Abstract
HMGN proteins promote chromatin unfolding, enhance access to nucleosomes, and modulate transcription from chromatin templates. It is not known whether they act indiscriminately as general modulators of transcription or whether they regulate specific gene expression. Here, we investigated the role of HMGN3, a recently discovered HMGN family member, in transcription in vivo. We created cell lines overexpressing HMGN3a or its splice variant, HMGN3b, and analyzed their gene expression profiles using microarrays and reverse transcriptase PCR. We found that ectopic expression of HMGN3a alters the expression of approximately 0.8% of genes. Both HMGN3a and HMGN3b upregulate the expression of the glycine transporter 1 gene (Glyt1). Glyt1 encodes a membrane transporter that regulates the glycine concentration in synaptic junctions. Both GLYT1 and HMGN3 are highly expressed in glia cells and the eye, and we show that both proteins are coexpressed in the retina. Chromatin immunoprecipitation assays showed that HMGN3 protein is recruited to a region of the Glyt1 gene encompassing the Glyt1a transcriptional start site. These results suggest that HMGN3 regulates Glyt1 expression and demonstrate that members of the HMGN family can regulate the transcription of specific genes.
In eukaryotes, all of the DNA is complexed with histone proteins and packaged into a highly folded, well-ordered, and dynamic structure called chromatin. This packaging modulates the ability of regulatory factors to access their DNA targets and plays a major role in regulating various nuclear activities, including transcription (18, 48). Chromatin folding is modulated by numerous nuclear factors including nucleosome remodeling complexes, histone-modifying enzymes, and architectural proteins such as linker histones and HMG proteins. Members of the HMG superfamily interact with chromatin and DNA and affect a wide range of DNA-dependent activities such as transcription, replication, and recombination (9).
One of the HMG families, the HMGN family, is comprised of small, basic proteins that bind specifically to nucleosomes (8). HMGN proteins are highly conserved and found only in vertebrates. The two founding members of the family, HMGN1 and HMGN2 (formerly named HMG-14 and HMG-17) (8), have been studied extensively. They contain a highly conserved nucleosome binding domain, a bipartite nuclear localization signal, and a C-terminal chromatin-unfolding domain (12, 47). When incorporated into minichromosomes, HMGN proteins confer a more open chromatin structure that is more sensitive to nucleases and that is transcribed and replicated more efficiently (13, 14, 35, 46, 50). Their ability to unfold chromatin also enhances the rate of DNA repair, as recently demonstrated in mice lacking HMGN1 (5).
Although HMGN proteins display little or no DNA sequence specificity when binding to nucleosomes (45), several lines of evidence indicate that HMGN binding within the nucleus is nonrandom. Immunofluorescence studies have shown that HMGN proteins are localized in many foci within the nucleus and that the foci contain either HMGN1 or HMGN2 (38). They have a slight preference for binding to transcriptionally active genes (16, 17, 20, 39), and it has also been shown that they tend to bind in clusters on arrays of approximately six contiguous nucleosomes (38). However, the organization of HMGN proteins is highly dynamic in live cells, and their association with any specific nucleosome is temporary (37). It is conceivable that HMGNs are targeted to specific regions by their association with other nuclear proteins, and indeed, biochemical studies suggest that HMGN proteins form multiple metastable complexes with a number of as-yet-unidentified nuclear proteins (29).
An additional member of the HMGN family, HMGN3, was discovered more recently in a yeast two-hybrid screen for interaction partners of the thyroid hormone receptor (26). The structure of HMGN3 is very similar to those of HMGN1 and HMGN2 in that it contains domains homologous to the nucleosome binding domain, the bipartite nuclear localization signal, and the chromatin-unfolding domain. HMGN3 is expressed as two splice variants, HMGN3a and HMGN3b, and the latter lacks most of the C-terminal chromatin-unfolding domain (53). HMGN3b interacts with TR-RXR in a ligand-dependent manner and can promote thyroid hormone-dependent transcription from chromatin templates (2). Thyroid hormone can induce HMGN3b expression during tadpole development, and this induction is highest in tissues undergoing differentiation or remodeling (2). Studies of HMGN2 expression during mouse development also revealed highest expression in tissues undergoing differentiation (27, 28). However, the expression pattern of mouse HMGN3a/b is distinct from those of HMGN1 and HMGN2, being highly expressed in the eye and brain (4, 23, 53). Taken together, the data raise the possibility that HMGNs function as coactivators in tissue-specific gene expression. One of the major questions in the field is whether HMGN proteins act indiscriminately as general facilitators of transcription during processes such as differentiation or whether they act specifically to regulate the expression of particular target genes.
To determine whether HMGN proteins can regulate specific gene expression, we generated several cell lines expressing either HMGN3a or HMGN3b and performed microarray and reverse transcriptase PCR (RT-PCR) analyses to study the gene expression profiles within these cells. The results show that the two splice forms regulate the expression of distinct subsets of genes. We focused our attention on one of the gene targets identified by this screen: the glycine transporter 1 gene (Glyt1). Glyt1 is expressed in the eye and in glia cells, tissues that also have a high content of HMGN3. GLYT1 regulates the extracellular glycine concentration at synaptic junctions in the central nervous system. Glycine is an inhibitory neurotransmitter, and its concentration within the synapse is critical for the appropriate processing of motor and sensory information. Very little is known about the molecular mechanisms regulating Glyt1 expression. Here we demonstrate that both HMGN3a and HMGN3b upregulate Glyt1 expression. We show that HMGN3 binds the Glyt1 gene in vivo and that HMGN3 and GLYT1 proteins are coexpressed in the mouse retina. Our study shows that HMGN3 is a potential regulator of Glyt1 gene expression in vivo and indicates that HMGN proteins can specifically regulate individual gene expression.
MATERIALS AND METHODS
Plasmid construction.
The insulator sequences from plasmid pBAW3 (a kind gift from G. Felsenfeld, National Institutes of Health [NIH]) were inserted on either side of the expression cassette of plasmid pCI (Promega) to minimize chromatin position effects after random integration into the genome. Two tandem copies of the 250-bp core insulator from HS4 of the chicken β-globin locus (11) were inserted in the same orientation in the BglII and SpeI sites of pCI to create pCI-INS2. The open reading frames of mouse HMGN3a and -b were amplified by PCR and inserted into the Mlu I and SalI sites of pCI-INS2 to create pCI-INS2-N3a and pCI-INS2-N3b, respectively.
Creation of stable cell lines.
Hepa-1 cells were transfected with linearized pTK-Hyg and either pCI-INS2, pCI-INS2-N3a, or pCI-INS2-N3b and then selected in the presence of 400 μg of hygromycin (BD Biosciences) per ml. Colonies were expanded and screened by Western blot analysis of 5% perchloric acid extracts using antibody 2752 against HMGN3 (36, 53). The number of integrated plasmid copies in the clones selected for further study was estimated by Southern analysis to be between 1 and 10.
