Abstract
Background
Most traditional genome sequencing projects involving viruses include the culture and purification of the virus particles. However, purification of virions may yield insufficient material for traditional sequencing. The electrophoretic method described here provides a strategy whereby the genomic DNA of the Korean isolate of Pieris rapae granulovirus (PiraGV-K) could be recovered in sufficient amounts for sequencing by purifying it directly from total host DNA by pulse-field gel electrophoresis (PFGE).
Methodology/Principal Findings
The total genomic DNA of infected P. rapae was embedded in agarose plugs, treated with restriction nuclease and methylase, and then PFGE was used to separate PiraGV-K DNA from the DNA of P. rapae, followed by mapping of fosmid clones of the purified viral DNA. The double-stranded circular genome of PiraGV-K was found to encode 120 open reading frames (ORFs), which covered 92% of the sequence. BLAST and ORF arrangement showed the presence of 78 homologs to other genes in the database. The mean overall amino acid identity of PiraGV-K ORFs was highest with the Chinese isolate of PiraGV (∼99%), followed up with Choristoneura occidentalis ORFs at 58%. PiraGV-K ORFs were grouped, according to function, into 10 genes involved in transcription, 11 involved in replication, 25 structural protein genes, and 15 auxiliary genes. Genes for Chitinase (ORF 10) and cathepsin (ORF 11), involved in the liquefaction of the host, were found in the genome.
Conclusions/Significance
The recovery of PiraGV-K DNA genome by pulse-field electrophoretic separation from host genomic DNA had several advantages, compared with its isolation from particles harvested as virions or inclusions from the P. rapae host. We have sequenced and analyzed the 108,658 bp PiraGV-K genome purified by the electrophoretic method. The method appears to be generally applicable to the analysis of genomes of large viruses.
Introduction
Baculoviruses represent a diverse group of viruses with covalently closed, double-stranded, circular, supercoiled genomes, with sizes varying from 80 to 180 kb, encoding between 90 and 180 genes. The DNA genome is packaged in rod-shaped nucleocapsids that are 230–385 nm in length and 40–60 mm in diameter. The virions occur in two types- occluded virions (ODV) and budded virus particles (BV). Baculoviridae are divided into four genera, Alphabaculovirus [lepidopteran-specific nuclear polyhedrosis virus (NPVs)], Betabaculovirus [lepidopteran-specific granulosis virus (GVs)], Gammabaculovirus (hymenopteran-specific NPVs) and Deltabaculovirus (dipteran-specific NPVs) [1], [2]. Viruses belonging to the order Hymenoptera contain the smallest genomes, at >80 kb, which has been explained as a result of their restricted life cycle, confined to replication in insect gut cells [3]. Group I alphabaculoviruses cluster ∼130 kb, whereas Group II shows a high degree of diversity, varying from ∼130 to 170 kb. The larger genomes of the Group II alphabaculoviruses can be attributed to a combination of repeated genes that are not found in the smaller genomes. This is in contrast to the betabaculoviruses genomes, varying from 101 kb in the case of Plutella xylostella granulovirus (PlxyGV) [4] to 178 kb in Xestia c-nigrum granulovirus (XecnGV) [5]. Despite the large difference in gene content in betabaculovirus genomes, as reflected in this range of sizes, their genomes are surprisingly collinear, compared with alphabaculoviruses, which show a high degree of variation [6], [7]. The first dipteran-specific deltabaculovirus, the Culex nigripalpus nucleopolyhedrovirus (CunniNPV), was isolated and sequenced from the mosquito Culex nigripalpus [8]. A phylogenetic analysis showed its distinctive form, making it a member of a new genus within the family Baculoviridae [9]. Compared to alphabaculoviruses family members, betabaculoviruses have been investigated to a lesser degree, because of the limitations of permissive cell lines [10]. Currently, 60 complete genomes are known in the Baculoviridae family; 45 genomes from NPV (41 alphabaculoviruses, 3 gammabaculoviruses, and 1 deltabaculovirus), 14 genomes from GV, and 1 unclassified Hemileuca sp. NPV (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=10442).
The small cabbage white butterfly, Pieris rapae (P. rapae) is a serious pest of cultivated cabbages and other mustard family crops worldwide. A serious infestation can lead to the death of the plant due to reduced photosynthesis. P. rapae granulovirus (PiraGV) infects P. rapae in nature and functions as an important biological agent in controlling the population of P. rapae in the ecosystem. Although PiraGV is now a registered biocontrol agent for the control of P. rapae, research on the genetic and molecular information of the virus is still limited, apart from a recent study on occlusion-derived virus (ODV)-associated proteins of the betabaculovirus [11]. Sequencing of the complete genome of the Chinese isolate of P. rapae granulovirus (PiraGV-C) showed a size of 108,592 bp and predicted 120 open reading frames (GenBank, GQ884143) [12]. Although sequencing efforts have been significant, more detailed information about a wide range of isolates inhabiting different geographical regions would provide a more comprehensive overview of baculoviruses and further establish their candidature as pest control agents.
This study is unique, as we have taken advantage of the large-sized genome and high titer of infection of P. rapae granulovirus (Korean isolate) to purify the viral genome away from host DNA by pulse-field gel electrophoresis. The viral DNA is recovered in amounts sufficient for its classical genome sequencing. The procedure requires less starting material than would be necessary if starting with the purification of virus particles from inclusion bodies. The genome sequence produced in this work was through a subcloning approach, without recourse to the use of automated high-throughput next-generation sequencing (NGS) technology.
Materials and Methods
Separation of Nuclei from P. rapae
Larvae of P. rapae were obtained from a mass rearing facility at Hampyeong Insect Institute (Hampyeong, Korea) and were reared in the laboratory on kale leaf at 25±3oC with 60±5% relative humidity, under a 12/12 hr natural light/dark cycle for a short duration. The final instar larvae were dissected to remove the gut and were subsequently ground and centrifuged (5,000 rpm, 10 min, 4oC) to separate the nuclei and remove the cell debris from the solution.
Chemicals
All chemicals used were of analytical grade, and were obtained from Sigma Chemical Co. (St. Louis, MO, USA) until indicated otherwise.
Preparation of High Molecular Weight (HMW) DNA Plugs Embedded in Agarose
HMW DNA is considered vulnerable to mechanical shearing forces and suffers frequent double-stranded breaks. It is thus not suited to large-insert cloning. To prevent HMW DNA from being damaged in the nucleus lysis process, the separated nuclei were embedded in agarose gel. The nuclei were warmed for 5 min at 45oC and were mixed with 1% InCert agarose. The mixture was subsequently poured into a plug mold (BioRad, Hercules, CA), kept on ice and allowed to solidify for 1–2 hr. The agarose plugs were then put into 50 ml of proteinase K lysis buffer (0.5 M EDTA, 1% N-lauroylascosine, 1 mg of proteinase K/ml) and incubated for 24 hr at 50oC. After the subsequent removal of proteinase K lysis buffer from the agarose plugs, the lysis process was repeated, for a further 24 hr. After 2–3 washes in deionized water, the plugs were placed in 50 ml of TE50 buffer (10 mM Tris-HCl, 50 mM EDTA, pH 8.0) and washed for 12 hr. Additional washing was performed for another 12 hr after replacing with TE50 buffer. Subsequently, the plugs were incubated for 2 hr in 0.1 mM phenylmethylsulfonylfluoride (PMSF) buffer at 4oC to inactivate proteinase K, followed by another subsequent wash in TE50 buffer for 24 hr, and were stored in 0.5 M EDTA at 4oC.
Pre-electrophoresis of Agarose Plugs
Next, the agarose plugs were placed in 0.5× TBE buffer (45 mM Tris-base, 1 mM EDTA, 45 mM boric acid) and dialyzed for 3 hr. Subsequently, they were inserted into the preparative slot of 1% pulse- field certified agarose gel, and PFGE was conducted using 0.5× TBE buffer and the CHEF DR-II apparatus (Bio-Rad, Hercules, CA) with a pulse time of 5 s for 10 hr at 12oC and a voltage of 4V/cm. After the electrophoresis, the plugs were removed from the slot, stored in 50 ml of 0.5 M EDTA buffer, and dialyzed overnight at 4oC.
Partial Digestion of Plugs
HMW DNA embedded plugs (n = 10) were placed in 500 µl of an enzyme mixture, consisting of 1 µl EcoRI at a concentration of 2 U/µl, 1 µl EcoRI methylase at a concentration of 40 U/µl (New England Biolabs, Ipswich, MA), 25 µl of 100× Bovine Serum Albumin (10 mg/ml), 5 µl of polyamine (100×), 50 µl of methylase buffer (10×) in 394 µl of DW and equilibrated for 2 hr at 4oC, followed by a 4 hr incubation at 37oC. After digestion, the plugs were treated with 150 µl of 0.5 M EDTA, 37.5 µl of 20% N-lauroylsarcosine and 15 µl of proteinase K (20 mg/ml), and incubated for 1 hr at 37oC to inactivate the endonuclease. Subsequently, PFGE was conducted with a CHEF DR-II apparatus (Bio-Rad) with a pulse time between 0.1 and 40 s for 16 hr at a voltage of 6 V/cm to check the partially digested plugs.
Separation of PiraGV-K DNA from P. rapae Genomic DNA
PiraGV-K DNA was separated by PFGE with an initial pulse time of 0.1 s, a final pulse time of 40 s, a temperature of 12oC and a voltage of 6 V/cm for 14 hr. Furthermore, a lambda ladder PFG marker (New England Biolabs, Ipswich, MA) was used as a size marker to enable the band of PiraGV-K at ∼125 kb to be eluted selectively.
After the PFGE treatment, the edge of the gel, including a size marker, was cut and put into ethidium bromide staining buffer to mark the location of the 125 kb band of PiraGV-K. Subsequently, the eluted portion was placed into a dialysis bag to recover the PiraGV-K DNA using PFGE with a pulse time between 0.1 and 40 s and a voltage of 6 V/cm for 14 hr.
Construction and Characterization of PiraGV-K Fosmid Library
Randomly sheared PiraGV-K DNA was cloned into the Eco72I blunt-end site of the CopyControl pCC1FOS fosmid vector (Epicentre Biotechnologies, Madison, WI). The fosmids were packaged using ultra-high efficiency MaxPlax lambda packaging extracts and plated on TransforMax EPI300 E. coli (Epicentre Biotechnologies, Madison, WI). The quality of the constructed fosmid library was assessed using standard techniques. Of a total of 6,000 clones, 96 were picked randomly and the fosmids were end sequenced from both directions using the primers (forward sequencing primer 5′– GGATGTGCTGCAAGGCGATTAAGTTGG –3′ and reverse sequencing primer 5′– CTCGTATGTTGTGGAATTGTGAGC –3′) to the vector. Stand-alone BLAST was performed for the nucleotide sequences against a locally curated viral sequence database (http://edunabi.com/~prgv/).
Whole Genome Shotgun Sequencing
Based on the mapping data in the locally curated viral sequence database (http://edunabi.com/~prgv/), a minimum tiling path was prepared and four fosmid library clones were selected to construct a shotgun library. The selected fosmid clones were named as NB-FOS-1-1-F40_A05A02 (27 kb), NB-FOS-1-1-F40_A23B06 (33 kb), NB-FOS-1-1-F40_C07D02 (32 kb) and NB-FOS-1-1-F40_E13E04 (37 kb). Equivalent volumes of fosmid DNA clones were digested with NotI to obtain 3-7 kb DNA pieces that were then ligated into a purified pUC118 BamHI/BAP ready vector (Takara Bio Inc., Shiga, Japan) [13]. Ligated products were transformed into E. coli DH5α cells by electroporation and spread on LB (ampicillin, 100 µg/ml) plates. The quality of the library was checked for E. coli genomic DNA contamination and empty vector contamination by cross-match. Plasmid clones that were eight times larger than each of the selected clones were randomly picked for plasmid preparation and sequencing with M13 forward and reverse universal primers using an Applied Biosystems 3730 XL DNA analyzer (Applied Biosystems, Carlsbad, CA) using the cycle sequencing method with fluorescent dye terminators and AmpliTaq DNA polymerase (ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction, Perkin Elmer, Waltham, MA). Applied Biosystems sequencing software was used for lane tracking, trace extraction and data were transferred to UNIX workstations for further processing.
