Abstract
HOXA9 expression is a common feature of acute myeloid leukemia, and high-level expression is correlated with poor prognosis. Moreover, HOXA9 overexpression immortalizes murine marrow progenitors that are arrested at a promyelocytic stage of differentiation when cultured and causes leukemia in recipient mice following transplantation of HOXA9 expressing bone marrow. The molecular mechanisms underlying the physiologic functions and transforming properties of HOXA9 are poorly understood. This study demonstrates that HOXA9 is phosphorylated by protein kinase C (PKC) and casein kinase II and that PKC mediates phosphorylation of purified HOXA9 on S204 as well as on T205, within a highly conserved consensus sequence, in the N-terminal region of the homeodomain. S204 in the endogenous HOXA9 protein was phosphorylated in PLB985 myeloid cells, as well as in HOXA9-immortalized murine marrow cells. This phosphorylation was enhanced by phorbol ester, a known inducer of PKC, and was inhibited by a specific PKC inhibitor. PKC-mediated phosphorylation of S204 decreased HOXA9 DNA binding affinity in vitro and the ability of the endogenous HOXA9 to form cooperative DNA binding complexes with PBX. PKC inhibition significantly reduced the phorbol-ester induced differentiation of the PLB985 hematopoietic cell line as well as HOXA9-immortalized murine bone marrow cells. These data suggest that phorbol ester-induced myeloid differentiation is in part due to PKC-mediated phosphorylation of HOXA9, which decreases the DNA binding of the homeoprotein.
HOXA9 is a member of the HOX homeobox gene family, a family of genes which regulate body pattern formation and tissue identity during embryogenesis (15, 24, 36). In addition to their vital role in vertebrate morphogenesis, HOX genes are also expressed during adult hematopoiesis (31, 35). HOX gene expression is tightly regulated during the process of hematopoietic proliferation and differentiation, and in general, HOX gene expression is observed in primitive progenitors and down-regulated as cells mature.
There is a substantial body of data demonstrating a role for the HOXA9 protein in both normal hematopoiesis and leukemic transformation. HOXA9-deficient mice show defects in myeloid, erythroid, and lymphoid compartments (20, 29). Overexpression of the HOXA9 protein immortalizes and blocks the differentiation of growth factor-dependent myeloid progenitors (7, 47), eventually leading to malignant transformation after a latency period, indicating that secondary events are required (26, 54). MEIS1, a member of the TALE (3-amino-acid loop extension) homeodomain (HD) family (39), is a leukemic collaborator that shortens the latency period of HOXA9 induced myeloid leukemias (54). HOXA9 is overexpressed in a majority of human acute myeloid leukemia (AML) cases and is frequently coexpressed with MEIS1 (30), and high expression of HOXA9 has been shown to correlate with poor patient prognosis (18). HOXA9 is overexpressed in a subset of human AMLs as a NUP98-HOXA9 chimeric protein resulting from a t(7;11) chromosomal translocation (3, 41). The NUP98-HOXA9 fusion protein also immortalizes myeloid progenitors and arrests growth factor-induced differentiation (8). Mice overexpressing the NUP98-HOXA9 fusion protein develop a myeloproliferative disorder, with a phenotype that is similar to the human disease, and it is accelerated to AML with MEIS1 coexpression (27). The chromosomal translocation-generated oncoprotein E2A-PBX1a also has been shown to collaborate with HOXA9 to transform mouse bone marrow cells (55). HOXA9 as well as other HOX genes have been proposed to be common distal oncogenes which are indirectly activated in many myeloid leukemias by a variety of chromosomal translocations (32).
A number of recent studies have begun to define the molecular mechanism of HOXA9 function. Like other HOX proteins, HOXA9 binds DNA through its HD (40). HOXA9 forms a heterodimeric DNA binding complex with PBX on the consensus sequence TGATTTA(C/T), which increases its DNA binding affinity (50). The PBX interaction site was shown to consist of an ANW sequence that is conserved in HOX9 and 10 paralog proteins (50), and this interaction was recently confirmed by X-ray structure analysis (28). In addition, HOXA9 has been shown to interact indirectly with MEIS1 in myeloid cells, as part of a cooperative triple complex with PBX (51). HOXA9 is thought to function as a transcription factor, and we have recently identified putative HOXA9 gene targets by DNA microarray analysis and have shown that HOXA9 can act both as an activator and repressor of transcription (13).
