Abstract
p150Sal2, a vertebrate homologue of the Drosophila melanogaster homeotic transcription factor Spalt, has previously been shown to be a binding target of the polyomavirus large T antigen. p150Sal2 acts as an inhibitor of viral DNA synthesis, and the binding of p150Sal2 by large T overcomes this inhibition. The present study focuses on the effects of p150Sal2 on the growth and survival of ovarian carcinoma (OVCA) cells that are deficient in expression of p150Sal2 and of normal established human ovarian surface epithelial (HOSE) cells which abundantly express the protein. Transient expression of exogenous p150Sal2 in OVCA cells that show little or no endogenous expression resulted in inhibition of DNA synthesis and colony formation and in increased apoptosis. OVCA cells stably transfected and expressing physiological levels of p150Sal2 showed reduced tumorigenicity accompanied by increased expression of p21WAF1/CIP1 (p21) and BAX. Conversely, reduction of endogenous levels of p150Sal2 in HOSE resulted in reduced p21 expression and increased DNA synthesis. p150Sal2 bound to the p21 promoter adjacent to the known p53 binding sites and stimulated transcription in the absence of p53. We propose that p150Sal2, acting in part as a p53-independent regulator of p21 and BAX, can function in some cell types as a regulator of cell growth and survival.
Mammalian p150Sal2 (Sal2, SALL2) belongs to the SALL family of proteins homologous to the region-specific homeotic transcription factor Spalt in Drosophila melanogaster (2, 17, 18). SALL proteins contain multiple zinc fingers frequently present as C2H2 pairs with a conserved linker motif (18). In humans, mutations in SALL genes have been linked to developmental abnormalities (2, 7, 13, 15). Mice carrying a homozygous knockout of SALL1 die in the early postnatal period following failure in kidney development (26). Mice lacking p150Sal2 appear to develop normally (32), although recent observations indicate a defect in myeloid stem cell maturation in these mice (Y. Ma, unpublished observation) similar to that seen in those lacking p21 (10). p150Sal2 is differentially expressed in mouse tissues, with particularly high levels found in the ovary (20). Several signaling pathways upstream of SALL genes have been recognized, and these have implications in human cancers (3, 8, 16, 23, 29). The specific biological functions and downstream target genes of Sal2 remain unknown.
p150Sal2 was identified independently through investigations of the oncogenic murine polyomavirus. Using a “tumor host range” selection procedure designed to uncover cellular factors which become targets for inactivation by the virus, p150Sal2 was found to be a binding partner of the viral large T antigen (20). In normal mouse cells, p150Sal2 has an inhibitory effect on polyoma viral DNA synthesis. The binding of p150Sal2 by large T overcomes this inhibition and is an essential step in virus replication and tumor induction (20). The large T proteins of polyomavirus and simian virus 40 (SV40), like oncoproteins of other DNA tumor viruses, function in part by inactivating tumor suppressor genes. Interestingly, while the large T antigens of both viruses bind and inactivate the retinoblastoma tumor suppressor protein pRb (9, 19a, 22), polyomavirus large T fails to bind and inactivate p53 in the manner of SV40 large T (4), and conversely, SV40 large T fails to bind p150Sal2 (D. Li, unpublished observation). To better understand the molecular and biological functions of p150Sal2, we studied both ovarian carcinoma (OVCA)-derived cells which are deficient in p150Sal2 expression and established human ovarian surface epithelial (HOSE) cells as the normal precursors which abundantly express the protein.
MATERIALS AND METHODS
Cells.
SKOV-3 and P19 cells were obtained from the American Type Culture Collection. HOSE and RUMG-S cells (12a) were gifts from Samuel Mok, Brigham and Women's Hospital, Harvard Medical School. IGR-OV-1 was a gift from Jean Feunteun, Institut Gustav-Roussy, Villejuif, France.
Antibody production.
Monoclonal and polyclonal antibodies against p150Sal2 have been described previously (20). Antisera were purified using protein A (Pierce).
p150Sal2 expression constructs.
p150Sal2 cDNAs were amplified from whole mouse embryo mRNA (Clontech) or human fetal brain mRNA (Clontech) and cloned into pcDNA3 (Invitrogen). Deletions of the DNA binding triple zinc fingers (designated Δ3; deletion of amino acids [aa] 631 to 711) and C-terminal large T-binding zinc finger pairs (designated Δ2; deletion of aa 911 to 956) were generated using a QuikChange site-directed mutagenesis kit (Stratagene). The p150 stable expression construct was generated using human p150 cDNA cloned into pTet-Splice (Invitrogen).
p21 promoter luciferase constructs were derived from p21waf1-Luc (31). Deletions were produced by either restriction digestion or PCR cloning of the desired p21 promoter region into pGL3 basic (Promega).
In vitro translation.
In vitro translation was performed using TNT T7 Quick.
Colony reduction assay.
SKOV-3 or HOSE cells were transfected with mouse or human p150Sal2 expression construct as described previously (20) After 48 h, the cells were seeded in 10-cm-diameter plates and selected in 400 μg of geneticin (Invitrogen)/ml for 2 weeks. The colonies were stained with crystal violet and photographed.
Assessment of apoptosis.
SKOV-3 cells were treated with actinomycin D (5 ng/ml) for 48 h. Cells with characteristic apoptosis DAPI (4′,6′-diamidino-2-phenylindole) staining were confirmed by use of TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) staining (Intergen). The apoptotic DAPI staining pattern was later used to score apoptosis for SKOV-3 cells in culture. For tumor sections, TUNEL staining was performed using ApopRed (Intergen) according to the manufacturer's suggestions.
BrdU incorporation assay.
SKOV-3 cells were transfected with p150 expression constructs and pEGFP-N1 (Clontech) at a ratio of 4 to 1. The cells were then growth arrested in 0.5% fetal bovine serum (FBS) for 48 h and stimulated with medium containing 15% FBS and 100 μM bromodeoxyuridine (BrdU; Sigma) for 20 h. Cells were stained using an anti-BrdU kit (Amersham-Pharmacia). The percentage of BrdU-negative cells among 200 green fluorescent protein (GFP)-positive cells in four random fields was plotted.