Genomic DNA Assembly
Contigs were prepared using the software Pregap4, including PHRED [14], [15], PHRAP (www.phrap.org), and vector masking on the average read length, clustering and assembling a repeated sequence. The primer walking procedure was used to close remaining gaps. The map of the first clone selected from PiraGV-K was constructed and a clone capable of covering 60 k to 85 k was also screened.
Sequence Analysis
Putative coding regions of PiraGV-K genome was predicted using the Genemark [16]; Glimmer [17] and AMIgene [18] open reading frame (ORF) finding software. ORFs of more than 150 bp were designated as putative genes; the overlap between any two ORFs was set to a maximum of 25 amino acids (aa); otherwise, the longer one was selected. Gene annotations and comparison of the sequences with those in public databases were carried out using the BLAST at National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequence analysis was performed using Clustal X and GeneDoc (2.7.0). The PiraGV-K genomic DNA sequence was deposited in GenBank under the accession number JX968491.
Twelve betabaculovirus genomes were used to identify gene conservation in PiraGV-K. These genomes were from Adoxophyes orana GV (AdorGV; NC_005038) [19], Agrotis segetum GV (AgseGV; NC_005839), Choristoneura occidentalis GV (ChocGV; NC_008168) [20], Cryptophlebia leucotreta GV (CrleGV; NC_005068) [7], Cydia pomonella GV (CypoGV; NC_002816) [21], Helicoverpa armigera GV (HearGV; NC_010240) [22], Phthorimaea operculella GV (PhopGV; NC_004062), P. rapae GV-Chinese isolate (PiraGV-C; NC_013797), Plutella xylostella GV (PlxyGV; NC_002593 [4], Xestia c-nigrum GV (XecnGV; NC_002331) [5], Pseudaletia unipuncta GV (PsunGV; NC_013772) and Spodoptera litura GV (SpliGV; NC_009503) [23]. Detailed descriptions of the putative PiraGV-K ORFs, including their positions in the genome, length, and their relationship with AdorGV, AgseGV, ChocGV, CrleGV, CypoGV, HearGV, PhopGV, PiraGV-C, PlxyGV, PsunGV, SpliGV, and XecnGV are presented in Table S1.
Data Access
The whole-genome data of PiraGV-K and relevant sequence information has been maintained in a database at ‘http://edunabi.com/~prgv/’ for ready reference. The PiraGV-K whole genome sequence is registered under GenBank accession number JX968491.
Results and Discussion
The Electrophoretic Separation Method for PiraGV-K Whole-genome Sequencing
Today, most genome sequencing projects rely on the whole-genome shotgun (WGS) method, which uses the Sanger technique to sequence genomic libraries over conventionally mapped clones using bacterial artificial chromosome (BAC), cosmid or fosmid libraries [24]–[26]. Although WGS strategy has provided rapid access to new gene models from diverse organisms with continued improvements in the assemblers, read lengths and mate pair technologies, the resulting assemblies still remain highly fragmented with an incomplete genomic representation [27], [28]. This has helped the focus on BAC-based physical map construction and its integration with high-density genetic maps that have benefited from next-generation sequencing (NGS) platforms and high-throughput array platforms [29], [30]. In this context, fosmids, with a narrower insert range (average of 40 kb), stable maintenance, and easy production, have found wide applications in studies related to structural variation and the organization of genomes [30]–[32].
The selection of target substances from the environment is the most critical component for the implementation of suitable approaches for whole-genome sequencing. In the case of infectious viruses, the study of the genome is more cumbersome because these agents are difficult to culture and purify. Conventional methods for the purification of genomic DNA fragments present the drawback of obtaining a large number of populations from multiple locations to acquire sufficient high-quality DNA samples for sequence analysis.
The genome sequencing method (Fig. 1), detailed here for the first time, was used to construct fosmid library clones of double-stranded PiraGV-K genome, generating a library size of 100–150 kb corresponding to the genome size of the virus. This approach was successful in the analysis of the PiraGV-K genome, without the need for purifying PiraGV-K from P. rapae, thus simplifying sampling and reducing labor time. This approach provides a significant advantage over traditional protocols for the sequencing of dsDNA genomes and could potentially be used for circular DNA genomes of viruses, although its wider application needs to be further validated. Recently, a report highlighted the importance of sequencing small genomes without the need for standard library preparation using the Pacific Biosciences RS sequencer (the “PacBio”) with as little as 1 ng of DNA [33]. That our method can be performed without the specialized expertise required for virus culturing and purification from their hosts, coupled with its requirement for little time and reliable precision, makes it particularly useful for laboratories lacking sophisticated viral culturing facilities. The limitations of the genome sequencing method purified by the electrophoretic method may lie in the sequencing of RNA viruses, because they are less stable than DNA in nature and may require the maintenance of cultured viral isolates, unlike our approach. A new system for rapid determination of viral RNA sequence (RDV) uses small amounts of RNA to synthesize first- and second- strand cDNAs for library construction and direct sequencing using optimized primers [34]. Although reverse transcription followed by polymerase chain reaction is commonly used for deciphering RNA viral genomes, low-copy number viral samples remain a challenge; sequence-independent methods provide attractive solutions [35], [36].
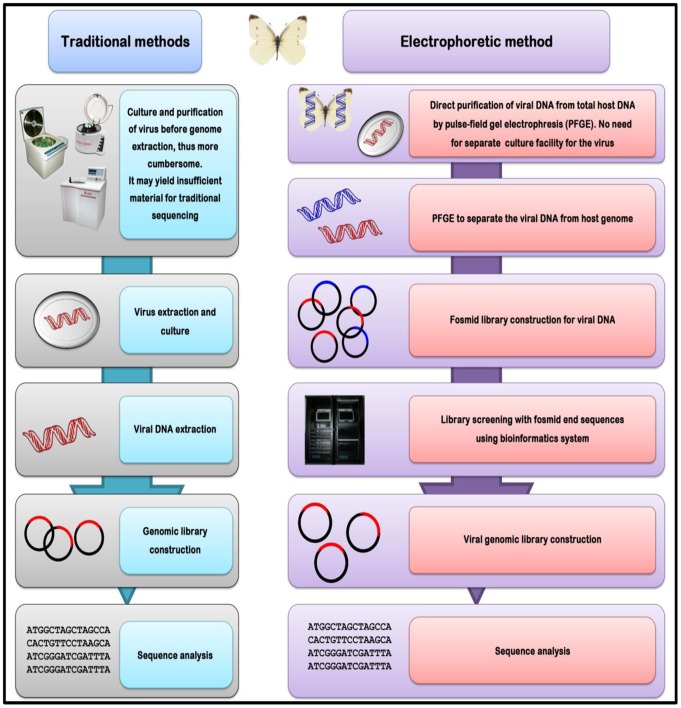
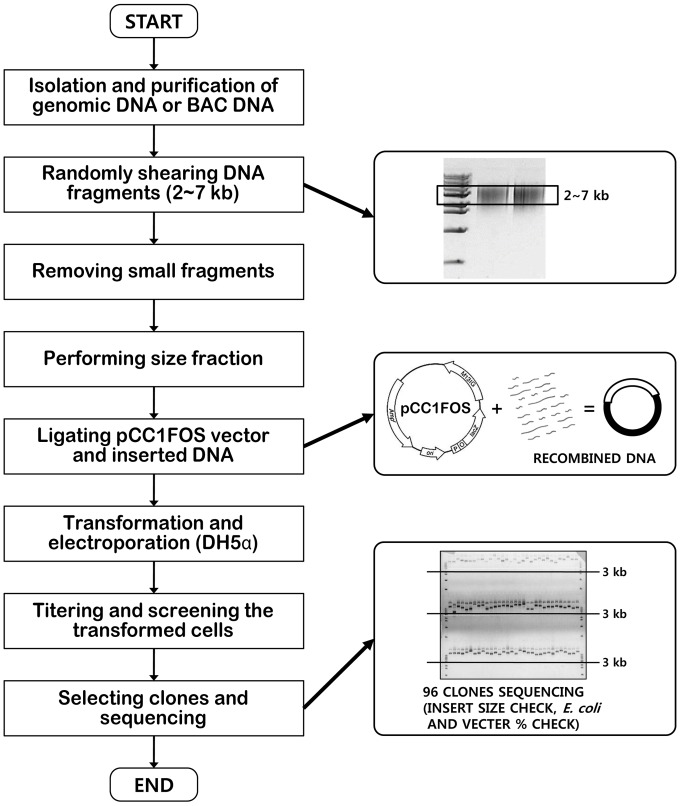
Figure 1. Comparative depiction of the electrophoretic and traditional methods for separation of viral genomic DNA.
Flow chart showing the electrophoretic method for purification of the virus from the host genome for the construction of fosmid library of PiraGV-K and its significance in comparison with the traditional methods.
In the method described here, HMW DNA embedded agarose plugs of P. rapae were digested with EcoRI, before confirmation of the potential PiraGV-K DNA at 125 kb by PFGE analysis (Fig. 2). The potential PiraGV-K DNA was found readily when EcoRI (8 U) and methylase (20 U) were used after a 2 hr pre-electrophoresis step. The partial digestion step is considered critical for both the construction of the host BAC library, and also converting the viral genome into a family of circularly-permuted linear molecules of genome length. The linear form of the viral genome, thus obtained from the digestion step facilitates efficient separation of the genomic DNA in PFGE. Subsequently, PCR was conducted with different primers, designed to provide variable sizes from the nucleotide sequence of PiraGV-K, to check the validity of the potential PiraGV-K DNA. The PCR product size in all cases was found to be the same as expected for the PiraGV-K DNA sequence (Fig. 3). Subsequently, for effective separation of PiraGV-K DNA, pre-electrophoresis and partial digestion of agarose plugs was repeated with PFGE. Following the PFGE run, the DNA band of 125 kb corresponding to PiraGV-K DNA was eluted, eventually separating PiraGV-K DNA from P. rapae embedded agarose molds (Fig. 4A). The eluted DNA (20 ng) was subsequently electrophoresed in parallel with a 1 kb ladder to validate the separation process (Fig. 4B). The eluted and end-repaired PiraGV-K DNA was ligated into the pCC1FOS vector and the purified products were checked for quality by titering. In total, approximately 6,000 clones resulted, out of which 96 were selected and end-sequenced. To effectively map the fosmid-end sequences, we performed a stand-alone BLAST against a locally constructed viral sequence database. Based on the mapping data from the databases, a minimum tiling path (MTP) was prepared, leading to the selection of four fosmid library clones for the construction of a PiraGV-K shotgun library. The sizes of the four selected fosmid clones, (NB-FOS-1-1-F40_C07D02, NB-FOS-1-1-F40_E13E04, NB-FOS-1-1-F40_A05A02, and NB-FOS-1-1-F40_A23B06), measured by NotI restriction digestion were approximately 32, 37, 27, and 33 kb, respectively (Fig. 5). The shotgun library resulted in a total of 20,000 clones, of which 96 were selected and sequenced (Fig. 6).
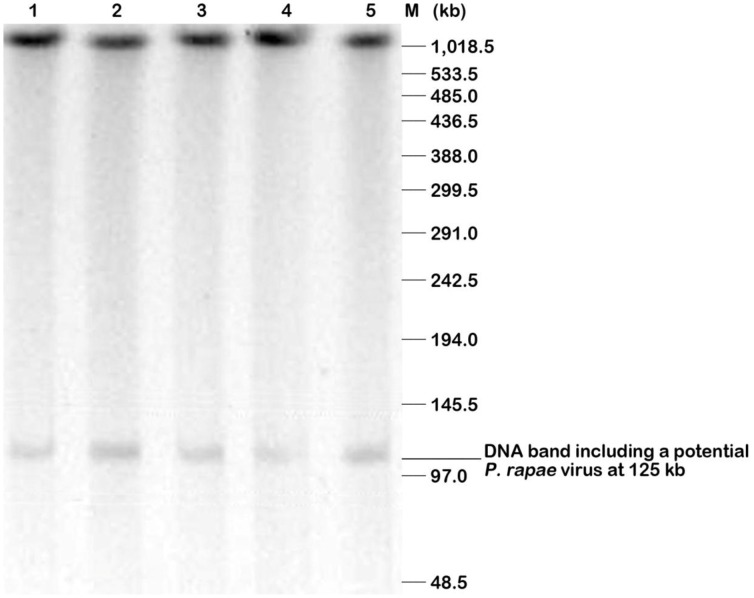
Figure 2. Pulse-field gel electrophoresis (PFGE) confirmation of the potential PiraGV-K DNA.
