Abstract
The general transcription factor TFIIB is required for accurate initiation, although the mechanism by which RNA polymerase II (RNAP II) identifies initiation sites is not well understood. Here we describe results from genetic and biochemical analyses of an altered form of yeast TFIIB containing an arginine-78 → cysteine (R78C) replacement in the “B-finger” domain. TFIIB R78C shifts start site selection downstream of normal and confers a cold-sensitive growth defect (Csm−). Suppression of the R78C Csm− phenotype identified a functional interaction between TFIIB and the Rpb2 subunit of RNAP II and defined a novel role for Rpb2 in start site selection. The rpb2 suppressor encodes a glycine-369 → serine (G369S) replacement, located in the “lobe” domain of Rpb2 and near the Rpb9 subunit, which was identified previously as an effector of start site selection. The Rpb2-Rpb9 “lobe-jaw” region of RNAP II is downstream of the catalytic center and distal to the site of RNAP II-TFIIB interaction. A TFIIB R78C mutant extract was defective for promoter-specific run-on transcription but yielded an altered pattern of abortive initiation products, indicating that the R78C defect does not preclude initiation. The sua7-3 rpb2-101 double mutant was sensitive to 6-azauracil in vivo and to nucleoside triphosphate substrate depletion in vitro. In the context of the recent X-ray structure of the yeast RNAP II-TFIIB complex, these results define a functional interaction between the B-finger domain of TFIIB and the distal lobe-jaw region of RNAP II and provide insight into the mechanism of start site selection.
Promoter-dependent transcription by RNA polymerase II (RNAP II) requires a set of general transcription factors that include the TATA-binding protein (TBP), TFIIB, TFIIE, TFIIF, and TFIIH (20, 36). Order-of-addition experiments with purified general transcription factors and RNAP II defined a stepwise model for assembly of the transcription preinitiation complex (PIC). Accordingly, TBP binds to the TATA element of promoter DNA to nucleate PIC assembly, followed by binding of TFIIB both upstream and downstream of TATA. The DNA-TBP-TFIIB ternary complex forms the binding site for RNAP II, which enters the complex in association with TFIIF. TFIIE and TFIIH complete PIC assembly and are required for ATP-dependent promoter melting by RNAP II (27). An additional factor, TFIIA, is not essential for accurate initiation in vitro but stabilizes formation of the DNA-TBP-TFIIB ternary complex and stimulates recruitment of TFIID (TBP plus TBP-associated factors) to promoter DNA (1, 28, 44). Although the pathway for PIC assembly in vivo might be different from that defined in vitro, PIC formation has been shown to proceed via formation of structural intermediates in vivo (44).
RNAP II comprises 12 subunits, encoded by the RPB1 to RPB12 genes in Saccharomyces cerevisiae (53, 54). The two largest subunits, Rpb1 and Rpb2, are homologs of the β′ and β subunits, respectively, of bacterial RNA polymerase, and the Rpb3/Rpb11 heterodimer is structurally and functionally related to the α2 dimer. Accordingly, Rpb1, Rpb2, Rpb3, and Rpb11 form the catalytic core of RNAP II and the counterpart of the bacterial α2ββ′ RNAP core enzyme. Rpb6 facilitates RNAP II assembly and stability and is the counterpart of the bacterial ω subunit (31). High-resolution X-ray structures of 10-subunit (lacking Rpb4/Rpb7) and 12-subunit yeast RNAP II complexes have been described previously (4, 10, 13, 14), as have yeast RNAP II-DNA-RNA transcribing complexes (19, 51a). These structures, in combination with high-resolution structures of bacterial RNAPs (32), offer insight into the mechanism of transcription and a framework for the interpretation of a wealth of genetic and biochemical data.
The RNAP II transcription cycle involves several distinct steps, including PIC assembly, open complex formation (promoter melting), initiation, promoter escape, elongation, termination, and reinitiation (19a, 51). Promoter escape is a transition phase characterized by functional instability of the RNAP II transcription complex and the synthesis of short, abortive transcripts. Commitment to promoter escape is dependent upon the length of the nascent transcript and is facilitated by TFIIE and TFIIH (15, 24, 29, 37). As RNAP II clears the promoter, numerous protein-protein and protein-DNA contacts established in the open complex must be disrupted, allowing RNAP II to move forward into the elongation phase. After initiation, a subcomplex of the PIC that includes TFIIA, TFIID, TFIIE, TFIIH, and Mediator subunits, but neither TFIIB nor TFIIF, remains assembled at the promoter to enhance subsequent rounds of transcription (55).
TFIIB plays a key role in transcription initiation. The SUA7 gene, which encodes yeast TFIIB, was initially identified based on mutations that alter start site selection (40). The sua7-1 and sua7-3 alleles shift initiation at the CYC1 and ADH1 promoters downstream of normal and encode, respectively, glutamate-62 → lysine (E62K) and arginine-78 → cysteine (R78C) replacements (41). TFIIB E62 replacements completely abolish transcription in vitro but do not affect PIC assembly (12, 44). Structural analyses of TFIIB have defined distinct domains: the C-terminal core domain, which consists of two α-helical repeats that interact with TBP and promoter DNA (5, 35); an N-terminal region that forms a zinc ribbon and interacts with TFIIF and the “dock” domain of RNAP II (11, 57); and a phylogenetically conserved domain downstream of the zinc ribbon that is critical for accurate start site selection in yeast (6, 12, 18, 39, 41, 44, 56), humans (17, 21), and Archaea (7). The recent high-resolution X-ray structure of a yeast RNAP II-TFIIB complex defined the conserved region as a “B-finger” (residues 55 to 88) that projects into the RNAP II active center via the “saddle” between the “clamp” and “wall” domains (10a). This structure suggests that a steric clash between the B-finger and nascent RNA on the saddle accounts for abortive initiation (10a).
In an effort to further define the mechanism of initiation by RNAP II, we have isolated and characterized suppressors of TFIIB start site defects. Earlier work from our laboratory identified mutations in the genes encoding the larger subunit of TFIIF (ssu71/TFG1) and the Rpb9 subunit (ssu73/RPB9) of RNAP II as suppressors of TFIIB E62K (49, 50). Here we report the identification of Rpb2 as a suppressor of the TFIIB R78C replacement and examine the effects of R78C and the Rpb2 suppressor in vitro. We interpret our results in the context of RNAP II and RNAP II-TFIIB tertiary structures.
MATERIALS AND METHODS
Yeast strains.
The yeast strains used in this study were derived from strain T16 (MATα cyc1-5000 cyc7-67 ura3-52 leu2-3,112 cyh2) (40). YIP363 (sua7-3) is a spontaneous Cyc+, cold-sensitive (Csm−) revertant of T16 (40); sua7-3 encodes an arginine-78 → cysteine (R78C) replacement in TFIIB (41). YBC14 (sua7-3 rpb2-101) is a UV-exposed Csm+ revertant of YIP363. YMH183 (MATa his1 lys2 trp2 sua7-3) is a meiotic segregant derived from a cross of YIP363 with D311-3A (MATa his1 lys2 trp2). YBC112 (sua7-3 ura3 RPB2-URA3) was tagged at the RPB2 locus by site-specific integration of URA3 with plasmid pDP39.
