Abstract
The HRC gene encodes the histidine-rich calcium-binding protein, which is found in the lumen of the junctional sarcoplasmic reticulum (SR) of cardiac and skeletal muscle and within calciosomes of arterial smooth muscle. The expression of HRC in cardiac, skeletal, and smooth muscle raises the possibility of a common transcriptional mechanism governing its expression in all three muscle cell types. In this study, we identified a transcriptional enhancer from the HRC gene that is sufficient to direct the expression of lacZ in the expression pattern of endogenous HRC in transgenic mice. The HRC enhancer contains a small, highly conserved sequence that is required for expression in all three muscle lineages. Within this conserved region is a consensus site for myocyte enhancer factor 2 (MEF2) proteins that we show is bound efficiently by MEF2 and is required for transgene expression in all three muscle lineages in vivo. Furthermore, the entire HRC enhancer sequence lacks any discernible CArG motifs, the binding site for serum response factor (SRF), and we show that the enhancer is not activated by SRF. Thus, these studies identify the HRC enhancer as the first MEF2-dependent, CArG-independent transcriptional target in smooth muscle and represent the first analysis of the transcriptional regulation of an SR gene in vivo.
Skeletal, cardiac, and smooth muscle represent the three major muscle cell types in vertebrates. These three types of muscle share the property of being contractile and have overlapping, but distinct, patterns of gene expression. Cardiac, skeletal, and smooth muscle have distinct embryonic origins, and each has evolved to perform highly specialized functions in vivo. As such, these three major muscle types have important differences in their contractile properties and in their gene expression programs. The MADS (MCM1, agamous, deficiens, serum response factor [SRF]) box transcription factors, SRF and myocyte enhancer factor 2 (MEF2), play key roles in the regulation of muscle-specific gene expression. SRF binds to the consensus sequence CC(A/T6)GG, known as a CArG box, found in the cis-regulatory elements of nearly every smooth muscle gene defined to date (50). SRF bound at smooth muscle CArG elements recruits smooth muscle cell-specific transcriptional coregulators to the DNA. The cysteine-rich proteins, CRP1 and CRP2, have been shown to strongly potentiate transcription through DNA-bound SRF (10), and myocardin has also been shown to be a potent transcriptional cofactor for SRF in smooth muscle (57, 58, 60). In addition to its critical role in smooth muscle transcription, SRF has also been shown to activate the expression of a subset of cardiac and skeletal muscle genes (11, 30, 37, 44).
The MEF2 family comprises four vertebrate genes, mef2a to -d, and a single gene in Drosophila melanogaster (8). Inactivation of the Drosophila Mef2 gene results in a complete loss of muscle differentiation (9, 33, 53), and targeted disruption of the mouse mef2c gene leads to embryonic lethality due to cardiovascular defects (4, 34, 35). In addition, expression of a dominant-negative form of MEF2 in cultured skeletal muscle cells resulted in a failure of myoblasts to differentiate (49). MEF2 factors bind to a consensus A/T-rich sequence, YTA(A/T4)TAR, found in the control regions of nearly every skeletal or cardiac muscle gene analyzed in vivo (1, 8). MEF2 factors are also expressed in vertebrate smooth muscle cells (18); however, to date no transcriptional targets of MEF2 in smooth muscle have been identified in vivo.
The product of the HRC gene, the histidine-rich calcium-binding protein (HRCBP), is localized to the sarcoplasmic reticulum (SR) of cardiac and skeletal muscle and to calciosomes within arterial smooth muscle cells (20, 21, 51). HRCBP binds calcium in vitro with low affinity and high capacity (20, 52) and is present in the lumen of the junctional SR, the site of calcium release by the ryanodine receptor (15, 20, 29, 55). The function of HRCBP is not known, but its expression pattern, subcellular localization to the lumen of the SR, and association with components of the calcium release channel complex suggest a possible role in calcium release during excitation-contraction coupling (15, 20, 21, 25, 29). The expression of HRC in cardiac, skeletal, and smooth muscle suggests the possibility that HRC is the target of a common transcriptional program in the three muscle lineages.
In this study, we investigated the transcriptional regulation of the HRC gene in vivo using a transgenic approach. We identify the cis-regulatory promoter and enhancer sequences from the HRC gene and show that the HRC enhancer is dependent on an evolutionarily conserved, high-affinity MEF2 site for function in all three muscle lineages. Furthermore, the entire HRC enhancer sequence lacks any discernible CArG motifs and is not activated by SRF, suggesting that the HRC enhancer directs smooth muscle expression in an SRF-independent manner. Thus, these studies identify the HRC enhancer as the first example of a MEF2-dependent, CArG box-independent transcriptional target in vascular smooth muscle and represent the first analysis of the transcriptional regulation of an SR gene in vivo.
MATERIALS AND METHODS
Cloning, plasmids, and mutagenesis.
A 2,726-bp fragment of the human HRC gene encompassing the region from −2609 to +117 relative to the transcriptional start site was subcloned from a lambda GT10 genomic library as a SalI-Psp1406I fragment into SalI-ClaI-cleaved pBluescript SKII(+), using standard techniques. The resulting product was further subcloned into the promoterless lacZ reporter plasmid AUG-β-gal (40) to create the plasmid HRC-lacZ for generation and analysis of transgenic mice and for transfection analyses. The 2,726-bp product was also subcloned into pCAT-Basic (Promega) to create plasmid HRC-CAT for transfection analyses comparing expression in fibroblasts, myoblasts, and myotubes. The myogenin promoter and enhancer (−1565 to +18) cloned into plasmid pCAT-Basic to create plasmid pMYO1565CAT has been described elsewhere (17). A 3.8-kb fragment of the mouse smooth muscle α-actin (SMaa) promoter and enhancer, including 1.1 kb of upstream sequence and 2.7 kb from the first intron, was amplified from genomic DNA using the 5′ primer 5′-ACACCATAAAACAAGTGCATGAGC-3′ and the 3′ primer 5′-GCAGCGTCTCAGGGTTCTGCA-3′. This fragment was confirmed by sequencing and was cloned into plasmid AUG-β-gal for transfection analyses. This construct is nearly identical to the rat SMaa promoter and enhancer, which has been described previously (37). The expression plasmids pCDNA1.MEF2A and pCDNA1.MEF2C are also described elsewhere (6). Plasmid pCGN.SRF contains the mouse SRF cDNA under control of the cytomegalovirus promoter (57). The MEF2 mutation in the HRC enhancer was generated using the PCR mutagenesis technique of gene splicing by overlap extension (gene SOEing) (24) to create the following mutant sequence in the context of the full-length 2,726-bp HRC fragment: 5′-CCTCCGAGCTGGATCCTCCGCCCTGGCCTAG-3′. The entire sequence of the mutant fragment was confirmed by sequencing on both strands. The GenBank accession numbers for the sequences of the human and mouse HRC enhancers are AY321454 and AY321455, respectively.
Generation of transgenic mice.
Transgenic reporter fragments were digested and gel purified using standard techniques and were suspended in 5 mM Tris-Cl, 0.2 mM EDTA (pH 7.4) at a concentration of 2 ng/μl for pronuclear injection as described previously (23). Injected embryos were implanted into pseudopregnant CD-1 females, and embryos were collected at indicated time points for transient analysis or were allowed to develop to adulthood for establishment of stable transgenic lines. DNA was extracted from the yolk sac and amnion of embryos or from tail biopsies from mice by digestion in tail lysis buffer (100 mM NaCl, 25 mM EDTA, 1% sodium dodecyl sulfate, 10 mM Tris-Cl, 200 μg of proteinase K/ml; pH 8.0) at 56°C overnight. Digested samples were extracted once with phenol-chloroform and ethanol precipitated. DNA preparations were digested with EcoRV and analyzed by Southern blotting using a radiolabeled lacZ probe. All experiments using animals complied with federal and institutional guidelines and were reviewed and approved by the UCSF Institutional Animal Care and Use Committee.
X-Gal staining and immunohistochemistry.
