Abstract
The virion host shutoff protein (vhs) of herpes simplex virus triggers accelerated degradation of cellular and viral mRNAs while sparing other cytoplasmic RNA species. Previous work has shown that vhs forms a complex with translation initiation factor eIF4H, which displays detectable RNase activity in the absence of other viral or host proteins. However, the contributions of eIF4H and other host factors to the activity and mRNA targeting properties of vhs have not yet been directly examined. An earlier report from our laboratory demonstrated that rabbit reticulocyte lysate (RRL) contains one or more factors that strongly stimulate the RNase activity of vhs produced in Saccharomyces cerevisiae. We report here that such yeast extracts display significant vhs-dependent RNase activity in the absence of mammalian factors. This activity differs from that displayed by vhs generated in RRL in that it is not targeted to the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES). Activity was strongly enhanced by the addition of RRL, eIF4H, or the related translation factor eIF4B. RRL also reconstituted strong targeting to the EMCV IRES, resulting in a major change in the RNA cleavage pattern. In contrast, eIF4H and eIF4B did not reconstitute IRES-directed targeting. These data indicate that eIF4B and 4H stimulate the nuclease activity of vhs, and they provide evidence that additional mammalian factors are required for targeting to the EMCV IRES.
Herpes simplex virus (HSV) is a large, enveloped DNA virus that establishes a latent infection in neural cells. During lytic infection, viral genes are expressed in a coordinately and temporally regulated fashion through both transcriptional and posttranscriptional controls (51). HSV genes fall into three kinetic classes: immediate early (IE or α), early (E or β), and late (L or γ). The IE genes are transcribed in the absence of de novo protein synthesis and stimulate the expression of the E and L genes. The E genes encode proteins required for DNA synthesis, and the L genes encode structural proteins. The HSV virion contains a number of regulatory proteins that affect the earliest events of viral infection. These proteins are part of the tegument—an amorphous protein layer located between the capsid and the envelope. The tegument-associated regulatory proteins are delivered into the cytoplasm upon fusion of the virion envelope with the host cell membrane and can therefore act prior to the onset of viral gene expression. One of these regulatory proteins is the virion host shutoff (vhs) protein, encoded by the UL41 gene, which triggers rapid shutoff of host protein synthesis following HSV infection (12, 13, 19, 42, 45, 46, 55, 57).
Upon delivery into the host cytoplasm, the 58-kDa vhs protein directs the shutoff of host protein synthesis, disruption of preexisting polyribosomes, and the degradation of cellular mRNAs in the absence of de novo viral gene expression (11, 14-16, 27, 28, 40, 41, 46, 63). The destabilization of cellular mRNAs by vhs is thought to aid viral mRNAs in accessing the cellular translational machinery by alleviating competition from cellular mRNAs. Additionally, vhs also destabilizes viral mRNAs belonging to all kinetic classes (27, 40, 41, 63), which sharpens the transitions between sequential phases of viral gene expression by tightly coupling changes in transcription with mRNA levels (15, 27, 40, 41, 46, 63). Some evidence suggests that the activity of the vhs delivered by the infecting virion is downregulated later during infection by one or more newly synthesized viral proteins, allowing viral mRNAs to accumulate to high levels after cellular mRNAs have been degraded (12, 15, 57). The transactivator VP16 has been implicated in this negative regulation of vhs activity. VP16 binds directly to vhs (56). Moreover, a VP16 null mutant undergoes accelerated turnover of host and viral mRNA and arrest of all protein synthesis at intermediate times postinfection (29), an effect that is eliminated by inactivating the UL41 gene (29, 39). In addition, a mutant VP16 protein which is unable to interact with vhs is able to complement the growth of a VP16/vhs null mutant but not a VP16 null virus, suggesting that direct interaction between VP16 and vhs is required for efficient virus growth (23).
Although vhs is not essential for virus replication, vhs mutants do display a 5- to 10-fold reduction in virus yield in tissue culture infection (45, 46, 57). In addition, vhs plays a critical role in vivo in HSV pathogenesis. Viral isolates bearing inactivating mutations in the UL41 gene display a greatly impaired ability to replicate in mouse trigeminal ganglia, corneas, and brains and a severely impaired ability to establish latency (31, 60-62). The basis for this profound attenuation remains unclear. However, accumulating evidence suggests that vhs plays a key role in disarming host innate and adaptive immunity, and it is possible that these effects contribute to the attenuation of vhs mutants. Thus, vhs reduces the expression of major histocompatibility complex class I molecules, thereby contributing to the resistance of lysis of HSV-infected cells by cytotoxic T lymphocytes (66). vhs also aids in reducing the expression of major histocompatibility complex class II molecules (67) and prevents dendritic cell activation in response to viral infection (52), thus reducing the amount of antigen presentation to the immune system. vhs has also been found to suppress production of the cytokines interleukin-1β (IL-1β), IL-8, and macrophage inflammatory protein 1α in macrophages and IL-8 in HEL cells (64).
Strong evidence suggests that vhs is either an RNase or the subunit of a multicomponent RNase that contains the active site. First, extracts of HSV-infected mammalian cells and of partially purified virions display RNase activity, and this activity is eliminated by vhs mutations (26, 59, 68). Second, antibodies against vhs inhibit the RNase activity of virion extracts (68). Third, vhs induces endonucleolytic cleavage of a number of RNA substrates when it is expressed as the only HSV protein in the rabbit reticulocyte lysate (RRL) in vitro translation system (4, 5, 68). Fourth, vhs, along with homologues from other alphaherpesviruses, shares amino acid sequence similarity in two regions with a family of mammalian, yeast, bacterial, and phage nucleases involved in DNA replication and repair (3, 6, 7). These similarities appear to be functionally relevant, as mutations in vhs that alter conserved residues that are known to be located in the active site and essential for catalytic activity of cellular homologues inhibit the ability of the vhs mutants to inhibit the expression of a reporter gene (7). Lastly, Everly et al. partially purified a soluble complex of vhs and the eukaryotic translation initiation factor 4H (eIF4H) formed through coexpression in Escherichia coli and showed that it displays RNase activity (7). The RNase activity of the complex was eliminated by mutations that altered residues located in the nuclease motif of vhs (7), suggesting that vhs contains at least part of the active site. Taken together, these data indicate that vhs is an integral and required part of the vhs-dependent RNase. However, vhs has not yet been purified to homogeneity in a soluble and biologically active form. Thus, the possibility remains that the vhs-dependent RNase requires one or more cellular factors such as eIF4H for full activity.
Most if not all HSV and cellular mRNAs are targeted for degradation by vhs, while rRNAs and tRNAs are spared in both in vitro and in vivo systems (26, 27, 40, 68). Thus, mRNAs may be selected for degradation via a common feature not present in other RNAs. The 3′ poly(A) tail was originally proposed as a recognition site for the vhs-dependent RNase (68); however, in both the RRL system and in extracts of HSV-infected cells it has been observed that the poly(A) tail is not required for vhs-dependent degradation of an RNA substrate (4, 21). The 5′ cap was also found to be dispensable for vhs-dependent degradation of mRNAs in both RRL and extracts of partially purified virions (4, 68). Although the foregoing results might be taken to exclude roles for both the 5′ cap and 3′ poly(A) tail, it is important that neither modification is required for efficient translation in RRL. Thus, either or both features may be important for substrate recognition in vivo. Indeed, several lines of evidence suggest that vhs targets mRNAs via interactions with the translation apparatus. In HSV-infected cells, sequences at the 5′ end of HSV thymidine kinase mRNA are degraded before the sequences at the 3′ end of the transcript (21). Consistent with this observation, the initial cleavage sites of signal recognition particle α-subunit (SRP-α) mRNA are clustered around the 5′ end in RRL containing pretranslated vhs (4). As well, the internal ribosome entry sites (IRES) of the encephalomyocarditis virus (EMCV) and poliovirus serve as movable elements which direct vhs-dependent cleavage events to sequences immediately 3′ of the IRES (5).
In all of these cases, the initial cleavage sites are apparently clustered in the vicinity of regions of translation initiation, raising the possibility that vhs may initially contact mRNAs via interactions with a component(s) of the translation initiation machinery. Consistent with this hypothesis, vhs has been found to interact with eIF4H in mammalian cells (9). Some mutant forms of vhs which are unable to degrade RNA in vivo are also unable to interact with eIF4H (9), suggesting the interaction may be important for vhs activity. eIF4H is a 25-kDa protein which stimulates the translational activity of eIF4B and eIF4F (composed of eIF4E, eIF4A, and eIF4G) and stimulates the helicase activity of eIF4A by increasing its processivity (47-50). eIF4H is thought to work together with eIF4A and eIF4B to unwind the secondary structure in the 5′ untranslated regions of mRNAs. eIF4B is a sequence paralogue of eIF4H and displays similar functions. It stimulates the RNA binding and ATPase activity of eIF4A (reviewed in reference 50) and together with eIF4H modulates the helicase activity of eIF4A and eIF4F (50).
As noted above, Everly et al. (7) have shown that the vhs-eIF4H complex displays detectable RNase activity in the absence of other viral and host proteins. However, it is not yet clear if vhs has RNase activity in the absence of eIF4H. In addition, the potential role of eIF4H and other cellular factors in controlling vhs activity and targeting it to mRNAs has not yet been assessed. Possibly relevant to these issues, a previous report from our laboratory indicated that cell extracts of Saccharomyces cerevisiae engineered to express vhs display little if any vhs-dependent RNase activity (34). However, RNase activity was readily apparent when blank RRL was added to the extracts (34), suggesting that the vhs-dependent RNase requires one or more mammalian factors for efficient activity. Given that vhs interacts with eIF4H and the vhs-eIF4H complex has RNase activity, we speculated that eIF4H might be the relevant factor. We therefore sought to determine if eIF4H contributes to the RNase activity of vhs-dependent RNase and, if so, whether it suffices to target vhs to specific sites on RNA substrates. In order to address this question, we partially purified the translation initiation factors eIF4A, eIF4B, and eIF4H and assayed them for their abilities to enhance the RNase activity of extracts of yeast expressing vhs. The results indicated that both eIF4H and its sequence paralogue eIF4B interact with vhs and stimulate its RNase activity. However, none of the translation initiation factors tested could restore IRES-mediated targeting to vhs, indicating that one or more other cellular factors are required to restore targeting to vhs.
MATERIALS AND METHODS
Plasmids.