Microarray analysis.
RNA was purified from 90% confluent cells by using Trizol (Invitrogen) followed by RNeasy (Qiagen) as recommended by the manufacturers. The protocol for microarray hybridization was derived from that published by Hegde and coworkers (21). Fluorescently labeled cDNA was prepared by using the Cyscribe first-strand cDNA labeling kit according to the manufacturer's instructions (AP Biotech). Mouse expression arrays were manufactured by the Advanced Technology Center at the National Cancer Institute, NIH (Gaithersburg, Md.) and contained 2,704 cDNA spots. After hybridization, the microarrays were scanned and quantified, using a GenePix 4000A microarray scanner (Axon Instruments, Foster City, Calif.). Data were analyzed with GenePix Pro 3.0 software and mAdb online software run by the NIH Center for Information Technology in collaboration with the National Cancer Institute Center for Cancer Research. Two separate reverse-fluor hybridizations were performed for each experiment, such that RNA samples A and B were labeled with Cy3 and Cy5, respectively, in the first hybridization but were labeled with Cy5 and Cy3, respectively, in the second hybridization. For data analysis, data were normalized such that the median Cy5/Cy3 ratio was set to 1. Two selection criteria were used to identify genes altered by HMGN3a or -b expression. First, any gene which was altered by more than 1.5-fold in four of the six arrays (standard and reverse fluor for three clones) was selected. Second, Student’s t test had to show a significant (P < 0.05) difference between either the ratios from these six arrays and the four arrays from the two control cell lines or between the six arrays for HMGN3a and the six for HMGN3b. Hierarchical clustering was performed using the programs Cluster and Treeview (Michael Eisen, Lawrence Berkeley National Laboratory, University of California, Berkeley).
RT-PCR.
Reverse transcriptase reactions were carried out using Multiscribe reverse transcriptase (Applied Biosystems) and oligo(dT)16 according to the manufacturer's instructions. An aliquot of the cDNA was used in real-time PCR, using SYBR green (Applied Biosystems) in an ABI PRISM 7900HT sequence detection system according to the manufacturer's instructions. The PCR primers were designed by using Primer Express (Applied Biosystems), and sequences are available on request. Efficient amplification of each mRNA was confirmed using a cDNA dilution series. For each sample, the mean threshold cycle (Ct) from three replicate PCRs was taken. Expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the control sample using either the comparative ΔΔCt method (30) or by comparison with standard curves as described by the manufacturer (Applied Biosystems).
Transient transfection.
Hepa-1 cells (90 to 95% confluent) were transfected with PCI-INS2, PCI-INS2-N3a, or PCI-INS2-N3b, using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen), and RNA was harvested 48 h later. Transfection efficiency was 60%, as assayed by transfection with pEGFP.
Immunofluorescence. (i) Cultured cells.
Cells were grown in chamber slides, fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), and then permeabilized with 1% Triton X-100 in PBS. The primary antibody against GLYT1 (GLYT11a from Alpha Diagnostics International) was used at 10 μg/ml in PBS with 10% fetal bovine serum. The secondary antibody was AlexaFluor 488 conjugated anti-rabbit immunoglobulin G (IgG) (Molecular Probes, Eugene, Oreg.) diluted 1:500 in PBS with 10% fetal bovine serum. DNA was stained with Hoescht stain (1:3,000 dilution in PBS with 20 mM glycine; Molecular Probes). Confocal microscopy was performed with a Zeiss 510 LSM confocal microscope configured with an argon/krypton and a helium/neon laser (Carl Zeiss).
(ii) Mouse eyes.
Unfixed eyes from 3-week-old C57BL/6J mice were embedded in tissue-freezing medium, and 16-μm sections were generated as described previously (43). HMGN3 was detected with antibody 2752 raised in rabbit (53). GLYT1 protein was detected with an antibody raised in rabbit by using a synthetic peptide corresponding to the final 15 amino acids on the carboxyl terminus of GLYT1 (41). The primary antibody for HMGN3 was diluted 1:5,000 and the GLYT1 antibody was diluted 1:200 in PBS with 1% bovine serum albumin (BSA). The primary antibodies were visualized after incubation with the appropriate secondary antibody (AlexaFluor 568 conjugated anti-rabbit IgG, diluted 1:50 in PBS with 1% BSA; Molecular Probes). Cell nuclei were detected with To-Pro-3 (1:3,000 dilution in PBS with 1% BSA; Molecular Probes). Sections were mounted as described previously (43) and stored at −20°C. Confocal microscopy was performed as described above.
Chromatin immunoprecipitation analysis.
Chromatin immunoprecipitations were performed by using a modification of the protocol described by Orlando et al. (34). Cells were grown to 70 to 80% confluence, harvested with trypsin, and resuspended in growth medium at 5 × 106 cells/ml. Formaldehyde was added to a final concentration of 0.35%, and the suspension was rocked at room temperature for up to 240 s. Approximately 1 × 108 cells were used for each time point. Reactions were stopped with glycine (final concentration, 0.125 M). Cells were washed at 4°C in PBS, in TEG (1 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl [pH 8]) containing 0.25% Triton X-100, and then in TEG containing 0.2 M NaCl and resuspended in 2 ml of TEG. Cells were sonicated in the presence of 0.5- mm-diameter microglass beads (Sigma). Sodium lauryl sarcosine was added to 0.5%, and the suspension was incubated at room temperature for 15 min and then centrifuged at 15,000 × g for 15 min. Each sample was made up to 13 ml in TEG plus 0.5% sarcosyl containing 1.42 g of cesium chloride/ml and adjusted to a refractive index of 1.3735. Samples were centrifuged in an SW40Ti rotor for 99 h at 40,000 rpm, and fractions were collected. The chromatin fragment length was assayed by electrophoresis after reversal of cross-links and purification of DNA. Chromatin fragments used for immunoprecipitation were shorter than 2 kb. Additional sonication was carried out to reduce fragment size if necessary.
Chromatin was dialyzed into ChIP buffer (0.01% sodium dodecyl sulfate, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.1], 167 mM NaCl). Aliquots of 100 μl of chromatin were diluted into 1 ml of ChIP buffer and precleared at 4°C with 4 μg of rabbit IgG for 2 h, followed by 2 h with 50 μl of protein A-agarose slurry blocked with salmon sperm DNA (Upstate Biotech). After the protein A-agarose was collected, immunoprecipitations (IPs) were performed overnight at 4°C using 5 μg each of two anti-HMGN3 antibodies, 2751 and 2752 (53). A “no-antibody” control was also set up. Protein A-agarose (50 μl) was added for 2 h, and antibody complexes were collected by centrifugation. The supernatant of the no-antibody control was collected as the input DNA. Complexes were washed and eluted, cross-links were reversed, and DNA was purified as described by Upstate Biotech. DNA was quantified with Picogreen as described by the manufacturer (Molecular Probes).
Real-time PCR.
