Abstract
The herpes simplex virus type 1 (HSV-1) alkaline nuclease, encoded by the UL12 gene, plays an important role in HSV-1 replication, as a null mutant of UL12 displays a severe growth defect. Although the precise in vivo role of UL12 has not yet been determined, several in vitro activities have been identified for the protein, including endo- and exonuclease activities, interaction with the HSV-1 single-stranded DNA binding protein ICP8, and an ability to promote strand exchange in conjunction with ICP8. In this study, we examined a naturally occurring N-terminally truncated version of UL12 called UL12.5. Previous studies showing that UL12.5 exhibits nuclease activity but is unable to complement a UL12 null virus posed a dilemma and suggested that UL12.5 may lack a critical activity possessed by the full-length protein, UL12. We constructed a recombinant baculovirus capable of expressing UL12.5 and purified soluble UL12.5 from infected insect cells. The purified UL12.5 exhibited both endo- and exonuclease activities but was less active than UL12. Like UL12, UL12.5 could mediate strand exchange with ICP8 and could also be coimmunoprecipitated with ICP8. The primary difference between the two proteins was in their intracellular localization, with UL12 localizing to the nucleus and UL12.5 remaining in the cytoplasm. We mapped a nuclear localization signal to the N terminus of UL12, the domain absent from UL12.5. In addition, when UL12.5 was overexpressed so that some of the enzyme leaked into the nucleus, it was able to partially complement the UL12 null mutant.
The alkaline nuclease of herpes simplex virus type 1 (HSV-1) is encoded by the UL12 gene (6, 7, 25). The enzyme exhibits both endonuclease and 5′-3′ exonuclease activities, with a pH optimum in the alkaline range (12, 13, 18, 33). Although the in vitro nuclease activity has been well characterized, the role of UL12 in the HSV-1 life cycle is not well understood. Null mutants incapable of expressing UL12 exhibit a 100- to 1,000-fold reduction in viral titers, suggesting that while not essential, UL12 plays an important role in efficient virus production (37). In cells infected with AN-1, a virus with the UL12 gene deleted, replicating HSV-1 DNA exhibits an aberrant structure (23). The AN-1 mutant virus packages DNA into capsids, but most of these fail to exit the nucleus (30), leading to low titers. The exonuclease activity of UL12 is required for its role in vivo, since wild-type UL12 on a transfected plasmid complements the growth of the null virus while point mutations lacking exonuclease activity do not (10, 11).
The UL12 protein interacts with the HSV-1 single-stranded DNA binding protein, ICP8 (34, 36). Recently, it was shown that these two proteins work together to mediate a strand exchange reaction in vitro, demonstrating that the proteins have activities analogous to that of the Red recombinase from bacteriophage lambda (26). The Red recombinase is composed of two components, the 5′-3′ exonuclease Red alpha and the single-stranded DNA binding protein Red beta, which acts as a synaptase. The UL12 gene shows homology with lambda red α, the gene for the exonuclease partner of the two-part recombinase. In fact, every member of the herpesvirus family studied to date encodes a nuclease with homology to the red α product (R. S. Myers and K. E. Rudd, presented at the 1998 Miami Nature Biotechnology Winter Symposium, 1998). The conservation of these sequences from viruses of plants, animals, and bacteria (Myers and Rudd, presented at the 1998 Miami Nature Biotechnology Winter Symposium) suggests that recombination may play an important role in the biology of double-stranded DNA viruses with linear genomes. Our finding that the UL12 protein has in vitro recombinase activity like that of the Red recombinase is consistent with this hypothesis. Since HSV-1 DNA replication is associated with a high degree of homologous recombination (1, 8, 9, 28, 35) and replication intermediates exhibit a complex, possibly branched structure (29), we and others have proposed that recombination-dependent replication is important in HSV (reviewed in reference 38). The observation that UL12 and ICP8 can carry out a strand exchange reaction supports the notion that UL12 has a role in mediating homologous recombination. The exonuclease activity is required for the recombinase activity of UL12, as a mutant lacking exonuclease activity was shown to be unable to mediate strand exchange (26).
The UL12 gene is encoded by a 2.3-kb mRNA, and embedded within this gene is a subgenic 1.9-kb mRNA encoding an N-terminally truncated version of UL12, designated UL12.5 (6, 7, 24). Like UL12, UL12.5 is expressed with early or β kinetics, but the 1.9-kb transcript is expressed threefold less efficiently than the 2.3-kb transcript for full-length UL12 (7). The UL12.5 protein is found in much smaller amounts than UL12 (4). Comparisons of UL12 and UL12.5 activities have shown that the endo- and exonuclease activities of UL12.5 are similar to those of UL12 (4, 11). However, despite the similarity of activity, UL12.5 cannot efficiently complement the AN-1 virus when UL12.5 is introduced into cells via the transfection of expression plasmids (11, 24). Furthermore, a frameshift mutation, ANF-1, consisting of two nucleotides inserted 14 nucleotides downstream of the translational start site of full-length UL12, was generated, and it expresses UL12.5 at wild-type levels but is null for the full-length protein (24). ANF-1 has a phenotype identical to that of the null AN-1 virus, despite the fact that UL12.5 is present. The inability of UL12.5 to complement a UL12 null mutant thus poses a dilemma and suggests that an essential function of UL12 lies within the first 126 residues.
To further characterize essential functions in this genetic locus, we set out to compare in greater detail the biochemical activities of these two proteins. We constructed a recombinant baculovirus for the expression of UL12.5, and we purified UL12.5 in soluble form. We confirmed the earlier studies by finding that UL12.5 possesses nuclease activity similar to that of UL12. In addition, we found that UL12.5 could be coimmunoprecipitated with ICP8 as efficiently as UL12 and could also mediate strand exchange activity with ICP8. These results did not explain why UL12.5 was not able to efficiently complement AN-1. The answer to this apparent dilemma came with the observation that in transfected cells UL12.5 was cytoplasmic, while UL12 was strongly localized to the nucleus. Furthermore, a green fluorescent protein (GFP) fusion of the N-terminal 126 residues of UL12 localized to the nucleus, indicating that this domain contains a nuclear localization signal (NLS). The cytoplasmic localization of UL12.5 is most likely the reason for its inability to complement the UL12 null virus.
MATERIALS AND METHODS
Cells and viruses.
Spodoptera frugiperda (Sf9) cells were grown in Grace's insect medium (Invitrogen, Carlsbad, Calif.) containing 10% (vol/vol) fetal bovine serum (GIBCO), 0.1 mg of streptomycin/ml, and 100 U of penicillin (Invitrogen)/ml. African green monkey kidney (Vero) cells (American Type Culture Collection, Manassas, Va.) and the 6-5 cell line, which is permissive for the UL12 null virus, were propagated in Dulbecco's modified Eagle medium (Invitrogen) containing 5% (vol/vol) fetal calf serum, 0.1 mg of streptomycin/ml, and 100 U of penicillin/ml (30). The 6-5 cell line was maintained in the presence of 400 U of G418 (Invitrogen)/ml.
DNA.
