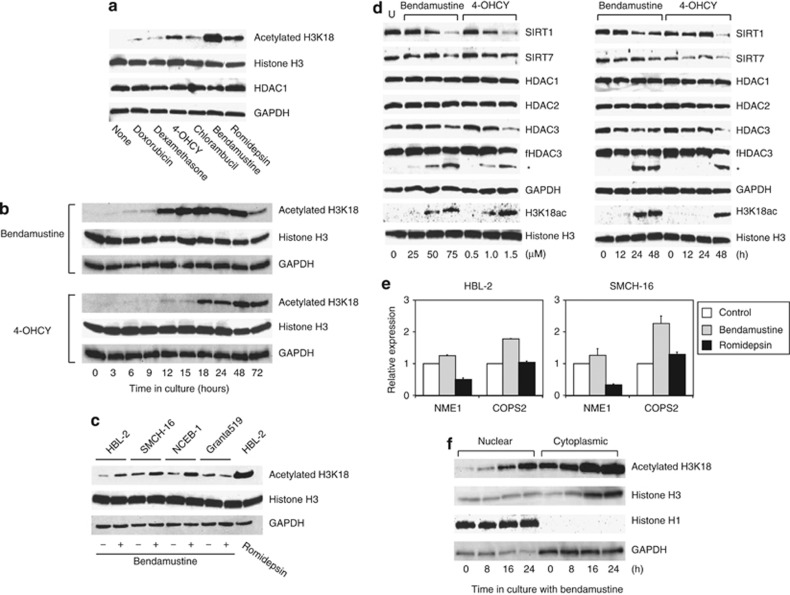
Histone modifications have crucial roles in diverse biological processes including development, differentiation and oncogenesis. Among them, acetylation of histone H3 at the lysine-18 residue (H3K18) is particularly important, because specific deacetylation of H3K18 is indispensable for oncogenic transformation by adenovirus1 and for host responses to bacterial infection.2 Regarding the former, it has also been demonstrated that H3K18 hypoacetylation is linked to the maintenance of malignant phenotypes3 and poor prognosis4, 5 in cancer.
Given the fundamental roles of H3K18 hypoacetylation in cancer biology, we investigated the possibility of therapeutic targeting of this modification in mantle cell lymphoma (MCL), in which epigenetic alterations, including aberrant hypomethylation of the HDAC1 gene,6 are central to the disease process and in which conventional therapies are usually not effective.7 Toward this end, we first screened for the effects of various anti-cancer drugs on the levels of H3K18 acetylation in the MCL cell line HBL-2. As shown in Figure 1a, in addition to the HDAC inhibitor romidepsin, two alkylating agents, bendamustine and 4-hydroperoxy-cyclophosphamide (4-OHCY; an active metabolite of cyclophosphamide), were able to reverse H3K18 hypoacetylation, whereas doxorubicin, dexamethasone and chlorambucil failed to do so at equitoxic concentrations. The increase in the acetylation levels of H3K18 was time dependent and considerably sustained until cell death (∼72 h; Figure 1b). This observation was reproducible in other MCL cell lines, except in Granta519, which is highly resistant to chemotherapeutic agents,7 upon treatment with bendamustine (Figure 1c). Moreover, we confirmed the induction of H3K18-specific acetylation in the bendamustine-sensitive Burkitt lymphoma cell line Namalwa but not in the bendamustine-resistant myeloid leukemia cell line K-562 (Supplementary Figure 1). It is of note that neither bendamustine nor 4-OHCY alone induced hyperacetylation at other sites including H3K9, H3K27, H4K5, H4K12 and H4K16 (Supplementary Figure 1).
Figure 1.
