Abstract
Small ubiquitin-related modifiers (SUMOs) are proteins that are posttranslationally conjugated to other cellular proteins, particularly those that localize and function in the nucleus. Enzymes regulating SUMO modification localize in part to nuclear pore complexes (NPCs), indicating that modification of some proteins may occur as they are translocated between the nucleus and the cytoplasm. Substrates that are regulated by SUMO modification at NPCs, however, have not been previously identified. Among the most abundant cargos transported through NPCs are the heterogeneous nuclear ribonucleoproteins (hnRNPs). HnRNPs are involved in various aspects of mRNA biogenesis, including regulation of pre-mRNA splicing and nuclear export. Here, we demonstrate that two subsets of hnRNPs, the hnRNP C and M proteins, are substrates for SUMO modification. We demonstrate that the hnRNP C proteins are modified by SUMO at a single lysine residue, K237, and that SUMO modification at this site decreases their binding to nucleic acids. We also show that Nup358, a SUMO E3 ligase associated with the cytoplasmic fibrils of NPCs, enhances the SUMO modification of the hnRNP C and M proteins. Based on our findings, we propose that SUMO modification of the hnRNP C and M proteins may occur at NPCs and facilitate the nucleocytoplasmic transport of mRNAs.
The heterogeneous nuclear ribonucleoproteins (hnRNPs) are a large family of highly conserved RNA-binding proteins that have important roles in regulating multiple steps in mRNA biogenesis and function (10, 11, 23). In the nucleus, the hnRNPs form large complexes with primary RNA polymerase II transcripts that contain more than 20 different hnRNPs ranging in size from 30 to 120 kDa (4, 31). HnRNPs affect mRNA transcription (16, 28), regulate mRNA translation in the cytoplasm (12-14, 18, 21), and are involved in the maintenance of the single-stranded DNA (ssDNA) extensions at chromosome telomeres (7, 12-14, 24). HnRNPs, however, are best known for their roles in regulating the nuclear posttranscriptional events involved in mRNA biogenesis, including regulation of pre-mRNA splicing, pre-mRNA polyadenylation, and 3′-end processing (1, 40) and mRNA nuclear export (10).
With regard to their role in nuclear export, most hnRNPs remain associated with the mRNAs as they are translocated through nuclear pore complexes (NPCs) and into the cytoplasm (32, 41). Once in the cytoplasm, these hnRNPs are released from the mRNAs by an unknown mechanism and shuttle back into the nucleus. Intriguingly, several hnRNPs, including the hnRNP C proteins, do not shuttle between the nucleus and the cytoplasm and are presumably released from newly formed mRNAs in the nucleus prior to export (32). Exactly where and how the nonshuttling hnRNPs are released from mRNAs in the nucleus is not currently known. Although significant progress has been made in identifying the factors and steps involved in targeting nuclear mRNPs to NPCs for export, very little is still understood about the exact mechanisms involved in their translocation through NPCs. The complexity of translocation through NPCs has been eloquently illustrated through analysis of the Balbiani ring mRNPs expressed in the salivary gland of Chironomus tentans. Balbiani ring mRNPs contain an ∼40-kb mRNA transcript whose biogenesis and export from the nucleus to the cytoplasm have been characterized through analysis by electron microscopy. These studies demonstrate that mRNP export through the NPC is a highly orchestrated event that involves dramatic changes in the conformation of the mRNP as it is translocated from the nucleoplasmic basket of NPCs to the cytoplasm (22, 38). The exact nature of these conformational changes and how they are induced and regulated are currently unknown.
SUMOs are small ubiquitin-like proteins that are posttranslationally conjugated to other proteins in the cell and thereby regulate a wide range of biological processes, including transcription, the cell cycle, apoptosis, chromatin integrity and dynamics, and nucleocytoplasmic transport (27). Unlike ubiquitination, SUMO modification does not lead to substrate degradation. Instead, its functions appear to be substrate dependent and can involve inducing changes in the target's subcellular localization, its protein-protein interactions, or its protein-DNA interactions. SUMO modification requires three types of enzymes and parallels the steps involved in protein ubiquitination (27). The first step of the modification process is ATP dependent and results in the formation of a high-energy thioester bond between the SUMO C-terminal glycine and a cysteine residue in the SUMO E1 activating enzyme, a heterodimer made up of Aos1 and Uba1. Following E1 activation, SUMO is transferred to the active-site cysteine residue of Ubc9, the SUMO E2 conjugating enzyme. Ubc9 recognizes and binds to a SUMO consensus sequence, ψKXE (where ψ is any hydrophobic amino acid, K is a lysine, X is any amino acid, and E is glutamic acid), present in most target proteins (3). Although the SUMO E1 and E2 enzymes are sufficient to modify most substrates in vitro, several SUMO E3-like factors have also been described (17, 19, 29). One recently identified SUMO E3-like factor, Nup358 (also referred to as RanBP2), is a component of the cytoplasmic fibrils of the NPC (29).
The discovery that Nup358 has SUMO E3-like activity is one of several factors suggesting that SUMO modification may occur at NPCs as proteins are transported between the nucleus and the cytoplasm. Other evidence includes the localization of Ubc9 to both the cytoplasmic and nucleoplasmic fibrils of the NPC (42). It is thought that Ubc9 is associated with Nup358 on the cytoplasmic fibrils and that together they may function to facilitate SUMO modification at this site. Also, SENP2, a SUMO protease that is able to remove SUMO from modified proteins, localizes to the nucleoplasmic face of NPCs (15, 42). Thus, SUMO demodification could also be coupled to the transport of proteins between the nucleus and the cytoplasm. Finally, in vitro experiments using digitonin-permeabilized cells have suggested that multiple nuclear envelope/NPC-associated proteins may be modified by SUMO at the NPC (29). With the exception of RanGAP1, however, substrates that are SUMO modified or demodified at NPCs have not been identified.
Among the most abundant substrates transported between the nucleus and the cytoplasm are the hnRNPs. Here, we demonstrate that a subset of hnRNPs can be modified by SUMO. We first identified the hnRNP M proteins as SUMO substrates that copurify with NPCs. Subsequent studies of immunopurified hnRNP complexes revealed that both the hnRNP M and the hnRNP C proteins are efficiently, and selectively, modified by SUMO. Further characterization of the hnRNP C proteins revealed that they are SUMO modified at a single lysine residue, K237, and that modification at this site decreases their binding to nucleic acids. We also found that Nup358 is able to act as an E3 ligase and stimulate the SUMO modification of the hnRNP M and C proteins. Based on these findings we propose a model in which SUMO modification of the hnRNP C and M proteins occurs at NPCs and regulates the conformation and/or composition of hnRNP complexes, thereby facilitating nucleocytoplasmic transport.