HMW DNA embedded agarose plugs of P. rapae confirmed by PFGE, wherein the plugs were partially digested by an enzyme mixture following pre-electrophoresis. ‘M’ represents PFG lambda marker (NEB) and lanes 1–5 depict EcoRI digested DNA molds. A potential PiraGV-K DNA band was seen approximately at 125 kb after PFGE of enzyme digested DNA. PFGE conditions included 1% pulsed field certified agarose gel, a pulse time between 0.1–40 sec for up to 16 hrs and a voltage of 6 V/cm to check for partially digested plugs.
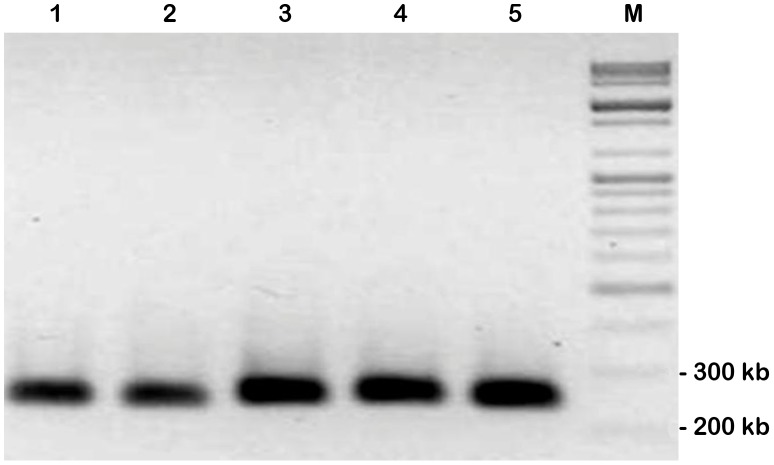
Figure 3. Confirmation of PiraGV-K DNA separated by PFGE.
PCR was conducted to check the identity of PiraGV-K with 5 primers designed from the nucleotide sequence of PiraGV-K. The size of the PCR product was the same as the expected size of the nucleotide sequence. Lane 1; primer 1: AY-519253-1 (expected size of 227 bp), lane 2; primer 2: AY-706575-1 (expected size of 223 bp), lane 3; primer 3: AY-428513-1 (expected size of 234 bp), lane 4; primer 4: AY-449794-2 (expected size of 212 bp), lane 5; primer 5: AY-519252-1 (expected size of 231 bp).
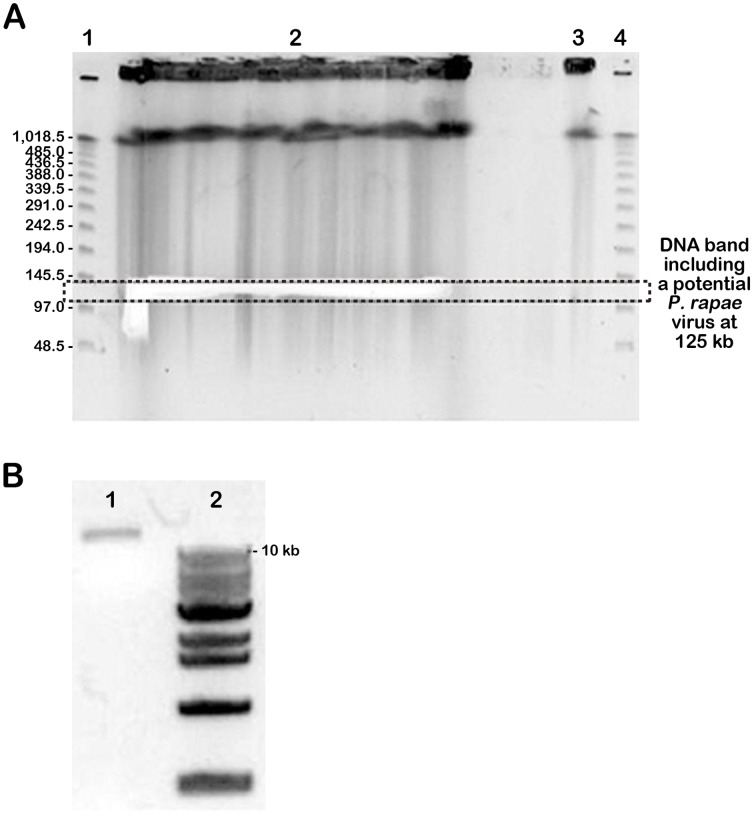
Figure 4. Separation of potential PiraGV-K DNA from agarose molds after PFGE.
(A) Elution of DNA band (approximately 125 kb) of potential PiraGV-K. This indicates that the DNA of PiraGV-K is separated from P. rapae DNA embedded agarose molds. Lanes 1 and 4 show PFG lambda marker (NEB) and lanes 2 and 3 depict EcoRI digested DNA molds. (B) This indicates the concentration of DNA that has been collected by PFGE as determined using a spectrophotometer. Lanes 1 and 2 show eluted DNA (20 ng loading) and a 1 kb ladder, respectively.
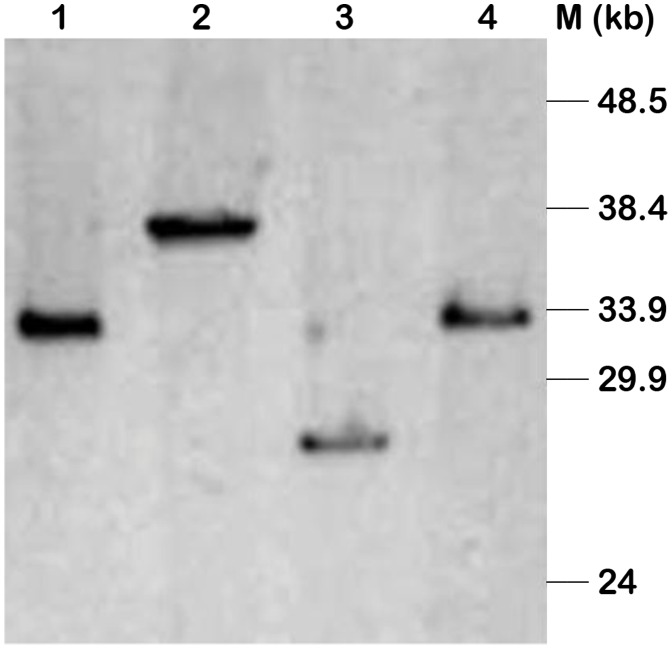
Figure 5. Restriction digestion of selected fosmid clone DNA by NotI enzyme.
Four fosmid clones were selected on the basis of minimum tiling path towards construction of shotgun library. Lane 1, fosmid clone NB-FOS-1-1-F40_C07D02 (approximately 32 kb); Lane 2, fosmid clone NB-FOS-1-1-F40_E13E04 (approximately 37 kb); Lane 3, fosmid clone NB-FOS-1-1-F40_A05A02 (approximately 27 kb); Lane 4, fosmid clone NB-FOS-1-1-F40_A23B06 (approximately 33 kb). Lane ‘M’ is represented by monocot lambda marker.
Figure 6. Flow chart depicting shotgun library construction.
Genomic DNA or BAC DNA isolation and purification was followed by size fractionation and ligation into a pUC118 ready vector for 4oC followed with transformation by electroporation into DH5α. The quality of thus constructed shotgun library was checked by titering (40 µl of cell stock, white: blue = 400∶100). The number of clones was approximately 20,000 in total. 96 clones were selected and sequenced including insert size check, E. coli and vector % check.
Characteristics of the PiraGV-K Genome Sequence
To date, whole-genome sequencing has been conducted successfully for 60 baculoviruses: 45 were NPVs (41 alphabaculoviruses, 3 gammabaculoviruses and 1 deltabaculovirus). Only 14 complete genomes have been sequenced of betabaculoviruses, including PiraGV-C [12]. The growing number of fully sequenced baculovirus genomes now allows some understanding of the evolutionary history of baculoviruses by comprehensive analyses of nucleotide/protein sequences, gene order, and content [37], [38]. We have sequenced and analyzed the 108,658 bp PiraGV-K genome purified by electrophoretic method. The approach allows for the determination of the viral sequence with multiple fold redundancy per base position. An 8x sequence of the PiraGV-K genome was compiled from the sequence data generated here. The size of the final draft sequence was 108,658 nt (Fig. 7). The length of the sequence obtained was consistent with the predicted size of PiraGV-C (108,592 nt), differing by only 66 nt. It can thus be categorized as one of the smaller betabaculoviruses sequences, with AdorGV (99,657 nt) being the smallest. XecnGV has a whole genome size of 178,733 nt [5], which is largest genome among the completely sequenced betabaculoviruses and is closely related to sequences studied from noctuid moths, including Autographa gamma GV, Hoplodrina ambigua GV, Euxoa ochrogaster GV, and Scotogramma trifolii GV [39]. These are closely followed by HearGV, with a genome size of 169,794 bp [40]. PiraGV-K coding sequences represent 92% of the genome, leaving very little noncoding DNA.
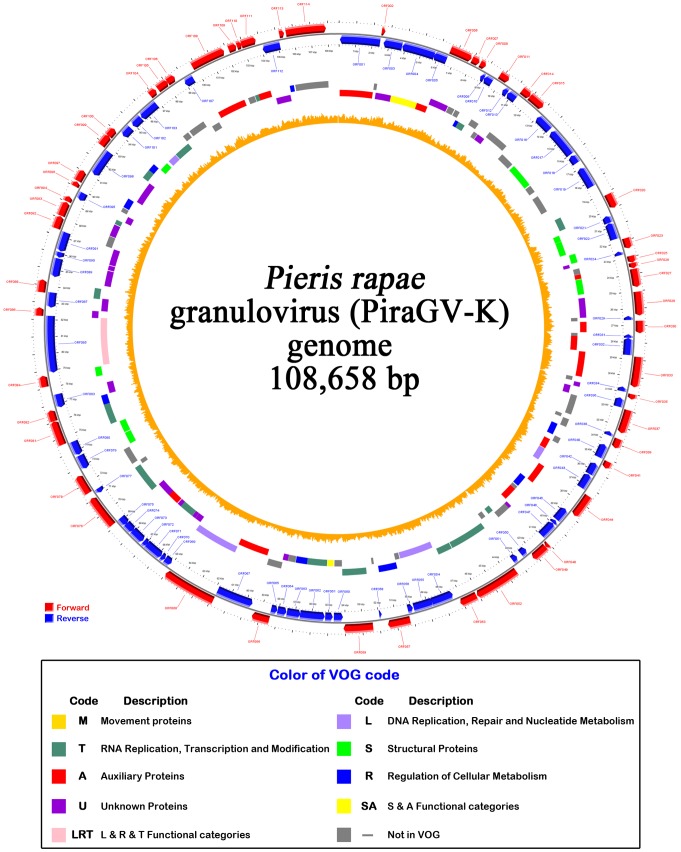
Figure 7. Circular representation of the PiraGV-K genome map.
ORFs are represented by arrows, with the position and direction of the arrow indicating ORF position and orientation. Red arrows and blue arrows represent forward and reverse strand ORFs in the circular map. VOG code and colors assigned indicate the grouping of the genes according to function.