Primer sets were created corresponding to 27 points across the Glyt1 gene locus. Sequences are available on request. PCRs contained SYBR green master mix and were carried out as described above. Results from the experimental IPs were normalized to the input DNA and then to a control primer set (set 1) using the ΔΔCt method (30).
RESULTS
Microarray analysis of cells constitutively overexpressing HMGN3a or HMGN3b.
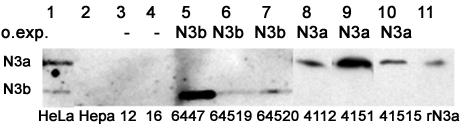
To test whether HMGN3 regulates the expression of specific genes, we generated stable cell lines overexpressing HMGN3a or HMGN3b. A mouse hepatoma cell line, Hepa-1, was used, as these cells have very low levels of endogenous HMGN3 (53). The expression of HMGN3a or -b in the transfected clones was assayed by Western blotting. Three stable lines constitutively expressing HMGN3a (4112, 4151, and 41515) and three lines expressing HMGN3b (6447, 64519, and 64520) were selected for further study. The level of HMGN3a or -b expression in these lines was comparable to that in HeLa cells (Fig. 1). Two lines containing an empty expression vector were selected as controls (lines 12 and 16).
FIG. 1.
Western blots showing expression of HMGN3a or -b in transgenic cell lines. The mobilities of HMGN3a (N3a) and HMGN3b (N3b) are indicated on the left. The recombinant protein expressed in each cell line is indicated above the respective lane. The cell type or clone number is indicated below each lane. Lane 1, HeLa cell extract, positive control. Lane 2, Hepa-1 cell extract, parent cell line. Lanes 3 and 4, extracts from control cells containing empty vector. Lanes 5 to 7, cells expressing recombinant HMGN3b. Lanes 8 to10, cells expressing recombinant HMGN3a. Lane 11, 5 μg of recombinant HMGN3a. Equal loading was confirmed by Coomassie blue staining of parallel gels (results not shown).
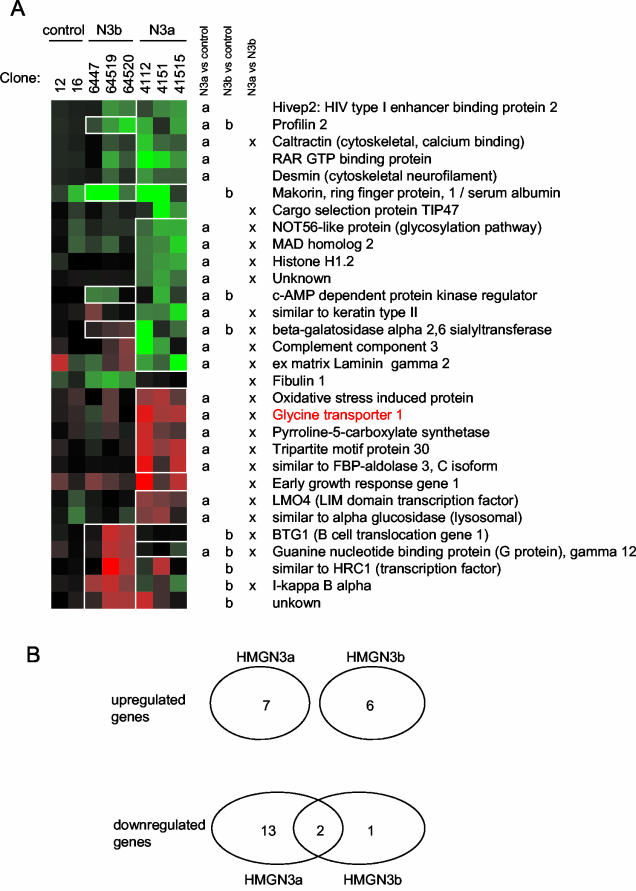
Two-color microarray analysis was performed with RNA from each clone, using RNA from the parent Hepa-1 cell line as the control in each hybridization. In cells expressing the full-length isoform, HMGN3a, the expression of 22 genes was altered (approximately 0.8% of the 2,704 cDNAs analyzed on the microarray chip) (Fig. 2). The microarray data was confirmed by real-time RT-PCR analysis of several of the genes with altered expression (data not shown). Seven of the genes were upregulated, and 15 were downregulated. In HMGN3b-expressing cells, six genes were upregulated and three genes were downregulated. There is no overlap between the genes upregulated by HMGN3a and HMGN3b, whereas 2 of the 15 genes downregulated by HMGN3a are also downregulated by HMGN3b. (Fig. 2). Overall, the results suggest that the two HMGN3 splice variants regulate the expression of distinct, but overlapping, subsets of genes. The 30 genes identified in total have a wide variety of functions within the cell, and there appears to be no selection for genes with related activities.
FIG. 2.
Changes in gene expression in cells expressing HMGN3a or HMGN3b. (A) Hierarchical clustering of genes whose expression was altered in HMGN3a or -b transgenic lines. Each vertical column represents a different transgenic cell line, and each horizontal row represents a gene. Red squares represent upregulation, green squares indicate downregulation, and black indicates no change in gene expression. The intensity of the color represents the magnitude of the change, which is calculated from the average of the two microarrays performed for each cell line. The results of Student's t tests for each gene are represented to the right of the cluster diagram. a or b indicates that the expression ratios for that gene in the microarrays for HMGN3a or HMGN3b lines, respectively, were significantly different from those in the microarrays for control lines (P < 0.05). The squares corresponding to these genes are outlined in white boxes on the cluster diagram for clarity. x indicates that the ratios for that gene in arrays for HMGN3a lines were significantly different from those for HMGN3b cell lines. The glycine transporter gene is marked in red. (B) Venn diagram to show intersections of gene subsets whose expression was altered in HMGN3a or -b transgenic lines.
Glyt1 expression is increased in cells expressing HMGN3a or -b.
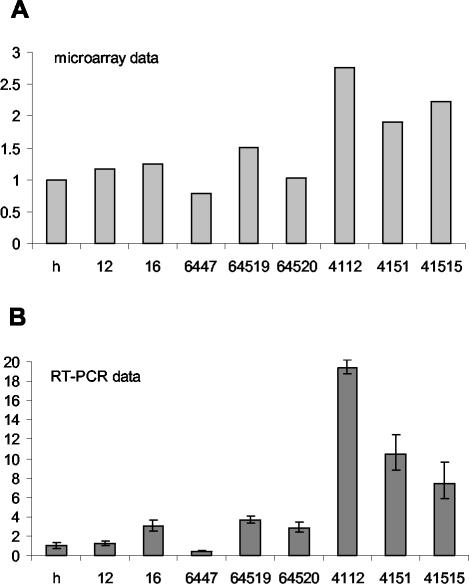
Among the genes with an altered expression pattern was the glycine transporter 1 gene (Glyt1). Glyt1 is known to be highly expressed in the brain and the eye, two tissues which also have high levels of HMGN3 expression (23, 53). Real-time RT-PCR analysis confirmed the microarray data. Expression of Glyt1 was increased in all three HMGN3a lines, with increases of between 1.9- and 2.8-fold as assayed by microarrays (Fig. 3A) and larger increases of between 7- and 19-fold when quantified more accurately by RT-PCR (Fig. 3B). In two of the three lines expressing the splice-form HMGN3b, Glyt1 expression was upregulated (2.9- to 3.7-fold increase assayed by RT-PCR), whereas it was downregulated in the third (2.4-fold decrease assayed by RT-PCR). This indicates that either HMGN3b does not affect Glyt1 expression or that its effect is weak and can be masked by variation between cell lines.
