Abstract
The Sleeping Beauty (SB) transposase is the most active transposase in vertebrate cells, and the SB transposon system has been used as a tool for insertional mutagenesis and gene delivery. Previous studies have indicated that the frequency of chromosomal transposition is considerably higher in mouse germ cells than in mouse embryonic stem cells, suggesting the existence of unknown mechanisms that regulate SB transposition. Here, we demonstrated that CpG methylation of the transposon region enhances SB transposition. The transposition efficiencies of a methylated transposon and an unmethylated transposon which had been targeted in the same genomic loci by recombination-mediated cassette exchange in mouse erythroleukemia cells were compared, and at least a 100-fold increase was observed in the methylated transposon. CpG methylation also enhanced transposition from plasmids into the genome. Chromatin immunoprecipitation assays revealed that histone H3 methylated at lysine-9, a hallmark of condensed heterochromatin, was enriched at the methylated transposon, whereas the unmethylated transposon formed a relaxed euchromatin structure, as evidenced by enrichment of acetylated histone H3 and reporter gene expression. Possible roles of heterochromatin formation in the transposition reaction are discussed. Our findings indicate a novel relationship between CpG methylation and transposon mobilization.
DNA transposons are mobile elements that can change location in the genome and have a major impact on the evolution of the genome (36). DNA transposons consist of two elements, a DNA element flanked by terminal inverted repeats (IRs) and a transposase that catalyzes the transposition reaction by a “cut-and-paste” mechanism. The DNA element flanked by IRs is excised from the genome in the “cut” step and is reintegrated into the genome in the “paste” step. After excision, several base pairs of additional nucleotides typically remain at the excised site and are known as a “footprint.” The DNA transposons can be classified into autonomous and nonautonomous transposons. The former encode the transposase gene within the transposon and can move by themselves, whereas the latter do not carry a functional transposase gene and can move only when transposases are supplied from other sources.
DNA transposons have been successfully engineered and utilized as tools for insertional mutagenesis or gene delivery in model organisms such as Drosophila (1, 33), Caenorhabditis elegans (13, 27), and plants (30). Although the vertebrate genomes contain numerous DNA transposon sequences, the vast majority of DNA transposons have been inactivated during evolution. Applications in vertebrates remained dormant until the recent emergence of the Sleeping Beauty (SB) transposon system (19). The SB transposase is a Tc1/mariner-like transposase that was reconstructed from the salmonid genome by restoring its activity through the correction of accumulated mutations. The SB transposon system was developed as a nonautonomous transposon system consisting of two parts: the SB transposon, containing IRs that flank the DNA element intended for mobilization, and the SB transposase gene, which is expressed from a separate expression vector (19). Each IR contains two copies of a direct repeat (DR) that function as binding sites for the SB transposase, and these structures are termed IR/DRs. SB transposases bound to the DRs need to interact with each other for the transposition reaction to occur (21). Since the SB transposase was found to show the most efficient transposition in vertebrate cells (10, 20), the SB system has been utilized as a tool for germ line mutagenesis (3, 17) and gene delivery (6) in vertebrates and has promising applications in gene therapy (38, 39).
Efficient chromosomal transposition of the SB transposon in mouse germ cells with a transposition frequency as high as 0.03 to 0.2 event per transposon per spermatid has been demonstrated (7, 10, 16). However, the frequency of chromosomal transposition in mouse embryonic stem (ES) cells has been reported to be several orders of magnitude lower than that in mouse germ cells, at most 3.5 × 10−5 events per cell per generation (25). This finding suggests that there are unknown mechanisms which regulate SB transposition. It is thought that the activities of mobile elements, such as DNA transposons, retrotransposons, and retroviruses, are regulated in living organisms in order to maintain genomic integrity. In Arabidopsis, the DNA transposon and retrotransposon were found to be activated in a mutant with DNA hypomethylation (15, 26). In other instances, the Tc1 transposon in C. elegans (22, 34) and the retrotransposon in Chlamydomonas (37) were reactivated in strains defective in RNA interference. Moreover, the endogenous retrovirus was transcriptionally activated in mouse embryos deficient in DNA methyltransferase-1 (35). In view of these various findings, in order to attain optimal use of the SB transposon as a tool for functional genomics and gene delivery, it is important to identify the regulatory mechanisms of SB transposition.
Here, we demonstrate that CpG methylation within the transposon sequence enhances the transposition frequency of the SB transposon. CpG methylation is a well-studied epigenetic modification of the genome and is associated with other epigenetic modifications, such as heterochromatin formation (2, 24). Our findings indicate a new aspect of the transposition mechanism.
MATERIALS AND METHODS
Construction and methylation of plasmids.
A targeting vector to introduce the SB transposon into the serine palmitoyltransferase long-chain base subunit 2 (Sptlc2) locus was generated by first cloning a 5-kb XhoI-KpnI fragment of the Sptlc2 gene containing exon 5 into the XhoI-KpnI site of pBluescript II (pBS; Stratagene), resulting in pBS-Sptlc2. A SalI-BamHI fragment of pCX-EGFP-PigA (16) and a BamHI-NotI fragment of a 150-bp synthetic splice donor site (a gift from M. Ikawa) were then cloned into the SalI-NotI site of pBS, followed by the deletion of the BamHI site, to generate pCX-EGFP-SD. A blunted SalI-NotI fragment of pCX-EGFP-SD was inserted into the blunted EcoRI-BamHI site of pBS-IR/DR(R,L) to generate pTransCX-EGFP-SD. An EcoRI-NotI fragment of pTransCX-EGFP:Neo (16) containing a PGK-neo cassette was cloned into pBS, after which the BamHI site was deleted to generate pBS-Neo. A NotI-KpnI fragment of pBS-Neo and a KpnI-XhoI fragment of pTransCX-EGFP-SD were inserted into the NotI-XhoI site of pBS-Sptlc2, resulting in the targeting vector.
A vector for recombinase-mediated cassette exchange (RMCE) was constructed in the following manner. To remove the loxP site downstream of the PGK-neo cassette of pTransCX-EGFP:Neo, a BamHI-AflIII fragment and a NotI-AflIII fragment were religated, resulting in pTransCX-EGFP:Neo-3′lox. After blunt ending and NotI linker ligation of all cleavage ends, a ClaI-BglII fragment of pL1CMVEGFP1L (8) was replaced with a PvuI fragment of pTransCX-EGFP:Neo-3′lox containing the SB transposon and a ∼200-bp flanking sequence on both sides, resulting in pL1TransCX-EGFP1L.
pTransSAβ-geo and pTc3/SAβ-geo were constructed by cloning a blunt-ended XhoI fragment of pROSAβ-geo (11) into the blunt-ended EcoRI-BamHI site of pTransCX-GFP (16) and the blunt-ended BspEI-NcoI site of pRP790 (9), respectively.
For the expression of a six-His-tagged 123-amino-acid N-terminal fragment of the SB transposase (N123), the N-terminal fragment was first PCR amplified from pCMV-SB (pSB10) (19) with primers 5′-CATGCCATGGGAAAATCAAAAGAAATC-3′ and 5′-CCGCTCGAGCAGTGGCTTCTTCCTTG-3′ and digested with NcoI, BsrGI, and XhoI. Then, the NcoI-BsrGI fragment and the BsrGI-XhoI fragment were cloned into NcoI- and XhoI-digested pET21d (Novagen), resulting in pET-N123.
For the DNA probe of IR/DR-L for the electrophoretic mobility shift assay (EMSA), the HindIII-KpnI fragment of pTransCX-EGFP:Neo was cloned into the HindIII-KpnI site of pBS, resulting in pBS-IR/DR-L.
Methylation of plasmid DNAs was performed with SssI CpG methylase (NEB) according to the manufacturer's protocol, followed by purification with a PCR purification kit (Qiagen). Complete methylation was confirmed by resistance to digestion with methylation-sensitive enzymes.
Cell culture and gene targeting.
Mouse ES cells were cultured in Dulbecco modified Eagle medium containing 20% fetal bovine serum, nonessential amino acids, sodium pyruvate, and 1,000 U of leukemia-inhibitory factor/ml on mitomycin C-treated mouse embryonic fibroblasts. Mouse erythroleukemia (MEL) cell clones RL5 and RL6 (8) were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum.
Targeted integration of the SB transposon at the Sptlc2 locus was performed by means of insertional-type homologous recombination. Briefly, 25 μg of the targeting vector was linearized at the BamHI site located in the homologous region and introduced into 1.0 × 107 ES cells by electroporation (240 V, 500 μF) with a Gene Pulser II (Bio-Rad). Cells were selected for 7 days with 150 μg of G418/ml, after which resistant clones were picked up, expanded, and analyzed with Southern blotting.