Stable p150-elevated cells.
SKOV-3 cells were transfected with human p150Sal2 cDNA in the vector pTet-splice (Invitrogen) and selected for 2 weeks in the presence of 400 μg of geneticin (Invitrogen)/ml until resistant colonies formed. The cells were pooled and cloned by limiting dilution. Single colonies were selected by testing the expression of p150 with Western blotting to generate SK-P150 clones. The empty vector-transfected cells (SK-vector) were pooled and used as controls.
Semiquantitative reverse transcription (RT)-PCR.
Total RNAs from SK-vector and SK-P150 clones were amplified using the primer pair 5′-CGT CAC CTG AGG TGA CAC AGC AAA GC-3′ and 5′-CGC TTC CAG GAC TGC AGG CTT CCT G-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPD) was amplified using 5′-CAG ACC CCA AAT CTG CAG ATA CTC AG-3′ and 5′-CAC TGG AAT TGG AAC TCT TCT GTC GAG-3′. Amplification mixtures from cycle numbers with linear amplification were used for comparison of relative quantities of transcripts. Amplified GAPD cDNA was used as an internal control to normalize the amount of p21 cDNA. The ratio of p21 to GAPD is the average of three linear amplification cycles.
Luciferase assays.
The p21-luciferase construct (0.3 μg) was cotransfected with 1.5 μg of p150Sal2 or p53 expression constructs and 0.3 μg of pCMV-β-Gal (Promega). For dosage response experiments, 0, 0.5, or 1.5 μg of p150 construct was used and empty vector was added to make up the different amounts of DNA. Cells were extracted 72 h posttransfection and assayed for luciferase and β-galactosidase activity. The relative luciferase activity of each sample was normalized by the respective β-galactosidase activity to assess the promoter activity.
Immunoprecipitation of p150 target DNA fragments.
The immunoprecipitation assay has been described previously (25). The restriction fragments were end-labeled with [α-32P]dCTP by Klenow fragment.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described previously (34). P19 cell extracts were used for ChIP. The bound chromatin fraction was amplified using mouse p21 promoter-specific primers 5′-GAA GTA GGA GTC ACC GTC CTG TTT ACC-3′ and 5′-GAT GTC TCT GTA TAG CCC TGG CTG TC-3′ for 45 cycles. As a nonspecific control, the GAPD (glyceraldehyde-3-phosphate dehydrogenase) gene was amplified with 5′-GCT GAA CGG GAA GCT CAC TGG CAT GG-3′ and 5′-GAG GTC CAC CAC CCT GTT GCT GTA GC-3′.
siRNA.
Two p150-specific small interfering RNA (siRNA) duplexes were made (5′-AAG GAG AUG GAC AGU AAU GAG-3′ and 5′-AAC CCC AUU ACC UCC AGA AUC-3′) and were transfected together into HOSE cells by using Oligofectamine (Invitrogen) according to the manufacturer's suggestions. The cells were serum starved (0.2% serum) for 48 h and stimulated with 10% FBS and 100 μM BrdU. After 18 h, cells were fixed and stained for BrdU and DAPI. p21 was detected by immunoprecipitation followed by Western blotting. p150 and tubulin were detected directly by Western blotting.
RESULTS
Exogenous p150Sal2 inhibits growth and induces apoptosis in p150Sal2-deficient OVCA cell lines.
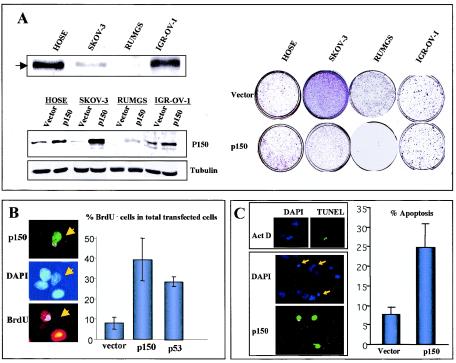
SKOV-3 is a human OVCA-derived cell line that is p53 null and produces tumors in nude mice with a histology similar to that of human ovarian carcinomas (36). Expression of p150Sal2 in SKOV-3 is greatly reduced compared to that in normal established HOSE cells. Two additional OVCA-derived cell lines were found to be either totally lacking (RUMGS) or essentially normal (IGR-OV-1) in p150Sal2 expression (Fig. 1A, top left). Transient expression of wild-type human p150Sal2 in SKOV-3 and RUMGS cells led to reduced colony formation. The growth of ovarian tumor line IGR-OV-1, which expresses endogenous p150Sal2 at a normal level, was unaffected by exogenous p150Sal2 (Fig. 1A, right). Western blots confirmed similar expression levels of p150Sal2 constructs following transfection (Fig. 1A, lower left).
FIG. 1.
Effects of p150Sal2 expression on the growth of SKOV-3 cells. (A) Colony reduction assay. Shown are a Western blot of HOSE cells and three other ovarian tumor cell lines with various levels of p150 expression (arrow) (top left panel) and a Western blot of p150 expression in various cells with transfected empty vector or a p150Sal2 expression construct (lower left panel). Tubulin was used as a loading control. Neomycin-resistant colonies were reduced in SKOV-3 and RUMGS cells transfected with a p150Sal2 expression construct (p150) or empty vector (Vector) but not in HOSE or IGR-OV-1 cells (right panel). (B) BrdU incorporation assay. SKOV-3 cells were transfected with either empty vector or constructs expressing p150Sal2 or p53 and incubated with BrdU-containing medium. Enhanced GFP construct was cotransfected in each experiment to identify the transfected cells. The cells were then stained for BrdU and DAPI. Results are presented as the percentage of BrdU-negative (BrdU−) cells among the total number of transfected cells (positive for cotransfected enhanced GFP). (C) Apoptosis analysis. The DAPI staining pattern of apoptotic SKOV-3 cells induced by actinomycin D was confirmed by TUNEL assay (top left panel). SKOV-3 cells were transiently transfected with p150 or empty vector together with GFP and stained with DAPI (bottom left panel). Arrows indicate cells that are p150 transfected and also apoptotic. Quantification is shown on the right as the percentage of apoptotic cells among all transfected cells.