M13mp18 replicative form was purified from infected Escherichia coli UT481 [Δ(lac-pro) hsdS(r−m−) lacIq lacZ] cells using the Qiagen (Valencia, Calif.) maxiplasmid kit. M13mp18 single-stranded DNA was purified from M13 phage-infected UT481 cells according to standard protocols (21). DNA fragments were purified from agarose gels using the GeneClean spin kit (Bio-101, La Jolla, Calif.).
Materials.
Restriction endonucleases and other DNA-modifying enzymes were purchased from New England Biolabs (Beverly, Mass.) or from Invitrogen. DNA primers were from Invitrogen.
Plasmids.
For expression in mammalian cells, the vector pSAK was used. This plasmid was derived from the plasmid pEGFP-C1 (Clontech-BD Biosciences, San Jose, Calif.), which expresses enhanced GFP (EGFP) from the cytomegalovirus (CMV) immediate-early promoter. The coding region for EGFP was deleted from this plasmid by cutting it with NheI and BglII. The ends were blunted with the Klenow fragment of DNA polymerase I and religated, creating the pSAK vector. The vector was used for expression of UL12 and UL12.5 in Vero cells. Vectors pSAKUL12.5 and pSAKUL12/12.5 were generated by PCR of the wild-type UL12 gene found on the previously described plasmid pUC119-AK (24). The plasmid construct expressing only UL12, and not UL12.5, called pSAKUL12, was generated by PCR of the plasmid pF1′-CMV-AK(M127F) (22). The PCR primers used in the preparation of these constructs were as follows: forward primer for UL12 constructs, 5′ GGAATTCCGCCACCATGGAGTCCACGGGAGGCCC; forward primer for UL12.5, 5′ GGAATTCCGCCACCATGTGGTCGGCGTCGGTGAT; reverse primer for UL12 and UL12.5 constructs, 5′ GGGGTACCTCAGCGAGACGACCTCCCCG. The restriction sites for EcoRI and KpnI are italicized, and the Kozak consensus sequence for initiation (19) is shown in boldface. The PCR-generated fragments were cloned into pSAK at the EcoRI and KpnI sites. The plasmids for expression of UL12, UL12.5, or the N terminus of UL12 fused to EGFP or to a hemagglutinin (HA) tag were constructed by first cloning full-length gene fragments with no stop codons (fusion ready) into pSAK. The forward primers for PCR were the same as those listed above, and the reverse primers were as follows: for full-length UL12 and UL12.5, 5′ GTGGATCCTCAGGTACCGCGAGACGACCTCCCCGTCG; for the N terminus of UL12, 5′ GGGGTACCAGAATCAAGGTCCGGGGAGTC, with the KpnI sites italicized. These PCR fragments were cloned into pSAK at the EcoRI and KpnI sites. HA tags were fused to the C termini of the various constructs by cutting the vectors with KpnI and BamHI and ligating annealed oligonucleotides encoding the tag and possessing the overhangs necessary for annealing to KpnI and BamHI. The oligonucleotides used were 5′ CTACCCATACGATGTTCCGGATTACGCTTGAG and 5′ GATCCTCAAGCGTAATCCGGAACATCGTATGGGTAGGTAC, with the annealing sequences for BamHI and KpnI italicized. To add EGFP to the C termini of UL12, UL12.5, or the N-terminal domain of UL12, a fragment encoding EGFP was generated by PCR using pEGFP-C1 as a template. The primers used were as follows: forward primer, 5′ CGGGGTACCGTGAGCAAGGGCGAGGAGCTG; reverse primer, 5′ CGCGGATCCTCACTTGTACAGCTCGTCCATGC, with the KpnI and BamHI sites underlined. The GFP-containing fragment was cloned into the fusion-ready vectors at the KpnI and BamHI sites. The resulting clones had EGFP fused in frame to the C termini of UL12 and UL12.5 or the N-terminal domain of UL12.
Antibodies.
The anti-UL12 BWp12 antibody was a generous gift from Joel Bronstein and Peter Weber (3, 24). The polyclonal anti-ICP8 antibody 3-83 was kindly provided by David Knipe (17). The monoclonal anti-ICP8 antibody 39-S was generously provided by M. Zweig (31). Goat anti-rabbit conjugated to Alexa Fluor 488 was from Molecular Probes (Eugene, Oreg.). The anti-HA antibody sc-805 was from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Protein expression and purification.
For preparation of a recombinant baculovirus expressing UL12.5, the Bac-to-Bac system (Invitrogen) was used. The UL12.5 gene was generated by PCR using pUC119AK as the template with the following primers: forward primer, 5′ CGGGATCCGCCACCATGTGGTCGGCGTCGGTGATCCCC; reverse primer, 5′ GGAATTCTCAGCGAGACGACCTCCC. The restriction sites for BamHI and EcoRI are italicized, and the Kozak consensus sequence for initiation (19) is shown in boldface. The PCR-generated fragment was digested with EcoRI and BamHI and cloned into the pFastBac vector at those sites. The resulting plasmid, pFastBacUL12.5L, was confirmed by DNA sequencing (Molecular Core, University of Connecticut Health Center). The transfer of the UL12.5 construct to the baculovirus bacmid by homologous recombination was done according to the manufacturer's protocols. The resulting baculovirus was named AcUL12.5. The UL12-expressing baculovirus AcAN was kindly provided by Fred L. Homa (Pharmacia & Upjohn, Kalamazoo, Mich.). The ICP8-expressing baculovirus AcUL29 was a generous gift from Nigel D. Stow (32).
The UL12.5 protein was purified according to the protocol that had been used for the purification of UL12, with minor modifications (10). In order to be able to compare the activities of the two proteins, both UL12 and UL12.5 were purified, using the same protocol. The UL12 protein, prepared by Josh Goldstein and described previously (10), was also used as a control. The new UL12 preparations had the same exonuclease activity as the UL12 protein described previously (reference 10 and data not shown). Briefly, Sf9 insect cells were grown in suspension and infected with recombinant baculovirus. The infected cells were collected after 50 h of incubation at 27°C. The cells were pelleted, washed with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4, pH 7.3), repelleted, and frozen at −80°C. Cells (∼5 g) were thawed and resuspended in 40 ml of cold buffer A (10 mM Tris-Cl, pH 7.5, 1 mM MgCl2, 80 mM KCl, 0.5 mM dithiothreitol [DTT], 0.2% NP-40). Protease inhibitors were added at the following concentrations: 13 μg of aprotinin/ml, 7 μg of leupeptin/ml, 7 μg of pepstatin A/ml, and 1 mM phenylmethylsulfonyl fluoride (PMSF). After 10 min of swelling on ice, the cells were disrupted by 10 strokes of a Dounce homogenizer. The nuclei were pelleted, and the supernatant was clarified by centrifugation at 100,000 × g in an SW28 rotor. The extract was brought to 20% saturation with ammonium sulfate, and the precipitate was removed by centrifugation. The UL12 or UL12.5 protein was then precipitated from the supernatant by bringing the ammonium sulfate saturation up to 55%. The precipitate was resuspended in 5 ml of buffer B (20 mM potassium phosphate, pH 8.0, 20% glycerol, 5 mM β-mercaptoethanol) and dialyzed overnight against buffer B. The dialysate was clarified by centrifugation and loaded on a 20-ml HiLoad 16/10 SP-Sepharose column (Pharmacia, Piscataway, N.J.) that had been equilibrated with buffer B. The proteins were eluted using a gradient from 20 to 500 mM potassium phosphate. UL12 and UL12.5 typically eluted at 0.1 to 0.2 M salt. Fractions containing the desired protein were pooled, concentrated using Biomax 50K concentrators (Millipore, Bedford, Mass.), and loaded onto a Superose 12 gel filtration column (Pharmacia) that had been equilibrated with buffer C (20 mM potassium phosphate, pH 8.0, 10% glycerol, 5 mM β-mercaptoethanol). Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The fractions used for biochemical analysis were determined to be >90% pure, as measured by densitometry of Coomassie blue-stained gels. Protein concentrations were determined by the Bradford method (2) using the reagent prepared by Bio-Rad (Hercules, Calif.).
