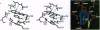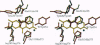Abstract
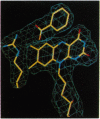
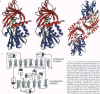
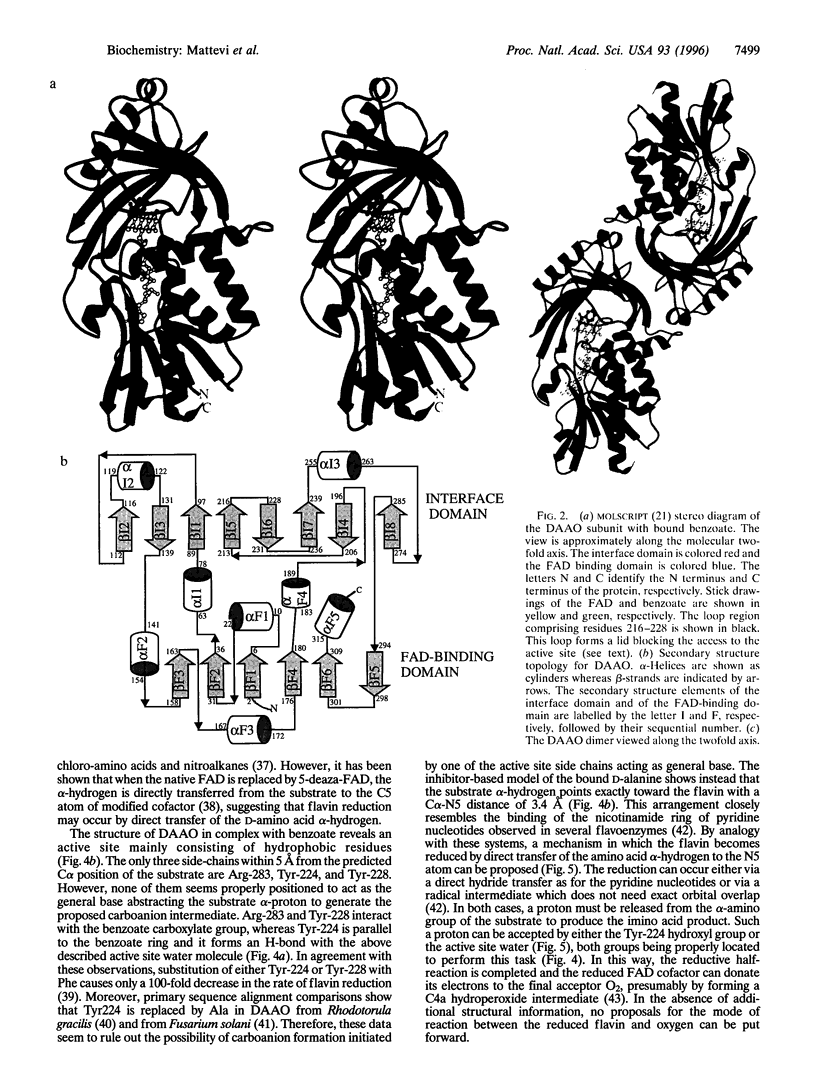
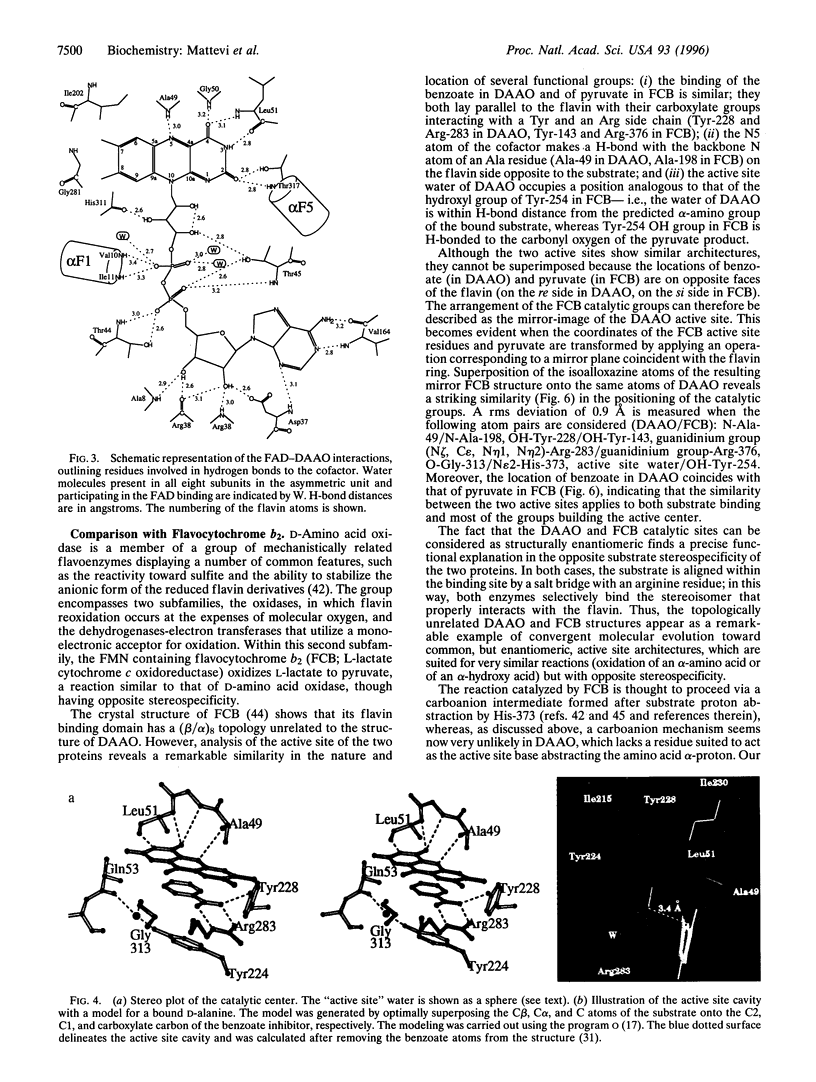
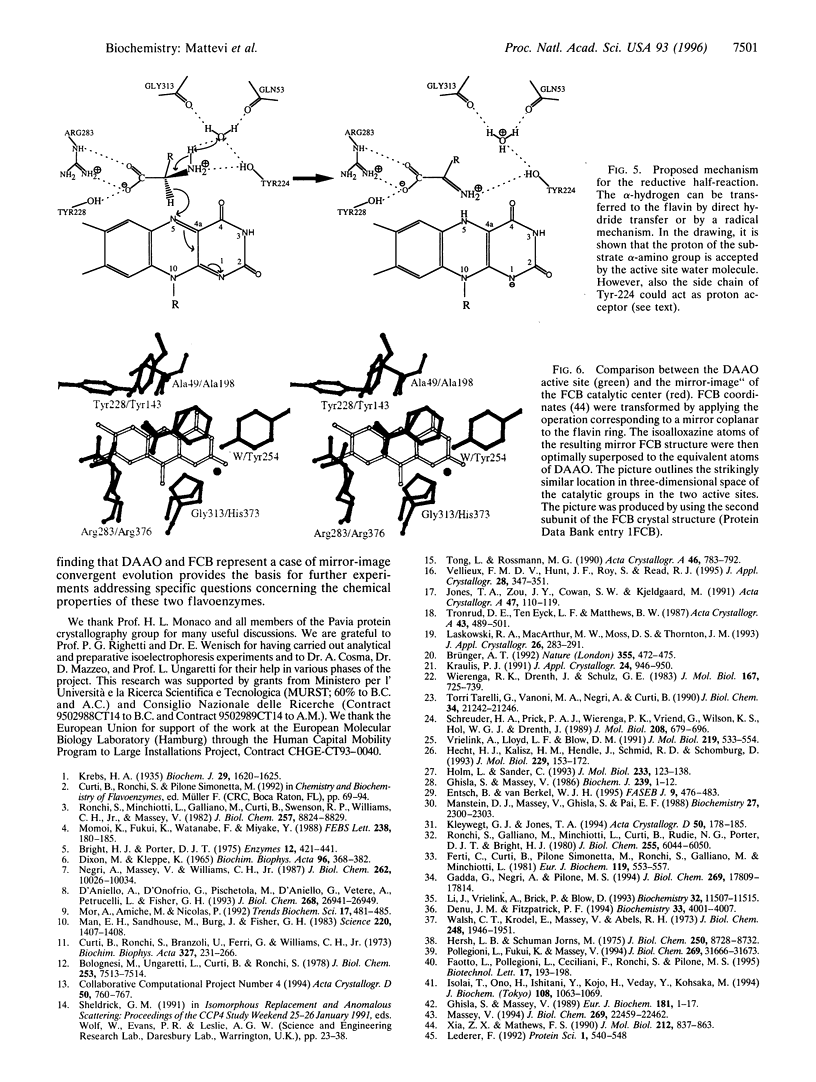
D-amino acid oxidase is the prototype of the FAD-dependent oxidases. It catalyses the oxidation of D-amino acids to the corresponding alpha-ketoacids. The reducing equivalents are transferred to molecular oxygen with production of hydrogen peroxide. We have solved the crystal structure of the complex of D-amino acid oxidase with benzoate, a competitive inhibitor of the substrate, by single isomorphous replacement and eightfold averaging. Each monomer is formed by two domains with an overall topology similar to that of p-hydroxybenzoate hydroxylase. The benzoate molecule lays parallel to the flavin ring and is held in position by a salt bridge with Arg-283. Analysis of the active site shows that no side chains are properly positioned to act as the postulated base required for the catalytic carboanion mechanism. On the contrary, the benzoate binding mode suggests a direct transfer of the substrate alpha-hydrogen to the flavin during the enzyme reductive half-reaction.The active site Of D-amino acid oxidase exhibits a striking similarity with that of flavocytochrome b2, a structurally unrelated FMN-dependent flavoenzyme. The active site groups (if these two enzymes are in fact superimposable once the mirror-image of the flavocytochrome b2 active site is generated with respect to the flavin plane. Therefore, the catalytic sites of D-amino acid oxidase and flavocytochrome b2 appear to have converged to a highly similar but enantiomeric architecture in order to catalvze similar reactions (oxidation of alpha-amino acids or alpha-hydroxy acids), although with opposite stereochemistry.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolognesi M., Ungaretti L., Curti B., Ronchi S. Crystallographic studies on D-amino acid oxidase. J Biol Chem. 1978 Oct 25;253(20):7513–7514. [PubMed] [Google Scholar]