β-Galactosidase expression from lacZ transgenic embryos, embryonic tissues, and adult tissues was detected by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining as described previously (16). Some embryos were dehydrated in ethanol and cleared for 1 to 3 h in a 1:1 mixture of benzyl alcohol and benzyl benzoate prior to photography for better visualization of staining under the skin. For transverse sections, embryos were collected at 11.5 days postcoitum (dpc), fixed, and stained with X-Gal. Following staining, the embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), rinsed, and dehydrated with a series of ethanol washes (70 to 100%) followed by three brief washes in xylene. Samples were then mounted in paraffin, and transverse sections were cut at a thickness of 5 μm using a Leica RM 2155 microtome and mounted on glass slides. Sections were counterstained with Nuclear Fast Red to visualize embryonic structures. For antibody staining, embryos were collected, fixed, and sectioned as described above. The sections were rehydrated through a series of ethanol washes (100 to 70%) and then were placed in PBS for 5 min. The sections were then blocked for 20 min in 3% normal goat serum diluted in PBS. Incubation in both primary antibodies was performed concurrently for 1 h at room temperature in a humid chamber. Mouse monoclonal anti-skeletal muscle myosin (MY-32; Sigma) and rabbit anti-β-galactosidase (ICN) were diluted 1:300 in 3% normal goat serum. Following incubation with the primary antibodies, the sections were washed three times for 10 min each with PBS. The secondary antibodies, Oregon Green-conjugated goat anti-rabbit (Molecular Probes) and tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-mouse (Sigma), were diluted 1:300 into 3% normal goat serum and incubated for 1 h at room temperature in a humid chamber in the dark, followed by three washes in PBS. Slides were mounted using a SlowFade Light antifade kit (Molecular Probes) and photographed on a fluorescence microscope.
Cell culture, transfections, and reporter assays.
C3H10T1/2 (10T1/2) cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum. C2C12 myoblasts were maintained in DMEM plus 15% fetal calf serum. For generation of myotubes, C2C12 cells were maintained in DMEM plus 2% horse serum as described previously for the transfection of myotubes (5). Transfections were performed by calcium phosphate precipitation in 60-mm-diameter dishes as described elsewhere (16). In transfections of the reporter plasmid only into 10T1/2 cells, C2C12 myoblasts, and C2C12 myotubes, 10 μg of the HRC-CAT reporter (−2609 to +117), the myogenin-CAT reporter (pMYO1565CAT), pCAT-Basic (Promega), or a constitutively active simian virus 40 (SV40)-CAT plasmid were used. Within each cell type, transfections were normalized as described previously (5). To account for differences in transfection efficiencies between the different cell types, the activity of SV40-CAT was set to 100% in each set of transfections for each cell type, and the data are expressed as a percentage of the activity obtained with SV40-CAT in that cell type. The activity of SV40-CAT is roughly equivalent among the three cell types used in these studies when normalized for transfection efficiency (5). For trans-activation analyses, 5 μg of HRC-lacZ or the SMaa-lacZ reporter was transfected along with either 5 μg of pCDNA1.MEF2A, 5 μg of pCDNA1.MEF2C, or 5 μg of pCDNA1.SRF expression plasmid by calcium phosphate precipitation. In samples where a cDNA expression plasmid was not transfected, an equal amount of the parental pCDNA1/amp expression vector (Invitrogen) was transfected. For chloramphenicol acetyltransferase (CAT) assays, transfected cells were harvested, and cellular extracts were prepared by sonication, heat inactivated, and normalized as described previously (14). CAT activity was determined as described previously (56). Reactions were conducted for 5 h at 37°C. Conversion to acetylated forms was analyzed by thin-layer chromatography and quantitated by phosphorimager analysis (Molecular Dynamics, Inc.). For β-galactosidase assays, transfected cells were harvested and cellular extracts were prepared by sonication and normalized as described previously (16). Chemiluminescent β-galactosidase assays were performed using the luminescent β-gal kit (Clontech) according to the manufacturer's recommendations, and relative light units were detected using a Tropix TR717 microplate luminometer (PE Applied Biosystems).
EMSAs.
DNA-binding reactions were performed as described previously (16). Briefly, double-stranded oligonucleotides for use in binding reactions were labeled with [32P]dCTP using Klenow to fill in overhanging 5′ ends and purified on a nondenaturing polyacrylamide-Tris-borate-EDTA gel. Binding reactions were preincubated at room temperature in 1× binding buffer (40 mM KCl, 15 mM HEPES [pH 7.9], 1 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol) containing 2 μg of reticulocyte lysate containing recombinant MEF2A or SRF protein or 2 μg of unprogrammed reticulocyte lysate, 1 μg of poly(dI-dC), and competitor DNA (100-fold excess where indicated) for 10 min prior to probe addition. Recombinant MEF2A and SRF proteins were generated from plasmid pCDNA1.MEF2A and plasmid pCDNA1.SRF by transcribing with T7 polymerase and translating in vitro using the TNT Quick coupled transcription-translation system as described in the manufacturer's directions (Promega). Reaction mixtures were incubated an additional 20 min at room temperature after probe addition and electrophoresed on a 6% nondenaturing polyacrylamide gel. The oligonucleotides for the myogenin MEF2 site and a mutant form of that site have been described previously (59). The oligonucleotides for the SMaa intronic CArG box and a mutant form of that site have also been described previously (37). The sense-strand sequences of the oligonucleotides used for electrophoretic mobility shift assays (EMSAs) were as follows: wild-type MEF2 site, 5′-TCCCAGCTGTATTTATAGCCCTGGCCTAGCCCA-3′; mutant MEF2 site, 5′-TCCCAGCTGGATCCTCCGCCCTGGCCTAGCCCA-3′.
RESULTS
Identification of a muscle-specific enhancer from the HRC gene.
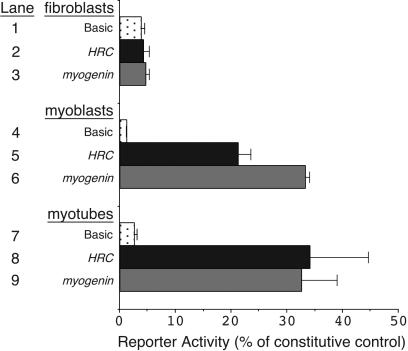
As a first step to define sequences regulating HRC transcription, we isolated a 2,726-bp fragment of the human HRC gene extending from −2609 to +117, relative to the transcriptional start site (Fig. 1A). This region of the HRC gene encompassed 141 bp of conserved noncoding sequence extending from −608 to −468 (Fig. 1B). This region is more than 80% conserved between the mouse and human HRC genes and contains a consensus MEF2-binding site, suggesting that this region might represent a muscle-restricted transcriptional enhancer. As an initial analysis to determine if the 2,726-bp HRC gene fragment contained functional promoter and enhancer sequences, we cloned this region into a CAT reporter plasmid such that transcription would be initiated at the transcriptional start site from the HRC gene (22) and translation would be initiated at the start codon in the CAT reporter mRNA. This HRC-CAT reporter plasmid was transfected into 10T1/2 fibroblasts, proliferating C2C12 skeletal myoblasts, and differentiating C2C12 skeletal myotubes to determine if this region of upstream sequence from the HRC gene was sufficient to direct transcription and whether expression would occur with muscle specificity (Fig. 2). We compared the expression of the HRC upstream region to that of the well-studied myogenin promoter, which is known to direct high levels of expression only in skeletal muscle cells (13, 17, 59), and to a promoterless parent CAT reporter gene construct (Basic). The results presented in Fig. 2 show that the HRC enhancer directed robust expression in myoblasts (Fig. 2, lane 5) and myotubes (Fig. 2, lane 8), but not in fibroblasts (Fig. 2, lane 2), where activity was similar to the background level of CAT activity directed by the promoterless Basic construct (Fig. 2, lane 1). This pattern of expression in transfected cells was nearly identical to the expression directed by the myogenin promoter, which also directed robust expression to myoblasts and myotubes (Fig. 2, lanes 6 and 9, respectively) but did not direct expression to nonmuscle fibroblasts (Fig. 2, lane 3).
FIG. 1.
An evolutionarily conserved noncoding sequence resides in the upstream region of the HRC gene. (A) Schematic representation of the human HRC upstream region. The HRC upstream region (−2609 to +117) was cloned as a BamHI-Psp1406I fragment into CAT and β-galactosidase reporter plasmids such that transcription would start from the HRC transcriptional start site (red arrow) and translation would initiate in the reporter cDNA. Green box, evolutionarily conserved region; black arrow, HRC translational start site; B, BamHI; N, NcoI; Bg, BglII; P, Psp1406I. (B) Sequence alignment of the evolutionarily conserved element from the HRC upstream region from −608 to −468, relative to the transcriptional start site, in the human sequence. The box denotes the evolutionarily conserved MEF2 site.
FIG. 2.