Growth media, genetic methods, and phenotypes.
Growth media were prepared by using standard recipes (47). 6AU was added either to synthetic complete medium at a concentration of 200 μg/ml or to synthetic complete medium lacking uracil at 30 μg/ml. Standard yeast genetic methods were used for making crosses, selecting diploids, inducing sporulation, and dissecting tetrads (48). The following designations denote phenotypes: Tsm− and Csm−, impaired growth relative to a wild-type strain on yeast extract-peptone-dextrose (YPD) medium at 37 or 38°C and 16°C, respectively; Cyc+, growth on medium containing 2% lactic acid as the sole carbon source; Ino−, impaired growth on inositol-lacking (−Ino) medium at 30°C relative to growth on +Ino control medium; and 6AUs and 6AUr, impaired or normal growth on 6AU medium at 30°C relative to growth on either synthetic or rich medium.
Identification of the rpb2-101 suppressor of sua7-3.
A total of 6 × 107 cells of strain YIP363 were spread on YPD medium and irradiated with UV light to 50% cell survival. Eighty-three independent revertants were isolated. One of these strains, YBC14, exhibited pleiotropic phenotypes, including Tsm−, Ino−, and 6AUs, and was chosen for further analysis. YBC14 was backcrossed with an sua7-3 mutant of the opposite mating type (YMH183), and the resulting diploid strain was phenotypically identical to the primary mutant YIP363 (Csm− Tsm+ Ino+ 6AUr), indicating that all revertant phenotypes are the result of recessive mutation(s). Following sporulation and tetrad dissection, the Tsm−, Ino−, and 6AUs phenotypes cosegregated with Csm+, thereby defining these phenotypes as markers for the sua7-3 suppressor.
To establish allelism between the suppressor and RPB2, we tagged the RPB2 locus with the URA3 marker to create strain YBC112 (sua7-3 ura3 RPB2::URA3), which was crossed with YBC14 (sua7-3 ura3 rpb2-101), sporulated, and dissected. The Csm−:Csm+ and Ura+:Ura− phenotypes segregated 2:2, and the Csm+/Ura− and Csm−/Ura+ phenotypes cosegregated among all four-spore progeny, thereby establishing that the suppressor is allelic to RPB2. Accordingly, we designated the sua7-3 suppressor rpb2-101.
Determination of transcription start sites.
Primer extension was performed as described previously, by using total RNA and the ADH1-specific primer oIP87 (40). DNA products were resolved in an 8% polyacrylamide DNA sequencing gel and visualized by autoradiography.
Isolation and sequence analysis of rpb2-101.
The rpb2-101 allele was isolated from the chromosomal DNA of strain YBC14 by gap repair as described previously (38). The resulting plasmid failed to rescue the 6AUs and Ino− phenotypes when introduced into YBC14, confirming the recovery of rpb2-101. Template DNA was prepared by using a commercial kit (QIAGEN), and the entire rpb2-101 open reading frame (ORF) was sequenced with an ABI Prism automated DNA sequencer by using a primer set that spans both strands of RPB2 DNA. The DNA sequence was compared with the published RPB2 sequence in the Saccharomyces Genome Database (www.yeastgenome.org).
Purification of recombinant TFIIB.
The plasmid construct for expression of recombinant TFIIB (rTFIIB) was acquired from Steve Buratowski (Harvard Medical School). The sua7-3 allele encoding TFIIB R78C was amplified by PCR, cloned into the vector pET-11d (Stratagene), and expressed in Escherichia coli strain BL21. The sua7-3 mutation encoding R78C in pET-11d was confirmed by DNA sequence analysis. The normal and R78C forms of rTFIIB were purified as described previously (12). Briefly, BL21 transformants were grown in 2× yeast extract-tryptone medium supplemented with ampicillin (100 μg/ml) at 37°C. When cell growth reached an A600 of 0.5, rTFIIB expression was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (0.4 mM) for 4 h. Cell extracts were prepared and passed through an S-Sepharose column (Amersham Pharmacia Biotech). rTFIIB was eluted with 0.5 M potassium acetate. The full-length rTFIIB fraction also contained a truncation product that arises from internal translation initiation at the methionine-119 codon. To separate the full-length protein from the truncation product, the S-Sepharose eluate was applied to a hydroxyapatite column (Bio-Rad) and equilibrated with 10 mM potassium phosphate (pH 7.6), 100 mM potassium acetate, 20% glycerol, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride. rTFIIB was eluted with a gradient (100 to 240 mM potassium acetate) of the same buffer. Proteins were monitored by Coomassie blue staining and confirmed by Western blotting with anti-TFIIB antibody.
Purification of yeast proteins.
RNAP II and TFIIF were purified from yeast strains SHY407B and SHY391A expressing TAP-tagged Rpb9 and Tfg1, respectively. These strains were a generous gift from Steve Hahn (Fred Hutchinson Cancer Research Center). Cells were cultured at 30°C in YPD medium, collected at late log phase, and lysed mechanically with glass beads in disruption buffer (0.2 M Tris-H2SO4, pH 7.9, 0.39 M ammonium sulfate, 10 mM magnesium sulfate, 20% glycerol, 1 mM EDTA). Proteins were purified as described previously (43).
In vitro transcription assays.
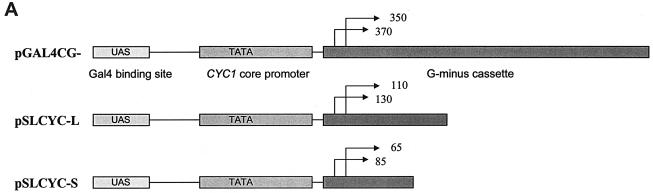
Whole-cell extracts from T16, YIP363, and YBC14 were prepared by using standard procedures (52). The 25-μl transcription reaction mixture contained 50 mM HEPES-KOH (pH 7.3), 100 mM glutamate, 15 mM magnesium acetate, 5 mM EGTA, 2.5 mM DTT, 10% glycerol, 4 mM phosphoenolpyruvate, 0.25 U of inhibit-ACE, 0.4 mM each ATP and CTP, 2 μM UTP, 5 μCi of [α-32P]UTP (3,000 Ci/mmol; NEN Life Science Products), 100 μg of whole-cell extract protein, and 300 ng of the template plasmids pGAL4CG−, pSLCYC-L, or pSLCYC-S (obtained from N. Woychik). Reaction mixtures were incubated at 25°C for 30 min without nucleotide triphosphates (NTPs). Initiation occurred upon addition of NTPs at 25°C for another 30 min and was stopped and processed with RNase T1 and proteinase K as described previously (30). RNA was extracted with phenol-chloroform, precipitated, resolved in a 6, 15, or 24% polyacrylamide gel containing 7 M urea, and visualized by autoradiography. For activated transcription, pGAL4CG− was incubated with GAL4-VP16 (obtained from D. Reinberg) for 5 min on ice before adding whole-cell extract. Reactions were supplemented, where indicated, with 80 ng of rTFIIB. For runoff transcription, the HindIII/SacII fragment of pSLCYC-S, which contains the CYC1 promoter and part of the G-less cassette, was used as a linear template (see Fig. 4A).