MATERIALS AND METHODS
In vitro protein expression and SUMO modification.
cDNAs coding for the indicated proteins were transcribed and translated in rabbit reticulocyte lysate in the presence of [35S]methionine according to the manufacturer's instructions (Promega, Madison, Wis.). In vitro SUMO modification reactions were carried out using two different assay conditions. Assays performed in the presence of high concentrations of E1 and E2 enzymes contained 2 μl of translation product in a 10-μl reaction mixture containing 20 mM HEPES (pH 7.3), 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM dithiothreitol (DTT), 0.5 μM recombinant Aos1/Uba2, 2 μM recombinant Ubc9, 10 μM recombinant SUMO-1, 1 mM ATP, 10 mM phosphocreatine (Sigma, St. Louis, Mo.), 40 U of creatine phosphokinase (Sigma)/ml, and 1.2 μg of inorganic pyrophosphatase (Sigma)/ml. Reactions using limiting amounts of E1 and E2 enzymes were carried out using similar conditions but contained 0.1 μM Aos1/Uba2, 0.25 μM Ubc9, and 0.05 μM Nup358 (amino acids 2596 to 2863 of human Nup358) or 0.3 μM PIAS1. Modification reactions were incubated at 37°C for 1 h and stopped by the addition of an equal volume of sodium dodecyl sulfate (SDS) sample buffer.
Proteins associated with isolated rat liver nuclear envelopes were SUMO modified using similar assay conditions. Heparin-extracted rat liver nuclear envelopes were prepared as previously described (25) and resuspended at a concentration of 100 U (1 U being equivalent to 3 × 106 nuclei) per ml in buffer containing 110 mM potassium acetate, 2 mM magnesium acetate, 20 mM HEPES-KOH (pH 7.3), 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride. Fifty microliters of suspended nuclear envelopes were incubated for 1 h at room temperature in the presence of 0.5 μM Aos1/Uba2, 2 μM Ubc9, 10 μM SUMO-1, 1 mM ATP, 10 mM phosphocreatine, 40 U of creatine phosphokinase/ml, and 1.2 μg of inorganic pyrophosphatase/ml. Following incubation, the nuclear envelopes were pelleted for 1 min at 8,000 × g, resuspended in SDS sample buffer, and analyzed by immunoblot analysis using the SUMO-1-specific monoclonal antibody 21C7 (25) and the hnRNP M protein-specific monoclonal antibody 1D8 (a gift of M. Swanson, University of Florida).
ssDNA binding assays.
Fifty microliters of ssDNA cellulose beads (Amersham Pharmacia Biotech, Piscataway, N.J.) were preequilibrated in four different binding buffers, each containing 10 mM Tris (pH 7.4), 2.5 mM MgCl2, and various concentrations of sodium chloride (100 mM, 500 mM, 1 M, or 2 M). Equivalent amounts of in vitro-translated hnRNP C1, before or after in vitro SUMO modification, were added to the DNA cellulose beads in the presence of 500 μl of binding buffer. Follow a 10-min incubation at room temperature, the beads were pelleted and washed four times with the respective binding buffer and once with binding buffer containing 100 mM NaCl. After the final wash, bound proteins were eluted with SDS sample buffer and analyzed SDS-polyacrylamide gel electrophoresis (PAGE) or by quantification using a scintillation counter. All binding reactions were performed in triplicate, and the averages of the three individual values were plotted together with their standard deviations.
Glutathione bead binding assays.
One hundred microliters of glutathione-Sepharose beads (Amersham Pharmacia Biotech) were preequilibrated in binding buffer containing 20 mM HEPES (pH 7.3), 110 mM potassium acetate, 2 mM magnesium acetate, and 1 mM DTT. The beads were then incubated with in vitro-translated hnRNP C1, before or after SUMO modification, in the presence of 500 μl of binding buffer for 1 h at 4°C. Following incubation, the beads were pelleted and washed four times with binding buffer. Bound proteins were eluted with SDS-PAGE sample buffer and analyzed by SDS-PAGE and autoradiography.
Cell culture.
HeLa cells were grown to 80% confluence in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 10 mM HEPES (pH 7.4). Cells were labeled with [35S]methionine as previously described (9).
Immunopurifications.
hnRNP complexes were immunopurified from the HeLa cell nucleoplasm by using the hnRNP C protein-specific monoclonal antibody 4F4 (a gift of G. Dreyfuss, University of Pennsylvania) as previously described (4). Following immunopurification, hnRNP complexes were either eluted from the protein A beads with SDS sample buffer or two-dimensional gel sample buffer or subjected to in vitro SUMO modification assays. Assays were performed in a total volume of 100 μl for 2 h at 37°C and contained similar concentrations of components as described above. Following incubation, the beads were washed four times with buffer containing 10 mM Tris (pH 7.4), 100 mM NaCl, and 2.5 mM MgCl2. Proteins were eluted with SDS sample buffer for SDS-PAGE analysis or by addition of two-dimensional sample buffer for two-dimensional PAGE analysis. SDS-PAGE and two-dimensional PAGE were performed as previously described (33).
RESULTS
Nuclear envelope-associated hnRNP M protein is a SUMO substrate.
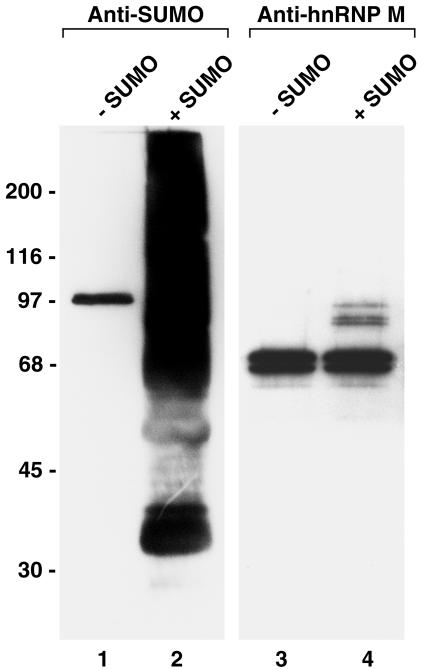
Several findings have suggested that SUMO modification may occur at NPCs as substrates are transported between the nucleus and the cytoplasm (30). However, the identities of those substrates that may be SUMO modified or demodified at NPCs are not known. A previous study using digitonin-permeabilized cells demonstrated that nuclear envelope- and/or NPC-associated proteins could be modified by SUMO in vitro (29). In order to identify potential substrates that could be regulated by SUMO modification at NPCs, we performed similar in vitro SUMO modification assays using isolated rat liver nuclear envelopes. Envelopes were incubated with SUMO E1 and E2 enzymes and ATP in the presence or absence of SUMO-1, and reactions were resolved by SDS-PAGE and analyzed by immunoblot analysis with an antibody specific for SUMO-1. In the absence of exogenously added SUMO-1, only a single protein with a relative molecular mass of ∼90 kDa was detected (Fig. 1, lane 1). This protein corresponds to the previously described SUMO-1-modified RanGAP1 that is associated with the cytoplasmic fibrils of the NPC (26). In contrast, we detected a general smear of high-molecular-mass SUMO conjugates associated with nuclear envelopes incubated in the presence of exogenously added SUMO (Fig. 1, lane 2).