The HRC upstream region contains promoter and enhancer sequences sufficient to direct muscle-specific expression. HRC-CAT (lanes 2, 5, and 8), myogenin-CAT (lanes 3, 6, and 9), and the promoterless CAT-Basic (lanes 1, 4, and 7) reporter plasmids were transfected into fibroblasts (lanes 1 to 3), myoblasts (lanes 4 to 6), or myotubes (lanes 7 to 9). The HRC and myogenin reporter plasmids exhibited no significant activity over background in nonmuscle fibroblasts (lanes 2 and 3), but both reporters were robustly active in myoblasts (lanes 5 and 6) and myotubes (lanes 8 and 9). Data are expressed as a percentage of the activity obtained using a constitutively active SV40-CAT plasmid in each cell type. The data shown represent the mean values obtained in five independent transfections and analyses. Error bars represent the standard errors of the means.
The HRC enhancer is sufficient to direct expression to skeletal, cardiac, and arterial smooth muscle in vivo.
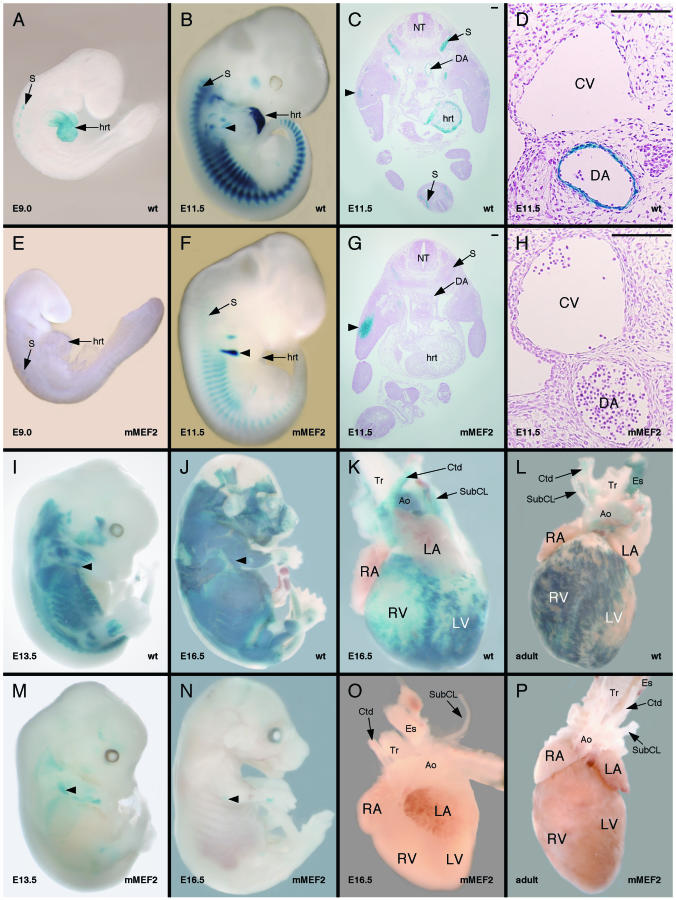
The results presented in Fig. 2 demonstrate that a fragment of the HRC gene from −2609 to +117 contains promoter and enhancer elements sufficient to direct strong expression in a muscle-specific fashion in cultured skeletal muscle cells. Based on these observations, we next wanted to determine if this region of the HRC gene was sufficient to direct expression in transgenic mice. We fused this fragment of HRC to the promoterless lacZ reporter AUG-β-gal and tested its ability to direct expression during embryonic development (Fig. 3). Expression of the HRC-lacZ transgene was clearly present in the developing myocardium and was faintly detectable within the myotomal compartment in rostral somites at 8.5 dpc (Fig. 3A). The expression of lacZ became much more robust in the heart and in the somitic myotome at 9.5 dpc (Fig. 3B) and 11.5 dpc (Fig. 3C). At 11.5 dpc, skeletal muscle expression was evident in both the hypaxial and epaxial compartments within the somites (Fig. 3C), and expression was present in all of the skeletal muscles in the embryo by 13.5 dpc (Fig. 3D). Cardiac expression of the transgene was clearly restricted to the ventricles by 11.5 dpc (Fig. 3E and F) and 13.5 dpc (Fig. 3H), and expression in the heart remained ventricle specific throughout development and in adulthood (see Fig. 6).
FIG. 3.
The HRC enhancer directs cardiac, skeletal, and arterial smooth muscle expression in transgenic mouse embryos. A 2,726-bp fragment of the human HRC gene was fused to a lacZ reporter plasmid and used to generate transgenic mice. This fragment of the HRC gene was sufficient to direct expression to all three muscle lineages in the mouse embryo in the pattern of endogenous HRC. (A to D) Representative X-Gal-stained transgenic embryos are shown at 8.5 dpc (A), 9.5 dpc (B), 11.5 dpc (C), and 13.5 dpc (D). The embryos in panels C and D have been cleared in a 1:1 mixture of benzyl alcohol and benzyl benzoate to help visualization of internal structures. Expression was evident at 8.5 dpc in the heart (hrt) and in the myotomal compartment of the somites (S) and by 11.5 dpc in arterial smooth muscle. No expression was observed in venous smooth muscle or in other smooth muscle cell types. (E to G) Transverse sections from X-Gal-stained transgenic embryos at 11.5 dpc. Expression was evident in somites, heart, and arterial vascular smooth muscle, including the dorsal aorta (DA) and branchial arch arteries (BAA). By contrast, expression was not observed in venous smooth muscle, including the cardinal veins (CV). No expression was observed in the smooth muscle of the trachea (Tr) or esophagus (Es) (G). Expression in the heart was restricted to the ventricles (E and F). Bar, 100 μm. (H) The heart and associated vasculature removed from an HRC-lacZ transgenic embryo at 13.5 dpc and stained with X-Gal. Expression in the heart at 13.5 dpc was restricted to the ventricles. Expression was also evident in arterial vascular smooth muscle, including the aorta (Ao) and subclavian (SubCL) and carotid arteries (Ctd). LA, left atrium; LV, left ventricle; NT, neural tube; RA, right atrium; RV, right ventricle. Seven independent transgenic lines all displayed nearly identical patterns of expression.
FIG. 6.
The MEF2 site in the HRC enhancer is required for expression in cardiac, skeletal, and arterial smooth muscle in vivo. Wild-type MEF2 site (A to D and I to L) and mutant MEF2 site (E to H and M to P) HRC-lacZ transgenic mice were analyzed for expression in vivo. Representative X-Gal-stained, transgenic embryos are shown at 9.0 dpc (A and E), 11.5 dpc (B and F), 13.5 dpc (I and M), and 16.5 dpc (J and N). X-Gal-stained hearts are shown from transgenic embryos dissected at 16.5 dpc (K and O) or transgenic adults dissected at 16 weeks of age (L and P). The embryos in panels J and N have been skinned to help visualize the underlying skeletal muscle. Panels C, D, G, and H show transverse sections of transgenic embryos collected and X-Gal stained at 11.5 dpc. Bar, 100 μm. The 2,726-bp wild-type (wt) HRC enhancer construct directed lacZ expression to the heart throughout embryonic development (A and B). By 11.5 dpc, expression was restricted to the ventricles (C), and the ventricle-restricted cardiac expression continued in the fetal (K) and adult (L) heart. The mutant MEF2 2,726-bp HRC enhancer construct (mMEF2) failed to direct expression to the heart at any stage in the embryo (E to G), fetus (O), or adult (P). The wild-type construct directed strong expression to arterial smooth muscle, including the dorsal aorta, beginning at 11.5 dpc (D). Arterial smooth muscle expression was also evident in the fetus (K) and adult (L). Smooth muscle expression was restricted to arteries. Note that the esophagus staining in the adult (L) represents skeletal muscle in the adult esophagus. The MEF2 mutant enhancer failed to drive lacZ expression in smooth muscle at all stages, including 11.5 dpc (G and H), 16.5 dpc (O), and adult (P). The wild-type transgene was robustly expressed in skeletal muscle in both the hypaxial and epaxial domains at 11.5 dpc (B and C). The MEF2 mutant transgene was also expressed in both hypaxial and epaxial myotomes, but the level of expression was much weaker at 11.5 dpc (F and G). Both the wild-type (B and C) and mutant MEF2 (F and G) enhancers directed expression to the dorsal limb muscles at 11.5 dpc. The wild-type enhancer drove strong lacZ expression in all skeletal muscles at 13.5 dpc (I) and 16.5 dpc (J). The mutant MEF2 enhancer directed only very weak skeletal muscle expression at 13.5 dpc (M), and expression was essentially absent by 16.5 dpc (N). Ao, aorta; Ctd, carotid artery; CV, cardinal vein; DA, dorsal aorta; Es, esophagus; hrt, heart; LA, left atrium; LV, left ventricle; NT, neural tube; S, somite (myotome); RA, right atrium; RV, right ventricle; SubCL, subclavian artery; Tr, trachea. Arrowheads denote expression in dorsal limb muscles.