The eIF4H open reading frame (ORF) was obtained by reverse transcription-PCR from HeLa cell RNA using the following PCR primers: OSW1 (5′-GACGGCATATGGCGGACTTCGACC-3′), used to engineer an NdeI site at the AUG start codon to clone eIF4H in frame into pET15b (Novagen), and OSW2 (5′-CTCGACTCCCTCCCAACCGCAGGCTCATTC-3′), used to engineer a XhoI site after the stop codon. The PCR products were ligated into the pGEM-T Easy vector (Promega). Both eIF4H and eIF4Hi were isolated, creating plasmids pSW1 and pSW2, respectively. The sequence of eIF4Hi was pristine; however, the sequence coding for eIF4H contained a deletion of one cytosine in a run of seven cytosines at position 531 to 537. To correct the deletion, oligonucleotide-directed mutagenesis was used. eIF4H was cut out of pSW1 using SphI and SstI and ligated into pSELECT-1 (Altered Sites mutagenesis kit; Promega). The mutagenic oligo (5′-GGCTGCCCATGGGGGGGCCTGTTCGCC-3′) was annealed to the recombinant single-stranded DNA template. T4 DNA polymerase was used to synthesize the mutant strand, and the ligated plasmid was transformed into BMH71-18 mutS. DNA was isolated and retransformed into HB101. Candidate plasmids were sequenced, and one with the eIF4H mutation corrected was designated pGC2.
The NdeI fragment of pGC2 bearing the eIF4H ORF was cloned into the NdeI site of pET15b (Novagen), creating pRD1. To clone eIF4Hi into pET15b, pSW2 was digested with SphI and then NdeI after the ends were made flush with T4 DNA polymerase. The SphI-NdeI fragment bearing the eIF4Hi ORF was cloned into the NdeI-BamHI site of pET15b (after repairing the BamHI site with the Klenow fragment of DNA polymerase I) to generate pRD2.
Gene Storm expression-ready clones that contained the ORF of eIF4A (accession no. D13748) or eIF4B (accession no. X55733) were purchased from Invitrogen. To clone eIF4A into pET15b, the eIF4A ORF was PCR amplified (PlatinumTaq High Fidelity; Invitrogen) from pCDNA3.1/GS/eIF4A (Invitrogen) using PCR primers ORD4 (5′-CATATGTCTGCGAGCCAGCAT-3′), used to engineer an NdeI site at the start codon, and ORD5 (5′-TCAGATGAGGTCAGCAACATTGAGG-3′), used to create a stop codon at the carboxy terminus of the protein. The eIF4A PCR product was ligated into pCR2.1-TOPO (Invitrogen) to generate pRD6. The eIF4B ORF was PCR amplified as described for eIF4B using PCR primers ORD1 (5′-CATATGGCGGCCTCAGCAAAAAAG-3′) and ORD14 (5′-TTACTATCATTCGGCATAATCTTCTCCCTC-3′) and cloned into pCR2.1-TOPO (Invitrogen), creating pRD9. The NdeI-BamHI fragments of pRD6 and pRD9 containing the ORFs of eIF4A and eIF4B, respectively, were cloned between the NdeI-BamHI sites of pET15b to generate pRD12 and pRD10.
DNA primers were synthesized and plasmids were sequenced at the Biochemistry DNA Services Laboratory, Faculty of Medicine, University of Alberta.
pcdc34Δ209His is a bacterial expression plasmid that expresses the yeast ubiquitin-conjugating enzyme cdc34Δ209 (44) and was generously supplied by Michael Ellison (University of Alberta).
The in vitro transcription vectors pCITE-1 and pSPSR19N, which were used to generate the reporter RNA substrates pCITE and SRP-α, respectively, and the vhs in vitro translation vector pSP6vhs have been described previously (4, 5).
The yeast expression vectors pYEX-BX (Clontech) and pYEX-BX2.1vhs, which encodes an 8xHis-hemagglutinin-tagged vhs ORF have been previously described (34). A derivative of pYEX-BX2.1vhs containing the inactivating D215N mutation (7) was created using the QuikChange site-directed mutagenesis kit (Stratagene) according to supplied protocols. Sequences of the complementary mutagenic oligonucleotide primers were as follows: 5′-CACCACGGACACTAACCTCCTGTTGATGGG and 5′-CCCATCAACAGGAGGTTAGTGTCCGTGGTG.
Bacterial strains and growth conditions.
All plasmids unless otherwise stated were maintained and amplified in E. coli strain HB101 (F− Δ[gpt-proA]62 leuB6 supE44 ara-14 galK2 lacY1 Δ[mcrC-mrr] rpsL20 [Strr] xyl-5 mtl-1 recA13) (36). Plasmids derived from pCR2.1-TOPO were maintained and amplified in strain TOP10 Electrocomp (Invitrogen) [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara.leu)7697 galU galK rpsL (Strr) endA1 nupG]. Plasmids derived from pcDNA3.1/GS were maintained and amplified in strain GeneHogs (Invitrogen) [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 endA1 recA1 deoR araD139 Δ(ara.leu)7697 galU galK rpsL nupG]. E. coli strains BMH17-18mutS (Promega) (thi supE Δ[lac-proAB] [mutS::Tn10] [F′ proA+B+ laqIqZΔM15]) and XL1-Blue (Stratagene) were used in oligonucleotide-directed mutagenesis. All protein expression was performed with strain BL21(DE3)/pLysS (Novagen) [F− dcm ompT hsdS (r8− m8−) gal λ(DE3)/pLysS Camr].
All strains transformed with recombinant plasmids were cultured at 37°C in Luria-Bertani medium (1.0% Bacto Tryptone, 0.5% yeast extract, 1.0% NaCl; pH 7.0) containing 100 μg of ampicillin per ml in a shaker incubator set at 225 rpm. Chloramphenicol (34 μg/ml) was also added when culturing strain BL21(DE3)/pLysS. For the GeneHogs strain, the salt concentration of the Luria-Bertani medium was reduced to 0.5%, the pH was increased to pH 7.5, and the antibiotic zeocin (25 μg/ml; Invitrogen) was used.
Yeast strain, growth conditions, and yeast transformation.
The S. cerevisiae strain W303-1A (MATa SUC3 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) (32) was used as the host for plasmids derived from pYEX-BX. Growth conditions and transformation of W303-1A have been described previously (34).
Preparation of yeast extracts for the vhs activity assay.
The method of Schultz et al. (53, 54) was used for the preparation of frozen cells and yeast extracts. Briefly, 1 liter of YNBG (0.67% yeast nitrogen base without amino acids, 2% galactose; supplemented with adenine, histidine, and tryptophan) was inoculated with an overnight culture of S. cerevisiae strain W303-1A/pYEX-BX, W303-1A/pYEX-BX2.1vhs, or W303-1A/pYEX-BX2.1vhs D215N and grown to an optical density at 600 nm (OD600) of ∼3 to 4. The culture was induced with 0.15 mM CuSO4 for 5 h. The cells were then harvested, washed, frozen in liquid nitrogen, and stored at −80°C. About 3 g of frozen cells was used to prepare the extracts. Frozen cells were ground in a coffee mill, thawed, and resuspended in 1.3 volumes of extraction buffer (100 mM HEPES-KOH [pH 7.9], 245 mM KCl, 5 mM EGTA, 2.5 mM dithiothreitol [DTT]) with protease inhibitors. The extract was centrifuged at 100,000 × g for 2 h. The supernatant was then collected and dialyzed into vhs assay buffer (1.6 mM Tris-acetate, 80 mM potassium acetate, 2.0 mM magnesium acetate, 0.1 mM DTT). Aliquots were frozen and stored at −80°C. ATP (0.25 mM) and 1 μl of RNaseOut (Invitrogen) per 50 μl sample were added upon thawing of aliquots.
Protein expression and purification.
Protocols were adapted from The QIAexpressionist (Qiagen). For the expression of eIF4H, eIF4Hi, and cdc34Δ209, 1 liter of culture was grown to an OD600 of ∼0.3 to 0.4 and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. The cells were then harvested, resuspended in 2 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 5 mM imidazole, 25% glycerol; pH 8.0) per g (wet weight), and frozen at −80°C. All subsequent purification steps were performed at 4°C or on ice. Cells were thawed for 30 min and lysed by three sonication cycles of 30 s each (550 Sonic Dismembrator; Fisher Scientific). The lysate was incubated with DNase I (5 μg/ml) and 8 mM MgCl2 for 20 min, and the insoluble material was removed by centrifugation for 25 min at 10,000 × g. The supernatant was incubated with 1 ml of Ni-nitrilotriacetic acid-agarose (Qiagen) per 4 ml of lysate for 1 h on a Nutator (Becton Dickinson). The resin was pelleted and washed sequentially with 10 ml of lysis buffer, wash buffer (lysis buffer plus 20 mM imidazole), and lysis buffer. The protein was batch eluted twice with a volume equivalent to the resin of elution buffer (lysis buffer plus 400 mM imidazole). Elution fractions were combined and dialyzed overnight into diethyl pyrocarbonate-treated vhs assay buffer with 25% glycerol or buffer B (20 mM Tris, 1 mM DTT, 0.1 mM EDTA, 100 mM KCl, 2.0 mM MgCl2) with 25% glycerol. Aliquots were frozen at −80°C. ATP (0.25 mM) and RNaseOut were added after thawing of aliquots. We obtained 3 to 4 mg of eIF4H and 1 to 1.5 mg of cdc34Δ209 per liter of culture. eIF4H was as active in buffer B as in vhs assay buffer.
eIF4A and eIF4B were expressed and purified in the same manner as eIF4H, with a few modifications. Two liters of culture per protein purified was grown to an OD600 of 1.0 and induced with 1 mM IPTG for 2 h. Cells expressing eIF4B were grown at 30°C instead of 37°C. Cells were harvested, resuspended in lysis buffer (10% glycerol), and lysed as described above. One milliliter of Ni-nitrilotriacetic acid-agarose was added per 6 ml of lysates expressing eIF4B. Bound protein was batch eluted with one-half the volume of resin of elution buffer (10% glycerol). Elution fractions were dialyzed overnight into diethyl pyrocarbonate-treated buffer B with 10% glycerol. Aliquots were frozen and stored at −80°C. ATP (0.25 mM) and 1 μl of RNaseOut per 50 μl of protein were added upon thawing of aliquots. Protein concentration was determined with the Bio-Rad protein assay using bovine serum albumin (BSA) as the standard.