FIG. 3.
Increase in Glyt1 expression in transgenic cell lines expressing HMGN3a or HMGN3b. (A) Glyt1 expression in transgenic cell lines was assayed by microarrays. The ratio of expression in each cell line compared to the parent Hepa-1 line is shown. h, parent Hepa-1 cell line; 12 and 16, empty vector control lines. 6447, 64519, and 64520 all express HMGN3b. 4112, 4151, and 41515 express HMGN3a. (B) Real-time RT-PCR quantification of Glyt1 expression in transgenic cell lines. RNA samples were the same as those used for microarray analysis in panel A. Glyt1 mRNA levels were normalized to GAPDH and to expression in the parental cell line (Hepa-1). The average and standard deviation of triplicate assays are shown. The data are representative of results of several experiments done with different RNA preparations.
Glyt1 is expressed as three splice forms, Glyt1a, Glyt1b, and Glyt1c, which differ at their 5′ ends. Glyt1a and Glyt1b/c are transcribed from two different promoters (see diagram in Fig. 7A). In Hepa-1 cells, Glyt1a is the predominant isoform and is expressed at approximately 10,000-fold-higher levels than Glyt1b (data not shown); the level of Glyt1b mRNA was too low to quantify accurately. Similar RT-PCR results were obtained with primers specific for Glyt1a and with primers that detect all splice forms.
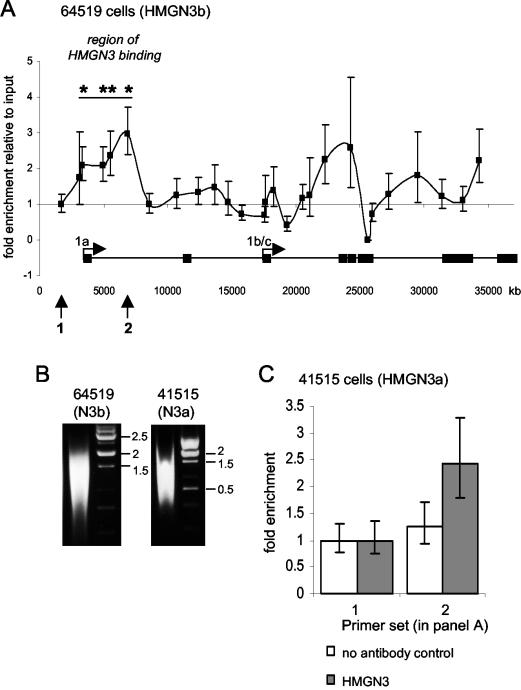
FIG. 7.
ChIP assays show that HMGN3 binds the Glyt1 gene in vivo. (A) ChIP assay for HMGN3 in 64519 cells expressing HMGN3b. Enrichment of each DNA sequence in the HMGN3 immunoprecipitate relative to the input DNA was normalized to the no-antibody control and is plotted against the position of the PCR primer pair within the Glyt1 gene locus. Enrichment at primer pair 1 (arrow) was set to 1. Error bars represent standard deviations between identical PCRs. Asterisks indicate primer sets where the HMGN3 IP is significantly different from that of the no-antibody control (P < 0.05). The positions of the two Glyt1 promoters (1a and 1b/c) and exons are shown at the bottom. Arrows 1 and 2 correspond to the primer pairs used in panel C. (B) Agarose gel showing the length of chromatin fragments prepared from cells expressing HMGN3b or HMGN3a. The fragments were prepared by sonication of formaldehyde-cross-linked chromatin. The cross-links were reversed and the DNA was purified prior to agarose gel electrophoresis. The same cross-linked chromatin populations were used for IP in panels A and C. (C) ChIP assays done with 41515 cells expressing HMGN3a. Enrichment in IP DNA versus input is shown for the no-antibody control (white bars) and anti-HMGN3 (grey bars) at two positions on the Glyt1 gene. The positions of primer sets 1 and 2 are shown at the bottom of panel A. Primer pair 1 is the control, and pair 2 corresponds to the region of HMGN3 enrichment. Error bars represent standard deviations of identical PCRs.
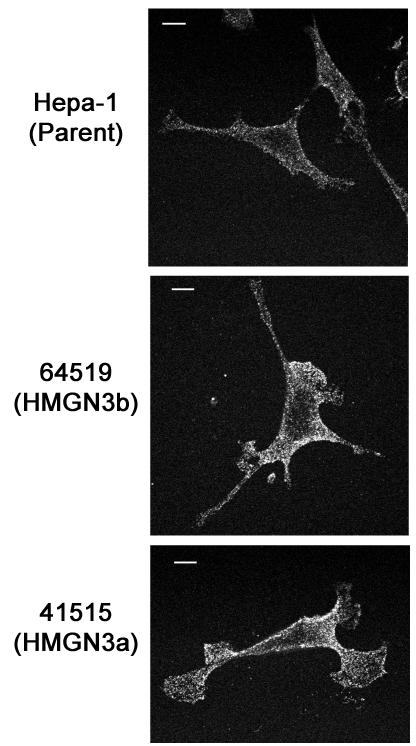
Immunofluorescence was used to investigate expression of GLYT1 protein. Increased GLYT1 expression can be seen in 64519 cells (Fig. 4, middle panel), which express HMGNb, as compared with the parent Hepa-1 cell line (Fig. 4, top panel). This is consistent with the 3.7-fold increase in Glyt1 mRNA observed in these cells. Increased GLYT1 protein was also observed in 41515 cells (Fig. 4, bottom panel), which express HMGN3a and have a 10-fold increase in Glyt1 mRNA.
FIG. 4.
Detection by immunofluorescence of GLYT1 protein in transgenic cell lines. GLYT1 immunofluorescence in the parent Hepa-1 line, 64519 cells (expressing HMGN3b), and 41515 cells (expressing HMGN3a) is shown. Scale bar = 2 μm. Each image is representative of 10 images of randomly selected cells. Cell line 4151 also had increased GLYT1 protein expression compared to Hepa-1 and empty vector controls (data not shown).
Glyt1 expression is altered in cells transiently expressing HMGN3a or HMGN3b.