FIG. 4.
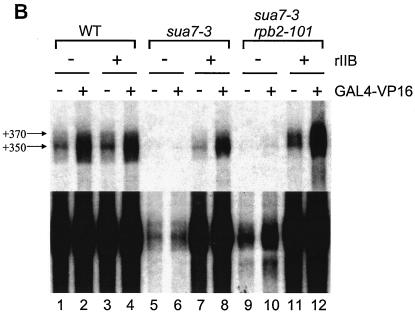
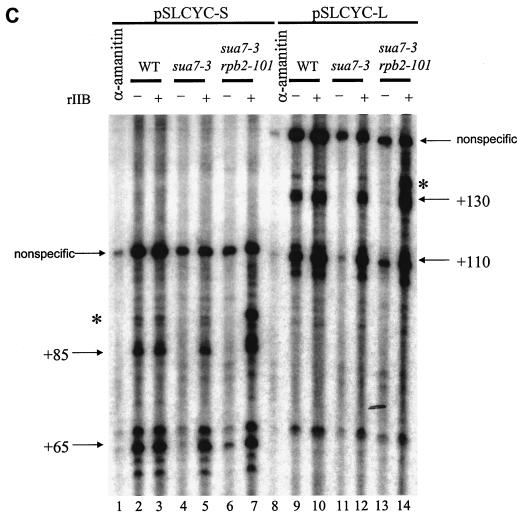
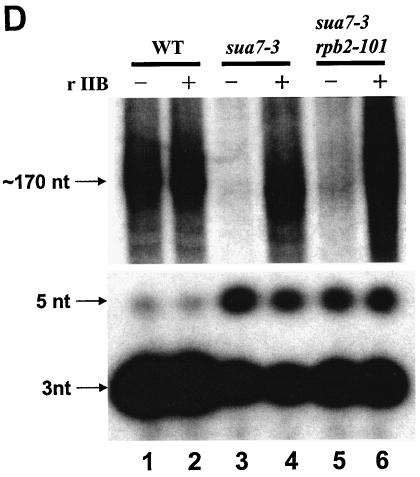
Effects of sua7 and rpb2 mutations on transcription in vitro. (A) Schematic depiction of the pGAL4CG−, pSLCYC-L, and pSLCYC-S templates. The pGAL4CG− template contains a single Gal4-binding site, a CYC1 TATA element, and a G-less cassette; following RNase T1 digestion, products of 350 and 370 nt corresponding to initiation at the indicated sites are produced. The templates pSLCYC-L and pSLCYC-S are identical to pGAL4CG− except that 110- and 130- or 85- and 65-nt transcripts are produced. (B) In vitro transcription was performed by using the pGAL4CG− template with whole-cell extracts from strains T16 (SUA7 RPB2), YIP363 (sua7-3 RPB2), and YBC14 (sua7-3 rpb2-101) in the presence (+) or absence (−) of the activator Gal4-VP16 or wild-type rTFIIB (rIIB). Reaction products were resolved in a 7 M urea−6% polyacrylamide gel. The lower panel is a longer exposure of the upper panel. (C) In vitro transcription was done as described for panel B, except with pSLCYC-L and pSLCYC-S as templates. Reaction products were resolved in a 7 M urea−6% polyacrylamide gel. For lanes 1 and 8, transcription was performed by using wild-type (WT) extracts incubated with 10 μg of α-amanitin/ml, which specifically inhibits RNAP II transcription. Lanes 7 and 14 exhibit an additional band (indicated with an asterisk) that corresponds to the upstream start site shift caused by rpb2-101. The bands flanking +65 (pSLCYC-S) and +110 (pSLCYC-L) represent products from minor initiation sites. The uppermost bands (nonspecific) correspond to promoter-independent transcripts from the G-less template whose 5′ and 3′ endpoints are defined by RNase T1 digestion. (D) In vitro transcription reactions were performed as described for panel C, by using the linear pSLCYC-S template. Runoff products (∼170 nt) were resolved with a 15% polyacrylamide gel (upper panel), whereas abortive initiation products (3 and 5 nt) were detected with a 24% polyacrylamide gel (lower panel).
Gel mobility shift assays.
The 20-μl binding reaction mixture contained 20 mM Tris-HCl (pH 7.9), 20 mM KCl, 40 mM potassium acetate, 5 mM MgCl2, 100 μg of bovine serum albumin/ml, 1 mM DTT, 6.25 μg of poly(dG-dC)/ml, 10% glycerol, and 5,000 cpm of probe. The XbaI/XhoI fragment of pDR27 (obtained from D. Reinberg), which contains the adenovirus major late promoter TATA element and initiator, was used as a probe. Recombinant or native yeast proteins were added to the binding reactions as follows: 5 ng of rTBP, 15 ng of rTFIIB, 60 ng of yeast RNAP II, and 15 ng of yeast TFIIF. The reaction mixture was incubated at 25°C for 40 min and loaded onto a 5% polyacrylamide gel containing 25 mM Tris-acetate (pH 8.3), 190 mM glycine, 0.5 mM DTT, and 2.5% glycerol.
RESULTS
Isolation of an rpb2 suppressor of the TFIIB R78C defect.
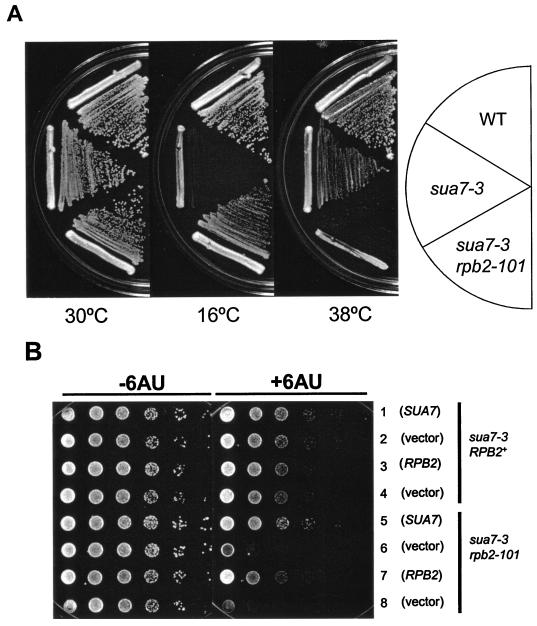
The sua7-3 allele of strain YIP363 encodes a TFIIB R78C replacement that shifts transcription start site selection downstream of normal and causes a cold-sensitive (Csm−) growth defect at 16°C (41). In an effort to further define the role of TFIIB in transcription, we sought suppressors of the sua7-3 mutation on YPD medium at 16°C. One Csm+ revertant, designated YBC14, exhibited multiple phenotypes, including heat sensitivity (Tsm−) and impaired growth on medium containing 6-azauracil (6AUs) (Fig. 1). By using a set of plasmid-borne genes that encode general transcription factors and RNAP II subunits, we found that RPB2, encoding the second-largest subunit of RNAP II, complemented all YBC14 phenotypes. An allelism test (see Materials and Methods) showed that the TFIIB R78C suppressor segregates opposite to a tagged RBP2::URA3 locus. Accordingly, we have designated the TFIIB R78C suppressor rpb2-101.