Examination of the HOXA9 amino acid sequence revealed potential consensus phosphorylation sites for protein kinase C (PKC) and casein kinase II (CK-II), including two PKC consensus sequences in the N-terminal region of the HD. The PKCα isoform has been shown to be ubiquitously expressed in hematopoietic lineages, including CD34+ primitive hematopoietic progenitors (44), and has been shown to promote myeloid cell differentiation (38). Among the biologic effects of phorbol esters is the capacity to induce myeloid cell differentiation (25, 33, 46), and previous studies have shown that phorbol esters often function by activation of PKC (34, 53, 59, 60). In this study, we have identified the HOXA9 residues phosphorylated by PKCα and CK-II and demonstrate that PKC-mediated phosphorylation of HOXA9 on S204 blocks DNA binding. Finally, we show that phorbol ester induction of myeloid cell differentiation is correlated with phosphorylation of HOXA9 on S204 and the loss of in vivo DNA binding activity, suggesting that PKC regulates the role of HOXA9 in myeloid cell proliferation and differentiation.
MATERIALS AND METHODS
Protein expression.
cDNAs encoding a full-length murine HOXA9 protein (accession number NM_010456) and murine HOXA9-HD, which comprises the 60 amino acid residues of the HD plus short N-terminal (S204) and C-terminal (K265-E271) tails (49), were subcloned into a derivative of the pET28a expression vector (Novagen) in which a FLAG epitope sequence (MDYKDDDDK) replaced the T7 epitope, downstream of the His-Tag sequence as previously described (51). By site-directed mutagenesis (Ex-site Mutagenesis; Stratagene), codons for S204 and T205 were replaced with codons for Ala in the full-length protein to make HOXA9-S204A/T205A and HOXA9-HD protein individually and in combination to make HOXA9-HD-S204A/T205A, HOXA9-HD-S204A, and HOXA9-HD-T205A. All mutations were confirmed by sequence analysis. The His-tagged proteins were grown and purified using Probond resin (Invitrogen) according to the standard protocol. HOXA9 proteins were synthesized using the TNT-coupled in vitro transcription-translation system (Promega) in the presence and absence of [35S]methionine, for gel-shift and in vitro kinase assays. Electrophoresis of the labeled proteins demonstrated synthesis of the appropriate length products (data not shown).
Antibodies.
Antibodies to HOXA9 were as follows: a goat antipeptide antibody (N20) from Santa Cruz Biotechnology (antibody 1), an affinity-purified chicken antibody to the full-length protein (51) (antibody 2), and an affinity-purified rabbit antibody to a peptide N terminal to the HD (ENESGGDKPPIDPNN) (antibody 3). Two different custom rabbit α-pS204 antibodies were generated using the phosphorylated HOXA9 peptide sequence ANWLHAR(pS)TRKKR, affinity purified against the phosphorylated peptide, and then cross-absorbed against the nonphosphorylated peptide (Invitrogen). Preliminary experiments established that both the resulting antibodies recognized PKC-phosphorylated HOXA9 but showed minimal residual binding to the native HOXA9 protein (data not shown). Rabbit antibody to PBX1a was from Santa Cruz Biotechnology. M2 antibody to FLAG was from Sigma. Rabbit antibody to pSer was from Zymed Laboratories.
Phosphorylation assays.