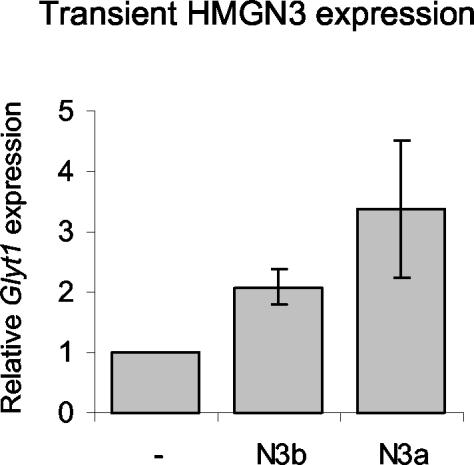
To further verify that HMGN3 enhances Glyt1 expression and to exclude the possibility that the elevated levels of Glyt1 are due to selection pressure during the lengthy process of clone selection, we transiently transfected the parent Hepa-1 cell line with expression vectors for HMGN3a or -b and harvested RNA 48 h later. RT-PCR revealed similar changes in gene expression in the transiently transfected cells and in the stable transgenic lines (Fig. 5). Glyt1 expression was increased by 3.4-fold in cells transiently expressing HMGN3a, and a significant increase of 2-fold was observed in cells expressing HMGN3b. The observation that Glyt1 expression is altered in cells both stably and transiently expressing HMGN3a strongly indicates that this gene is regulated, either directly or indirectly, by HMGN3a. The observations that HMGN3b increases Glyt1 expression in transient transfection assays and in two of three stable cell lines indicates that HMGN3b also regulates Glyt1 expression, although its effect is weaker than that of the full-length HMGN3a isoform.
FIG. 5.
Increase in Glyt1 expression after transient transfection with HMGN3a or HMGN3b. Cells were transfected with plasmids expressing HMGN3b or HMGN3a, and RNA was harvested 48 h later. mRNA was quantified by RT-PCR and was normalized to GAPDH and to expression in cells transfected with empty vector. Results show the averages and standard deviations from triplicate PCRs from two experiments.
HMGN3 and GLYT1 are coexpressed in the mouse retina.
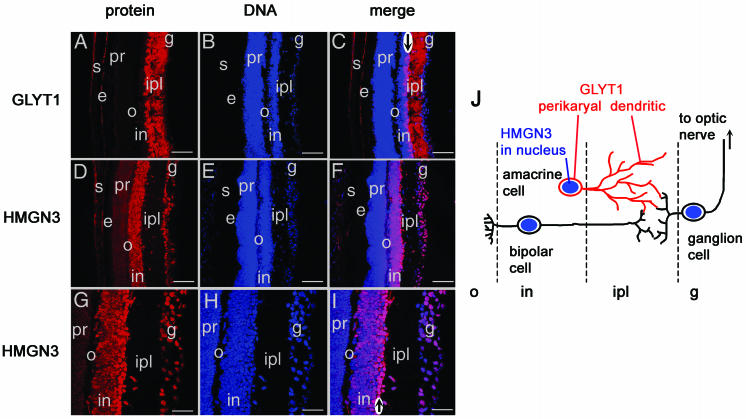
We have previously demonstrated that HMGN3 is expressed most highly in the mouse eye and brain (53) and that within the brain, expression is highest in glia cells expressing glial fibrillary acidic protein (23). GLYT1 is also expressed very highly in glia cells and in retinal neurons (1, 33, 54). We used immunofluorescence to test whether HMGN3 and GLYT1 are coexpressed in the same cells in the retina (Fig. 6). HMGN3 was localized to the nuclei of cells in the inner nuclear layer, which includes amacrine, bipolar, and horizontal neurons, and in the nuclei of ganglion neurons (Fig. 6D to I). In agreement with previous reports (33, 41, 42), the membrane protein GLYT1 was located predominantly in the inner plexiform layer (IPL) (Fig. 6A to C), corresponding to the location of the axons and dendrites projected by the amacrine, bipolar, and ganglion neurons. The absence of cell bodies in the IPL is demonstrated by lack of DNA staining in this region (Fig. 6B). Previous studies have shown that GLYT1 is expressed predominantly by amacrine neurons (33, 41, 42), which form the inner two rows of cell bodies of the inner nuclear layer next to the IPL. Indeed, colocalization of GLYT1 and DNA at the right-hand edge of the inner nuclear layer (see arrow in Fig. 6C) corresponds to the perikaryal localization of GLYT1 around these cell bodies. The strongest expression of HMGN3 also occurs in the same inner two rows of cell bodies (arrow in Fig. 6I) as the GLYT1 perikaryal staining. Thus, the immunolocalization studies demonstrate nuclear staining of HMGN3 and perikaryal GLYT1 staining at the edge of the inner nuclear layer and dendritic localization of GLYT1 in the IPL. While proof of exact colocalization would require double immunofluorescence, these data suggest that amacrine cells express both HMGN3 and GLYT1, as shown diagrammatically in Fig. 6J.
FIG. 6.
Colocalization of HMGN3 and GLYT1 in 3-week-old mouse retina. (A, B, and C) GLYT1 protein localization in the IPL, where the axons of bipolar and amacrine neurons connect to the dendrites of ganglion cells. (A) GLYT1, red; (B) To-Pro-3 DNA stain, blue; (C) merge of GLYT1 and DNA staining. (D, E, F, G, H, and I) HMGN3 protein localization in the nuclei of bipolar, amacrine, and ganglion cells. (D and G) HMGN3, red; (E and H) To-Pro-3 DNA stain, blue; (F and I) merge of HMGN3 and DNA staining, with the HMGN3/DNA overlap staining pink. Abbreviations: g, ganglion cells; ipl, inner plexiform layer; in, inner nuclear layer; o, outer plexiform layer; pr, photoreceptors; e, epithelial cells; s, sclera. Scale bars for panels A to F = 77 μm; scale bars for panels G to I = 38 μm. Arrows in panels C and I indicate cell bodies in the inner rows of the inner nuclear layer (see text for discussion). (J) Diagrammatic representation of the arrangement of cells within the retina. HMGN3 (blue) is located in the nuclei of bipolar, amacrine, and ganglion cells, whereas GLYT1 (red) is located on the cell membranes of amacrine cells.
HMGN3 binds to the Glyt1 gene in vivo.
Taken together with our analysis of the genes induced by stable and transient ectopic expression of HMGN3, the colocalization of GLYT1 and HMGN3 suggests a role for HMGN3 in regulating Glyt1 expression. Since HMGNs are nucleosome binding proteins, we used chromatin IP (ChIP) assays to determine whether HMGN3 binds directly to the Glyt1 gene. HMGN proteins do not bind chromatin with any sequence specificity (45), and although there is some evidence that they bind preferentially to active genes rather than inactive genes (39), it is not known whether they are targeted to promoters or transcribed regions. Therefore, we tested for the presence of HMGN3 at several sites in the 35-kb Glyt1 gene. Assays were carried out using cell line 64519, which expresses HMGN3b and has a 3.7-fold increase in Glyt1 expression. HMGN3 binding was assayed at 27 different regions across the Glyt1 gene locus by quantitative PCR (Fig. 7A). The average size of the chromatin fragments used was 1.2 kb, corresponding to approximately six nucleosomes (Fig. 7B).