The PiraGV-K genome has an AT content of 66%, identical to PiraGV-C (66%), and is closely related to CrleGV, having the highest known AT content of 67.6%. This result is consistent with previous findings that the sequenced betabaculovirus genomes are AT-rich, with the lowest AT content of 54.8% observed in case of CypoGV, with an overall average of 62.6%. The difference in AT content is due to the base composition at the third nucleotide position of the codon in the coding regions. It has been established previously that proteins encoded by more extreme AT and GC-rich genomes generally have lower compositional complexity than those of more typical organisms [41]. A consequence of this is that the overall amino acid composition of the peptides in such organisms is skewed. Peptides of AT-rich organisms have higher proportions of Phe, Leu, Ile, Met, Asn, Lys and Tyr that are relatively rare in the organisms with GC-rich genomes. Similar correlation has been noted with smaller data sets in earlier research [19], [42], [43]. The end result of this is that organisms with an extreme genome composition encode peptides of a lower complexity, as measured by the global complexity value [44]. It is known that the median global complexity value, G1 for AT-rich genes from a variety of cellular organisms is in the range of 0.72 to 0.78 [41]. Whereas most PiraGV-K ORFs had an AT composition (average 65%) close to the average AT composition of PiraGV-K genome (66%), granulin had an AT composition that was significantly lower at 56% (results not shown). It is to be noted that in case of extremely anchored proteins, such as granulin, it might be impossible for the virus to maintain its preferred nucleotide composition and codon usage and still encode a particular peptide. This observation has been confirmed in other annotated, AT-rich, viral genomes [19], [45] Also, it is understood that, although various ORF prediction methods have been used (Fig. 8), no one method can define all possible ORFs in compositionally extreme (AT or GC-rich) genomes, as is clearly illustrated in the PiraGV-K genome. PiraGV-K granulin had a subjective appearance of an “alien” gene, because the codon usage did not conform to the overall codon usage [46]. However, we believe that granulin represents a specific class of highly expressed, complex peptide that the virus encodes by sacrificing the constraints it maintains on other genes.
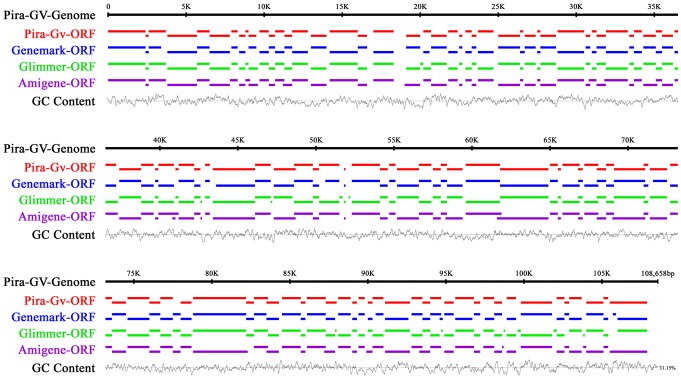
Figure 8. Predictive map of the putative coding regions of PiraGV-K genome.
The putative coding regions were predicted using GeneMark (Georgia Institute of Technology, Atlanta, Georgia), Glimmer-Microbial gene-finding system (University of Maryland) and AMIgene-an integrated computer environment for sequence annotation and analysis (Institut Pasteur, France) ORFs finding softwares.
The primary criteria used to identify potential ORFs on the PiraGV-K genome were a minimum of 50 aa in length, having minimal overlap with larger ORFs, and sharing significant sequence identity with previously characterized ORFs of betabaculoviruses (Table 1). Also, by convention, the first nucleotide of the methionine start codon of granulin was defined as the first nucleotide of the genome, and the sequence was numbered in the direction of transcription of the gene. As in the case of other baculovirus genomes, minimal overlaps were observed in the PiraGV-K genome sequence with 65 ORFs in the granulin-sense orientation and, 54 in the opposite orientation, clustering together according to expression or function. Homologous repeat regions (hrs), functioning as enhancers of transcription and origins of replication, were also found interspersed in the genome. These repeated sequences have been reported to be more variable in betabaculoviruses than in alphabaculoviruses, where they consist of repeated palindromes. The CypoGV genome includes 13 hrs, as do the XecnGV and HearGV genomes. The AdorGV genome includes nine repeated regions that are unlike typical hrs [19]. Six repeat regions, including one unique hrs, have also been identified in the EppoMNPV genome [47]. In the completely sequenced genome of SpltNPV, 17 hrs were identified [48]. In the AcMNPV, hrs consist of repeated units of about 70 bp with an imperfect 30 bp palindrome near their center, binding to the transcriptional activator ie1 (Ac147) [49]. Also, cAMP and 12-O-tetradecanoylphorbol 13-acetate (TPA) response elements (CRE and TRE)-like sequences, located between hrs palindromes have been found to be evolutionarily conserved in alphabaculoviruses, but were not found in betabaculoviruses.
Table 1. Analysis and annotation of PiraGV-K ORFs.
| QueryID | SubjectID | Annotation | Pid | Psi | Eval | Db | Best Hit Annotation | Source |
| GVORF001 | AAR06236.1 | granulin | 100 | 100 | 0 | gb | granulin | P. rapae granulovirus-Chinese strain |
| GVORF002 | YP_654423.1 | hypothetical protein COGV_gp002 | 61.9 | 73 | 6E-20 | ref | hypothetical protein COGV_gp002 | C. occidentalis granulovirus |
| GVORF003 | YP_654424.1 | pk-1 | 67 | 83.3 | 1E-141 | ref | pk-1 | C. occidentalis granulovirus |
| GVORF004 | NP_148788.1 | ORF4 similar to Cydia pomonella GV ORF4 | 65.4 | 81.4 | 7E-92 | ref | ORF4 similar to XcGV ORF7 | C. pomonella granulovirus |
| GVORF005 | YP_654427.1 | hypothetical protein COGV_gp006 | 60.3 | 74.4 | 1E-25 | ref | hypothetical protein COGV_gp006 | C. occidentalis granulovirus |
| GVORF006 | YP_654428.1 | ie-1 | 52.3 | 72.6 | 1E-144 | ref | ie-1 | C. occidentalis granulovirus |
| GVORF007 | YP_654429.1 | hypothetical protein COGV_gp008 | 58.1 | 75.3 | 1E-77 | ref | hypothetical protein COGV_gp008 | C. occidentalis granulovirus |
| GVORF008 | NP_148793.1 | ORF8 similar to Cydia pomonella GV ORF9 | 69.3 | 88.1 | 1E-55 | ref | ORF9 similar to AcMNPV ORF145 | C. pomonella granulovirus |
| GVORF009 | unknown | No hit | No hit | |||||
| GVORF010 | NP_148794.1 | chitinase | 65.3 | 78.2 | 0 | ref | ORF10 chitinase | C. pomonella granulovirus |
| GVORF011 | NP_148795.1 | cathepsin | 69.2 | 82.8 | 1E-180 | ref | ORF11 cathepsin | C. pomonella granulovirus |
| GVORF012 | NP_148796.1 | unknown | 60.4 | 77.4 | 1E-18 | ref | ORF12 | C. pomonella granulovirus |
| GVORF013 | NP_148797.1 | gp37 | 59.8 | 75.2 | 1E-111 | ref | ORF13 gp37 | C. pomonella granulovirus |
| GVORF014 | YP_001256961.1 | odv-e18 | 62.2 | 73 | 2E-21 | ref | odv-e18 | S. litura granulovirus |
| GVORF015 | YP_654434.1 | p49 | 63.8 | 80 | 0 | ref | p49 | C. occidentalis granulovirus |
| GVORF016 | YP_654435.1 | odv-e56 | 72.1 | 85.8 | 0 | ref | odv-e56 | C. occidentalis granulovirus |
| GVORF017 | NP_068221.1 | ORF17 similar to PxORF2 peptide | 37 | 51 | 6E-13 | ref | PxORF2 peptide | P. xylostella granulovirus |
| GVORF018 | NP_872468.1 | ORF18 similar to Adoxophyes orana granulovirus ORF14 | 31.8 | 48.5 | 7E-12 | ref | ORF_14 | A. orana granulovirus |
| GVORF019 | NP_148803.1 | ORF19 similar to Cydia pomonella GV ORF19 | 48.7 | 65.4 | 1E-18 | ref | ORF19 similar to AcMNPV ORF29 | C. pomonella granulovirus |
| GVORF020 | NP_148804.1 | pep1 | 51.7 | 63.7 | 2E-74 | ref | ORF20 similar to XcGV ORF17 | C. pomonella granulovirus |
| GVORF021 | NP_148806.1 | calyx/polyhedral envelope protein | 67.9 | 78.3 | 1E-170 | ref | ORF22 similar to XcGV ORF19 | C. pomonella granulovirus |
| GVORF022 | NP_148807.1 | pep2 | 66.7 | 76.4 | 3E-60 | ref | ORF23 similar to XcGV ORF18 | C. pomonella granulovirus |
| GVORF023 | NP_148808.1 | pe-38 | 28.9 | 47.8 | 1E-14 | ref | ORF24 PE-38 | C. pomonella granulovirus |
| GVORF024 | YP_654443.1 | hypothetical protein COGV_gp022 | 29.2 | 44 | 5E-32 | ref | hypothetical protein COGV_gp022 | C. occidentalis granulovirus |
| GVORF025 | NP_148814.1 | ORF25 similar to Cydia pomonella GV ORF30 | 41.5 | 63.6 | 7E-41 | ref | ORF30 similar to XcGV ORF26 | C. pomonella granulovirus |
| GVORF026 | NP_148815.1 | ORF26 similar to Cydia pomonella GV ORF31 | 64.1 | 80.4 | 0 | ref | ORF31 similar to AcMNPV ORF23 | C. pomonella granulovirus |
| GVORF027 | ZP_01774245.1 | ORF27 similar to Adoxophyes orana GV ORF24 | 33 | 40.9 | 1E-20 | ref | conserved hypothetical protein | G. bemidjiensis Bem |
| GVORF028 | NP_148817.1 | ORF28 similar to Cydia pomonella GV ORF33 | 37.5 | 55.1 | 1E-58 | ref | ORF33 similar to XcGV ORF29 | C. pomonella granulovirus |
| GVORF029 | NP_891880.1 | unknown | 63 | 77.2 | 8E-83 | ref | hypothetical protein ClgVgp033 | C. leucotreta granulovirus |
| GVORF030 | NP_148819.1 | ORF30 similar to Cydia pomonella GV ORF35 | 58.5 | 76.1 | 1E-83 | ref | ORF35 similar to AcMNPV ORF115 | C. pomonella granulovirus |
| GVORF031 | NP_148823.1 | ORF31 similar to Cydia pomonella GV ORF39 | 72.5 | 90.2 | 6E-55 | ref | ORF39 similar to XcGV ORF34 | C. pomonella granulovirus |
| GVORF032 | YP_003429356.1 | phosphohydrolase | 99 | 100 | 1E-141 | ref | phosphohydrolase | P. rapae granulovirus-Chinese strain |
| GVORF033 | YP_654450.1 | lef-2 | 61.3 | 78 | 3E-72 | ref | lef-2 | C. occidentalis granulovirus |
| GVORF034 | NP_148826.1 | hypothetical protein COGV_gp030 | 48.8 | 65.9 | 4E-20 | ref | ORF42 similar to XcGV ORF36 | C. pomonella granulovirus |
| GVORF035 | YP_654452.1 | hypothetical protein COGV_gp031 | 45.9 | 67.6 | 3E-29 | ref | hypothetical protein COGV_gp031 | C. occidentalis granulovirus |
| GVORF036 | YP_654453.1 | hypothetical protein COGV_gp032 | 55 | 73.8 | 3E-40 | ref | hypothetical protein COGV_gp032 | C. occidentalis granulovirus |
| GVORF037 | NP_891890.1 | metalloproteinase | 46.6 | 64.9 | 1E-137 | ref | metalloproteinase | C. leucotreta granulovirus |
| GVORF038 | NP_148831.1 | p13 | 59.7 | 75.4 | 1E-108 | ref | ORF47 p13 | C. pomonella granulovirus |
| GVORF039 | NP_148821.1 | odv-e66 | 31.2 | 50.8 | 1E-59 | ref | ORF37 odv-e66 | C. pomonella granulovirus |
| GVORF040 | YP_654456.1 | pif-2 | 71.1 | 83.5 | 0 | ref | pif-2 | C. occidentalis granulovirus |
| QueryID | SubjectID | Annotation | Pid | Psi | Eval | Db | Best Hit Annotation | Source |
| GVORF041 | NP_663210.1 | hypothetical protein PogVgp045 | 46.9 | 73.4 | 8E-17 | ref | hypothetical protein PogVgp045 | P. operculella granulovirus |
| GVORF042 | NP_148834.1 | ORF42 similar to Cydia pomonella GV ORF050 | 41.4 | 59.4 | 2E-51 | ref | ORF50 similar to XcGV ORF47 | C. pomonella granulovirus |