For in vitro kinase assays with purified enzymes, affinity-purified FLAG-HOXA9 and FLAG-HOXA9-HD proteins immobilized on beads or control beads were incubated with either PKCα (100 U; Panvera) in PKC assay buffer (20 mM morpholinepropanesulfonic acid, 25 mM β-glycerol phosphate, 1 mM sodium orthovanadate, 1 mM dithiothreitol, 1 mM CaCl2, 75 mM magnesium chloride, 500 μM ATP, 2 μM PKA inhibitor peptide, 20 μM CaM kinase inhibitor compound R24571, PKC lipid activators phosphatidyl serine [0.5 mg/ml] and diacylglycerol [0.05 mg/ml]) and 1 μl of [γ-32P]ATP (6,000 Ci/mmol; NEN Dupont) at 30°C for 10 min or CK-II in reaction buffer (20 mM HEPES, 20 mM MgCl2, 27 mM β-glycerol phosphate, 0.5 mM dithiothreitol, 0.4 mM sodium orthovanadate, 20 μM ATP) and 1 μl of [γ-32P]ATP at 30°C for 30 min.
For in vitro kinase assays with cell extracts, TNT-coupled in vitro transcription-translated FLAG-HOXA9 protein was immunoprecipitated overnight with anti-FLAG (α-FLAG) antibody on protein G beads. The beads were washed two times with wash buffer (75 mM NaCl, 15 mM Tris HCl, 1% bovine serum albumin [BSA]) and then incubated with U937 or THP-1 whole-cell lysates (lysis buffer-0.1% Triton X-100, 20 mM HEPES, 75 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, 25 mM β-glycerol phosphate, 0.5 mM dithiothreitol, 0.4 mM sodium orthovanadate, 50 mM phenylmethylsulfonyl fluoride [PMSF], aprotinin [25 μg/ml], leupeptin [25 μg/ml]) for 1 h at 4°C. The beads were washed with wash buffer (above) and then incubated in reaction buffer (defined above) and 1 μl of [γ-32P]ATP at 30°C for 30 min. The immunoprecipitated HOXA9 was also incubated with purified PKC and CK-II as described above. All reactions were terminated by adding sodium dodecyl sulfate (SDS) sample buffer and boiling for 5 min. Protein samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiography.
Phosphopeptide mapping and phosphoamino acid analysis.
PKC-phosphorylated 32P-labeled HOXA9-HD protein prepared as described above was subjected to SDS-PAGE and autoradiography. The labeled HOXA9-HD band was excised and subjected to in-gel trypsin (Promega) digestion, and the tryptic peptides were extracted with 60% acetonitrile-0.1% trifluoroacetic acid. The phosphopeptides (2,000 cpm) were subjected to electrophoresis followed by chromatography as described by Boyle et al. (5). For phosphoamino acid analysis, the phosphopeptides scraped off the TLC plate were eluted and hydrolyzed with 6 N HCl at 110°C for 60 min. The hydrolysate was dried, resuspended in 4 μl of buffer (pH 1.9) and then electrophoresed according to the protocol as described previously (5).
HPLC and Edman sequencing.
Gel-purified full-length PKC-phosphorylated 32P-labeled FLAG-HOXA9 protein was digested with chymotrypsin (Sigma), endoproteinase Glu-C (StaphA/V8) (Roche Biochemicals), or trypsin. CK-II-phosphorylated 32P-labeled FLAG-HOXA9 was digested with endoproteinase Glu-C or trypsin. The resulting peptides were dissolved in 0.1% trifluoroacetic acid and resolved on a C18 reverse-phase high-performance liquid chromatography (HPLC) column (The Separations Group) as described previously (48). Peptides were detected at 215 nm absorbance, and aliquots from each fraction were tested for radioactivity. The radioactive fractions contributing to each peak were pooled, dried, and used for phosphoamino acid analysis or sequence analysis by Edman degradation using a Procise sequencer (Perkin-Elmer). The radioactivity in each Edman cycle fraction, which corresponds to sequential N-terminal amino acids, was counted. By combining the observed cycles in which radioactivity was released with the enzymatic cleavage specificities, we were able to determine the PKC- and CK-II-phosphorylated residues.
Cytoplasmic and nuclear extraction, immunoprecipitation, Western blotting, and phosphatase treatment.