Immunoprecipitation.
Sf9 insect cells (9 × 106) were plated in 100-mm-diameter dishes 3 h prior to infection. The cells were infected with the recombinant baculoviruses and harvested 50 h later by scraping the cells from the dish. The cells were centrifuged at 2,000 × g and washed with PBS. The cell pellets were lysed in 1.2 ml of modified RIPA buffer (25 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 10 mM β-glycerophosphate, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 20 μg of aprotinin/ml), and were disrupted by sonication with a microtip (Misonix, Farmingdale, N.Y.) for 30 s. The insoluble fraction was removed by centrifugation at 100,000 × g for 30 min at 4°C. The protein concentrations of the lysates were determined by the Bradford method (2) using the reagent prepared by Bio-Rad. The lysates were analyzed by SDS-PAGE and immunoblotting (21).
For immunoprecipitation experiments, 150 μl of insect cell lysate (normalized for protein concentration so that the same amount of total protein was present in each sample) was combined with 150 μl of modified RIPA buffer containing 1 mM phenylmethylsulfonyl fluoride and 20 μg of aprotinin/ml. Each reaction mixture was incubated with 25 μl of mouse monoclonal anti-ICP8 antibody (39-S) for 1.5 h at 4°C. To precipitate the protein-antibody complexes, 15 μl of protein A-G-Sepharose (Amersham Pharmacia Biotech, Piscataway, N.J.) was added, and incubation was continued for 1 h. Immune complexes were pelleted by centrifugation at 14,000 × g for 10 s. The volume of each sample was reduced to 100 μl by aspiration, and each sample was washed three times in RIPA buffer. After the final wash, the volume was reduced to 15 μl, and 5 μl of 4× gel loading buffer was added. The samples were analyzed by SDS-PAGE and immunoblotting using the anti-UL12 BWp12 antibody (3, 24) and the 3-83 anti-ICP8 antibody (17). The immunoblots were developed using the ECL system (Amersham).
Transfection of Vero cells.
For transfection of Vero cells, 106 cells were plated in 35-mm-diameter six-well dishes 1 day prior to transfection. The cells were transfected with a total of 1 μg of plasmid DNA using Lipofectamine PLUS (Invitrogen). For experiments involving titration of the amount of protein-expressing vector, the empty parent vector was included in the transfection so that the total amount of DNA remained 1 μg. The cells were harvested 24 h posttransfection, washed in PBS, and lysed in modified RIPA buffer. The crude cell lysates were centrifuged at 14,000 × g and 4°C and used for SDS-PAGE and immunoblot analyses.
Strand exchange assay.
The reaction was carried out in a final volume of 20 μl as described previously (26). Each reaction mixture consisted of 100 ng of circular single-stranded M13mp18 DNA (2 nM), 100 ng of linear (PstI-cut) double-stranded M13mp18 (1 nM), 18.8 ng of UL12 (13.9 nM) or 15.3 ng of UL12.5 (13.9 nM), 4.5 μg of ICP8 (1.75 μM), 20 mM Tris-Cl (pH 7.5), 40 mM NaCl, 1 mM MgCl2, and 1 mM DTT. The reaction mixture was incubated at 37°C, and the reaction was stopped by adding 5 μl of 5× stop buffer (50% glycerol, 50 mM EDTA, 1% SDS, 0.2% bromphenol blue). Samples were electrophoresed on a 1% agarose gel with 0.7 μg of ethidium bromide/ml in TAE buffer (0.04 M Tris-acetate, 0.001 M EDTA). Adobe Photoshop (version 6.0) and Adobe Illustrator (version 10.0) were used in the preparation of figures.
Exonuclease assay.
Total unlabeled chromosomal DNA from E. coli was isolated from late-log-phase UT481 cells by phenol extraction and ethanol precipitation essentially as described previously (10). [Thymidine-methyl-3H]DNA (derived from E. coli) (NEN-Perkin Elmer, Boston, Mass.) was mixed with unlabeled chromosomal E. coli DNA to provide a substrate with the desired specific radioactivity. The nuclease assay was performed in a 20-μl volume, with 2.4 μg of [3H]DNA as the substrate (360 μM nucleotides). UL12 (6.75 ng; 5 nM) and UL12.5 (5.5 ng; 5 nM) were assayed for nuclease activity using the following buffer: 20 mM Tris-Cl, pH 8.8, 40 mM NaCl, 10 mM MgCl2, 1 mM DTT. Reaction mixtures were incubated at 37°C and then stopped with 5 μl of 0.6-mg/ml DNA and 25 μl of 20% (wt/vol) trichloroacetic acid. After 10 min on ice, samples were centrifuged for 10 min at 14,000 × g, and the radioactivity in 25 μl of the supernatant fraction was determined by scintillation counting.
Endonuclease assay.
UL12 or UL12.5 (200 nM) was incubated at 37°C in a 20-μl reaction volume with 225 ng of pEGFP-C1 plasmid in a buffer consisting of 20 mM Tris-Cl, pH 8.8, 40 mM NaCl, 10 mM MgCl2, and 1 mM DTT. The reaction was stopped by adding 5 μl of 5× stop buffer. Samples were electrophoresed on a 1% agarose gel with 0.7 μg of ethidium bromide/ml in TAE buffer.
IF microscopy.
Vero cells were plated on glass coverslips and transfected as described above. When indicated, the cells were superinfected with AN-1 virus (at a multiplicity of infection of 5) at 24 h posttransfection. At 24 h posttransfection, or 5 h postsuperinfection, the cells were processed for immunofluorescence (IF) microscopy. After a brief wash with PBS, the cells were fixed by incubating them with 4% (wt/vol) paraformaldehyde in PBS for 10 min at room temperature. The cells were then washed with PBS and permeabilized for 10 min in 1% Triton X-100 in PBS. Coverslips were blocked in 3% normal goat serum (NGS) in PBS overnight at 4°C and then incubated with primary antibody diluted in 3% NGS for 1.5 h at room temperature. For the detection of UL12 and UL12.5, the anti-UL12 antibody BWp12 was used (3, 24). For the detection of proteins with an HA tag, the anti-HA antibody sc-805 (Santa Cruz Biotechnology) was used. After extensive washing with PBS, cells were incubated for 1.5 h with Alexa Fluor 488 goat anti-rabbit secondary antibody (Molecular Probes) in 3% NGS. After a final wash in PBS, the coverslips were mounted in glycerol gelatin containing 2.5% 1,4-diazobicyclo-[2.2.2]octane to retard photobleaching. For the detection of GFP fusion proteins, cells were fixed in paraformaldehyde, washed with PBS, and mounted on slides as described above. The coverslips were mounted on slides and observed under oil immersion. A BX60 microscope (Olympus America Inc., Melville, N.Y.) was used for the visualization of samples.