RMCE was performed as described previously (32). Briefly, 25 μg of methylated or unmethylated plasmids was introduced by electroporation (250 V, 1,070 μF) into MEL cell clone RL5 or RL6 together with 20 μg of Cre expression vector pBS185 and 200 μg of salmon sperm DNA. Cells were selected for 10 days, starting 4 days after electroporation, with 10 μM ganciclovir. Limiting dilution was performed for the isolation of independent clones, which were expanded and prescreened by PCR with the primers EGFP-1U (5′-CACCCTCGTGACCACCCTGACCTAC-3′) and EGFP-1L (5′-CTTGATGCCGTTCTTCTGCTTGTCG-3′) for the detection of enhanced green fluorescent protein (EGFP) and with the primers HYG-1U (5′-CGGGCGTATATGCTCCCCATTGGTCTTGAC-3′) and TK-1L (5′-TGGTGTAGATGTTCGCGATTGTCTCGGAAG-3′) for the detection of HYTK. The orientation of the exchanged cassette was determined with the primers M13F (5′-ACGACGTTGTAAAACGACGGCCAGT-3′) and RMCE-DL1 (5′-GCATCGCCATGGGTCACGACGAGATCCTC-3′) for orientation D and with the primers M13F and RMCE-IL-1 (5′-AAGTGAGTTTAAATGTATTTGGCTAAGGTG-3′) for orientation I. Cassette exchange and methylation statuses were further confirmed by Southern blot analysis.
Isolation of male germ cells.
Testes of a mouse carrying donor transposons (16) were decapsulated and chopped into ∼1-mm2 fragments with a razor blade. Germ cells were released by repeated pipetting. After brief centrifugation to remove seminiferous tubules, the supernatant was collected and sedimented by further centrifugation at 1,500 × g. Then, DNA was extracted from the cell pellet and analyzed with Southern blotting.
Excision assay of transiently transfected transposon.
For the excision assay of the transiently transfected transposon, 1.0 × 106 MEL cells were transfected with 1.0 μg of methylated or unmethylated pTransCX-EGFP:Neo together with 1.0 μg of pSB10 or pBS by using TransFast (Promega). Genomic DNA was extracted with a DNeasy kit (QIAGEN) 48 h after transfection. Four hundred nanograms and 80 ng of HindIII-digested DNAs were used to amplify the excision product and the neo fragment, respectively, with the HotStarTaq system (QIAGEN). The neo fragment is present in the pTransCX-EGFP:Neo vector and was used as an internal control for transfection efficiency. Primers TgTP-1U (5′-GACCGCTTCCTCGTGCTTTACGGTATC-3′) and TgTP-2L (5′-ACACAGGAAACAGCTATGACCATGATTACG-3′) were used to detect the excision product, and primers neo-U1 (5′-GGGTGGAGAGGCTATTCGGCTATGA-3′) and neo-L1 (5′-TGGATACTTTCTCGGCAGGAGCAAG-3′) were used to amplify the neo fragment. Amplification of the excision product was carried out under the following conditions: 95°C for 15 min, followed by 35 cycles of 94°C for 1 min, 63°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 7 min. The same conditions were used for the amplification of the neo fragment except that the annealing temperature was 60°C and the number of cycles was 19.
The excision product and the neo fragment were also quantified by real-time PCR on a LightCycler instrument (Roche Diagnostics) from 40 ng of template DNA by using the LightCycler FastStart DNA Master Hybridization Probes kit (Roche Diagnostics) according to the manufacturer's instructions. Fluorescently labeled probes for the excision product were 5′-CGGCCGCTCTAGCGGTACCCTAC-FITC-3′ and 5′-LCRed640-GTAGGGGATCGACCTCGAGGGG-3′, and those for the neo fragment were 5′-GCTGTGCTCGACGTTGTCACTGAAG-FITC-3′ and 5′-LCRed640-GGGAAGGGACTGGCTGCTATTGGG-3′. PCR primers for the excision product were 5′-GTTGGGTCGTTTGTTCGGAT-3′ and 5′-CGCGCAATTAACCCTCACTA-3′, and those for the neo fragment were 5′-AATGAACTGCAGGACGAGGC-3′ and 5′-ATGGATACTTTCTCGGCAGG-3′. Four attograms to 2.0 pg and 0.2 pg to 2.0 ng of control plasmids carrying the target sequences were used as standards for the excision product and the neo fragment, respectively. Amplification conditions were 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 10 s. The excision product and the neo fragment were quantified on the basis of the standard curves, and the amount of the excision product was divided by the amount of the neo fragment for normalization.
Excision assay of stably integrated transposon.
For the excision assay of the stably integrated transposon, 2.0 × 105 targeted ES cell clones or 1.0 × 106 MEL cell clones were transfected with 2.0, 0.67, 0.22, 0.074, 0.025, and 0.0082 μg of pSB10, and genomic DNAs were extracted with a DNeasy kit 48 h posttransfection. One microgram of genomic DNA was analyzed by PCR with the primers LCB2XL2 (5′-TTCCAAAAGAAGTAGAGTGGAGAACCAGTG-3′) and PGK2 (5′-AGGCCACTTGTGTAGCGCCAAGT-3′) for the targeted ES cell clone and with the primers TgTP-1U and TgTP-2L for MEL cell clones. Nested PCR with 1 μl of the first PCR product as a template was performed with the primers LCB2XL1 (5′-CCAACCAAATACATTTAACATATTCTAGGT-3′) and PGK4 (5′-GCTGCTAAAGCGCATGCTCCAGACTG-3′) for the targeted ES cell clone and with the primers TgTP-2U (5′-TCTATCGCCTTCTTGACGAGTTCTTCTGAG-3′) and TgTP-3L (5′-CAAGCGCGCAATTAACCCTCACTAAAGG-3′) for MEL cell clones. PCR conditions were 95°C for 15 min, followed by 30 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 7 min. To quantitate the expression level of the SB transposase, total RNA was extracted by using TRIzol (Invitrogen) in accordance with the manufacturer's protocol and treated with RQ1 RNase-Free DNase (Promega). One microgram of RNA was reverse transcribed with SuperscriptII (Invitrogen) by using random hexamer primer in a total volume of 20 μl in accordance with the manufacturer's protocol, and 2 μl of the reaction product was analyzed by PCR. PCR primers were 5′-AATAGAACTGTTTGGCCATAATGACCATCG-3′ and 5′-ATCCACATAATTTTCCTTCCTCATG-3′ for the amplification of the SB transposase gene and 5′-CAGGGTGTGATGGTGGGAATGGGTCAGAAG-3′ and 5′-TACGTACATGGCTGGGGTGTTGAAGGTCTC-3′ for the amplification of the β-actin gene that was used as an internal control. PCR conditions were the same as those used for the amplification of the excision product except that the number of cycles was 30 for the SB transposase gene and 18 for the β-actin gene.
SB transposase- and Tc3-mediated integration of promoter trap transposon.
For SB transposase-mediated integration of the promoter trap transposon, 2.0 × 105 ES cells were transfected with 1.6 μg of methylated or unmethylated pTransSAβ-geo together with 0.4 μg of pSB10 by using TransFast and plated into 1 well of a 24-well plate. The transfected cells were then transferred to a 10-cm dish 48 h posttransfection and selected for 7 days with 150 μg of G418/ml, after which the colonies were Giemsa stained and counted. Several G418-resistant clones were expanded, and their DNAs and RNAs were extracted. The flanking sequences of integration sites were determined by ligation-mediated PCR as described previously (16) and were analyzed by using the Ensembl mouse genome database (version 16.30.1). Promoter trap events were verified by reverse transcription (RT)-PCR with the transposon-specific primer β-geo (5′-TGCCAGTTTGAGGGGACGACGACAGTATCG-3′) and the gene-specific primers 5′-TGGAGTGAGCTAGAATCAGAAAGATGACAC-3′ for M2S, 5′-GACTTTCAAGACCTTCGACGCACCGTTCAC-3′ for M2L, 5′-TCTTCAGCCACAGGCTCCCAGACATGACAG-3′ for M4, and 5′-GATATGAAGAGCTGTCAGTTTGTAGCAGTC-3′ for N1. PCR products were directly sequenced. For Tc3-mediated integration of the promoter trap transposon, 2.0 × 105 ES cells were transfected with 1.0 μg of methylated or unmethylated pTc3/SAβ-geo together with 1.0 μg of pRP2302 (10), an expression vector of Tc3 transposase, by using TransFast. Transfected cells were passaged and selected by G418 under the conditions described above.
Protein expression and EMSA.
The N123 peptide containing the IR/DR recognition domain was expressed and purified as described previously (19). Oligonucleotide sequences for oligonucleotide probes corresponding to the outer binding site of IR/DR-L were 5′-TACAGTTGAAGTCGGAAGTTTACATACACTTAAG-3′ (Unmet-U) and 5′-CTTAAGTGTATGTAAACTTCCGACTTCAACTGTA-3′ (Unmet-L) for unmethylated probes and 5′-TACAGTTGAAGTOGGAAGTTTACATACACTTAAG-3′ (Met-U) and 5′-CTTAAGTGTATGTAAACTTCOGACTTCAACTGTA-3′ (Met-L) (where “O” represents 5-methyl cytosine) for methylated probes. The oligonucleotides were first labeled with [γ-32P]ATP by using T4 polynucleotide kinase and then annealed. A DNA fragment of IR/DR-L was prepared by digestion of pBS-IR/DR-L with HindIII and KpnI following methylation with SssI methylase or no methylation, and this fragment was then treated with alkaline phosphatase. The digested fragment was extracted with phenol-chloroform, precipitated with ethanol, and finally labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Nucleoprotein complexes were formed in a buffer described previously (19) in the presence of 1.0 μg of poly(dI-dC), 0.2 pmol of the oligonucleotide probes or 100 pg of the IR/DR-L fragment, and 1.0 μl of N123. After 30 min of incubation at 25°C, samples were loaded onto 5% native polyacrylamide gels, which were run for 1.5 h for the oligonucleotide probe and for 2.5 h for the IR/DR-L fragment at a constant voltage of 150 V in 0.5× Tris-borate-EDTA. After the gel was dried, the nucleoprotein complexes were visualized by autoradiography.