FIG. 1.
Nuclear envelope-associated hnRNP M proteins are substrates for SUMO modification. Isolated rat liver nuclear envelopes were incubated with SUMO E1 and E2 conjugating enzymes in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of exogenously added SUMO-1. Following incubation, proteins were separated by SDS-PAGE, and SUMO conjugates (lanes 1 and 2) and the hnRNP M proteins (lanes 3 and 4) were identified by immunoblot analysis using specific monoclonal antibodies. Relative molecular masses are indicated on the left.
Previous proteomic analysis of proteins that copurify with isolated NPCs identified a subset of hnRNPs, including the hnRNP H, F, and M proteins (6). We considered the possibility that these hnRNPs may be among the proteins modified by SUMO in the assays performed on isolated nuclear envelopes. To test this possibility, we performed immunoblot analysis with a monoclonal antibody specific for the hnRNP M protein. Analysis of nuclear envelopes incubated with SUMO E1 and E2 enzymes but in the absence of SUMO revealed a doublet of ∼68 kDa, demonstrating that the hnRNP M proteins are associated with the isolated nuclear envelope fraction and that they are not obviously modified under these conditions (Fig. 1, lane 3). Similar analysis of nuclear envelopes incubated in the presence of exogenously added SUMO revealed the same ∼68-kDa doublet but also the appearance of a triplet of ∼90 kDa (Fig. 1, lane 4). The relative 20-kDa shift in mobility of these bands is consistent with their corresponding to the hnRNP M proteins modified by SUMO at a single lysine residue. These results, as well as those indicated below, indicate that the nuclear envelope-associated hnRNP M proteins are substrates for SUMO modification. Analysis with antibodies specific for the hnRNP H and F proteins indicated that these proteins, although copurifying with nuclear envelopes, are not modified by SUMO in the same assay (data not shown). Other hnRNPs, including the hnRNP C proteins, do not copurify with isolated nuclear envelopes.
SUMO modification of components of immunopurified hnRNP complexes.
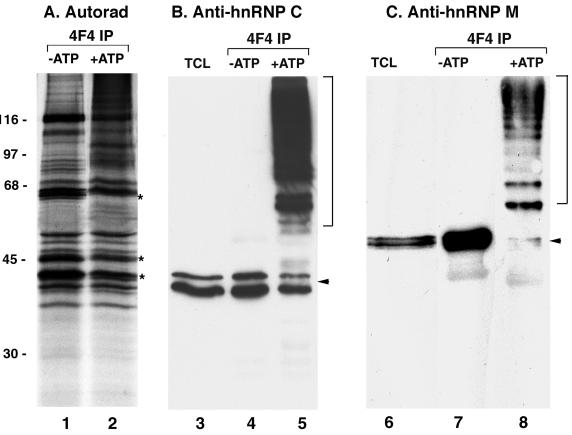
Evidence that the hnRNP M proteins are substrates for SUMO modification led us to examine whether additional hnRNPs may be subject to the same posttranslational modification. To that effect we performed SUMO modification assays on hnRNP complexes immunopurified from [35S]methionine-labeled cells. HnRNP complexes were immunopurified with a monoclonal antibody specific for the hnRNP C proteins, and the purified complexes (while still immobilized on the antibody-protein A beads) were then incubated in the presence of ATP, SUMO, and SUMO E1 and E2 enzymes. As a control, reactions were also performed in the absence of ATP, conditions that do not support SUMO modification. Following incubation, the immobilized hnRNP complexes were washed once with reaction buffer and subsequently eluted with SDS sample buffer. When control reactions were separated by SDS-PAGE and analyzed by autoradiography, we detected the predicted pattern of proteins corresponding to previously described hnRNP complexes (Fig. 2A, lane 1). However, several individual protein bands were noticeably reduced in intensity following incubation in reactions supporting SUMO modification, and there was a detectable smear of higher-molecular-mass proteins (Fig. 2A, lane 2). One of the protein bands that was noticeably reduced in intensity migrated at a position similar to that of the hnRNP M proteins. Immunoblot analysis of the reactions with an anti-hnRNP M antibody confirmed that the M proteins were SUMO modified under these assay conditions (Fig. 2C). Using these particular conditions, we detected hnRNP M proteins that appeared to be modified at single lysine residues (shifted by ∼20 kDa in mass) as well as higher-molecular-mass species possibly modified by SUMO at multiple lysine residues or by SUMO chains (Fig. 2C, lane 8). In control assays with incubation in the absence of ATP, only unmodified hnRNP M proteins migrating at ∼68 kDa were detected (Fig. 2C, lane 7).
FIG. 2.
The hnRNP M and C proteins are selectively modified by SUMO within immunopurified hnRNP complexes. HnRNP complexes were immunopurified from the HeLa cell nucleoplasm and incubated with SUMO E1 and E2 conjugating enzymes and SUMO-1 either in the presence or in the absence of ATP. (A) Reactions were resolved by SDS-PAGE and analyzed by autoradiography. Asterisks indicate bands whose intensities decreased following incubation in the presence of ATP (conditions supporting SUMO modification). Molecular mass markers are indicated on the left. (B) Reactions were resolved by SDS-PAGE and analyzed by immunoblot analysis with an antibody specific for the hnRNP M proteins. The arrow indicates unmodified hnRNP M proteins; the bracket indicates hnRNP M protein-SUMO conjugates. (C) Reactions were resolved by SDS-PAGE and analyzed by immunoblot analysis with an antibody specific for the hnRNP C proteins. The arrow indicates unmodified hnRNP C1 and C2, and the bracket indicates hnRNP C protein-SUMO conjugates. Equivalent amounts of protein were loaded in lanes 4 to 8.
In addition to the observed shift in the hnRNP M proteins in reactions incubated with SUMO and ATP, we also observed a decrease in the intensities of bands migrating at apparent molecular masses of 44 and 47 kDa, similar to the molecular masses of the hnRNP C proteins (Fig. 2A, lane 2). To determine whether the hnRNP C proteins were also modified by SUMO, we performed immunoblot analysis with an hnRNP C protein-specific monoclonal antibody. In reactions incubated in the absence of ATP, only unmodified hnRNP C proteins were detected (Fig. 2B, lane 4). In contrast, in reactions incubated in the presence of SUMO and ATP, a doublet shifted in molecular mass by approximately 20 kDa relative to unmodified hnRNP C proteins was detected, as well as a higher-molecular-mass smear of SUMO-hnRNP C conjugates (Fig. 2B, lane 5). Together, these results demonstrate that both the hnRNP M proteins and hnRNP C proteins can be modified by SUMO while present in intact hnRNP complexes.