Complementation assay.
Vero cells were plated in 35-mm-diameter dishes and transfected as described above. Twenty-four hours posttransfection, the cells were superinfected with the AN-1 virus at a multiplicity of infection of 5 and incubated for 1 h. The cells were then washed five times with PBS, and 2 ml of fresh medium was added. Eighteen hours later, the medium was collected and titers were determined by a plaque assay of the permissive cell line 6-5.
RESULTS
Purification of UL12.5.
In previous studies, UL12.5 activity was assayed using protein that was purified in insoluble form and subsequently denatured and refolded (4) or protein translated in an in vitro translation system (11). Thus, direct comparisons of the activity of UL12.5 with that of UL12, which can be purified in soluble form from baculovirus-infected insect cells (10, 16), have not been possible. In this study, we prepared a new construct for the expression of UL12.5 in insect cells and have successfully purified soluble UL12.5.
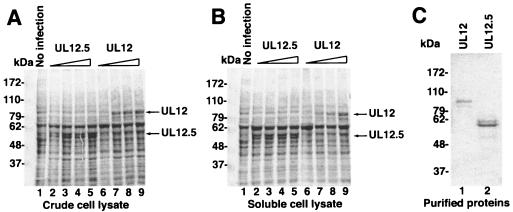
The UL12.5 gene was integrated into the baculovirus genome using the Bac-to-Bac system and was expressed from the polyhedrin promoter. In order to improve translation of the transcript, the sequence directly upstream of the first ATG in UL12.5 was changed to GCCACC, in accordance with the Kozak consensus sequences (19). Sf9 cells were infected with the recombinant baculovirus, and the UL12.5 protein was expressed, appearing as a 55-kDa band on SDS-PAGE (Fig. 1A, lanes 2 to 5). The amount of UL12.5 protein increased with the amount of baculovirus stock used to infect the insect cells. UL12 protein expressed in AcAN-infected cells was also clearly detectable (Fig. 1A, lanes 6 to 9). To determine whether the protein expressed was soluble, the extracts were centrifuged at 100,000 × g, and the supernatants were again analyzed by SDS-PAGE. Like UL12, which had been previously demonstrated to be soluble in this system, UL12.5 appears soluble by this analysis (Fig. 1B, lanes 2 to 5). The UL12.5 protein was purified from the baculovirus-infected insect cells and was estimated to be >90% pure by densitometry of Coomassie blue-stained gels (Fig. 1C). The identity of the UL12.5 protein was confirmed by immunoblot analysis using the anti-UL12 antibody BWp12 that also recognizes UL12.5 (24) (data not shown). The UL12 and UL12.5 proteins appear as doublets (Fig. 1C). This characteristic has been noted before for these proteins and may be due to posttranslational modification (4, 16).
FIG. 1.
Expression and purification of UL12.5. SF21 cells were infected with increasing amounts of AcAN (for UL12) or AcUL12.5 (for UL12.5) baculovirus stock. (A and B) Total (A) and soluble-fraction (B) cell lysates were subjected to SDS-PAGE, and the gels were stained by Coomassie brilliant blue. (C) Purified UL12 and UL12.5. Masses are expressed in kilodaltons (kDa).
Nuclease activity of UL12.5.
UL12 and UL12.5 have been shown to possess both endo- and exonuclease activities (3, 4, 11, 14). The endonuclease activity is weaker, requiring more enzyme to see appreciable activity than for exonuclease activity (3). We assayed the soluble preparations of UL12 and UL12.5 for both the exo- and endonuclease activities.
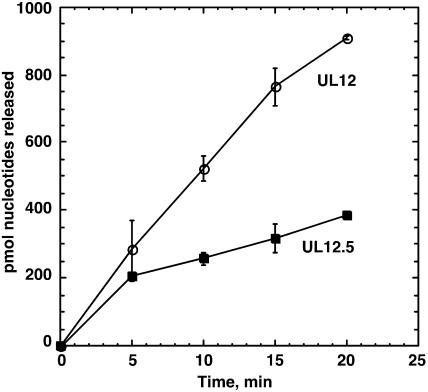
The exonuclease activities of the proteins were assayed on a substrate consisting of total E. coli chromosomal DNA. As shown in Fig. 2, both UL12 and UL12.5 possess nuclease activity, but UL12.5 appears to be slightly less active and the activity reaches a plateau quickly. This is in contrast to UL12, where the activity remains linear for at least 15 min. Thus, it appears that UL.12.5 retains significant nuclease activity. Although it is formally possible that a portion of the nuclease activity exhibited by UL12.5 is due to the presence of a contaminating nuclease in the preparation, we think it is unlikely that this contaminating nuclease contributes significantly to the activities seen. The UL12 and UL12.5 proteins are purified to >90% purity, indicating that any contaminant would have to have very high specific activity to account for the results seen in Fig. 2. Moreover, the observation that UL12.5 exhibits exonuclease activity is consistent with the results of previous studies with purified proteins (4, 11).
FIG. 2.
Exonuclease activity of UL12.5. Exonuclease activity was measured using 2.4 μg of 3H-labeled E. coli DNA (360 μM nucleotides) and 5 nM UL12 or UL12.5 as described in Materials and Methods. Open circles, UL12; solid squares, UL12.5.
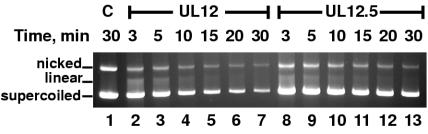
The endonuclease activities of the enzymes were tested by using a supercoiled plasmid substrate. The conversion of the supercoiled form to nicked and linear forms is an indicator of the endonucleolytic activity. UL12 and UL12.5 both had endonuclease activity, leading to the appearance of the linear form and the disappearance of the supercoiled form of the plasmid (Fig. 3). The nicked and linear forms of the plasmid did not accumulate during the assay, as the more robust exonucleolytic activity of the proteins rapidly degraded these forms. Although the qualitative nature of the endonuclease assay does not allow precise calculation of units of activity, Fig. 3 demonstrates that UL12 is more active than UL12.5, causing a more rapid disappearance of the supercoiled form of the plasmid. The endonuclease assay data was also consistent with an early plateau in the activity of UL12.5. The difference in activity between the two enzymes may be due to lower stability of UL12.5 under the assay conditions or an intrinsic difference between the activities of the two proteins. The concentrated stocks of the proteins appeared to be stable, as both enzymes were unchanged by seven freeze-thaw cycles, exhibiting the same exonuclease activity on the first and seventh thaws (data not shown). In summary, although UL12.5 exhibits slightly lower endo- and exonuclease activities than UL12, this difference does not seem to be sufficient to account for the inability of UL12.5 to complement the UL12 null mutant virus, indicating that UL12.5 likely lacks another essential function.
