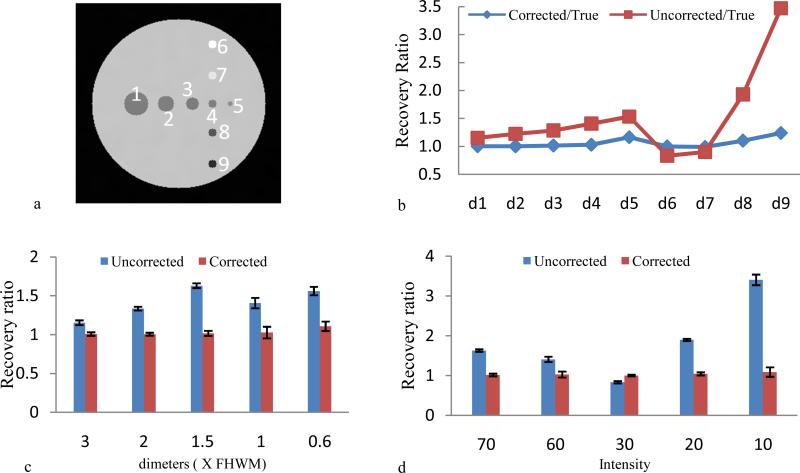
The circles are numbered from 1 to 9 as Fig .2a. Radioactivity recovery ratio is computed as mean activity within the circle divided with true activity in the region and the recovery ratio improvement from the correction is demonstrated in Fig. 2b. Expressing as percentage of true activity, the correction with respect to the circle diameters is shown is Fig. 2c that demonstrates the method restore 102 ± 7% of the true object activity for object having size equal or greater than FHWM of PSF. Fig. 2d shows the effects of object radioactivity to partial volume correction. 1%, 5%, 10%, 15% and 20% noise are added to PET and the correction results generate the error bar in Fig 2(c) and (d).
Fig. 2.
Quantitative evaluation of partial volume correction of simulated data. (a) Each circle was labeled with a number from 1 to 9. (b) The activity recovery ratio between the acquired PET data and the true activity for cases with and without correction. (c) The activity recovery verse object dimension related to FHWM of PSF in the objects (1,2,3,4,5). (d) The activity recovery verse object intensity for the circles (6,7,4,8,9).
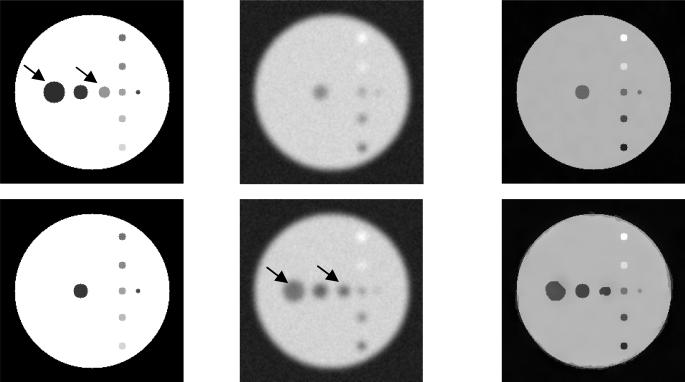
There are some situations MRI does not differentiate nearby structures which shows different radioactivity level. The true PET cannot be correctly restored in common used MRI guided method which assumes same MRI structures have exactly same radioactivity. Our method can preserve the edge present in PET if an appropriate threshold selected for PET. We purposely remove two circles in MRI in first row of Fig.3, but activity different background are present in these circle in true activity distribution. Our method can successfully restore the objects even though MRI does not detect the objects from background. Even we cannot select an appropriate threshold to preserving the edges, the method can correctly restore the activity detected by MRI which like a local restoration. The second row of Fig. 3 shows the local partial volume correction from MRI defined region.
Fig. 3.
Demonstration of the restoration of true activity. The columns are MRI, PET, and corrected PET, respectively. In the first row two circles are not visible in PET but in MRI (labeled with arrows) true PET is correctly restored although MRI and the edges of the two circles were restored. In second row, the two circles presences in PET (labeled with arrows), but not MRI. By selecting an appropriate threshold for PET, the two circles can be restored even MRI does not show the anatomical information.