Alkylating agents induce histone H3K18 hyperacetylation through SIRT7 downregulation and HDAC3 cleavage. (a) HBL-2 cells were cultured with the indicated drugs at IC50 (doxorubicin 20 nM, dexamethasone 100 nM, 4-OHCY 1 μM, chlorambucil 4 μM, bendamustine 50 μM and romidepsin 2 nM) for 24 h and subjected to immunoblotting with specific antibodies against the histone H3 acetylated at lysine-18 (Cell Signaling Technology, Beverly, MA, USA), total histone H3 (Cell Signaling Technology), HDAC1 (Sigma-Aldrich, St Louis, MO, USA) and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA). (b) HBL-2 cells were cultured with either bendamustine (provided by SymBio Pharmaceuticals, Tokyo, Japan) or 4-OHCY (provided by Shionogi, Osaka, Japan) at IC50 (50 and 1.0 μM, respectively) for the indicated periods and subjected to immunoblotting with specific antibodies against the histone H3 acetylated at lysine-18 (H3K18), histone H3 and GAPDH (loading controls). (c) We cultured four MCL cell lines in the absence (−) or presence (+) of 50 μM bendamustine for 12 h and subjected them to immunoblotting with specific antibodies against histone H3 acetylated at lysine-18 (H3K18), histone H3 and GAPDH (loading controls). HBL-2 cells were treated with 2 nM romidepsin (provided by Gloucester Pharmaceuticals, Cambridge, MA, USA) for 12 h to serve as a positive control for H3K18 hyperacetylation.14 (d) HBL-2 cells were cultured with either bendamustine or 4-OHCY at the indicated concentrations for 24 h (left panel) or at IC50 (50 and 1.0 μM, respectively) for the indicated periods (right panel) and subjected to immunoblotting with specific antibodies against SIRT1, SIRT7, HDAC2, acetylated H3K18, histone H3 (Cell Signaling Technology), HDAC1, HDAC3 (Upstate Biotechnology, Lake Placid, NY, USA) and GAPDH. We also used another anti-HDAC3 antibody (BD Transduction Laboratories, San Diego, CA, USA) to detect both full-length HDAC3 (fHDAC3) and cleaved HDAC3 (asterisk).13 (e) HBL-2 and SMCH-16 cells were cultured with either bendamustine or romidepsin at IC50 (50 μM and 2 nM, respectively) for 24 h and subjected to real-time quantitative RT-PCR using the TaqMan Expression Assays (Hs02621161 for NME1 and Hs00182826 for COPS2). The data were quantified with the 2−ΔΔCt method using simultaneously amplified GAPDH (Hs01922876) as a reference. The y axis indicates relative gene expression with the expression levels of untreated control cells being set at 1.0. (f) We cultured HBL-2 cells with 50 μM bendamustine for the indicated periods and separated nuclear and cytoplasmic fractions using the Nuclear Extraction Kit (Cayman Chemical Company, Ann Arbor, MI, USA) according to the manufacturer's instructions. We monitored the quality of separation using histone H1 and GAPDH as nuclear and cytoplasmic markers, respectively. The data shown are representative of multiple independent experiments.
Next, we sought to clarify the mechanisms by which bendamustine and 4-OHCY specifically enhanced H3K18 acetylation in target cells. A recent study by Barber et al.3 indicated that SIRT7, a class III HDAC, is responsible for site-specific deacetylation at H3K18 in cancer cells. Consistent with their finding, both bendamustine and 4-OHCY were seen to reduce the expression levels of SIRT7 in a dose- and time-dependent manner in HBL-2 cells (Figure 1d). Another class III HDAC, SIRT1, was also markedly downregulated under the same condition (Figure 1d), whereas there were no changes in the expression of SIRT2 and SIRT6 (data not shown). In addition, the two drugs decreased the abundance of full-length HDAC3 (∼49 kDa) concomitantly with the appearance of the cleaved form (∼42 kDa), whereas they did not affect the expression and activities of HDAC1 and HDAC2 (Figure 1d and data not shown). It is worth noting that the abundance of cleaved HDAC3 correlated precisely with the increase in H3K18 acetylation (Figure 1d).
Bendamustine-mediated H3K18 hyperacetylation might cause upregulation of a unique subset of target genes. Indeed, Barber et al.3 reported that SIRT7-mediated hypoacetylation of H3K18 contributes to cancer development and maintenance through silencing of tumor suppressor genes such as NME1 and COPS2. As anticipated, bendamustine increased the expression of these genes stronger than did romidepsin, which could not inhibit SIRT7 activity, in HBL-2 and SMCH-16 cells in association with the induction of H3K18 hyperacetylation (Figure 1e).
It is well known that core histones are acetylated immediately after translation in the cytoplasm and deacetylation allows their nuclear import by facilitating the binding of specific transporters to positively charged N-termini.8 It is possible that, as a reciprocal process, H3K18 hyperacetylation causes cytoplasmic translocation of nuclear histone H3. Immunoblot analyses of nuclear and cytoplasmic fractions of bendamustine-treated HBL-2 cells clearly demonstrated that this was the case (Figure 1f), which may contribute to growth inhibition because a ready supply of histones is required for the assembly of newly replicated DNA to complete S-phase progression. This is fully consistent with a recent report by Qian et al.,9 in which core histones are acetylated upon DNA damage and transferred to the cytoplasm, where they are degraded by the PA200/Bim 10-containing proteasomes, to facilitate DNA repair. Taken together, these results suggest that the repression of SIRT7 and HDAC3 is mainly responsible for H3K18 hyperacetylation induced by the two alkylating agents and that downregulation of SIRT1, a nuclear and cytoplasmic deacetylase for H3K9 and H4K16 as well as multiple non-histone cytosolic proteins including p53, may be implicated in further modification of translocated histone H3 in the cytoplasm.