| GVORF043 | NP_891897.1 | ORF43 similar to Cryptophlebia leucotreta GV ORF50 | 78 | 88 | 1E-119 | ref | hypothetical protein ClgVgp050 | C. leucotreta granulovirus |
| GVORF044 | NP_047452.1 | odv-e66 | 70.1 | 83.2 | 0 | ref | odv-e66 | B. mori NPV |
| GVORF045 | YP_654460.1 | UBQ | 89.2 | 93.5 | 6E-59 | ref | UBQ | C. occidentalis granulovirus |
| GVORF046 | YP_654461.1 | hypothetical protein COGV_gp040 | 65.8 | 80.8 | 1E-176 | ref | hypothetical protein COGV_gp040 | C. occidentalis granulovirus |
| GVORF047 | NP_148840.1 | ORF47 similar to Cydia pomonella GV ORF56 | 53.1 | 76.6 | 5E-21 | ref | ORF56 similar to XcGV ORF54 | C. pomonella granulovirus |
| GVORF048 | YP_654463.1 | 39K | 48.7 | 67 | 1E-75 | ref | 39K | C. occidentalis granulovirus |
| GVORF049 | YP_654464.1 | lef-11 | 65.2 | 79.3 | 5E-40 | ref | lef-11 | C. occidentalis granulovirus |
| GVORF050 | NP_663219.1 | superoxide dismutase | 67.3 | 82.7 | 3E-78 | ref | superoxide dismutase | P. operculella granulovirus |
| GVORF051 | NP_148844.1 | ORF51 similar to p74 (Baculoviridae p74 conserved region | 61.7 | 76.2 | 0 | ref | ORF60 p74 | C. pomonella granulovirus |
| GVORF052 | YP_610994.1 | p22.2 | 34.3 | 49.7 | 8E-29 | ref | p22.2 | A. pernyi nucleopolyhedrovirus |
| GVORF053 | YP_743425.1 | dehydrogenase catalytic domain-containing protein | 71.7 | 77.4 | 3E-12 | ref | dehydrogenase catalytic protein | A. ehrlichei MLHE-1 |
| GVORF054 | YP_654469.1 | hypothetical protein COGV_gp048 | 61.6 | 77.8 | 4E-88 | ref | hypothetical protein COGV_gp048 | C. occidentalis granulovirus |
| GVORF055 | NP_891907.1 | ORF55 similar to Cryptophlebia leucotreta GV ORF60 | 71.4 | 88.9 | 7E-26 | ref | hypothetical protein ClgVgp060 | C. leucotreta granulovirus |
| GVORF056 | YP_654471.1 | p47 | 71.1 | 83.2 | 0 | ref | p47 | C. occidentalis granulovirus |
| GVORF057 | NP_891909.1 | Nudix_Hydrolase | 77.8 | 90.5 | 1E-129 | ref | hypothetical protein ClgVgp062 | C. leucotreta granulovirus |
| GVORF058 | YP_654473.1 | p24 capsid protein | 60 | 79.4 | 5E-78 | ref | p24 capsid protein | C. occidentalis granulovirus |
| GVORF059 | YP_654475.1 | 38.7KD protein | 44.5 | 63 | 3E-35 | ref | 38.7KD protein | C. occidentalis granulovirus |
| GVORF060 | YP_654476.1 | lef-1 | 68.7 | 82.4 | 1E-126 | ref | lef-1 | C. occidentalis granulovirus |
| GVORF061 | YP_654477.1 | pif-1 | 63.7 | 79.1 | 0 | ref | pif-1 | C. occidentalis granulovirus |
| GVORF062 | YP_654478.1 | fgf-1 | 58.9 | 77.2 | 6E-98 | ref | fgf-1 | C. occidentalis granulovirus |
| GVORF063 | YP_654479.1 | COGV_gp058 contains chitin binding domain | 42 | 63 | 1E-18 | ref | hypothetical protein COGV_gp058 | C. occidentalis granulovirus |
| GVORF064 | NP_872520.1 | ORF64 similar to Adoxophyes orana GV ORF66 | 35.3 | 52.7 | 6E-28 | ref | ORF_66 | A. orana granulovirus |
| GVORF065 | YP_654481.1 | lef-6 | 52.5 | 72.5 | 4E-27 | ref | lef-6 | C. occidentalis granulovirus |
| GVORF066 | YP_654482.1 | DBP | 66.8 | 82.8 | 1E-136 | ref | DBP | C. occidentalis granulovirus |
| GVORF067 | YP_654484.1 | hypothetical protein COGV_gp063 | 35 | 52.9 | 2E-34 | ref | hypothetical protein COGV_gp063 | C. occidentalis granulovirus |
| GVORF068 | NP_148867.1 | ORF68 similar to Cydia pomonella GV ORF83 | 80.9 | 90.7 | 0 | ref | ORF83 similar to AcMNPV ORF103 | C. pomonella granulovirus |
| GVORF069 | NP_148868.1 | ORF69 similar to Cydia pomonella GV ORF84 | 65.7 | 79.4 | 5E-43 | ref | ORF84 similar to AcMNPV ORF102 | C. pomonella granulovirus |
| GVORF070 | YP_654487.1 | hypothetical protein COGV_gp066 | 70.6 | 85.6 | 0 | ref | hypothetical protein COGV_gp066 | C. occidentalis granulovirus |
| GVORF071 | YP_654488.1 | p6.9 | 81 | 87.9 | 2E-17 | ref | p6.9 | C. occidentalis granulovirus |
| GVORF072 | NP_148871.1 | lef-5 | 71.8 | 83.7 | 1E-129 | ref | ORF87 lef-5 | C. pomonella granulovirus |
| GVORF073 | YP_654490.1 | hypothetical protein COGV_gp069 | 70.8 | 84.2 | 1E-161 | ref | hypothetical protein COGV_gp069 | C. occidentalis granulovirus |
| GVORF074 | NP_148873.1 | 19KD | 67.5 | 81.9 | 4E-72 | ref | ORF89 similar to AcMNPV ORF96 | C. pomonella granulovirus |
| GVORF075 | YP_654492.1 | helicase-1 | 64.5 | 80 | 0 | ref | helicase-1 | C. occidentalis granulovirus |
| GVORF076 | NP_148875.1 | odv-e25 | 81.3 | 93.9 | 1E-131 | ref | ORF91 odv-e25 | C. pomonella granulovirus |
| GVORF077 | NP_148876.1 | ORF77 similar to Cydia pomonella GV ORF92 | 60.4 | 79.9 | 2E-66 | ref | ORF92 similar to AcMNPV ORF93 | C. pomonella granulovirus |
| GVORF078 | NP_148877.1 | ORF78 similar to Cydia pomonella GV ORF93 | 68.5 | 88 | 1E-142 | ref | ORF93 similar to AcMNPV ORF92 | C. pomonella granulovirus |
| GVORF079 | NP_891932.1 | iap | 37 | 60.2 | 1E-48 | ref | iap | C. leucotreta granulovirus |
| GVORF080 | NP_148879.1 | lef-4 | 56.9 | 73.5 | 0 | ref | ORF95 lef-4 | C. pomonella granulovirus |
| QueryID | SubjectID | Annotation | Pid | Psi | Eval | Db | Best Hit Annotation | Source |
| GVORF081 | YP_654497.1 | vp39 capsid | 73.4 | 84.3 | 1E-158 | ref | vp39 capsid | C. occidentalis granulovirus |
| GVORF082 | YP_654498.1 | odv-e27 | 71.4 | 86.4 | 1E-148 | ref | odv-e27 | C. occidentalis granulovirus |
| GVORF083 | YP_654499.1 | hypothetical protein COGV_gp078 | 51.9 | 71.1 | 1E-129 | ref | hypothetical protein COGV_gp078 | C. occidentalis granulovirus |
| GVORF084 | NP_891938.1 | ORF084 similar to Cryptophlebia leucotreta ORF91 | 59 | 73.8 | 2E-20 | ref | hypothetical protein ClgVgp091 | C. leucotreta granulovirus |
| GVORF085 | NP_891939.1 | vp91 capsid | 46.5 | 67.2 | 0 | ref | vp91 capsid | C. leucotreta granulovirus |
| GVORF086 | NP_891940.1 | tlp20 | 39 | 54.2 | 9E-25 | ref | tlp20 | C. leucotreta granulovirus |
| GVORF087 | NP_891941.1 | hypothetical protein ClgVgp094 | 79.2 | 94 | 1E-112 | ref | hypothetical protein ClgVgp094 | C. leucotreta granulovirus |
| GVORF088 | NP_148888.1 | Structural glycoprotein p40/gp41 conserved | 73.8 | 88.1 | 1E-164 | ref | ORF104 GP41 | C. pomonella granulovirus |
| GVORF089 | YP_654506.1 | hypothetical protein COGV_gp085 | 51.6 | 64.2 | 6E-23 | ref | hypothetical protein COGV_gp085 | C. occidentalis granulovirus |
| GVORF090 | NP_891944.1 | vlf1 (very late expression factor 1) | 73.8 | 87.4 | 0 | ref | vlf-1 | C. leucotreta granulovirus |
| GVORF091 | NP_148891.1 | ORF91 similar to Cydia pomonella GV ORF107 | 73.8 | 89.3 | 2E-43 | ref | ORF107 similar to AcMNPV ORF76 | C. pomonella granulovirus |
| GVORF092 | ABC67291.1 | unknown | 100 | 100 | 1e-104, | gb | unknown | P. rapae granulovirus-Chinese strain |
| GVORF093 | ACZ63579.1 | DNA polymerase | 99.7 | 99.9 | 0.0, | gb | DNA polymerase | P. rapae granulovirus-Chinese strain |
| GVORF094 | ACZ63580.1 | desmoplakin | 100 | 100 | 0.0, | gb | desmoplakin | P. rapae granulovirus-Chinese strain |
| GVORF095 | ACZ63581.1 | lef-3 | 100 | 100 | 0.0, | gb | lef-3 | P. rapae granulovirus-Chinese strain |
| GVORF096 | ABC67295.1 | unknown | 100 | 100 | 6e-89, | gb | unknown | P. rapae granulovirus-Chinese strain |
| GVORF097 | ABC67296.1 | unknown | 99.4 | 100 | 1e-123, | gb | unknown | P. rapae granulovirus-Chinese strain |
| GVORF098 | ABC67297.1 | iap-5 | 99.6 | 100 | 0.0, | gb | iap-5 | P. rapae granulovirus |
| GVORF099 | ABC67298.1 | lef-9 | 100 | 100 | 0.0, | gb | lef-9 | P. rapae granulovirus |
| GVORF100 | ABC67299.1 | fp | 100 | 100 | 1e-110, | gb | fp | P. rapae granulovirus |
| GVORF101 | ABC67300.1 | unknown | 100 | 100 | 1e-126, | gb | unknown | P. rapae granulovirus |
| GVORF102 | NP_148904.1 | DNA ligase | 66.1 | 80.5 | 0 | ref | ORF120 DNA LIGASE | C. pomonella granulovirus |
| GVORF103 | YP_004376313.1 | hypothetical protein ClanGV_gp105 | 55.3 | 68.4 | 0.000001 | ref | hypothetical protein ClanGV_gp105 | C. anachoreta granulovirus |
| GVORF104 | YP_654522.1 | hypothetical protein COGV_gp101 | 60.9 | 79.7 | 3E-22 | ref | hypothetical protein COGV_gp101 | C. occidentalis granulovirus |
| GVORF105 | NP_891960.1 | fgf-2 | 44.7 | 66.6 | 1E-106 | ref | fgf | C. leucotreta granulovirus |
| GVORF106 | NP_663280.1 | ORF106 similar to Phthorimaea operculella GV ORF115 | 63 | 85.9 | 2E-42 | ref | hypothetical protein PogVgp115 | P. operculella granulovirus |
| GVORF107 | YP_654525.1 | ALK-EXO | 60.8 | 77.4 | 1E-177 | ref | ALK-EXO | C. occidentalis granulovirus |
| GVORF108 | NP_148910.1 | HELICASE-2 | 62.5 | 78.4 | 0 | ref | ORF126 HELICASE-2 | C. pomonella granulovirus |
| GVORF109 | YP_654527.1 | hypothetical protein COGV_gp106 | 38.2 | 59.2 | 5E-41 | ref | hypothetical protein COGV_gp106 | C. occidentalis granulovirus |
| GVORF110 | NP_148915.1 | lef-8 | 74.3 | 85.9 | 0 | ref | ORF131 lef-8 | C. pomonella granulovirus |
| GVORF111 | AAT77801.1 | unknown | 42.5 | 64.2 | 2E-16 | gb | unknown | C. anachoreta granulovirus |
| GVORF112 | NP_148917.1 | ORF112 similar to Cydia pomonella GV ORF133 | 52.4 | 76.2 | 4E-13 | ref | ORF133 similar to XcGV ORF170 | C. pomonella granulovirus |
| GVORF113 | NP_148918.1 | ORF113 similar to Cydia pomonella GV ORF134 | 71.4 | 86.5 | 4E-73 | ref | ORF134 similar to AcMNPV ORF53 | C. pomonella granulovirus |
| GVORF114 | YP_654531.1 | hypothetical protein COGV_gp110 | 37.1 | 58.5 | 8E-56 | ref | hypothetical protein COGV_gp110 | C. occidentalis granulovirus |
| GVORF115 | YP_654532.1 | hypothetical protein COGV_gp111 | 57.4 | 72.2 | 3E-14 | ref | hypothetical protein COGV_gp111 | C. occidentalis granulovirus |
| GVORF116 | AAT67151.1 | vp1054 | 68.9 | 83.7 | 1E-140 | gb | unknown | C. anachoreta granulovirus |
| GVORF117 | unknown | No hit | No hit | |||||
| GVORF118 | NP_891974.1 | fgf-3 | 57.5 | 73.3 | 1E-103 | ref | hypothetical protein ClgVgp127 | C. leucotreta granulovirus |
| GVORF119 | YP_654536.1 | Ecdysteroid UDP-glucosyltransferase | 59 | 74.9 | 0 | ref | egt | C. occidentalis granulovirus |
| GVORF120 | NP_148927.1 | ME53-like protein | 57.9 | 78.1 | 1E-136 | ref | ORF143 ME53 | C. pomonella granulovirus |
PiraGV-K ORFs are represented by the Query ID and the source ORFs are represented by Subject ID. Proteins from viral complete genomes were clustered by sequence similarity based on BLASTP pairwise alignments using the viral clusters of orthologous groups (VOG) approach. Pid - percent identity; Psi - percent similarity; Eval - Best hit E-value; Db - Databases.