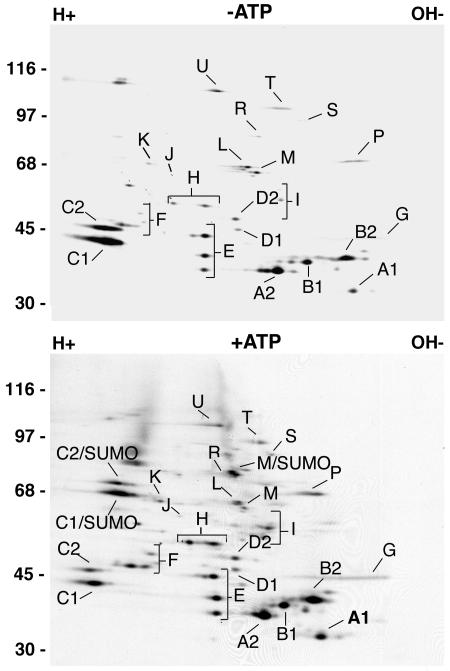
To determine whether other components of immunopurified hnRNP complexes could be modified by SUMO, we also analyzed the above-described reactions using two-dimensional gel electrophoresis. Complexes incubated in the absence of ATP revealed the expected pattern of hnRNPs (Fig. 3, top panel). Several additional spots corresponding to SUMO-modified hnRNP C and M proteins, however, were detected in complexes incubated with SUMO and ATP (Fig. 3, bottom panel). Significantly, no other proteins appeared to be modified by SUMO, indicating that only a specific subset of hnRNPs are substrates for SUMO modification.
FIG. 3.
SUMO modification of the hnRNP M and C proteins does not significantly disrupt immunopurified hnRNP complexes. hnRNP complexes were immunopurified from the [35S]methionine-labeled HeLa cell nucleoplasm and incubated with SUMO E1 and E2 conjugating enzymes in the presence (bottom panel) and in the absence (top panel) of ATP. Reactions were resolved by two-dimensional gel electrophoresis. Individual hnRNPs are labeled according to the method of Piñol-Roma et al. (31). Molecular mass markers are indicated on the left.
HnRNP C1 is modified by SUMO at lysine 237.
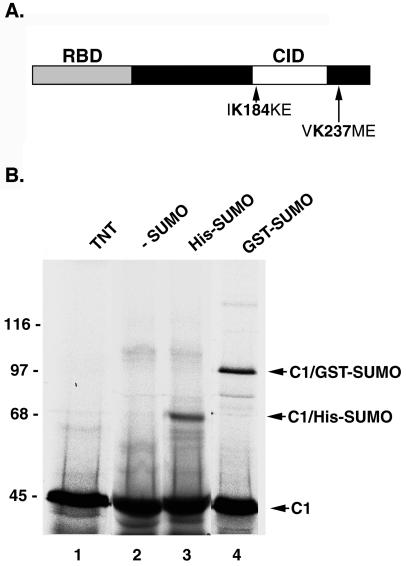
Because the hnRNP C proteins are among the most highly characterized hnRNPs, we focused our attention on further characterizing their SUMO modification. We first investigated whether hnRNP C1 produced by translation in rabbit reticulocyte lysate could be modified by SUMO using previously established in vitro assays (35, 36). HnRNP C1 was translated in rabbit reticulocyte lysate in the presence of [35S]methionine and found to migrate at its predicted molecular mass of ∼45 kDa by SDS-PAGE (Fig. 4B, lane 1). When incubated in the presence of SUMO E1 and E2 enzymes and histidine-tagged SUMO, a more slowly migrating band of ∼65 kDa appeared (Fig. 4B, lane 3), consistent with the predicted size of hnRNP C1 modified by a single molecule of SUMO. To provide further evidence that the higher-molecular-mass band corresponds to SUMO-modified hnRNP C1, we also performed modification reactions using glutathione S-transferase (GST)-tagged SUMO. Using these conditions, a more slowly migrating band of ∼95 kDa was observed (Fig. 4B, lane 4), consistent with the predicted molecular mass of hnRNP C1 modified by a single molecule of GST-SUMO. Neither of these more slowly migrating bands was detected in reactions where exogenous SUMO was not included (Fig. 4B, lane 2). Together, these findings further demonstrate that the hnRNP C proteins can be modified by SUMO.
FIG. 4.
hnRNP C1 contains two SUMO modification consensus sequences and can be expressed and modified by SUMO in vitro. (A) A schematic representation of the hnRNP C1 protein structure is depicted. SUMO modification consensus sequences surrounding lysines 184 and 237 are indicated. RBD, RNA-binding domain; CID, C-protein interaction domain. (B) hnRNP C1 was translated in rabbit reticulocyte lysate in the presence of [35S]methionine (lane 1) and incubated with SUMO E1 and E2 conjugating enzymes either in the absence of exogenously added SUMO-1 (lane 2) or in the presence of His-tagged SUMO-1 (lane 3) or GST-tagged SUMO-1 (lane 4). Reactions were separated by SDS-PAGE and analyzed by autoradiography. Molecular mass standards are indicated on the left. Unmodified hnRNP C1, His-SUMO-modified hnRNP C1, and GST-SUMO-modified hnRNP C1 are indicated on the right.
The hnRNP C proteins have two functional domains: an RNA-binding domain located in the amino terminus and an oligomerization domain located near the carboxyl terminus (Fig. 4A). Upon inspection of the hnRNP C-protein sequence, two potential modification sites were identified that correspond to the SUMO consensus sequence. This consensus sequence defines the modification site of many known SUMO substrates and conforms to the sequence “ΨKXE,” where Ψ is a hydrophobic amino acid, K is the lysine to which SUMO is conjugated, X can be any residue, and E is glutamic acid (36). The first SUMO consensus sequence surrounds lysine 184 of hnRNP C1, a residue within the oligomerization domain, while the second sequence surrounds lysine 237 near the carboxyl terminus (Fig. 4A).