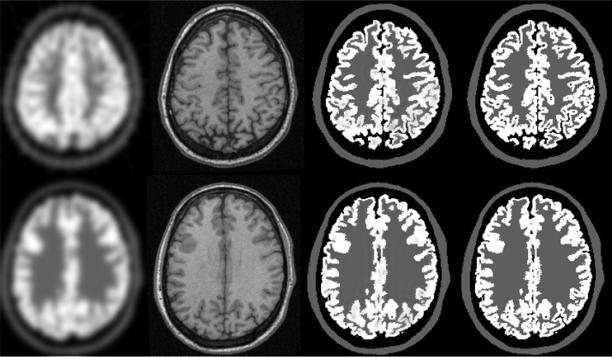
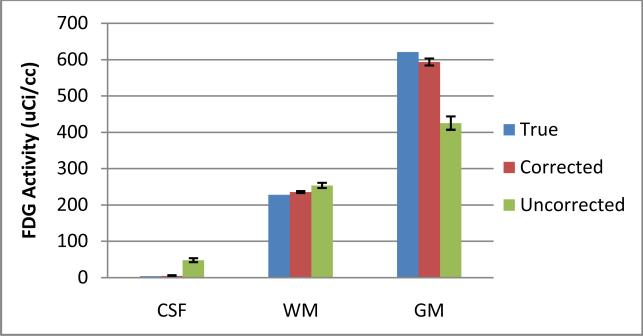
Publicly available dataset of 16 Monte Carlo dynamic FDG brain PET [9] are used to evaluate the proposed method. The dataset was simulated by the SORTTEO Monte Carlo simulated software modeling the Exact ECAT HR+ scanner operating in 3D mode. The simulation accounts for most of the phenomena encountered during PET acquisition including scatter, random and system dead-time, Poisson nature of the emission and positron range in tissues. Realistic anatomical models were derived from human brain images. For FDG activity model, constant activity of 8.45kBq/cc and 22.99kBq/cc were assigned to tissues classes of WM and GM respectively. The PSF was approximated by 3-D Gaussian function. The full-width at half maximum parameters was computed from imaging a point source located at 5 cm from the center of field of view of the simulated scanner. The FWHM for FDG were 9.2 mm in axial rand radial directions. True activity, PET images and registered MRI provide ideal dataset for quantitative evaluation of the method. Results on brain FDG PET image from PET-SORTEO database is shown in Fig 4. Quantitative comparison of activity recovery for CSF, WM and GM is shown in Fig. 5, which including average activity in each region from the true, PET images and corrected images, and the activity standard deviation within each region for each image.
Fig. 4.
Partial volume correction of human brain images. The first column is the interpolated PET images. The second column is the corrected PET. The third column is the MRI images and the last column is the ground true of PET FDG distribution. Two axial slices are demonstrated.
Fig. 5.
Recovery of FDG activity in the brain after partial volume correction. FDG radioactivity within CSF, WM and GM from five PET images are computed from the true, corrected image and uncorrected PET images. The true activity of CSF is zero.
CONCLUSIONS
We developed an MRI-guided partial volume correction method for PET images. The method can improve PET image quality and the quantification of radioactivity concentration. The method does not require prior information about tracer activity within tissue regions. It can offer a partial volume correction method for various applications in PET imaging. It can be particularly useful for combined PET/MRI studies.
REFERE1CES
- 1.Chen CH, Muzic RF, Jr., Nelson AD, Adler LP. A nonlinear spatially variant object-dependent system model for prediction of partial volume effects and scatter in PET. IEEE Trans Med Imaging. 1998;17:214–227. doi: 10.1109/42.700733. [DOI] [PubMed] [Google Scholar]
- 2.Fazio F, Perani D. Importance of partial-volume correction in brain PET studies. J Nucl.Med. 2000;41:1849–1850. [PubMed] [Google Scholar]
- 3.Chen CH, Muzic RF, Jr., Nelson AD, Adler LP. Simultaneous recovery of size and radioactivity concentration of small spheroids with PET data. J Nucl.Med. 1999;40:118–130. [PubMed] [Google Scholar]
- 4.Baete K, Nuyts J, Van Laere K, Van Paesschen W, Ceyssens S, De Ceuninck L, Gheysens O, Kelles A, Van den EJ, Suetens P, Dupont P. Evaluation of anatomy based reconstruction for partial volume correction in brain FDG-PET. Neuroimage. 2004;23:305–317. doi: 10.1016/j.neuroimage.2004.04.041. [DOI] [PubMed] [Google Scholar]
- 5.Frouin V, Comtat C, Reilhac A, Gregoire MC. Correction of partial-volume effect for PET striatal imaging: fast implementation and study of robustness. J Nucl.Med. 2002;43:1715–1726. [PubMed] [Google Scholar]
- 6.Rousset OG, Ma Y, Evans AC. Correction for partial volume effects in PET: principle and validation. J Nucl.Med. 1998;39:904–911. [PubMed] [Google Scholar]
- 7.Soret M, Bacharach SL, Buvat I. Partial-volume effect in PET tumor imaging. J Nucl.Med. 2007;48:932–945. doi: 10.2967/jnumed.106.035774. [DOI] [PubMed] [Google Scholar]
- 8.Hager WW, Zhang H. Algorithm 851: CG_DESCENT, a conjugate gradient method with guaranteed descent. ACM transcations on Mathmatical Softwares. 2006;32:113–137. [Google Scholar]
- 9.Reilhac A, Lartizien C, Costes N, Sans S, Comtat C, Gunn R, Evans A. PET-SORTEO: A Monte Carlo-based simulator with high count rate capabilities. IEEE Trans on Nuclear Science. 2004;51:46–52. [Google Scholar]