Genomic sequence identity of PiraGV-K was studied against other known betabaculoviruses genomes, with a maximum identity of 99% with PiraGV-C (Table 2). The 1% difference was thought to be related to the presence of extra nucleotides in the intronic sequences of the PiraGV-K genome and not corresponded to any known ORF. The identity with other genomes was in the order of 42–58%, with greater identity with ChocGV (58.5%), CrleGV (55.78%) and CypoGV (55.6%) genome sequences. Of a total of 120 ORFs, only ORFs 9, 32, and 117 were considered unique to the PiraGV genome sequences of the Korean and Chinese strains. This represents 1.7% of the whole genome sequence. Also, 78 ORFs found in all betabaculoviruses sequences studied, have been called “core GV genes”. Based on gene function, PiraGV-K ORFs have been grouped into four functional categories (Table-3): transcription (10 genes), replication (11 genes), structural (25 genes), and auxiliary (15 genes), with 59 unrepresented in the annotation. The most conserved among the core set of genes was granulin, with 100% identity with PiraGV-C. We compared the identified PiraGV-C ODV associated proteins [11], with the structural proteins found in PiraGV-K and found that the ORFs complemented and matched each other. PiraGV-K-ORF 1 (granulin), ORF-14 (odv-e18), ORF-15 (p49), ORF-16 (odv-e56), ORF-17 (p10), ORF-39 (odv-e66), ORF-44 (odv-e66a), ORF-45 (ubiquitin), ORF-51 (p74), ORF-61 (pif-1), ORF-71 (p6.9), ORF-75 (helicase-1), ORF-81 (vp39), ORF-82 (odv-e27), ORF-85 (vp91 capsid), ORF-88 (gp-41), ORF-90 (vlf-1), ORF-93 (DNA pol), ORF-95 (lef-3), ORF-118 (fgf-3) and ORF-120 (ME-53) were also among the reported proteins in PiraGV-C. Other proteins common to both the PiraGV genomes were found to be hypothetical or unknown proteins.
Table 2. The granuloviruses genome used for the characterization of PiraGV-K.
| GV | Refseq | GenBank | Length (nt) | GC (%) | Genes | Identity (%) | Completed | Nation | Ref. |
| AdorGV | NC_005038 | AF547984 | 99,657 | 34 | 119 | 49 | 7/15/2003 | UK | 27 |
| AgseGV | NC_005839 | AY522332 | 131,680 | 37 | 132 | 46 | 4/9/2004 | China | NCBI |
| ChocGV | NC_008168 | DQ333351 | 104,710 | 32 | 116 | 58 | 6/19/2006 | Canada | 28 |
| CrleGV | NC_005068 | AY229987 | 110,907 | 32 | 129 | 56 | 8/13/2003 | Germany | 13 |
| CypoGV | NC_002816 | U53466 | 123,500 | 45 | 143 | 56 | 4/2/2001 | UK | 29 |
| HearGV | NC_010240 | EU255577 | 169,794 | 40 | 179 | 43 | 1/9/2008 | USA | 47 |
| PhopGV | NC_004062 | AF499596 | 119,217 | 35 | 130 | 52 | 7/1/2002 | France | NCBI |
| PlxyGV | NC_002593 | AF270937 | 100,999 | 40 | 121 | 44 | 10/29/2000 | Japan | 10 |
| PsunGV | NC_013772 | EU678671 | 176,677 | 39 | 183 | 45 | 1/30/2010 | China | NCBI |
| SpliGV | NC_009503 | DQ288858 | 124,121 | 38 | 136 | 42 | 5/30/2010 | Korea | 31 |
| XecnGV | NC_002331 | AF162221 | 178,733 | 40 | 181 | 43 | 6/7/2000 | USA | 11 |
| PiraGV-C | NC_013797 | GQ884143 | 108,592 | 33 | 120 | 99 | 2/11/2010 | China | 20 |
Table 3. PiraGV-K genes grouped according to function.
| Functional category | PiraGV-K (ORF) |
| Transcription | pe-38 (23), 39K (48), lef-11 (49), p47 (56), lef-6 (65), lef-5 (72), lef-4 (80), vlf-1 (90), lef-9 (99), lef-8 (110) |
| Replication | ie-1 (6), lef-2 (33), lef-1 (60), dbp (66), 38.7 k (59), helicase-1 (75), DNA polymerase (93), lef-3 (95), DNA ligase (102) helicase-2 (108), me53 (120) |
| Structural | granulin (1), pk-1 (3), odv-e18 (14), odv-e56 (16), p10 (17), pep-1 (20), calyx/pep (21), pep-2 (22), p13 (38), odv-e66 (39), pif-2 (40), odv-e66a (44), p74 (51), vp24 (58), pif-1 (61), p6.9 (71), odv-e25 (76), vp39 (81), odv-e27 (82), vp91 (85), tlp20 (86), gp41 (88), desmoplakin (94), fp (100), vp1054 (116) |
| Auxilliary | chitinase (10), cathepsin (11), gp37 (13), p49 (15), phosphohydrolase (32), metalloproteinase (37), ubiquitin (45), superoxide dismutase (50), Nudix hydrolase (57), fgf-1 (62), iap (79), fgf-2 (105), alk-exo (107), fgf-3 (118), ecdysteroid UDP-glucosyl transferase (119) |
| Unknown | ORF2, ORF4, ORF5, ORF7, ORF8, ORF9, ORF12, ORF18, ORF19, ORF24, ORF25, ORF26, ORF27, ORF28, ORF29, ORF30, ORF31, ORF34, ORF35, ORF36, ORF41, ORF42, ORF43, ORF46, ORF47, ORF51, ORF52, ORF53, ORF54, ORF55, ORF63, ORF64, ORF67, ORF68, ORF69, ORF70, ORF73, ORF74, ORF77, ORF78, ORF83, ORF84, ORF87, ORF89, ORF91, ORF92, ORF96, ORF97, ORF101, ORF103, ORF104, ORF106, ORF109, ORF111, ORF112, ORF113, ORF114, ORF115, ORF117 |
PiraGV-K ORF 98 encoded an inhibitor of apoptosis (iap-5) that seems to be betabaculovirus specific [21]. Also, PiraGV-K ORF 37 (homologous to Cypo46, Xecn40, and Plxy35) is likely a member of the stromelysin family within the matrix metalloproteinase (MMP) superfamily. It has been observed that this peptide is retained within infected cells until death, and subsequently is released into the body of the insect, causing proteolysis of tissues [4], [50]. The most conserved baculovirus gene is polyhedrin/granulin, the major component of occlusion bodies. Another conserved PiraGV-K structural gene was odv-e25 (PiraGV-K, ORF 76), showing 80% amino acid identity to betabaculovirus homologs. In contrast, p24 capsid (PiraGV-K-58, ORF 58), which encodes a protein associated with both ODV and BV [51], was found to be poorly conserved (60% average amino acid identity to other betabaculoviruses). The p80/p87-capsid gene was absent from the PiraGV-K genome, as with other betabaculovirus genomes. The putative p10 (PiraGV-K, ORF17) gene showed similarities to three XecnGV ORFs (Xecn ORF 5, Xecn ORF 19, and Xecn ORF 83). Homologs of these three ORFs were found in PlxyGV (Plxy ORF 2, Plxy ORF 21, and Plxy ORF 50) and they were thus suggested to be p10 homologs [4]. p10 is implicated in occlusion body morphogenesis and disintegration of the nuclear body matrix, resulting in dissemination of OBs [52]. In NPV-infected cells, p10 forms fibrillar structures in the nucleus and cytoplasm. PiraGV-K ORF 17 showed a significantly low identity 14%, with AcMNPV p10, and was smaller than its counterpart (104 vs 336 amino acids). A high sequence identity of 48% was noted with ClanGV p10, having 101 amino acid residues in relation to other betabaculoviruses.
The PiraGV-K genome did not encode the glycoprotein gp64 that constitutes a major envelope fusion protein in AcMNPV, BmNPV, OpMNPV, and EppoMNPV [53], [54]. This protein thus appears to be unique to group I NPVs [55], [56]. Also, 19 lef genes have been found in AcMNPV genomes, and have been implicated in DNA replication and transcription [57]. Early baculovirus genes are transcribed by the host cell RNA polymerase II, but these are often transactivated by genes such as ie-0, ie-1, ie-2, and pe38 [58]. Of these early baculovirus genes, the PiraGV-K genome contained only ie-1 and it was found to be poorly conserved in comparison with other betabaculovirus genomes, except PiraGV-C. These genes have previously been reported to be poorly conserved among baculoviruses. The CypoGV and PhopGV genomes have been reported to have a pe38, consistent with PiraGV-K genome [21].
Six genes have been described as essential for baculovirus DNA replication: lef-1, lef-2, lef-3, dnapol, helicase and ie-1 [59]. Homologs for all these necessary genes were found in the whole-genome of PiraGV-K with moderately conserved sequences. A PiraGV-K genome-wide scan suggested the absence of a lef-7 homolog. Earlier reports suggested that lef-7 was a group I NPV-specific gene, and stimulated transient DNA replication in AcMNPV and BmNPV [60], [61]. The PiraGV-K ORFs also encode a DNA ligase (PiraGV-K ORF 102) and a helicase-2 (PiraGV-K ORF 108), in common with LdMNPV and other betabaculovirus genomes. The LdMNPV DNA ligase displays catalytic properties of a type-III DNA ligase [62]. Because the homologs of helicase-2 and DNA ligase are involved in DNA repair and recombination [63], the PiraGV-K genes likely have similar functions. The PiraGV-K genome lacks large (rr1) and small (rr2) subunits of ribonucleotide reductase and deoxyuridyltriphosphate (dUTPase) genes, that may account for the loss of enzymatic functions during facilitation of virus replication in non-dividing cells, where dNTP pathways are inactive. The lack of these genes has also been noted in alphabaculoviruses, such as AcMNPV, BmNPV, HaSNPV, HzSNPV, and EppoMNPV and other betabaculoviruses, such as PlxyGV and XecnGV [4], [5], [22], [51], [63], [64]. Late transcription genes, including lef 4–6, 8–11, 39K, p47, and vlf-1 [65] have been found among the PiraGV-K ORFs, except a lef-10 homolog. The most conserved PiraGV-K lef homolog was lef-8, while lef-6 was the most poorly conserved. It has been understood that the GV lef-6 genes are smaller than the NPV lef-6 genes (86–102 amino acids vs 138–187 amino acids) and were reported in the XecnGV genome [5].