- Curti B., Ronchi S., Branzoli U., Ferri G., Williams C. H., Jr Improved purification, amino acid analysis and molecular weight of homogenous D-amino acid oxidase from pig kidney. Biochim Biophys Acta. 1973 Dec 19;327(2):266–273. doi: 10.1016/0005-2744(73)90409-9. [DOI] [PubMed] [Google Scholar]
- D'Aniello A., D'Onofrio G., Pischetola M., D'Aniello G., Vetere A., Petrucelli L., Fisher G. H. Biological role of D-amino acid oxidase and D-aspartate oxidase. Effects of D-amino acids. J Biol Chem. 1993 Dec 25;268(36):26941–26949. [PubMed] [Google Scholar]
- Denu J. M., Fitzpatrick P. F. Intrinsic primary, secondary, and solvent kinetic isotope effects on the reductive half-reaction of D-amino acid oxidase: evidence against a concerted mechanism. Biochemistry. 1994 Apr 5;33(13):4001–4007. doi: 10.1021/bi00179a029. [DOI] [PubMed] [Google Scholar]
- Entsch B., van Berkel W. J. Structure and mechanism of para-hydroxybenzoate hydroxylase. FASEB J. 1995 Apr;9(7):476–483. doi: 10.1096/fasebj.9.7.7737455. [DOI] [PubMed] [Google Scholar]
- Ferti C., Curti B., Simonetta M. P., Ronchi S., Galliano M., Minchiotti L. Reactivity of D-amino acid oxidase with 1,2-cyclohexanedione: evidence for one arginine in the substrate-binding site. Eur J Biochem. 1981 Oct;119(3):553–557. doi: 10.1111/j.1432-1033.1981.tb05643.x. [DOI] [PubMed] [Google Scholar]
- Gadda G., Negri A., Pilone M. S. Reaction of phenylglyoxal with arginine groups in D-amino-acid oxidase from Rhodotorula gracilis. J Biol Chem. 1994 Jul 8;269(27):17809–17814. [PubMed] [Google Scholar]