PLB985 and U937 cells were cultured in RPMI supplemented with 10% fetal calf serum (FCS). Prior to cell stimulation the medium was changed to RPMI containing 0.5% serum. PLB985 cells or murine bone marrow cell lines infected with FLAG-HOXA9 retrovirus were stimulated with either 12-O-tetradecanoylphorbol-13-acetate (TPA) (100 nM; Sigma) or dimethyl sulfoxide control for 30 min or pretreated with a specific PKC inhibitor (bisindolylmaleamide1) (5 μM; Calbiochem) for 5 min prior to stimulation with TPA. Following stimulation of PLB985 cells, cytoplasmic and nuclear extracts were prepared by standard method (1). HOXA9 protein was immunoprecipitated from precleared cytoplasmic and nuclear extracts with goat α-HOXA9 antibody overnight at 4°C on protein G beads. Whole-cell lysates of murine bone marrow-HOXA9 cells were prepared by solubilizing cells briefly in SDS lysis buffer (0.5% [wt/vol] SDS, 0.05 M Tris Cl [pH 8], 1 mM dithiothreitol), followed by RIPA correction buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.01 M sodium phosphate [pH 7.2], 2 mM EDTA, 50 mM sodium fluoride, 0.2 mM sodium vanadate, 1× EDTA-free protease inhibitor cocktail). Following incubation with α-FLAG antibody overnight at 4°C, the immunoprecipitated proteins were washed with RIPA buffer containing 3% BSA and eluted in SDS sample buffer. Equal amounts of the immunoprecipitated samples were subjected to SDS-PAGE and Western blotting with chicken α-HOXA9 antibody or α-pS204 antibody. For phosphatase treatment, immunoprecipitated proteins on protein G beads from cells stimulated with TPA were incubated with protein phosphatase (λ-PPase; New England BioLabs) in phosphatase buffer (50 mM Tris-HCl [pH 7.5], 0.1 mM Na2EDTA, 5 mM dithiothreitol, 0.01% Brij 35, supplemented with 2 mM MnCl2) or control buffer at 30°C for 30 min. The immunoprecipitates were then washed with RIPA buffer supplemented with 3% BSA, eluted in SDS sample buffer, and processed as indicated above. To demonstrate that the phosphatase was active, phosphatase incubation was performed on purified PKCα-phosphorylated 32P-labeled HOXA9 protein. The phosphorylated protein was completely dephosphorylated after the phosphatase treatment (data not shown).
EMSA.
An oligonucleotide containing a consensus heterodimeric binding site for PBX1a and HOXA9 (ctgcgATGATTTACGACcgc) was used for gel shift assays, performed as described previously (49). Prior to electrophoretic mobility shift assay (EMSA), 3 μl of reticulocyte lysate reaction mixture, containing HOXA9 or HOXA9-S204A/T205A mutant, was incubated in the presence or absence of PKCα under conditions described earlier, and then 5 μl of the PKC assay reaction mixture (containing 1 μl of the original lysate) was incubated with the 32P-labeled oligonucleotide probe and subjected to EMSA. Nuclear extracts from PLB985 cells stimulated with control or with TPA were prepared as described earlier, and 5-μl aliquots of the extracts were used in the gel shift reaction. In some cases the extracts were preincubated with antibodies against HOXA9 (chicken) or PBX1a at 4°C for 30 min prior to EMSA.
Cell lines.
HOXA9 and HOXA9-S204A/T205A immortalized cell lines were prepared by a modification of a previously described method (7). Briefly, murine bone marrow progenitors were isolated from femurs and tibias of 8- to 10-week-old C57BL/6 mice 5 days after treatment with 5-fluorouracil (150 mg/kg of body weight) by intraperitoneal injection and incubated in “prestim” medium (DME-H21 medium supplemented with 15% heat-inactivated (HI) FCS, 1× penicillin-streptomycin (P/S), stem cell factor (100 ng/ml), interleukin-6 (IL-6) (50 ng/ml), and IL-3 (10 ng/ml) (all cytokines from StemCell Technologies) for 48 h. High-titer retrovirus was generated by transient transfection of 293-T cells with the desired retroviral vector and the pCL ecotrophic packaging plasmid (42). Marrow infections were accomplished by two “spinoculations” performed on consecutive days; marrow progenitors were incubated with retroviral supernatants in the presence of Polybrene (2 μg/ml) while spinning at 1,000 × g for 90 min at room temperature. Marrow cells were then transferred to growth medium (RPMI, 10% HI FCS, 1× P/S, and either IL-3 [5 ng/ml] or 1% B16 granulocyte-macrophage cell-stimulating factor [GM-CSF] conditioned medium). Nonadherent cells were transferred to new plates every 7 days. PLB 985 cells were grown in RPMI-10% HI FCS-1× P/S.