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed with the ChIP assay kit (Upstate) according to the manufacturer's protocol. Antibodies used in this study were anti-acetyl H3 (Upstate) and anti-trimethylated H3K9 (Abcam). Precipitated DNAs were analyzed at multiple loci by PCR with the primers 5′-CCTTGTACGGGTTGGTGGAGGTCAC-3′ and 5′-CGCCACTCGAACAGGTGGACAATAG-3′ for the amylase 2.1 gene, 5′-TGCGAGGATAAGAACAGACACTAC-3′ and 5′-ACAGACTCAGAAGCAAACGTAAGA-3′ for the β-minor globin gene, EGFP-1U and EGFP-1L for the EGFP gene; 5′-GCACGGGTGTTGGGTCGTTTGTTC-3′ and 5′-CTTCTAAAGCCATGACATCATTTTCTG-3′ for IR/DR-L, and 5′-GAAGGCTACTCGAAATGTTTGACCCAAG-3′ and 5′-CAAGCGCGCAATTAACCCTCACTAAAGG-3′ for IR/DR-R. PCR conditions consisted of 95°C for 15 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 7 min with the HotStarTaq system. To allow semiquantitative analysis, input DNA was used at 1, 0.2, 0.04, and 0.008 μl per reaction and precipitated DNAs were used at 1 and 0.2 μl per reaction except for EGFP, IR/DR-L, and IR/DR-R in the fraction of anti-trimethylated H3K9, for which amounts of 5 and 1 μl per reaction were used.
RESULTS
Correlation of transposition efficiency with SB transposon methylation.
To find an explanation for the difference in transposition efficiencies between mouse germ cells and ES cells, we considered two possible lines of investigation, one related to transposase and the other related to the transposon. The first possibility is that the expression level of the SB transposase was not optimal in ES cells. In fact, it was shown recently that an optimal expression level of the SB transposase is required to achieve a high frequency of transposition and that overexpression of the SB transposase reduces transposition efficiency (12, 38). The second possibility is that the SB transposon region undergoes different epigenetic modifications in mouse germ cells and ES cells, resulting in more efficient transposition of the SB transposon in mouse germ cells.
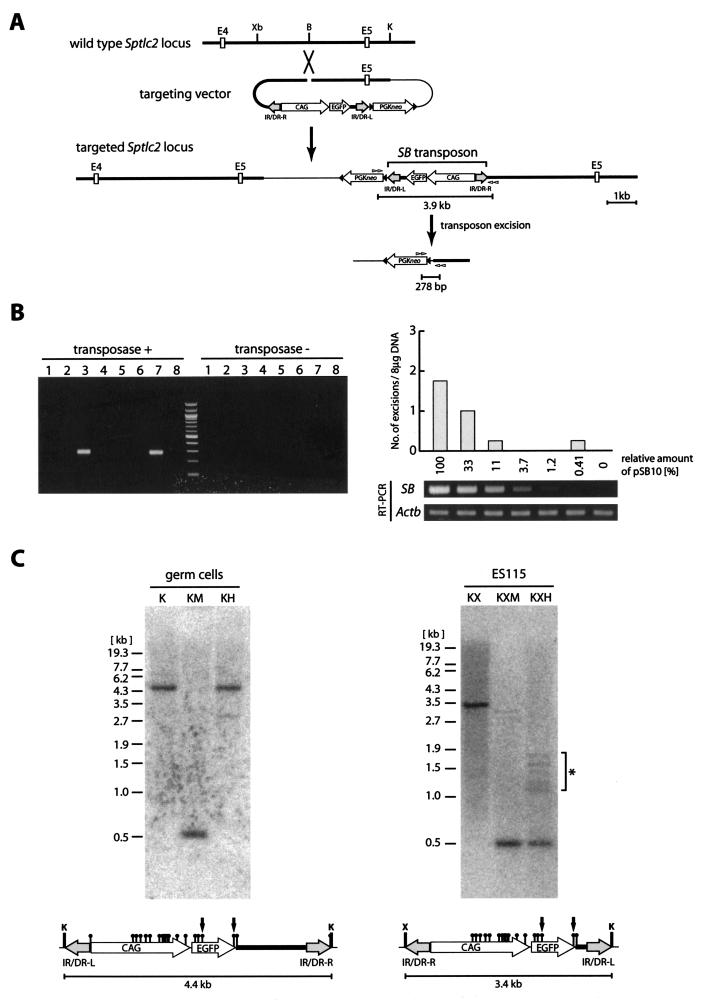
To test the first possibility, we generated an ES cell clone designated ES115 containing a single copy of the SB transposon at the Sptlc2 locus by gene targeting (Fig. 1A). Targeted ES cells (2.0 × 105 cells) were transfected with the SB transposase expression vector pSB10 (19). We routinely transfected more than 80% of the ES cells, as measured by transient enhanced green fluorescent protein (EGFP) expression. Forty-eight hours after transfection, transposon excision was analyzed by nested PCR. With 1 μg of genomic DNA as a template for each reaction, two out of eight PCRs were shown to be positive (Fig. 1B). Since a single molecule of the excision product could be detected under the PCR conditions used in this study (data not shown) and since 1 μg of genomic DNA corresponds to approximately 1.5 × 105 cells, this result indicates that only about two excision events occurred in approximately 1.2 × 106 cells. This frequency did not increase when the amount of pSB10 was serially reduced to 0.41% by using threefold decrements (Fig. 1B). This result demonstrates that the low frequency of transposon excision in ES cells, which is consistent with the results of a previous study (25), is not caused by an inadequate SB transposase expression level.
FIG.1.
High frequency of transposition is related to CpG methylation. (A) Introduction of a single copy of the SB transposon into the Sptlc2 locus by insertional-type homologous recombination and detection of transposon excision by PCR. Open boxes, exons; black triangles, loxP sites; gray and white arrowheads, nested PCR primers; thin line, plasmid backbone sequence; CAG, CAG promoter; IR/DR-R and IR/DR-L, right and left IR/DRs, respectively; Xb, XbaI; B, BamHI; K, KpnI. (B) Excision of the SB transposon at the Sptlc2 locus in ES cells. The ES115 clone with targeted integration of the SB transposon at the Sptlc2 locus (shown in panel A) was transfected with serially diluted pSB10 expressing the wild-type SB transposase (transposase +) or pSB10-ΔDDE (19) expressing the inactive SB transposase with the DDE box deleted (transposase −) and screened by nested PCR as shown in panel A. Four independent transfections were performed for each dilution factor of pSB10, and 8 μg of genomic DNA was screened in each transfection by using 1 μg of genomic DNA per reaction in eight independent PCRs. A representative PCR result with the maximum amount of pSB10 (2 μg) is shown in the left panel. The average number of positive PCRs for each transfection and results of RT-PCR analysis of the expression level of the SB transposase are shown in the right panel. Actb, β-actin. (C) Southern blot analysis of methylation status within transposon sequences in male mouse germ cells and ES cells. EGFP was used as a probe. Genomic DNA from germ cells was not digested with HpaII, indicating that the sites were methylated. On the other hand, the genomic DNA from the ES115 clone was almost completely digested. The presence of faint bands, shown by an asterisk, indicates that a small fraction of HpaII sites have been methylated in ES cells. Black circle, HpaII (H) or MspI (M) site; X, XhoI site. The 0.5-kb bands in the KM, KXM, and KXH lanes come from the HpaII-MspI fragment indicated by arrows.
To test the second possibility, epigenetic modification of the SB transposon region was assessed. We reasoned that the SB transposon DNA might be heavily methylated in mice because a previous study demonstrated that the expression of the EGFP reporter carried by the transposon is repressed prior to transposition (16). Methylation status within the transposon sequence in male germ cells and ES cells was examined by using the methylation-sensitive restriction enzyme HpaII. As shown in Fig. 1C, double digestion of germ cell DNA with KpnI and HpaII yielded a 4.4-kb fragment which corresponded to the 4.4-kb KpnI fragment, while triple digestion of ES115 DNA with KpnI, XhoI, and HpaII produced a 0.5-kb fragment, which was also produced when MspI was used instead of HpaII. This finding indicates that the HpaII sites within the transposon region were nearly completely methylated in mouse germ cells but not in ES cells. Our results suggest that there is a correlation between transposition frequency and CpG methylation of the transposon sequence and a possibility that CpG methylation may enhance transposition frequency.
Effect of methylation on excision of transiently transfected transposon DNA.