- Ghisla S., Massey V. Mechanisms of flavoprotein-catalyzed reactions. Eur J Biochem. 1989 Apr 15;181(1):1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- Ghisla S., Massey V. New flavins for old: artificial flavins as active site probes of flavoproteins. Biochem J. 1986 Oct 1;239(1):1–12. doi: 10.1042/bj2390001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht H. J., Kalisz H. M., Hendle J., Schmid R. D., Schomburg D. Crystal structure of glucose oxidase from Aspergillus niger refined at 2.3 A resolution. J Mol Biol. 1993 Jan 5;229(1):153–172. doi: 10.1006/jmbi.1993.1015. [DOI] [PubMed] [Google Scholar]
- Hersh L. B., Jorns M. S. Use of 5-deazaFAD to study hydrogen transfer in the D-amino acid oxidase reaction. J Biol Chem. 1975 Nov 25;250(22):8728–8734. [PubMed] [Google Scholar]
- Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993 Sep 5;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Isogai T., Ono H., Ishitani Y., Kojo H., Ueda Y., Kohsaka M. Structure and expression of cDNA for D-amino acid oxidase active against cephalosporin C from Fusarium solani. J Biochem. 1990 Dec;108(6):1063–1069. doi: 10.1093/oxfordjournals.jbchem.a123306. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kleywegt G. J., Jones T. A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994 Mar 1;50(Pt 2):178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- Krebs H. A. Metabolism of amino-acids: Deamination of amino-acids. Biochem J. 1935 Jul;29(7):1620–1644. doi: 10.1042/bj0291620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer F. Extreme pKa displacements at the active sites of FMN-dependent alpha-hydroxy acid-oxidizing enzymes. Protein Sci. 1992 Apr;1(4):540–548. doi: 10.1002/pro.5560010409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Vrielink A., Brick P., Blow D. M. Crystal structure of cholesterol oxidase complexed with a steroid substrate: implications for flavin adenine dinucleotide dependent alcohol oxidases. Biochemistry. 1993 Nov 2;32(43):11507–11515. [PubMed] [Google Scholar]
