Abstract
Haemophilus influenzae is a Gram negative bacterium that belongs to the family Pasteurellaceae, causes bacteremia, pneumonia and acute bacterial meningitis in infants. The emergence of multi-drug resistance H. influenzae strain in clinical isolates demands the development of better/new drugs against this pathogen. Our study combines a number of bioinformatics tools for function predictions of previously not assigned proteins in the genome of H. influenzae. This genome was extensively analyzed and found 1,657 functional proteins in which function of 429 proteins are unknown, termed as hypothetical proteins (HPs). Amino acid sequences of all 429 HPs were extensively annotated and we successfully assigned the function to 296 HPs with high confidence. We also characterized the function of 124 HPs precisely, but with less confidence. We believed that sequence of a protein can be used as a framework to explain known functional properties. Here we have combined the latest versions of protein family databases, protein motifs, intrinsic features from the amino acid sequence, pathway and genome context methods to assign a precise function to hypothetical proteins for which no experimental information is available. We found these HPs belong to various classes of proteins such as enzymes, transporters, carriers, receptors, signal transducers, binding proteins, virulence and other proteins. The outcome of this work will be helpful for a better understanding of the mechanism of pathogenesis and in finding novel therapeutic targets for H. influenzae.
Introduction
Haemophilus influenzae strain Rd KW20 is a Gram-negative bacterium frequently isolated from the lower respiratory tract of patients with chronic bronchitis [1], [2] which is the “fourth-most-common” cause of death in the United States [1]. Due to comparatively small genome size and its phylogenetic closeness to Escherichia coli, H. influenzae is a very convenient model organism for genomic and proteomic findings [3], [4], [5]. The genome of H. influenzae was successfully sequenced [6], and it consists of 1,830,140 base pairs in a single circular chromosome that contains 1740 protein-coding genes, 2 transfer RNA genes, and 18 other RNA genes [6]. Due to successful sequencing of whole genome, H. influenzae serve as a model organism for whole-genome annotation, computational analysis and cross-genome comparisons [7]. Furthermore, genome-scale model of metabolic fluxes construction [8], [9], [10] and whole-genome transposon mutagenesis analysis [11], [12] was first implemented in H. influenzae. Moreover, in this study it is also used as a test genome to evaluate the performance of various bioinformatics approaches for proteome analysis, with the ultimate aim of determining the in silico properties of the protein set expressed by the bacterium under certain conditions.
Genomic analysis of 102 bacterial genomes shows that the respective genomic pool contain 45,110 proteins organized in 7853 orthologous groups with unknown function [13]. Proteins with unknown function may be termed as Hypothetical Proteins (HPs) or putative conserved proteins because these proteins are showing limited correlation to known annotated proteins [14], [15]. The HPs have not been functionally characterized and described at biochemical and physiological level [15]. Nearly half of the proteins in most genomes belong to HPs, and this class of proteins presumably have their own importance to complete genomic and proteomic information [16], [17]. We have been working on structure based rational drug design where we always need a selective target for drug design [18], [19], [20]. A precise annotation of HPs of particular genome leads to the discovery of new structures as well as new functions, and helps in bringing out a list of additional protein pathways and cascades, thus completing our fragmentary knowledge on the mosaic of proteins [17]. Furthermore, novel HPs may also serve as markers and pharmacological targets for drug design, discovery and screen [21], [22].
The use of advanced bioinformatics tools for sequence analysis and comparison is an initial step to identify homologue for only a part of the region shared between proteins, which could lead to a robust function prediction. Most commonly used method for functional prediction of gene products is by identification of related well-characterized homologues using sequence-based search procedures such as BLAST [23]. Multiple sequence alignment of homologues of a family is a suitable method to obtain structurally/functionally important positions and structurally conserved domains. We have considered functional domains as the basis to infer the biological role of HPs. Motif analysis is an obligatory step in the identification and characterization of HPs. Detection of common motifs among proteins in particular with absent or low sequence identities (e.g. less than 30%) may provide important clues for function or classification of HPs into appropriate families [24]. A series of signature databases are publically available, and are used for motif finding including GenomeNet [25] (contains PROSITE [26], PRINTS [27], Pfam [28], ProDom [29], BLOCKS [30]) and InterPro [31] using InterProScan [32]. A potent method for motif searches represents the use of MEME suite [33], a resource for investigating candidate's functional and structural motifs/sites in HPs ( Table 1 ). Furthermore, study of protein interactions using STRING database [34] is crucial to understand the functional role of individual proteins in a well-organized biological network.
Table 1. List of bioinformatics tools and databases used for sequence based function annotation.
| S. No. | Software name | URL | Remark |
| 1) Sequence similarity search | |||
| 1. | BLAST: Basic Local Alignment Search Tool | http://www.ncbi.nlm.nih.gov/BLAST/ | BLASTp is used for finding similar sequences in protein databases |
| 2. | HHpred | ftp://toolkit.genzentrum.lmu.de/pub/HH-suite/ | Protein homology detection by HMM-HMM comparison |
| 2) Physicochemical characterization | |||
| 3. | ExPASy – ProtParam tool | http://web.expasy.org/protparam/ | Used for computation of various physical and chemical parameters |
| 3) Sub-cellular localization | |||
| 4. | PSORT B | http://www.psort.org/psortb | PSORTb attained an overall precision of 97% |
| 5. | PSLpred | http://www.imtech.res.in/raghava/pslpred/ | The overall accuracy of PSLpred is 91.2%. |
| 6. | CELLO | http://cello.life.nctu.edu.tw | The overall accuracy of CELLO is 91%. |
| 7. | SignalP | http://www.cbs.dtu.dk/services/SignalP/ | Predict signal peptide cleavage sites |
| 8. | SecretomeP | http://www.cbs.dtu.dk/services/SecretomeP/ | Predict bacterial non-classical secretion |
| 9. | TMHMM | http://www.cbs.dtu.dk/services/TMHMM/. | Predict membrane topology |
| 10. | HMMTOP | http://www.enzim.hu/hmmtop/ | Predict transmembrane topology |
| 4) Sequence alignment | |||
| 11. | PRALINE (PRofile ALIgNEment) | http://ibivu.cs.vu.nl/programs/pralinewww/ | Integrates homology-extended and secondary structure information for multiple sequence alignment |
| 5) Protein classification | |||
| 12. | Pfam | http://pfam.sanger.ac.uk/. | Collection of multiple protein-sequence alignments and HMMs |
| 13. | CATH (Class, Architecture, Topology, Homology) | http://www.cathdb.info/ | Hierarchical domain classification of PDB structures |
| 14. | SUPERFAMILY | http://supfam.cs.bris.ac.uk/SUPERFAMILY | Based on SCOP database |
| 15. | SYSTERS | http://systers.molgen.mpg.de | - |
| 16. | SVMProt | http://jing.cz3.nus.edu.sg/cgi-bin/svmprot.cgi. | SVM based classification with accuracy of 69.1–99.6% |
| 17. | CDART (The Conserved Domain Architecture Retrieval Tool) | http://www.ncbi.nlm.nih. gov/Structure/Lexington/Lexington.cgi. | NCBI Entrez Protein Database search of domain architecture |
| 18. | PANTHER (Protein Analysis THrough Evolutionary Relationships) | http://www.pantherdb.org | Classification based on HMM-HMM search |
| 19. | ProtoNet | http://www.protonet.cs.huji.ac.il | Based on automatic hierarchical clustering of the protein sequences |
| 20. | SMART (Simple Modular Architecture Research Tool) | http://smart.embl.de/ | Identification and annotation of protein domains |
| 6) Motif Discovery | |||
| 21. | InterProScan | http://www.ebi.ac.uk/InterProScan/ | Searches InterPro for motif discovery |
| 22. | MOTIF | http://www.genome.jp/tools/motif/ | Japanese GenomeNet service for motif discovery |
| 23. | MEME Suite | http://meme.nbcr.net | - |
| 7) Clustering | |||
| 24. | CLUSS | http://prospectus.usherbrooke.ca/cluss/ | Clustering on the basis of Substitution Matching Similarity (SMS) |
| 8) Virulence factor analysis | |||
| 25. | VirulentPred | http://bioinfo.icgeb.res.in/virulent/ | Accomplish an accuracy of 81.8% |
| 26. | VICMpred | http://www.imtech.res.in/raghava/vicmpred/ | Attain accuracy of 70.75%. |
| 9) Protein-protein interaction | |||
| 27. | STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) | http://string-db.org | Version –9.05 |
Here we have used recent bioinformatics tools to assign function to all HPs encoded by H. influenzae genome. The Receiver Operating Characteristic (ROC) analysis [35] is used for evaluating the performance of used bioinformatics tools. We also measured the confidence level of the function prediction on the basis of used bioinformatics tools [36]. The function prediction has high confidence level if more than three tools indicate the same functions. While if there is less than three tools then it is less confidently predicted function [36]. So, we have successfully assigned functions to all 296 HPs of H. influenzae genome with high confidence. We have performed an extensive sequence analysis of proteins associated with virulence using tools like Virulentpred [37] and VICMpred [38], because H. influenzae is the causative agent of infection in respiratory tract.