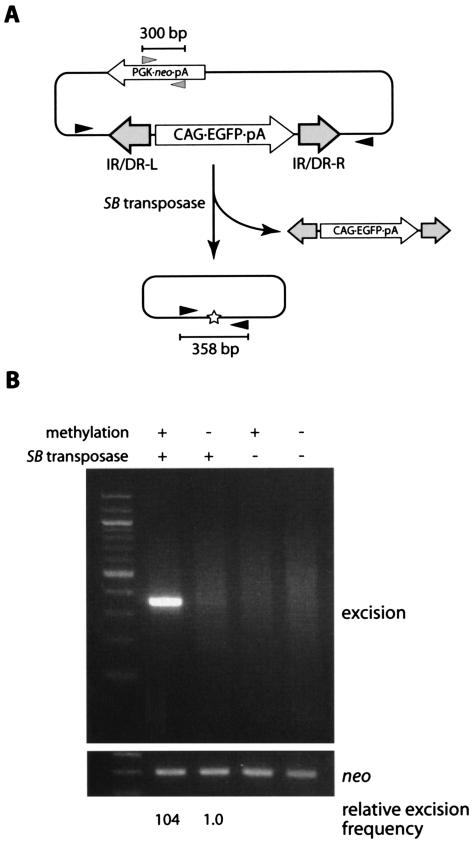
In order to investigate whether CpG methylation enhances transposition efficiency, we first compared the excision frequencies of transiently transfected transposon DNAs with and without methylation. The transposon vector pTransCX-EGFP:Neo, which showed a high transposition frequency in mice in a previous study (16), was methylated in vitro with SssI CpG methylase. Methylation was verified by complete resistance to digestion with CpG methylation-sensitive restriction enzymes (data not shown). Methylated or unmethylated transposon vectors were then introduced into 1.0 × 106 MEL cells together with the SB transposase expression vector or pBS as a negative control. The MEL cell line was chosen in order to test the effect of methylation at defined chromosomal loci, as described below. Cells transfected with the unmethylated vector showed the GFP signal under fluorescence microscopy, but cells transfected with the methylated vector did not, indicating that the activity of the CAG promoter was blocked by CpG methylation (data not shown). Total DNAs were extracted 48 h after transfection and used as PCR templates to detect excision. A 358-bp PCR band can be expected when the transposon is excised from the plasmid (Fig. 2A). A larger quantity of the PCR product was obtained from cells transfected with the methylated plasmid and the SB transposase expression vector (Fig. 2B), indicating that the methylated SB transposon was excised from plasmid DNA at a higher frequency. Quantification of the excision product by real-time PCR (see Materials and Methods) indicated that the enhancement of the excision reaction was approximately 100-fold (Fig. 2B).
FIG. 2.
Effect of methylation on the excision of the transiently transfected transposon. (A) Schematic representation of excision assay. Methylated or unmethylated transposon plasmids were cotransfected with the SB transposase expression vector or pBluescript as a negative control. After 48 h, whole DNA was extracted and PCR was performed. Excised plasmids yielded a 358-bp PCR product. Black and gray arrowheads, PCR primers; star, footprint; pA, polyadenylation signal. (B) High excision frequency of the methylated SB transposon. While the unmethylated plasmid with the transposase yielded a faint 358-bp band, the methylated plasmid with transposase produced strong amplification. The neo fragment was amplified as an internal control for transfection efficiency. The first lane contains the 100-bp ladder. The relative frequency of excision was estimated by real-time PCR (see Materials and Methods).
Effect of methylation on excision of stably integrated transposon DNA.
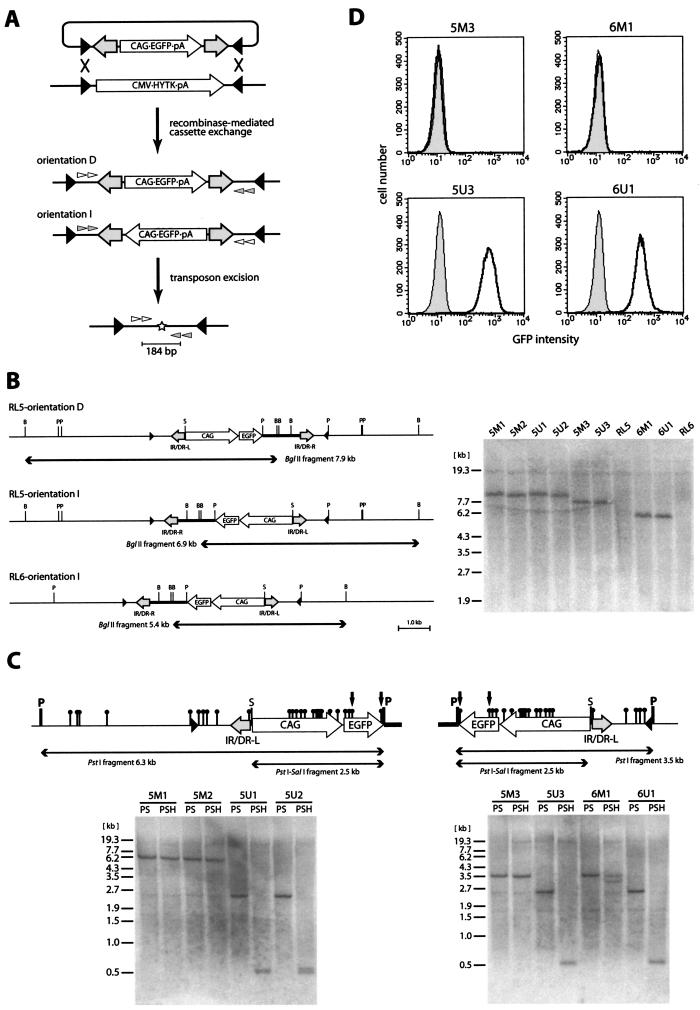
Although the excision assay of the transiently transfected transposon demonstrated that excision was enhanced by CpG methylation, the same effect could not be directly extrapolated to a stably introduced transposon. In fact, it was shown that the frequency of SB transposition was affected by the chromosomal location of the SB transposon (25). The elimination of the position effect is thought to be critical for determining transposition efficiency in a chromosomal context. To study the effect of methylation on a stably introduced transposon, we used RMCE to generate cells bearing a methylated or unmethylated transposon at the same locus (8, 32). RMCE makes it possible to introduce various DNA cassettes flanked by inverted lox511 sites into the defined genomic loci harboring a HYTK fusion gene flanked by the same configuration of inverted lox511 sites (Fig. 3A) and therefore eliminates the position effect on transposition efficiency. MEL cell clones RL5 and RL6, containing a recipient locus at different chromosomes, were used because it was previously demonstrated that the methylation status of the plasmid DNA methylated in vitro was maintained at both loci (32).
FIG.3.
Generation of cells carrying the methylated or unmethylated SB transposon at the defined genomic loci. (A) Schematic representation of the procedure to investigate the effect of CpG methylation on transposon excision at the defined genomic loci. The transposon flanked by inverted lox511 sites was either methylated or unmethylated and introduced with the Cre expression vector into MEL cell clone RL5 or RL6 in order to carry out RMCE. Targeted cells could be selected with ganciclovir due to a loss of HYTK gene expression. After ganciclovir selection, exchanged clones were isolated and then transfected with the SB transposase expression vector. Finally, extracted genomic DNAs were analyzed by nested PCR. The SB transposon was integrated in two different orientations, orientation D (direct) and orientation I (inverted). Black triangles, lox511 sites; white and gray arrowheads, nested PCR primers; star, footprint; HYTK, hygromycin phosphotransferase and HSV-thymidine kinase fusion gene. (B) Southern blot analysis of cassette exchange. The restriction maps are shown on the left for each combination of parental cell lines (RL5 and RL6) and transposon orientations (orientations D and I). EGFP was used as a probe. B, BglII; P, PstI; S, SalI. Clones 5M1 to 5M3 and clones 5U1 to 5U3 were obtained by transfection of RL5 with the methylated and unmethylated transposons, respectively. Clones 6M1 and 6U1 were obtained by transfection of RL6 with the methylated and unmethylated transposons, respectively. (C) Southern blot analysis of methylation status. Detailed restriction maps within the PstI fragment (see panel B) are shown above the panels. The left panel shows the result of RL5-derived orientation D, and the right panel shows the results of RL5- and RL6-derived orientation I. EGFP was used as a probe. Black circles, HpaII sites. Resistance to SalI and HpaII digestion demonstrates the methylation of corresponding restriction sites. The 0.5-kb bands in the PSH lanes come from the HpaII fragment indicated by arrows. (D) Fluorescence-activated cell sorter analysis of EGFP expression in the targeted clones. Gray area, wild-type cells; thick lines, targeted clones.
pL1TransCX-GFP1L, an RMCE vector containing the transposon flanked by the inverted lox511 sites, was introduced into RL5 and RL6 in either the methylated or the unmethylated form together with the Cre expression vector. Ganciclovir-resistant clones were isolated by limiting dilution and prescreened by PCR (see Materials and Methods). Some of the PCR-positive clones were further analyzed by means of Southern blotting. All clones analyzed had either of the expected two orientations of transposon integration shown in Fig. 3B. In order to examine the methylation status of transposon DNA, genomic DNAs were analyzed with the CpG methylation-sensitive enzymes HpaII and SalI (Fig. 3C). In both RL5- and RL6-derived clones transfected with the methylated transposon, neither HpaII nor SalI sites were digested, indicating that the methylation status had been maintained. In contrast, both HpaII and SalI sites were completely digested in all clones transfected with the unmethylated transposon, demonstrating that the unmethylated state had been maintained. Consistent with these results, the expression of the EGFP reporter located within the transposon vector was suppressed in cells containing the methylated transposon but not in those containing the unmethylated transposon (Fig. 3D). Both the methylated and the unmethylated statuses of the transposon regions were maintained for at least 10 weeks after the RMCE reaction.