FIG. 1.
Phenotypes associated with sua7-3 and its rpb2-101 suppressor. (A) Growth of strains T16 (SUA7 RPB2), YIP363 (sua7-3 RPB2), and YBC14 (sua7-3 rpb2-101) is shown on rich (YPD) medium at 16, 30, and 38°C. YBC14 was selected as a Csm+ revertant of YIP363 on YPD medium at 16°C. (B) Synthetic 6AUs phenotype associated with the sua7-3 rpb2-101 mutant. Rows 1 to 4 are strain YIP363 transformed with plasmid DNA carrying either SUA7+ (pM269), RPB2+(pN1002), or the respective vectors (pRS316 or YCp50). Rows 5 to 8 are strain YBC14 transformed with the same set of plasmids as in rows 1 to 4, as indicated. Cells were grown on −Ura medium without 6AU (−6AU) or supplemented with 30 μg of 6AU/ml (+6AU). For both panels, cell growth was photographed after 2 days of incubation, except for the 16°C plate, which was photographed after 7 days of incubation.
The rpb2-101 suppressor confers a synthetic 6AUs phenotype.
We were surprised to find a strong 6AUs phenotype associated with a suppressor of a TFIIB defect. 6AU reduces intracellular GTP and UTP levels by inhibiting IMP dehydrogenase and orotidylate decarboxylase, enzymes that catalyze rate-limiting steps in NTP biosynthesis (16). Sensitivity to 6AU can be a consequence of defective expression of the IMD2 gene, which requires optimally functioning elongation machinery for induction (46). Accordingly, 6AUs is often regarded as a phenotype indicative of transcription elongation defects. This idea is supported by the fact that rpb1, rpb2, rpb6, and rpb9 mutants are 6AUs and exhibit diminished rates of elongation in vitro (3, 23, 26, 42).
In an effort to further characterize this effect, we asked whether the YBC14 6AUs phenotype is conferred by rpb2-101 alone or if the sua7-3 primary mutation is also required. We introduced plasmid-borne SUA7+ (pM269) or RPB2+ (pN1002) into YBC14 and scored the resulting transformants for complementation of 6AUs. Either gene alone was sufficient to rescue the 6AUs phenotype (Fig. 1B, cf. rows 5 and 7 with 6 and 8). Thus, 6AUs is a synthetic growth defect, dependent upon both the sua7-3 and rpb2-101 alleles. These results define a functional interaction between TFIIB and the Rpb2 subunit of RNAP II and suggest that this interaction occurs at a step in the transcription cycle that is sensitive to NTP levels.
The rpb2-101 mutation partially restores normal start site selection in the sua7-3 background.
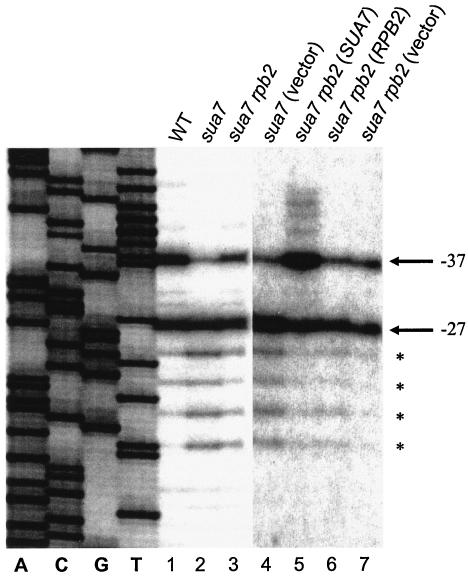
In addition to conferring a Csm− growth defect, the sua7-3 mutation shifts start site selection at the CYC1 and ADH1 genes downstream of normal. We therefore asked whether the rpb2-101 suppressor, in addition to suppressing the sua7-3 Csm− phenotype, would restore the normal initiation pattern. Transcription start sites were mapped at the ADH1 gene by primer extension (Fig. 2). In the wild-type strain, transcription initiates with equal efficiency at two major sites located at positions −27 and −37 relative to the ATG start codon (lane 1). Consistent with previous results, the sua7-3 mutation shifted start sites downstream of normal, diminishing initiation at −37 and enhancing initiation at multiple sites downstream of −27 (lane 2). The rpb2-101 mutation partially suppressed the downstream shift, producing an initiation pattern intermediate between that of the wild-type and sua7-3 strains (lane 3). Initiation was quantified at position −37 relative to all ADH1 start sites in each strain. Results revealed 40% initiation at −37 in the wild-type strain, 10% in the sua7-3 mutant, and 25% in the sua7-3 rpb2-101 revertant.
FIG. 2.
Primer extension analyses of ADH1 transcription start sites. Lanes: 1, T16 (SUA7 RPB2); 2, YIP363 (sua7-3 RPB2); 3, YBC14 (sua7-3 rpb2-101); 4, YIP363/pRS316 (sua7-3 RPB2); 5, YBC14/pM1553 (sua7-3/SUA7 rpb2-101); 6, YBC14/pM1764 (sua7-3 rpb2-101/RPB2); 7, YBC14/pRS315 (sua7-3 rpb2-101). Lanes A, C, G, and T are molecular size markers and correspond to a sequence ladder of SUA7 DNA. The principal ADH1 transcription start sites in the wild-type strain (lane 1) are located at positions −37 and −27 (A of the ATG start codon is designated +1).
We next asked whether the rpb2-101 mutation affected start site selection in a SUA7 wild-type background (Fig. 2). In this case, primer extension was performed by using RNA isolated from the sua7-3 rpb2-101 revertant (YBC14) that had been transformed with plasmid-borne SUA7 (pM1553), RPB2 (pM1764), or vector alone (pRS315). Whereas the sua7-3 mutation shifted initiation downstream of normal (lane 4) and the rpb2-101 mutation compensated for this defect (lane 7), the rpb2-101 mutation alone shifted initiation upstream of normal, enhancing initiation at −37 relative to −27 and promoting the use of novel start sites upstream of −37 (lane 5). Thus, the altered forms of TFIIB and Rpb2 encoded by the sua7-3 and rpb2-101 mutations exert opposite effects on start site selection. These effects are comparable to those of the sua7-1-encoded TFIIB E62K replacement and its rpb9 suppressor (50).