Materials and Methods
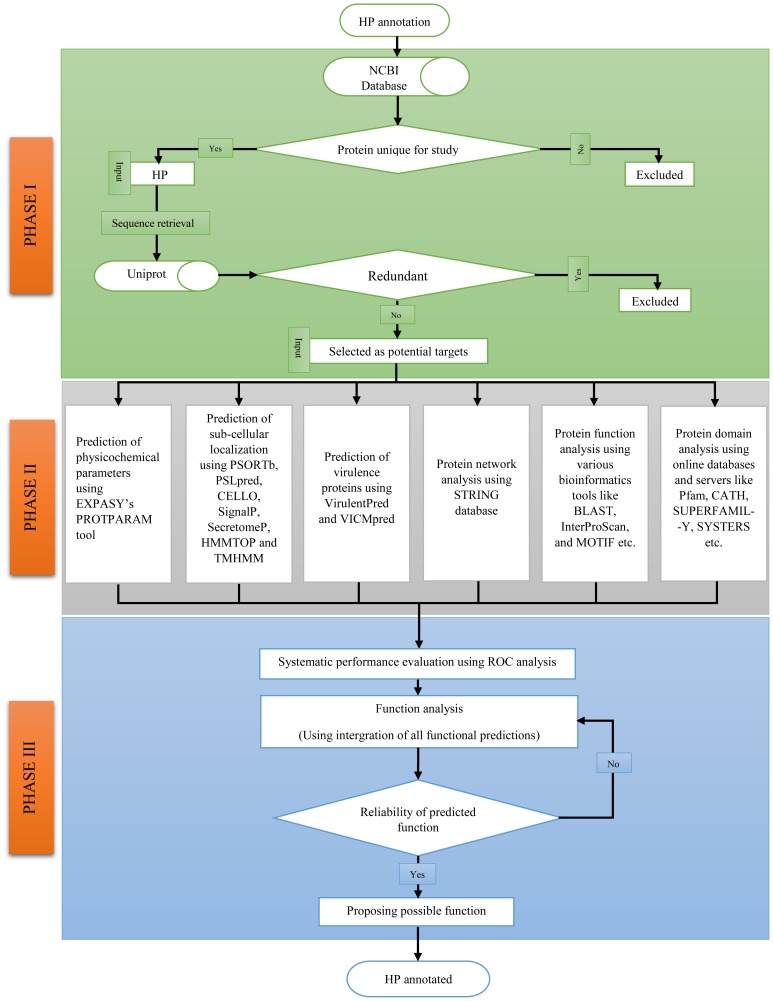
The computational framework used for functional annotation of HPs is given in Figure 1 , is divided into three phases namely, Phase I, II and III. The Phase I include the characterization and sequence retrieval of HPs by analyzing the genome of H. influenzae. The Phase II comprises the automated annotation of various functional parameters using various online servers. In Phase III, the systematic performance evaluation of various bioinformatics tools by using H. influenzae protein sequences with known function by performing ROC analysis. The probable functions of the characterized HPs were predicted by the integration of various functional predictions made in PHASE II. In latter phase expert knowledge is used for performing ROC analysis and for confidently annotating the HPs functional properties.
Figure 1. Computational framework used for annotating function of 429 HPs from H. influenzae.
Methodology is divided into three phases: PHASE I. H. influenzae HP characterization and sequence retrieval from online databases. PHASE II. The extensive analysis of sub-cellular localization, physicochemical parameters, virulence, function and domain present in HPs. PHASE III. This phase include assessment of predicted functions using the protein with known function from H. influenzae and reliable prediction of possible functions of HPs.
Sequence retrieval
We have analyzed the genome of H. influenzae and found 1,657 proteins present in it (http://www.ncbi.nlm.nih.gov/genome/). The 429 proteins are characterized as HPs and their fasta sequences were retrieved from UniProt (http://www.uniprot.org/) using the primary accession number of all HPs.
Physicochemical characterization
Expasy's ProtParam server [39] has been used for theoretical measurements of physiochemical properties such as molecular weight, isoelectric point, extinction coefficient [40], instability index [41], aliphatic index [42] and grand average of hydropathicity (GRAVY) [43]. These predicted parameters are listed in Table S1.
Sub-cellular localization
A protein can be characterized as drug or vaccine target by utilizing the knowledge of sub-cellular localization. The proteins localized in cytoplasm can act as possible drug targets, while surface membrane proteins are considered as potent vaccine targets [44]. Databases like UniProt provide valuable information about sub-cellular location of proteins [45]. If experimental information about HP localization is absent, then we have used sub-cellular localization prediction tools like PSORTb [46], PSLpred [47] and CELLO [48], [49]. CELLO (version 2.0) two-level support vector machine based system, which comprises 1444 and 7589 protein sequences as standard datasets for the prediction of bacterial and eukaryotic protein localization, respectively [48], [49]. The PSLpred is used only for predicting sub-cellular localization of Gram negative bacteria. We have used SignalP 4.1 [50] for predicting signal peptide and SecretomeP [51] for identifying protein involvement in non-classical secretory pathway. TMHMM [52] and HMMTOP [53] have been used for predicting the propensity of a protein to be a membrane protein. The sub-cellular localization predictions of 429 HPs are listed in Table S2.
Sequence comparisons
The first step towards predicting the functionality of a protein is generally a sequence similarity search in various available gene and protein databases. We have used BLASTp [23] and HHpred [54] for searching similar sequences with known function. BLAST is a popular bioinformatics tool, most frequently used for calculating sequence similarity by performing local alignments. The BLASTp search against the non-redundant protein sequences (nr) database returns 100 homologs of each HP, and proteins with low query coverage (<50%) or low sequence identity (<20%) are excluded. Proteins showing high sequence identities (>40%) and e-value (<0.005) are referred to as close homologs of HPs and those with low identities (<26%) are considered as remote homologues. The search with the highest value of the respective parameters considered as probable function of the given HP. The BLASTp also used for checking the availability of structural homologs in Protein Data Bank (PDB). Whereas, HHpred utilizes pair wise comparison of profile hidden Markov models (HMMs) for remote protein homology detection by searching various protein databases like PDB [55], [56], SCOP [57], CATH [58], etc. is also used for detection of structural homologs. We have used BLASTp for determining the sequence identity between two proteins sequences and PRALINE [59] for multiple sequences comparison (Table S3).
Function prediction
We have used various tools for precise functional assignments to all 429 HPs from H. influenzae are described in Table 1 . The functional domain of a protein is predicted by using various publically available databases such as Pfam, SUPERFAMILY [60], CATH, PANTHER [61], SYSTERS [62], SVMProt [63], CDART [64], SMART [65], and ProtoNet [66] (Table S4). The database SYSTERS was used for clustering proteins on the basis of their functions. We used BLASTp for searching SYSTERS database and the output is obtained in the form of clusters of functionally related proteins. The clusters with e-value (<0.005) are considered as a proper classification of HP. SVMProt was used for the SVM based classification of proteins into 54 functional families from its primary sequences. The significance level of classification is measured in the form of R-value and P-value (%), classification with R-value (>2.0) and P-value (>60%) are considered as significant. CDART and SMART were used for similarity search based on domain architecture and profiles rather than by direct sequence similarity. The Simple modular architecture research tool (SMART) search for similar domain in Swiss-Prot [67], SP-TrEMBL [68] and stable Ensembl [69] proteomes in normal mode. The search with e-value (<0.005) was considered as a significant match for the given HP.
Similarly, PANTHER is a comprehensively organized database of protein families, trees and subfamilies, used to develop evolutionary relationships to infer the functions of HPs. The HMM- based search is performed on PANTHER database for functional annotation of HPs and important hits with e-value greater than 1e-3 are reported in the output. ProtoNet (Version 6.0) tree provided an automatic hierarchical clustering of the protein sequences. The “Classify your protein” option in ProtoNet is used for assignment of a biological function to HPs.
Protein sequence motifs are signatures of protein families and can often be used as tools for the prediction of protein function, particularly in enzymes, in which motifs are associated with catalytic functions. We used InterProScan which combines different protein signature recognition methods from the InterPro consortium which is the integration of several large databases, including PANTHER, Pfam, SMART, ProSite and SUPERFAMILY etc. for motif discovery. The output generated by InterProScan is presented in the form of the checksum of the protein sequence which is supposed to be unique, e-value of the match which should be less than 0.005 and status of the match in the form of true (T) or unknown (?), indicative of reliability of the generated result. The MOTIF and MEME suite have been used to perform motif- sequence database searching and assignment of function. The MOTIF tool generates a very large set of output and to identify the probable function of the HP we check whether the SCOP database predicted fold in HP is also present in the MOTIF generated functional annotations. While in motif discovery using MEME suite we first cluster the protein sequences of HPs into clusters using CLUSS [70], [71] online server and then submit the clustered sequences in the MEME suite server. MEME suite server identified three motif sites in the clustered HPs by default. The MAST [33] module of MEME suite then perform database searching for assigning function to the discovered motifs in the HPs.
Virulence factors analysis
Virulence factors (VFs) are described as potent targets for developing drugs because it is essential for the severity of infection [72]. For identifying these VFs we have used VICMpred and Virulentpred. Both are SVM based method to predict bacterial VFs from protein sequences with an accuracy of 70.75% and 81.8%, respectively. Both methods use five-fold cross-validation technique for the evaluation of various prediction strategies.
Functional protein association networks
The function and activity of a protein are often modulated by other proteins with which it interacts. Therefore, understanding of protein-protein interactions serve as valuable information for predicting the function of a protein. We have used STRING (version–9.05) [34] to predict protein interactions partners of HPs. The interactions include direct (physical) and indirect (functional) associations, experimental or co-expression. STRING quantitatively integrates interaction data from these sources for a large number of organisms, and transfers information between these organisms wherever applicable.
Performance assessment
The statistical estimation of diagnostic accuracy is considered as an important step towards the validation of the predicted outcome of the adopted pipeline [73]. There are various available conventional methods for comparing the accuracy of various predicted models but ROC analysis is an extensively used method for analyzing and comparing the diagnostic accuracy [74], provides the most comprehensive explanation of diagnostic accuracy available till date [74]. We used six levels at which diagnostic efficacy can be evaluated. The two binary numerals “0” or “1” used to classify the prediction as true positive (“1”) or true negative (“0”). The integers (2, 3, 4 and 5) are used as confidence rating for each case. The ROC analysis is carried out for sequences of 100 proteins with known function from H. influenzae. We used the above explained in silico pipeline for the function prediction these known proteins using various online bioinformatics tools. We further classified the predicted function of proteins using already known function (Table S5 and S6). The classification results are submitted to “ROC Analysis: Web-based Calculator for ROC Curves” [75] in format 1 form as required by the software. This online software automatically calculates the ROC using the submitted data and generates the result in the form of accuracy, sensitivity, specificity and the ROC area. These generated parameters are utilized for validating the predicted functions of HPs. The average accuracy of used pipeline is 96.25% (Table S7) and indicates that outcomes of functional annotation of HPs are reliable that can be further utilized for other experimental research.
Results and Discussion
Sequence analysis
We have extensively analyzed sequences of 429 HPs using BLAST, Pfam, PANTHER, CATH, CDART, and SVMProt. Tools like InterProScan, MOTIF, and MEME suite were used for discovering functional motifs in the HPs. We have successfully assigned a proposed function to each of 429 HPs present in H. influenzae (Table S3 and Table S4) and discovered motif in 420 HPs using MEME suite using 208 predicted clusters of CLUSS [70], [71] online software tool (Table S8), among which 296 HPs are characterized with high confidence and are listed in Table 2 , and less confident annotated proteins are listed in Table S9. All sequence analyses were compiled. It was observed that in HPs present in H. influenzae, there are 139 enzymes, 57 transporters, 32 binding proteins, 21 bacteriophage related proteins, 15 lipoproteins and the rest are involved in various cellular process like transcription, translation, replication, etc. ( Figure 2 ). These analyses suggest a possible role of HPs in the development and pathogenesis of the organism, and identified groups are described here separately.