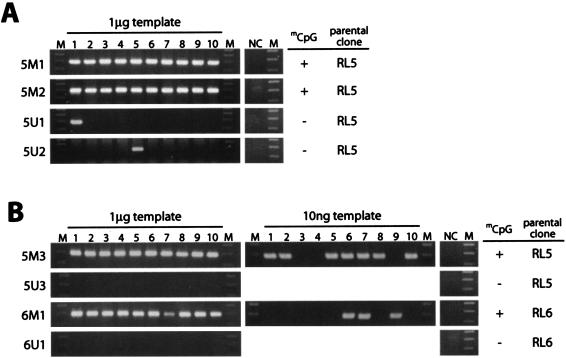
To determine the effect of CpG methylation on transposon excision, we transfected the clones with the SB transposase expression vector and performed PCR to detect excision 48 h after transfection (Fig. 4A and B). In clones containing the unmethylated transposon, excision was detected in 1 out of 10 PCRs (clones 5U1 and 5U2) or in none of the PCRs (clones 5U3 and 6U1) when 1 μg of genomic DNA was used as a template. This frequency was similar to that observed for the ES cells containing the unmethylated transposon at the Sptlc2 locus (Fig. 1B). On the other hand, in clones containing the methylated transposon, excision was detected in all 10 reactions (clones 5M1, 5M2, 5M3, and 6M1). When genomic DNA for the first PCR was diluted, 10 ng of genomic DNA from clones containing the methylated transposon still yielded positive signals (Fig. 4B, clones 5M3 and 6M1). The methylated transposon consistently showed a higher frequency of excision when the amount of the SB transposase expression vector was serially reduced to about 10% by using threefold decrements (data not shown), indicating that the expression level of the SB transposase is not responsible for the difference in excision frequencies. This result indicates that the efficiency of transposon excision is at least 100-fold higher in the methylated transposon. This result is also consistent with the ∼100-fold increase in excision efficiency in the transiently transfected methylated transposon (Fig. 2B).
FIG. 4.
High frequency of excision of the methylated SB transposon at the defined genomic loci. One microgram or 10 ng of genomic DNAs from the clones shown in Fig. 3 was used as a template for nested PCR. Ten PCRs were carried out for each clone. (A) Orientation D; (B) orientation I. NC, nontransfected genomic DNA as a negative control; M, 100-bp ladder. Methylation statuses of the transposon regions and parental clones are shown on the right.
Effect of methylation on overall transposition.
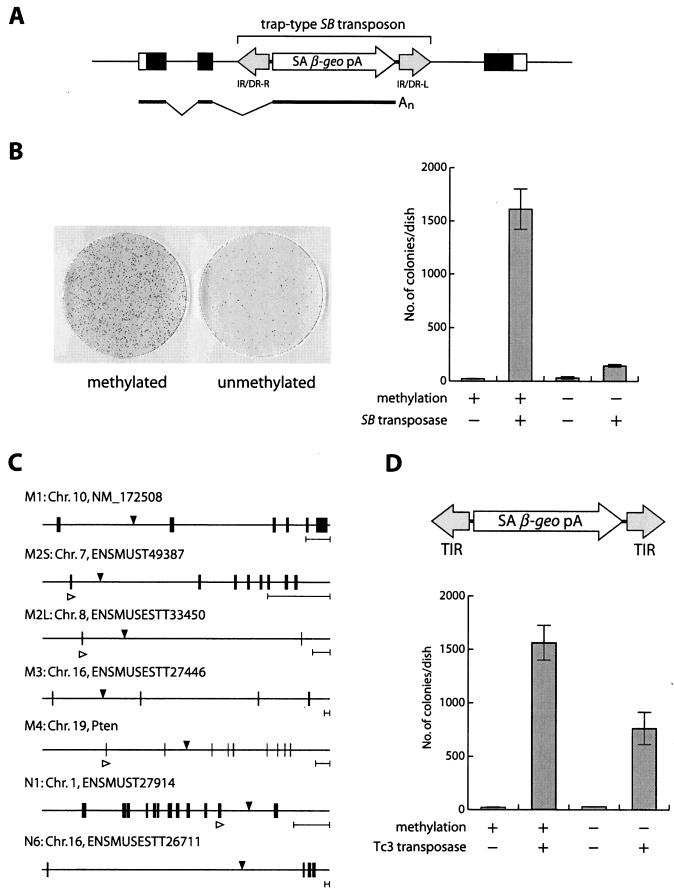
As described above, excision of the transposon from genomic loci, the first step of the transposition reaction, was shown to be enhanced by CpG methylation. The excised transposon fragment needs to be reintegrated into the genome to complete the transposition reaction. We therefore compared methylated and unmethylated transposons in terms of the number of transposons integrated into the genome. Since CpG methylation inhibits promoter activity, the expression cassette of selection markers within a transposon could not be utilized to detect transposition into the genome. Instead, we employed a gene trap scheme (11) by constructing a gene trap-type transposon vector, pTransSAβ-geo, which contained a splice acceptor site upstream of β-geo (Fig. 5A). When the transposon is integrated into an active gene, a chimeric fusion transcript consisting of an endogenous gene and β-geo is thought to be generated. If β-geo is in frame with the trapped gene, cells become resistant to G418 due to the expression of β-geo (Fig. 5A). The methylated or unmethylated transposon vector was introduced into ES cells together with the SB transposase expression vector and selected with G418. The methylated transposon produced 1,600 colonies, while the unmethylated transposon yielded 140 colonies (Fig. 5B). This result indicates that the efficiency of overall transposition, which includes both excision and integration, is enhanced 11-fold in the methylated transposon. Transposon insertion sites were determined by ligation-mediated PCR, and gene trap events were verified in randomly selected clones by RT-PCR analysis (Fig. 5C), and the results obtained indicated that vector methylation did not affect gene trap selection.
FIG. 5.
Integration of methylated SB and Tc3 transposons. (A) Schematic representation of the gene trap with the SB transposon. Open boxes, untranslated regions; closed boxes, coding regions; thick line, mRNA; SA, splice acceptor site; β-geo, β-galactosidase and neomycin phosphotransferase fusion gene. (B) Effect of CpG methylation on SB transposition. Giemsa staining of ES colonies is shown on the left. Average numbers of G418-resistant colonies per dish for three independent transfections are presented on the right. Error bars indicate standard deviations. (C) Structures of trapped genes and transposon integration sites. Integration sites are indicated with black arrowheads. Clones M1 to M4 were derived by transfection with the methylated transposon, and clones N1 and N6 were derived from the unmethylated transposon. Chromosome numbers, Ensembl gene designators, and gene names are also shown. One integration site was identified in each clone except clone M2, in which two integration sites, designated M2S and M2L, were characterized. Accurate splicing in some clones (M2S, M2L, M4, and N1) was confirmed by RT-PCR with primers for the upper exon (white arrowheads) and transposon. Black boxes, exons. Five-kilobase scale bars are shown on the right. (D) Effect of methylation on Tc3 transposition. The structure of the trap vector is shown at the top. TIR, terminal inverted repeat. Average numbers of G418-resistant colonies per dish for three independent transfections are presented at the bottom.
To test the effect of methylation in a different transposon system, we constructed another gene trap-type vector, pTc3/SAβ-geo, by using the Tc3 transposon derived from C. elegans (9, 10) and introduced this vector with or without methylation into ES cells together with the Tc3 transposase expression vector. CpG methylation increased gene trap events twofold (Fig. 5D). This result indicates that the enhancement of transposition by CpG methylation is seen in other transposon systems, although the extent of enhancement varies.
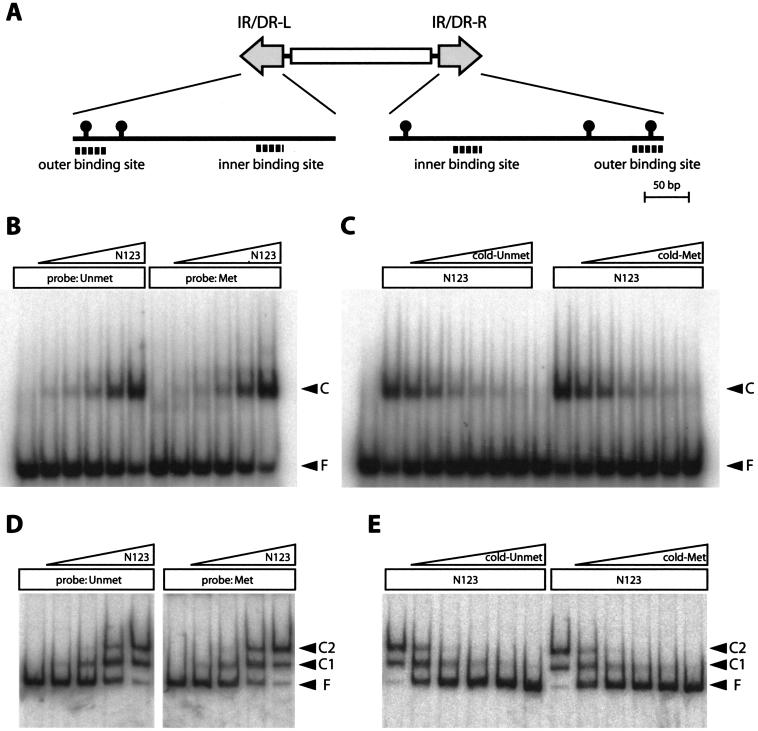
Unaltered affinity of the SB transposase DNA binding domain to naked methylated IR/DR.