We find that HMGN3b binding is enriched relative to the input and the no-antibody control over a 3.5-kb region around the start of Glyt1a transcription (see marked region in Fig. 7A). This region spans bases −328 to + 3170 relative to the Glyt1a transcription start site. ChIP assays were then performed on cells expressing HMGN3a to determine whether the longer splice form also binds to this region. Enrichment of HMGN3a was observed at primer set 2 relative to the no-antibody control and the control primer set 1 (Fig. 7C). These data demonstrate that HMGN3a also binds the Glyt gene in the vicinity of the Glyt1a start site, in agreement with the ChIP data for HMGN3b. These results show that both HMGN3a and HMGN3b bind to the Glyt1 gene in vivo, supporting the possibility that HMGN3 directly affects Glyt1 expression.
DISCUSSION
Identification of Glyt1 as a specific gene target for HMGN3.
HMGN proteins unfold chromatin and promote transcription and replication from chromatin templates in vitro (13, 35, 46, 49, 50). They are associated with sites of transcription within the nucleus (22) and have been shown to bind to and increase the nuclease sensitivity of chromatin regions containing active genes (39, 51, 52). However, it is still not known whether they are indiscriminate transcriptional regulators, designed to stimulate overall gene expression during a particularly active period, such as differentiation, or whether they specifically target individual genes. Because HMGN3 has a tissue-specific expression pattern (53) and binds to a hormone receptor (2, 26), we chose to investigate whether this member of the HMGN family regulates the expression of individual genes. Microarray analysis of transgenic cell lines show that HMGN3a and HMGN3b do indeed regulate the expression of individual genes, rather than having a broader, more general effect on transcription.
The changes in gene expression that we observed were in the range of 2- to 11-fold, depending on the cells used and the gene studied. These values are in the same range as those obtained in previous studies of enhancement of transcription or replication from minichromosomes by HMGN proteins, which varied between 2.2- and 9-fold, depending on the system used (2, 13, 15, 46, 47, 49, 50). This is consistent with the role of HMGN proteins as architectural elements, unfolding chromatin to improve access by other regulatory factors, and thus differs from the greater levels of transcriptional activation observed for sequence-specific transcriptional activators, for example. The access of both transcriptional activators and repressors to the DNA will be equally improved, and the overall effect of HMGN3 on the transcription of a particular gene is likely to depend on the DNA sequence elements within the gene and how the balance of activator and repressor binding is altered. The ability of HMGN3 to promote both gene activation and repression is analogous to that of linker histone H1. Ablation of histone H1 results in activation and repression of different genes in Tetrahymena (44). Similarly, the chromatin remodeling enzymes Swi/Snf and Rsc have also been shown to both activate and repress transcription (3, 32). Not all the observed changes in gene expression are likely to be a direct consequence of HMGN3 binding to those genes, however. HMGN3-dependent changes in the expression levels of regulatory factors will have knock-on effects on the genes those factors regulate, resulting in increased or decreased expression depending on the factors involved.
Glyt1 was of particular interest among the genes identified in the microarray screen, since its expression pattern is similar to that of HMGN3. RNA dot blots, Western blotting, and immunohistochemistry studies have shown that HMGN3 is highly expressed in the mouse eye and brain (23, 53). Glyt1 is known to be predominantly expressed in glia cells in the central nervous system and in retinal neurons (1, 7, 19, 33, 54). The Glyt1a splice form has also been detected in the liver and the lung, which is consistent with our results showing expression of Glyt1a but not Glyt1b in the hepatoma cell line used here. RT-PCR analysis of cells stably and transiently expressing HMGN3 showed that both splice forms upregulate Glyt1a expression.
HMGN3 binds the Glyt1 gene in vivo.
To gain insights into the molecular mechanism whereby HMGN3 affects Glyt1 expression, we used ChIP assays to determine whether HMGN3 was bound to the Glyt1 gene in vivo. Glyt1 is expressed in glia cells as three splice forms, Glyt1a, Glyt1b, and Glyt1c. Glyt1a and Glyt1b/c are transcribed from two different promoters, 1a and 1b/c. GLYT1a and GLYT1b proteins differ from each other by 10 and 15 amino acids, respectively, at the N terminus, while GLYT1c has an extra exon encoding 40 amino acid residues inserted after the first 15 residues of GLYT1b (6, 7, 31, 54). Glyt1a is expressed in grey matter in the brain, whereas Glyt1b/c is expressed in white matter (7). Very little is known about how these promoters are regulated, although it has been shown that a 2.5-kb fragment from the 5′-flanking region of Glyt1a can drive transcription from a transiently transfected reporter plasmid (6).
The high-resolution ChIP results presented here show HMGN3 binding to a 3.5-kb region encompassing the start site of Glyt1a transcription, supporting a direct role for HMGN3 in Glyt1a expression. Although we have not shown that the profile of HMGN3a is identical to that of HMGN3b throughout the Glyt1 gene, we expect the two to be very similar, as they both contain the nucleosome binding domain. The nucleosome binding domain is an independent functional domain, and its interaction with chromatin is identical to that of the corresponding full-length protein in all respects (12, 40, 47). The specific recruitment of HMGN3 to a relatively small region of the Glyt1 gene is intriguing, and we are investigating whether particular protein-protein interactions or histone tail modifications are responsible.
The binding of HMGN3 over part of the Glyt1a promoter and for up to 3 kb downstream suggests that HMGN3 may be important for transcription initiation and/or elongation. Previous studies have shown that HMGN proteins can increase the efficiency of either transcription initiation (35) or elongation (14, 15), depending on the system studied. HMGN proteins also increase the initiation of replication and the rate of replication fork movement (49). The splice variant HMGN3b lacks most of the C-terminal chromatin-unfolding domain (53), which is required for HMGN proteins to unfold chromatin (14, 47). Truncation of HMGN2 at the same point abolishes its ability to open up chromatin or to compete with histone H1, although its affinity for nucleosomes is unaffected (10, 14, 47). However, both HMGN3b and HMGN2 are able to interact with TR/RXR and to stimulate thyroid hormone- and TR/RXR-dependent transcription from chromatin templates (2, 26). HMGN3b can thus promote transcription through mechanisms other than direct unfolding of chromatin. HMGN3b could also regulate transcription by influencing modifications of the core histone N-terminal tails; for example, HMGN1 inhibits phosphorylation of histone H3 on serine 10 (J. H. Lim and M. Bustin, unpublished results). Modulation of the histone code (24) by HMGN3b could influence the recruitment of other factors and thus alter the level of gene expression.