HRC transgene expression became evident in smooth muscle at 11.5 dpc (Fig. 3C), when lacZ expression could easily been seen in larger arteries, such as the dorsal aorta (Fig. 3G) and the branchial arch arteries (Fig. 3F). Transgene expression could also be seen in smaller arterial vessels, but the level of expression was weaker in those vessels. Expression in smooth muscle was restricted to arterial vascular smooth muscle and was not detected in other smooth muscle cell types. For example, no expression was observed in the smooth muscle components of the trachea or esophagus at this stage (Fig. 3G). Expression of the transgene in venous smooth muscle, such as the cardinal vein (Fig. 3G), was also essentially absent at this and all other stages, although weak, patchy expression was rarely observed in very large veins such as the vena cava (data not shown). Similarly at 13.5 dpc, expression of the transgene was evident in arterial smooth muscle, including the aorta and the subclavian and carotid arteries (Fig. 3H). Arterial smooth muscle expression was maintained in the fetus and adult (see Fig. 6). These observations demonstrate that the HRC enhancer is sufficient to direct expression to smooth muscle in an artery-specific fashion. This is consistent with the expression of the endogenous gene product in smooth muscle, where it is restricted to arteries (51). Overall, the results presented in Fig. 3 demonstrate that the 2,726-bp fragment from the HRC gene is sufficient to direct expression to cardiac, skeletal, and smooth muscle during embryonic development in vivo.
An evolutionarily conserved upstream region is required for HRC function in vivo.
As a first step towards defining the cis-acting elements within the HRC enhancer required to direct cardiac, skeletal, and arterial smooth muscle-specific transcription in vivo, we performed deletional analyses of the HRC upstream region to identify the minimal sequence required for expression of lacZ (Fig. 4). Deletion from −2609 to −770 had no effect on expression in skeletal muscle, but this deletion had a dramatic effect on cardiac and smooth muscle expression, which was only faintly detectable (Fig. 4, constructs 1 and 2). Deletion of the upstream region to −510 had a more dramatic effect, completely eliminating expression in all three muscle lineages (Fig. 4, construct 3). Importantly, deletion of this 261-bp region (Δ510-770) in the context of the entire −2609 to +117 HRC enhancer fragment also completely eliminated expression of lacZ in transgenic mouse embryos (Fig. 4, construct 4). Since the region of the HRC enhancer between −770 and −510 was required for expression, we tested whether this region was also sufficient for expression in vivo. We cloned this 261-bp region of the HRC gene into plasmid HSP68-lacZ (HSP68/510-770) such that if functional enhancer sequences were present, lacZ would be transcribed due to the presence of the heterologous, minimal HSP68 promoter (27). This small region from the HRC gene was sufficient to drive strong expression in skeletal muscle, but it directed only very weak expression in cardiac and smooth muscle (Fig. 4, construct 5). These results, combined with the observation that deletion from −2600 to −770 had the same effect on transgene function, indicate that sequences upstream of −770 are required for the full expression of HRC in cardiac and smooth muscle. Taken together, all of the results summarized in Fig. 4 showed that the 261-bp region between −770 and −510 was absolutely required for HRC enhancer function in all three muscle lineages in vivo.
FIG. 4.
Deletional analysis of the HRC upstream region identified a conserved region required for expression in vivo. Schematic representations of the various deletion constructs of the HRC upstream region analyzed in these studies are shown in the center. Red arrow, HRC transcriptional start site; green arrow, transcriptional start site directed by the heterologous HSP68 promoter; blue arrow, lacZ translational start site; B, BamHI; N, NcoI; Bg, BglII; P, Psp1406I. Construct number and nucleotides, relative to the HRC transcriptional start site at +1, are indicated on the left. Expression of lacZ in somites, limbs, heart, and arteries is noted in the columns to the right. +++, very robust, easily detectable expression; +, very weak expression; −, a complete lack of detectable expression. The column on the far right indicates the number of independent transgenic lines or F0 transgenic embryos that expressed lacZ in the indicated pattern as a fraction of the total number of transgene-positive F0 embryos or lines examined. For HSP68/510-770 (construct 5), three of the five lines examined expressed lacZ in the indicated pattern. The other two F0 transgenic embryos showed no expression of lacZ.
The HRC enhancer contains an evolutionarily conserved, functional MEF2 site.
The region between −770 and −510 was required for expression in all three muscle cell types in vivo (Fig. 4). This region of the HRC enhancer contained the majority of a 141-bp evolutionarily conserved noncoding sequence, which contained a completely conserved, consensus MEF2 sequence (Fig. 1B). To determine if this potential MEF2 site in the HRC enhancer represented a bona fide binding site for MEF2, we tested its ability to bind to MEF2A by EMSA (Fig. 5A). The MEF2 consensus sequence in the HRC enhancer represented a strong binding site for MEF2 in vitro (Fig. 5A, lane 2). MEF2A bound specifically to the HRC MEF2 site, since binding was efficiently competed by an excess of unlabeled probe (Fig. 5A, lane 3) but was not competed by a 100-fold excess of unlabeled mutant MEF2 site from the HRC enhancer (Fig. 5A, lane 4). In addition, a 100-fold excess of an unlabeled MEF2 site from the myogenin promoter (59) also efficiently competed for MEF2 binding to the HRC MEF2 site (Fig. 5A, lane 5), but a mutant version of the myogenin MEF2 site (59) failed to compete (Fig. 5A, lane 6). The mutant form of the HRC MEF2 site used for the competition in lane 4 did not show any detectable binding to MEF2A (Fig. 5A, lane 8).
FIG. 5.
The conserved region of the HRC enhancer contains a high-affinity, functional MEF2 site. (A) MEF2 binds specifically to the HRC MEF2 site in vitro. MEF2A was transcribed and translated in vitro and used in EMSA analyses with radiolabeled double-stranded oligonucleotides representing the HRC MEF2 site (lanes 1 to 6) or a mutant version of the HRC MEF2 site (lanes 7 and 8). MEF2 efficiently bound to the HRC MEF2 site (lane 2) but failed to bind to the mutant MEF2 site (lane 8). Binding of MEF2 to the HRC MEF2 site was specific, since a 100-fold excess of unlabeled HRC MEF2 site efficiently competed for binding (lane 3) but a mutant version of the HRC MEF2 site (mHRC) failed to compete for binding even at a 100-fold excess (lane 4). Likewise, an unlabeled control MEF2 site from the myogenin gene (My) efficiently competed for binding (lane 5), but a 100-fold excess of a mutant myogenin MEF2 site (mMy) did not compete for binding (lane 6). In samples where in vitro-translated proteins were absent (lanes 1 and 7), an equal amount of unprogrammed reticulocyte lysate was included. Lysate-derived, nonspecific mobility shifts are noted. (B) The HRC enhancer is activated directly by MEF2 factors through the MEF2 site in the enhancer. MEF2A expression plasmid (lanes 3 and 4), MEF2C expression plasmid (lanes 5 and 6), or parental expression vector (lanes 1 and 2) was cotransfected with a full-length HRC-lacZ reporter plasmid (lanes 1, 3, and 5) or a mutant version of that reporter containing a disrupted MEF2 site (lanes 2, 4, and 6) into 10T1/2 fibroblasts. The parental expression vector failed to significantly activate the HRC enhancer (lane 1). MEF2A and MEF2C were each able to significantly trans-activate the HRC-dependent reporter (lanes 3 and 5, respectively). Neither MEF2A nor MEF2C activated the MEF2 mutant enhancer (lanes 4 and 6, respectively). The data shown represent the mean values obtained in three independent transfections and analyses. Error bars represent the standard errors of the means. (C) The HRC MEF2 site is not bound by SRF. Either MEF2A (lanes 2 to 4) or SRF (lanes 5 to 7 and 9 to 11) was transcribed and translated in vitro and used in EMSA analyses with radiolabeled double-stranded oligonucleotides representing the HRC MEF2 site (lanes 1 to 7) or the SMaa intronic CArG box (lanes 8 to 11). MEF2 efficiently bound to the HRC MEF2 site (lane 2), whereas SRF was completely unable to bind to the HRC MEF2 site (lane 5) under conditions in which it efficiently bound to the SMaa CArG box (lane 9). In samples where in vitro-translated proteins were absent (lanes 1 and 8), an equal amount of unprogrammedreticulocyte lysate was included. Lysate-derived, nonspecific mobility shifts are noted. Wild-type (HRC and CArG) and mutant (mHRC and mCArG) competitors were used at a 100-fold excess where indicated. (D) The HRC enhancer is not trans activated by SRF. Expression plasmids for MEF2C (lane 2), SRF (lanes 3 and 5), or the parental expression vector (lanes 1 and 4) were cotransfected with a full-length HRC-lacZ reporter plasmid (lanes 1 to 3) or a SMaa-lacZ reporter plasmid (lanes 4 and 5) into 10T1/2 fibroblasts. SRF failed to activate the HRC reporter (lane 3) under conditions in which MEF2C activated the HRC reporter more than 10-fold (lane 2) over the background level of activation indicated by parental expression vector cotransfection (lane 1). By contrast, SRF activated the SMaa reporter in the same experiment more than 16-fold (lane 5) over the background level of activation indicated by parental expression vector cotransfection (lane 4). The data shown represent the mean values obtained in three independent transfections and analyses. Error bars represent the standard errors of the means.