Transient expression of exogenous p150Sal2 in SKOV-3 cells resulted in reduced DNA synthesis as indicated by the roughly fourfold increase in cells that failed to incorporate BrdU. A similar degree of inhibition of DNA synthesis was found upon reintroduction of p53 (Fig. 1B). SKOV-3 cells transfected with p150Sal2 also showed a three- to fourfold increase in apoptotic cells (Fig. 1C). These results demonstrate that p150Sal2, when restored to p150Sal2-deficient OVCA cells, acts in a p53-independent manner to reduce cell replication and viability.
Stable expression of p150Sal2 in SKOV-3 cells reduces tumorigenicity.
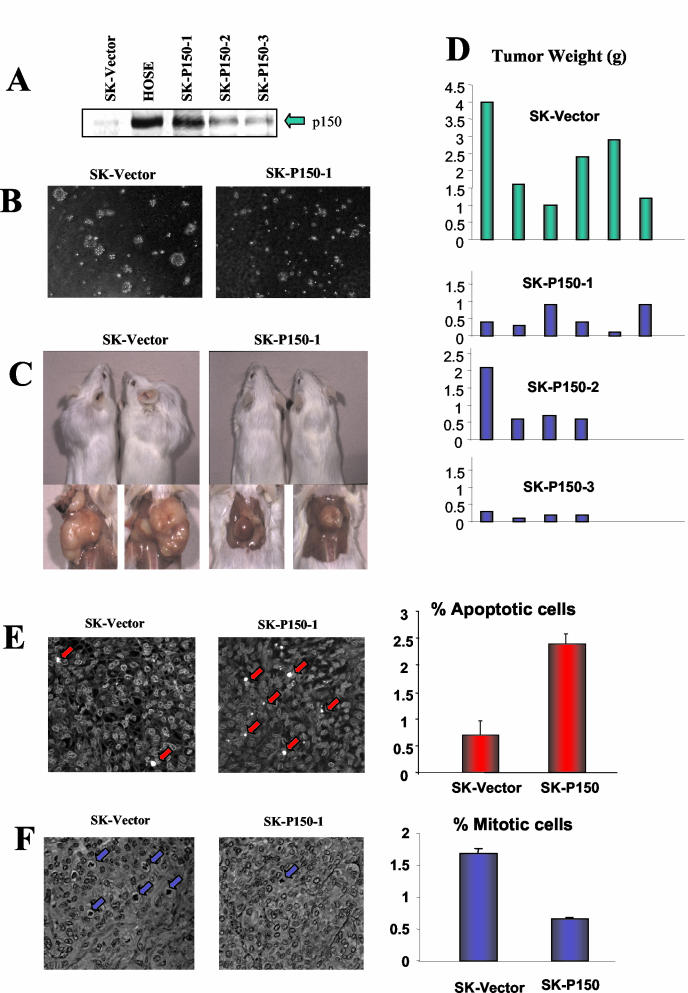
Attempts were made to isolate viable clones of SKOV-3 cells in which expression of p150Sal2 was stably restored to approximately normal levels. Three independent clones (SK-P150-1, -2, and -3) showing increased levels of p150Sal2 expression that were roughly in the range of, but not exceeding, that of normal HOSE cells were isolated (Fig. 2A). These SK-P150 cells showed diminished cloning efficiency and reduced colony size in soft agar compared to what was seen with the empty vector-transfected controls cells (SK-vector) (Fig. 2B). When inoculated subcutaneously into SCID mice, SK-vector cells produced fast-growing tumors weighing 2.2 g on average after 2 months. In contrast, SK-P150 cells produced slower-growing tumors averaging 0.6 g in weight (Fig. 2C and D). Apoptotic cells were found at three- to fourfold-higher frequency and mitotic cells were found at two- to threefold-lower frequency in SK-P150 compared to SK-vector tumors (Fig. 2E and F). The smaller sizes of SK-P150 tumors therefore reflect both proapoptotic and growth-suppressive effects of p150Sal2.
FIG. 2.
Effect of p150 expression on tumorigenicity. (A) Western blot comparison of p150Sal2 expression in normal HOSE cells, pooled neomycin-resistant SKOV-3 cells transfected with empty vector (SK-vector), and three independently derived SKOV-3 clones with various p150Sal2 expression levels (SK-P150-1, -2, and -3). (B) Representative figure showing the reduction of anchorage-independent growth in SK-P150 cells with restored p150Sal2 expression compared to SK-vector cells. (C) SCID mice inoculated by SKOV-3 cells with stably integrated empty vector (SK-vector) and SKOV-3 cells with stably integrated p150 expression construct (SK-P150-1). (D) Weights of tumors from SK-vector cells and from three independent SKOV-3 clones with human p150 (SK-P150-1, SK-P150-2, and SK-P150-3). (E) Apoptosis in SK-vector- and SK-P150-induced tumors. TUNEL staining of sections of SK-vector and SK-P150-1 tumors is shown at left (arrows indicate TUNEL-positive cells). Quantification of apoptotic cells in SK-vector and SK-P150-1 tumors is shown at right. Results are the averages for two independent tumors of each type, two sections of different areas, and three random fields of each section. (F) Mitosis in SK-vector and SK-P150 tumors. Hematoxylin and eosin staining of sections of SK-vector and SK-P150 tumors is shown at left (arrows indicate mitotic cells). Quantification of mitotic cells in SK-vector and SK-P150 tumors is shown at right. The sampling was the same as described for panel E.
p150Sal2 is a p53-independent activator of p21 and BAX.