Table 2. List of annotated HPs from H. influenzae.
| S. NO. | PROTEIN NAME | GENE ID | UNIPROT ID | Protein Function |
| 1. | HP HI0020 | 950917 | Q57048 | Sodium/sulphate symporter |
| 2. | HP HI0034 | 950928 | P44471 | Protein Iojap ribosomal silencing factor RsfS |
| 3. | HP HI0035 | 950933 | P44472 | K+ uptake protein TrkA |
| 4. | HP HI0044 | 950935 | P44477 | Bax inhibitor-1 like protein |
| 5. | HP HI0051 | 950946 | P44484 | TRAP-type transporter system, small permease component |
| 6. | HP HI0052 | 950947 | P71336 | TRAP type C4 dicarboxylate transport system, periplasmic component |
| 7 | HP HI0056 | 950954 | P43932 | Integral membrane protein TerC |
| 8. | HP HI0065 | 950963 | P44492 | P-loop containing nucleoside triphosphate hydrolases |
| 9. | HP HI0077 | 950975 | P43935 | Ferritin- like protein |
| 10. | HP HI0080 | 950976 | P43936 | PemK-like family protein |
| 11. | HP HI0081 | 950980 | P44500 | TatD related DNase |
| 12. | HP HI0082 | 950979 | P43937 | Acyl-CoA dehydrogenase |
| 13. | HP HI0090 | 950992 | P44506 | Alanine racemase |
| 14. | HP HI0091 | 950989 | P44507 | Glycerate kinase |
| 15. | HP HI0092 | 950987 | Q57493 | Gluconate transporter |
| 16. | HP HI0093 | 950994 | P44509 | Putative sugar diacid recognition |
| 17. | HP HI0094 | 950995 | P43939 | GntP family permease |
| 18. | HP HI0095 | 950997 | Q57060 | Methyltransferase type II |
| 19. | HP HI0103 | 951002 | P44515 | Arsenate reductase (ArsC protein) |
| 20. | HP HI0105 | 951007 | Q57354 | NIF3-like protein (metal-binding protein) |
| 21. | HP HI0112 | 951016 | P71339 | Transposase |
| 22. | HP HI0118 | 951021 | Q57097 | Ubiquitin activating enzyme |
| 23. | HP HI0125 | 951038 | P44530 | xanthine/uracil/vitamin C permease |
| 24. | HP HI0134 | 951034 | P43952 | sugar transporter (AsmA-like C-terminal domain protein) |
| 25. | HP HI0143 | 951052 | P44540 | HTH-type transcriptional regulator |
| 26. | HP HI0146 | 951056 | P44542 | sialic acid transporter, TRAP-type C4-dicarboxylate transport system, periplasmic component |
| 27. | HP HI0147 | 951057 | P44543 | C4-dicarboxylate ABC transporter permease |
| 28. | HP HI0149 | 951059 | P43953 | protein-S-isoprenylcysteinemethyltransferase |
| 29. | HP HI0150 | 951060 | P44545 | Band 7 protein/HflC protease |
| 30. | HP HI0152 | 951063 | P43954 | 4′-phosphopantetheinyl transferase |
| 31. | HP HI0175 | 951085 | P44552 | multi-copper polyphenol oxidoreductase laccase |
| 32. | HP HI0177 | 951089 | P44553 | Tetratricopeptide repeat like |
| 33. | HP HI0178 | 951088 | P43961 | Prokaryotic membrane protein lipid attachment site profile |
| 34. | HP HI0217 | 951128 | P43965 | transposase IS200-family protein |
| 35. | HP HI0220.2 | 951123 | O86222 | Uracil-DNA glycosylase |
| 36. | HP HI0223 | 951139 | P44579 | DMT superfamily drug/metabolite transporter RarD |
| 37. | HP HI0228 | 951145 | P43966 | glycosyltransferase family 8 |
| 38. | HP HI0242 | 949384 | P44593 | SulfurtransferaseTusA family |
| 39. | HP HI0243 | 949380 | P43971 | Hemerythrin HHE cation binding domain protein |
| 40. | HP HI0246 | 949373 | P43972 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 41. | HP HI0257 | 949379 | P71346 | S30EA ribosomal protein/Sigma 54 modulation protein |
| 42. | HP HI0270 | 950625 | P44606 | tRNA-dihydrouridine synthase C |
| 43. | HP HI0275 | 949970 | P43975 | Sulphatases EC 3.1.6. |
| 44. | HP HI0277 | 949404 | P44609 | SEC-C motif domain-containing protein |
| 45. | HP HI0315 | 949441 | P44634 | DNA-binding regulatory protein, YebC |
| 46. | HP HI0318 | 949431 | P43984 | isoprenylcysteine carboxyl methyltransferase family protein |
| 47. | HP HI0325 | 950706 | P44640 | sodium:protonantiporter |
| 48. | HP HI0326 | 949439 | P43987 | primosomal replication protein N |
| 49. | HP HI0329 | 949459 | P44641 | Lysine 2,3-aminomutase |
| 50. | HP HI0352 | 949950 | P24324 | CMP-neu5Ac-lipooligosaccharide alpha 2–3 sialyltransferase |
| 51. | HP HI0367 | 949469 | Q57065 | transcriptional regulator with an N-terminal xre-type HTH domain |
| 52. | HP HI0370 | 949833 | P43989 | TPR-like (Tetratricopeptide repeat) |
| 53. | HP HI0371 | 949472 | P44668 | Fe-S cluster related protein IscX |
| 54. | HP HI0374 | 950642 | P44670 | histidyl-tRNA synthetase |
| 55. | HP HI0376 | 950630 | P44672 | iron-binding protein IscA |
| 56. | HP HI0379 | 949480 | P44675 | Rrf2 family transcriptional regulator |
| 57. | HP HI0380 | 949482 | P44676 | tRNA/rRNAmethyltransferase |
| 58. | HP HI0386 | 950554 | P44679 | acyl-CoA thioesterase |
| 59. | HP HI0388 | 950019 | P43990 | O-Sialoglycoproteinendopeptidase |
| 60. | HP HI0391 | 949488 | P43992 | Rhamnogalacturonanacetylesterase -like domain family protein |
| 61. | HP HI0395 | 949524 | P43994 | RnfH family Ubiquitin |
| 62. | HP HI0396 | 950708 | P44683 | RmlC-like cupins |
| 63. | HP HI0398 | 949499 | P44684 | ADP-ribose pyrophosphatase |
| 64. | HP HI0407 | 949507 | P44691 | ABC transporter involved in vitamin B12 uptake, BtuC family protein |
| 65. | HP HI0409 | 949412 | P44693 | Endopeptidases (Peptidase, M23/M37 family) |
| 66. | HP HI0414 | 949402 | Q57392 | Porin, opacity type |
| 67. | HP HI0420 | 949520 | P43995 | Ribbon-helix-helix superfamily protein |
| 68. | HP HI0423 | 949527 | P44702 | tRNA (adenine-N6)-methyltransferase |
| 69. | HP HI0441 | 949523 | P31777 | S-adenosyl-L-methionine-dependent methyltransferases |
| 70. | HP HI0442 | 950773 | P44711 | YbaB/EbfC DNA-binding protein |
| 71. | HP HI0449 | 949746 | P43997 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 72. | HP HI0452 | 949660 | P44717 | cystathionine-beta-synthase CBS domain protein |
| 73. | HP HI0454 | 949545 | P44718 | TatD type deoxyribonuclease |
| 74. | HP HI0457 | 950653 | P44720 | aminodeoxychorismate lyase |
| 75. | HP HI0466 | 949552 | P44000 | Aminomethyltransferase folate-binding domain family protein |
| 76. | HP HI0467 | 949553 | P44726 | YICC alpha Helix stress-induced protein |
| 77. | HP HI0487 | 950695 | P44003 | PTS-regulatory domain, PRD |
| 78. | HP HI0489 | 949626 | P44005 | SNARE associated Golgi protein |
| 79. | HP HI0493 | 949783 | O05023 | Transposase/integrase |
| 80. | HP HI0500 | 949635 | P44733 | DNA recombination protein RmuC |
| 81. | HP HI0510 | 949577 | P44740 | tRNA (adenine(37)-N6)-methyltransferase |
| 82. | HP HI0520 | 949583 | P44743 | Radical SAM protein |
| 83. | HP HI0521 | 950665 | P44744 | glycine radical enzyme, YjjI family |
| 84. | HP HI0526 | 949589 | P44012 | Ribonuclease T2 |
| 85. | HP HI0552 | 949603 | P44013 | Glucose-6-phosphate 1-dehydrogenase |
| 86. | HP HI0554 | 949606 | P44014 | Transposase IS200-like |
| 87. | HP HI0561 | 950224 | P44016 | oligopeptide transporter, OPT family |
| 88. | HP HI0562 | 949610 | P44754 | S4 RNA-binding domain |
| 89. | HP HI0573 | 949619 | P44759 | DNA-binding domain/SlyX like |
| 90. | HP HI0575 | 950683 | P44761 | YheO DNA-binding (transcription regulator) |
| 91. | HP HI0577 | 949622 | P44017 | SulfurtransferaseTusD -like domain family protein |
| 92. | HP HI0585 | 949628 | P44018 | C4-dicarboxylate anaerobic carrier |
| 93. | HP HI0586 | 950596 | P44019 | C4-dicarboxylate anaerobic carrier |
| 94. | HP HI0594 | 949632 | P44023 | C4-dicarboxylate anaerobic carrier |
| 95. | HP HI0597 | 950123 | P44771 | Cof protein like hydrolase |
| 96. | HP HI0617 | 950684 | P44782 | 23S rRNA/tRNApseudouridine synthase A |
| 97. | HP HI0627 | 950813 | P44025 | Succinate dehydrogenase assembly factor 2, -like domain family |
| 98. | HP HI0633 | 950781 | P44026 | Voltage gated chloride channel |
| 99. | HP HI0638 | 950538 | P44796 | High frequency lysogenization protein HflD |
| 100. | HP HI0650 | 949696 | P44028 | Prokaryotic membrane lipoprotein lipid attachment site profile protein |
| 101. | HP HI0656 | 950161 | P44807 | tRNAthreonylcarbamoyladenosine biosynthesis protein RimN |
| 102. | HP HI0656.1 | 949423 | P46494 | Topoisomerase DNA binding C4 zinc finger |
| 103. | HP HI0660 | 950644 | P44031 | Phage derived protein Gp49-like |
| 104. | HP HI0665 | 949704 | P44033 | HipA-like N-terminal domain |