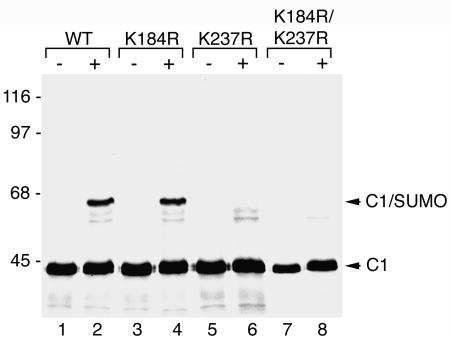
To determine whether either of these two lysines could serve as acceptor sites for SUMO modification, we mutated lysines 184 and 237 to arginine. Mutant proteins were translated in rabbit reticulocyte lysate and tested for their ability to be modified by SUMO using reactions similar to those described above. The lysine 184-to-arginine mutant (Fig. 5, lanes 3 and 4) was found to be modified by SUMO as efficiently as wild-type hnRNP C1 (Fig. 5, lanes 1 and 2), as indicated by the appearance of the shifted band of ∼65 kDa following incubation in the presence of SUMO and SUMO E1 and E2 enzymes. SUMO modification of the lysine 237-to-arginine mutant (Fig. 4, lanes 5 and 6), however, was almost completely inhibited. A similar inhibition was also observed with a double mutant in which both lysines 184 and 237 were mutated to arginines (Fig. 5, lanes 7 and 8). Weak bands running below the major SUMO-modified hnRNP C1 protein were detected both in the lysine 237 single mutant and in the lysine 184/237 double mutant. The origin of these bands is not known; however, they could possibly correspond to SUMO modification at less efficiently recognized sites. In summary, these results indicate that lysine 237 is the major SUMO modification site in hnRNP C1.
FIG. 5.
hnRNP C1 is modified by SUMO at lysine residue 237. Wild-type hnRNP C1 and hnRNP C1 mutants containing lysine-to-arginine substitutions at the two predicted SUMO modification sites (lysines 184 and 237) were translated in rabbit reticulocyte lysate in the presence of [35S]methionine (lanes 1, 3, 5, and 7). The proteins were incubated with SUMO E1 and E2 conjugating enzymes and SUMO-1 (lanes 2, 4, 6, and 8), and their ability to be modified was determined by SDS-PAGE followed by autoradiography. Molecular mass markers are indicated on the left, and unmodified and SUMO-modified hnRNP C1 proteins are indicated on the right.
Nup358 functions as an E3-like factor to enhance SUMO modification of hnRNP C1.
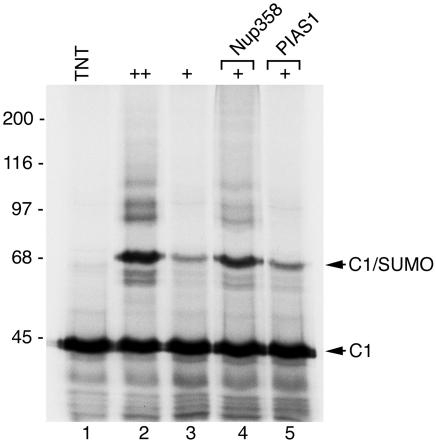
SUMO E3-like proteins are not strictly required for SUMO modification in vitro. However, when limiting concentrations of E1 and E2 enzymes are present, SUMO E3-like factors can enhance the efficiency of substrate modification (19, 29). To determine whether two known SUMO E3-like factors, Nup358 or PIAS1, could enhance the SUMO modification of hnRNP C1, we altered the above in vitro assay conditions by reducing the concentrations of SUMO E1 and E2 enzymes. Using conditions in which SUMO E1 and E2 enzymes were limiting, we observed a decrease in the level of hnRNP C1 modification (Fig. 6, lane 3) compared to results under the more optimal assay conditions (Fig. 6, lane 2). When a domain of Nup358 containing SUMO E3-like activity was included in the assays with limiting concentrations of E1 and E2 enzymes, modification was dramatically enhanced (Fig. 6, lane 4), approaching levels observed using optimal concentrations of E1 and E2 alone (Fig. 6, lane 2). The addition of PIAS1 to the assays, however, had little if any effect on hnRNP C1 SUMO modification (Fig. 6, lane 5). PIAS1, however, did enhance the SUMO modification of the androgen receptor under similar assay conditions (data not shown). These findings demonstrate that Nup358, but not PIAS1, is able to function as an E3-like factor to enhance the SUMO modification of hnRNP C1.
FIG. 6.
Nup358 functions as an E3-like factor to enhance the SUMO modification of hnRNP C1. hnRNP C1 was translated in rabbit reticulocyte lysate in the presence of [35S]methionine (lane 1) and incubated with SUMO-1 and high concentrations of SUMO E1 and E2 conjugating enzymes (lane 2) or low concentrations of E1 and E2 enzymes (lanes 2 to 5). Reactions containing low concentrations of E1 and E2 conjugating enzymes were supplemented with a fragment of Nup358 known to have SUMO E3-like activity (lane 4) or with PIAS1 (lane 5). Molecular mass markers are indicated on the left, and unmodified and SUMO-modified hnRNP C1 is indicated on the right.
SUMO modification decreases the interaction between hnRNP C1 and ssDNA.
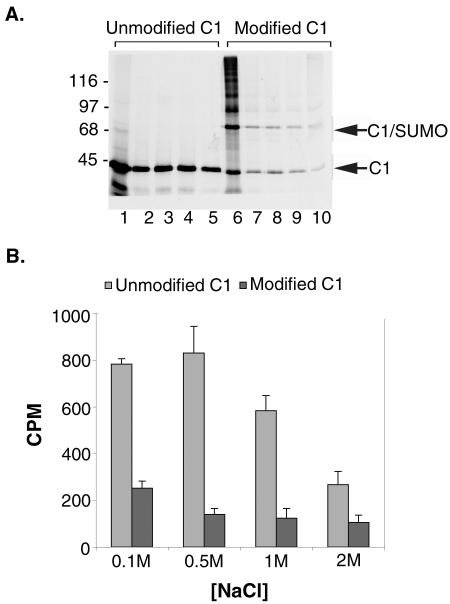
To begin to address the effects of SUMO modification on hnRNP C1 function, we analyzed the binding of unmodified and SUMO-modified hnRNP C1 to ssDNA. ssDNA was chosen for these studies due to its stability and because all of the major hnRNPs have previously been shown to interact equally well with ssDNA and RNA (33). hnRNP C1 was translated in rabbit reticulocyte lysate in the presence of [35S]methionine and incubated with ssDNA-agarose beads in buffers of increasing ionic strength. Bound proteins were eluted with SDS-PAGE sample buffer and analyzed visually by SDS-PAGE, or quantitatively by counting bound radioactive counts using a scintillation counter. As expected, hnRNP C1 bound to the ssDNA at all of the tested salt conditions, including 2 M NaCl (Fig. 7A, lanes 1 to 5). Based on quantitative analysis, binding was most robust at 500 mM NaCl (Fig. 7B).
FIG. 7.
SUMO modification influences the nucleic acid binding activity of the hnRNP C proteins. hnRNP C1 was translated in rabbit reticulocyte lysate in the presence of [35S]methionine and incubated with ssDNA-cellulose before or following modification by SUMO-1. Equal numbers of radioactive protein counts were added to all binding reactions. Binding reactions were performed in buffers containing 0.1, 0.5, 1, and 2 M NaCl. (A) Proteins bound to the ssDNA-cellulose beads were eluted with SDS sample buffer and analyzed by SDS-PAGE followed by autoradiography. Molecular mass markers are indicated on the left, and unmodified and SUMO-modified hnRNP C1 proteins are indicated on the right. (B) Proteins bound to the ssDNA-cellulose beads were eluted with SDS sample buffer, and binding was quantified by determining bound radioactive counts using a scintillation counter. Binding reactions were carried out in triplicate, and average values and standard deviations were plotted with bound counts per minute (CPM) on the y axis and salt concentrations on the x axis.