In vitro transcription and RNA labeling.
vhs mRNA to be used for in vitro translation was produced using the method of Elgadi et al. (4). Uncapped, internally labeled reporter RNAs were generated in the same manner, except that the cap primer was omitted, the GTP concentration was reduced to 0.25 mM, and the length of the reaction was shortened to 40 min (5, 58). To generate pCITE reporter RNA, EcoNI-linearized template DNA (pCITE-1) was transcribed with T7 RNA polymerase to yield a runoff transcript of 2.3 kb (5). SRP-α reporter RNA was transcribed using an EcoRV-linearized template DNA (pSPSR19N) and SP6 RNA polymerase to yield a runoff transcript of 2.4 kb (4). Unlabeled pCITE reporter RNA was generated using the MEGAscript transcription kit (Ambion) according to the manufacturer's protocol.
In vitro translation.
In vitro translation of vhs using RRL has been previously described (4, 58).
vhs activity assay.
The vhs activity assay has been described elsewhere (34). Briefly, yeast extracts were mixed with various test proteins, blank RRL, or buffer. The amount of total protein present in the volume of yeast extract used per time point was 20 μg before the addition of test proteins. Reporter RNAs (4,000 Cerenkov cpm per time point) were added to the indicated samples, and all reaction mixtures were adjusted to the same final volume with vhs assay buffer or buffer B. The reactions were incubated at 30°C, and aliquots were removed at the indicated time points. RNA was recovered with the RNeasy Mini kit (Qiagen) using the clean-up protocol following the manufacturer's instructions and then precipitated with 1 volume of isopropanol and a 1/10 volume of 3 M sodium acetate. The RNA pellet was washed sequentially with 70 and 95% ethanol and then resuspended in 25 μl of RNA sample buffer (1× morpholinepropanesulfonic acid [MOPS] buffer [200 mM MOPS, 50 mM sodium acetate, 5 mM EDTA, adjusted pH to 7.0 with NaOH], 50% deionized formamide, 16.7% formaldehyde).
Agarose gel electrophoresis and Northern blot analysis.
Samples were incubated at 55°C for 10 to 15 min and then transferred to ice for 2 min. RNA samples were combined with 2 μl of RNA loading buffer (50% glycerol, 1 mM EDTA, 10 mg of xylene cyanol/ml, and 10 mg of bromophenol blue/ml) and loaded onto a 1.2 to 1.4% agarose gel containing 2% formaldehyde. Electrophoresis was carried out in 1× MOPS buffer at 130 V for 2 h until the gel had run ∼7 to 8 cm. RNA was transferred to a GeneScreen Plus nylon membrane (NEN Life Sciences Products) in 10× SSC (1× SSC is 1.5 M sodium chloride and 150 mM sodium citrate). Following UV cross-linking (Stratalinker 2400; Stratagene), 32P-labeled RNA fragments were detected by exposure to Kodak BioMax MS film at −80°C.
Unlabeled RNA fragments were detected by hybridization. After UV cross-linking, the membranes were rinsed in 2× SSC and prehybridized in 15 ml of modified Westneat solution (6.6% sodium dodecyl sulfate [SDS], 250 mM MOPS [pH 7.0], 5× Denhardt's solution, 1 mM EDTA) at 48°C for 1 h. Membranes were hybridized to either ORD17 (5′-GCCTTATTCCAAGCGGCTTCGGCCAGTC-3′), complementary to residues 45 to 72 of the pCITE transcript, or JPP-E (5′-GCCGGAGTTGGATGATGACCCGACG-3′), complementary to the extreme 3′ end of the pCITE RNA. Oligonucleotides were 5′ 32P labeled using T4 polynucleotide kinase (Invitrogen). The entire kinase reaction mixture, after a 10-min incubation at 95°C, was added to the prehybridization Westneat solution. Hybridization was carried out overnight at 48°C. The membrane was then washed twice in 2× SSC-0.2% SDS at room temperature for 5 min and twice in 0.2× SSC-0.2% SDS at 48°C for 20 min before being subjected to autoradiography.
Far Western analysis.
The far Western protocol was performed following the method of Guichet et al. (17). Briefly, 8 μg of each test protein was run on an SDS-12% polyacrylamide gel (SDS-PAGE) gel and transferred to a nitrocellulose membrane (Hybond ECL membrane; Amersham Biosciences). Human protein phosphatase 1 (PP1) was kindly provided by Kathleen Perrault (University of Alberta). After the transfer was complete, the blot was blocked with 2% milk powder in AC buffer (10% glycerol, 100 mM NaCl, 20 mM Tris, 0.5 mM EDTA, 0.1% Tween 20) for 1 h at 4°C. To prepare the probe, RRL containing 35S-labeled vhs generated by in vitro translation was spun through a 1-ml syringe packed with G-25 resin (Amersham Pharmacia) to remove unincorporated amino acids. The probe was mixed with 10 ml of 2% milk powder in AC buffer with 1 mM DTT. After the blocking step, the probe mix was added to the blot and incubated for 2 h at 4°C. The blot was washed with 2% skim milk in AC buffer for 15 min and four times for 20 min in AC buffer. The blot was dried and visualized by autoradiography
RESULTS
Expression of vhs in S. cerevisiae induces an RNase activity that is stimulated by one or more mammalian factors.
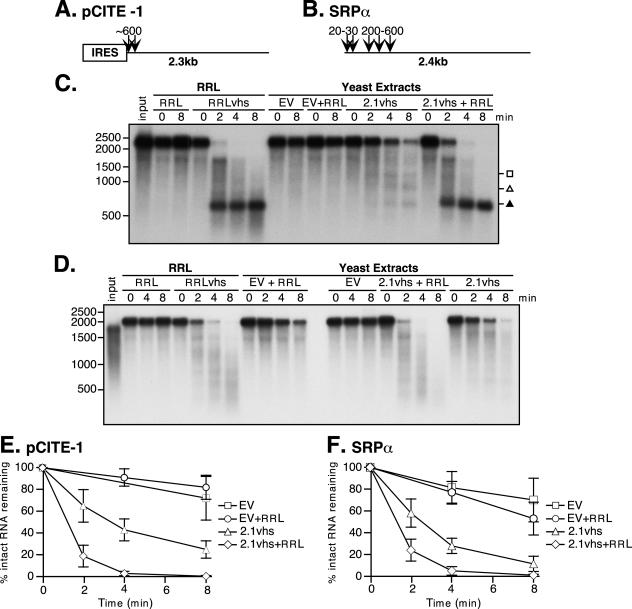
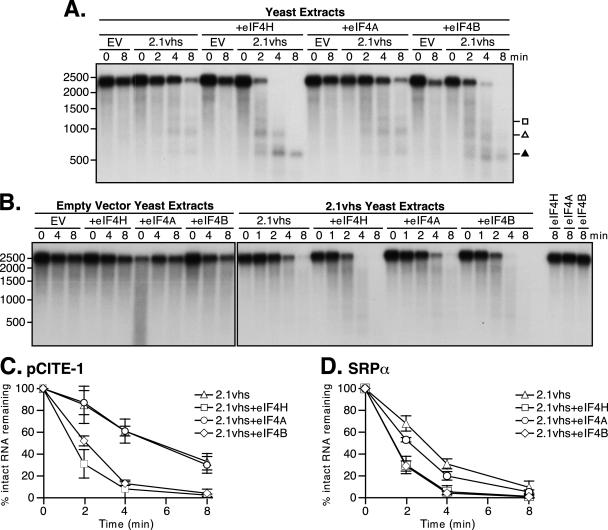
A previous study from this laboratory reported that cell extracts prepared from yeast engineered to express vhs display little if any RNase activity when assayed under conditions that readily detect the activity of vhs translated in RRL (34). In contrast, vhs-dependent activity was easily detected when RRL was added to the yeast extracts. These observations suggested that vhs produced in yeast requires one or more mammalian factors for efficient activity in vitro. Our more recent trials of this experiment confirm the stimulatory effect of RRL (Fig. 1). However, in contrast to the previous study, we are now able to reproducibly detect vhs-dependent RNase activity in the yeast extracts in the absence of RRL, as shown in the following representative experiments.
FIG. 1.
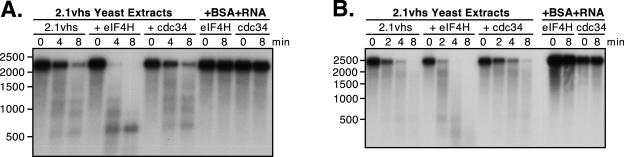
Extracts of yeast expressing 2.1vhs contain endoribonuclease activity that is enhanced by one or more mammalian cofactors. (A and B) pCITE-1 and SRP-α RNA substrates, respectively, showing the sites of initial cleavage events by vhs translated in RRL. The EMCV IRES on pCITE-1 is indicated. (C and D) Analysis of nuclease activity on pCITE-1 and SRP-α RNA substrates, respectively. Internally labeled pCITE-1 and SRP-α RNA was added to control RRL (RRL), RRL pretranslated with vhs (RRLvhs), extracts of yeast harboring the empty expression vector either alone (EV) or mixed with RRL (EV+RRL), and extracts of yeast expressing 2.1vhs either alone (2.1vhs) or mixed with RRL (2.1vhs+RRL). RNA was extracted at the indicated time points, resolved on a 1.2% agarose-2% formaldehyde gel, and transferred to a Gene Screen Plus membrane, and the RNA signal was detected by autoradiography. The solid triangle indicates the previously described 600-nt degradation product corresponding to the EMCV IRES. The position and size of the markers are indicated in nucleotides at the left. (E and F) Quantification of nuclease activity on pCITE-1 and SRP-α RNAs, respectively. The quantity of full-length RNA in panels C and D and in at least two other experiments was determined using phosphorimager analysis and plotted against the time points indicated. The error bars indicate the standard deviation of each time point calculated from at least three independent experiments.