Chitinase [66] and cathepsin were present as auxiliary genes in the PiraGV-K genome. These genes have been identified in almost all the baculoviruses completely sequenced to date, except PlxyGV [4] and AdorGV [19]. The protein products encoded by these genes provide selective advantages in the breakdown of insect tissues at the end of infection and the release of OBs to the environment, which then spread horizontally [67]. The lack of the same in the cases of the PlxyGV and AdorGV genomes may account for the infected larvae not lysing at the end of infection; this may lead to the spread of viral infection by discharging large amounts of virus from their posterior ends. PiraGV-K ORF 50 corresponded to superoxide dismutase (sod), a well-conserved gene in baculoviruses. Among the betabaculoviruses, it was not reported in the SpliGV genome, although it is known in other betabaculoviruses. Although, SOD functions as an endogenous antioxidant, its proper function in baculoviruses remains unknown. Gene deletion studies conducted in AcMNPV did not show any deleterious effect [68], although it may be predicted that SOD may protect OBs from superoxide radicals generated by exposure to sunlight in the environment.
PiraGV-K ORF 45 corresponded to a ubiquitin protein, which have been identified in all baculoviruses sequenced to date, although it was found fused to gp37 as a single ORF in SpltMNPV [48]. Apart from polyhedrin and granulin [69], it is also one of the most highly conserved genes in the baculovirus genome, with 73% average amino acid identity to betabaculovirus homologs. Interestingly, the homolog of viral ubiquitin has not been reported in AcMNPV-ODV or HearNPV-ODV, but is known in AcMNPV-BV [70]. Per os infectivity factors (pif), another highly conserved gene, involved in oral infectivity of baculovirus ODV, has been characterized from almost all baculovirus genomes sequenced so far. We identified ORF 61, corresponding to pif-1, and ORF 16, corresponding to ODV-E56, also known as pif-5 [71] in the PiraGV-K genome. Although pif-1 and p74 (ORF 51 in the PiraGV-K genome) have been proposed to form structural components of the ODV envelope and may regulate infectivity of OBs, pif-5 is not an essential protein for binding and fusion of ODV or virus replication [72], [73]. Additionally, the PiraGV-K genome was found to contain three putative fibroblast growth factors (fgf), represented by ORFs 62, 105, and 118. These fgfs contained the fgf superfamily domains, as determined by a conserved family domain search with the BLAST program. No enhancin homolog was found in PiraGV-K genome and is consistent with the absence of the same in the AdorGV, CypoGV and PlxyGV genomes. In contrast to the above betabaculovirus genomes, four enhancin homologs were reported in XecnGV, two in LdMNPV, and one in MacoNPV. Enhancin functions in disrupting the insect peritrophic membrane, and facilitates the initiation of infection [74]. PiraGV-K ORF 13 corresponded to the gp37 homolog (spindling acting as enhancing factor) that was shown to be absent from the AdorGV, AgseGV, ChocGV, CrleGV, PhopGV, PlxyGV, and SpliGV genomes, although the ORF was reported in the CypoGV, HearGV, PsunGV, XecnGV, and PiraGV-C genomes.
Furthermore, PiraGV-K was found to lack a conotoxin-like (ctl) homolog, as reported in the BmNPV, SeMNPV, HaSNPV, AdorGV, CypoGV, and PlxyGV genomes, although a ctl homolog has been identified in the genome of XecnGV. The ORF contains a six-cysteine motif similar to that in chitin-binding proteins [75]. A gene encoding protein kinase 1 (pk-1; PiraGV-K ORF 3) was also identified in the whole-genome sequence of PiraGV-K; this may be involved in the regulation of the phosphorylation status of viral and host proteins during infection. Two members of the iap genes, corresponding to iap (PiraGV-K ORF 79) and iap5 (PiraGV-K ORF 98), were also identified in the PiraGV-K genome. Although the p35 with antiapoptotic activity has been identified previously in the AcMNPV, BmNPV, and SpltMNPV genomes, it is absent from betabaculovirus genomes. The iap homologs generally contain two baculovirus IAP repeats (BIP) [76], that are associated with binding to apoptosis-inducing proteins [77], and a C-terminal zinc finger-like (RING) Cys/His motif [78]. The iap-5 appears to be GV-specific, and all betabaculoviruses sequenced to date have iap-5. PiraGV-K ORF 94 is a homolog of Plxy ORF 94, named desmoplakin because it shows similarity to an internal region of a human desmoplakin, an essential constituent of intracellular junctions [4]. Baculovirus-repeated ORFs (bro) have not been seen in the PiraGV-K genome, although truncated versions have been observed in CpGV [21]. These repeats are more conspicuously present in many baculoviruses (1 and 16 copies), although their function is unclear, with the possibility of binding to DNA.
Two uncharacterized ORFs were also identified in the whole genome sequence of PiraGV-K and PiraGV-C, indicated as PiraGV-K ORF 9 and PiraGV-K ORF 117.
Conclusions
There has been a significant increase in the number of whole-genome sequencing projects using the shotgun method, but traditional mapped clone methods using BAC, cosmid, and fosmid libraries remain an important intermediate layer for hybrid sequencing strategies. With a view towards advancing the whole-genome sequencing strategies of infectious viruses, we adopted a method for the construction of a fosmid library of virus mixed with the infected host and further screening only the viral genomic library. The method overcomes the often-difficult need to culture and purify viruses by traditional methods of genome analysis and reduces the difficulties in obtaining starting material than would be necessary if starting with the purification of virus particles from inclusion bodies. The viral DNA is recovered in amounts sufficient for classical genome sequencing, without recourse to the use of automated high-throughput NGS technology. Thus, the analysis of the genome of PiraGV-K by the novel method of electrophoretic separation provides significant advances towards analysis of other infectious viruses.
Supporting Information
Analysis and homology search of PiraGV-K ORFs. The PiraGV-K ORFs have been analyzed for homology using representative granulovirus genomes such as Adoxophyes orana granulovirus (AdorGV), Agrotis segetum granulovirus (AgseGV), Choristoneura occidentalis granulovirus (ChocGV), Cryptophlebia leucotreta granulovirus (CrleGV), Cydia pomonella granulovirus (CypoGV), Helicoverpa armigera granulovirus (HearGV), Phthorimaea operculella granulovirus (PhopGV), Pieris rapae granulovirus-Chinese isolate (PiraGV-C), Plutella xylostella granulovirus (PlxyGV), Pseudaletia unipuncta granulovirus (PsunGV), Spodoptera litura granulovirus (SpliGV) and Xestia c-nigrum granulovirus (XecnGV). Pid and Psi refers to percent identity and percent similarity.
(DOCX)
Acknowledgments
This work was supported in part by Soonchunhyang University.
Funding Statement
This study was supported by Mid-career researcher program through NRF grant funded by the MEST (No. 2010-0027694). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Garcia-Maruniak A, Maruniak JE, Zanatto PMA, Doumbouya AE, Liu JC, et al. (2004) Sequence analysis of the genome of the Neodiprion sertifera Nucleopolyhedrovirus. J Virol 78: 7036–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jehle JA, Lange M, Wang H, Zhihong H, Wang Y, et al. (2006) Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 346: 180–193. [DOI] [PubMed] [Google Scholar]
- 3. Lauzon HA, Garcia-Maruniak A, Zanatto PM, Clemente JC, Herniou EA, et al. (2006) Genomic comparison of Neodiprion sertifera and Neodiprion lecontei nucleopolyhedroviruses and identification of potential hymenopteran baculovirus-specific open reading frames. J Gen Virol 87: 1477–1489. [DOI] [PubMed] [Google Scholar]
- 4. Hashimoto Y, Hayakawa T, Ueno Y, Fugita T, Sano Y, et al. (2000) Sequence analysis of the Plutella xylostella granulovirus genome. Virology 275: 358–372. [DOI] [PubMed] [Google Scholar]
- 5. Hayakawa T, Ko R, Okano K, Seong S, Goto C, et al. (1999) Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 262: 277–297. [DOI] [PubMed] [Google Scholar]
- 6. Hayakawa T, Rohrmann GF, Hashimoto Y (2000) Patterns of genome organization and content in lepidopteran baculoviruses. Virology 278: 1–12. [DOI] [PubMed] [Google Scholar]
- 7. Lange M, Jehle JA (2003) The genome of the Cryptophlebia leucotreta granulovirus. Virology 317: 220–236. [DOI] [PubMed] [Google Scholar]
- 8. Afonso CL, Tulman ER, Lu Z, Balinsky CA, Moser BA, et al. (2001) Genome sequence of a baculovirus pathogenic for Culex nigripalpus . J Virol 75: 11157–11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moser BA, Becnel JJ, White SE, Alfonso C, Kutish G, et al. (2001) Morphological and molecular evidence that Culex nigripalpus baculovirus is an unusual member of the family Baculoviridae. J Gen Virol 82: 283–297. [DOI] [PubMed] [Google Scholar]
- 10. Winstanley D, Crook NE (1993) Replication of Cydia pomonella granulosis virus in cell cultures. J Gen Virol 74: 1599–1609. [DOI] [PubMed] [Google Scholar]
- 11. Wang XF, Zhang BQ, Xu HJ, Cui YJ, Xu YP, et al. (2011) ODV-associated proteins of the Pieris rapae granulovirus. J Prot Res 10: 2817–2827. [DOI] [PubMed] [Google Scholar]
- 12. Zhang BQ, Cheng RL, Wang XF, Zhang CX (2012) The genome of Pieris rapae granulovirus (Genome announcement). J Virol 86: 9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yanisch-Perron CJ, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequencing of the M13mp18 and pUC9 vectors. Gene 33: 103–119. [DOI] [PubMed] [Google Scholar]
- 14. Ewing B, Green P (1998) Base-calling of automated sequencer traces using PHRED. II. Error probabilities. Genome Res 8: 186–194. [PubMed] [Google Scholar]
- 15. Ewing B, Hillier L, Wendl M, Green P (1998) Base-calling of automated sequencer traces using PHRED. I. Accuracy assessment. Genome Res 8: 175–185. [DOI] [PubMed] [Google Scholar]
- 16. Borodovsky M, McIninch JD (1993) GeneMark: Parallel gene recognition for both DNA strands. Comp Chem 17: 123–133. [Google Scholar]
- 17. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27: 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bocs S, Cruveiller S, Vallenet D, Nuel G, Medique C (2003) AMIGene: Annotation of microbial genes. Nucleic Acids Res 31: 3723–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wormleaton S, Kuzio J, Winstanley D (2003) The complete sequence of the Adoxophyes orana granulovirus genome. Virology 311: 350–365. [DOI] [PubMed] [Google Scholar]
- 20. Escasa SR, Lauzon HAM, Mathur AC, Krell PJ, Arif BM (2006) Sequence analysis of the Choristoneura occidentalis granulovirus genome. J Gen Virol 87: 1917–1933. [DOI] [PubMed] [Google Scholar]
- 21. Luque T, Finch R, Crook N, O’Reilly, Winstanley D (2001) The complete sequence of the Cydia pomonella granulovirus genome. J Gen Virol 82: 2531–2547. [DOI] [PubMed] [Google Scholar]
- 22. Chen XW, Ijkel WFJ, Tarchini R, Sun XL, Sandbrink H, et al. (2001) The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J Gen Virol 82: 241–257. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y, Choi JY, Roh JY, Liu Q, Tao XY, et al. (2011) Genomic sequence analysis of granulovirus isolated from the tobacco cutworm, Spodoptera litura . PloS ONE 6(11): e28163 Doi:10:1371/journal.pone.0028163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park TH, Park BS, Kim JA, Hong JK, Jin M, et al. (2011) Construction of random sheared fosmid library from Chinese cabbage and its use for Brassica rapa genome sequencing project. J Gent Genomics 38: 47–53. [DOI] [PubMed] [Google Scholar]