Differentiation assays.
PLB985 cells or HOXA9- or HOXA9-S204A/T205A-immortalized murine bone marrow cells were washed twice with phosphate-buffered saline and plated in triplicate in six-well plates at a density of 0.5 × 106 cells/well containing RPMI, 10% HI FCS, and 1× P/S. To induce differentiation the cells were treated with 100 nM TPA. In some cases, the cells were pretreated with 2 μM PKC inhibitor 5 min prior to stimulation with TPA. At 24 h after cell stimulation nonadherent cells were collected by aspiration and gentle rinsing of culture dishes two times with phosphate-buffered saline. Adherent cells were then dislodged with a rubber policeman and collected. Viable adherent and nonadherent cells were counted based on trypan blue dye exclusion. Cytospins of pooled adherent and nonadherent cells were stained with Wright-Giemsa stain. Monocytic differentiation was assessed by adherence to tissue culture plates and morphological features, including nuclear morphology and cytoplasmic characteristics. Immature cells were defined as those with high nuclear/cytoplasmic ratios and open (noncondensed) chromatin with or without prominent nucleoli. Features consistent with monocytic differentiation included a decreased nuclear/cytoplasmic ratio, condensed chromatin, indented or folded nucleus, and eccentrically placed nucleus. For all histologic analyses, a minimum of 300 cells from more than 10 distinct 100× fields were examined. A minimum of three experiments was performed with each cell line.
RESULTS
HOXA9 protein is phosphorylated in vitro by PKC and CK-II.
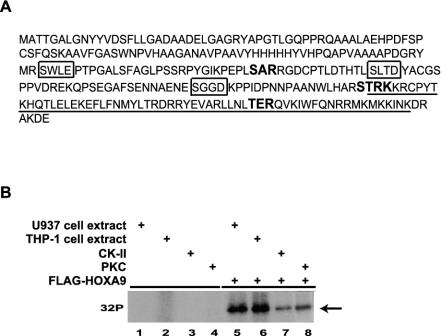
The consensus PKC and CK-II phosphorylation sequences, as determined by the Prosite sequence analysis program, in the HOXA9 protein are shown in Fig. 1A. Since HOXA9 is broadly expressed in myeloid cells, total cell extracts from two commonly used myeloid cells, U937 and THP-1, were used to determine if the HOXA9 protein could be phosphorylated. Reticulocyte lysate-synthesized FLAG-HOXA9, consisting of a full-length 260-amino-acid protein fused to an N-terminal FLAG epitope tag (Fig. 1A), was incubated with either U937 or THP-1 cell extracts or purified PKC and CK-II enzymes in the presence of [γ-32P]ATP. As shown in Fig. 1B, FLAG-HOXA9 was phosphorylated by U937 and THP-1 cell extracts, as well as by PKC, and CK-II. As shown in Fig. 2A, purified FLAG-HOXA9-HD protein, consisting of the 60-amino-acid HD plus S204 at the N terminus and the short 6-amino-acid C-terminal tail, was also phosphorylated by U937 cell extract and PKC but was only weakly phosphorylated by CK-II. These results suggested that (i) HOXA9 protein was a substrate for PKC and CK-II, (ii) at least some phosphorylation sites are within the HD, and (iii) kinases that phosphorylate HOXA9 are present in U937 and THP-1 myeloid leukemia cells. In these experiments it is difficult to assess the relative contribution of PKC or CK-II to total phosphorylation activity in the lysates, since the kinase profiles of the lysates are unknown. Nevertheless, the most-parsimonious interpretation of the phosphorylation intensities in Fig. 1B is that additional kinases act on the HOXA9 protein. A similar analysis would suggest that PKC phosphorylates the HOXA9 HD as effectively as the cell lysate (Fig. 2A), and PKC may be the major kinase acting on the HD.