FIG. 3.
Endonuclease activity of UL12.5. pEGFP-C1 plasmid (100 ng) was incubated with 100 pmol of UL12.5 or UL12 protein for the times indicated, as described in Materials and Methods. The samples were run on a 0.7% agarose gel in TAE buffer and stained with ethidium bromide. C, control (no protein added).
Interaction of UL12 with the HSV-1 single-stranded DNA binding protein, ICP8.
UL12 has been reported to interact with the HSV-1 single-stranded DNA binding protein, ICP8 (34, 36). The N terminus of UL12 is rich in proline, and proline-rich sequences are common in protein-protein interaction domains (15). We considered that this domain might be responsible for the interaction with ICP8. The UL12.5 protein, which lacks the first 126 amino acids of UL12, is missing this proline-rich domain. We therefore tested whether UL12.5 is able to interact with ICP8.
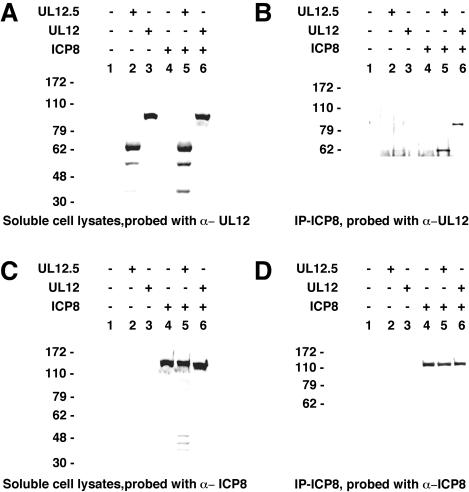
The interaction between UL12.5 (or UL12) and ICP8 was analyzed by coimmunoprecipitation. Immunoprecipitations were carried out using lysates of insect cells infected with recombinant baculoviruses, either alone or in combination. Figure 4 shows that the soluble fractions of the lysates expressed similar amounts of UL12 and UL12.5, as detected by Western blotting with the anti-UL12 antibody (Fig. 4A). Likewise, cells expressing ICP8 either alone or in combination with UL12 or UL12.5 contained equal amounts of this protein (Fig. 4C). For the immunoprecipitation, the monoclonal anti-ICP8 antibody was used, and the potential interaction partners, UL12.5 and UL12, were detected by Western blot analysis of the precipitated proteins. Both UL12.5 and UL12 were coimmunoprecipitated with ICP8 (Fig. 4B, lanes 5 and 6, respectively), indicating that both UL12.5 and UL12 interact with ICP8. Figure 4D is a control showing that the same amount of ICP8 was immunoprecipitated in each lane. Therefore, the inability of UL12.5 to complement the UL12 null mutant is not due to an inability of UL12.5 to interact with ICP8. Furthermore, this experiment shows that the UL12-ICP8 interaction domain is not found in the first 126 amino acids of UL12.
FIG. 4.
Coimmunoprecipitation of UL12, UL12.5, and ICP8. UL12, UL12.5, and ICP8 were expressed in Sf21 insect cells through infection with recombinant baculoviruses. The anti-ICP8 antibody 39-S was used for immunoprecipitation of proteins from soluble cell lysates. Protein A-G agarose was used to bind the antibody-protein complexes. Soluble cell lysates (A and C) and anti-ICP8 immunoprecipitates (B and D) were separated by SDS-PAGE, blotted to nitrocellulose, and probed with anti-UL12 Bwp12 antibody (A and B) or with anti-ICP8 antibody 3-83 (C and D). Lanes 1, mock infection; lanes 2, infection with AcUL12.5; lanes 3, infection with AcAN; lanes 4, infection with AcUL29; lanes 5, infection with AcUL29 and AcUL12.5; lanes 6, infection with AcUL29 and AcAN. +, present; −, absent.
Strand exchange activity of UL12.5.
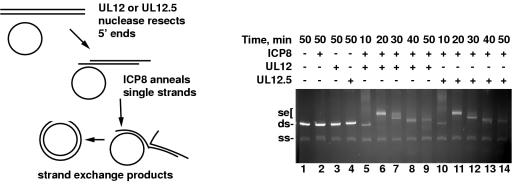
It was recently shown that UL12 and ICP8 can work together to mediate a strand exchange reaction, much like the Red system of bacteriophage lambda (26). This activity shows that these proteins function like a recombinase, and this activity could be important for the replication of HSV-1 DNA, which is associated with a high level of homologous recombination. Therefore, we tested whether UL12.5 retained the ability to mediate strand exchange. In the in vitro reaction, UL12 digests linear double-stranded M13 DNA from the 5′ ends, and ICP8 binds to the 3′ single-stranded tail and pairs it with homologous single-stranded circular M13 DNA. A schematic diagram of this assay and the expected products is shown in Fig. 5, left panel. We compared the strand exchange activity of UL12 with that of UL12.5. Figure 5 shows that UL12.5 can mediate strand exchange with ICP8 as efficiently as UL12, producing slowly migrating strand exchange products with the same kinetics seen with UL12. Although we have previously shown that UL12.5 has a weaker nuclease activity than UL12 (Fig. 2), in this assay the difference does not affect the overall strand exchange activity, probably because it is carried out with saturating amounts of UL12 and UL12.5 for the reaction. This experiment shows that the inability of UL12.5 to complement the UL12 null virus is not due to an inability to mediate strand exchange.
FIG. 5.
Strand exchange activities of UL12.5 and ICP8. (Left) Schematic representation of the strand exchange reaction. (Right) Strand exchange with UL12, UL12.5, and ICP8. Linear double-stranded and circular single-stranded M13 DNA substrates were incubated with 13.9 nM UL12 or UL12.5 and 1.75 μM ICP8 at 37°C for the times indicated. Samples were run on a 0.7% agarose gel in TAE buffer and stained with ethidium bromide. ds, double-stranded DNA; ss, single-stranded DNA; se, strand exchange products; +, present; −, absent.
Expression of UL12.5 in mammalian cells.