- 25. Huang SW, Lin YY, You EM, Liu TT, Shu HY, et al. (2011) Fosmid library end sequencing reveals a rarely known genome structure of marine shrimp Penaeus monodon . BMC Genomics 12: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ariyadasa R, Stein N (2012) Advances in BAC-based physical mapping and map integration strategies in plants. J Biomed Biotech doi: 10.1155/2012/184854. [DOI] [PMC free article] [PubMed]
- 27. Feuillet C, Leach JE, Rogers J, Schnable PS, Eversole K (2011) Crop genome sequencing: lessons and rationales. Trends Plant Sci 16: 77–88. [DOI] [PubMed] [Google Scholar]
- 28. Alkan C, Sajjadian S, Eichler EE (2011) Limitations of next generation genome sequence assembly. Nature Methods 8: 61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schatz MC, Delcher AL, Salzberg SL (2010) Assembly of large genomes using second-generation sequencing. Genome Res doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed]
- 30. Kim CG, Fujiyama A, Saitou N (2003) Construction of a gorilla fosmid library and its PCR screening system. Genomics 82: 571–574. [DOI] [PubMed] [Google Scholar]
- 31. Meyer JDF, Deleu W, Garcia-Mas J, Havey MJ (2008) Construction of a fosmid library of cucumber (Cucumis sativus) and comparative analyses of the Eif4E and eIF(iso)4E regions from cucumber and melon (Cucumis melo). Mol Genet Genomics 279: 473–480. [DOI] [PubMed] [Google Scholar]
- 32.Hao DC, Ge G, Xiao P, Zhang Y, Yang L (2011) The first insight into the tissue specific Taxus transcriptome via Illumina second generation sequencing. PLoS one 6, doi:10.1371/journal.pone.0021220. [DOI] [PMC free article] [PubMed]
- 33. Coupland P, Chandra T, Quail M, Reik W, Swerdlow H (2012) Direct sequencing of small genomes on the Pacific Biosciences RS without library preparation. BioTechniques 53: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mizutani T, Endoh D, Okamoto M, Shirato K, Shimizu H, et al. (2007) Rapid genome sequencing of RNA Viruses. Emerging Infectious Diseases 13: 322–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malboeuf CM, Yang X, Charlebois P, Qu J, Berlin AM, et al. (2013) Complete viral RNA genome sequencing of ultra-low copy samples by sequence-independent amplification. Nucleic Acids Res 41(1): e13 Doi:10.1093/nar/gks794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ninomiya M, Ueno Y, Funayama R, Nagashima T, Nishida Y, et al. (2012) Use of illumina deep sequencing technology to differentiate hepatitis C virus variants. J Clin Microbiol 50: 857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L, Donly C, Li Q, Willis LG, Keddie BA, et al. (2002a) Identification and genomic analysis of a second species of nucleopolyhedrovirus isolated from Mamestra configurata . Virology 297: 226–244. [DOI] [PubMed] [Google Scholar]
- 38. Li Q, Donly C, Li L, Willis LG, Theilmann DA, et al. (2002b) Sequence and organization of the Mamestra configurata nucleopolyhedrovirus genome. Virology 294: 106–121. [DOI] [PubMed] [Google Scholar]
- 39. Lange M, Wang H, Zhihong H, Jehle JA (2004) Towards a molecular identification and classification system of lepidopteran-specific baculovirus. Virology 325: 36–47. [DOI] [PubMed] [Google Scholar]
- 40. Harrison RL, Popham HJ (2008) Genomic sequence analysis of a granulovirus isolated from the Old World bollworm, Helicoverpa armigera . Virus Genes 36: 565–581. [DOI] [PubMed] [Google Scholar]
- 41. Wan H, Wootton JC (2000) A global compositional complexity measure for biological sequences: AT-rich and GC-rich genomes encode less complex proteins. Comput Chem 24: 71–94. [DOI] [PubMed] [Google Scholar]
- 42. Jukes TH, Bhushan V (1986) Silent nucleotide substitutions and G+C content of some mitochondrial and bacterial genes. J Mol Evol 24: 39–44. [DOI] [PubMed] [Google Scholar]
- 43. D’Onofrio G, Mouchiroud D, Aissani B, Gautier C, Bernardi G (1991) Correlations between the compositional properties of human genes, codon usage, and amino acid composition of proteins. J Mol Evol 32: 504–510. [DOI] [PubMed] [Google Scholar]
- 44.Mount DW (2001) Bioinformatics: Sequence and Genome analysis. Cold Spring Harbor, NY, p.308.
- 45. Hilton S, Winstanley D (2008) Genomic sequence and biological characterization of a nucleopolyhedrovirus isolated from the summer fruit tortrix, Adoxophyes orana . J Gen Virol 89: 2898–2908. [DOI] [PubMed] [Google Scholar]
- 46. Karlin S (1998) Global dinucleotide signatures and analysis of genomic heterogeneity. Curr Opin Microbiol 1: 598–610. [DOI] [PubMed] [Google Scholar]
- 47. Hyink O, Dellow RA, Olsen MJ, Caradoc-Davies KMB, Drake K, et al. 2002. Whole genome analysis of the Epiphyas postvittana nucleopolyhedrovirus. J Gen Virol 83: 957–971. [DOI] [PubMed] [Google Scholar]
- 48. Pang Y, Yu J, Wang L, Hu X, Li W, et al. (2001) Sequence analysis of the Spodoptera litura multicapsid nucleopolyhedrovirus genome. Virology 287: 391–404. [DOI] [PubMed] [Google Scholar]
- 49. Leisy DJ, Rasmussen C, Kim HT, Rohrmann GF (1995) The Autographa californica nuclear polyhedrosis virus homologous region 1a: identical sequences are essential for DNA replication activity and transcriptional enhancer function. Virology 208: 742–752. [DOI] [PubMed] [Google Scholar]
- 50. Ko R, Okano K, Maeda S (2000) Structural and functional analysis of the Xestia c-nigrum granulovirus matrix metalloproteinase. J Virol 74: 11240–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wolgamot GM, Gross CH, Russell RLQ, Rohrmann GF (1993) Immunocytochemical characterization of p24, a baculovirus capsid-associated protein. J Gen Virol 74: 103–107. [DOI] [PubMed] [Google Scholar]
- 52. van Oers MM, Vlak JM (1997) The baculovirus 10-kDa protein. J Invertebr Pathol 70: 1–17. [DOI] [PubMed] [Google Scholar]
- 53. Monsma SA, Oomens AG, Blissard GW (1996) The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J Virol 70: 4607–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pearson MN, Groten C, Rohrmann GF (2000) Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J Virol 74: 6126–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ijkel WFJ, Westenberg M, Goldbach RW, Blissard GW, Vlak JM, et al. (2000) A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 275: 30–41. [DOI] [PubMed] [Google Scholar]
- 56. Rapp JC, Wilson JA, Miller LK (1998) Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J Virol 72: 10197–10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friesen PD (1997) Regulation of baculovirus early gene expression. In: L.K. Miller (ed). The Baculoviruses, Plenum Press. 141–170.
- 58.Lu A, Miller LK (1997) Regulation of baculovirus late and very late gene expression. In: L.K. Miller (ed). The Baculoviruses, Plenum Press. 193–216.
- 59. Gomi S, Zhou CE, Yih W, Majima K, Maeda S (1997) Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology 230: 35–47. [DOI] [PubMed] [Google Scholar]
- 60. Morris TD, Todd JW, Fisher B, Miller LK (1994) Identification of lef-7: a baculovirus gene affecting late gene expression. Virology 200: 360–369. [DOI] [PubMed] [Google Scholar]
- 61. Pearson MN, Rohrmann GF (1998) Characterization of a baculovirus encoded ATP-dependent DNA ligase. J Virol 72: 9142–9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kuzio J, Pearson MN, Harwood SH, Funk CJ, Evans JT, et al. (1999) Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar . Virology 253: 17–34. [DOI] [PubMed] [Google Scholar]
- 63. Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD (1994) The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202: 586–605. [DOI] [PubMed] [Google Scholar]
- 64. Gomi S, Majima K, Maeda S (1999) Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol 80: 1323–1337. [DOI] [PubMed] [Google Scholar]
- 65.Lu A, Miller LK (1997) Regulation of baculovirus late and very late gene expression. In: L.K. Miller (ed.) The Baculoviruses, Plenum Press. 193–216.
- 66.Oh S, Kim DH, Patnaik BB, Jo YH, Noh MY, et al. (2013) Molecular and immunohistochemical characterization of the chitinase gene from Pieris rapae granulovirus. Arch Virol DOI 10.1007/s00705-013-1649-z. [DOI] [PubMed]
- 67. Hawtin RE, Zarkowska T, Arnold K, Thomas CK, Gooday GW, et al. (1997) Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238: 243–253. [DOI] [PubMed] [Google Scholar]
- 68. Tomalski MD, Eldridge R, Miller LK (1991) A baculovirus homolog of a Cu/Zn superoxide dismutase gene. Virology 184: 149–161. [DOI] [PubMed] [Google Scholar]
- 69. Oh S, Kim DH, Patnaik BB, Jo YH, Noh MY, et al. (2013) Molecular and immunohistochemical characterization of granulin gene encoded in Pieris rapae granulovirus genome. J Invertebr Pathol 113: 7–17. [DOI] [PubMed] [Google Scholar]
- 70. Wang RR, Deng F, Hou DH, Zhao Y, Guo L, et al. (2010) Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J Virol 84: 7233–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xiang XW, Chen L, Guo AQ, Yu SF, Yang R, et al. (2011) The Bombyx mori nucleopolyhedrovirus (BmNPV) ODV-E56 envelope protein is also a per os infectivity factor. Virus Res 155: 69–75. [DOI] [PubMed] [Google Scholar]
- 72. Song JJ, Wang RR, Deng F, Wang HL, Hu ZH (2008) Functional studies of per os infectivity factors of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. J Gen Virol 89: 2331–2338. [DOI] [PubMed] [Google Scholar]
- 73. Wang P, Grandos RR (1998) Observations on the presence of the peritrophic membrane in larval Trichoplusia ni and its role in limiting baculovirus infection. J Invertebr Pathol 72: 57–62. [DOI] [PubMed] [Google Scholar]
- 74. Jiang H, Liu S, Zhao P, Pope C (2009) Recombinant expression and biochemical characterization of the catalytic domain of acetylcholinesterase-1 from the African malaria mosquito, Anopheles gambiae. . Insect Biochem Mol Biol 39: 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dall D, Luque T, O’Reilly D (2001) Insect-virus relationships: sifting by informatics. Bioessays 23: 184–193. [DOI] [PubMed] [Google Scholar]
- 76. Birnbaum MJ, Clem RJ, Miller LK (1994) An apoptosis inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol 68: 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vucic D, Kaiser WJ, Harvey AJ, Miller LK (1997) Inhibition of Reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc Natl Acad Sci USA 94: 10183–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Crook NE, Clem RJ, Miller LK (1993) An apoptosis-inhibiting baculovirus gene with a zinc-finger motif. J Virol 67: 2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis and homology search of PiraGV-K ORFs. The PiraGV-K ORFs have been analyzed for homology using representative granulovirus genomes such as Adoxophyes orana granulovirus (AdorGV), Agrotis segetum granulovirus (AgseGV), Choristoneura occidentalis granulovirus (ChocGV), Cryptophlebia leucotreta granulovirus (CrleGV), Cydia pomonella granulovirus (CypoGV), Helicoverpa armigera granulovirus (HearGV), Phthorimaea operculella granulovirus (PhopGV), Pieris rapae granulovirus-Chinese isolate (PiraGV-C), Plutella xylostella granulovirus (PlxyGV), Pseudaletia unipuncta granulovirus (PsunGV), Spodoptera litura granulovirus (SpliGV) and Xestia c-nigrum granulovirus (XecnGV). Pid and Psi refers to percent identity and percent similarity.
(DOCX)