The results of the EMSA analyses presented in Fig. 5A demonstrated that the MEF2 site in the HRC enhancer was efficiently bound by MEF2 in vitro. To determine whether MEF2 factors could bind to the HRC MEF2 site in vivo and trans-activate the enhancer independent of other muscle-specific factors, we transfected the HRC-lacZ reporter plasmid with expression plasmids for either MEF2A or MEF2C (Fig. 5B). The transfection results presented in Fig. 2 showed that the HRC enhancer exhibited minimal activity in 10T1/2 fibroblasts. Thus, these cells provided a silent background to test the ability of MEF2 factors to activate the HRC enhancer. MEF2A and MEF2C each trans-activated the full-length HRC enhancer in 10T1/2 fibroblasts (Fig. 5B, lanes 3 and 5), and this activation was dependent on the presence of an intact MEF2 binding site in the enhancer (Fig. 5B, lanes 4 and 6). The results of these trans-activation experiments demonstrated that MEF2 factors can independently activate the HRC enhancer. Taken together with the results of the EMSA in Fig. 5A, which showed that MEF2 could bind directly to the HRC MEF2 site, these results strongly suggest that HRC is a direct target of MEF2 transcription factors via the conserved MEF2 site in its enhancer.
The data presented in Fig. 3 demonstrated that the HRC enhancer functions in arterial smooth muscle cells. Because the vast majority of smooth muscle genes described to date depend on the MEF2-related transcription factor SRF for activation in vivo (50), we wanted to test whether the HRC enhancer might also be a target of SRF. Even though the MEF2 site in the HRC enhancer meets the consensus sequence constraints for MEF2 binding (1) and does not match the CArG consensus for SRF binding (50), we wanted to exclude the possibility that the HRC MEF2 site might represent a noncanonical SRF-binding site. Under conditions in which MEF2 bound efficiently to the HRC MEF2 site (Fig. 5C, lane 2), SRF exhibited no detectable binding to the site (Fig. 5C, lane 5). The failure of SRF to exhibit any detectable binding to the HRC MEF2 site occurred in the same experiment in which SRF bound very robustly to the bona fide CArG box from the SMaa intronic enhancer (Fig. 5C, lane 9). Thus, the data presented in Fig. 5C further demonstrate that the HRC MEF2 site represents a bona fide MEF2 site and that it does not represent a binding site for SRF.
As an additional test for a potential role for SRF in the activation of HRC, we tested the ability of SRF to trans-activate the HRC enhancer (Fig. 5D). SRF cotransfection failed to cause any detectable activation over background (Fig. 5D, compare lanes 1 and 3) in the same experiment in which MEF2C trans-activated the HRC enhancer greater than 10-fold (Fig. 5D, compare lanes 1 and 2). Furthermore, SRF resulted in greater than 16-fold activation of the SMaa enhancer, a bona fide SRF target, in the same experiment in which it failed to activate HRC at all (Fig. 5D, lane 5). These results, taken together with the observation that the HRC enhancer lacks any recognizable CArG motifs, strongly suggest that HRC is not an SRF target gene.
The MEF2 site in the HRC enhancer is required for transgene expression in all three muscle lineages in vivo.
To test the function of the HRC MEF2 site in vivo, we introduced a mutation in the MEF2 site in the context of the 2,726-bp HRC-lacZ transgene (Fig. 4, construct 1) and generated transgenic embryos. Mutation of the HRC MEF2 site completely abolished cardiac and smooth muscle expression (Fig. 6). Cardiac expression directed by the wild-type transgene was apparent throughout development and in adulthood (Fig. 6A to C, K, and L). Cardiac expression directed by the wild-type HRC enhancer was robust during embryonic development (Fig. 6B) and was reduced but remained easily detectable in the fetal and adult heart (Fig. 6K and L). Adult cardiac expression directed by the wild-type enhancer was somewhat variable among individual animals but was consistently present in the ventricles in every case. Very weak expression in the adult atria was rarely observed (data not shown). In contrast to the expression directed by the wild-type enhancer, no expression of the MEF2 mutant transgene was detected in the heart at any stage (Fig. 6E to G, O, and P). The wild-type transgene was expressed in arterial smooth muscle beginning at 11.5 dpc (Fig. 6C and D), and expression could also be detected easily in arterial smooth muscle at later stages in development (Fig. 6K) and in adulthood (Fig. 6L). Mutation of the HRC MEF2 site completely disrupted lacZ expression in smooth muscle at all stages. No smooth muscle expression of the MEF2 mutant transgene was observed in the embryo (Fig. 6G and H), fetus (Fig. 6O), or adult (Fig. 6P). Six independent MEF2 mutant lines were examined. None displayed any expression in cardiac or arterial smooth muscle. Seven independent transgenic lines were examined for the wild-type HRC enhancer construct. All displayed nearly identical patterns of expression. Taken together, the results presented in Fig. 6 clearly indicate that the MEF2 site in the HRC enhancer is required for function in cardiac and smooth muscle in vivo.
The wild-type HRC enhancer directed easily detectable lacZ expression to skeletal muscle by 9.0 dpc (Fig. 6A), and this expression was maintained throughout development and adulthood (Fig. 6). Activity of the MEF2 mutant enhancer could be detected at 9.0 dpc in somites (Fig. 6E), although expression was slightly weaker than that observed with the wild-type enhancer (Fig. 6A). By 11.5 dpc, the difference in the activities of the wild-type and mutant enhancers became more pronounced in skeletal muscle. The wild-type HRC enhancer directed robust lacZ expression to myoblasts in the hypaxial and epaxial somites and to the muscles of the forelimb bud (Fig. 6B and C). Expression of the MEF2 mutant transgene at 11.5 dpc could also be observed in epaxial and hypaxial muscles within the somites and in the myoblasts of the forelimb bud (Fig. 6F and G). However, the overall expression level of lacZ directed by the MEF2 mutant transgene was severely reduced in the somites, such that expression was nearly undetectable in rostral somites (Fig. 6F and G). Interestingly, expression of the mutant transgene was quite robust in dorsal limb muscles (Fig. 6F and G) compared to that of the wild-type HRC-lacZ transgene (Fig. 6B and C). The disparity in skeletal muscle expression directed by the wild-type versus the mutant transgenes continued to become more pronounced throughout development, such that the wild-type transgene was expressed at very high levels at 13.5 dpc (Fig. 6I) and at 16.5 dpc (Fig. 6J). The MEF2 mutant transgene was only very weakly expressed by 13.5 dpc (Fig. 6M) and was essentially inactive by 16.5 dpc (Fig. 6N) and in the adult (data not shown).