To investigate the effects of SUMO modification on hnRNP C1, we repeated the binding assays with SUMO-modified protein produced using the in vitro assay described above. Under these assay conditions, approximately 50% of the expressed hnRNP C1 is modified by SUMO (Fig. 7A, lane 6), and as indicated below, modified and unmodified proteins are present in the extracts as hetero-oligomers. Analysis of the binding reactions by SDS-PAGE demonstrated that both unmodified and SUMO-modified hnRNP C1 bound to the ssDNA resin at all of the assayed salt concentrations, indicating that SUMO modification does not qualitatively prevent interactions between hnRNP C1 and nucleic acids (Fig. 7A, lanes 6 to 10). However, analysis of the binding assays by scintillation counting of bound protein demonstrated that SUMO modification had a significant quantitative effect on the binding of hnRNP C1 to ssDNA (Fig. 7B). This analysis revealed a consistent reduction in the binding of hnRNP C1 to ssDNA at all of the tested salt concentrations, with the greatest effect (∼80% reduction in binding) seen at 500 mM NaCl. Comparable quantitative data was also obtained by phosphorimage analysis of the gel shown in Fig. 7A (data not shown). These results indicate that SUMO modification decreases the affinity between hnRNP C1 and single-stranded nucleic acids.
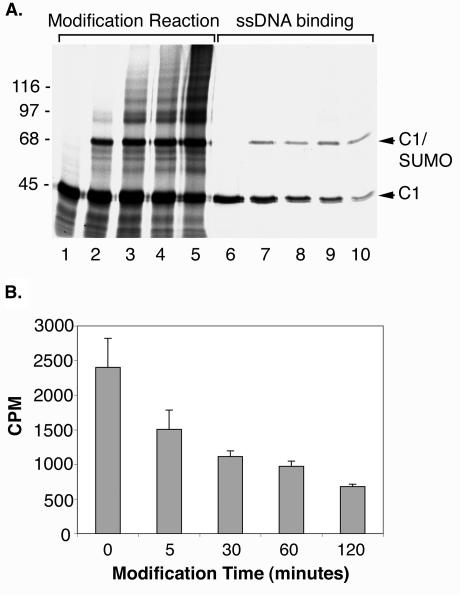
To further examine the effects of SUMO modification on hnRNP C1, we also performed ssDNA-binding assays in which the ratio of SUMO-modified protein to unmodified protein varied. hnRNP C1 was expressed in rabbit reticulocyte lysate (Fig. 8A, lane 1) and incubated for various lengths of time in an in vitro SUMO modification assay (Fig. 8A, lanes 2 to 5). At the earliest time point (Fig. 8A, lane 2), approximately one-fourth of the expressed hnRNP C1 protein was modified by SUMO, whereas approximately one-half of the protein was SUMO modified at the latest time point (Fig. 8A, lane 5). Upon incubation of each of the hnRNP C1 SUMO modification reactions with ssDNA, we detected a direct correlation between the extent of SUMO modification and the effect on ssDNA binding (Fig. 8A, lanes 6 to 10). Quantitative analysis of the binding revealed an approximately 1.6-fold reduction in ssDNA binding for reactions containing the lowest levels of SUMO-modified hnRNP C1 and an approximately threefold reduction in binding for reactions containing the highest levels of SUMO-modified hnRNP C1 (Fig. 8B).
FIG. 8.
Increased SUMO modification of hnRNP C1 correlates with increased reduction in ssDNA binding. (A) hnRNP C1 was translated in rabbit reticulocyte lysate in the presence of [35S]methionine and incubated in an in vitro SUMO modification assay for increasing lengths (lane 1, 0 min; lane 2, 5 min; lane 3, 30 min; lane 4, 60 min; lane 5, 120 min) of time to generate samples with increasing ratios of SUMO-modified to unmodified hnRNP C1. The resulting samples were incubated with ssDNA-cellulose in the presence of 0.5 M NaCl, and bound proteins were eluted with SDS sample buffer and examined by SDS-PAGE (lanes 6 to 10). Molecular mass markers are indicated on the left, and unmodified and SUMO modified hnRNP C1 proteins are indicated on the right. (B) Proteins bound to the ssDNA-cellulose beads were eluted with SDS sample buffer, and binding was quantified by determining bound radioactive counts using a scintillation counter. Binding reactions were performed in triplicate, and average values and standard deviations were plotted with bound counts per minute (CPM) on the y axis and incubation times for each SUMO modification reaction on the x axis.
SUMO-modified and unmodified hnRNP C1 exist as hetero-oligomers in vitro.
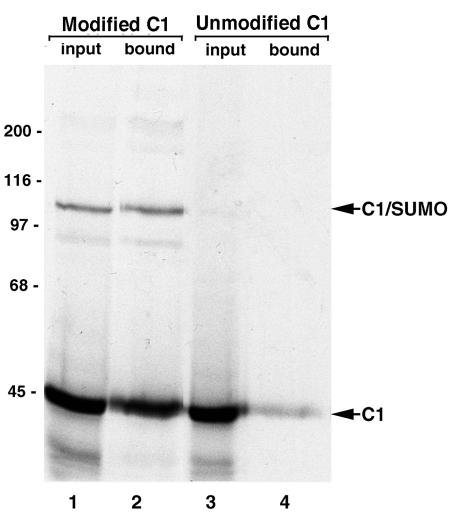
hnRNP C1 forms oligomers with its alternatively spliced isoform C2 in vivo and with itself in vitro, and it has been suggested that the C proteins bind to RNA as a tetramer (2, 34). Because only ∼50% of the hnRNP C1 produced in rabbit reticulocyte lysate could be modified by SUMO, we examined whether SUMO-modified and unmodified proteins might be present as hetero-oligomers in our extracts. hnRNP C1 was translated in rabbit reticulocyte lysate in the presence of [35S]methionine and was modified using GST-tagged SUMO (Fig. 9, lane 1) as described above. SUMO-modified hnRNP C1 was then purified using glutathione-Sepharose beads, and bound proteins were analyzed by SDS-PAGE. Using these conditions, we detected both SUMO-modified and unmodified hnRNP C1 copurifying on the glutathione beads (Fig. 9, lane 2). A control experiment using only unmodified hnRNP C1 indicated that the C1 protein itself has relatively little affinity for the glutathione beads (Fig. 9, lane 4). Together, these findings indicate that SUMO-modified and unmodified hnRNP C1 proteins are present as hetero-oligomers in the in vitro extracts. These findings indicate that SUMO modification does not disrupt the oligomerization of the hnRNP C1 protein. These findings also indicate that the in vitro ssDNA-binding assay described above measure interactions between hnRNP C1/SUMO-hnRNP C1 protein hetero-oligomers and ssDNA.