To elucidate the mechanism of enhanced transposition by CpG methylation, we compared the affinities of the recombinant SB transposase peptide to methylated and unmethylated IR/DRs by EMSA. Both the right and the left IR/DRs contain two binding sites for SB transposase (Fig. 6A). Since only the outer binding sites contain the CpG sequence (Fig. 6A), the left outer binding site was used as a probe and competitor. The first 123 amino acids of the SB transposase (N123), which was previously reported to contain an IR/DR recognition domain (19), was expressed in Escherichia coli, purified via a C-terminal histidine tag, and used to make a nucleoprotein complex. The intensities of the shifted bands when unmethylated or methylated binding sequences were used as probes were comparable (Fig. 6B). No difference was observed when the unmethylated site or the methylated site was used as a competitor against a labeled unmethylated probe (Fig. 6C). The same experiments were performed with the entire region of the left IR/DR fragment as a probe and competitor, and no effect of methylation was observed (Fig. 6D and E). These results indicate that CpG methylation of IR/DR does not alter the direct binding of the N123 peptide to naked IR/DR, suggesting that the native chromatin structure at the transposon region may need to be considered to understand the enhancement of transposition.
FIG. 6.
Unaltered affinity of the SB transposase DNA binding domain to naked methylated IR/DR. (A) Schematic representation of CpG sites and transposase binding sites in IR/DRs. Black circles, CpG sites. (B to E) EMSA with the recombinant SB transposase peptide (N123). (B and C) Results obtained with 34-bp outer binding site of IR/DR-L; (D and E) results obtained with 300-bp IR/DR-L fragment. (B) The unmethylated or methylated outer binding site was mixed with increasing concentrations of N123 peptide (1,600- to 100-fold dilutions of the purified peptide) for nucleoprotein complex formation. (C) The unmethylated outer binding site was labeled as a probe, and the unmethylated or methylated outer binding site at increasing concentrations (1- to 50-fold molar excess of the probe) was used as a competitor. (D) An unmethylated or methylated IR/DR-L fragment was mixed with increasing concentrations of N123 peptide (5,100- to 160-fold dilutions of the purified peptide) for nucleoprotein complex formation. (E) An unmethylated IR/DR-L fragment was labeled as a probe, and the unmethylated or methylated unlabeled IR/DR-L fragment at increasing concentrations (500- to 8,000-fold molar excess of the probe) was used as a competitor. Unmet, unmethylated; Met, methylated; F, free probe; C, complex. Complex 1 (C1) and C2 represent the binding of one and two molecules of N123 peptide per IR/DR, respectively (19).
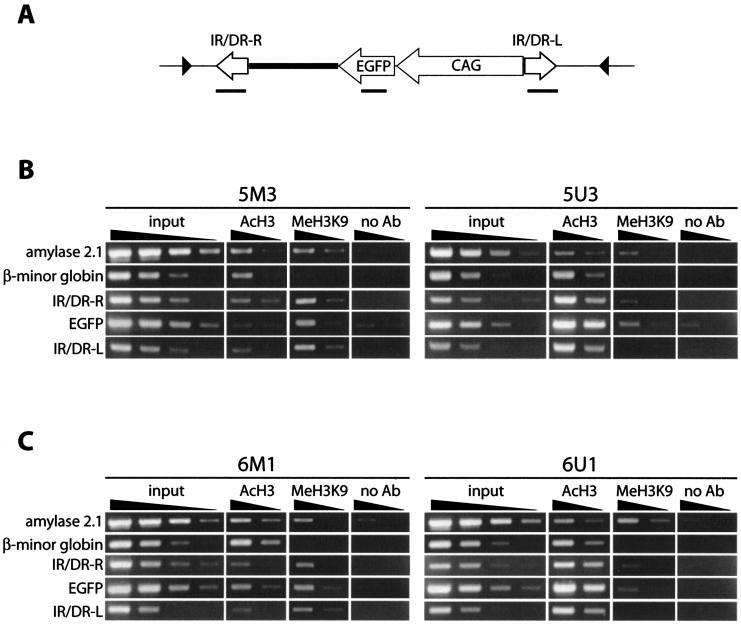
Chromatin structure at the methylated and unmethylated transposons.
DNA recombination reactions can be affected by conformational changes of the DNA substrate. For example, the E. coli protein HU stimulates recombination efficiency in bacteriophage Mu by bringing together transposase binding sites (23). A similar effect of the high-mobility group B1 (HMGB1) protein on SB transposition was reported recently (42). We therefore used the ChIP assay to investigate the chromatin structure at the methylated and unmethylated transposon regions in MEL cell clones. It has been demonstrated that modifications of the histone tail play crucial roles in the organization of higher-order chromatin structures and the regulation of gene expression. Recent studies have shown that acetylated histone H3 is located on euchromatin, in which DNA is loosely packed, and that histone H3 methylated at the lysine-9 residue (H3K9) is localized on heterochromatin, in which DNA is condensed (14). We therefore used antibodies recognizing acetyl H3 or trimethylated H3K9 for the ChIP assay and compared the chromatin structures of methylated and unmethylated transposon regions at two different loci by using RL5- and RL6-derived clones (Fig. 7). As internal controls for the ChIP assay, the pancreatic amylase 2.1 gene was used as a heterochromatic marker and the β-minor globin gene was used as a euchromatic marker (5, 31). As shown in Fig. 7, the quantity of the amylase 2.1 gene in the fraction enriched for trimethylated H3K9 relative to that enriched for acetyl H3 was significantly higher than that of the β-minor globin gene, consistent with the expectation that the amylase 2.1 gene and the β-minor globin gene are located in the heterochromatin and the euchromatin, respectively. The enrichment of transposon sequences (IR/DR-R, EGFP, and IR/DR-L) in the fraction of acetyl H3 was observed in clones containing the unmethylated transposon (5U3 and 6U1). In contrast, the enrichment in the fraction of trimethylated H3K9 was observed in clones containing the methylated transposon (5M3 and 6M1). This enrichment closely correlates with the expression pattern of the GFP reporter (Fig. 3D). These results show that the methylated and unmethylated transposon regions formed heterochromatin and euchromatin, respectively.
FIG. 7.
ChIP assay of the methylated or unmethylated transposon at the defined loci. (A) Region of the SB transposon analyzed in the ChIP assay. PCR-amplified regions are shown as thick lines below each component. Black triangles, lox511 sites. (B and C) PCR analysis of precipitated DNAs. Clones 5M3 and 5U3 (B) and clones 6M1 and 6U1 (C) were derived from parental clones RL5 and RL6, respectively, as shown in Fig. 3 and 4. Fivefold serial dilutions of input and precipitated DNAs were analyzed. AcH3, anti-acetyl H3; MeH3K9, anti-trimethylated H3K9; no Ab, no-antibody control. Amylase 2.1 and β-minor globin were used as respective controls for the heterochromatic and euchromatic regions. Although the intensities of the bands of amylase 2.1 and β-minor globin are almost the same in the anti-acetyl H3 fraction of 5M3, the higher amplification efficiency of the amylase 2.1 sequence in the input DNA indicates that the β-minor globin sequence is enriched in this fraction compared to the amylase 2.1 sequence. Faint bands seen in the lanes with the no-antibody control come from nonspecific binding of genomic DNA to the protein A agarose beads used for precipitation. Quantification was performed twice, and a representative result is presented.
DISCUSSION
In order to identify gene functions in mammalian contexts, a large number of mutant mice have been generated by means of gene targeting in the last decade. Although this reverse-genetics approach has provided direct information about gene functions, a large-scale phenotype-driven genetic screen is also required to understand the complex mammalian genome. The recent development of N-ethyl-N-nitrosourea mutagenesis has produced many mutant mice with a wide range of phenotypes (18, 29). However, the identification of genes responsible for these phenotypes is often difficult and time-consuming. Transposon-tagged mutagenesis is another approach to produce mutants and has proven to be an effective method with model organisms. The SB transposon system was recently developed as an active transposon system for vertebrates (19) and has been used for mice as a tool for germ line mutagenesis (3, 17) and transgenesis (6) and as a promising vehicle in gene therapy (38, 39). For such applications, however, a highly active transposon is apparently required. Efforts to improve the SB transposon system have so far resulted in a three- to fourfold increase in transposition efficiency compared to that of the original system (4, 12). We assumed that the difference in transposition efficiency between mouse germ cells (7, 10, 16) and ES cells (25) implies the presence of regulatory mechanisms of SB transposition and that an understanding of this mechanism would provide a key to the further improvement of transposition efficiency.
Chromosomal transposition was not affected by overexpression of the SB transposase.
We first examined the possibility that the expression level of the SB transposase was not adequate in ES cells but was optimal in germ cells. In fact, a recent study clearly showed that overexpression of the SB transposase reduced transposition frequency (12, 38). To attain the expression of the SB transposase at different concentrations, we transfected an ES cell clone carrying a single copy of the transposon with the serially diluted SB transposase expression vector. However, no significant increase in excision frequency was observed (Fig. 1B). The difference between the assay used in our study and that used in the above-mentioned study for the detection of the transposition reaction may account for this finding; our assay was designed to detect the excision of the transposon on the chromosome, whereas the effect of the overexpression of the SB transposase was observed in the transposition from plasmids into the genome. We therefore concluded that the low efficiency of chromosomal transposition in ES cells was not caused by the overexpression of the SB transposase.