| 105. | HP HI0666 | 949708 | P44034 | HipA-like N-terminal |
| 106. | HP HI0666.1 | 949707 | O86228 | HTH-type transcriptional regulator |
| 107. | HP HI0668 | 949710 | P44812 | cell division protein ZapB |
| 108. | HP HI0677 | 950735 | P44036 | N-acetyl transferase, NAT family |
| 109. | HP HI0687 | 949720 | P71356 | Multidrug resistance efflux transporter EmrE family |
| 110 | HP HI0694 | 950211 | P44827 | ribosomal large subunit pseudouridine synthase E |
| 111. | HP HI0698 | 950204 | P44038 | bacterial surface antigen protein |
| 112. | HP HI0700 | 949725 | P44831 | Regulator of ribonuclease activity B |
| 113. | HP HI0704 | 949730 | P44040 | outer membrane antigenic lipoprotein B |
| 114. | HP HI0710 | 950711 | P71357 | bifunctional antitoxin/transcriptional repressor RelB |
| 115. | HP HI0711 | 949734 | P44041 | Plasmid stabilisation system protein RelE/ParE |
| 116 | HP HI0719 | 949739 | P44839 | Endoribonuclease L-PSP |
| 117. | HP HI0722 | 949742 | P44842 | Translation elongation factor EFG, V domain |
| 118. | HP HI0725 | 949753 | P44043 | coproporphyrinogen III oxidase |
| 119. | HP HI0744 | 949771 | P44854 | rhodanese-related sulfurtransferase |
| 120. | HP HI0755 | 949515 | P44863 | Polysaccharide deacetylase |
| 121. | HP HI0756 | 950697 | P44864 | peptidase M23 family protein |
| 122. | HP HI0760 | 949979 | P44048 | Fe(2+)-trafficking protein |
| 123. | HP HI0762 | 949781 | P44050 | Calcineurin-like phosphoesterase |
| 124. | HP HI0767 | 949786 | P44869 | 16S rRNA m(2)G966 methyltransferase |
| 125. | HP HI0804 | 950170 | P44053 | cAMP-dependent protein kinase regulatory subunit -like domain ½ family |
| 126. | HP HI0806 | 949820 | P44054 | Sulfite exporter TauE/SafE family protein |
| 127. | HP HI0827 | 949716 | P44886 | acyl-CoA thioester hydrolase |
| 128. | HP HI0841 | 949855 | P44898 | Sulphatases EC 3.1.6. |
| 129. | HP HI0842 | 949857 | P44058 | N-isopropylammelide isopropyl amidohydrolase |
| 130. | HP HI0852 | 949865 | P44903 | Drug resistance transporter EmrB/QacA |
| 131. | HP HI0857 | 950666 | P44062 | BolA family transcriptional regulator |
| 132. | HP HI0858 | 949870 | P44905 | 5-formyltetrahydrofolate cyclo-ligase |
| 133. | HP HI0866 | 950756 | P44063 | lipopolysaccharide biosynthesis protein WzzE |
| 134 | HP HI0868 | 949464 | Q57022 | glycosyl transferase family A protein |
| 135. | HP HI0869 | 949879 | P44064 | Glycosyltransferase |
| 136. | HP HI0874 | 949882 | P44067 | O-antigen ligase WaaL |
| 137. | HP HI0878 | 949421 | P71360 | multidrug resistance efflux transporter EmrE |
| 138. | HP HI0902 | 949698 | P44070 | Sulfite exporter TauE/SafE |
| 139 | HP HI0906 | 949908 | P44931 | Cytidinedeaminase |
| 140. | HP HI0912 | 950836 | P44074 | SAM dependent methyltransferase |
| 141. | HP HI0918 | 949920 | P44936 | Peptidase M50 (metalloendopeptidase) |
| 142. | HP HI0920 | 950624 | P44938 | Undecaprenyl pyrophosphate synthetase |
| 143. | HP HI0925 | 950812 | P44075 | type I restriction enzyme M protein |
| 144. | HP HI0926 | 949651 | P44076 | glutaredoxin-like protein (electron transport) |
| 145. | HP HI0929 | 949927 | P44940 | Bifunctionalglutathionylspermidine synthetase/amidase |
| 146. | HP HI0930 | 949932 | P44077 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 147. | HP HI0933 | 949936 | P44941 | FAD/NAD(P)-binding oxidoreductase |
| 148. | HP HI0938 | 949906 | P44079 | Type II secretory pathway, pseudopilin |
| 149 | HP HI0948 | 949840 | Q57120 | Antidote-toxin recognition MazE |
| 150. | HP HI0960 | 950757 | P44084 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 151. | HP HI0966 | 950444 | P44085 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 152. | HP HI0973 | 949511 | Q57133 | transferrin-binding protein |
| 153. | HP HI0976 | 949977 | Q57147 | EamA-like transporter family protein |
| 154. | HP HI0976.1 | 949978 | O86230 | Multidrug resistance efflux transporter EmrE |
| 155. | HP HI0979 | 949982 | P44965 | tRNA-dihydrouridine synthase |
| 156. | HP HI0983 | 949986 | P43907 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 157. | HP HI0984 | 949993 | P43908 | Peroxide stress response protein YAAA |
| 158. | HP HI1005 | 949997 | P44974 | Sulphatases EC 3.1.6. |
| 159. | HP HI1008 | 950002 | Q57134 | competence protein ComE |
| 160. | HP HI1011 | 950004 | P44093 | D-Tagatose-1,6-bisphosphate aldolase |
| 161. | HP HI1013 | 950733 | Q57151 | hydroxypyruvate isomerase |
| 162. | HP HI1014 | 950006 | P44094 | Nucleoside-diphosphate-sugar epimerase |
| 163. | HP HI1016 | 949991 | P44095 | cyclase family protein |
| 164. | HP HI1028 | 949528 | P44992 | TRAP dicarboxylate transporter subunit DctP |
| 165. | HP HI1029 | 949652 | P44993 | C4-dicarboxylate ABC transporter permease |
| 166. | HP HI1030 | 950014 | P44994 | C4-dicarboxylate ABC transporter permease |
| 167. | HP HI1037 | 950020 | P44098 | glutamine amidotransferase |
| 168. | HP HI1038 | 950021 | P44099 | AAA+ superfamily ATPase |
| 169. | HP HI1048 | 949536 | P44103 | transglutaminase family protein |
| 170. | HP HI1053 | 950030 | Q57498 | Carboxymuconolactone decarboxylase |
| 171. | HP HI1054 | 950034 | P44104 | Type III restriction-modification system restriction enzyme |
| 172. | HP HI1058 | 949400 | P44106 | type III restriction/modification enzyme methylation subunit |
| 173. | HP HI1064 | 950040 | P71367 | Sulphatases EC 3.1.6. |
| 174. | HP HI1082 | 949428 | P45026 | BolA family transcriptional regulator |
| 175. | HP HI1099 | 950069 | P44112 | Prokaryotic membrane lipoprotein lipid attachment site |
| 176. | HP HI1146 | 950109 | P45071 | P-loop containing ATPase protein |
| 177. | HP HI1152 | 950115 | P45077 | TldD/PmbA, Putative modulator of DNA gyrase |
| 178. | HP HI1161 | 950121 | P45083 | Thioesterase |
| 179. | HP HI1162 | 950122 | P44116 | Restriction endonuclease type II-like |
| 180. | HP HI1163 | 950119 | Q57252 | FAD-linked oxidoreductase |
| 181. | HP HI1165 | 949810 | P45085 | Glutaredoxin (electron carrier) |
| 182. | HP HI1173 | 950125 | P44119 | Zinc metal-binding SPRT metallopeptidase |
| 183. | HP HI1189 | 950138 | P45097 | Methyltransferase (radical SAM protein) |
| 184. | HP HI1191 | 950043 | P44124 | 7-cyano-7-deazaguanine synthase(QueC) |
| 185. | HP HI1192 | 950139 | P44125 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 186. | HP HI1198 | 950741 | P45103 | Sua5/YciO/YrdC/YwlC family protein (Double stranded RNA binding) |
| 187. | HP HI1199 | 950150 | P45104 | ribosomal large subunit pseudouridine synthase B |
| 188. | HP HI1202 | 950140 | P44126 | Smr protein/MutS2 |
| 189. | HP HI1208 | 950157 | P71373 | Amidophosphoribosyltransferase (Epimerase) |
| 190. | HP HI1246 | 950184 | P44135 | Sulphatases EC 3.1.6. |
| 191. | HP HI1248 | 950186 | P44136 | Nickel/cobalt transporter(ABC-type transport system) |
| 192. | HP HI1250 | 950243 | P44138 | plasmid maintenance system killer protein (Toxin-antitoxin system) |
| 193. | HP HI1253 | 950692 | P44139 | invasion protein expression up-regulator SirB |
| 194. | HP HI1254 | 950259 | P44140 | tRNA(Met) cytidineacetyltransferase |
| 195. | HP HI1265 | 950187 | P44144 | YcaO protein (Involved in beta-methylthiolation of ribosomal protein S12) |
| 196. | HP HI1273 | 950164 | P44150 | S-adenosyl-L-methionine-dependent methyltransferases |
| 197. | HP HI1282 | 950221 | P45138 | ribosome maturation protein RimP |
| 198. | HP HI1292 | 949593 | P44154 | Zn-ribbon-containing protein (DNA binding protein) |
| 199. | HP HI1293 | 950226 | P44156 | SufE protein probably involved in Fe-S center assembly |
| 200. | HP HI1297 | 950233 | P45145 | LrgA like protein (Export murein hydrolases) |
| 201. | HP HI1298 | 950227 | P45146 | murein hydrolase regulator LrgB |
| 202. | HP HI1307 | 950239 | Q57320 | Lysine-type exporter protein (LYSE/YGGA) |
| 203. | HP HI1309 | 950234 | P45154 | 2Fe-2S ferredoxin-type domain (elctron carrier) |
| 204. | HP HI1315 | 950581 | P71375 | Sodium/solute symporter |
| 205. | HP HI1317 | 950209 | P44160 | Aldose 1-epimerase |
| 206. | HP HI1323 | 950258 | P44161 | MacrodomainTer protein, MatP |