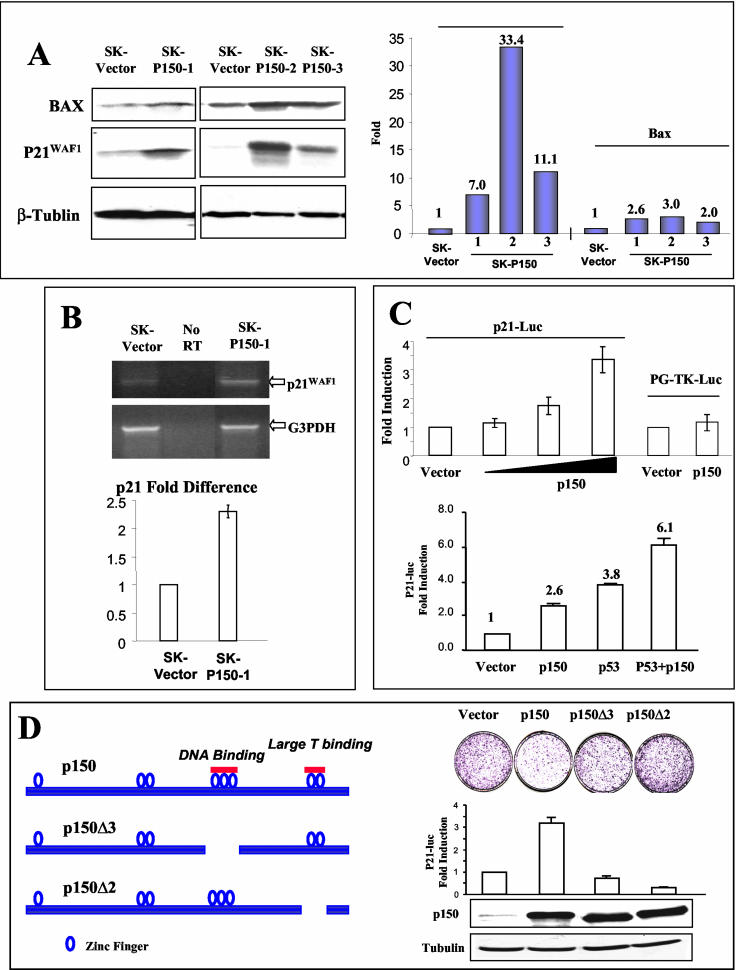
SKOV-3 cells are p53 deficient (36) and can be growth arrested by exogenous p53 (11, 12). Growth inhibition results largely from p53-dependent transcriptional activation of p21, causing cell cycle arrest, and from increased apoptosis mediated by Bax (30, 33). To determine whether the reduction of tumorigenicity by p150Sal2 is the result of cell growth-inhibitory and proapoptotic pathways similar to those induced by p53, we compared the expression levels of p53-regulated genes p21 and Bax in the three SK-P150 clones and SK-vector cells by Western blotting (Fig. 3A). Bax protein was elevated two- to threefold in the SK-P150 clones compared to the control cells. The p21 protein levels were increased more dramatically, in the range of 7- to 30-fold. Levels of P21 transcripts in SK-P150 cells were more than twofold higher in SK-P150 than in SK-vector cells as determined by RT-PCR (Fig. 3B), indicating that increased transcription contributes to the increased p21 levels in SK-P150 cells.
FIG. 3.
p150Sal2 regulates p21Cip1/Waf1 expression in SKOV-3 and HOSE cells. (A) Western blots for p21 in empty vector-transfected SKOV-3 cells (SK-vector) and three independent SKOV3-p150Sal2 clones (SK-P150-1, SK-P150-2, and SK-P150-3) is shown at left. α-Tubulin was used as a loading control. Quantification of p21 levels in SK-vector and SK-P150 clones is shown at right. (B) Semiquantitative RT-PCR comparison of p21 RNA levels using either SK-vector or SK-P150 total RNA. GAPD transcript (G3PDH) was used as an internal control. Relative levels of p21 transcripts of SK-vector and SK-P150 were assessed by comparing the 0.6-kb p21 RT-PCR products of the same cycle numbers within the linear range of amplification (34 to 40 cycles). The quantification was normalized by the levels of transcripts of GAPD within its own linear range of amplification (28 to 34 cycles). (C) p150Sal2 stimulates p21 promoter activity (top). Increasing amounts of a human p150 cDNA expression construct (p150) were cotransfected into SKOV-3 cells along with a luciferase reporter driven by a 2.7-kb human p21 promoter (P21-Luc). An empty vector (Vector) and an unrelated (thymidine kinase) promoter-luciferase construct (PG-TK-Luc) were used as controls. Stimulation of p21 promoter activity by p150Sal2 is presented as the increase (n-fold) in luciferase activity over that of the vector control normalized by the activity of cotransfected β-galactosidase. Also shown is the effect of p150 and p53 on P21-Luc induction (bottom). An additive effect was seen when p150Sal2 and p53 were cotransfected (P53+p150) compared to what was seen with either p150 or p53 alone. (D) At left is a schematic presentation of p150Sal2 protein showing the location of deleted zinc finger motifs in p150Δ3 (deletion of aa 631 to 711) and p150Δ2 (deletion of aa 911 to 956). Shown at top right are the results of a colony reduction assay using empty vector (Vector), p150Sal2 (p150), and zinc finger domain deletions of the DNA binding triple zinc finger (p150Δ3) or the T-antigen-interacting last zinc finger pair (p150Δ2) in SKOV-3 cells. The graph at middle right shows induction of p21-Luc activities by vector, p150, p150Δ3, and p150Δ2 in SKOV-3 cells. Shown at bottom right are the expression levels of p150Sal2 in SKOV-3 cells transfected with vector, p150, p150Δ3, or p150Δ2.