- Man E. H., Sandhouse M. E., Burg J., Fisher G. H. Accumulation of D-aspartic acid with age in the human brain. Science. 1983 Jun 24;220(4604):1407–1408. doi: 10.1126/science.6857259. [DOI] [PubMed] [Google Scholar]
- Manstein D. J., Massey V., Ghisla S., Pai E. F. Stereochemistry and accessibility of prosthetic groups in flavoproteins. Biochemistry. 1988 Apr 5;27(7):2300–2305. doi: 10.1021/bi00407a009. [DOI] [PubMed] [Google Scholar]
- Massey V. Activation of molecular oxygen by flavins and flavoproteins. J Biol Chem. 1994 Sep 9;269(36):22459–22462. [PubMed] [Google Scholar]
- Momoi K., Fukui K., Watanabe F., Miyake Y. Molecular cloning and sequence analysis of cDNA encoding human kidney D-amino acid oxidase. FEBS Lett. 1988 Sep 26;238(1):180–184. doi: 10.1016/0014-5793(88)80252-7. [DOI] [PubMed] [Google Scholar]
- Mor A., Amiche M., Nicolas P. Enter a new post-translational modification: D-amino acids in gene-encoded peptides. Trends Biochem Sci. 1992 Dec;17(12):481–485. doi: 10.1016/0968-0004(92)90333-5. [DOI] [PubMed] [Google Scholar]
- Negri A., Massey V., Williams C. H., Jr D-aspartate oxidase from beef kidney. Purification and properties. J Biol Chem. 1987 Jul 25;262(21):10026–10034. [PubMed] [Google Scholar]
- Pollegioni L., Fukui K., Massey V. Studies on the kinetic mechanism of pig kidney D-amino acid oxidase by site-directed mutagenesis of tyrosine 224 and tyrosine 228. J Biol Chem. 1994 Dec 16;269(50):31666–31673. [PubMed] [Google Scholar]
- Ronchi S., Galliano M., Minchiotti L., Curti B., Rudie N. G., Porter D. J., Bright H. J. An active site-tyrosine-containing heptapeptide from D-amino acid oxidase. J Biol Chem. 1980 Jul 10;255(13):6044–6046. [PubMed] [Google Scholar]