Cultures of S. cerevisiae harboring a previously described vhs expression vector (2.1vhs) (34) or empty vector were induced with copper sulfate, and whole-cell extracts were prepared as described in Materials and Methods. The extracts were then assayed for vhs-dependent RNase activity in the presence and absence of added RRL, as previously described (34). Two RNA substrates were used for this assay: pCITE-1 RNA, which bears the IRES of EMCV at its 5′ end, and SRP-α RNA (Fig. 1A and B). Internally labeled RNAs generated by in vitro transcription were added to the extracts, and samples withdrawn at various times were analyzed by formaldehyde-agarose gel electrophoresis followed by autoradiography (Fig. 1C and D). As controls, the substrate RNAs were also incubated in RRL containing pretranslated vhs (RRLvhs) and in blank RRL. Previous studies have shown that pCITE-1 RNA is initially cleaved immediately 3′ of the IRES in the RRL in vitro assay system (Fig. 1A), giving rise to 5′ and 3′ products of 600- and 1,800-nucleotides (nt), respectively (5). The 600-nt fragment containing the IRES is stable throughout the course of the reaction, while the 1,800-nt fragment is subjected to further decay. As shown in Fig. 1C, we readily detected the stable 600-nt fragment in the reaction mixture with RRL containing pretranslated vhs; however, we could not unambiguously detect the predicted 1,800-nt 3′ fragment in this experiment because the input RNA substrate contained small amounts of an RNA species that migrated at this position in the gel (see Fig. 7A for clearer evidence for production of the 1,800-nt 3′ fragment). The pCITE-1 RNA was quite stable in the extract prepared from yeast harboring the empty expression vector, both in the absence and presence of added RRL (Fig. 1C, EV and EV+RRL). However, it was significantly less stable in the extract prepared from yeast induced to express vhs (Fig. 1C, 2.1vhs, and data quantified in E). Moreover, several discrete degradation products could be detected in the 2.1vhs (but not control) extract (Fig. 1C). These effects, observed in multiple experiments with several independently prepared extracts, indicated that expression of vhs in yeast is associated with the appearance of a novel RNase activity in cell extracts. The degradation profile observed with the 2.1vhs extracts differed from that observed in RRLvhs in that substantially reduced levels of 600-nt product were detected and 900- and 1,200-nt fragments were also produced (Fig. 1C). These findings raised the possibility that the vhs-dependent RNase detected in the yeast extracts is not targeted to the RNA substrate in the same fashion as in RRL, a hypothesis that is explored in further detail below (see Fig. 7). Addition of RRL enhanced the RNase activity of the 2.1vhs extracts and altered the overall pattern, such that substantial amounts of the 600-nt fragment accumulated. In contrast, RRL had no effect when added to the empty vector extracts (Fig. 1C and 2). The addition of RRL may enhance the recovery of a stable 600-nt fragment by targeting cleavage to just 3′ of the IRES (thus causing the nuclease to bypass the IRES), by providing factors that bind the IRES and protect it from degradation, or by a combination of both mechanisms. Broadly similar results were obtained with SRP-α RNA, with the exception that in this case discrete reaction products were not readily detected in any of the reactions (Fig. 1D and data quantified in F). Thus, the transcript was significantly less stable in extracts of yeast expressing vhs than in control extracts, and the activity of these extracts was enhanced by the addition of RRL.
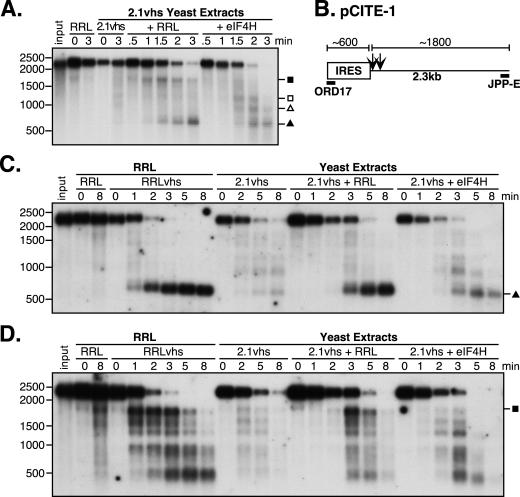
FIG. 7.
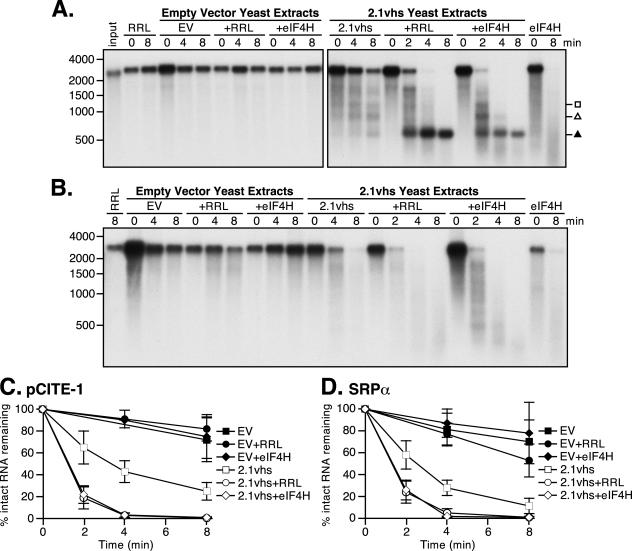
eIF4H does not restore efficient IRES-directed targeting to yeast-expressed vhs. (A) Analysis of nuclease activity on pCITE-1 RNA. Internally labeled pCITE-1 RNA was added to control RRL (RRL) and yeast extracts containing 2.1vhs in the presence or absence of RRL or eIF4H (2.1vhs, 2.1vhs+RRL, and 2.1vhs+eIF4H). Samples were processed as described in the legend for Fig. 1. (B) Diagram of pCITE-1 RNA, indicating the positions of hybridization of oligonucleotides ORD17 and JPP-E specific for the 5′ and 3′ ends of the RNA, respectively. Also indicated are the RNA fragments produced upon initial cleavage of the RNA by vhs translated in RRL. (C and D) Northern blot analysis of nuclease activity on pCITE-1 RNA using oligonucleotide probes for the 5′ and 3′ ends, respectively, of pCITE-1 RNA. Unlabeled pCITE-1 RNA was added to control RRL (RRL), RRL containing pretranslated vhs (RRLvhs), and yeast extracts containing 2.1vhs, either alone (2.1vhs), mixed with RRL (2.1vhs+RRL), or mixed with eIF4H (2.1vhs+eIF4H). RNA was extracted at the indicated time points, resolved on an agarose-formaldehyde gel, transferred to a Gene Screen Plus membrane, and analyzed by Northern blot analysis. Symbols are as described in the legend for Fig. 3. The closed square indicates the position of the 1,800-nt fragment. The positions and sizes of the RNA markers in nucleotides are at the left.
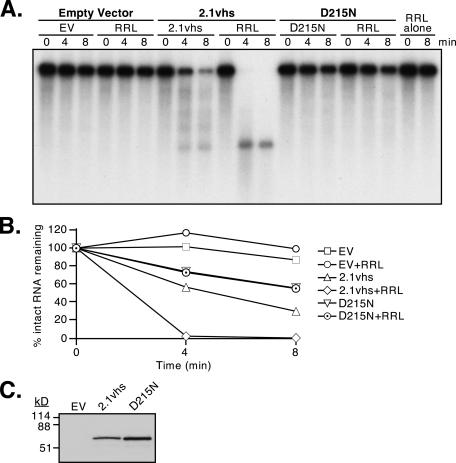
FIG. 2.
Effect of the D215N vhs mutation on the RNase nuclease activity of yeast extracts. (A) Nuclease activity of cell extracts. Internally labeled pCITE-1 RNA was added to extracts of yeast harboring the empty expression vector (EV), 2.1 vhs expression vector (2.1vhs), or 2.1 expression vector bearing the D215N mutation (D215N), either alone or supplemented with RRL. In addition, a portion of the RNA was incubated in RRL (RRL). Samples were withdrawn at the indicated times, and RNA was extracted and analyzed as described in the legend for Fig. 1. (B) The data obtained in the experiment shown in panel A were quantified by phosphorimager analysis. (C) Western blot analysis of yeast extracts. Aliquots of the extracts used for panel A (each containing 20 μg of protein) were subjected to electrophoresis through an SDS-10% polyacrylamide gel, transferred to a nitrocellulose membrane, and then analyzed by Western blotting using a polyclonal rabbit anti-vhs antiserum (AE328).
The simplest explanation for the enhanced RNase activity observed in 2.1vhs extracts is that it stems from the nuclease activity of vhs. Alternatively, it is possible that expression of vhs in yeast alters the levels of one or more cellular RNases. As one approach to discriminate between these possibilities, we examined extracts of yeast expressing a mutant form of vhs that lacks nuclease activity. To this end, we introduced the D215N single amino acid substitution into the 2.1vhs expression vector. Everly et al. (7) have previously shown that this mutation lies in the nuclease domain of vhs and eliminates vhs activity without affecting binding of eIF4H. We found that the D215N extracts displayed significantly less overall RNase activity than 2.1vhs extracts, although perhaps more than that in extracts prepared from cells harboring the empty expression vector (Fig. 2A and 6A [data are quantified in Fig. 2B and 6B]). Moreover, the 600-, 900-, and 1,200-nt degradation products characteristic of the 2.1vhs extracts could not be detected. In addition, RRL had no effect on the D215N extracts. Inasmuch as the D215N extract contained at least as much intact vhs protein as the 2.1vhs extract (Fig. 2C), these data suggest that the discrete degradation products produced in the 2.1vhs extracts arise through the action of the vhs nuclease and that this activity comprises a significant fraction of the total RNase activity in these extracts.
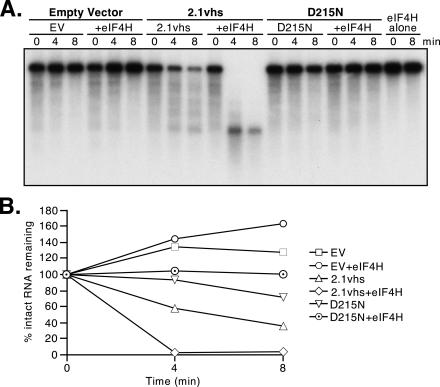
FIG. 6.
eIF4H does not enhance the RNase activity of yeast expressing inactive (D215N) vhs. Internally labeled pCITE-1 RNA was added to extracts of yeast harboring the empty expression vector (EV), 2.1 vhs expression vector (2.1vhs), or 2.1 expression vector bearing the D215N mutation (D215N), either alone or supplemented with eIF4H. In addition, a portion of the RNA was incubated with eIF4H alone. Samples were withdrawn at the indicated times, and the RNA was extracted and analyzed as described in the legend for Fig. 1. (B) The data obtained in the experiment shown in panel A were quantified by phosphorimager analysis.
Taken in combination, these results indicate that expression of vhs in yeast induces a novel vhs-dependent RNase activity that is enhanced, and perhaps altered, by one or more factors present in RRL.
eIF4H stimulates the endoribonuclease activity of vhs produced in yeast.