To examine whether p150Sal2 functions directly as a transcriptional activator of p21, p150Sal2 expression constructs and a luciferase reporter driven by a 2.7-kb human p21 promoter (31) were cotransfected into SKOV-3 cells. p150Sal2 activated the p21 promoter two- to threefold in a dosage-dependent manner, consistent with the increase in p21 mRNA in SK-P150 cells. p21 promoter activation by p150Sal2 is specific since the luciferase reporter driven by a thymidine kinase promoter was not activated by p150Sal2 (Fig. 3C, top). Transfection of exogenous wild-type p53 stimulated the p21 promoter 3.8-fold compared to 2.6-fold stimulation by p150Sal2 (Fig. 3C, bottom). When these same amounts of p150 and p53 plasmids were added together, a 6.1-fold induction was observed, indicating independent and additive effects on the p21 promoter.
Both the putative DNA binding (see Fig. 5) and polyomavirus large T interaction domains of p150Sal2 are essential for transactivation of the p21 promoter. This was shown by transfection of two mutant p150Sal2 constructs lacking the triple zinc finger (p150Δ3) and C-terminal double zinc finger (p150Δ2). Though efficiently expressed, neither of these mutant constructs was able to transactivate the p21-Luc reporter (Fig. 3D). As expected, these mutant constructs also failed to inhibit colony formation (Fig. 3D).
FIG. 5.
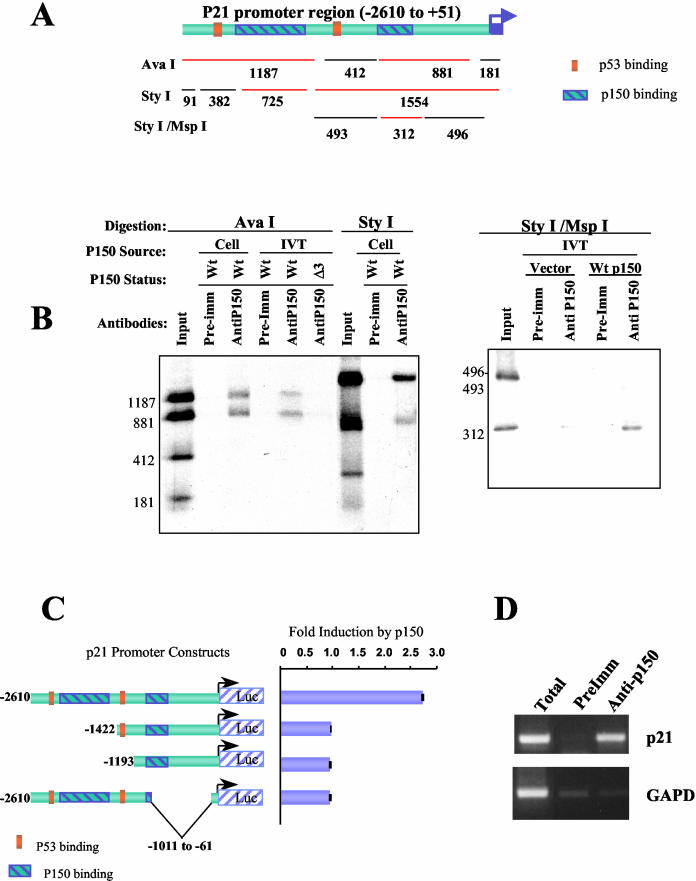
p150Sal2 directly interacts with p21 promoter and regulates p21 transcription through a cis-acting element. (A) Schematic representation of the 2.7-kb human p21 promoter region and its digestion fragments used in the protein DNA immunoprecipitation assay (25) (sizes in base pairs are given under each fragment). (B) Immunoprecipitation of AvaI-digested, StyI-digested, or StyI/MspI double-digested fragments of p21 promoter by p150 antibody (AntiP150) with preimmune serum (Pre-Imm) as a control. Wild-type (Wt) p150 or p150 with a deletion of the putative DNA binding domain (Δ3) was from either P19 cell extract (Cell) or the in vitro translation (IVT) product of cloned human cDNA. Empty cloning vector was used as a negative control for in vitro-translated p150 in the immunoprecipitation of StyI/MspI fragments. (C) A p150-responsive cis-acting element is located 1.4 kb upstream of the human p21 promoter corresponding to the distal p150Sal2 binding region. A luciferase assay was conducted in SKOV-3 cells to assess the responsiveness of various p21 promoter deletion constructs. Fold induction is the corresponding luciferase activity with cotransfected p150Sal2 over that with cotransfected empty vector. The p53 binding sites are shown as landmarks. (D) p150 binds p21 promoter region in vivo. ChIP was performed using serum against p150 (Anti-p150) to precipitate chromatin cross-linked with p150 in P19 cells and amplified using p21 promoter-specific primers (p21) and primers for GAPD. The same sets of primers were also used for PCR amplification from preimmune serum-precipitated chromatin (PreImm) and total input chromatin extract (Total) as controls.
Partial reduction of p150Sal2 in HOSE cells leads to increased DNA synthesis and reduced levels of p21.
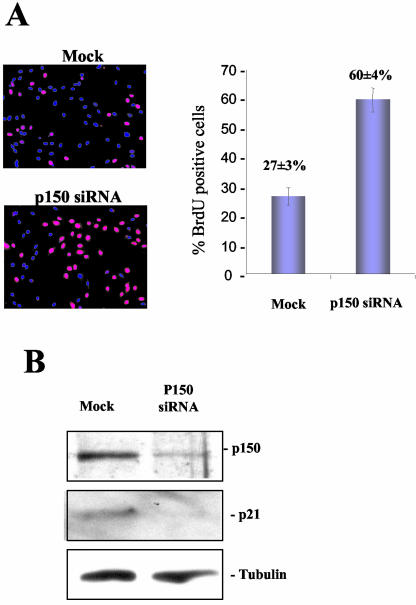
To determine whether the maintenance of high levels of p150Sal2 is important in the growth regulation of normal HOSE cells, a targeted reduction of p150Sal2 was carried out using siRNAs (Fig. 4). Roughly 68 h after addition of siRNAs to HOSE cells, the level of p150Sal2 was reduced three- to fourfold compared to that in control cells. This reduction in p150Sal2 level was accompanied by a substantial increase in the number of cells synthesizing DNA (Fig. 4A). A roughly eightfold reduction in p21 level was seen in the same siRNA-treated HOSE cells (Fig. 4B). These results suggest that p150Sal2, acting through p21, can function as a negative regulator of ovarian surface epithelial cell growth.