FIG. 9.
Unmodified and SUMO-modified hnRNP C1 form hetero-oligomers. hnRNP C1 was translated in rabbit reticulocyte lysate in the presence of [35S]methionine and modified with GST-tagged SUMO-1 (lane 1). Modification reactions were incubated with glutathione-Sepharose beads, and bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE followed by autoradiography (lane 2). Binding reactions were also performed using unmodified hnRNP C1 as a control for nonspecific interactions between hnRNP C1 and glutathione Sepharose (lanes 3 and 4). Molecular mass markers are indicated on the left, and modified and unmodified hnRNP C1 proteins are indicated on the right.
DISCUSSION
Several lines of evidence suggest that SUMO modification may occur at NPCs as proteins are transported between the nucleus and the cytoplasm (30). However, what role SUMO modification plays at NPCs is currently not known, in part because specific protein targets that are modified during their nucleocytoplasmic transport have not been identified. We have demonstrated by a combination of in vitro assays that two groups of hnRNPs, which are among the most abundant cargo transported through NPCs, are substrates for SUMO modification. First, we identified the hnRNP M proteins as SUMO substrates using an assay designed to identify proteins modified by SUMO at the nuclear envelope. Apart from the hnRNP M proteins this assay also detected the presence of additional SUMO substrates, whose identities remain to be determined. Although the smear of high-molecular-mass SUMO conjugates detected in these experiments may suggest the presence of a large number of possible substrates (Fig. 1), short exposures of these immunoblots reveal the presence of a relatively small number of major SUMO conjugates in addition to hnRNP M proteins (data not shown). We had previously identified the hnRNP M proteins as one of only a subset of hnRNPs that copurify with isolated nuclear envelopes and NPCs (6). Although the exact nature of the interaction of hnRNP M proteins with the NPC is not known, we hypothesize that they interact directly with specific protein components of the NPC. Their selective association with the NPC suggests that they may play a distinct role in the transport of mRNPs from the nucleus to the cytoplasm. Unlike the hnRNP M proteins, most other hnRNPs, including the hnRNP C proteins, do not copurify with isolated nuclear envelopes.
Following the identification of the hnRNP M proteins as SUMO substrates, we investigated whether other hnRNPs could be modified by SUMO using alternative approaches. Using immunopurified hnRNP complexes and in vitro SUMO modification assays, we found that only two subsets of hnRNPs are modified by SUMO. Consistent with our nuclear envelope findings, we again identified the hnRNP M proteins as SUMO substrates by using this assay. In addition, we also identified the hnRNP C1 and C2 proteins as SUMO substrates. Both the hnRNP M and C proteins were modified in intact hnRNP complexes, indicating their ability to be recognized by SUMO conjugating enzymes while being part of large protein-RNA complexes. Why these two specific subsets of hnRNPs are targets for SUMO modification is of great interest, and we propose that they may have a similar function in mRNA biogenesis that involves a common regulatory mechanism dependent on SUMO. Importantly, the selective SUMO modification of just this subset of hnRNPs demonstrates the specificity of the in vitro reactions.
The hnRNP C proteins bind selectively to poly(U)-rich mRNA sequences (39) and have direct roles in pre-mRNA splicing (5). Of particular interest is that the hnRNP C proteins are among the very few hnRNPs that do not accompany mRNAs during translocation into the cytoplasm (32). Because the hnRNP C proteins do not shuttle between the nucleus and the cytoplasm, they are apparently released from newly matured mRNAs within the nucleus or at the NPC. The hnRNP M proteins bind preferentially to poly(G)- and poly(U)-rich mRNA sequences (8, 39), and like the C proteins they have also been implicated in regulating pre-mRNA splicing (20). It is currently unknown whether the hnRNP M proteins exit the nucleus with mRNPs or whether they are released from newly matured mRNAs in the nucleus or at the NPC. Based on their apparent regulatory roles in pre-mRNA splicing, SUMO modification of the hnRNP C and M proteins could play a role in mediating the dynamic changes in protein-protein and protein-RNA interactions that occur during the stages of spliceosome assembly, intron excision, and exon joining. In addition, SUMO modification of the hnRNP M and C proteins could also play a role in mediating changes in mRNP structure and/or composition during transport from the nucleus to the cytoplasm. SUMO modification of a number of proteins regulates their subnuclear localization (37), and it is therefore possible that the modification of hnRNPs targets them to specific nuclear domains. Our preliminary results, however, have revealed no obvious differences in the subnuclear localization of wild-type and mutant (K237R) hnRNP C1 (data not shown).
Analysis of the interactions between the hnRNP C proteins and ssDNA indicates that SUMO modification influences their associations with nucleic acids. Our results also demonstrate that the SUMO-modified and unmodified hnRNP C proteins are present as hetero-oligomers in our in vitro reactions and that SUMO modification of only one or two subunits of an oligomer may be sufficient to have a dramatic effect on its nucleic acid binding. Moreover, we observed a parallel decrease in ssDNA-binding activity as the ratio of SUMO-modified to unmodified hnRNP C1 was increased in the binding reactions. To what extent the hnRNP C proteins exist as oligomers in vivo is not clear; however, our findings suggest a possible mechanism whereby the function of the hnRNP C proteins could be differentially regulated through SUMO modification of one or more subunits within an oligomer.
Based on our present observations, we propose a model in which SUMO modification and rapid demodification of the hnRNP C and M proteins occurs at the NPC and facilitates the transport of mRNPs from the nucleus to the cytoplasm. This model is supported by the finding that Nup358 acts as an E3 ligase and enhances the SUMO modification of both the hnRNP C and M proteins (Fig. 6; also data not shown). In addition to Nup358, both Ubc9 (the SUMO E2 conjugating enzyme) and SENP2 (a SUMO deconjugating enzyme) localize to the NPC (42), further suggesting that SUMO modification and demodification of substrates may be linked to nucleocytoplasmic transport. We propose that SUMO modification of the hnRNP C and M proteins could occur at the NPC (either at the cytoplasmic filaments, assisted by Nup358, or at the nucleoplasmic basket, where Ubc9 has also been detected) and mediate a change in mRNP structure or composition that facilitates translocation into the cytoplasm. This model is consistent with the observations that Balbiani ring mRNP transcripts undergo dramatic conformational changes at the NPC as they are translocated through the nucleoplasmic basket and into the cytoplasm (22, 38). As part of our model, SUMO modification of the hnRNPs would be very transient, with both modification and rapid demodification occurring at the NPC. SUMO modification of only the small fraction of hnRNP M and C proteins engaged at the NPC, as well as the transient nature of the modification, would suggest that only a very low level of SUMO-modified hnRNP C and M proteins would be present in the cell at any given time, consistent with our present findings. Since this is only a working model, there are also other possible functions for SUMO modification of the hnRNP C and M proteins. Apart from having a role in mediating the nucleocytoplasmic transport of mRNPs, SUMO modification could also influence other steps in mRNA biogenesis, including pre-mRNA splicing. We hope to address the question of how SUMO modification of the hnRNP C and M proteins specifically affects their roles in mRNA biogenesis in future studies.