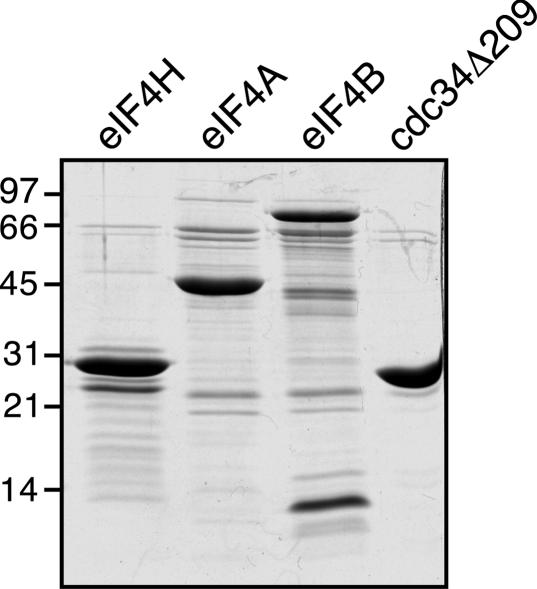
As reviewed in the introduction, the initial cleavage events induced by vhs translated in RRL occur near areas of translation initiation (4, 5). vhs has been shown to interact with the translation initiation factor eIF4H (9), and a complex of vhs-eIF4H has RNase activity (7). We therefore sought to determine whether eIF4H is the mammalian factor that stimulates the RNase activity of vhs expressed in yeast and/or serves to target vhs to specific sites on mRNAs. The eIF4H cDNA was cloned into a bacterial expression vector to express eIF4H as an N-terminally His6-tagged protein which was partially purified as described in Materials and Methods. Both eIF4H and eIF4Hi, an alternatively spliced version of eIF4H which contains an insertion of 20 amino acids after residue 137 (9), were used in preliminary assays. We did not detect any differences in the activities of the two proteins, so subsequent experiments were performed using only the shorter version of eIF4H. As a negative control, we also expressed a His6-tagged version of the yeast ubiquitin-conjugating enzyme cdc34Δ209, which is truncated at amino acid 209 (44), a presumably irrelevant protein. Translation initiation factors eIF4A and eIF4B were also expressed and partially purified as described in Materials and Methods and were used in later experiments (see Fig. 9 and 10). Figure 3 shows a Coomassie-stained gel of the partially purified proteins. The extra bands seen below the intact eIF4H on the Coomassie-stained gel are degradation products that contain the amino terminus, as an antibody directed against the His6 tag reacted with the majority of the degradation products (data not shown).
FIG. 9.
eIF4B interacts in vitro with vhs. (A) Far Western analysis of interactions between vhs and the proteins indicated. Eight micrograms of each protein was resolved on an SDS-12% PAGE gel, transferred to a nitrocellulose membrane, and incubated with RRL containing pretranslated 35S-labeled vhs, and the interaction was detected by autoradiography. (B) Analysis of purified proteins. After transfer of proteins to the membrane, the SDS-PAGE gel was stained for residual protein with Coomassie brilliant blue. The positions and sizes of the protein markers (in kilodaltons) are indicated on the left.
FIG. 10.
eIF4B stimulates the endoribonuclease activity of the vhs protein produced in yeast. (A and B) Analysis of nuclease activity on pCITE-1 and SRP-α RNAs, respectively. Internally labeled pCITE-1 and SRP-α RNAs were added to extracts of yeast containing the empty expression vector either alone (EV), mixed with eIF4H (+eIF4H), mixed with eIF4A (+eIF4A), or mixed with eIF4B (+eIF4B) and yeast extracts containing 2.1vhs either alone (2.1vhs), mixed with eIF4H (+eIF4H), mixed with eIF4A (+eIF4A), or mixed with eIF4B (+eIF4B). The concentration of purified proteins used was as follows: eIF4H, 2.1 μM; eIF4B, 2.1 μM; eIF4A, 7.6 μM. Purified proteins were mixed with buffer B and 20 μg of BSA and 1 μg of total yeast RNA per time point. Symbols are as described in the legend for Fig. 3. The positions and sizes of the RNA markers in nucleotides are at the left. (C and D) Quantification of nuclease activity on pCITE-1 and SRP-α RNA substrates, respectively. The quantities of full-length RNA in panels A and B and two other experiments were determined using phosphorimager analysis and plotted against the time points indicated. The error bars indicate the standard deviation of each time point calculated from at least three independent experiments.
FIG. 3.
Analysis of purified proteins. Partially purified eIF4H, eIF4A, eIF4B, and cdc34Δ209 (5 μg of each) were resolved on an SDS-12% PAGE gel and stained with Coomassie brilliant blue. The positions and sizes of the protein markers are indicated in kilodaltons on the left.
Many of the recombinant protein preparations described above (eIF4A, eIF4B, eIF4H, and cdc34Δ209) contained trace amounts of contaminating RNase activity that could be detected when a 1,000-fold mass excess of the partially purified protein was incubated alone with radiolabeled substrate (Fig. 4A and B [for eIF4H] and data not shown [for other proteins]), although the amount of nuclease activity varied between preparations (data not shown). However, the partially purified proteins had no effect on the stability of the RNA substrate when they were added to control yeast extract (Fig. 4 and 10, EV+eIF4H, EV+eIF4A, and EV+eIF4B). This observation suggested that the large excess of yeast protein or (more likely) RNA present in the control extracts served to suppress this trace nuclease activity. Consistent with this interpretation, when eIF4H and cdc34Δ209 were mixed with RNase-free BSA and total RNA was purified from control extracts to the same concentration as in control extracts, no appreciable nuclease activity was observed (Fig. 5, eIF4H and cdc34Δ209). Thus, we judged that these preparations were suitable for testing the effects of the various partially purified proteins on the vhs-dependent nuclease in yeast extracts.
FIG. 4.
eIF4H stimulates the endoribonuclease activity of vhs produced in yeast. (A and B) Analysis of nuclease activity on pCITE-1 and SRP-α RNA substrates, respectively. Internally labeled pCITE-1 and SRP-α RNAs were added to control RRL (RRL), eIF4H mixed with vhs assay buffer (eIF4H), control extracts alone, mixed with RRL (+RRL), or mixed with eIF4H (+eIF4H), and yeast extracts containing 2.1vhs alone, mixed with RRL (+RRL), or mixed with eIF4H (+eIF4H). eIF4H was used at a concentration of 7.7 μM. Samples were processed as described in the legend for Fig. 1. Symbols and brackets are as described in the legend for Fig. 1. The solid triangle indicates the 600-nt fragment. The open square and triangles indicate additional ca. 900- and 1,200-nt degradation products, respectively. The positions and sizes of the RNA markers are indicated in nucleotides at the left. (C and D) Quantification of nuclease activity on pCITE-1 and SRP-α RNAs, respectively. The quantity of full-length RNA in panels A and B and at least two other experiments was determined using phosphorimager analysis and plotted against time points indicated. The error bars indicate the standard deviation of each time point calculated from at least three independent experiments.
FIG. 5.
The enhancement of the RNase activity of vhs expressed in yeast is specific to eIF4H. (A and B) Analysis of nuclease activity on pCITE-1 and SRP-α RNAs, respectively. Internally labeled pCITE-1 and SRP-α RNAs were added to yeast extracts containing 2.1vhs either alone, mixed with eIF4H (+eIF4H), or mixed with cdc34Δ209 (+cdc34). The RNA substrates were also added to eIF4H or cdc34Δ209 mixed with 20 μg of BSA and 1 μg of total yeast RNA per time point. eIF4H and cdc34Δ209 were used at a concentration of 7.7 μM. Samples were processed as described in the legend for Fig. 1. The positions and sizes of the RNA markers in nucleotides are indicated at the left.
Partially purified eIF4H was used to investigate whether eIF4H affects the activity of vhs produced in yeast. As described above, eIF4H had no detectable effect on the stability of the RNA substrates in extracts prepared from yeast harboring the empty expression vector (Fig. 4A and B, EV+eIF4H). In marked contrast, addition of eIF4H to the 2.1vhs extracts significantly accelerated the decay of both pCITE-1 and SRP-α RNAs (Fig. 4A and B, 2.1vhs+eIF4H; data are quantified in C and D). This effect appeared to be specific, as a presumably irrelevant protein (yeast cdc34Δ209) purified in the same manner had no such effect (Fig. 5, 2.1vhs+cdc34). In addition, eIF4H altered the pattern of degradation products of pCITE-1 RNA compared to that observed in 2.1vhs extracts alone. Specifically, more of the 600-, 900-, and 1,200-nt degradation products were produced, and the 600-nt fragment was quite stable even after the disappearance of the 900- and 1,200-nt fragments (Fig. 3A, 2.1vhs+eIF4H). The 600-nt product comigrated with the 600-nt 5′ fragment produced in RRLvhs and, as documented below, at least some of the material migrating at this position arose by cleavage just downstream of the IRES (see Fig. 7). While the predicted 1,800-nt 3′ fragment diagnostic of IRES targeting was not observed with eIF4H, the detection of this fragment may have been compromised because the input RNA contained RNA species that migrated at the same position (Fig. 1). Although eIF4H altered the pattern of degradation products, it did not have the same effect as adding RRL to the 2.1vhs extracts. With the addition of RRL, the degradation product profile was much more similar to that observed in the intact RRL assay, in that substantial amounts of the 600- and 1,800-nt fragments and much less (if any) of the 900- and 1,200-nt fragments were produced. Thus, although eIF4H accelerated the cleavage of the RNA substrates, these data raise the possibility that it does not efficiently reconstitute preferential cleavage to downstream of the EMCV IRES. This point is examined further below (see Fig. 7).
As noted above, the preparation of eIF4H used in the experiments shown in Fig. 4 and 5 contained trace amounts of RNase activity that could be detected when a 1,000-fold excess of the protein was incubated alone with the radiolabeled RNA substrate. The level of nuclease contamination varied between eIF4H preparations, and we were able to generate some preparations that were devoid of detectable RNase activity (Fig. 6). Such nuclease-free preparations of eIF4H retained the ability to enhance the activity of 2.1vhs (but not D215N) extracts (Fig. 6). These data confirmed that the stimulatory activity of eIF4H does not stem from contaminating bacterial RNases.
Effects of RRL and eIF4H on targeting of vhs activity to the EMCV IRES.
Elgadi and Smiley provided evidence that the EMCV IRES strongly targets the initial vhs-induced cleavage events to sequences located just downstream of the IRES in the RRL assay system (5). Two of the key observations that led to this conclusion were as follows. First, when internally labeled pCITE-1 RNA was used as the substrate, 600- and 1,800-nt degradation products were simultaneously produced early during the reaction, with no evidence of prior intermediates (5). These two products roughly sum to yield the length of the intact substrate. Second, the 600- and 1,800-nt fragments were shown to correspond to the 5′ and 3′ ends of the transcript, respectively, using direct and indirect end-labeling techniques (5). The experiments depicted in Fig. 1 and 4 did not provide definitive information about the targeting properties of vhs expressed in yeast (either in the presence or absence of mammalian factors) because we could not unambiguously detect the 1,800-nt 3′ degradation fragment and the origin of the various degradation intermediates was not examined in detail.