FIG. 4.
RNA interference analysis with HOSE cells. (A) Mock-transfected or p150 siRNA-transfected HOSE cells were cultured in BrdU-containing medium for 20 h (left panels). Nuclei with BrdU incorporation were stained with anti-BrdU antibody (red). Nuclei were also stained with DAPI (blue). The results were quantified by counting BrdU-positive cells in four random fields as the percentage of total cells (right). (B) Western blot analysis showing reduced p150Sal2 levels after addition of siRNA (P150 siRNA) and reduced p21 levels in HOSE cells compared to what was seen with mock-transfected cells. The same blot was stripped and reprobed for α-tubulin as the loading control.
p150Sal2 binds to the p21 promoter in vitro and in vivo.
To determine whether p150Sal2 activates p21 transcription by binding directly to the p21 promoter, antibody to p150Sal2 was used to immunoprecipitate labeled DNA restriction fragments incubated with p150Sal2 (25). Extracts of P19 cells, which express high levels of endogenous p150Sal2, and in vitro translated p150 cDNA were used as sources of protein. Antibody to p150Sal2 but not preimmune serum immunoprecipitated two noncontiguous DNA fragments from the p21 promoter. The fragments that were bound by p150Sal2 flank but do not overlap the two known p53 binding sites (6) (Fig. 5A and B). In vitro-translated p150 bound the same fragments as endogenous p150Sal2 from the P19 cell extract. p150Sal2 contains a triple zinc finger motif (20) similar to the DNA binding motif of the transcription factor SP1 (27). An in vitro-translated p150Sal2 with a deletion of this triple zinc finger region (Δ3; aa 631 to 711) was unable to bind the p21 promoter fragments (Fig. 5B). Thus, p150Sal2 binds to the p21 promoter directly and the triple zinc finger motif is essential for binding.
To validate the p150Sal2 binding regions as functional elements in the p21 promoter, a series of promoter constructs with truncations and internal deletions were transfected into SKOV-3 cells along with p150Sal2 cDNA. Removal of the distal p150Sal2 binding region totally abolished inducibility, indicating a p150Sal2-responsive element in this region (Fig. 5C). Similarly, an internal deletion of sequences containing the proximal binding region also abolished inducibility. The latter construct responded well to p53, demonstrating that core elements in the promoter proximal region were still present (data not shown). In this cell context, retention of both p150Sal2 binding regions appears to be essential for a p150Sal2-mediated response.
Further evidence for binding of p150Sal2 to the p21 promoter in vivo was sought by ChIP using anti-p150Sal2 antibody and chromatin from P19 murine embryonal carcinoma cells (24) which express high levels of the protein. Specific amplification of sequences from the p21 promoter was seen when anti-p150Sal2 antibody, but not preimmune serum, was used (Fig. 5D).
DISCUSSION
SALL genes are conserved in evolution from flies to humans and play important roles in development, yet little is known at the molecular or cellular level about their functions. A clue as to the possible molecular function(s) of the murine SALL2 gene emerged in the course of studies of a tumor host range mutant of polyomavirus in which p150Sal2 was shown to inhibit viral DNA replication (20). This inhibition is overcome by the viral large T antigen which binds to p150Sal2. A mutant virus unable to bind p150Sal2 is unable to replicate or induce a broad spectrum of tumors in the mouse. The selection procedure used to isolate this mutant was designed to identify cellular factors that must be altered or inhibited by the virus in order to replicate. Since polyomavirus replication requires that the virus be able to promote G1-to-S progression as well as to block apoptosis, it may be expected that p150Sal2 would function as a regulator of cell cycle progression and perhaps of apoptosis as well.
Among a number of normal mouse tissues tested for p150Sal2 expression, the ovary was found to have the highest level. The SALL2 gene has been mapped to human chromosome 14q11-12 (14), adjacent to and possibly overlapping a region of loss of heterozygosity in ovarian cancer (1). For these reasons, the present study focused on cells derived from human ovarian carcinomas which are deficient in p150Sal2 and on ovarian surface epithelial cells as the normal precursors which express high levels of the protein.
p150Sal2 is shown here to be a transcriptional activator of the cyclin-Cdk inhibitor p21, a key factor in G1 checkpoint control (5). Though the level of induction by p150Sal2 of a p21 promoter-luciferase reporter construct was in the range of two- to threefold in OVCA cells restored to normal levels of p150Sal2 expression, the level of p21 protein achieved in these cells was much higher (7- to 30-fold). Furthermore, the magnitude of the effects of added p150Sal2 were comparable to those of added p53 in terms of p21 induction (Fig. 3C) and reduction in BrdU incorporation (Fig. 1B). Similar levels of induction of p21 with significant biological effects have been reported for transforming growth factor β (19, 28) and retinoic acid (21). p150Sal2 also induces apoptosis accompanied by modestly elevated levels of Bax. Whether the proapoptotic effect of p150Sal2 is mediated entirely or only in part by Bax is unclear. Additional targets of p150Sal2 most likely remain to be identified.
The growth-suppressive and proapoptotic functions of p150Sal2 are expressed in the absence of p53. p150Sal2 binds to the p21 promoter in vitro and in vivo and stimulates transcription. DNA binding by p150Sal2 depends on a triple zinc finger motif, and transactivation requires retention of the C-terminal zinc finger pair which is also essential for binding to polyomavirus large T antigen (20). These functions of p150Sal2 are accompanied by negative effects on cell growth and survival. They are consistent with predictions of the tumor host range selection procedure and also with the general roles SALL genes are thought to play in processes of cell fate determination and terminal differentiation during embryonic development.