Acknowledgments
We acknowledge Gideon Dreyfuss for the generous gift of the 4F4 monoclonal antibody to hnRNP C1 and Maurice Swanson for hnRNP C1 and hnRNP M cDNAs and hnRNP M monoclonal antibodies. We also thank members of the Matunis lab for their advice and support during course of this work.
This work was supported by grants from the National Institutes of Health (GM60980 to M.J.M.) and the American Cancer Society (RSG-01-064-01-CSM to M.J.M.).
REFERENCES
- 1.Arhin, G. K., M. Boots, P. S. Bagga, C. Milcarek, and J. Wilusz. 2002. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 30:1842-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, S. F., D. L. Friedman, and W. M. LeStourgeon. 1989. The C proteins of HeLa 40S nuclear ribonucleoprotein particles exist as anisotropic tetramers of (C1)3 C2. Mol. Cell. Biol. 9:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. [DOI] [PubMed] [Google Scholar]
- 4.Choi, Y. D., and G. Dreyfuss. 1984. Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc. Natl. Acad. Sci. USA 81:7471-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, Y. D., P. J. Grabowski, P. A. Sharp, and G. Dreyfuss. 1986. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science 231:1534-1539. [DOI] [PubMed] [Google Scholar]
- 6.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dallaire, F., S. Dupuis, S. Fiset, and B. Chabot. 2000. Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J. Biol. Chem. 275:14509-14516. [DOI] [PubMed] [Google Scholar]
- 8.Datar, K. V., G. Dreyfuss, and M. S. Swanson. 1993. The human hnRNP M proteins: identification of a methionine/arginine-rich repeat motif in ribonucleoproteins. Nucleic Acids Res. 21:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreyfuss, G., Y. D. Choi, and S. A. Adam. 1984. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol. Cell. Biol. 4:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 11.Dreyfuss, G., M. J. Matunis, S. Pinol-Roma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 12.Eversole, A., and N. Maizels. 2000. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol. 20:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford, L. P., J. M. Suh, W. E. Wright, and J. W. Shay. 2000. Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol. Cell. Biol. 20:9084-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford, L. P., W. E. Wright, and J. W. Shay. 2002. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene 21:580-583. [DOI] [PubMed] [Google Scholar]
- 15.Hang, J., and M. Dasso. 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277:19961-19966. [DOI] [PubMed] [Google Scholar]
- 16.Hay, D. C., G. D. Kemp, C. Dargemont, and R. T. Hay. 2001. Interaction between hnRNPA1 and IκBα is required for maximal activation of NF-κB-dependent transcription. Mol. Cell. Biol. 21:3482-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochstrasser, M. 2001. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107:5-8. [DOI] [PubMed] [Google Scholar]
- 18.Holcik, M., B. W. Gordon, and R. G. Korneluk. 2003. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 23:280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735-744. [DOI] [PubMed] [Google Scholar]
- 20.Kafasla, P., M. Patrinou-Georgoula, J. D. Lewis, and A. Guialis. 2002. Association of the 72/74-kDa proteins, members of the heterogeneous nuclear ribonucleoprotein M group, with the pre-mRNA at early stages of spliceosome assembly. Biochem J. 363:793-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J. H., K. Y. Paek, K. Choi, T. D. Kim, B. Hahm, K. T. Kim, and S. K. Jang. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23:708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiseleva, E., M. W. Goldberg, B. Daneholt, and T. D. Allen. 1996. RNP export is mediated by structural reorganization of the nuclear pore basket. J. Mol. Biol. 260:304-311. [DOI] [PubMed] [Google Scholar]
- 23.Krecic, A. M., and M. S. Swanson. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 11:363-371. [DOI] [PubMed] [Google Scholar]
- 24.LaBranche, H., S. Dupuis, Y. Ben-David, M. R. Bani, R. J. Wellinger, and B. Chabot. 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19:199-202. [DOI] [PubMed] [Google Scholar]
- 25.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matunis, M. J., J. Wu, and G. Blobel. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 28.Michelotti, E. F., G. A. Michelotti, A. I. Aronsohn, and D. Levens. 1996. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 16:2350-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 30.Pichler, A., and F. Melchior. 2002. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3:381-387. [DOI] [PubMed] [Google Scholar]
- 31.Pinol-Roma, S., Y. D. Choi, M. J. Matunis, and G. Dreyfuss. 1988. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 2:215-227. [DOI] [PubMed] [Google Scholar]
- 32.Pinol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730-732. [DOI] [PubMed] [Google Scholar]
- 33.Pinol-Roma, S., M. S. Swanson, M. J. Matunis, and G. Dreyfuss. 1990. Purification and characterization of proteins of heterogeneous nuclear ribonucleoprotein complexes by affinity chromatography. Methods Enzymol. 181:326-331. [DOI] [PubMed] [Google Scholar]
- 34.Rech, J. E., W. M. LeStourgeon, and P. F. Flicker. 1995. Ultrastructural morphology of the hnRNP C protein tetramer. J. Struct. Biol. 114:77-83. [DOI] [PubMed] [Google Scholar]
- 35.Rogers, R. S., C. M. Horvath, and M. J. Matunis. 2003. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J. Biol. Chem. 278:30091-30097. [DOI] [PubMed] [Google Scholar]
- 36.Sampson, D. A., M. Wang, and M. J. Matunis. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664-21669. [DOI] [PubMed] [Google Scholar]
- 37.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 38.Skoglund, U., K. Andersson, B. Bjorkroth, M. M. Lamb, and B. Daneholt. 1983. Visualization of the formation and transport of a specific hnRNP particle. Cell 34:847-855. [DOI] [PubMed] [Google Scholar]
- 39.Swanson, M. S., and G. Dreyfuss. 1988. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol. 8:2237-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veraldi, K. L., G. K. Arhin, K. Martincic, L. H. Chung-Ganster, J. Wilusz, and C. Milcarek. 2001. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 21:1228-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visa, N., A. T. Alzhanova-Ericsson, X. Sun, E. Kiseleva, B. Bjorkroth, T. Wurtz, and B. Daneholt. 1996. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell 84:253-264. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, H., H. Saitoh, and M. J. Matunis. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]