The rpb2-101 allele encodes a G369S replacement.
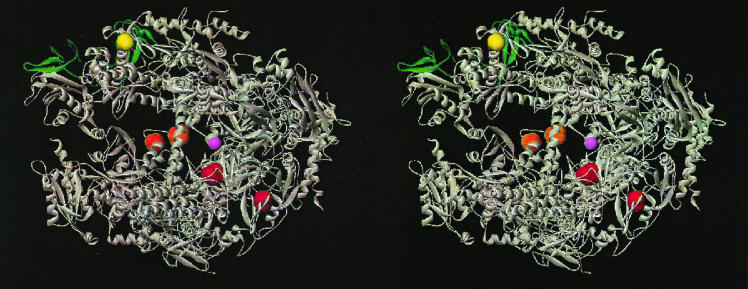
The high-resolution X-ray structures of RNAP II offer an opportunity to interpret the rpb2-101 defect in the context of the holoenzyme. The rpb2-101 allele was cloned by gap repair (see Materials and Methods), and the entire ORF was sequenced with a set of RPB2-specific primers that span both strands of the ORF. A single base pair substitution at position 1105 (G1105A) was identified, encoding a glycine-to-serine replacement at amino acid position 369 (G369S). G369 is located within the “lobe” domain of Rpb2, immediately preceding the α10 helix (Fig. 3) (14). This region of RNAP II is distal to the dock that interacts with TFIIB but proximal to the Rpb9 subunit, which with Rpb2 forms the “jaw-lobe” module downstream of the active center (11, 14). Consistent with a role for the jaw-lobe in start site selection, deletion of Rpb9 was previously identified as a suppressor of a TFIIB E62K start site defect (50). Thus, altered forms of two different subunits of RNAP II suppress related TFIIB defects, and the normal subunits are in physical proximity to each other and are distal to the docking site for TFIIB.
FIG. 3.
Stereo image of the 10-subunit RNAP II structure. Positions where residue replacements affect start site selection are indicated in color: the G369S (rbp2-101) and G369D (rpb2-504 and rpb2-505) replacements (yellow) are located in the lobe domain of Rpb2; the T1080I (rpb1-501) and S1096F (rpb1-502) replacements (orange) are located in the cleft domain of Rpb1; and the A402R (sua8-3) and N445S (sua8-1 and sua8-2) replacements (red) are located in the Rpb1 dock and active site domains, respectively. The dock domain is the binding site for TFIIB (10a, 11). The active center Mg2+ is shown in magenta. The Rpb9 subunit is depicted in green. The Rpb4 and Rpb7 subunits are not included in this structure. This figure was generously provided by Richard H. Ebright (Waksman Institute, Rutgers University).
Effects of sua7-3 and rpb2-101 on transcription in vitro.
To characterize the effects of sua7-3 and rpb2-101 on transcription in vitro, whole-cell extracts were prepared from wild-type (T16), sua7-3 primary mutant (YIP363), and sua7-3 rpb2-101 revertant (YBC14) strains. The GALUAS-CYC1TATA promoter fused with the G-less cassette was utilized as the template (pGAL4CG−) (Fig. 4A), which in our system yields a predominant transcript of 350 nucleotides (nt) (Fig. 4B). Transcript levels were induced about 10-fold in the presence of the Gal4-VP16 activator protein. The addition of rTFIIB did not affect either basal or activated transcription in the wild-type extract.
The sua7-3 mutation nearly eliminated basal transcription in vitro and abolished the response to the Gal4-VP16 activator (Fig. 4B, lanes 5 and 6, top and bottom panels). Basal and activated transcription were restored when the mutant extract was supplemented with rTFIIB (lanes 7 and 8), demonstrating that TFIIB R78C is directly responsible for diminished transcription in the mutant extract. The rpb2-101 suppressor of sua7-3 partially restored transcription, enhancing basal transcription (cf. lanes 5 and 9) and the response to the Gal4-VP16 activator (cf. lanes 6 and 10). Again, normal transcription was restored by rTFIIB (lanes 11 and 12). Thus, the TFIIB R78C replacement effectively abolished basal transcription as well as the response to an activator in vitro, and these effects were partially compensated by the RNAP II suppressor.
The transcription results shown in Fig. 4B monitor run-on transcription from the full-length G-less cassette. Because the 6AUs phenotype is often indicative of transcription elongation defects, we considered the possibility that sua7-3 is defective at a stage in the transcription cycle subsequent to initiation. To address this possibility, we repeated the in vitro transcription reactions with two truncated versions of the G-less cassette, one (pSLCYC-S) producing 65- and 85-nt transcripts, the other (pSLCYC-L) producing 110- and 130-nt transcripts (Fig. 4A). Results comparable to those with the full-length template were observed: near elimination of transcription by the sua7-3 extract (Fig. 4C, cf. lanes 2 and 4 and lanes 9 and 11) and partial restoration by rpb2-101 (cf. lanes 4 and 6 and lanes 11 and 13). Rescue by rpb2-101 was limited to initiation at the downstream site yielding the +65 (cf. lanes 4 and 6) and +110 (cf. lanes 11 and 13) transcripts, with little or no effect at the upstream site (+85 and +130). An additional transcript was made apparent by using the sua7-3 rpb2-101 extract in the presence of rTFIIB (lanes 7 and 14), presumably reflecting upstream initiation associated with rpb2-101 in vivo (Fig. 2). Despite the implications of the 6AUs phenotype, these results demonstrate that the sua7-3 defect is manifest prior to the entry of RNAP II into the elongation phase of transcription.
We next used the in vitro transcription reactions to assay abortive initiation products. In this case, products from the pSLCYC-S template with wild-type, sua7-3, and sua7-3 rpb2-1-101 extracts were resolved on a 24% polyacrylamide gel. The results revealed the presence of abortive initiation products in all three strains (Fig. 4D). Whereas the wild-type extract yielded primarily an abortive 3-nt product, the sua7-3 extract resulted in diminished levels of the 3-nt product and enhanced formation of a 5-nt product, and the sua7-3 rpb2-1-101 extract showed intermediate levels of the two abortive products (Fig. 4D, cf. lanes 1, 3, and 5). These results demonstrate that TFIIB R78C does not preclude transcription initiation but adversely affects the transition from abortive to productive initiation.
The sua7-3 rpb2-101 mutant is sensitive to NTP depletion in vitro.