The action of p150Sal2 in regulating p21 and Bax partially reverses the tumorigenic properties of OVCA cells lacking p53. Introduction of exogenous p150Sal2 into OVCA cells led to a sharp reduction in replication and to an increase in the number of apoptotic cells. Clones of p53-negative OVCA cells in which p150Sal2 was stably restored to approximately normal levels expressed elevated levels of p21 and Bax and showed substantially reduced growth as tumors in SCID mice. The decreased net growth of tumors expressing exogenous p150Sal2 was accompanied by a lower mitotic index and an increased apoptotic index. In normal ovarian surface epithelial cells, targeted reduction of the endogenous p150Sal2 level led to decreased p21 expression and a concomitant increase in the number of cells synthesizing DNA. Thus, the maintenance of p150Sal2 levels appears to play a role in regulating the growth of these progenitor cells of OVCA.
p150Sal2 and p53 appear to function similarly in inhibiting cell replication and in inducing apoptosis. Loss of p53 is common in human ovarian cancer (35). It is possible that p150Sal2 serves as a backup to p53 in protecting against development of this important type of cancer. This model predicts that expression of SALL2 may be lost or reduced in primary OVCA and that mutations that diminish the ability of p150Sal2 to activate the p21 promoter may be found in ovarian tumors. The possibility that loss of SaLL2 may constitute an additional risk factor synergizing with p53 loss in OVCA should be evaluated.
Acknowledgments
This work was supported by a grant from the National Cancer Institute (RO1 CA-92520). T.B. is a D. K. and Virginia Ludwig Professor of cancer research and teaching.
We thank Jean Dahl, Peter Howley, and Raymond Erikson for constructive suggestions and reading of the manuscript, John Carroll and Yingxin Kong for their expert technical assistance, and Sam Mok and Jean Feunteun for gifts of ovarian tumor cell lines.
REFERENCES
- 1.Bandera, C. A., H. Takahashi, K. Behbakht, P. C. Liu, V. A. LiVolsi, I. Benjamin, M. A. Morgan, S. A. King, S. C. Rubin, and J. Boyd. 1997. Deletion mapping of two potential chromosome 14 tumor suppressor gene loci in ovarian carcinoma. Cancer Res. 57:513-515. [PubMed] [Google Scholar]
- 2.Barrio, R., J. F. de Celis, S. Bolshakov, and F. C. Kafatos. 1999. Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev. Biol. 215:33-47. [DOI] [PubMed] [Google Scholar]
- 3.Carl, M., and J. Wittbrodt. 1999. Graded interference with FGF signalling reveals its dorsoventral asymmetry at the mid-hindbrain boundary. Development 126:5659-5667. [DOI] [PubMed] [Google Scholar]
- 4.Dey, D. C., R. P. Bronson, J. Dahl, J. P. Carroll, and T. L. Benjamin. 2000. Accelerated development of polyoma tumors and embryonic lethality: different effects of p53 loss on related mouse backgrounds. Cell Growth Differ. 11:231-237. [PubMed] [Google Scholar]
- 5.Dotto, G. P. 2000. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim. Biophys. Acta 1471:M43-M56. [DOI] [PubMed] [Google Scholar]
- 6.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 7.Engels, S., J. Kohlhase, and J. McGaughran. 2000. A SALL1 mutation causes a branchio-oto-renal syndrome-like phenotype. J. Med. Genet. 37:458-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrell, E. R., and A. E. Munsterberg. 2000. csal1 is controlled by a combination of FGF and Wnt signals in developing limb buds. Dev. Biol. 225:447-458. [DOI] [PubMed] [Google Scholar]
- 9.Freund, R., R. T. Bronson, and T. L. Benjamin. 1992. Separation of immortalization from tumor induction with polyoma large T mutants that fail to bind the retinoblastoma gene product. Oncogene 7:1979-1987. [PubMed] [Google Scholar]
- 10.Gartel, A. L., and A. L. Tyner. 1999. Transcriptional regulation of the p21(WAF1/CIP1) gene. Exp. Cell Res. 246:280-289. [DOI] [PubMed] [Google Scholar]
- 11.Jin, X., W. Burke, K. Rothman, and J. Lin. 2002. Resistance to p53-mediated growth suppression in human ovarian cancer cells retain endogenous wild-type p53. Anticancer Res. 22:659-664. [PubMed] [Google Scholar]
- 12.Johnson, P., D. Gray, M. Mowat, and S. Benchimol. 1991. Expression of wild-type p53 is not compatible with continued growth of p53-negative tumor cells. Mol. Cell. Biol. 11:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Kim, J. H., S. J. Skates, T. Uede, K. K. Wong, J. O. Schorge, C. M. Feltmate, R. S. Berkowitz, D. W. Cramer, and S. C. Mok. 2002. Osteopontin as a potential diagnostic biomarker for ovarian cancer. JAMA 287:1671-1679. [DOI] [PubMed]
- 13.Kohlhase, J., M. Heinrich, L. Schubert, M. Liebers, A. Kispert, F. Laccone, P. Turnpenny, R. M. Winter, and W. Reardon. 2002. Okihiro syndrome is caused by SALL4 mutations. Hum. Mol. Genet. 11:2979-2987. [DOI] [PubMed] [Google Scholar]
- 14.Kohlhase, J., R. Schuh, G. Dowe, R. P. Kuhnlein, H. Jackle, B. Schroeder, W. Schulz-Schaeffer, H. A. Kretzschmar, A. Kohler, U. Muller, M. Raab-Vetter, E. Burkhardt, W. Engel, and R. Stick. 1996. Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene spalt. Genomics 38:291-298. [DOI] [PubMed] [Google Scholar]