The disparity in the expression directed by the wild-type and mutant transgenes at 11.5 and 13.5 dpc in the somites and limbs prompted us to examine lacZ transgene expression in skeletal muscle in more detail (Fig. 7). At 11.5 dpc, the wild-type enhancer directed strong expression within the myotomal compartment of the somites (Fig. 7A and B) but directed only weak expression of lacZ to the forelimb bud (Fig. 7C). By contrast, activity of the MEF2 mutant enhancer was barely detectable in the somites (Fig. 7D and E), but it directed very robust expression in the forelimb bud (Fig. 7F). We compared β-galactosidase expression in the somites to the expression of skeletal muscle myosin heavy chain by immunofluorescence using antibodies directed against both proteins (Fig. 7). β-Galactosidase protein expression in caudal somites at 11.5 dpc (Fig. 7G) completely overlapped with the expression of myosin (Fig. 7H and I) and appeared to be restricted to mononucleated muscle cells. Expression in more rostral somites at 11.5 dpc or in caudal somites at later times in development was present in mononucleated myosin-positive cells and in myosin-positive multinucleated myotubes (data not shown). Somitic expression of β-galactosidase protein directed by the MEF2 mutant transgene at 11.5 dpc was much weaker than that of the wild-type transgene, although it could be detected (Fig. 7J). As with the wild-type transgene, expression overlapped the expression of myosin and was restricted to mononucleated cells in caudal somites, although many myosin-positive cells were not positive for β-galactosidase at this stage (Fig. 7K and L).
FIG. 7.
The HRC enhancer directs β-galactosidase expression to myosin-positive skeletal muscle cells in the somites and limbs. Wild-type MEF2 site (A to C, G to I, and M to O) and mutant MEF2 site (D to F, J to L, and P to R) HRC-lacZ transgenic mice were analyzed for enhancer activity by X-Gal staining (A to F) or by immunohistochemistry using an anti-β-galactosidase polyclonal antibody and anantimyosin monoclonal antibody to detect protein expression (G to R). Antimyosin was detected using a TRITC-conjugated rabbit anti-mouse antibody (red), and anti-β-galactosidase was detected using an Oregon Green-conjugated goat anti-rabbit antibody (green). In panels I, L, O, and R, red and green digital photographs of the same section costained with the two markers were merged in Adobe Photoshop by overlaying the two images. The 2,726-bp wild-type (wt) HRC enhancer construct directed lacZ expression to the myotomal compartment of the somites (my) at 11.5 dpc (A, B, and G). This expression overlapped with the expression of myosin (H). Weak expression directed by the wild-type transgene could be observed in the limb bud at 11.5 dpc by X-Gal staining (C). By 13.5 dpc, the wild-type enhancer directed strong expression of β-galactosidase (M and O) to myosin-positive, multinucleated myotubes (N and O) in the forelimb. The mutant MEF2 2,726-bp HRC enhancer construct (mMEF2) directed weak but detectable expression to the myotomal compartment of the somites at 11.5 dpc (D, E, and J), and this expression overlapped with the expression of myosin (K). In mutant transgenic embryos, all cells expressing β-galactosidase in the myotome were myosin positive, but there were numerous myosin-positive cells that did not express detectable levels of β-galactosidase (L), which is in sharp contrast to the overlap observed from wild-type transgenic embryos (I). Also in contrast to the wild-type enhancer, the MEF2 mutant enhancer directed robust expression of lacZ to the forelimb bud at 11.5 dpc (F). However, by 13.5 dpc, β-galactosidase expression directed by the mutant enhancer in the muscles of the limb was barely detectable (P) in spite of the presence of numerous multinucleated, myosin-positive muscle fibers (Q). The weak expression directed by the mutant transgene was completely overlapping with the expression of myosin in the limbs (R). Sections through the somites at 11.5 dpc were cut at hind limb level. In all panels, dorsal is to the top, except for panels C, F, and M to R, in which dorsal is to the right. In all panels, the bar equals 100 μm. DRG, dorsal root ganglia; LB, limb bud; my, myotome; NT, neural tube.
By 13.5 dpc, lacZ expression in the limb muscles of MEF2 mutant transgenic mice (Fig. 6M) was noticeably weaker than expression directed by the wild-type transgene (Fig. 6I). β-Galactosidase expression directed by the wild-type transgene was clearly evident in multinucleated myotubes in the forelimb, although some mononucleated β-galactosidase cells could still be seen (Fig. 7M). The expression of β-galactosidase completely overlapped the expression of myosin (Fig. 7N and O), indicating that the transgene was exclusively expressed in muscle cells at this stage (Fig. 7O). The MEF2 mutant transgene was also exclusively expressed in muscle cells at this stage (Fig. 7R), but many myosin-positive cells in the limb (Fig. 7Q) had low or undetectable levels of β-galactosidase (Fig. 7P). Taken together, the results presented in Fig. 6 and 7 demonstrate that mutation of the MEF2 site in the HRC enhancer had a profound effect on transgene expression in skeletal muscle, where it appears to be required to maintain expression of the transgene after initial activation. This was evident in a subset of limb muscles, where the initial expression of the transgene was strong in the absence of a functional MEF2 site (Fig. 6F and 7F), but eventually expression was lost in those muscles (Fig. 6N and 7P). These observations suggest that MEF2 may not be required for the initial expression of HRC in skeletal myoblasts but may play a critical role in the maintenance of expression in those cells.
DISCUSSION
The HRC gene is expressed in skeletal, cardiac, and arterial smooth muscle (20, 21, 51). In this study, we have defined promoter and enhancer sequences that are sufficient to recapitulate that expression pattern in vivo (Fig. 3). Our results demonstrate that the function of the HRC enhancer is dependent on an evolutionarily conserved, high-affinity MEF2 site (Fig. 6). Furthermore, the HRC enhancer can be trans-activated by MEF2 factors independently of other tissue-restricted transcription factors (Fig. 5), strongly suggesting that it is a direct target of MEF2 in all three muscle lineages in vivo. The observation that the function of the HRC enhancer in arterial smooth muscle is dependent on MEF2 is consistent with earlier studies that used a multimerized MEF2 reporter in transgenic mice to show that functional MEF2 proteins are present in vascular smooth muscle during development (47). However, despite the evidence for a role for MEF2 proteins in smooth muscle differentiation and gene expression, no vertebrate MEF2-dependent smooth muscle target genes have previously been defined in vivo. Thus, these studies establish HRC as the first bona fide in vivo direct target of MEF2 factors in smooth muscle.
Several other genes expressed in vascular smooth muscle have had the cis-regulatory regions controlling their expression in vivo defined in transgenic analyses. In nearly every smooth muscle gene described to date, expression is dependent on the presence of one or more SRF-binding CArG boxes in the promoter or enhancer (26, 30, 32, 37-39, 44, 50), and a number of smooth muscle enhancers are able to discriminate arterial from venous smooth muscle expression in vivo, including the SM22α, CRP1, and desmin enhancers (31, 32, 44). In each of these cases, smooth muscle expression is largely restricted to arteries and is dependent on one or more conserved CArG boxes in the enhancer (26, 30, 32, 44).
While most smooth muscle genes defined to date are SRF dependent in vivo, there have been a few exceptions to this model. A recent paper by Chang et al. showed that the cysteine-rich protein 2 (CRP2) enhancer directed lacZ expression in smooth muscle in a pattern that was restricted to arteries in vivo through a CArG-independent pathway (12). Notably, the CRP2 enhancer also does not contain any MEF2 sites (12). The mouse aortic carboxypeptidase-like protein (ACLP) gene promoter directs expression to both venous and arterial smooth muscle in vivo, and it also lacks any discernible CArG or MEF2 elements (28). Importantly, the promoter and enhancer elements from the HRC gene are sufficient to direct expression to arterial smooth muscle in vivo, yet the HRC enhancer lacks any CArG motifs and cannot be trans-activated by SRF (Fig. 5D), suggesting that the HRC enhancer functions in smooth muscle via an SRF-independent pathway.
The cis-acting elements controlling the expression of several cardiac and skeletal muscle genes with products restricted to the SR have been analyzed in cell culture studies. These include the sarcoplasmic endoplasmic reticulum calcium ATPase 1 and 2 genes, the ryanodine receptor 1 and 2 genes, the phospholamban gene, and the calsequestrin gene (2, 3, 19, 43, 48, 54). The majority of these promoters contain at least one consensus MEF2 site, although no direct role for MEF2 has been demonstrated for any SR gene in vivo prior to the present study. It will be interesting to determine if other SR genes are also dependent on MEF2 for expression in vivo and if the genes are coordinately regulated at the transcriptional level.