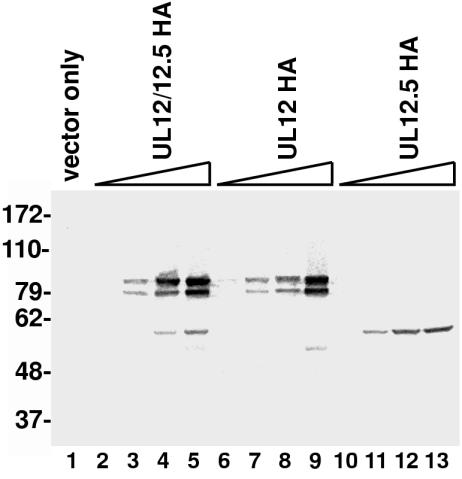
We have shown that the in vitro activities of purified UL12.5 do not differ appreciably from that of UL12. However, previous studies have shown that UL12.5 expressed from a plasmid or expressed from the ANF-1 virus is unable or only partially able to compensate for the loss of UL12 (11, 24). One possible reason for the inability of UL12.5 to complement is that the level of expression of UL12.5 was too low. The natural promoter of UL12.5 is much weaker than that of UL12, and the 1.9-kb UL12.5 transcript is transcribed threefold less efficiently than the 2.3-kb transcript for UL12 (7). The sequence upstream of the start codon of UL12.5 also was not a favored sequence for efficient translation. We therefore constructed a more efficient expression vector for UL12.5, designated pSAKUL12.5, in which the UL12.5 gene can be expressed from the CMV promoter and the translational start sequence has been optimized for translation. The wild-type UL12 gene was also cloned in this vector for direct comparison with UL12.5. Because the natural UL12.5 promoter and start site are contained within the wild-type UL12 gene, this plasmid expresses both UL12 and UL12.5, although the UL12.5 expression is much lower. The plasmid is therefore designated pSAKUL12/12.5. A third plasmid was generated in which M127 of UL12, which is the start site of the UL12.5 gene, has been changed to phenylalanine. Previous work from our laboratory has shown that this mutation eliminates the production of UL12.5 but does not appear to alter UL12 activity, as this construct is fully capable of complementing the UL12 null mutant (22). This M127F UL12 mutant plasmid is therefore called pSAKUL12. In order to improve the quantification of the amounts of expressed proteins, we also prepared vectors for the expression of the UL12 and UL12.5 proteins fused to an HA tag. This tag adds 11 residues to the C termini of the proteins and does not appear to affect the function of UL12 (see below). Since the HA-tagged UL12 and UL12.5 each have only one copy of the HA tag, using the anti-HA antibody to detect and quantify these proteins should give a more accurate assessment of the amount of the protein expressed in the cells than using the polyclonal anti-UL12 antibody, which recognizes several epitopes. Vero cells were transfected with plasmids for the expression of HA-tagged and native UL12 and UL12.5 proteins, and bands of the expected sizes were observed (Fig. 6 and data not shown). The wild-type UL12 construct, pSAKUL12/12.5HA, expresses both UL12 and UL12.5, as mentioned above, although the amount of UL12.5 produced is much smaller than that of UL12 (Fig. 6, lanes 2 to 5). The constructs pSAKUL12HA and pSAKUL12.5HA express only the UL12 or UL12.5 protein, respectively (Fig. 6, lanes 6 to 9 and 10 to 13). By increasing the amount of plasmid used for the transfection, we achieved greater levels of expression of UL12 or UL12.5. These increased amounts of protein represent true per-cell increases and not an increase in the number of transfected cells, as the percentages of UL12- and UL12.5-expressing cells were comparable, as seen by indirect IF (data not shown). Although both proteins were expressed from plasmid constructs with identical promoters and upstream sequences, the amount of soluble UL12.5 was smaller than that of UL12, with the amount of soluble UL12.5 obtained using 600 ng of plasmid being comparable to the amount of soluble UL12 obtained using 40 to 200 ng of plasmid (Fig. 6, compare lane 13 with lanes 3 and 4). Therefore, it appears that the inability of the original UL12.5 constructs to complement the AN-1 virus may have been partly due to an inability to produce sufficient quantities of soluble protein.
FIG. 6.
Expression of UL12.5 in transfected Vero cells. Vero cells were transfected with the plasmids pSAK, pSAKUL12/12.5HA, pSAKUL12HA, and pSAKUL12.5HA as described in Materials and Methods. The amounts of expressing plasmids used were 5, 40, 200, and 600 ng, although the empty parent vector pSAK was added to each transfection so that each plate was transfected with a total of 1 μg of DNA. Lane 1, control, with transfection of 1 μg of pSAK only. Cells were collected 24 h posttransfection, and extracts were analyzed by SDS-PAGE. Proteins were immunoblotted, and the anti-HA antibody sc-805 was used for detection. Numbers on left refer to kilodaltons.
Complementation of the UL12 null virus with UL12.5.
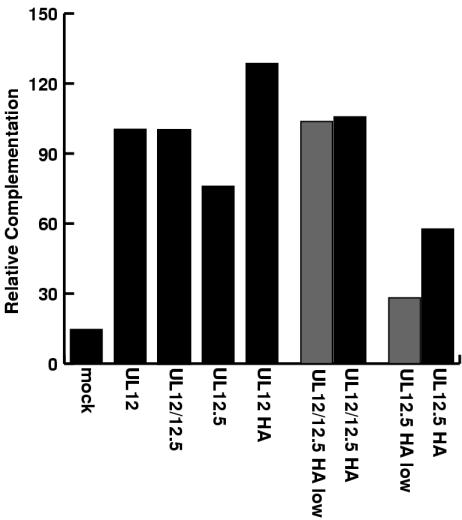
The UL12- and UL12.5-expressing plasmids were used to test complementation of the UL12 null virus, AN-1. Vero cells were transfected with the expression plasmids and were subsequently superinfected with AN-1. The supernatants were collected, and titers were determined on the complementing cell line, 6-5. Both the native and tagged versions of UL12 were able to complement the null virus, indicating that the tag did not appear to affect the activity of UL12 (Fig. 7). UL12 could fully complement AN-1, even when a small amount of plasmid (40 ng) was used. UL12.5 was also able to complement AN-1, but not to the same extent as UL12. Complementation by UL12.5 never exceeded 75%, even when 600 ng of expression plasmid was used, which produces appreciable amounts of soluble UL12.5 (Fig. 6 and 7). This indicated that the inability of UL12.5 to complement AN-1 was not due solely to the lower levels of expression of soluble UL12.5.
FIG. 7.
Complementation of the AN-1 mutant virus by transfection with UL12-expressing plasmids. Vero cells were transfected with the plasmids pSAK, pSAKUL12, pSAKUL12/12.5, pSAKUL12.5, pSAKUL12HA, pSAKUL12/12.5HA, and pSAKUL12.5HA and superinfected with the AN-1 virus 24 h posttransfection. Cell supernatants were collected 18 h later, and titers were determined on the complementing cell line 6-5. Relative complementation was calculated by setting the number of plaques obtained with 600 ng of pSAKUL12/12.5 (wild-type) transfection at 100%. Solid bars, 600 ng of plasmid; shaded bars, 40 ng of plasmid; mock, transfected with control plasmid.
Intracellular localization of UL12.5.