FIG. 1.
HOXA9 is phosphorylated in vitro. (A) Sequence of the HOXA9 full-length protein with the PKC consensus sequences (in boldface type) and CK-II consensus sequences (boxed) as predicted by the Prosite sequence analysis program. The HD sequence is underlined. (B) His-tagged FLAG-HOXA9 protein synthesized using the TNT reticulocyte lysate system or control lysate was subjected to kinase assays with U937 cell extracts, THP-1 cell extracts, purified CK-II, or purified PKC. Radiolabeled bands migrating with the mobility of HOXA9 were detected in the kinase reactions with cell extracts as well as purified enzymes.
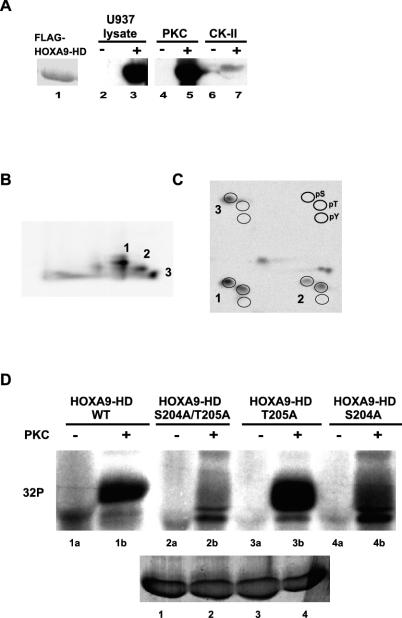
FIG. 2.
FLAG-HOXA9-HD protein is phosphorylated in vitro by PKC at S204 and T205. (A) FLAG-HOXA9-HD (consisting of the 60-amino-acid HD and 1 N-terminal residue [S204] and 6 C-terminal amino acids [K265-E271]) expressed from the pET28a expression vector was purified as a His-tagged protein on Probond resin and visualized by Coomassie blue staining (lane 1). Lanes 2 to 6 represent autoradiographs of His-tagged FLAG-HOXA9-HD immobilized on the resin (lanes 3, 5, and 7) or control resin that were subjected to kinase assays with U937 cell extracts (lanes 2 and 3), purified PKC (lanes 4 and 5) and purified CK-II (lanes 6 and 7) in the presence of [γ-32P]ATP, followed by elution and electrophoresis. (B) Tryptic peptides of PKC phosphorylated, 32P-labeled FLAG-HOXA9-HD were separated by thin-layer electrophoresis with pH 1.9 buffer in the first dimension and chromatography with n-butanol-pyridine-acetic acid-H2O in the second dimension. Three strong phosphopeptides were detected. (C) Phosphoamino acid analysis was performed on the three phosphopeptides. Phosphorylated serine, threonine, and tyrosine residues migrate in a standard pattern indicated at the top right. (D) Affinity-purified FLAG-HOXA9-HD wild-type (#1) and mutant proteins; S204A/T205A (#2), T205A (#3), S204A (#4), immobilized on beads were subjected to in vitro kinase assay in the absence (lanes 1a to 4a) or presence (lanes 1b to 4b) of purified PKC. Phosphorylated bands for FLAG-HOXA9-HD-WT and -T205A proteins were detected in the PKC-treated reactions, whereas very weak or no phosphorylated bands were detected for FLAG-HOXA9-HD-S204A/T205A and -S204A mutant proteins. The bottom gel represents Coomassie blue-stained bands demonstrating equal amounts of wild-type, and mutant proteins were used for the labeling experiment.
PKC phosphorylates HOXA9-HD protein at the STRK site.