HMGN3 and GLYT1 are coexpressed in vivo.
To confirm whether GLYT1 and HMGN3 were indeed coexpressed in the same cells, we performed immunofluorescence on mouse retinas. In the retina, bipolar and amacrine cells transfer information from activated photoreceptors to the IPL, where the signal is transferred to ganglion cells. The cell bodies of amacrine neurons lie in a narrow layer between the bipolar cells and the IPL, and their dendrites extend into the IPL. Our results are consistent with previous studies showing that GLYT1 is predominantly located on nerve fibers in the IPL and on plasma membranes surrounding the cell bodies of amacrine neurons (33, 41, 42). We show that the strongest HMGN3 expression is in the innernuclear layer, where the cell bodies of bipolar and amacrine neurons are located, and in the ganglion cell layer. HMGN3 expression is particularly strong in a narrow layer of cells where the cell bodies of the amacrine neurons are located (33). These data strongly suggest that HMGN3 and GLYT1 are coexpressed in the same cell types. The antibody used detects both HMGN3a and HMGN3b, and previous work has shown that HMGN3a and HMGN3b are always expressed together (53). In mouse tissues, the level of HMGN3b is always greater than that of HMGN3a, and this ratio does not change between the different tissues examined (53). Thus, the immunolocalization most likely reflects the expression patterns of both HMGN3a and HMGN3b. The coexpression of HMGN3 and GLYT1 in the retina supports the conclusion that HMGN3 regulates Glyt1 expression in vivo.
Examination of previous expression studies reveals that Hmgn3 and Glyt1 may be coexpressed in some other tissues within the body, including the kidney and the pancreas (25, 53). However, it is clear that their expression patterns do not always overlap, as can be seen from the expression of HMGN3 but not GLYT1 in bipolar and ganglion cells in this study. The high expression of Glyt1 in amacrine and glia cells is likely to be regulated by several sequence-specific transcription factors with their own tissue-specific patterns of expression. As a chromatin-unfolding protein, HMGN3 is likely to modulate the abilities of the sequence-specific factors to activate Glyt1 expression and thus refine its tissue-specific expression pattern.
In summary, the use of a microarray screen followed by RT-PCR analysis identified several target genes for HMGN3. Our data support the possibility that HMGN proteins can act specifically to regulate the expression of individual genes. We have shown that both HMGN3 splice forms upregulate Glyt1 expression and that they are bound to the endogenous Glyt1 gene in vivo. We have also demonstrated that both HMGN3 and GLYT1 are expressed in the same cell types in the mouse retina. Taken together, our data show that HMGN3 is a potential regulator of Glyt1 expression in vivo, although further investigations will be required to prove this relationship in mice. The Glyt1 locus will now provide a model system for the study of how HMGN proteins are recruited to specific genes and how they influence the different stages in transcription initiation and elongation. Given the known role of GLYT1 in neurotransmission, an understanding of the molecular mechanisms regulating Glyt1 expression may also have important implications for several disease conditions, including schizophrenia (31).
Acknowledgments
We are grateful to David Pow (University of Queensland, Brisbane, Australia) for providing one of the GLYT1 antibodies, to Gary Felsenfeld (NIH, Bethesda, Md.) for providing plasmid pBAW3, containing the HS4 insulator, and to Adam West (University of Glasgow) for providing protocols and advice concerning ChIP assays.
REFERENCES
- 1.Adams, R. H., K. Sato, S. Shimada, M. Tohyama, A. W. Puschel, and H. Betz. 1995. Gene structure and glial expression of the glycine transporter GlyT1 in embryonic and adult rodents. J. Neurosci. 15:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano, T., K. Leu, K. Yoshizato, and Y. B. Shi. 2002. Thyroid hormone regulation of a transcriptional coactivator in Xenopus laevis: implication for a role in postembryonic tissue remodeling. Dev. Dyn. 223:526-535. [DOI] [PubMed] [Google Scholar]
- 3.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 4.Birger, Y., Y. Ito, K. L. West, D. Landsman, and M. Bustin. 2001. HMGN4, a new nucleosomal binding protein encoded by an intronless gene. DNA Cell Biol. 20:257-264. [DOI] [PubMed] [Google Scholar]
- 5.Birger, Y., K. L. West, Y. V. Postnikov, J. H. Lim, T. Furusawa, J. P. Wagner, C. S. Laufer, K. H. Kraemer, and M. Bustin. 2003. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 22:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowsky, B., and B. J. Hoffman. 1998. Analysis of a gene encoding two glycine transporter variants reveals alternative promoter usage and a novel gene structure. J. Biol. Chem. 273:29077-29085. [DOI] [PubMed] [Google Scholar]
- 7.Borowsky, B., E. Mezey, and B. J. Hoffman. 1993. Two glycine transporter variants with distinct localization in the CNS and peripheral tissues are encoded by a common gene. Neuron 10:851-863. [DOI] [PubMed] [Google Scholar]
- 8.Bustin, M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 26:431-437. [DOI] [PubMed] [Google Scholar]
- 9.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catez, F., D. T. Brown, T. Misteli, and M. Bustin. 2002. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3:760-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, J. H., A. C. Bell, and G. Felsenfeld. 1997. Characterization of the chicken beta-globin insulator. Proc. Natl. Acad. Sci. USA 94:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crippa, M. P., P. J. Alfonso, and M. Bustin. 1992. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J. Mol. Biol. 228:442-449. [DOI] [PubMed] [Google Scholar]
- 13.Crippa, M. P., L. Trieschmann, P. J. Alfonso, A. P. Wolffe, and M. Bustin. 1993. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO J. 12:3855-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding, H. F., M. Bustin, and U. Hansen. 1997. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol. Cell. Biol. 17:5843-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding, H. F., S. Rimsky, S. C. Batson, M. Bustin, and U. Hansen. 1994. Stimulation of RNA polymerase II elongation by chromosomal protein HMG-14. Science 265:796-799. [DOI] [PubMed] [Google Scholar]
- 16.Dorbic, T., and B. Wittig. 1987. Chromatin from transcribed genes contains HMG17 only downstream from the starting point of transcription. EMBO J. 6:2393-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorbic, T., and B. Wittig. 1986. Isolation of oligonucleosomes from active chromatin using HMG17-specific monoclonal antibodies. Nucleic Acids Res. 14:3363-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 19.Goebel, D. J. 1996. Quantitative gene expression of two types of glycine transporter in the rat central nervous system. Brain Res. Mol. Brain Res. 40:139-142. [DOI] [PubMed] [Google Scholar]