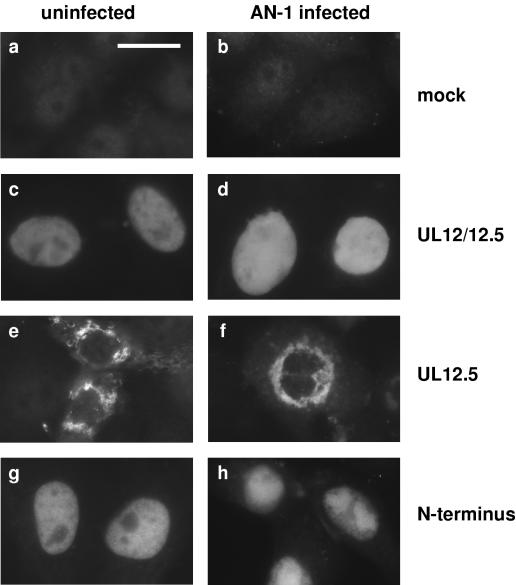
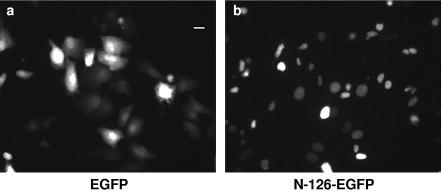
Previous studies have shown that UL12 localizes to the nucleus (34). Since HSV-1 replication occurs in the nucleus and UL12 is proposed to play a part in this process, localization to the nucleus could be an important characteristic of UL12. Because even increased levels of UL12.5 were found to be insufficient to completely complement the UL12 null virus, we hypothesized that UL12.5 may not properly localize to the nucleus. To test this possibility, we used indirect IF to determine the cellular localization of UL12 and UL12.5 in transfected cells. The results obtained with IF using both native (Fig. 8) and HA-tagged (not shown) versions of the proteins were the same. With all constructs, UL12 clearly localized to the nucleus, with or without superinfection with the UL12 null virus, AN-1 (Fig. 8C and D). In contrast, UL12.5 remained in the cytoplasm, primarily located in a ring around the nucleus, in what appeared to be aggregates (Fig. 8E and F). This is consistent with previous studies, in which UL12.5 was found to be insoluble in mammalian cells. There does appear to be some faint nuclear staining of UL12.5 in the cells transfected with 600 ng of plasmid (Fig. 8E and F), which does not appear when smaller amounts of plasmid are used (data not shown). The faint nuclear staining could be the result of nonspecific leakage of UL12.5 into the nucleus. The localization patterns are consistent with the data for complementation of the UL12 null virus shown in Fig. 7. When small amounts of plasmid were used, complementation was also low, correlating with small amounts of soluble protein and undetectable nuclear staining, but with greater amounts of plasmid, a higher level of complementation and some nuclear staining were observed. These observations explain the inability of UL12.5 to complement AN-1; at low levels of UL12.5 expression, the protein is found in the cytoplasm and is unable to participate in nuclear events, while at high levels of expression, some UL12.5 leaks into the nucleus in order to partially complement the UL12 null virus. These results also suggest that the NLS of UL12 resides in the first 126 amino acids of UL12, the region that is absent from UL12.5. PredictNLS Online was used to screen the UL12 protein sequence for possible NLSs (5). This program located a strong potential NLS at residues 35 to 39 (KRPRP) of UL12. The prediction was tested experimentally by fusing the N terminus of UL12 to GFP. The native GFP protein is distributed over the nucleus and cytoplasm of the cell (Fig. 9a), but when GFP was fused to the N terminus of UL12, the fusion protein localized strictly to the nucleus (Fig. 9b). Thus, the N-terminal region of UL12 possesses an NLS that is capable of directing GFP to the nucleus and is likely to be responsible for the nuclear localization of full-length UL12.
FIG. 8.
UL12 localizes to the nucleus and UL12.5 does not. Vero cells were transfected with 600 ng of plasmids pSAK, pSAKUL12/12.5, pSAKUL12.5, and pSAKN126 for the analysis of mock, wild-type UL12, UL12.5, and the N terminus of UL12, respectively. In the panels on the left, cells were fixed and processed for IF 24 h posttransfection. In the panels on the right, cells were superinfected with AN-1 virus 24 h posttransfection and then fixed and processed 5 h postinfection. The anti-UL12 antibody Bwp12 was used as the primary antibody, and Alexa Fluor 488-conjugated goat-anti-rabbit was used as the secondary antibody. Bar, 40 μm.
FIG. 9.
Localization of GFP fusion proteins. Vero cells were transfected with 600 ng of the plasmid pEGFP-C1 (a) or pSAKN126-GFP (b) for the expression of EGFP and the UL12-N terminus-EGFP fusion protein, respectively. The cells were fixed 24 h posttransfection. Bar, 50 μm.
DISCUSSION
The HSV-1 alkaline nuclease plays an important role in the life cycle of the virus. Although the precise role of UL12 in vivo has not yet been elucidated, the recent observation that it can function as part of a viral recombinase suggests a role for UL12 in recombination-dependent replication (26). In this paper, we have addressed the domain structure of UL12 in an attempt to correlate the functional roles played by the full-length UL12, the UL12.5 protein lacking the N-terminal 126 residues of UL12, and the N-terminal domain itself. Various activities, including exo- and endonuclease activities, ability to carry out strand exchange, interaction with ICP8, and intracellular localization, were examined in order to determine the minimal activities required for in vivo function. We conclude that the enzymatic and ICP8 interaction domains, as well as the ability to carry out strand exchange, reside in the C-terminal 500 amino acids of UL12, while the NLS lies within the N-terminal 126 amino acids.
Proline-rich domains are found in many sites of protein-protein interaction (15). The N terminus of UL12 is rich in prolines, and this suggested that the problem with UL12.5 was that it was unable to contact a protein important for its in vivo activity. Of special interest was the HSV-1 single-stranded DNA binding protein, ICP8, which had been shown to interact with UL12 (34, 36). Thomas et al. have mapped domains of ICP8 responsible for interaction with UL12. These include the region of amino acids 325 to 585, as well as the C-terminal 167 residues (34). However, the corresponding regions of UL12 responsible for interaction with ICP8 have not yet been mapped. Our study shows that the ICP8 interaction domain of UL12 is not located within the first 126 amino acids of UL12. It will be interesting to see if a mutant of UL12 can be found that is nuclease active but unable to interact with ICP8 and whether such a mutant would be able to mediate strand exchange and complement AN-1.
Although the exact in vivo role of UL12 has not yet been defined, all of the suggested possibilities for UL12 activity, from processing of replicated DNA for packaging into capsids (37) to a role in recombination associated with replication (26), take place in the nucleus. Therefore, it is expected that UL12 would need to localize to the nucleus. Thomas et al. found that UL12 localized to the nucleus even in cells infected with viruses encoding mutant forms of ICP8 which did not localize to the nucleus, implying that UL12 possesses its own NLS (34). Kehm et al. also demonstrated that UL12 and a C-terminal 123-amino-acid truncated form of UL12 both localized to the nucleus in the absence of any other herpesvirus protein (16). Here, we confirm that UL12 does possess its own NLS and that it is contained within the N-terminal 126 residues of UL12. The NLS is most likely to be amino acids 35 to 39 (KRPRP), which was identified as an NLS by the PredictNLS program (5). The EBNA-2 protein from Epstein-Barr virus and mouse polyomavirus T antigen also possess identical sequences, which have been experimentally confirmed to be functional NLSs (20, 27). Our results demonstrate experimentally that the N terminus of UL12 contains an effective NLS, as fusion of this region to GFP, which normally is distributed over the entire cell, caused GFP to localize strictly to the nucleus. The UL12.5 protein, which lacks this domain, was found to localize to the cytoplasm.
These results do not explain why HSV-1 has evolutionarily retained the ability to transcribe a subgenic version of UL12. We do know that it is not essential for viral growth in culture (22). As has been suggested previously (22), it may be required for growth in animals or for latency or it may represent an earlier version on the evolutionary path to full-length UL12. Further work will be necessary to investigate these possibilities.