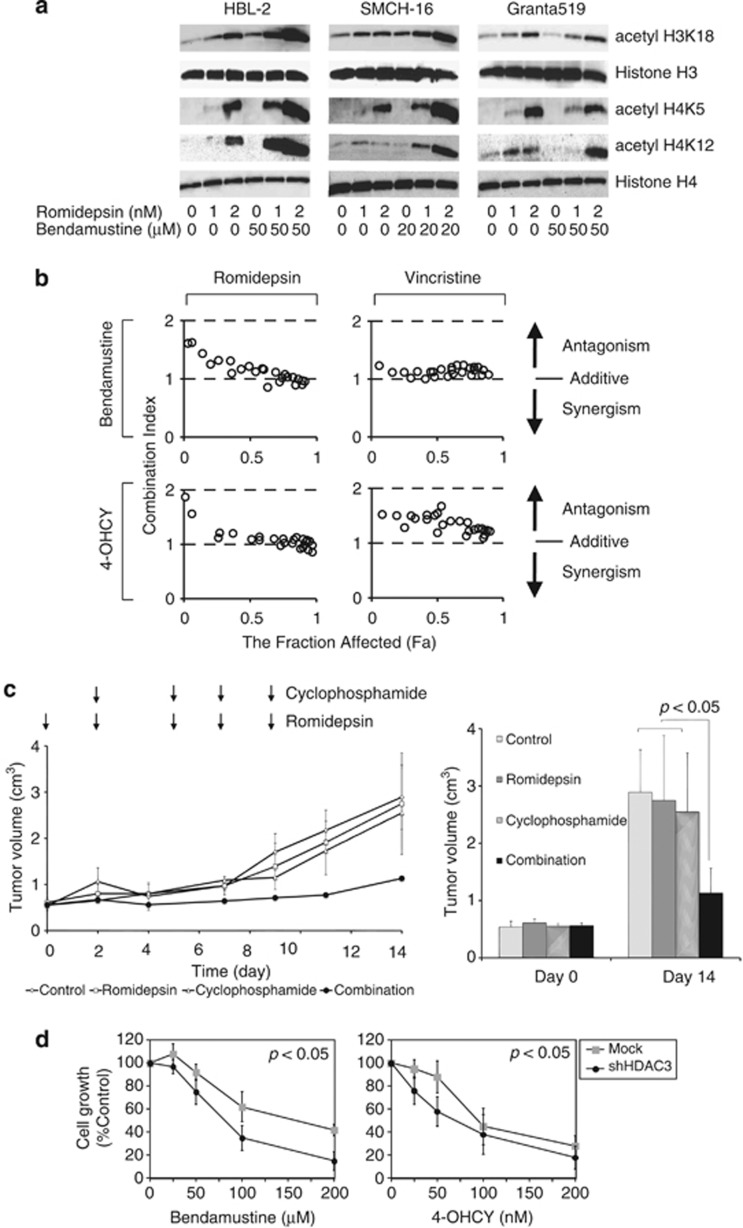
As bendamustine and 4-OHCY appear to suppress the expression of SIRT1, SIRT7 and HDAC3, the combination with authentic HDAC inhibitors, which primarily target the catalytic activity of class I HDACs especially HDAC1 and HDAC2,10 may produce synergistic effects in terms of histone acetylation and overall cytotoxicity. To substantiate this hypothesis, we first checked the acetylation status of histone tails in MCL cell lines treated with romidepsin and bendamustine simultaneously. As anticipated, romidepsin enhanced bendamustine-induced H3K18 acetylation in HBL-2 and SMCH-16 but not in Granta519 cells (Figure 2a). In addition, the two drugs synergistically induced hyperacetylation of other sites such as histones H3K9, H4K5, H4K12 and H4K16 (Figure 2a and data not shown). Drug combination analysis with isobolograms revealed that bendamustine and 4-OHCY were additive to synergistic cytotoxicity in combination with romidepsin against HBL-2 cells, whereas the combination with vincristine was rather antagonistic in vitro (Figure 2b). Next, we confirmed the synergistic effects of alkylating agents and HDAC inhibitors in vivo using a mouse xenograft model of MCL. As shown in Figure 2c, the combination of cyclophosphamide and romidepsin significantly retarded the growth of HBL-2 cells inoculated subcutaneously into immunodeficient mice at concentrations that did not affect tumor growth as single agents. Because romidepsin almost equally inhibits the enzymatic activity of HDAC1, 2 and 3,10 we attempted to clarify the direct target(s) responsible for the synergism with alkylating agents using knockdown approaches. shRNA against HDAC3 but not HDAC1 and 2 showed favorable effects in combination with bendamustine and 4-OHCY in HBL-2 and SMCH-16 cells (Figure 2d and data not shown), consistent with our finding that alkylating agents downregulated HDAC3 without affecting the expression and activities of HDAC1 and 2.