Enhancement of SB transposition by DNA methylation and relationship with heterochromatin formation.
We next focused on the relationship between the epigenetic status of transposon DNA and transposition efficiency and clearly demonstrated that CpG methylation of the SB transposon increases transposition efficiency. A lack of enhancement of the direct binding of the N123 peptide to a naked methylated IR/DR fragment (Fig. 6) suggested that the mechanism of enhanced transposition needed to be studied in the context of native chromatin structure. We therefore performed a ChIP assay and found that the methylated transposon was located in the heterochromatic region. Heterochromatin is thought to function in gene regulation and higher-order chromatin structures (14). Heterochromatin and DNA methylation were also thought to suppress the mobilization of transposable elements, both DNA transposons and retrotransposons, by inhibiting the expression of catalytic enzymes for transposition (15, 26, 35, 41). In contrast, our observations indicate that heterochromatin formation facilitates the transposition reaction when transposase is supplied in trans. This finding suggests the possibility that transposition may be enhanced in heterochromatic regions even in organisms that do not have CpG methylation. In this respect, it is interesting that enhanced transposition was also observed in the Tc3 transposon system of C. elegans, whose genome does not contain CpG methylation (Fig. 5D). Our findings may imply that Tc3 transposition is enhanced in C. elegans when Tc3 is located in heterochromatic regions. Our observation may also have the evolutionary implication that the host genome was protected from transposon-induced insertional mutations partly by the repression of transposase expression through heterochromatinization, whereas this effect was counterbalanced by the enhancement of transposition efficiency as shown in our study, allowing the persistence of the transposon sequence in the genome. Our findings may also explain the following observation: in our study of mice, although the SB transposition occurs frequently at donor sites in which the transposon DNA is heavily methylated, loses its GFP reporter expression, and probably forms heterochromatin, it is difficult to remobilize the transposon once it has been transposed, possesses GFP expression, and probably forms euchromatin (K. Yae, K. Horie, and J. Takeda, unpublished observation).
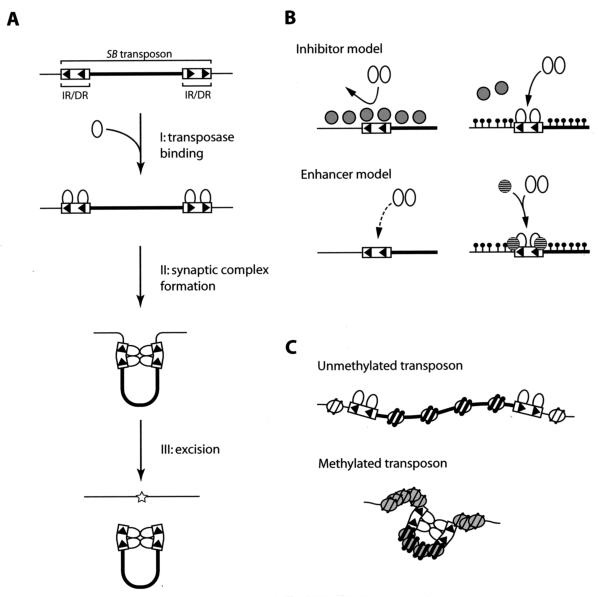
How does heterochromatin formation enhance SB transposition?
We hypothesized that heterochromatin plays a role during either the transposase binding step, the synaptic complex formation step, or both (Fig. 8A). In the model proposed for the transposase binding step, the presence of an inhibitor(s) or an enhancer(s) for the binding of the SB transposase to IR/DR is assumed (Fig. 8B). The putative inhibitor has high affinity to euchromatic DNA but low affinity to heterochromatic DNA, which is highly methylated. Thus, the SB transposase can bind to heterochromatic IR/DRs more efficiently, resulting in a higher frequency of transposition (Fig. 8B, top). On the other hand, the putative enhancer specifically binds to heterochromatin and recruits the SB transposase to the binding sites, thus promoting transposition (Fig. 8B, bottom). In the model proposed for the synaptic complex formation step (Fig. 8C), chromatin conformation is assumed to be the determinant for the efficiency of synaptic complex formation. Several reports have indicated that the efficiency of synaptic complex formation determines transposition efficiency. One observation concerns the effect of the distance between IR/DRs: the shorter the distance, the higher the transposition efficiency (12, 20); in other words, the shorter distance facilitates the interaction of the SB transposases binding to each of the IR/DRs. Another finding is that HMGB1 is required for efficient SB transposition (42). HMGB1 bends the DNA strand, and it has been posited that it juxtaposes two IR/DRs as well as two DRs in the same IR closer, thus resulting in the efficient interaction of the SB transposases binding to each of the DRs (42). In the present case, the synaptic complex formation may have been influenced by the different conformations of the transposon DNA in heterochromatin and in the euchromatin. It is difficult for the transposon in euchromatin to form the synaptic complex because of its relaxed chromatin structure (Fig. 8C, top), whereas both IR/DRs are located close together in a condensed heterochromatin, causing the SB transposases which binds to the IR/DRs to interact more frequently to form a stable synaptic complex (Fig. 8C, bottom). This model shows a significant resemblance to the centromeric heterochromatin structure. Among the centromere proteins, Cenp-B, which is similar to the Tc1/mariner family transposase, can specifically bind to the Cenp-B box within the minor satellite in the mouse genome, and Cenp-B is thought to dimerize and form higher-order compacted structures (28, 40). Based on this proposed structure, accessibility of the SB transposase to the heterochromatic region may not be inhibited.
FIG. 8.
Proposed models of enhancement of transposition reaction by CpG methylation and involvement of the heterochromatin structure. (A) Schematic illustration of transposition reaction. After the binding of the transposase to DRs (step I), the transposon region folds to form the synaptic complex (step II) and is excised from the genome (step III). Arrowhead, DR; thin line, genomic sequence; thick line, transposon sequence; oval, transposase; star, footprint. (B) Models of the regulation of transposition reaction during the transposase binding step (step I in panel A). Two models are proposed. In the inhibitor model (top), cellular factors that inhibit the binding of the transposase to DRs dissociate from the genome when the transposon region is methylated. In the enhancer model (bottom), the accessibility of the transposase to IR/DRs is enhanced in the methylated transposon. The transposase may be recruited by a cellular factor that has high affinity to the methylated region. In either model, it is possible that heterochromatinization is not required and that DNA methylation alone is sufficient. Gray circles, inhibitors; striped circles, enhancers; black circles, CpG methylation. (C) A model of the regulation of the transposition reaction during the synaptic complex formation step (step II in panel A). In the unmethylated transposon (top), the transposon region forms a relaxed euchromatin structure and transposases that are bound to each of the IR/DRs interact infrequently. In the methylated transposon (bottom), the transposon region forms a condensed heterochromatin structure and transposases on IR/DRs have more opportunities to interact with each other to form a synaptic complex because of the reduced distance between the IR/DRs. White and gray ovals coiled with DNA represent histones in the euchromatic and heterochromatic regions, respectively.
The enhancement of the efficiency of overall transposition, which includes both excision and integration, was lower than that observed in the excision assay (11-fold [Fig. 5B] versus ∼100-fold [Fig. 2B and 4B]). This result may indicate that the efficiency of integration decreases when transposon DNA is methylated. Alternatively, transcriptional machinery such as RNA polymerases or transcription factors may function as inhibitors (Fig. 8B, top), because the vectors used in the excision assay contain the strong CAG promoter (Fig. 2A and 3A) but the gene trap vector used to measure overall transposition efficiency does not have a transcriptional unit (Fig. 5A). However, the presence of the enhancement even in the gene trap vector lacking a transcriptional unit (Fig. 5A and B) suggests that mechanisms shown in Fig. 8B (bottom) and C also exist.
Other possibilities for efficient transposition in germ cells.
Methylation of the transposon does not fully explain the difference in transposition frequency between mouse germ cells and ES cells. As shown in Fig. 4B, when 10 ng of genomic DNA was used as a template for each of the PCRs, 7 out of 10 PCRs were positive for clone 5M3. According to the Poisson distribution, approximately 6 to 7 reactions out of 10 can be expected to be positive if the target molecule exists at a frequency of 1 molecule per reaction. It can therefore be assumed that one excision event is predicted to exist in 10 ng of genomic DNA in clone 5M3. Since 10 ng of genomic DNA corresponds to approximately 1,500 cells and the transfection efficiency of MEL cells is about 15% based on the transfection of the EGFP reporter, excision frequency is estimated as 1/(1500 × 0.15) = ∼4 × 10−3. This frequency is even higher than that in ES cells but is still much lower than that observed in mouse germ cells (i.e., 0.03 to 0.2 transposition events per transposon per spermatid) (7, 10, 16). A possible explanation for this discrepancy is that there is a specific cellular factor(s) in mouse germ cells that regulates transposition. Another possibility is that the specialized chromatin structure and DNA methylation status in germ cells are more appropriate for the transposition reaction. The chromatin structure changes dynamically in germ cells, as evidenced by the replacement of histones with protamines, and the methylation status of germ cells is also quite different from that of somatic cells (24). Identification and clarification of the mechanism affecting transposition efficiency will be essential for the optimal use of the SB system.