- Ronchi S., Minchiotti L., Galliano M., Curti B., Swenson R. P., Williams C. H., Jr, Massey V. The primary structure of D-amino acid oxidase from pig kidney. II. Isolation and sequence of overlap peptides and the complete sequence. J Biol Chem. 1982 Aug 10;257(15):8824–8834. [PubMed] [Google Scholar]
- Schreuder H. A., Prick P. A., Wierenga R. K., Vriend G., Wilson K. S., Hol W. G., Drenth J. Crystal structure of the p-hydroxybenzoate hydroxylase-substrate complex refined at 1.9 A resolution. Analysis of the enzyme-substrate and enzyme-product complexes. J Mol Biol. 1989 Aug 20;208(4):679–696. doi: 10.1016/0022-2836(89)90158-7. [DOI] [PubMed] [Google Scholar]
- Tarelli G. T., Vanoni M. A., Negri A., Curti B. Characterization of a fully active N-terminal 37-kDa polypeptide obtained by limited tryptic cleavage of pig kidney D-amino acid oxidase. J Biol Chem. 1990 Dec 5;265(34):21242–21246. [PubMed] [Google Scholar]
- Tong L. A., Rossmann M. G. The locked rotation function. Acta Crystallogr A. 1990 Oct 1;46(Pt 10):783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- Vrielink A., Lloyd L. F., Blow D. M. Crystal structure of cholesterol oxidase from Brevibacterium sterolicum refined at 1.8 A resolution. J Mol Biol. 1991 Jun 5;219(3):533–554. doi: 10.1016/0022-2836(91)90192-9. [DOI] [PubMed] [Google Scholar]
- Walsh C. T., Krodel E., Massey V., Abeles R. H. Studies on the elimination reaction of D-amino acid oxidase with -amino- -chlorobutyrate. Further evidence for abstraction of substrate -hydrogen as a proton. J Biol Chem. 1973 Mar 25;248(6):1946–1955. [PubMed] [Google Scholar]
- Wierenga R. K., Drenth J., Schulz G. E. Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J Mol Biol. 1983 Jul 5;167(3):725–739. doi: 10.1016/s0022-2836(83)80106-5. [DOI] [PubMed] [Google Scholar]
- Xia Z. X., Mathews F. S. Molecular structure of flavocytochrome b2 at 2.4 A resolution. J Mol Biol. 1990 Apr 20;212(4):837–863. doi: 10.1016/0022-2836(90)90240-M. [DOI] [PubMed] [Google Scholar]