- 20.Goodwin, G. H., C. G. Mathew, C. A. Wright, C. D. Venkov, and E. W. Johns. 1979. Analysis of the high mobility group proteins associated with salt-soluble nucleosomes. Nucleic Acids Res. 7:1815-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-550, 552-554, 556. [DOI] [PubMed] [Google Scholar]
- 22.Hock, R., F. Wilde, U. Scheer, and M. Bustin. 1998. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 17:6992-7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, Y., and M. Bustin. 2002. Immunohistochemical localization of the nucleosome-binding protein HMGN3 in mouse brain. J. Histochem. Cytochem. 50:1273-1275. [DOI] [PubMed] [Google Scholar]
- 24.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 25.Kim, K. M., S. F. Kingsmore, H. Han, T. L. Yang-Feng, N. Godinot, M. F. Seldin, M. G. Caron, and B. Giros. 1994. Cloning of the human glycine transporter type 1: molecular and pharmacological characterization of novel isoform variants and chromosomal localization of the gene in the human and mouse genomes. Mol. Pharmacol. 45:608-617. [PubMed] [Google Scholar]
- 26.Lee, J. W., H. S. Choi, J. Gyuris, R. Brent, and D. D. Moore. 1995. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with thyroid hormone receptor. Mol. Endocrinol. 9:243-254. [DOI] [PubMed] [Google Scholar]
- 27.Lehtonen, S., and E. Lehtonen. 2001. HMG-17 is an early marker of inductive interactions in the developing mouse kidney. Differentiation 67:154-163. [DOI] [PubMed] [Google Scholar]
- 28.Lehtonen, S., V. M. Olkkonen, M. Stapleton, M. Zerial, and E. Lehtonen. 1998. HMG-17, a chromosomal non-histone protein, shows developmental regulation during organogenesis. Int. J. Dev. Biol. 42:775-782. [PubMed] [Google Scholar]
- 29.Lim, J. H., M. Bustin, V. V. Ogryzko, and Y. V. Postnikov. 2002. Metastable macromolecular complexes containing high mobility group nucleosome-binding chromosomal proteins in HeLa nuclei. J. Biol. Chem. 277:20774-20782. [DOI] [PubMed] [Google Scholar]
- 30.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Corcuera, B., A. Geerlings, and C. Aragon. 2001. Glycine neurotransmitter transporters: an update. Mol. Membr. Biol. 18:13-20. [PubMed] [Google Scholar]
- 32.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 33.Menger, N., D. V. Pow, and H. Wassle. 1998. Glycinergic amacrine cells of the rat retina. J. Comp. Neurol. 401:34-46. [DOI] [PubMed] [Google Scholar]
- 34.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 35.Paranjape, S. M., A. Krumm, and J. T. Kadonaga. 1995. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 9:1978-1991. [DOI] [PubMed] [Google Scholar]
- 36.Pash, J., N. Popescu, M. Matocha, S. Rapoport, and M. Bustin. 1990. Chromosomal protein HMG-14 gene maps to the Down syndrome region of human chromosome 21 and is overexpressed in mouse trisomy 16. Proc. Natl. Acad. Sci. USA 87:3836-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 38.Postnikov, Y. V., J. E. Herrera, R. Hock, U. Scheer, and M. Bustin. 1997. Clusters of nucleosomes containing chromosomal protein HMG-17 in chromatin. J. Mol. Biol. 274:454-465. [DOI] [PubMed] [Google Scholar]
- 39.Postnikov, Y. V., V. V. Shick, A. V. Belyavsky, K. R. Khrapko, K. L. Brodolin, T. A. Nikolskaya, and A. D. Mirzabekov. 1991. Distribution of high mobility group proteins 1/2, E and 14/17 and linker histones H1 and H5 on transcribed and non-transcribed regions of chicken erythrocyte chromatin. Nucleic Acids Res. 19:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postnikov, Y. V., L. Trieschmann, A. Rickers, and M. Bustin. 1995. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J. Mol. Biol. 252:423-432. [DOI] [PubMed] [Google Scholar]
- 41.Pow, D. V., and A. E. Hendrickson. 1999. Distribution of the glycine transporter glyt-1 in mammalian and nonmammalian retinae. Vis. Neurosci. 16:231-239. [DOI] [PubMed] [Google Scholar]
- 42.Pow, D. V., and A. E. Hendrickson. 2000. Expression of glycine and the glycine transporter glyt-1 in the developing rat retina. Vis. Neurosci. 17:1-9. [DOI] [PubMed] [Google Scholar]
- 43.Reed, N. A., D. J. Oh, K. J. Czymmek, and M. K. Duncan. 2001. An immunohistochemical method for the detection of proteins in the vertebrate lens. J. Immunol. Methods 253:243-252. [DOI] [PubMed] [Google Scholar]
- 44.Shen, X., and M. A. Gorovsky. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86:475-483. [DOI] [PubMed] [Google Scholar]
- 45.Shirakawa, H., J. E. Herrera, M. Bustin, and Y. Postnikov. 2000. Targeting of high mobility group-14/-17 proteins in chromatin is independent of DNA sequence. J. Biol. Chem. 275:37937-37944. [DOI] [PubMed] [Google Scholar]
- 46.Trieschmann, L., P. J. Alfonso, M. P. Crippa, A. P. Wolffe, and M. Bustin. 1995. Incorporation of chromosomal proteins HMG-14/HMG-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 14:1478-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trieschmann, L., Y. V. Postnikov, A. Rickers, and M. Bustin. 1995. Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional enhancement domain distinct from the nucleosomal binding domain. Mol. Cell. Biol. 15:6663-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 49.Vestner, B., M. Bustin, and C. Gruss. 1998. Stimulation of replication efficiency of a chromatin template by chromosomal protein HMG-17. J. Biol. Chem. 273:9409-9414. [DOI] [PubMed] [Google Scholar]
- 50.Weigmann, N., L. Trieschmann, and M. Bustin. 1997. Enhancement of the transcription potential of nascent chromatin by chromosomal proteins HMG-14/-17 is coupled to nucleosome assembly and not DNA synthesis. DNA Cell Biol. 16:1207-1216. [DOI] [PubMed] [Google Scholar]
- 51.Weisbrod, S., M. Groudine, and H. Weintraub. 1980. Interaction of HMG 14 and 17 with actively transcribed genes. Cell 19:289-301. [DOI] [PubMed] [Google Scholar]
- 52.Weisbrod, S., and H. Weintraub. 1981. Isolation of actively transcribed nucleosomes using immobilized HMG 14 and 17 and an analysis of alpha-globin chromatin. Cell 23:391-400. [DOI] [PubMed] [Google Scholar]
- 53.West, K. L., Y. Ito, Y. Birger, Y. Postnikov, H. Shirakawa, and M. Bustin. 2001. HMGN3a and HMGN3b, two protein isoforms with a tissue-specific expression pattern, expand the cellular repertoire of nucleosome-binding proteins. J. Biol. Chem. 276:25959-25969. [DOI] [PubMed] [Google Scholar]
- 54.Zafra, F., C. Aragon, L. Olivares, N. C. Danbolt, C. Gimenez, and J. Storm-Mathisen. 1995. Glycine transporters are differentially expressed among CNS cells. J. Neurosci. 15:3952-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]