Our results reveal several factors that contribute to the inability of UL12.5 to complement the UL12 null virus, AN-1. The nuclease activity of the UL12.5 protein was lower than that of UL12, possibly due to lower stability of the UL12.5 protein. The UL12.5 protein was less soluble than UL12 and tended to aggregate in vivo, lowering its effective concentration in the cell. Furthermore, in previous studies, when complementation was studied using constructs and viruses employing the natural promoter of UL12.5, the total amount of UL12.5 expressed was much smaller than that of UL12 (24). Since the natural promoter of UL12.5 provides threefold less expression of UL12.5 than UL12 (7), this probably explains our previous failure to observe complementation of AN-1 by UL12.5. These various factors all contribute to the deficiency of UL12.5. However, we find it significant that the NLS resides in the N terminus of UL12, which is not found in the N-terminally truncated UL12.5. The localization of UL12.5 in the cytoplasm, prevented by the nuclear boundary from taking part in HSV-1 replication, would logically be primarily responsible for its inability to complement AN-1.
Acknowledgments
We thank Ping Bai for her excellent technical assistance in the preparation of the UL12 and UL12.5 proteins and Joshua N. Goldstein for the preparation of the UL12 protein. We are grateful to members of our laboratory for helpful comments on the manuscript.
This work was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship, DRG-1625 (N.B.R.), and Public Health Service grants AI21747 and AI37549 (S.K.W.).
REFERENCES
- 1.Bataille, D., and A. L. Epstein. 1995. Herpes simplex virus type 1 replication and recombination. Biochimie 77:787-795. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Bronstein, J. C., and P. C. Weber. 1996. Purification and characterization of herpes simplex virus type 1 alkaline exonuclease expressed in Escherichia coli. J. Virol. 70:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronstein, J. C., S. K. Weller, and P. C. Weber. 1997. The product of the UL12.5 gene of herpes simplex virus type 1 is a capsid-associated nuclease. J. Virol. 71:3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa, R. H., K. G. Draper, L. Banks, K. L. Powell, G. Cohen, R. Eisenberg, and E. K. Wagner. 1983. High-resolution characterization of herpes simplex virus type 1 transcripts encoding alkaline exonuclease and a 50,000-dalton protein tentatively identified as a capsid protein. J. Virol. 48:591-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draper, K. G., G. Devi-Rao, R. H. Costa, E. D. Blair, R. L. Thompson, and E. K. Wagner. 1986. Characterization of the genes encoding herpes simplex virus type 1 and type 2 alkaline exonucleases and overlapping proteins. J. Virol. 57:1023-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutch, R. E., V. Bianchi, and I. R. Lehman. 1995. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J. Virol. 69:3084-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutch, R. E., R. C. Bruckner, E. S. Mocarski, and I. R. Lehman. 1992. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J. Virol. 66:277-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein, J. N., and S. K. Weller. 1998. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology 244:442-457. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, J. O., L. J. Ball-Goodrich, and D. S. Parris. 1998. Structure-function analysis of the herpes simplex virus type 1 UL12 gene: correlation of deoxyribonuclease activity in vitro with replication function. Virology 243:247-259. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, P. J. 1981. Mechanism of degradation of duplex DNA by the DNase induced by herpes simplex virus. J. Virol. 38:1005-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann, P. J., and Y.-C. Cheng. 1978. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. J. Biol. Chem. 253:3557-3562. [PubMed] [Google Scholar]
- 14.Hoffmann, P. J., and Y.-C. Cheng. 1979. DNase induced after infection of KB cells by herpes simplex virus type 1 or type 2. J. Virol. 32:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 16.Kehm, E., M. Goksu, S. Bayer, and C. W. Knopf. 1998. Herpes simplex virus type 1 DNase: functional analysis of the enzyme expressed by recombinant baculovirus. Intervirology 41:110-119. [DOI] [PubMed] [Google Scholar]
- 17.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knopf, C. W., and K. Weisshart. 1990. Comparison of exonucleolytic activities of herpes simplex virus type-1 DNA polymerase and DNase. Eur. J. Biochem. 191:263-273. [DOI] [PubMed] [Google Scholar]
- 19.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 20.Ling, P. D., J. J. Ryon, and S. D. Hayward. 1993. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J. Virol. 67:2990-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Martinez, R., J. N. Goldstein, and S. K. Weller. 2002. The product of the UL12.5 gene of herpes simplex virus type 1 is not essential for lytic viral growth and is not specifically associated with capsids. Virology 298:248-257. [DOI] [PubMed] [Google Scholar]
- 23.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez, R., L. Shao, J. C. Bronstein, P. C. Weber, and S. K. Weller. 1996. The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology 215:152-164. [DOI] [PubMed] [Google Scholar]
- 25.McGeoch, D. J., A. Dolan, and M. C. Frame. 1986. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the exonuclease gene and neighbouring genes. Nucleic Acids Res. 14:3435-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuven, N. B., A. E. Staire, R. S. Myers, and S. K. Weller. 2003. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 77:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson, W. D., B. L. Roberts, and A. E. Smith. 1986. Nuclear location signals in polyoma virus large-T. Cell 44:77-85. [DOI] [PubMed] [Google Scholar]
- 28.Schaffer, P. A., M. J. Tevethia, and M. Benyesh-Melnick. 1974. Recombination between temperature-sensitive mutants of herpes simplex virus type 1. Virology 58:219-228. [DOI] [PubMed] [Google Scholar]
- 29.Severini, A., D. G. Scraba, and D. L. Tyrrell. 1996. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 70:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao, L., L. M. Rapp, and S. K. Weller. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196:146-162. [DOI] [PubMed] [Google Scholar]
- 31.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stow, N. D. 1992. Herpes simplex virus type 1 origin-dependent DNA replication in insect cells using recombinant baculoviruses. J. Gen. Virol. 73:313-321. [DOI] [PubMed] [Google Scholar]
- 33.Strobel-Fidler, M., and B. Francke. 1980. Alkaline deoxyribonuclease induced by herpes simplex virus type 1: composition and properties of the purified enzyme. Virology 103:493-501. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, M. S., M. Gao, D. M. Knipe, and K. L. Powell. 1992. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J. Virol. 66:1152-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umene, K. 1985. Intermolecular recombination of the herpes simplex virus type 1 genome analysed using two strains differing in restriction enzyme cleavage sites. J. Gen. Virol. 66:2659-2670. [DOI] [PubMed] [Google Scholar]
- 36.Vaughan, P. J., L. M. Banks, D. J. Purifoy, and K. L. Powell. 1984. Interactions between herpes simplex virus DNA-binding proteins. J. Gen. Virol. 65:2033-2041. [DOI] [PubMed] [Google Scholar]
- 37.Weller, S. K., M. R. Seghatoleslami, L. Shao, D. Rowse, and E. P. Carmichael. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 71:2941-2952. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson, D. E., and S. K. Weller. 2003. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life 55:451-458. [DOI] [PubMed] [Google Scholar]