Figure 2.
Alkylating agents potentiate HDAC inhibitor-mediated global histone hyperacetylation and cytotoxicity in vitro and in vivo. (a) MCL cell lines were cultured with romidepsin and bendamustine at the indicated concentrations for 12 h, and were subjected to immunoblotting using specific antibodies against acetylated histones H3K18, H4K5 and H4K12 (Cell Signaling Technology) and histone H3 and histone H4 (loading controls). (b) We treated HBL-2 cells with variable concentrations of bendamustine or 4-OHCY in combination with romidepsin or vincristine for 48 h and determined cell viability by the reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) using the Cell Counting Kit-8 (Wako Biochemicals, Osaka, Japan). We calculated the combination index of two anti-cancer drugs using CompuSyn software and generated isobolograms according to the manufacturer's instructions (www.combosyn.com). The overall effects of drug combination were analyzed by the method of Chou and Talalay as described previously in detail.15 Combination index <1.0 means synergism of the two drugs. The fraction affected (Fa) means the fraction of the population affected by drug treatment. (c) Non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice aged 6 to 8 weeks were inoculated subcutaneously in the right thigh with 3 × 106 HBL-2 cells in 0.5 ml IMDM medium mixed with 0.5 ml Matrigel basement membrane matrix (Becton Dickinson, Franklin Lakes, NJ, USA).14 When tumors were measurable (day 0), mice were assigned to four treatment groups: those receiving the vehicle alone (the control group), those receiving romidepsin alone, those receiving cyclophosphamide alone and those receiving cyclophosphamide plus romidepsin (the combination group) (n=3–6 animals/group). Romidepsin was given intravenously through the tail vein at 0.1 mg/kg on days 0, 2, 5, 7 and 9.14 Cyclophosphamide was given intraperitoneally at 50 mg/kg on days 2, 5, 7 and 9. The control group received the vehicle (0.9% NaCl) alone on the same schedule. (Left panel) Caliper measurements of the longest perpendicular tumor diameters were taken every alternate day to estimate the tumor volume using the following formula: 4/3π × (width/2)2 × (length/2), which represents the three-dimensional volume of an ellipse. (Right panel) The y axis shows the average tumor volume of each group at day 0 and 14. The means±s.d. (bars) are shown (n=3–4). P-values were calculated by one-way ANOVA with Tukey's multiple comparison test. (d) We transfected the lentiviral short-hairpin RNA/small-interfering RNA (shRNA/siRNA) expression vector pLL3.7-HDAC3 shRNA or empty vector into 293FT cells with packaging plasmids (Invitrogen, Carlsbad, CA, USA) to produce infective lentiviruses in culture supernatants. Lentiviruses were added to HBL-2 cell suspensions in the presence of 8 μg/ml polybrene and transduced for 24 h. The transduced cells were washed with fresh medium and cultured for an additional 24 h, followed by collection of GFP-positive cells using a FACS Aria II flow cytometer (Becton Dickinson). shRNA-treated HBL-2 cells were resuspended in fresh medium and cultured with the indicated concentrations of bendamustine or 4-OHCY. Cell growth was determined by the MTT assay after 48 h. The y axis indicates the relative growth of drug-treated cells against untreated cells (Control) transduced with empty vector (Mock) or shHDAC3 vector. The means±s.d. (bars) of three independent experiments are shown. P-values were calculated using one-way analysis of variance with the Student–Newman–Keuls multiple comparison test.