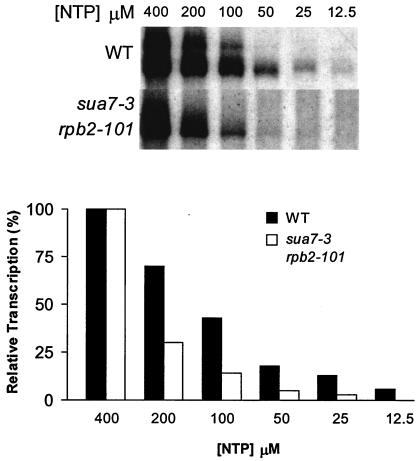
Promoter escape can be regarded as a competition between a stable PIC and a dynamic, initiation-competent RNAP II (7). If TFIIB R78C is defective for promoter escape, then wild-type and mutant extracts are likely to show differing sensitivities to limiting NTP substrate concentrations in an in vitro transcription assay. Precedent for this idea comes from the sensitivity to limiting NTP concentrations associated with an archaeal TFB E46K replacement (counterpart of TFIIB E62K) that blocks initiation between promoter melting and promoter escape (7). We tested the sensitivity to limiting NTP concentrations by the in vitro transcription assay using serial dilutions of NTPs, ranging from 400 to 12.5 μM. Transcript levels were normalized in each case to the signal obtained in the presence of 400 μM NTP. The results demonstrated that the sua7-3 rpb2 extract was significantly more sensitive than the wild-type extract to diminishing NTP levels (Fig. 5). As this experiment was performed using extract from the sua7-3 rpb2-101 suppressor strain (the sua7-3 primary mutant yields no appreciable run-on product), the effect of TFIIB R78C on promoter escape is probably underrepresented by these data. These results support the conclusion that TFIIB R78C affects promoter escape. Furthermore, enhanced sensitivity to limiting NTP concentrations in vitro is likely to account for the 6AUs phenotype of the sua7-3 rpb2-101 mutant in vivo.
FIG. 5.
The sua7 rpb2 mutant is sensitive to NTP depletion in vitro. In vitro transcription reactions were performed as described for Fig. 4, except that NTP substrate concentrations ranged from 25 to 400 μM, as indicated in the upper panel. The lower panel shows the quantitative results of transcript levels in the upper panel. Transcript abundance is expressed as a percentage of the levels at 400 μM NTP. Because of the decreased levels of transcription in the double mutant extract, we adjusted the exposure time of the films to observe approximately equal product signals in the presence of saturating NTP (400 μM).
Effect of TFIIB R78C on PIC formation.
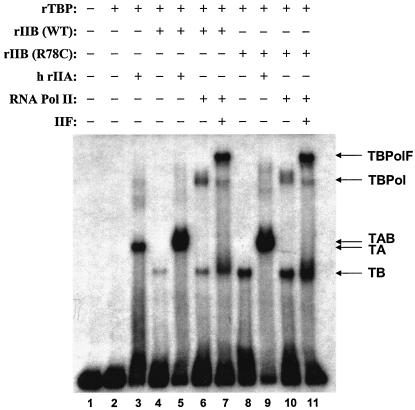
TFIIB binds the DNA-TBP complex, and the resulting ternary complex is a prerequisite for assembly of RNAP II into the PIC (9). Accordingly, we asked whether the TFIIB R78C replacement affects PIC assembly. We performed native gel electrophoresis assays (electrophoretic mobility shift assays) to monitor PIC assembly with purified transcription factors, including either normal TFIIB or its R78C derivative. The results are presented in Fig. 6. We observed a marked increase in DNA-TBP-TFIIB ternary complex formation associated with R78C (Fig. 6, cf. lanes 4, 6, and 7 with lanes 8, 10, and 11). However, there was no difference in accumulation of the higher-order DNA-TBP-TFIIB-RNAP II and DNA-TBP-TFIIB-TFIIF-RNAP II complexes (cf. lanes 6 and 7 and lanes 10 and 11). This observation is consistent with a previous report showing that TFIIB R64E stabilized the DNA-TBP-TFIIB ternary complex (6). Thus, formation of the DNA-TBP-TFIIB ternary complex, normally very weak in the absence of TFIIA (lanes 4 and 5), is enhanced significantly by alteration in the B-finger region of TFIIB. These results define a role for the B-finger prior to assembly of RNAP II into the PIC, perhaps reflecting intramolecular interaction between the TFIIB N- and C-terminal domains (45). Furthermore, the failure of R78C to enhance formation of higher-order complexes indicates that PIC assembly is minimally a two-step process with the second step unaffected by R78C.
FIG. 6.
TFIIB R78C effect on PIC assembly. PIC assembly was assayed by native gel electrophoresis. The AdML TATA element and initiator sequence, isolated as a XbaI-XhoI DNA fragment from plasmid pDR27 (from D. Reinberg), was used as the probe. Recombinant or native yeast proteins were added to the binding reactions as follows: 5 ng of rTBP, 15 ng of rTFIIB, 5 ng of TFIIA (rIIA), 60 ng of yeast RNAP II, and 15 ng of yeast TFIIF. Human (h) TFIIA, which is functionally interchangeable with yeast TFIIA for transcription in vitro (9), was purified from HeLa cells (a gift from S. Kim and D. Reinberg). WT and R78C denote the wild-type and mutant forms, respectively, of rTFIIB. Reaction mixtures and gel electrophoresis conditions are described in Materials and Methods. TA, DNA-TBP-TFIIA; TB, DNA-TBP-TFIIB; TAB, DNA-TBP-TFIIA-TFIIB; TBPol, DNA-TBP-TFIIB-RNAP II; TBPolF, DNA-TBP-TFIIB-RNAP II-TFIIF.
DISCUSSION
In this study, we used an altered form of TFIIB (R78C) to investigate the mechanism of transcription initiation by RNAP II. We draw three significant conclusions. First, in vitro experiments demonstrate that R78C impairs transcription subsequent to PIC formation, resulting in accumulation of abortive initiation products. This result implies that the altered start site selection associated with R78C and other TFIIB defects is coupled to the mechanism by which RNAP II overcomes abortive initiation and clears the promoter. Second, the growth defect associated with R78C is suppressed by an amino acid replacement (G369S) in the Rpb2 subunit of RNAP II, and this replacement is located in a region of the enzyme distal to the site of interaction between TFIIB and RNAP II. This result defines a functional interaction between TFIIB and the Rpb2 lobe and has implications for the mechanism of start site selection in yeast. Finally, our observation that the TFIIB R78C Rpb2 G369S double mutant exhibits a 6AUs phenotype yet is defective at a step in the transcription cycle prior to promoter escape offers a cautionary note about 6AUs as a hallmark of elongation defects. 6AUs appears to be indicative not only of defects in elongation but also of defects at other stages of transcription that are dependent upon NTP substrate concentrations, including promoter escape.