| 207. | HP HI1327 | 950255 | P44163 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 208. | HP HI1333 | 949671 | P71376 | RNA-binding, CRM domain |
| 209. | HP HI1338 | 950260 | P44164 | phosphohistidine phosphatase SixA |
| 210. | HP HI1339 | 950818 | P71378 | Late embryogenesis abundant protein |
| 211. | HP HI1340 | 950814 | P44165 | Outer membrane efflux porinTdeA |
| 212. | HP HI1343 | 949643 | P71379 | cysteine desulfurase, catalytic subunit CsdA |
| 213. | HP HI1349 | 950182 | P45173 | DNA-binding ferritin-like protein |
| 214. | HP HI1351 | 950443 | P44167 | tRNAmo(5)U34 methyltransferase, SAM-dependent |
| 215. | HP HI1361 | 950286 | P45180 | Glycosyl transferase, family 35 |
| 216. | HP HI1369 | 950892 | P45182 | TonB-dependent receptor |
| 217. | HP HI1376 | 950804 | P44170 | Multidrug resistance efflux transporter EmrE |
| 218. | HP HI1388.1 | 950703 | O86237 | Tautomerase/MIF |
| 219. | HP HI1394 | 950304 | P44172 | RNA binding domain (ASCH) |
| 220. | HP HI1395 | 950305 | P44173 | zeta toxin family protein |
| 221. | HP HI1400 | 950717 | P44176 | Polymerase and histidinol phosphatase like |
| 222. | HP HI1413 | 949414 | P44185 | Prokaryotic membrane lipoprotein lipid attachment site profile |
| 223. | HP HI1415 | 950713 | P44187 | Lysozyme-like superfamily protein |
| 224. | HP HI1416 | 950758 | P44188 | Phage holin, lambda family |
| 225. | HP HI1418 | 950323 | P44189 | BRO family, N-terminal domain |
| 226. | HP HI1419 | 949900 | P44190 | Phage derived protein Gp49-like |
| 227. | HP HI1420 | 950760 | P44191 | Helix-turn-helix protein |
| 228. | HP HI1422 | 949966 | P44193 | antA/AntBantirepressor family protein |
| 229. | HP HI1434 | 949657 | P45202 | Cys-tRNAPro/Cys-tRNACysdeacylaseybaK |
| 23.0 | HP HI1435 | 950339 | P44197 | tRNApseudouridine synthase C |
| 231. | HP HI1436 | 950784 | Q57152 | RNA pseudouridine synthase C |
| 232. | HP HI1454 | 950340 | P44202 | Cytochrome C biogenesis protein transmembrane region |
| 233. | HP HI1462 | 950787 | P45217 | Outer membrane efflux porinTdeA |
| 234. | HP HI1469 | 949595 | P44205 | molybdenum ABC transporter substrate-binding protein |
| 235. | HP HI1475 | 950353 | Q57380 | molybdate ABC transporter, permease |
| 236. | HP HI1479 | 950355 | P44208 | Transposase |
| 237. | HP HI1493 | 950360 | P44218 | N-acetylmuramoyl-L-alanine amidase |
| 238. | HP HI1497 | 950363 | P44221 | Zinc finger, DksA/TraR C4-type |
| 239. | HP HI1498.1 | 950365 | O86242 | Ribonuclease R winged-helix domain protein |
| 240. | HP HI1499 | 950366 | P44223 | Mu-like phage gp27 |
| 241. | HP HI1500 | 950367 | P44224 | Mu-like prophageFluMu protein gp28 |
| 242. | HP HI1501 | 950368 | P44225 | Mu-like prophageFluMu protein gp29 |
| 243. | HP HI1502 | 950369 | P44226 | F protein, phage head morphogenesis, SPP1 gp7 family domain protein |
| 244. | HP HI1505 | 950373 | P44227 | Mu-like prophageFluMu major head subunit |
| 245. | HP HI1508 | 950376 | P44230 | Mu-like prophage protein GP36 |
| 246. | HP HI1509 | 950377 | P44231 | Mu-like prophageFluMu protein gp37 |
| 247. | HP HI1510 | 950834 | P44232 | Mu-like prophageFluMu protein gp38 |
| 248. | HP HI1512 | 950378 | P44234 | Mu-like prophageFluMu tail tube protein |
| 249 | HP HI1513 | 950379 | P44235 | Mu-like prophageFluMu protein gp41 |
| 250. | HP HI1518 | 950383 | P44238 | Mu-like prophageFluMu protein gp45 |
| 251. | HP HI1519 | 950384 | P44239 | Mu-like prophageFluMu protein gp46 |
| 252. | HP HI1520 | 950385 | P44240 | Mu-like prophageFluMu protein gp47 |
| 253. | HP HI1521 | 950386 | P44241 | Mu-like prophageFluMu protein gp48 |
| 254. | HP HI1522 | 950387 | P44242 | Mu-like prophageFluMu defective tail fiber protein |
| 255. | HP HI1522.1 | 950388 | P71390 | Mu-like prophage protein Com |
| 256. | HP HI1523 | 949672 | P44243 | D12 class N6 adenine-specific DNA methyltransferase |
| 257. | HP HI1534 | 950396 | P44246 | tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein MnmC |
| 258. | HP HI1536 | 950398 | P44247 | TRNA U-34 5-methylaminomethyl-2-thiouridine biosynthesis protein MnmC, C-terminal |
| 259. | HP HI1542 | 950405 | P45244 | NAD(P)H nitroreductase |
| 26. | HP HI1555 | 949639 | P44252 | Outer membrane-specific lipoprotein ABC transporter, permease component LolE |
| 261. | HP HI1558 | 950418 | P45252 | Tetratricopeptide repeat (TPR) like |
| 262. | HP HI1559 | 950419 | P45253 | N5-glutamine S-adenosyl-L-methionine-dependent methyltransferase |
| 263. | HP HI1560 | 950420 | P44253 | RDD domain-containing protein |
| 264. | HP HI1562 | 950422 | P44254 | TPR repeat, Sel1 subfamily protein (key negative regulator of the Notch pathway) |
| 265. | HP HI1564 | 950424 | P44256 | DNA polymerase IV |
| 266. | HP HI1571.1 | 950429 | Q4QKT3 | bacteriophage replication protein A |
| 267. | HP HI1581 | 950440 | P44262 | Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyldioxygenase |
| 268. | HP HI1598 | 950454 | P45267 | adenylatecyclase |
| 269. | HP HI1600 | 950455 | P44268 | Xylose isomerase-like, TIM barrel domain |
| 270. | HP HI1602 | 950457 | P44270 | TQO small subunit DoxD family protein (subunit of the terminal quinol oxidase) |
| 271. | HP HI1605 | 950458 | P44272 | SH3 domain-containing protein |
| 272. | HP HI1625 | 950478 | P44277 | Sel1 repeat domain |
| 273. | HP HI1627 | 950462 | P71394 | Endoribonuclease L-PSP |
| 274. | HP HI1629 | 950844 | P45280 | SNARE associated Golgi protein |
| 275. | HP HI1632 | 950850 | Q57525 | Aspartokinase |
| 276. | HP HI1637 | 950851 | P44280 | P-loop containing nucleoside triphosphate hydrolases |
| 277. | HP HI1650 | 950489 | P44281 | DEAD/DEAH box helicase/type I restriction endonuclease subunit R |
| 278. | HP HI1651 | 950855 | P44282 | Signal transduction histidine kinase |
| 279. | HP HI1654 | 950491 | P45298 | S-adenosylmethionine-dependent methytransferase |
| 280. | HP HI1656 | 950807 | P45300 | Restriction endonuclease type II-like |
| 281. | HP HI1657 | 950796 | P52606 | Sedoheptulose 7-phosphate isomerase |
| 282. | HP HI1658 | 950803 | P45301 | Transport-associated and nodulation domain, bacteria (BON domain) (ion transport) |
| 283. | HP HI1663 | 950497 | Q57544 | Metallo-beta-lactamase |
| 284. | HP HI1664 | 950504 | P45305 | TatD-related deoxyribonuclease |
| 285. | HP HI1665 | 950493 | P44283 | Hedgehog signalling/DD-peptidase zinc-binding domain/Peptidase_M15_2 |
| 286. | HP HI1666 | 950486 | P44284 | Hedgehog signalling/DD-peptidase zinc-binding domain/Peptidase_M15_2 |
| 287. | HP HI1667 | 950498 | P44285 | L, D-transpeptidase |
| 288. | HP HI1671 | 950860 | P44287 | Paraquat-inducible protein A/Multihaem cytochrome (electron transport) |
| 289. | HP HI1672 | 950502 | P44288 | Mammalian cell entry (MCE) related protein |
| 290. | HP HI1680 | 950508 | P44289 | MFS general substrate transporter superfamily |
| 291. | HP HI1709 | 950526 | P44293 | Viral OB-fold, YgiW |
| 292. | HP HI1718 | 950877 | P44296 | trimericautotransporteradhesin |
| 293. | HP HI1720 | 950873 | Q57066 | Transposase |
| 294. | HP HI1728 | 950517 | O05087 | Mn2+ and Fe2+ transporter of the NRAMP family |
| 295. | HP HI1730 | 950540 | P44298 | allophanate hydrolase subunit 2 |
| 296. | HP HI1731 | 950880 | P44299 | allophanate hydrolase subunit 1 |
Figure 2. Classification of 429 HPs into various groups by utilizing the functional annotation result of various bioinformatics tools.
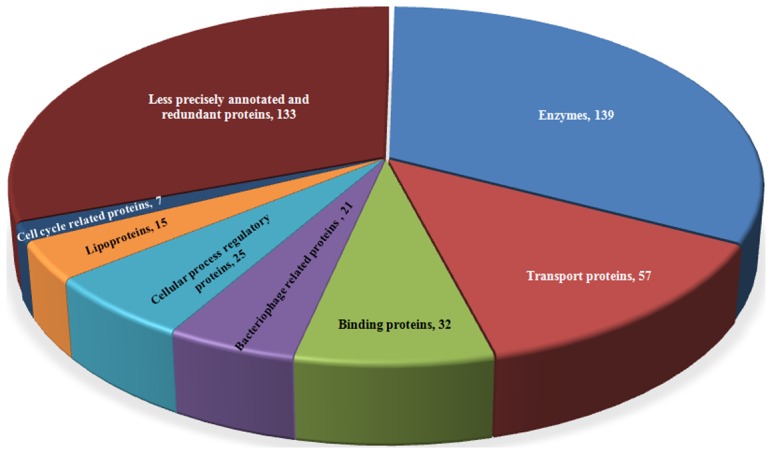
The chart shows that there are 41% are enzymes, 20% proteins involve in transportation, 12% binding proteins, 7% bacteriophage related proteins and rest are proteins involved in cellular processes like transcription, translation, replication etc., among 429 HPs from H. influenzae.
Enzymes
Enzymes produced by bacteria are key player for the survival of organism in their host because they provide nutrient for growth and responsible for pathogenesis of organism, for enzymes modify the local environment for favorable growth inside the host and metabolism of compounds inside the host [76]. We characterized 139 enzymes. Knowledge of these enzymes is important for understanding the host-pathogen interaction as well.