Acknowledgments
We acknowledge E. Bouhassira for providing the RL5 and RL6 cell lines and the pL1CMVEGFP1L vector; S. Osuka for providing the Sptlc2 genomic clone; R. Plasterk for providing the pRP790 and pRP2302 vectors; R. Feil, K. Ura, and V. Keng for comments on the manuscript; and J. Katahira, G. Kondoh, H. Koike, and R. Ikeda for discussions, helpful advice, and technical assistance.
This work was supported by a grant from the New Energy and Industrial Technology Development Organization of Japan, a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and a grant from the Preventure Program, Japan Science and Technology Agency.
REFERENCES
- 1.Bellen, H. J., C. J. O'Kane, C. Wilson, U. Grossniklaus, R. K. Pearson, and W. J. Gehring. 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3:1288-1300. [DOI] [PubMed] [Google Scholar]
- 2.Bird, A. 2001. Molecular biology. Methylation talk between histones and DNA. Science 294:2113-2115. [DOI] [PubMed] [Google Scholar]
- 3.Carlson, C. M., A. J. Dupuy, S. Fritz, K. J. Roberg-Perez, C. F. Fletcher, and D. A. Largaespada. 2003. Transposon mutagenesis of the mouse germline. Genetics 165:243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui, Z., A. M. Geurts, G. Liu, C. D. Kaufman, and P. B. Hackett. 2002. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J. Mol. Biol. 318:1221-1235. [DOI] [PubMed] [Google Scholar]
- 5.Dhar, V., A. I. Skoultchi, and C. L. Schildkraut. 1989. Activation and repression of a beta-globin gene in cell hybrids is accompanied by a shift in its temporal replication. Mol. Cell. Biol. 9:3524-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupuy, A. J., K. Clark, C. M. Carlson, S. Fritz, A. E. Davidson, K. M. Markley, K. Finley, C. F. Fletcher, S. C. Ekker, P. B. Hackett, S. Horn, and D. A. Largaespada. 2002. Mammalian germ-line transgenesis by transposition. Proc. Natl. Acad. Sci. USA 99:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupuy, A. J., S. Fritz, and D. A. Largaespada. 2001. Transposition and gene disruption in the male germline of the mouse. Genesis 30:82-88. [DOI] [PubMed] [Google Scholar]
- 8.Feng, Y. Q., M. C. Lorincz, S. Fiering, J. M. Greally, and E. E. Bouhassira. 2001. Position effects are influenced by the orientation of a transgene with respect to flanking chromatin. Mol. Cell. Biol. 21:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, S. E., H. G. van Luenen, and R. H. Plasterk. 1999. Cis requirements for transposition of Tc1-like transposons in C. elegans. Mol. Gen. Genet. 262:268-274. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, S. E., E. Wienholds, and R. H. Plasterk. 2001. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513-1523. [DOI] [PubMed] [Google Scholar]
- 12.Geurts, A. M., Y. Yang, K. J. Clark, G. Liu, Z. Cui, A. J. Dupuy, J. B. Bell, D. A. Largaespada, and P. B. Hackett. 2003. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol. Ther. 8:108-117. [DOI] [PubMed] [Google Scholar]
- 13.Greenwald, I. 1985. lin-12, a nematode homeotic gene, is homologous to a set of mammalian proteins that includes epidermal growth factor. Cell 43:583-590. [DOI] [PubMed] [Google Scholar]
- 14.Grewal, S. I., and S. C. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178-187. [DOI] [PubMed] [Google Scholar]
- 15.Hirochika, H., H. Okamoto, and T. Kakutani. 2000. Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horie, K., A. Kuroiwa, M. Ikawa, M. Okabe, G. Kondoh, Y. Matsuda, and J. Takeda. 2001. Efficient chromosomal transposition of a Tc1/mariner-like transposon Sleeping Beauty in mice. Proc. Natl. Acad. Sci. USA 98:9191-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horie, K., K. Yusa, K. Yae, J. Odajima, S. E. Fischer, V. W. Keng, T. Hayakawa, S. Mizuno, G. Kondoh, T. Ijiri, Y. Matsuda, R. H. Plasterk, and J. Takeda. 2003. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23:9189-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrabe de Angelis, M. H., H. Flaswinkel, H. Fuchs, B. Rathkolb, D. Soewarto, S. Marschall, S. Heffner, W. Pargent, K. Wuensch, M. Jung, A. Reis, T. Richter, F. Alessandrini, T. Jakob, E. Fuchs, H. Kolb, E. Kremmer, K. Schaeble, B. Rollinski, A. Roscher, C. Peters, T. Meitinger, T. Strom, T. Steckler, F. Holsboer, T. Klopstock, F. Gekeler, C. Schindewolf, T. Jung, K. Avraham, H. Behrendt, J. Ring, A. Zimmer, K. Schughart, K. Pfeffer, E. Wolf, and R. Balling. 2000. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 25:444-447. [DOI] [PubMed] [Google Scholar]
- 19.Ivics, Z., P. B. Hackett, R. H. Plasterk, and Z. Izsvak. 1997. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91:501-510. [DOI] [PubMed] [Google Scholar]
- 20.Izsvak, Z., Z. Ivics, and R. H. Plasterk. 2000. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 302:93-102. [DOI] [PubMed] [Google Scholar]
- 21.Izsvak, Z., D. Khare, J. Behlke, U. Heinemann, R. H. Plasterk, and Z. Ivics. 2002. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 277:34581-34588. [DOI] [PubMed] [Google Scholar]
- 22.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 23.Lavoie, B. D., and G. Chaconas. 1990. Immunoelectron microscopic analysis of the A, B, and HU protein content of bacteriophage Mu transpososomes. J. Biol. Chem. 265:1623-1627. [PubMed] [Google Scholar]
- 24.Li, E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3:662-673. [DOI] [PubMed] [Google Scholar]
- 25.Luo, G., Z. Ivics, Z. Izsvak, and A. Bradley. 1998. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 95:10769-10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miura, A., S. Yonebayashi, K. Watanabe, T. Toyama, H. Shimada, and T. Kakutani. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411:212-214. [DOI] [PubMed] [Google Scholar]
- 27.Moerman, D. G., G. M. Benian, and R. H. Waterston. 1986. Molecular cloning of the muscle gene unc-22 in Caenorhabditis elegans by Tc1 transposon tagging. Proc. Natl. Acad. Sci. USA 83:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muro, Y., H. Masumoto, K. Yoda, N. Nozaki, M. Ohashi, and T. Okazaki. 1992. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J. Cell Biol. 116:585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolan, P. M., J. Peters, M. Strivens, D. Rogers, J. Hagan, N. Spurr, I. C. Gray, L. Vizor, D. Brooker, E. Whitehill, R. Washbourne, T. Hough, S. Greenaway, M. Hewitt, X. Liu, S. McCormack, K. Pickford, R. Selley, C. Wells, Z. Tymowska-Lalanne, P. Roby, P. Glenister, C. Thornton, C. Thaung, J. A. Stevenson, R. Arkell, P. Mburu, R. Hardisty, A. Kiernan, A. Erven, K. P. Steel, S. Voegeling, J. L. Guenet, C. Nickols, R. Sadri, M. Nasse, A. Isaacs, K. Davies, M. Browne, E. M. Fisher, J. Martin, S. Rastan, S. D. Brown, and J. Hunter. 2000. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet. 25:440-443. [DOI] [PubMed] [Google Scholar]
- 30.Osborne, B. I., and B. Baker. 1995. Movers and shakers: maize transposons as tools for analyzing other plant genomes. Curr. Opin. Cell Biol. 7:406-413. [DOI] [PubMed] [Google Scholar]
- 31.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 32.Schubeler, D., M. C. Lorincz, D. M. Cimbora, A. Telling, Y. Q. Feng, E. E. Bouhassira, and M. Groudine. 2000. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 20:9103-9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spradling, A. C., D. M. Stern, I. Kiss, J. Roote, T. Laverty, and G. M. Rubin. 1995. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92:10824-10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 35.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 36.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 37.Wu-Scharf, D., B. Jeong, C. Zhang, and H. Cerutti. 2000. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science 290:1159-1162. [DOI] [PubMed] [Google Scholar]
- 38.Yant, S. R., A. Ehrhardt, J. G. Mikkelsen, L. Meuse, T. Pham, and M. A. Kay. 2002. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat. Biotechnol. 20:999-1005. [DOI] [PubMed] [Google Scholar]
- 39.Yant, S. R., L. Meuse, W. Chiu, Z. Ivics, Z. Izsvak, and M. A. Kay. 2000. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 25:35-41. [DOI] [PubMed] [Google Scholar]
- 40.Yoda, K., K. Kitagawa, H. Masumoto, Y. Muro, and T. Okazaki. 1992. A human centromere protein, CENP-B, has a DNA binding domain containing four potential alpha helices at the NH2 terminus, which is separable from dimerizing activity. J. Cell Biol. 119:1413-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, F., N. Zingler, G. Schumann, and W. H. Stratling. 2001. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 29:4493-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zayed, H., Z. Izsvak, D. Khare, U. Heinemann, and Z. Ivics. 2003. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]