To more rigorously determine if degradation of pCITE-1 RNA proceeds in the same manner as in the intact RRL system in 2.1vhs extracts following reconstitution with RRL or eIF4H, we first repeated the experiment using internally labeled substrate. The experiment was modified in two ways to maximize the likelihood of detecting the unstable 1,800-nt 3′ fragment: a more homogenous substrate was used, and samples were taken at earlier time points (Fig. 7A). The degradation pattern observed in the 2.1vhs extract supplemented with RRL closely resembled that previously observed in the intact RRL system (5), in that the 1,800- and 600-nt products were clearly evident at early times and these were the only obvious discrete degradation fragments detected (Fig. 7A, 2.1vhs+RRL). Moreover, the 600-nt fragment was quite stable and accumulated throughout the reaction, while the 1,800-nt fragment was subject to further decay. However, this was not the case in reactions supplemented with eIF4H. By 1.5 min, possibly up to four degradation products could be detected (Fig. 7A, 2.1vhs+eIF4H). Thus, 600-, 900-, 1,200-, and possibly 1,800-nt fragments were produced, although the 1,800-nt fragment was not one of the most predominant and was rapidly degraded. The 900- and 1,200-nt fragments were initially the most predominant but were subject to further decay, while the 600-nt fragment accumulated at later time points. These observations suggest that in the presence of eIF4H, vhs activity is not as strongly targeted by the EMCV IRES as in the presence of RRL.
We next used a slightly modified version of the vhs activity assay to track the fate of the 5′ and 3′ ends of the RNA substrate over the course of the reaction. Unlabeled pCITE-1 RNA was incubated with variously supplemented extracts, followed by Northern blot analysis using oligonucleotide probes complementary to the extreme 5′ and 3′ ends of the RNA (Fig. 7B). As previously described (5), the earliest products detected in RRLvhs were a stable 5′ 600-nt fragment and a predominant 3′ 1,800-nt fragment (Fig. 7C and D, RRLvhs). The 600-nt fragment was essentially the only product detected with the 5′ probe, and it arose through cleavage just downstream of the IRES. While the 1,800-nt fragment was the predominant 3′ product detected at early times, it was not the only 3′ degradation product present. Over the time course, the 1,800-nt fragment was subject to further decay and a shift from larger to smaller products occurred, with discrete intermediates. The sizes of these further degradation products mapped to the positions of preferential cleavage sites, from 5′ to 3′, as the initial 3′ product was further degraded. The overall pattern of degradation products observed in RRLvhs was consistent with a model where vhs cleaves the RNA in an overall 5′-to-3′ direction. Indeed, additional experiments using oligonucleotides complementary to multiple sites along the length of the RNA confirm this interpretation (J. Perez-Parada and J. R. Smiley, unpublished results). The 2.1vhs extracts showed a comparable loss of intact substrate, but comparably abundant discrete degradation products were not detected (Fig. 7C and D, 2.1vhs). We have reproducibly seen faint bands that comigrate with some of the degradation products in RRLvhs; however, they were found at a much lower molar yield. These observations demonstrated that 2.1vhs is not strongly targeted by the EMCV IRES. In contrast, RRL altered the pattern of degradation products to one almost identical to that seen with RRLvhs. A dominant 5′ 600-nt fragment containing the IRES was detected, although faint larger 5′ fragments were also observed. Furthermore, the 3′ 1,800-nt fragment was the most abundant product at early times, although other smaller 3′ products were also seen. However, unlike in RRLvhs, the 3′ degradation products were not obviously shifted from larger to smaller fragments over the course of the reaction. Further testing is required to determine if vhs cleaves the RNA in a 5′-to-3′ direction in yeast extracts supplemented with RRL. eIF4H also altered the degradation profile of the 2.1vhs extracts; however, the pattern was quite different from that seen with RRL (Fig. 7C and D, 2.1vhs+eIF4H). The 600-nt 5′ fragment bearing the IRES was detected; however, the molar yield was substantially lower and additional larger 5′ products were also produced at early times. Furthermore, the 3′ 1,800-nt fragment was barely detectable; rather, smaller 3′ products were more abundant. These observations indicate that in the presence of eIF4H, the sites of initial cleavage of pCITE-1 RNA are distributed throughout the length of the RNA, rather than focused immediately downstream of the IRES as in reactions containing RRL. Taken in combination, these results demonstrate that although RRL restores IRES-mediated targeting to vhs expressed in yeast, eIF4H does not.
eIF4B interacts directly with vhs.
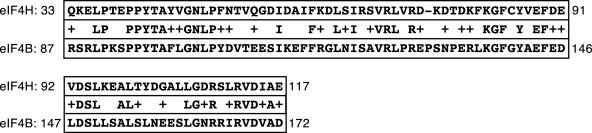
We decided to examine two other translation initiation factors for their ability to interact with and stimulate the RNase activity of vhs produced in yeast. The first translation initiation factor chosen was eIF4B, which is a sequence paralogue of eIF4H. These two proteins work together to modulate the helicase activity of eIF4A (47, 49, 50). They have 62% similarity and 39% identity in their amino acid sequences (48). By using the Blast 2 sequences alignment program (65), the region of greatest similarity was found between residues 33 to 117 of eIF4H and 87 to 147 of eIF4B (Fig. 8). This region of strong similarity overlaps with the region of eIF4H that interacts with vhs (residues 90 to 137), raising the possibility that eIF4B may also interact with vhs. The second candidate protein we investigated was eIF4A. eIF4A is an RNA helicase which is a component of eIF4F, a translation initiation factor that binds the cap of mRNAs and is known to interact functionally with both eIF4H and eIF4B in translation initiation (47, 49, 50). Both eIF4A and eIF4B were expressed as His6-tagged proteins and purified in exactly the same manner as eIF4H (Fig. 2, eIF4A and eIF4B). The eIF4B preparation contained both the full-length protein as well as a series of smaller polypeptides, some of which were quite abundant. The smaller species were presumably degradation products of eIF4B, as an anti-His tag antibody reacted with the majority of the polypeptides (data not shown). The degradation of eIF4B in E. coli has been previously noted by Methot et al. (37), who also reported that the degradation products do not interfere with the ability of eIF4B to stimulate the helicase activity of eIF4A. To compensate for the presence of these degradation products, we used equimolar amounts of full-length proteins in the assays described below.
FIG. 8.
Region of greatest similarity between eIF4H and eIF4B. Sequence similarity between residues 33 to 117 of eIF4H (accession no. NP_114381) and residues 87 to 147 of eIF4B (accession no. CAA39265) was determined using the Blast 2 sequence alignment program (65). The residues shown between the two sequences are conserved, and the residues from the same family are indicated with a +. There are 43% identity and 68% similarity between eIF4H and eIF4B in this region.
Potential interactions between vhs and eIF4B and eIF4A were analyzed by far Western blot analysis using radiolabeled vhs as a probe. Partially purified translation factors were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with RRL containing 35S-labeled vhs. eIF4H acted as a positive control, as it had already been shown to interact with vhs. cdc34Δ209, BSA, and PP1 were used as negative control proteins, and an interaction with vhs was not detected (Fig. 9). eIF4H and eIF4B were both found to interact with vhs, as seen by the appearance of bands on the far Western blot corresponding to the positions of the proteins on the Coomassie-stained gel (Fig. 9). While the signal intensity obtained with eIF4B was lower than with eIF4H, it is unclear whether this was due to a lower affinity for vhs or the consequence of a lower quantity of intact eIF4B compared to eIF4H present on the blot. An interaction between vhs and eIF4A was not detected (Fig. 9). The purified eIF4A protein retained RNA helicase activity (data not shown), suggesting that the protein preparation was functional. While we did not observe an interaction between eIF4A and vhs, the possibility of detecting an interaction by other methods cannot be ruled out. These data showed that both eIF4H and eIF4B can directly interact with vhs in vitro.
eIF4B stimulates the endoribonuclease activity of vhs expressed in yeast.
We next investigated whether eIF4B or eIF4A could stimulate the RNase activity or reconstitute IRES-directed targeting to vhs produced in yeast. vhs activity assays were performed as described for Fig. 4, adding either eIF4A or eIF4B to yeast extracts. The concentration of eIF4H and eIF4B used in these assays was 2.1 μM, as this was the highest concentration of full-length eIF4B that we were able to achieve. Both pCITE-1 and SRP-α RNA substrates were very stable in control extracts in the presence of eIF4A and eIF4B (Fig. 10A and B, EV+eIF4A and EV+eIF4B). eIF4B enhanced the RNase activity of vhs on pCITE-1 RNA, and the degradation product profile was similar to that seen when eIF4H was added to 2.1vhs extracts, with the exception that the 600-nt product was perhaps less stable (Fig. 10A, 2.1vhs+eIF4B; data are quantified in C). Similar results were seen with SRP-α RNA, with the exception that discrete degradation products were not observed (Fig. 10B, 2.1vhs+eIF4B; data are quantified in D). The ability of eIF4B to enhance the activity of vhs produced in yeast may vary with the substrate used, as eIF4B did not enhance the decay of pCITE-1 RNA as much as SRP-α RNA, relative to the effect of eIF4H (Fig. 10C versus D, 2.1vhs+eIF4B). While eIF4A did not affect the nuclease activity of the 2.1vhs extracts on pCITE-1 RNA, a slight stimulation was observed with SRP-α RNA (Fig. 10B, 2.1vhs+eIF4A; data are quantified in D). The concentration of eIF4A used in these experiments was 7.6 μM, to match that of eIF4H used in the experiments shown in previous figures. However, when the concentration of eIF4A was lowered to 2.1 μM to match that of eIF4H and eIF4B used in these particular experiments, an enhancement of nuclease activity of vhs could not be detected (data not shown).
As was the case for eIF4H, both eIF4A and eIF4B were unable to restore IRES-directed targeting to 2.1vhs (data not shown). In addition, eIF4H and eIF4A in combination were also unable to reconstitute targeting to 2.1vhs (data not shown). Taken together, these results demonstrate that while eIF4B and eIF4H stimulate the activity of 2.1vhs, one or more other additional mammalian factors are required to fully reconstitute IRES-mediated targeting to 2.1vhs.