We identified 14 oxidoreductase enzymes, which are critically important for bacterial virulence and pathogenesis. It is well understood that the disulfide bonds are important for the stability and/or structural rigidity of many extracellular proteins, including bacterial virulence factors. Bond formation is catalyzed by thiol-disulfide oxidoreductases (TDORs). Oxidoreductases like SdbA is required for disulfide bond formation in S. gordonii, which is required for autolytic activity [77]. Protein P45154 contain 2Fe-2S ferredoxin-type domain. Many bacteria produce protein antibiotics known as bacteriocins to kill competing strains of the same or closely related bacterial species. We identified protein P44743 as a radical SAM (S-adenosylmethionine) protein, it is understood that radical SAM proteins play a significant role in pathogenesis of an organism and is also validated that the inhibition of these enzymes is effective in preventing the lethal diseases [78].
Similarly, we identified 39 transferase enzymes which are required for the efficient spore germination and full virulence of bacteria like Bacillus anthracis. Transferase enzymes are essential for biosynthesis of lipoprotein, and bacterial lipoproteins play an important role in virulence of bacteria [79]. Proteins Q57022, P44064 and P45180 are glycosyl transferase, and on mutation it affects extracellular polysaccharide (EPS) and lipopolysaccharide (LPS) biosynthesis, cell motility, and reduces the development of disease symptoms [80], [81]. We have characterized protein P44256 as DNA polymerase IV and it is observed that virulent strains contain increased level of activity of DNA polymerase than non-virulent strains, indicating its role in virulence [82].
The protein Q57544 is found to be a β-lactamase. The enzyme responsible for generation of resistance against β-Lactam antibiotics like penicillin, cephalosporins, etc. [83]. We annotated 56 hydrolase enzymes having an established role in virulence of bacteria, e.g. Kdo hydrolase is the main cause of virulence in Francisella tularensis, which is classified as a bioterrorism agent [84]. Similarly, nudix hydrolase encoded by nudA gene in Bacillus anthracis is important for the complete virulence [85].
There are 8 lyase enzymes. These are important for the virulence of pathogen in host [76]. The P44717 protein is a cystathionine β-lyase, an enzyme which forms the cystathionine intermediate in cysteine biosynthesis, may be considered as the target for pyridiamine anti-microbial agents [86]. Similarly, isocitrate lyase is an enzyme of glyoxylate cycle, which catalyzes the cleavage of isocitrate to succinate and glyoxylate together with malate synthase. This enzyme bypasses two decarboxylation steps of TCA cycle. It is found to up-regulate glyoxylate cycle during pathogenesis, and therefore, this pathway is used by bacteria, fungi, etc., for survival in their hosts [87].
The isomerase enzyme catalyze changes within one molecule by structural rearrangement [88] and isomerases like peptidylprolyl cis/trans isomerases (PPIases) involved in protein folding. These isomerases are considered as surface-exposed proteins which are important for virulence and resistance to NaCl [88]. We identified 13 isomerases and 5 ligases in a group of 139 enzymes. Ligase enzymes are also part of virulence in the hosts. It is found that E3 ligase activity associated with the C-terminal region of XopL, a type III effectors, which specifically interacts with plant E2 ubiquitin conjugating enzyme that induce plant cell death and subvert plant immunity [89]. There are also 4 HPs with kinase activity, which play a significant role in growth, differentiation, metabolism and apoptosis in response to external and internal stimuli [90]. Thus, such enzymes are important for the survival of pathogen and may serve as a target for drug design and discovery [91].
Transport
Transport process plays a pivotal role in cellular metabolism, e.g., for the uptake of nutrients or the excretion of metabolic waste products, etc. We successfully predicted 50 transporters, 3 carriers, 3 receptors and 1 signal transduction proteins among HPs. It is recently identified that these proteins may be involved in virulence and essential for intracellular survival of pathogens [92]. The protein P44691 was predicted to be a member of ABC 3 transporter family, presumably involved in virulence because they are associated with the uptake of metal ions, such as iron, zinc, and manganese [93]. This protein also helps in the attachment of pathogenic bacteria to the mucosal surfaces of host cells, which is a critical step in bacterial pathogenesis, thereby present as a putative drug target [93].
We found protein P44005 and P45280 as SNARE associated Golgi protein. The soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) proteins play an essential role in the compartment fusion in eukaryotic cells [94]. They share a conserved motif, known as SNARE motif, and have been classified as glutamine containing SNAREs (Q-SNAREs) and arginine containing SNAREs (R-SNAREs) on the basis of favorably conserved residue at the center of this motif [95]. These proteins are central regulators of membrane fusion, so they are potential targets for intracellular organisms, which frequently rely on destabilizing the host intracellular traffic. This finding helps us to conclude that by mimicking SNAREs some inclusion proteins can control intracellular trafficking.
Bacteriocins proteins contain an N-terminal domain with an extensive resemblance to a [2Fe-2S] plant ferredoxin and a C-terminal colicin M-like catalytic domain and to gain entry into vulnerable cells. These proteins parasitize an existing iron uptake pathway by using a ferredoxin-containing receptor binding domain [96]. Protein Q57133 is a transferrin-binding protein. Transferrins are a group of non-haem iron-binding glycoproteins, widely distributed in the physiological fluids and cells of vertebrates. These proteins are involved in iron transport within the circulatory system of the vertebrates. Transferrins is important for bacterial virulence but their role in virulence is still not fully understood [97]. The membrane transferrin receptor-mediated endocytosis is a major route of cellular iron uptake and the efficient cellular uptake of transferrin pathway has shown potential in the delivery of anticancer drugs, proteins, and therapeutic genes into primarily proliferating malignant cells over expressed transferrin receptors [98], [99].
Binding Proteins
32 HPs are annotated as binding proteins in which 15 are DNA binding, 5 RNA binding, 9 metal binding and 3 ATP/coenzyme binding proteins. We have identified a tetratricopeptide repeat (TPR), a structural motif involved in the assembly of various multi-protein complexes in many HPs. TPR-containing proteins often play important roles in cell processes, and involved in virulence-associated functions [100].
HPs function as DNA-binding proteins also contribute to the virulence. The winged-helix-turn-helix (wHTH) motif in sarZ proteins in Staphylococcus aureus contributes to virulence by binding to cvf gene that encodes for alpha hemolysin [101]. In complex regulatory system of group A Streptococcus (GAS), there is the streptococcal regulator of virulence (Srv) which is the member of the CRP/FNR family of transcriptional regulators, and members of this family possess a characteristic C-terminal helix-turn-helix motif (HTH) that facilitates binding to DNA targets. Point mutation in this motif alters protein-DNA interaction [102], indicate that DNA binding motifs are regulatory factors of the virulence of bacteria. The RNA binding proteins are also contributing to the survival of the organism and control the virulence factors of the pathogens [103].
Lipoprotein
Lipoproteins identified in bacteria are formed by lipid modification of proteins that facilitate the anchoring of hydrophilic proteins to hydrophobic surfaces through hydrophobic interactions of the attached acyl groups to the cell wall phospholipids. This process has a considerable significance in many cellular and virulence phenomena. We found 15 lipoproteins from the group of HPs because they play crucial roles in adhesion to host cells, variation of inflammatory processes and translocation process of virulence factors into host cells. It is also discovered that lipoproteins may function as vaccines. The knowledge of these facts may be utilized for the generation of novel countermeasures to bacterial diseases [104].
Other Proteins
Structural motifs like helix-turn-helix are conserved in various organisms. A detection of these common patterns in a sequence refers that such proteins are mainly involved in the regulation of transcription. The transcription regulators like HilC and HilD also showed DNA binding activities and contributes to the virulence of Salmonella enterica, where these are involved in the invasion to the host cells [105]. We found 18 transcriptional regulatory, 3 translation regulatory, 1 replication regulatory, 3 cell cycle regulatory enzyme/protein. The regulatory protein RfaH is found in E. coli and enhances the expression of different factors that are supposed to play a role in the bacterial virulence. Furthermore, inactivation of rfaH decreases the virulence of uropathogenic E. coli strain [106]. Similarly, the RNA-binding protein Hfq has emerged as an important regulatory factor in varieties of physiological processes, including stress resistance and virulence in various Gram-negative bacteria such as E. coli. Hfq modulates the stability or translation of mRNAs and interacts with numerous small regulatory RNAs [107]. The cell cycle and related protein P44063, is involved in lipopolysaccharide biosynthesis and are important in understanding the virulence of H. influenzae, as proteins involved in this particular biosynthesis are considered as primary virulence factors [108].
Virulent proteins
We use the consensus of VICMpred and VirulentPred for predicting the virulence factors among the 429 HPs and found 40 HPs that give positive virulence score in both servers, and can be used as potent drug targets for drug design. These are listed in Table 3 . In this group of virulent proteins we observed that protein P43936 is a PemK superfamily toxin of the ChpB-ChpS toxin-antitoxin system protein involved in plasmid maintenance [109]. We have also identified 30 bacteriophage related proteins among HPs. It is known that SuMu protein 1a, a bacteriophage related protein, has shown homology to IgA metalloproteinase and IgA1 protease which are described as virulence factors in non-typeable H. influenzae [110]. So, SuMu proteins are considered as highly virulent proteins.