As noted above, mutation of the MEF2 site in the HRC enhancer had a dramatic impact on expression in all three muscle lineages in vivo, but the initial activation in skeletal muscle did not appear to require the MEF2 site. The MEF2 mutant HRC enhancer was activated in somites almost as robustly as the wild-type enhancer at the earliest stages of transgene expression, but it failed to continue to express lacZ at levels comparable to the wild-type transgene (Fig. 6 and 7). Likewise, expression directed by the HRC enhancer in the dorsal limb muscles appeared to be independent of the MEF2 site at the time of initial activation (Fig. 6F and 7F). However, at later stages in development and in adulthood, the MEF2 mutant HRC-lacZ transgene was not expressed in any muscles, including those in the limbs (Fig. 6 and 7). These observations support a model for HRC expression in which the initial activation of the enhancer in skeletal muscle is MEF2 independent but in which maintenance of expression is MEF2 dependent. Furthermore, our data suggest that MEF2 factors may actually be playing an early repressive role on the HRC enhancer in skeletal muscle, since the initial activation in dorsal limb muscles is more robust when the MEF2 site is mutated (Fig. 6F and G and 7F). The observation that limb muscle expression in the MEF2 site mutant is more robust and appears to be activated precociously is consistent with a potential role for a MEF2-dependent recruitment of histone deacetylases (HDACs) to the enhancer (42). In this model, a MEF2-HDAC complex bound at the MEF2 site would repress activation by myogenic bHLH factors bound at a nearby E box (36, 42) until HDAC proteins were displaced and shuttled out of the nucleus (41, 42).
A hallmark of MEF2 transcription factor function is the potential ability to serve as either an activator or repressor of transcription in skeletal muscle due to the recruitment of either positive or negative transcriptional coregulators (42). MEF2 recruitment of HDAC proteins results in transcriptional repression, while displacement of HDACs by MyoD interaction with MEF2 results in strong activation of transcription (41, 42). Mutation of the MEF2 site in the HRC enhancer appears to disrupt an early repressive effect of MEF2 on the enhancer in skeletal muscles, particularly in the limbs (Fig. 6 and 7). In contrast to its initial robust activation in skeletal muscle, the MEF2 mutant enhancer was never expressed in cardiac or smooth muscle, even at the earliest times of HRC expression (Fig. 6). This observation suggests that MEF2 may play an essential role in the initial activation of HRC in cardiac and smooth muscle. While MEF2 may be crucial for the initiation of HRC transcription in these lineages, we consider it unlikely that MEF2 is the only critical regulator of HRC in cardiac and smooth muscle. Instead, we favor a model in which MEF2 cooperates with other lineage-specific or -restricted transcription factors. This type of cooperative model for MEF2 factors has been demonstrated in skeletal muscle transcription, where MEF2 proteins cooperate with myogenic bHLH proteins to activate transcription (7, 45). Similarly, the cardiac-restricted transcription factor GATA4 has been shown to recruit MEF2 proteins to target promoters, resulting in transcriptional synergy (46). A cooperative model for transcriptional activation in smooth muscle has been demonstrated for SRF (10) but, to date, no coregulators of MEF2 activity have been identified in smooth muscle. It seems likely that additional MEF2 transcriptional coregulators function in the activation of HRC and other genes in each muscle lineage, and it will be interesting to determine whether previously unidentified MEF2 cofactors are responsible for the activation of the HRC enhancer in cardiac and smooth muscle.
Acknowledgments
We thank Sandy Hofmann (UT-Southwestern) for the original human HRC genomic clone.
E.J.J. and E.D. were supported in part by fellowships from the American Heart Association, Western States Affiliate. A.B.H. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship. This work was supported by the D.W. Reynolds Center for Clinical Cardiovascular Research to E.N.O., by a grant from the American Heart Association, Western States Affiliate, to B.L.B., and by grants from the National Institutes of Health to E.N.O. and B.L.B.
REFERENCES
- 1.Andres, V., M. Cervera, and V. Mahdavi. 1995. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J. Biol. Chem. 270:23246-23249. [DOI] [PubMed] [Google Scholar]
- 2.Baker, D. L., V. Dave, T. Reed, S. Misra, and M. Periasamy. 1998. A novel E box/AT-rich element is required for muscle-specific expression of the sarcoplasmic reticulum Ca2+-ATPase (SERCA2) gene. Nucleic Acids Res. 26:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, D. L., V. Dave, T. Reed, and M. Periasamy. 1996. Multiple Sp1 binding sites in the cardiac/slow twitch muscle sarcoplasmic reticulum Ca2+-ATPase gene promoter are required for expression in Sol8 muscle cells. J. Biol. Chem. 271:5921-5928. [DOI] [PubMed] [Google Scholar]
- 4.Bi, W., C. J. Drake, and J. J. Schwarz. 1999. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev. Biol. 211:255-267. [DOI] [PubMed] [Google Scholar]
- 5.Black, B. L., J. Lu, and E. N. Olson. 1997. The MEF2A 3′ untranslated region functions as a cis-acting translational repressor. Mol. Cell. Biol. 17:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, B. L., J. F. Martin, and E. N. Olson. 1995. The mouse MRF4 promoter is trans-activated directly and indirectly by muscle-specific transcription factors. J. Biol. Chem. 270:2889-2892. [DOI] [PubMed] [Google Scholar]
- 7.Black, B. L., J. D. Molkentin, and E. N. Olson. 1998. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol. Cell. Biol. 18:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black, B. L., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14:167-196. [DOI] [PubMed] [Google Scholar]
- 9.Bour, B. A., M. A. O'Brien, W. L. Lockwood, E. S. Goldstein, R. Bodmer, P. H. Taghert, S. M. Abmayr, and H. T. Nguyen. 1995. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 9:730-741. [DOI] [PubMed] [Google Scholar]
- 10.Chang, D. F., N. S. Belaguli, D. Iyer, W. B. Roberts, S. P. Wu, X. R. Dong, J. G. Marx, M. S. Moore, M. C. Beckerle, M. W. Majesky, and R. J. Schwartz. 2003. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell 4:107-118. [DOI] [PubMed] [Google Scholar]
- 11.Chang, P. S., L. Li, J. McAnally, and E. N. Olson. 2001. Muscle specificity encoded by specific serum response factor-binding sites. J. Biol. Chem. 276:17206-17212. [DOI] [PubMed] [Google Scholar]
- 12.Chang, Y. F., J. Wei, X. Liu, Y. H. Chen, M. D. Layne, and S. F. Yet. 2003. Identification of a CArG-independent region of the cysteine-rich protein 2 promoter that directs expression in the developing vasculature. Am. J. Physiol. Heart Circ. Physiol. 285:H1675-H1683. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, T. C., M. C. Wallace, J. P. Merlie, and E. N. Olson. 1993. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science 261:215-218. [DOI] [PubMed] [Google Scholar]
- 14.Cripps, R. M., B. L. Black, B. Zhao, C. L. Lien, R. A. Schulz, and E. N. Olson. 1998. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 12:422-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damiani, E., and A. Margreth. 1991. Subcellular fractionation to junctional sarcoplasmic reticulum and biochemical characterization of 170 kDa Ca2+- and low-density-lipoprotein-binding protein in rabbit skeletal muscle. Biochem. J. 277:825-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodou, E., S. M. Xu, and B. L. Black. 2003. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech. Dev. 120:1021-1032. [DOI] [PubMed] [Google Scholar]
- 17.Edmondson, D. G., T. C. Cheng, P. Cserjesi, T. Chakraborty, and E. N. Olson. 1992. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol. Cell. Biol. 12:3665-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firulli, A. B., J. M. Miano, W. Bi, A. D. Johnson, W. Casscells, E. N. Olson, and J. J. Schwarz. 1996. Myocyte enhancer binding factor-2 expression and activity in vascular smooth muscle cells. Association with the activated phenotype. Circ. Res. 78:196-204. [DOI] [PubMed] [Google Scholar]