DISCUSSION
As reviewed in the introduction, previous work has provided strong evidence that vhs is an integral part of the vhs-dependent RNase, an mRNA-specific nuclease that requires one or more cellular factors for efficient activity. In addition, vhs interacts with translation initiation factor eIF4H (10), and a complex of vhs-eIF4H has RNase activity (7). However, the role of eIF4H in the activity and selective targeting of the vhs-dependent RNase had remained unclear. In the present study, we demonstrated that cell extracts of yeast expressing vhs display vhs-dependent RNase activity in the absence of mammalian factors. This activity differed from that previously described for vhs produced in RRL in that it was not strongly targeted to sequences located immediately downstream of the EMCV IRES. RRL enhanced the activity of the extracts and reconstituted IRES-directed targeting of the nuclease. We also showed that the eIF4H paralogue eIF4B binds to vhs and that eIF4B and eIF4H each stimulate the activity of vhs produced in yeast. However, neither factor was able to fully restore targeting to the EMCV IRES, indicating that one or more other mammalian factors are required.
The data presented in this study clearly demonstrate that cell extracts prepared from yeast expressing vhs contain a novel RNase activity that can be detected in the absence of added mammalian factors. This activity has been detected in repeated experiments and with several independent preparations of yeast extracts, and it was reduced if not eliminated by the D215N mutation. Thus, we consider it likely that this activity stems from the RNase activity of vhs. However, a previous study from this laboratory did not detect appreciable vhs-dependent RNase activity in extracts of yeast expressing vhs until RRL was added to the extracts (34). The basis for this discrepancy remains unclear. Perhaps the RNase activity in extracts is near the lower detection limit of the assay and our ability to detect it has improved over time. It may also be that currently unknown variations in growth conditions affect the level of activity in the extracts. Possibly relevant in this context, Lu et al. found that the severity of the growth inhibitory effect of vhs in yeast varies depending on the carbon source used in the medium (34). It is unknown at this time whether vhs expressed in yeast has activity on its own or instead requires interactions with one or more yeast factors. Indeed, S. cerevisiae contains a homologue of eIF4H and eIF4B known as Tif3 or Stm1 (1, 2). Tif3/Stm1 plays an important role in translation initiation and was found to have 25% identity and 53% similarity to the sequence of eIF4H (48). Thus, it is possible that Tif3/Stm1 or other yeast proteins can interact with vhs and form a partially competent RNase. Additional studies are required to determine the subunit composition of the vhs-dependent RNase that we have detected in yeast extracts.
The vhs-dependent nuclease activity observed in yeast extracts may explain the previous finding that vhs strongly inhibits the growth of yeast on certain carbon sources, an effect that correlates with the mammalian host shutoff activity of vhs in mutational studies (34). However, this hypothesis is not obviously consistent with the observation that vhs does not induce global turnover of yeast mRNAs in vivo (34). Perhaps the RNase acts only on a subset of yeast mRNAs in vivo, or the effect is too weak to be detected by Northern blot analysis. An interesting alternative possibility is that vhs inhibits the growth of yeast by binding the eIF4H homologue Tif3/Stm1 and blocking its activity, rather than by (or in addition to) destabilizing mRNAs. Indeed, TIF3/STM1 gene disruptions cause a slow growth phenotype and a defect in translation initiation (1, 2). However, in this context it is worth noting that the D215N mutation abolishes the growth inhibitory effect of vhs (data not shown) yet does not prevent the interaction between vhs and human eIF4H (7). Further studies are required to determine if Tif3/Stm1 contributes to the growth inhibitory effect of vhs in yeast.
The RNase activity detected in yeast extracts differed in several respects from that of vhs translated in RRL. Previous work has shown that the activity in RRL is nonrandomly targeted relative to the structure of the substrate mRNA. Thus, the initial sites of cleavage on SRP-α mRNA are clustered over the 5′ quadrant of the transcript (4), and the EMCV IRES present in pCITE-1 RNA targets the initial cleavage events to 3′-flanking sequences (5). In addition, as noted above, our unpublished data strongly suggest that the subsequent degradation of pCITE-1 RNA (mediated by vhs [34]) proceeds in an overall 5′-to-3′ direction in the RRL system (Perez-Parada and Smiley, unpublished). We have not yet examined the detailed mode of decay of SRP-α mRNA in the yeast extract system and, therefore, we do not know whether the nuclease activity is initially targeted to the 5′ end of this RNA. However, our data provide a clear indication that the RNase is largely untargeted relative to the EMCV IRES, with no obvious strong clustering of preferred initial cleavage sites immediately downstream of the IRES in the pCITE-1 transcript. In addition, while discrete degradation products were produced from pCITE-1 RNA at relatively low molar yield, they did not appear in the defined temporal order indicative of initial IRES targeting and subsequent 5′-to-3′ decay as observed in RRL (Fig. 7). Moreover, the 600-nt IRES-containing fragment was relatively unstable in the yeast extracts. It is unclear why the molar yield of the discrete degradation intermediates of pCITE-1 RNA was reduced in the yeast extracts relative to that of reaction mixtures supplemented with RRL, eIF4H, or eIF4B. It is possible that the vhs-dependent nuclease cleaves the substrate at many more sites in the absence of mammalian factors, or it acts primarily as an exonuclease. Alternatively, the degradation intermediates may be more susceptible to yeast nucleases.
Our data demonstrate that mammalian factors significantly alter the characteristics of the nuclease activity of vhs expressed in yeast. Thus, RRL restored IRES-directed targeting and (as previously reported [34]) enhanced the RNase activity of yeast extracts containing vhs. In contrast, eIF4B and eIF4H stimulated the activity of vhs without fully restoring IRES-mediated targeting. Further investigation is required to determine if RRL, eIF4B, or eIF4H influences the overall polarity of RNA degradation by vhs.
It is perhaps not surprising that vhs requires mammalian factors to target to immediately downstream of the EMCV IRES, as this IRES cannot direct translation initiation in yeast (8). Although the basis for this inactivity is unclear, a likely possibility is that yeast translation initiation factors cannot functionally interact with the IRES. RRL fully reconstituted IRES-directed targeting to vhs expressed in yeast while eIF4B and eIF4H did not, suggesting that distinct, or multiple, mammalian factors are required for the targeting reaction. The combination of eIF4A and eIF4H was also inactive, and a role for eIF4H in IRES-directed targeting is perhaps unlikely, given that a mutant form of vhs (vhs1, T214→I [28, 46]) that no longer interacts with eIF4H (9) retains the ability to perform IRES-directed cleavage (35). However the combination of eIF4A and eIF4B was not tested. It will be important to do so in future studies, as eIF4A and eIF4B together greatly enhance the binding of each other to the EMCV IRES (43). eIF4G also plays a strong role in recruiting translation factors and promoting ribosomal attachment to the IRES (see the references listed in reference 25). The binding of eIF4A, eIF4B, and eIF4G to the EMCV IRES is strongly stimulated by the presence of all three factors (33, 43), and the IRES has evolved to bind the eIF4G/eIF4A complex with high affinity rather than eIF4G alone (33). Thus, it is reasonable to suggest that a combination of all three factors may be required for IRES-directed targeting by vhs. Moreover, additional factors may also be required, such as other canonical translation initiation factors and/or IRES trans-acting factors (ITAFs), such as pyrimidine tract-binding protein (20, 24), La protein (22), and ITAF45 (18). One potentially relevant canonical initiation factor may be eIF3, as it binds to both eIF4B (38) and eIF4G (30), and there is some evidence to suggest that it may be able to interact with the IRES directly without the need for eIF4G (33). Further studies are required to determine the precise interplay of factors required for IRES-directed targeting by vhs.
Our data establish that eIF4B and eIF4H are each able to stimulate the nuclease activity of vhs expressed in yeast. However, the mechanism of this enhancement remains to be defined. It seems likely that the stimulatory effect is a consequence of the direct interactions between the translation factors and vhs (see below, however). In addition, it seems plausible that eIF4B and eIF4H stimulate vhs activity through similar mechanisms, given the functional and sequence similarities between these proteins. We can think of three distinct ways that the physical interaction between vhs and eIF4B/eIF4H could enhance the nuclease activity of vhs. First, by directly binding to vhs, eIF4H and eIF4B may induce a conformational change in either vhs or the complex to create a more active nuclease. The active site, contained at least in part by vhs (7), may not be in a favorable conformation or may be partially hidden and require the binding of eIF4H or eIF4B to assume an optimal configuration. Second, the factors may enhance the affinity of vhs or the complex for the RNA substrate, either by inducing a conformational change in vhs or by directly contributing to RNA binding. Indeed, eIF4B and eIF4H each contain a conserved RNA recognition motif (see, for example, references 37 and 48) that might direct the complex to particular RNA sequences and/or enhance its general affinity for RNA. Third, the factors might act to alter the overall pattern of RNase activity, for example, by triggering a shift from a distributive to a processive mode of RNA degradation (or vice versa).
In this latter context, it is interesting that vhs is required both for the initial cleavage of mRNAs and the subsequent degradation of the decay products in the RRL assay system (35). Moreover, as noted above, the subsequent decay of the initial 3′ cleavage product of pCITE-1 mRNA appears to proceed 5′ to 3′ (Perez-Parada and Smiley, unpublished). Both observations are consistent with a model in which vhs is a processive enzyme that, once loaded, tracks along the RNA, cleaves it at particular sites, and remains associated with the 3′ cleavage product. Alternatively, vhs may be a distributive enzyme with a preference for the 5′ ends of the mRNA degradation intermediates. Some indirect evidence is consistent with a role of eIF4H (and by analogy, perhaps eIF4B) in the decay of the first products of vhs action. Specifically, as noted above, the vhs1 mutant version of vhs (T214→I), which is defective in host shutoff in vivo (28, 46) and unable to degrade SRP-α RNA in vitro (4), does not detectably interact with eIF4H (9). The vhs1 protein nevertheless retains the ability to perform IRES-directed cleavage of pCITE-1 RNA in the RRL system (35). However, the resulting 3′ 1,800-nt fragment is not subject to further decay, suggesting that the mutation severely impairs the ability of vhs to continue degradation of the RNA after the first cleavage reaction (35). Thus, considered in the light of the foregoing, eIF4H may enhance the decay of the initial degradation products by increasing the processivity of vhs or by promoting its distributive cycling on and off the substrate.
The three scenarios described above all assume that eIF4B and eIF4H stimulate vhs activity in yeast extracts solely through their direct interactions with vhs. However, it is important to stress that this is not necessarily the case. Mammalian eIF4B can functionally replace yeast Tif3/Stm1 in in vitro translation reactions (1) and is thus capable of interacting with other components of the yeast translational apparatus. The same may also be so for eIF4H. Therefore, it possible that some or all of the effects of eIF4B and eIF4H that we have observed stem from the recruitment of additional yeast proteins into the vhs nuclease complex.