Table 3. List of HPs with virulence factors in H. influenzae.
| S No. | UNIPROT ID | Virulent proteins | |
| Virulentpred | VICMpred | ||
| 1. | P71336 | Yes | Yes |
| 2. | P43936 | Yes | Yes |
| 3. | P44553 | Yes | Metabolism molecule |
| 4. | P44609 | Yes | Yes |
| 5. | P44670 | Yes | Yes |
| 6. | P44675 | Yes | Cellular process |
| 7. | P43990 | Yes | Cellular process |
| 8. | P44693 | Yes | Cellular process |
| 9. | Q57144 | Yes | Cellular process |
| 10. | P44733 | Yes | Cellular process |
| 11. | P44740 | Yes | Yes |
| 12. | P44023 | Yes | Yes |
| 13. | Q57523 | Yes | Yes |
| 14. | P44038 | Yes | Cellular process |
| 15. | P44041 | Yes | Information and storage |
| 16. | P44863 | Yes | Yes |
| 17. | P44054 | Yes | Yes |
| 18. | P44063 | Yes | Cellular process |
| 19. | Q57120 | Yes | Cellular process |
| 20. | Q57133 | Yes | Yes |
| 21. | P43907 | Yes | Cellular process |
| 22. | P44972 | Yes | Cellular process |
| 23. | P45074 | Yes | Cellular process |
| 24. | P45077 | Yes | Cellular process |
| 25. | P71373 | Yes | Yes |
| 26. | P44132 | Yes | Metabolism molecule |
| 27. | P44138 | Yes | Cellular process |
| 28. | P44140 | Yes | Yes |
| 29. | P44165 | Yes | Yes |
| 30. | P45182 | Yes | Yes |
| 31. | P44169 | Yes | Yes |
| 32. | P44183 | Yes | Yes |
| 33. | P56507 | Yes | Yes |
| 34. | P45217 | Yes | Yes |
| 35. | P44242 | Yes | Cellular process |
| 36. | P44246 | Yes | Yes |
| 37. | P44288 | Yes | Metabolism molecule |
| 38. | P44293 | Yes | Yes |
| 39. | P44296 | Yes | Metabolism molecule |
| 40. | P44298 | Yes | Yes |
Conclusions
Using an innovative in silico approach we have analyzed all 429 HPs from H. influenzae. Using the ROC analysis and confidence level measurements of the predicted results, we precisely predict the function of 296 HPs with confidence and successfully characterized them. We did not find enough evidences for functional prediction of 124 proteins, and hence these sequences require further analysis. The sub-cellular localization and physicochemical parameters prediction are useful in distinguishing the HPs with transporter activity from the rest of the protein. The protein-protein interaction also helps to find out the involvement of such proteins in various metabolic pathways. Further, we are able to detect the 40 virulence proteins essential for the survival of pathogen, particularly protein Q57523 showing highest virulence score in VICMpred which is known to be the most virulent HP among the listed virulence proteins. Our results could facilitate in developing drugs/vaccines, specifically targeting the pathogen's system without causing any allergic or side effect to the host. This in silico approach for functional annotation of HPs can be further utilized in drug discovery for characterizing putative drug targets for other clinically important pathogens.
Supporting Information
List of predicted physicochemical parameters by Expasy's ProtParam tool of 429 HP from H. influenzae.
(DOCX)
List of predicted sub-cellular localization of 429 HPs from H. influenzae.
(DOCX)
List of annotated functions of 429 HPs from H. influenzae using BLASTp, STRING, SMART, INTERPROSCAN and MOTIF.
(DOCX)
List of functionally annotated domains of 429 HPs from H. influenzae by CATH, SUPERFAMILY, PANTHER, Pfam, SYSTERS, CDART SVMProt and ProtoNet.
(DOCX)
List of annotated functions of 100 proteins with known function from H. influenzae using BLASTp, SMART, INTERPROSCAN and MOTIF for ROC analysis.
(DOCX)
List of functionally annotated domains of 100 proteins with known function from H. influenzae by CATH, SUPERFAMILY, PANTHER, Pfam, SYSTERS, CDART SVMProt and ProtoNet for ROC analysis.
(DOCX)
List of accuracy, sensitivity, specificity and ROC area of various bioinformatics tools used for predicting function of HPs from H. influenzae obtained after ROC analysis.
(DOCX)
List of clusters formed by CLUSS online tool and predicted motif sequence site and sequence by MEME Suite in 429 HPs from H. influenzae.
(DOCX)
List of annotated HPs at low confidence from H. influenzae.
(DOCX)
Funding Statement
The authors sincerely thank Indian Council of Medical Research for financial assistance (Grant No. BIC/12(04)/2012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sethi S, Murphy TF (2001) Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin Microbiol Rev 14: 336–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murphy TF, Sethi S (1992) Bacterial infection in chronic obstructive pulmonary disease. Am Rev Respir Dis 146: 1067–1083. [DOI] [PubMed] [Google Scholar]
- 3. Ball P (1996) Infective pathogenesis and outcomes in chronic bronchitis. Curr Opin Pulm Med 2: 181–185. [DOI] [PubMed] [Google Scholar]
- 4. Cash P, Argo E, Langford PR, Kroll JS (1997) Development of a Haemophilus two-dimensional protein database. Electrophoresis 18: 1472–1482. [DOI] [PubMed] [Google Scholar]
- 5. Evers S, Di Padova K, Meyer M, Fountoulakis M, Keck W, et al. (1998) Strategies towards a better understanding of antibiotic action: folate pathway inhibition in Haemophilus influenzae as an example. Electrophoresis 19: 1980–1988. [DOI] [PubMed] [Google Scholar]
- 6. Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496–512. [DOI] [PubMed] [Google Scholar]
- 7. Wong SM, Akerley BJ (2008) Identification and analysis of essential genes in Haemophilus influenzae. Methods Mol Biol 416: 27–44. [DOI] [PubMed] [Google Scholar]
- 8. Edwards JS, Palsson BO (1999) Systems properties of the Haemophilus influenzae Rd metabolic genotype. J Biol Chem 274: 17410–17416. [DOI] [PubMed] [Google Scholar]
- 9. Papin JA, Price ND, Edwards JS, Palsson BB (2002) The genome-scale metabolic extreme pathway structure in Haemophilus influenzae shows significant network redundancy. J Theor Biol 215: 67–82. [DOI] [PubMed] [Google Scholar]
- 10. Schilling CH, Palsson BO (2000) Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis. J Theor Biol 203: 249–283. [DOI] [PubMed] [Google Scholar]
- 11. Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, et al. (2002) A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc Natl Acad Sci U S A 99: 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herbert MA, Hayes S, Deadman ME, Tang CM, Hood DW, et al. (2002) Signature Tagged Mutagenesis of Haemophilus influenzae identifies genes required for in vivo survival. Microb Pathog 33: 211–223. [DOI] [PubMed] [Google Scholar]
- 13. Doerks T, von Mering C, Bork P (2004) Functional clues for hypothetical proteins based on genomic context analysis in prokaryotes. Nucleic Acids Res 32: 6321–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hawkins T, Kihara D (2007) Function prediction of uncharacterized proteins. J Bioinform Comput Biol 5: 1–30. [DOI] [PubMed] [Google Scholar]
- 15. Galperin MY, Koonin EV (2004) ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res 32: 5452–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loewenstein Y, Raimondo D, Redfern OC, Watson J, Frishman D, et al. (2009) Protein function annotation by homology-based inference. Genome Biol 10: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nimrod G, Schushan M, Steinberg DM, Ben-Tal N (2008) Detection of functionally important regions in “hypothetical proteins” of known structure. Structure 16: 1755–1763. [DOI] [PubMed] [Google Scholar]
- 18. Hassan MI, Kumar V, Somvanshi RK, Dey S, Singh TP, et al. (2007) Structure-guided design of peptidic ligand for human prostate specific antigen. J Pept Sci 13: 849–855. [DOI] [PubMed] [Google Scholar]
- 19. Hassan MI, Kumar V, Singh TP, Yadav S (2007) Structural model of human PSA: a target for prostate cancer therapy. Chem Biol Drug Des 70: 261–267. [DOI] [PubMed] [Google Scholar]
- 20. Thakur PK, Kumar J, Ray D, Anjum F, Hassan MI (2013) Search of potential inhibitor against New Delhi metallo-beta-lactamase 1 from a series of antibacterial natural compounds. J Nat Sci Biol Med 4: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minion FC, Lefkowitz EJ, Madsen ML, Cleary BJ, Swartzell SM, et al. (2004) The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J Bacteriol 186: 7123–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lubec G, Afjehi-Sadat L, Yang JW, John JP (2005) Searching for hypothetical proteins: theory and practice based upon original data and literature. Prog Neurobiol 77: 90–127. [DOI] [PubMed] [Google Scholar]
- 23. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 24. Rost B, Valencia A (1996) Pitfalls of protein sequence analysis. Curr Opin Biotechnol 7: 457–461. [DOI] [PubMed] [Google Scholar]
- 25. Kanehisa M (1997) Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem Sci 22: 442–444. [DOI] [PubMed] [Google Scholar]
- 26. Sigrist CJ, Cerutti L, de Castro E, Langendijk Genevaux PS, Bulliard V, et al. (2010) PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 38: D161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Attwood TK (2002) The PRINTS database: a resource for identification of protein families. Brief Bioinform 3: 252–263. [DOI] [PubMed] [Google Scholar]
- 28. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bru C, Courcelle E, Carrere S, Beausse Y, Dalmar S, et al. (2005) The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res 33: D212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Henikoff JG, Henikoff S (1996) Blocks database and its applications. Methods Enzymol 266: 88–105. [DOI] [PubMed] [Google Scholar]
- 31. Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, et al. (2011) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40: D306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, et al. (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39: D561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Metz CE (1978) Basic principles of ROC analysis. Semin Nucl Med 8: 283–298. [DOI] [PubMed] [Google Scholar]
- 36. Shanmughavel SAaP (2008) Computational Annotation for Hypothetical Proteins of Mycobacterium Tuberculosis. Journal of Computer Science & Systems Biology 1: 50–62. [Google Scholar]
- 37. Garg A, Gupta D (2008) VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinformatics 9: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saha S, Raghava GP (2006) VICMpred: an SVM-based method for the prediction of functional proteins of Gram-negative bacteria using amino acid patterns and composition. Genomics Proteomics Bioinformatics 4: 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, et al. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182: 319–326. [DOI] [PubMed] [Google Scholar]
- 41. Guruprasad K, Reddy BV, Pandit MW (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng 4: 155–161. [DOI] [PubMed] [Google Scholar]
- 42. Ikai A (1980) Thermostability and aliphatic index of globular proteins. J Biochem 88: 1895–1898. [PubMed] [Google Scholar]
- 43. Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132. [DOI] [PubMed] [Google Scholar]
- 44. Vetrivel U, Subramanian G, Dorairaj S A novel in silico approach to identify potential therapeutic targets in human bacterial pathogens. Hugo J 5: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, et al. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32: D115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, et al. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bhasin M, Garg A, Raghava GP (2005) PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics 21: 2522–2524. [DOI] [PubMed] [Google Scholar]
- 48. Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64: 643–651. [DOI] [PubMed] [Google Scholar]
- 49. Yu CS, Lin CJ, Hwang JK (2004) Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 13: 1402–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971. [DOI] [PubMed] [Google Scholar]
- 51. Bendtsen JD, Kiemer L, Fausboll A, Brunak S (2005) Non-classical protein secretion in bacteria. BMC Microbiol 5: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580. [DOI] [PubMed] [Google Scholar]
- 53. Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850. [DOI] [PubMed] [Google Scholar]
- 54. Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bernstein FC, Koetzle TF, Williams GJ, Meyer EF Jr, Brice MD, et al. (1977) The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur J Biochem 80: 319–324. [DOI] [PubMed] [Google Scholar]
- 56. Bernstein FC, Koetzle TF, Williams GJ, Meyer EF Jr, Brice MD, et al. (1978) The Protein Data Bank: a computer-based archival file for macromolecular structures. Arch Biochem Biophys 185: 584–591. [DOI] [PubMed] [Google Scholar]
- 57. Hubbard TJ, Ailey B, Brenner SE, Murzin AG, Chothia C (1999) SCOP: a Structural Classification of Proteins database. Nucleic Acids Res 27: 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, et al. (2013) New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Res 41: D490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Simossis VA, Heringa J (2005) PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 33: W289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313: 903–919. [DOI] [PubMed] [Google Scholar]
- 61. Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Meinel T, Krause A, Luz H, Vingron M, Staub E (2005) The SYSTERS Protein Family Database in 2005. Nucleic Acids Res 33: D226–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cai CZ, Han LY, Ji ZL, Chen X, Chen YZ (2003) SVM-Prot: Web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res 31: 3692–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Geer LY, Domrachev M, Lipman DJ, Bryant SH (2002) CDART: protein homology by domain architecture. Genome Res 12: 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40: D302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rappoport N, Karsenty S, Stern A, Linial N, Linial M (2012) ProtoNet 6.0: organizing 10 million protein sequences in a compact hierarchical family tree. Nucleic Acids Res 40: D313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gasteiger E, Jung E, Bairoch A (2001) SWISS-PROT: connecting biomolecular knowledge via a protein database. Curr Issues Mol Biol 3: 47–55. [PubMed] [Google Scholar]
- 68. Bairoch A, Apweiler R (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res 28: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hubbard T, Barker D, Birney E, Cameron G, Chen Y, et al. (2002) The Ensembl genome database project. Nucleic Acids Res 30: 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kelil A, Wang S, Brzezinski R (2008) CLUSS2: an alignment-independent algorithm for clustering protein families with multiple biological functions. Int J Comput Biol Drug Des 1: 122–140. [DOI] [PubMed] [Google Scholar]
- 71. Kelil A, Wang S, Brzezinski R, Fleury A (2007) CLUSS: clustering of protein sequences based on a new similarity measure. BMC Bioinformatics 8: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Baron C, Coombes B (2007) Targeting bacterial secretion systems: benefits of disarmament in the microcosm. Infect Disord Drug Targets 7: 19–27. [DOI] [PubMed] [Google Scholar]
- 73. Zou KH, Warfield SK, Fielding JR, Tempany CM, William MW 3rd, et al. (2003) Statistical validation based on parametric receiver operating characteristic analysis of continuous classification data. Acad Radiol 10: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Swets JA, Dawes RM, Monahan J (2000) Better decisions through science. Sci Am 283: 82–87. [DOI] [PubMed] [Google Scholar]
- 75.Eng J (2013) ROC analysis: web-based calculator for ROC curves. Baltimore, Maryland, USA: Johns Hopkins University.