The most interesting of the Prosite-identified PKC sites in HOXA9 are the overlapping sites (STRK) (PKC sites are shown in boldface type) located at the extreme N terminus of the HD. This sequence is highly conserved among HOXA9 proteins from a range of species and other HOX proteins. T205, which represents the first residue of the HD, and S204, referred to as ser −1, form part of a highly structured linker between the ANW PBX interaction motif and the HD N-terminal amino acids that bind in the DNA minor groove, as recently described in the structure of the HOXA9 HD and PBX1 bound to DNA (28). This region is not only immediately adjacent to DNA binding residues (16, 28) but is also part of a highly basic region RSTRKKR, which is similar to motifs reported in other HD proteins to be nuclear localization signals (17, 19, 22, 52). We therefore initially focused on identifying the specific residues in the HOXA9-HD protein that were phosphorylated by PKC.
The PKCα-phosphorylated FLAG-tagged HD protein band (Fig. 2A) was digested with trypsin, and the resulting tryptic phosphopeptide map showed three strong phosphopeptides (Fig. 2B). Phosphopeptides 1 and 2 were phosphorylated on serine and threonine residues, while phosphopeptide 3 was phosphorylated only on the serine residue, as indicated by phosphoamino acid analysis (Fig. 2C). The observation that the His-tagged FLAG-HOXA9-HD sequence contains a single predicted tryptic peptide containing a Ser and Thr residue strongly implicated the S204 and T205 residues within the overlapping PKC consensus sites (STRK) as PKC phosphorylation sites. Densitometric analysis of the phosphorylated serine and threonine residues of the three peptides as well as phosphoamino acid analysis of the entire FLAG-HOXA9-HD protein (data not shown) indicated that greater than 70% of the label was associated with the serine residue.
To confirm these results the S204 and T205 in HOXA9-HD were replaced with alanines by site-directed mutagenesis. Purified HOXA9-HD (wild type), HOXA9-HD-S204A/T205A, HOXA9-HD-S204A, and HOXA9-HD-T205A FLAG-tagged proteins were incubated with PKCα in vitro. HOXA9-HD-S204A/T205A was not phosphorylated by PKC, and HOXA9-HD-S204A was phosphorylated very weakly (Fig. 2D). However, HOXA9-T205A was phosphorylated by PKC, as was the HOXA9-HD wild-type protein. These results supported the conclusion that the serine residue within the PKC consensus sequence, STRK, in HOXA9-HD protein was the primary site of phosphorylation by PKC.
Full-length HOXA9 protein is phosphorylated on S204 by PKCα.
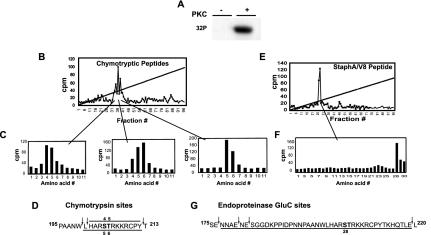
To confirm the PKCα-mediated phosphorylation of S204 within the full-length protein, and identify other possible sites of phosphorylation in the N terminus, purified full-length FLAG-HOXA9 was phosphorylated by PKC in vitro (Fig. 3A). For identification of phosphorylation sites, the phosphorylated protein was digested with either chymotrypsin or endoproteinase Glu-C. HPLC and Edman sequencing analysis of the chymotryptic peptides revealed three partially resolved radioactive peaks (Fig. 3B) that together incorporated the labeled phosphate at residues 4, 5, and 6 (Fig. 3C). Inspection of the predicted HOXA9 chymotryptic peptide map suggested that these peaks were generated, due to an incomplete chymotrypsin digestion, by a mixture of two overlapping HOXA9 chymotryptic peptides containing phosphorylated S204 and T205, (Fig. 3D). This phosphorylation site assignment was confirmed using endoproteinase Glu-C digestion (Fig. 3E to G). Phosphoamino acid analysis of PKCα-phosphorylated FLAG-HOXA9 digested with trypsin revealed that >65% of the label was associated with the serine residue on very small peptides, consistent with tryptic digestion of a labeled RSTR sequence (data not shown). These data confirmed our in vitro analysis of the HOXA9-HD polypeptide showing S204 and T205 within the overlapping consensus sites (STRK) as the sites of PKC phosphorylation. Furthermore, it appeared that this site was the only PKC phosphorylation site in the full-length protein and that S204 was the primary site of PKC phosphorylation.
FIG. 3.