- 19.Frank, K. F., L. Mesnard-Rouiller, G. Chu, K. B. Young, W. Zhao, K. Haghighi, Y. Sato, and E. G. Kranias. 2001. Structure and expression of the mouse cardiac calsequestrin gene. Basic Res. Cardiol. 96:636-644. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann, S. L., M. S. Brown, E. Lee, R. K. Pathak, R. G. Anderson, and J. L. Goldstein. 1989. Purification of a sarcoplasmic reticulum protein that binds Ca2+ and plasma lipoproteins. J. Biol. Chem. 264:8260-8270. [PubMed] [Google Scholar]
- 21.Hofmann, S. L., J. L. Goldstein, K. Orth, C. R. Moomaw, C. A. Slaughter, and M. S. Brown. 1989. Molecular cloning of a histidine-rich Ca2+-binding protein of sarcoplasmic reticulum that contains highly conserved repeated elements. J. Biol. Chem. 264:18083-18090. [PubMed] [Google Scholar]
- 22.Hofmann, S. L., M. Topham, C. L. Hsieh, and U. Francke. 1991. cDNA and genomic cloning of HRC, a human sarcoplasmic reticulum protein, and localization of the gene to human chromosome 19 and mouse chromosome 7. Genomics 9:656-669. [DOI] [PubMed] [Google Scholar]
- 23.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 24.Horton, R. M. 1997. In vitro recombination and mutagenesis of DNA: SOEing together tailor-made genes, p. 141-149. In B. A. White (ed.), PCR cloning protocols, vol. 67. Humana Press, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 25.Kim, E., D. W. Shin, C. S. Hong, D. Jeong, H. Kim Do, and W. J. Park. 2003. Increased Ca2+ storage capacity in the sarcoplasmic reticulum by overexpression of HRC (histidine-rich Ca2+ binding protein). Biochem. Biophys. Res. Commun. 300:192-196. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S., H. S. Ip, M. M. Lu, C. Clendenin, and M. S. Parmacek. 1997. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 17:2266-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kothary, R., S. Clapoff, S. Darling, M. D. Perry, L. A. Moran, and J. Rossant. 1989. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development 105:707-714. [DOI] [PubMed] [Google Scholar]
- 28.Layne, M. D., S. F. Yet, K. Maemura, C. M. Hsieh, X. Liu, B. Ith, M. E. Lee, and M. A. Perrella. 2002. Characterization of the mouse aortic carboxypeptidase-like protein promoter reveals activity in differentiated and dedifferentiated vascular smooth muscle cells. Circ. Res. 90:728-736. [DOI] [PubMed] [Google Scholar]
- 29.Lee, H. G., H. Kang, D. H. Kim, and W. J. Park. 2001. Interaction of HRC (histidine-rich Ca2+-binding protein) and triadin in the lumen of sarcoplasmic reticulum. J. Biol. Chem. 276:39533-39538. [DOI] [PubMed] [Google Scholar]
- 30.Li, L., Z. Liu, B. Mercer, P. Overbeek, and E. N. Olson. 1997. Evidence for serum response factor-mediated regulatory networks governing SM22α transcription in smooth, skeletal, and cardiac muscle cells. Dev. Biol. 187:311-321. [DOI] [PubMed] [Google Scholar]
- 31.Li, L., J. M. Miano, B. Mercer, and E. N. Olson. 1996. Expression of the SM22α promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J. Cell Biol. 132:849-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lilly, B., E. N. Olson, and M. C. Beckerle. 2001. Identification of a CArG box-dependent enhancer within the cysteine-rich protein 1 gene that directs expression in arterial but not venous or visceral smooth muscle cells. Dev. Biol. 240:531-547. [DOI] [PubMed] [Google Scholar]
- 33.Lilly, B., B. Zhao, G. Ranganayakulu, B. M. Paterson, R. A. Schulz, and E. N. Olson. 1995. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267:688-693. [DOI] [PubMed] [Google Scholar]
- 34.Lin, Q., J. Lu, H. Yanagisawa, R. Webb, G. E. Lyons, J. A. Richardson, and E. N. Olson. 1998. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development 125:4565-4574. [DOI] [PubMed] [Google Scholar]
- 35.Lin, Q., J. Schwarz, C. Bucana, and E. N. Olson. 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 37.Mack, C. P., and G. K. Owens. 1999. Regulation of smooth muscle alpha-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ. Res. 84:852-861. [DOI] [PubMed] [Google Scholar]
- 38.Mack, C. P., M. M. Thompson, S. Lawrenz-Smith, and G. K. Owens. 2000. Smooth muscle alpha-actin CArG elements coordinate formation of a smooth muscle cell-selective, serum response factor-containing activation complex. Circ. Res. 86:221-232. [DOI] [PubMed] [Google Scholar]
- 39.Manabe, I., and G. K. Owens. 2001. CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J. Clin. Investig. 107:823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFadden, D. G., J. Charite, J. A. Richardson, D. Srivastava, A. B. Firulli, and E. N. Olson. 2000. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development 127:5331-5341. [DOI] [PubMed] [Google Scholar]
- 41.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11:497-504. [DOI] [PubMed] [Google Scholar]
- 43.McTiernan, C. F., C. S. Frye, B. H. Lemster, E. A. Kinder, M. L. Ogletree-Hughes, C. S. Moravec, and A. M. Feldman. 1999. The human phospholamban gene: structure and expression. J. Mol. Cell. Cardiol. 31:679-692. [DOI] [PubMed] [Google Scholar]
- 44.Mericskay, M., A. Parlakian, A. Porteu, F. Dandre, J. Bonnet, D. Paulin, and Z. Li. 2000. An overlapping CArG/octamer element is required for regulation of desmin gene transcription in arterial smooth muscle cells. Dev. Biol. 226:192-208. [DOI] [PubMed] [Google Scholar]
- 45.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 46.Morin, S., F. Charron, L. Robitaille, and M. Nemer. 2000. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 19:2046-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naya, F. J., C. Wu, J. A. Richardson, P. Overbeek, and E. N. Olson. 1999. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development 126:2045-2052. [DOI] [PubMed] [Google Scholar]
- 48.Nishida, K., K. Otsu, M. Hori, T. Kuzuya, and M. Tada. 1996. Cloning and characterization of the 5′-upstream regulatory region of the Ca2+-release channel gene of cardiac sarcoplasmic reticulum. Eur. J. Biochem. 240:408-415. [DOI] [PubMed] [Google Scholar]
- 49.Ornatsky, O. I., J. J. Andreucci, and J. C. McDermott. 1997. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 272:33271-33278. [DOI] [PubMed] [Google Scholar]
- 50.Parmacek, M. S. 2001. Transcriptional programs regulating vascular smooth muscle cell development and differentiation. Curr. Top. Dev. Biol. 51:69-89. [DOI] [PubMed] [Google Scholar]
- 51.Pathak, R. K., R. G. Anderson, and S. L. Hofmann. 1992. Histidine-rich calcium binding protein, a sarcoplasmic reticulum protein of striated muscle, is also abundant in arteriolar smooth muscle cells. J. Muscle Res. Cell Motil. 13:366-376. [DOI] [PubMed] [Google Scholar]
- 52.Picello, E., E. Damiani, and A. Margreth. 1992. Low-affinity Ca2+-binding sites versus Zn2+-binding sites in histidine-rich Ca2+-binding protein of skeletal muscle sarcoplasmic reticulum. Biochem. Biophys. Res. Commun. 186:659-667. [DOI] [PubMed] [Google Scholar]
- 53.Ranganayakulu, G., B. Zhao, A. Dokidis, J. D. Molkentin, E. N. Olson, and R. A. Schulz. 1995. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev. Biol. 171:169-181. [DOI] [PubMed] [Google Scholar]
- 54.Schmoelzl, S., T. Leeb, H. Brinkmeier, G. Brem, and B. Brenig. 1996. Regulation of tissue-specific expression of the skeletal muscle ryanodine receptor gene. J. Biol. Chem. 271:4763-4769. [DOI] [PubMed] [Google Scholar]
- 55.Suk, J. Y., Y. S. Kim, and W. J. Park. 1999. HRC (histidine-rich Ca2+ binding protein) resides in the lumen of sarcoplasmic reticulum as a multimer. Biochem. Biophys. Res. Commun. 263:667-671. [DOI] [PubMed] [Google Scholar]
- 56.Verzi, M. P., J. P. Anderson, E. Dodou, K. K. Kelly, S. B. Greene, B. J. North, R. M. Cripps, and B. L. Black. 2002. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev. Biol. 249:174-190. [DOI] [PubMed] [Google Scholar]
- 57.Wang, D. Z., S. Li, D. Hockemeyer, L. Sutherland, Z. Wang, G. Schratt, J. A. Richardson, A. Nordheim, and E. N. Olson. 2002. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. USA 99:14855-14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Z., D. Z. Wang, G. C. Pipes, and E. N. Olson. 2003. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. USA 100:7129-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yee, S. P., and P. W. Rigby. 1993. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 7:1277-1289. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida, T., S. Sinha, F. Dandre, B. R. Wamhoff, M. H. Hoofnagle, B. E. Kremer, D. Z. Wang, E. N. Olson, and G. K. Owens. 2003. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ. Res. 92:856-864. [DOI] [PubMed] [Google Scholar]