Our data demonstrate that mammalian factors alter both the RNase activity and IRES-targeting properties of vhs. The in vitro assay described here will facilitate future studies designed to identify the factors required for IRES-directed targeting and illuminate the mechanisms that regulate vhs activity.
Acknowledgments
We thank Michael Ellison for the gift of the cdc34Δ209 expression vector; Duncan Wilson for the AE328 vhs antiserum; Kathleen Perault for the gift of purified PP1; Shawn Wasilenko and Gary Chan for construction of plasmids pSW1 and -2 and pGC2, respectively; Kim Ellison for technical advice; and Rob Maranchuk for help with figure preparation.
R.C.D. was supported by a studentship from the Alberta Heritage Foundation for Medical Research. This research was supported by a grant from the Canadian Institutes of Health Research to J.R.S.
REFERENCES
- 1.Altmann, M., P. P. Muller, B. Wittmer, F. Ruchti, S. Lanker, and H. Trachsel. 1993. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 12:3997-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coppolecchia, R., P. Buser, A. Stotz, and P. Linder. 1993. A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J. 12:4005-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doherty, A. J., L. C. Serpell, and C. P. Ponting. 1996. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 24:2488-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elgadi, M. M., and J. R. Smiley. 1999. Picornavirus IRES elements target RNA cleavage events induced by the herpes simplex virus vhs protein. J. Virol. 73:9222-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everly, D. N., Jr., and G. S. Read. 1997. Mutational analysis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): characterization of HSV type 1 (HSV-1)/HSV-2 chimeras. J. Virol. 71:7157-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everly, D. N. J., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 76:8560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evstafieva, A. G., A. V. Beletsky, A. V. Borovjagin, and A. A. Bogdanov. 1993. Internal ribosome entry site of encephalomyocarditis virus RNA is unable to direct translation in Saccharomyces cerevisiae. FEBS Lett. 335:273-276. [DOI] [PubMed] [Google Scholar]
- 9.Feng, P., D. N. J. Everly, and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng, W. C., C. M. Southwood, and J. J. Bieker. 1994. Analysis of β-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J. Biol. Chem. 269:1493-1500. [PubMed] [Google Scholar]
- 11.Fenwick, M. L., and J. Clark. 1982. Early and delayed shut-off of host protein synthesis in cells infected with herpes simplex virus. J. Gen. Virol. 61:121-125. [DOI] [PubMed] [Google Scholar]
- 12.Fenwick, M. L., and R. D. Everett. 1990. Inactivation of the shutoff gene (UL41) of herpes simplex virus types 1 and 2. J. Gen. Virol. 71:2961-2967. [DOI] [PubMed] [Google Scholar]
- 13.Fenwick, M. L., and R. D. Everett. 1990. Transfer of UL41, the gene controlling virion-associated host shutoff, between different strains of herpes simplex virus. J. Gen. Virol. 71:411-418. [DOI] [PubMed] [Google Scholar]
- 14.Fenwick, M. L., and M. M. McMenamin. 1984. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J. Gen. Virol. 65:1225-1228. [DOI] [PubMed] [Google Scholar]
- 15.Fenwick, M. L., and S. A. Owen. 1988. On the control of immediate early (alpha) mRNA survival in cells infected with herpes simplex virus. J. Gen. Virol. 69:2869-2877. [DOI] [PubMed] [Google Scholar]
- 16.Fenwick, M. L., and M. J. Walker. 1978. Suppression of the synthesis of cellular macromolecules by HSV. J. Gen. Virol. 22:37-51. [DOI] [PubMed] [Google Scholar]
- 17.Guichet, A., J. W. Copeland, M. Erdelyi, D. Hlousek, P. Zavorszky, J. Ho, S. Brown, A. Percival-Smith, H. M. Krause, and A. Ephrussi. 1997. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature 385:548-552. [DOI] [PubMed] [Google Scholar]
- 18.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 19.Jones, F. E., C. A. Smibert, and J. R. Smiley. 1995. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J. Virol. 69:4863-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaminski, A., and R. J. Jackson. 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karr, B. M., and G. S. Read. 1999. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology 264:195-204. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y. K., and S. K. Jang. 1999. La protein is required for efficient translation driven by encephalomyocarditis virus internal ribosomal entry site. J. Gen. Virol. 80:3159-3166. [DOI] [PubMed] [Google Scholar]
- 23.Knez, J., P. T. Bilan, and J. P. Capone. 2003. A single amino acid substitution in herpes simplex virus type 1 VP16 inhibits binding to the virion host shutoff protein and is incompatible with virus growth. J. Virol. 77:2892-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolupaeva, V. G., C. U. Hellen, and I. N. Shatsky. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2:1199-1212. [PMC free article] [PubMed] [Google Scholar]
- 25.Kolupaeva, V. G., I. B. Lomakin, T. V. Pestova, and C. U. Hellen. 2003. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 23:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krikorian, C. R., and G. S. Read. 1991. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J. Virol. 65:112-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong, A. D., J. A. Kruper, and N. Frenkel. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 30.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 31.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livingstone-Zatchej, M., A. Meier, B. Suter, and F. Thoma. 1997. RNA polymerase II transcription inhibits DNA repair by photolyase in the transcribed strand of active yeast genes. Nucleic Acids Res. 25:3795-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomakin, I. B., C. U. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, P., F. E. Jones, H. A. Saffran, and J. R. Smiley. 2001. The herpes simplex virus virion host shutoff protein requires a mammalian factor for efficient in vitro endoribonuclease activity. J. Virol. 75:1172-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, P., H. A. Saffran, and J. R. Smiley. 2001. The vhs1 mutant form of the herpes simplex virus virion host shutoff protein retains significant internal ribosome entry site-directed RNA cleavage activity. J. Virol. 75:1072-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Methot, N., A. Pause, J. W. Hershey, and N. Sonenberg. 1994. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol. Cell. Biol. 14:2307-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Methot, N., M. S. Song, and N. Sonenberg. 1996. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 16:5328-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oroskar, A. A., and G. S. Read. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oroskar, A. A., and G. S. Read. 1987. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J. Virol. 61:604-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pak, A. S., D. N. Everly, K. Knight, and G. S. Read. 1995. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology 211:491-506. [DOI] [PubMed] [Google Scholar]
- 43.Pestova, T. V., I. N. Shatsky, and C. U. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ptak, C., J. A. Prendergast, R. Hodgins, C. M. Kay, V. Chau, and M. J. Ellison. 1994. Functional and physical characterization of the cell cycle ubiquitin-conjugating enzyme CDC34 (UBC3). Identification of a functional determinant within the tail that facilitates CDC34 self-association. J. Biol. Chem. 269:26539-26545. [PubMed] [Google Scholar]
- 45.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptides. J. Virol. 67:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Read, G. S., and N. Frenkel. 1983. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate early) viral polypeptides. J. Virol. 46:498-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter, N. J., G. W. Rogers, Jr., J. O. Hensold, and W. C. Merrick. 1999. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J. Biol. Chem. 274:35415-35424. [DOI] [PubMed] [Google Scholar]
- 48.Richter-Cook, N. J., T. E. Dever, J. O. Hensold, and W. C. Merrick. 1998. Purification and characterization of a new eukaryotic protein translation factor, eukaryotic initiation factor 4H. J. Biol. Chem. 273:7579-7587. [DOI] [PubMed] [Google Scholar]
- 49.Rogers, G. W., Jr., N. J. Richter, and W. C. Merrick. 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274:12236-12244. [DOI] [PubMed] [Google Scholar]
- 50.Rogers, G. W. J., N. J. Richter, W. F. Lima, and W. C. Merrick. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276:30914-30922. [DOI] [PubMed] [Google Scholar]
- 51.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. H. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 52.Samady, L., E. Costigliola, L. MacCormac, Y. McGrath, S. Cleverley, C. E. Lilley, J. Smith, D. S. Latchman, B. Chain, and R. S. Coffin. 2003. Deletion of the virion host shutoff protein (vhs) from herpes simplex virus (HSV) relieves the viral block to dendritic cell activation: potential of vhs- HSV vectors for dendritic cell-mediated immunotherapy. J. Virol. 77:3768-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schultz, M. C., S. Y. Choe, and R. H. Reeder. 1991. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc. Natl. Acad. Sci. USA 88:1004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz, M. C., D. J. Hockman, T. A. Harkness, W. I. Garinther, and B. A. Altheim. 1997. Chromatin assembly in a yeast whole-cell extract. Proc. Natl. Acad. Sci. USA 94:9034-9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smibert, C. A., D. C. Johnson, and J. R. Smiley. 1992. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J. Gen. Virol. 73:467-470. [DOI] [PubMed] [Google Scholar]
- 56.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smibert, C. A., and J. R. Smiley. 1990. Differential regulation of endogenous and transduced β-globin genes during infection of erythroid cells in herpes simplex virus type 1 recombinant. J. Virol. 64:3882-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smiley, J. R., M. M. Elgadi, and H. A. Saffran. 2001. Herpes simplex virus vhs protein. Methods Enzymol. 342:440-451. [DOI] [PubMed] [Google Scholar]
- 59.Sorenson, C. M., P. A. Hart, and J. Ross. 1991. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 19:4459-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strelow, L., T. Smith, and D. Leib. 1997. The virion host shutoff function of herpes simplex virus type 1 plays a role in corneal invasion and functions independently of the cell cycle. Virology 231:28-34. [DOI] [PubMed] [Google Scholar]
- 61.Strelow, L. I., and D. A. Leib. 1996. Analysis of conserved domains of UL41 of herpes simplex virus type 1 in virion host shutoff and pathogenesis. J. Virol. 70:5665-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strelow, L. I., and D. A. Leib. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strom, T., and N. Frenkel. 1987. Effects of herpes simplex virus on mRNA stability. J. Virol. 61:2198-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzutani, T., M. Nagamine, T. Shibaki, M. Ogasawara, I. Yoshida, T. Daikoku, Y. Nishiyama, and M. Azuma. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 81:1763-1771. [DOI] [PubMed] [Google Scholar]
- 65.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174:247-250. [DOI] [PubMed] [Google Scholar]
- 66.Tigges, M. A., S. Leng, D. C. Johnson, and R. L. Burke. 1996. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-γ or when virion host shutoff functions are disabled. J. Immunol. 156:3901-3910. [PubMed] [Google Scholar]
- 67.Trgovcich, J., D. Johnson, and B. Roizman. 2002. Cell surface major histocompatibility complex class II proteins are regulated by the products of the γ134.5 and UL41 genes of herpes simplex virus 1. J. Virol. 76:6974-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zelus, B. D., R. S. Stewart, and J. Ross. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 70:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]