The recent yeast RNAP II-TFIIB X-ray structure offers insight into the mechanism of transcription initiation and provides a framework for the interpretation of TFIIB start site defects (10a). A remarkable feature of this structure is the B-finger, which encompasses TFIIB residues 55 to 88 and protrudes into the active center of RNAP II (10a). This structure is comparable to the σ-factor 3.2 loop, which follows a similar path into the active center and out the RNA exit channel of bacterial RNAP (33). The paths of the B-finger and the σ3.2 loop suggest that both structures should inhibit nascent RNA synthesis, potentially accounting for abortive initiation and the failure of RNAP (II) to clear the promoter prior to displacement of TFIIB or σ-factor (10a, 33).
Our observation that R78C affects the pattern of abortive initiation products in vitro (Fig. 4D) provides experimental support for this idea. Moreover, the effects of R78C on start site selection in vivo and abortive initiation in vitro define a functional connection between these two processes. The R78C defect was identified in the same genetic screen that yielded the E62K replacement, and these replacements cause similar growth phenotypes and start site defects (41). Other replacements at the phylogenetically conserved positions E62, W63, R64, F66, and R78 also shift initiation sites in yeast, human, and archaeal systems (6, 7, 12, 17, 18, 21, 39, 44, 56). All of these sites are located in the B-finger domain, with E62 and R78 situated on opposite sides of the finger where they are likely to form a salt bridge (8, 10a).
Specific Rpb1 amino acid replacements can affect start site selection in a manner similar to that of B-finger defects. The sua8 alleles of RPB1 were identified in the same genetic selection that uncovered the sua7-encoded E62K and R78C replacements, and both sets of mutants exhibit similar start site defects (8). Furthermore, sua7 sua8 double mutants are inviable, underscoring the relationship between Rpb1 and TFIIB in selecting initiation sites (8). The sua8 alleles encode an A402R replacement (sua8-3) in the RNAP II dock domain that binds TFIIB and an N445S replacement (sua8-1 and sua8-2) in the “active site” domain (Fig. 3). A subset of sit1 alleles of RPB1 also affects start site selection and includes a different replacement at position 445 (N445T) (2). These altered forms of TFIIB and Rpb1 should serve as valuable reagents to further define the relationship between abortive initiation and start site selection.
In contrast to the upstream Rpb1 and B-finger replacements, the Rpb2 G369S suppressor is located in the lobe domain, downstream of the active center and distal to the TFIIB-binding dock domain (Fig. 3). A different amino acid replacement at the same site, G369D, was identified in a selection for spt mutants that affect core promoter function (23). Although it was not reported whether G369D affects start site selection, the same spt selection yielded amino acid replacements in the “cleft” domain of Rpb1 (T1080I and S1096F) (Fig. 3), and these replacements shift initiation upstream of normal (22). Furthermore, deletion or truncation of the Rpb9 subunit of RNAP II compensates for the E62K start site defect and, like Rpb2 G369S, shifts initiation upstream in a TFIIB wild-type background (25, 50).
The similar genetic interactions of Rpb2 and Rpb9 with TFIIB, and their identical effects on start site selection, define a functional relationship between these two subunits of RNAP II. Rpb2 and Rpb9 might affect initiation indirectly, a consequence of their effects on RNAP II conformation. High-resolution X-ray structures of two different crystal forms of the 10-subunit yeast RNAP II core complex revealed four mobile modules (13, 14). One is the “upper jaw,” composed of the Rpb2-Rpb9 lobe-jaw region and a domain of Rpb1; a second module is the clamp, which in the 12-subunit RNAP II structure adopts a closed conformation resembling its position in the 10-subunit core elongation complex (4, 10). Rpb2 and Rpb9 might affect initiation indirectly, for example, by altering the position of the lobe-jaw relative to the clamp. An alternative possibility is suggested by a model for the bacterial RNAP-DNA promoter-open complex (RPo), one that is based on E. coli RNAP-DNA photo-cross-linking data and the X-ray structure of Thermus aquaticus RNAP (34). Extrapolation of these results to yeast RNAP II raises the possibility of a direct interaction between the Rpb2 lobe and the transcription bubble nontemplate strand. Whether direct or indirect, these results define an important role for RNAP II downstream of the active center and distal to the interaction of TFIIB in the mechanism of start site selection.
Acknowledgments
We are grateful to Subhrangsu S. Mandal and Qian Tan for technical advice; to Steve Buratowski, Steve Hahn, Danny Reinberg, and Nancy Woychik for reagents; to Richard Ebright, D. Reinberg, Krishnamurthy Shankarling, and Nancy Woychik for valuable comments on the manuscript; and to R. Ebright for providing Fig. 3.
This work was supported by NIH grant GM39484.
REFERENCES
- 1.Andel, F., III, A. G. Ladurner, C. Inouye, R. Tjian, and E. Nogales. 1999. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286:2153-2156. [DOI] [PubMed] [Google Scholar]
- 2.Archambault, J., D. B. Jansma, J. H. Kawasoe, K. T. Arndt, J. Greenblatt, and J. D. Friesen. 1998. Stimulation of transcription by mutations affecting conserved regions of RNA polymerase II. J. Bacteriol. 180:2590-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archambault, J., F. Lacroute, A. Ruet, and J. D. Friesen. 1992. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12:4142-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armache, K. J., H. Kettenberger, and P. Cramer. 2003. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 100:6964-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagby, S., S. J. Kim, E. Maldonado, K. I. Tong, D. Reinberg, and M. Ikura. 1995. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell 82:857-867. [DOI] [PubMed] [Google Scholar]
- 6.Bangur, C. S., T. S. Pardee, and A. S. Ponticelli. 1997. Mutational analysis of the D1/E1 core helices and the conserved N-terminal region of yeast transcription factor IIB (TFIIB): identification of an N-terminal mutant that stabilizes TATA-binding protein-TFIIB-DNA complexes. Mol. Cell. Biol. 17:6784-6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, S. D., and S. P. Jackson. 2000. The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J. Biol. Chem. 275:12934-12940. [DOI] [PubMed] [Google Scholar]
- 8.Berroteran, R. W., D. E. Ware, and M. Hampsey. 1994. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol. Cell. Biol. 14:226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratowski, S., S. Hahn, L. Guarente, and P. A. Sharp. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56:549-561. [DOI] [PubMed] [Google Scholar]
- 10.Bushnell, D. A., and R. D. Kornberg. 2003. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. USA 100:6969-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Bushnell, D. A., K. D. Westover, R. E. Davis, and R. D. Kornberg. 2004. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science 303:983-988. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H. T., and S. Hahn. 2003. Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell 12:437-447. [DOI] [PubMed] [Google Scholar]