- 76. Bjornson HS (1984) Enzymes associated with the survival and virulence of gram-negative anaerobes. Rev Infect Dis 6 Suppl 1S21–24. [DOI] [PubMed] [Google Scholar]
- 77. Davey L, Ng CK, Halperin SA, Lee SF (2013) Functional analysis of paralogous thiol-disulfide oxidoreductases in Streptococcus gordonii. J Biol Chem 288: 16416–16429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Parveen N, Cornell KA (2011) Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol 79: 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Okugawa S, Moayeri M, Pomerantsev AP, Sastalla I, Crown D, et al. (2012) Lipoprotein biosynthesis by prolipoprotein diacylglyceryl transferase is required for efficient spore germination and full virulence of Bacillus anthracis. Mol Microbiol 83: 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McQuiston JR, Vemulapalli R, Inzana TJ, Schurig GG, Sriranganathan N, et al. (1999) Genetic characterization of a Tn5-disrupted glycosyltransferase gene homolog in Brucella abortus and its effect on lipopolysaccharide composition and virulence. Infect Immun 67: 3830–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li Q, Zhang Y, Sheng Y, Huo R, Sun B, et al. (2012) Large T-antigen up-regulates Kv4.3 K(+) channels through Sp1, and Kv4.3 K(+) channels contribute to cell apoptosis and necrosis through activation of calcium/calmodulin-dependent protein kinase II. Biochem J 441: 859–867. [DOI] [PubMed] [Google Scholar]
- 82. Makioka A, Ohtomo H (1995) An increased DNA polymerase activity associated with virulence of Toxoplasma gondii. J Parasitol 81: 1021–1022. [PubMed] [Google Scholar]
- 83. Poole K (2004) Resistance to beta-lactam antibiotics. Cell Mol Life Sci 61: 2200–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Okan NA, Chalabaev S, Kim TH, Fink A, Ross RA, et al. (2013) Kdo hydrolase is required for Francisella tularensis virulence and evasion of TLR2-mediated innate immunity. MBio 4: e00638–00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Edelstein PH, Hu B, Shinzato T, Edelstein MA, Xu W, et al. (2005) Legionella pneumophila NudA Is a Nudix hydrolase and virulence factor. Infect Immun 73: 6567–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ejim LJ, D'Costa VM, Elowe NH, Loredo Osti JC, Malo D, et al. (2004) Cystathionine beta-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect Immun 72: 3310–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dunn MF, Ramirez Trujillo JA, Hernandez Lucas I (2009) Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155: 3166–3175. [DOI] [PubMed] [Google Scholar]
- 88. Reffuveille F, Connil N, Sanguinetti M, Posteraro B, Chevalier S, et al. (2012) Involvement of peptidylprolyl cis/trans isomerases in Enterococcus faecalis virulence. Infect Immun 80: 1728–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Huang J, Huang Q, Zhou X, Shen MM, Yen A, et al. (2004) The poxvirus p28 virulence factor is an E3 ubiquitin ligase. J Biol Chem 279: 54110–54116. [DOI] [PubMed] [Google Scholar]
- 90. Engh RA, Bossemeyer D (2002) Structural aspects of protein kinase control-role of conformational flexibility. Pharmacol Ther 93: 99–111. [DOI] [PubMed] [Google Scholar]
- 91. Stephenson K, Hoch JA (2002) Histidine kinase-mediated signal transduction systems of pathogenic microorganisms as targets for therapeutic intervention. Curr Drug Targets Infect Disord 2: 235–246. [DOI] [PubMed] [Google Scholar]
- 92. Freeman ZN, Dorus S, Waterfield NR (2013) The KdpD/KdpE two-component system: integrating K(+) homeostasis and virulence. PLoS Pathog 9: e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Garmory HS, Titball RW (2004) ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72: 6757–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jahn R, Scheller RH (2006) SNAREs – engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643. [DOI] [PubMed] [Google Scholar]
- 95. Fasshauer D, Sutton RB, Brunger AT, Jahn R (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A 95: 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Grinter R, Milner J, Walker D (2012) Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PLoS One 7: e33033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cheng Y, Zak O, Aisen P, Harrison SC, Walz T (2004) Structure of the human transferrin receptor-transferrin complex. Cell 116: 565–576. [DOI] [PubMed] [Google Scholar]
- 98. Kratz F, Beyer U, Roth T, Tarasova N, Collery P, et al. (1998) Transferrin conjugates of doxorubicin: synthesis, characterization, cellular uptake, and in vitro efficacy. J Pharm Sci 87: 338–346. [DOI] [PubMed] [Google Scholar]
- 99. Singh M (1999) Transferrin As A targeting ligand for liposomes and anticancer drugs. Curr Pharm Des 5: 443–451. [PubMed] [Google Scholar]
- 100. Kondo Y, Ohara N, Sato K, Yoshimura M, Yukitake H, et al. (2010) Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect Immun 78: 2846–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kaito C, Morishita D, Matsumoto Y, Kurokawa K, Sekimizu K (2006) Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol Microbiol 62: 1601–1617. [DOI] [PubMed] [Google Scholar]
- 102. Doern CD, Holder RC, Reid SD (2008) Point mutations within the streptococcal regulator of virulence (Srv) alter protein-DNA interactions and Srv function. Microbiology 154: 1998–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ariyachet C, Solis NV, Liu Y, Prasadarao NV, Filler SG, et al. (2013) SR-like RNA-binding protein Slr1 affects Candida albicans filamentation and virulence. Infect Immun 81: 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kovacs Simon A, Titball RW, Michell SL (2011) Lipoproteins of bacterial pathogens. Infect Immun 79: 548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Olekhnovich IN, Kadner RJ (2002) DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J Bacteriol 184: 4148–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nagy G, Dobrindt U, Schneider G, Khan AS, Hacker J, et al. (2002) Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect Immun 70: 4406–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH (2004) The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol 186: 3355–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang L, Vinogradov EV, Bogdanove AJ (2013) Requirement of the lipopolysaccharide O-chain biosynthesis gene wxocB for type III secretion and virulence of Xanthomonas oryzae pv. Oryzicola. J Bacteriol 195: 1959–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bukowski M, Lyzen R, Helbin WM, Bonar E, Szalewska-Palasz A, et al. (2012) A regulatory role for Staphylococcus aureus toxin-antitoxin system PemIKSa. Nat Commun 4: 2012. [DOI] [PubMed] [Google Scholar]
- 110. Zehr ES, Tabatabai LB (2012) Bayles (2012) DO Genomic and proteomic characterization of SuMu, a Mu-like bacteriophage infecting Haemophilus parasuis. BMC Genomics 13: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of predicted physicochemical parameters by Expasy's ProtParam tool of 429 HP from H. influenzae.
(DOCX)
List of predicted sub-cellular localization of 429 HPs from H. influenzae.
(DOCX)
List of annotated functions of 429 HPs from H. influenzae using BLASTp, STRING, SMART, INTERPROSCAN and MOTIF.
(DOCX)
List of functionally annotated domains of 429 HPs from H. influenzae by CATH, SUPERFAMILY, PANTHER, Pfam, SYSTERS, CDART SVMProt and ProtoNet.
(DOCX)
List of annotated functions of 100 proteins with known function from H. influenzae using BLASTp, SMART, INTERPROSCAN and MOTIF for ROC analysis.
(DOCX)
List of functionally annotated domains of 100 proteins with known function from H. influenzae by CATH, SUPERFAMILY, PANTHER, Pfam, SYSTERS, CDART SVMProt and ProtoNet for ROC analysis.
(DOCX)
List of accuracy, sensitivity, specificity and ROC area of various bioinformatics tools used for predicting function of HPs from H. influenzae obtained after ROC analysis.
(DOCX)
List of clusters formed by CLUSS online tool and predicted motif sequence site and sequence by MEME Suite in 429 HPs from H. influenzae.
(DOCX)
List of annotated HPs at low confidence from H. influenzae.
(DOCX)