Among class I HDACs, HDAC3 has distinct properties despite evolutionarily conserved structures with other members.10 For instance, it has been reported that HDAC3 has critical roles in tumor cell viability, chromosomal stability, S-phase progression and the DNA damage response.11 Another unique feature of HDAC3 is conditional cleavage by extrinsic stimuli. Xia et al.12 demonstrated that HDAC3 cleaved by osmotic stress is more active than full-length HDAC3 and decreases histone acetylation at promoter regions of the c-Jun gene. On the other hand, cleaved HDAC3 is exported to the cytoplasm during the apoptotic process because of the loss of nuclear localization signals, resulting in increased histone acetylation at promoter regions of apoptotic genes such as Fas and Bax, whereas global histone acetylation decreases probably because of concurrent inactivation of histone acetyltransferases.13 In the present study, alkylator-cleaved HDAC3 remained in the nucleus (data not shown) but could not retain the ability of H3K18 deacetylation, implying that HDAC3 is inactivated by alkylating agents upon cleavage. These data suggest that alkylating agents cleave HDAC3 via distinct mechanisms from osmotic stress12 and apoptotic insults such as Fas ligand and ultraviolet exposure.13 The underlying mechanisms are currently under investigation in our laboratory.
In summary, this study uncovered a novel unexpected function of alkylating agents targeting cancer-specific histone modification (H3K18 hypoacetylation), which provides a rationale for the combination with HDAC inhibitors to potentiate each anti-tumor activity against intractable malignancies by epigenetic means, as exemplified here in MCL.
Acknowledgments
We thank Professor Martin JS Dyer (MRC Toxicology Unit, Leicester University, Leicester, UK) for providing NCEB-1 and Granta519 cell lines. This work was supported in part by the High-Tech Research Center Project for Private Universities: Matching Fund Subsidy from MEXT, a grant-in-aid for Scientific Research from JSPS (to YF and JK), and research grants from The Naito Foundation, The Yasuda Medical Foundation, The Uehara Memorial Foundation (to YF), Japan Leukemia Research Fund and Takeda Science Foundation (to JK). NH is a winner of the Young Scientist Award of Jichi Medical University.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandarian HA, Impens F, Nahori M-A, Soubigou G, Coppée J-Y, Cossart P, et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341:1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Manuyakorn A, Paulus R, Farrell J, Dawson NA, Tze S, Cheung-Lau G, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol. 2010;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchenko VV, Kuo P-Y, Shaknovich R, Yang DT, Gellen T, Petrich A, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116:1025–1034. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vose JM. Mantle cell lymphoma: 2012 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2012;87:605–609. doi: 10.1002/ajh.23176. [DOI] [PubMed] [Google Scholar]
- Blackwell JS, Wilkinson ST, Mosammaparast N, Pemberton LF. Mutational analysis of H3 and H4 N termini reveals distinct roles in nuclear import. J Biol Chem. 2007;282:20142–20150. doi: 10.1074/jbc.M701989200. [DOI] [PubMed] [Google Scholar]
- Qian M-X, Pang Y, Liu CH, Haratake K, Du B-Y, Ji D-Y, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers AR, Fischer MA, Stengel KR, Zhao Y, Kaiser JF, Wells CE, et al. Hdac3 is essential for DNA replication in hematopoietic progenitor cells. J Clin Invest. 2013;123:3112–3123. doi: 10.1172/JCI60806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Wang J, Liu T-J, Yung WKA, Hunter T, Lu Z. c-Jun downregulation by HDAC3-dependent transcriptional repression promotes osmotic stress-induced cell apoptosis. Mol Cell. 2007;25:219–232. doi: 10.1016/j.molcel.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaffit F, Vaute O, Chevillard-Briet M, Segui B, Takami Y, Nakayama T, et al. Cleavage and cytoplasmic relocalization of histone deacetylase 3 are important for apoptosis progression. Mol Cell Biol. 2007;27:554–567. doi: 10.1128/MCB.00869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu R, Kikuchi J, Wada T, Ozawa K, Kano Y, Furukawa Y. HDAC inhibitors augment cytotoxic activity of rituximab by upregulating CD20 expression on lymphoma cells. Leukemia. 2010;24:1760–1768. doi: 10.1038/leu.2010.157. [DOI] [PubMed] [Google Scholar]
- Kikuchi J, Yamada S, Koyama D, Wada T, Nobuyoshi M, Izumi T, et al. The novel orally active proteasome inhibitor K-7174 exerts anti-myeloma activity in vitro and in vivo by down-regulating the expression of class I histone deacetylases. J Biol Chem. 2013;288:25593–25602. doi: 10.1074/jbc.M113.480574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.