- 12.Cho, E. J., and S. Buratowski. 1999. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem. 274:25807-25813. [DOI] [PubMed] [Google Scholar]
- 13.Cramer, P., D. A. Bushnell, J. Fu, A. L. Gnatt, B. Maier-Davis, N. E. Thompson, R. R. Burgess, A. M. Edwards, P. R. David, and R. D. Kornberg. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640-649. [DOI] [PubMed] [Google Scholar]
- 14.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1876. [DOI] [PubMed] [Google Scholar]
- 15.Dvir, A., R. C. Conaway, and J. W. Conaway. 1997. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc. Natl. Acad. Sci. USA 94:9006-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Exinger, F., and F. Lacroute. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22:9-11. [DOI] [PubMed] [Google Scholar]
- 17.Fairley, J. A., R. Evans, N. A. Hawkes, and S. G. Roberts. 2002. Core promoter-dependent TFIIB conformation and a role for TFIIB conformation in transcription start site selection. Mol. Cell. Biol. 22:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faitar, S. L., S. A. Brodie, and A. S. Ponticelli. 2001. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Mol. Cell. Biol. 21:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876-1882. [DOI] [PubMed] [Google Scholar]
- 19a.Hahn, S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol., in press. [DOI] [PMC free article] [PubMed]
- 20.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawkes, N. A., and S. G. Roberts. 1999. The role of human TFIIB in transcription start site selection in vitro and in vivo. J. Biol. Chem. 274:14337-14343. [DOI] [PubMed] [Google Scholar]
- 22.Hekmatpanah, D. S., and R. A. Young. 1991. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol. Cell. Biol. 11:5781-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemming, S. A., D. B. Jansma, P. F. Macgregor, A. Goryachev, J. D. Friesen, and A. M. Edwards. 2000. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J. Biol. Chem. 275:35506-35511. [DOI] [PubMed] [Google Scholar]
- 24.Holstege, F. C., and H. T. Timmers. 1997. Analysis of open complex formation during RNA polymerase II transcription initiation using heteroduplex templates and potassium permanganate probing. Methods 12:203-211. [DOI] [PubMed] [Google Scholar]
- 25.Hull, M. W., K. Mckune, and N. A. Woychik. 1995. RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev. 9:481-490. [DOI] [PubMed] [Google Scholar]
- 26.Ishiguro, A., Y. Nogi, K. Hisatake, M. Muramatsu, and A. Ishihama. 2000. The Rpb6 subunit of fission yeast RNA polymerase II is a contact target of the transcription elongation factor TFIIS. Mol. Cell. Biol. 20:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, T. K., R. H. Ebright, and D. Reinberg. 2000. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288:1418-1422. [DOI] [PubMed] [Google Scholar]
- 28.Kraemer, S. M., R. T. Ranallo, R. C. Ogg, and L. A. Stargell. 2001. TFIIA interacts with TFIID via association with TATA-binding protein and TAF40. Mol. Cell. Biol. 21:1737-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kugel, J. F., and J. A. Goodrich. 2000. A kinetic model for the early steps of RNA synthesis by human RNA polymerase II. J. Biol. Chem. 275:40483-40491. [DOI] [PubMed] [Google Scholar]
- 30.Liao, S. M., I. C. Taylor, R. E. Kingston, and R. A. Young. 1991. RNA polymerase II carboxy-terminal domain contributes to the response to multiple acidic activators in vitro. Genes Dev. 5:2431-2440. [DOI] [PubMed] [Google Scholar]
- 31.Minakhin, L., S. Bhagat, A. Brunning, E. A. Campbell, S. A. Darst, R. H. Ebright, and K. Severinov. 2001. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA 98:892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami, K. S., and S. A. Darst. 2003. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13:31-39. [DOI] [PubMed] [Google Scholar]
- 33.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 34.Naryshkin, N., A. Revyakin, Y. Kim, V. Mekler, and R. H. Ebright. 2000. Structural organization of the RNA polymerase-promoter open complex. Cell 101:601-611. [DOI] [PubMed] [Google Scholar]
- 35.Nikolov, D. B., H. Chen, E. D. Halay, A. A. Usheva, K. Hisatake, D. K. Lee, R. G. Roeder, and S. K. Burley. 1995. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377:119-128. [DOI] [PubMed] [Google Scholar]
- 36.Orphanides, G., T. LaGrange, and D. Reinberg. 1996. The general initiation factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 37.Pal, M., D. McKean, and D. S. Luse. 2001. Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Mol. Cell. Biol. 21:5815-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pappas, D. L., Jr., and M. Hampsey. 2000. Functional interaction between Ssu72 and the Rpb2 subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:8343-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pardee, T. S., C. S. Bangur, and A. S. Ponticelli. 1998. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J. Biol. Chem. 273:17859-17864. [DOI] [PubMed] [Google Scholar]
- 40.Pinto, I., D. E. Ware, and M. Hampsey. 1992. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell 68:977-988. [DOI] [PubMed] [Google Scholar]
- 41.Pinto, I., W.-H. Wu, J. G. Na, and M. Hampsey. 1994. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 269:30569-30573. [PubMed] [Google Scholar]
- 42.Powell, W., and D. Reines. 1996. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J. Biol. Chem. 271:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 44.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, S. G. E., and M. R. Green. 1994. Activator-induced conformational change in general transcription factor TFIIB. Nature 371:717-720. [DOI] [PubMed] [Google Scholar]
- 46.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 48.Sherman, F., and J. Hicks. 1991. Micromanipulation and dissection of asci. Methods Enzymol. 194:21-37. [DOI] [PubMed] [Google Scholar]
- 49.Sun, Z. W., and M. Hampsey. 1995. Identification of the gene (SSU71/TFG1) encoding the largest subunit of transcription factor TFIIF as a suppressor of a TFIIB mutation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92:3127-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, Z. W., A. Tessmer, and M. Hampsey. 1996. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 24:2560-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66:117-172. [DOI] [PubMed] [Google Scholar]
- 51a.Westover, K. D., D. A. Bushnell, and R. D. Kornberg. 2004. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303:1014-1016. [DOI] [PubMed] [Google Scholar]
- 52.Woontner, M., and J. A. Jaehning. 1990. Accurate initiation by RNA polymerase II in a whole cell extract from Saccharomyces cerevisiae. J. Biol. Chem. 265:8979-8982. [PubMed] [Google Scholar]
- 53.Woychik, N. A. 1998. Fractions to functions: RNA polymerase II thirty years later. Cold Spring Harbor Symp. Quant. Biol. 63:311-317. [DOI] [PubMed] [Google Scholar]
- 54.Woychik, N. A., and M. Hampsey. 2002. The RNA polymerase II machinery: structure illuminates function. Cell 108:453-463. [DOI] [PubMed] [Google Scholar]
- 55.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, D. Y., D. J. Carson, and J. Ma. 2002. The role of TFIIB-RNA polymerase II interaction in start site selection in yeast cells. Nucleic Acids Res. 30:3078-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, W. L., Q. D. Zeng, C. M. Colangelo, L. M. Lewis, M. F. Summers, and R. A. Scott. 1996. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat. Struct. Biol. 3:122-124. [DOI] [PubMed] [Google Scholar]