- 15.Kohlhase, J., A. Wischermann, H. Reichenbach, U. Froster, and W. Engel. 1998. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat. Genet. 18:81-83. [DOI] [PubMed] [Google Scholar]
- 16.Koster, R., R. Stick, F. Loosli, and J. Wittbrodt. 1997. Medaka spalt acts as a target gene of hedgehog signaling. Development 124:3147-3156. [DOI] [PubMed] [Google Scholar]
- 17.Kuhnlein, R. P., G. Bronner, H. Taubert, and R. Schuh. 1997. Regulation of Drosophila spalt gene expression. Mech. Dev. 66:107-118. [DOI] [PubMed] [Google Scholar]
- 18.Kuhnlein, R. P., G. Frommer, M. Friedrich, M. Gonzalez-Gaitan, A. Weber, J. F. Wagner-Bernholz, W. J. Gehring, H. Jackle, and R. Schuh. 1994. spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 13:168-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurisaki, K., A. Kurisaki, U. Valcourt, A. A. Terentiev, K. Pardali, P. Ten Dijke, C. H. Heldin, J. Ericsson, and A. Moustakas. 2003. Nuclear factor YY1 inhibits transforming growth factor β- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 23:4494-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Larose, A., L. St-Onge, and M. Bastin. 1990. Mutations in polyomavirus large T affecting immortalization of primary rat embryo fibroblasts. Virology 176:98-105. [DOI] [PubMed] [Google Scholar]
- 20.Li, D., K. Dower, Y. Ma, Y. Tian, and T. L. Benjamin. 2001. A tumor host range selection procedure identifies p150sal2 as a target of polyoma virus large T antigen. Proc. Natl. Acad. Sci. USA 98:14619-14624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, M., A. Iavarone, and L. P. Freedman. 1996. Transcriptional activation of the human p21WAF1/CIP1 gene by retinoic acid receptor. Correlation with retinoid induction of U937 cell differentiation. J. Biol. Chem. 271:31723-31728. [DOI] [PubMed] [Google Scholar]
- 22.Ludlow, J. W., J. A. DeCaprio, C. M. Huang, W. H. Lee, E. Paucha, and D. M. Livingston. 1989. SV40 large T antigen binds preferentially to an underphosphorylated member of the retinoblastoma susceptibility gene product family. Cell 56:57-65. [DOI] [PubMed] [Google Scholar]
- 23.Ma, Y., D. Li, L. Chai, A. M. Luciani, D. Ford, J. Morgan, and A. L. Maizel. 2001. Cloning and characterization of two promoters for the human HSAL2 gene and their transcriptional repression by the Wilms tumor suppressor gene product. J. Biol. Chem. 276:48223-48230. [DOI] [PubMed] [Google Scholar]
- 24.McBurney, M. W., and B. J. Rogers. 1982. Isolation of male embryonal carcinoma cells and their chromosome replication patterns. Dev. Biol. 89:503-508. [DOI] [PubMed] [Google Scholar]
- 25.McKay, R., and D. DiMaio. 1981. Binding of an SV40 T antigen-related protein to the DNA of SV40 regulatory mutants. Nature 289:810-813. [DOI] [PubMed] [Google Scholar]
- 26.Nishinakamura, R., Y. Matsumoto, K. Nakao, K. Nakamura, A. Sato, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, S. Scully, D. L. Lacey, M. Katsuki, M. Asashima, and T. Yokota. 2001. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development 128:3105-3115. [DOI] [PubMed] [Google Scholar]
- 27.Pabo, C. O., and R. T. Sauer. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61:1053-1095. [DOI] [PubMed] [Google Scholar]
- 28.Pardali, K., A. Kurisaki, A. Moren, P. ten Dijke, D. Kardassis, and A. Moustakas. 2000. Role of Smad proteins and transcription factor Sp1 in p21Waf1/Cip1 regulation by transforming growth factor-β. J. Biol. Chem. 275:29244-29256. [DOI] [PubMed] [Google Scholar]
- 29.Raftery, L. A., and D. J. Sutherland. 1999. TGF-β family signal transduction in Drosophila development: from Mad to Smads. Dev. Biol. 210:251-268. [DOI] [PubMed] [Google Scholar]
- 30.Ramirez, P. T., D. M. Gershenson, G. Tortolero-Luna, L. M. Ramondetta, D. Fightmaster, J. T. Wharton, and J. K. Wolf. 2001. Expression of cell-cycle mediators in ovarian cancer cells after transfection with p16INK4a, p21WAF1/Cip-1, and p53. Gynecol. Oncol. 83:543-548. [DOI] [PubMed] [Google Scholar]
- 31.Sambucetti, L. C., D. D. Fischer, S. Zabludoff, P. O. Kwon, H. Chamberlin, N. Trogani, H. Xu, and D. Cohen. 1999. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J. Biol. Chem. 274:34940-34947. [DOI] [PubMed] [Google Scholar]
- 32.Sato, A., Y. Matsumoto, U. Koide, Y. Kataoka, N. Yoshida, T. Yokota, M. Asashima, and R. Nishinakamura. 2003. Zinc finger protein Sall2 is not essential for embryonic and kidney development. Mol. Cell. Biol. 23:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengupta, P. S., A. T. McGown, V. Bajaj, F. Blackhall, R. Swindell, M. Bromley, J. H. Shanks, T. Ward, C. H. Buckley, K. Reynolds, R. J. Slade, and G. C. Jayson. 2000. p53 and related proteins in epithelial ovarian cancer. Eur. J. Cancer 36:2317-2328. [DOI] [PubMed] [Google Scholar]
- 34.Weinmann, A. S., and P. J. Farnham. 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26:37-47. [DOI] [PubMed] [Google Scholar]
- 35.Wenham, R. M., J. M. Lancaster, and A. Berchuck. 2002. Molecular aspects of ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 16:483-497. [DOI] [PubMed] [Google Scholar]
- 36.Yaginuma, Y., and H. Westphal. 1992. Abnormal structure and expression of the p53 gene in human ovarian carcinoma cell lines. Cancer Res. 52:4196-4199. [PubMed] [Google Scholar]