Identification of PKCα phosphorylation sites in the full-length FLAG-HOXA9 protein. (A) Affinity-purified His-tagged FLAG-HOXA9 was subjected to in vitro kinase assay with [γ-32]ATP in the absence or presence of purified PKC. (B) Chymotryptic peptides of PKCα-phosphorylated 32P-labeled FLAG-HOXA9, were separated by reverse-phase HPLC. (C) The radioactive fractions contributing to each peak were analyzed by Edman degradation. Sequencing of peak 1 released radioactivity in Edman cycles 4 and 5; similarly, sequencing of peaks 2 and 3 released radioactivity in cycles 4, 5, and 6 and cycles 5 and 6, respectively. (D) Sequence of the two overlapping FLAG-HOXA9 chymotryptic peptides, H201-Y212 and L200-Y212, which have serine and threonine residues in positions 4 and 5 and positions 5 and 6, respectively. Peak 1 was generated by chymotryptic peptide H201-Y212, peak 3 was generated by chymotryptic peptide L200-Y212, and peak 2 was generated by a mixture of the two overlapping chymotryptic peptides (E) Similarly, endoproteinase Glu-C peptides of 32P-labeled FLAG-HOXA9 were separated by reverse-phase HPLC and generated a single peak. (F) Edman sequencing of the endoproteinase Glu-C peak released the radioactivity in cycle 28. (G) Sequence of FLAG-HOXA9 with the predicted endoproteinase Glu-C cleavage sites, peptide N177-E219 has S204 at position 28.
HOXA9 is phosphorylated by CK-II within a region N terminal to the HD.
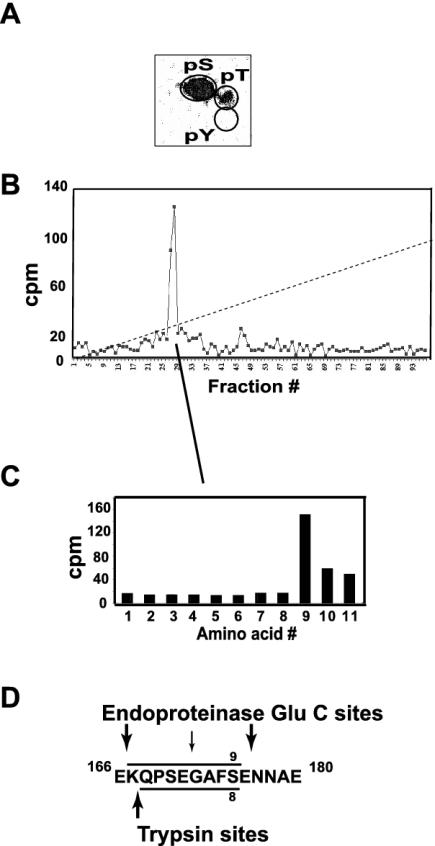
A similar HPLC fractionation coupled with Edman sequence analysis was performed on 32P-labeled, purified FLAG-HOXA9 following in vitro phosphorylation by CK-II. Phosphoamino acid analysis showed that all of the label was incorporated into serine (Fig. 4A). A single radioactive peptide was observed from an endoproteinase Glu-C digest (Fig. 4B). Edman analysis was consistent with phosphorylation of S175 (Fig. 4C). Similar analysis of the tryptic peptide digest confirmed the incorporation of label at this position (not shown). S175 is followed directly by E176. Although this sequence was not predicted by the Prosite program, this arrangement represents an unusual, but previously observed, CK-II consensus recognition site, in which there is no spacer amino acid between the target serine and the requisite acidic residue (37, 45, 57). Since the location of the lone CK-II phosphorylation site did not present any obvious regulatory implications for HOXA9 activity, we chose to focus on the PKC phosphorylation site within the HD, to explore its possible role in HOXA9 function.
FIG. 4.
Identification of a single CK-II phosphorylation site in the full-length FLAG-HOXA9 protein. (A) Phosphoamino acid analysis of the CK-II-phosphorylated 32P-labeled His-tagged FLAG-HOXA9 protein indicated that the label was predominantly associated with serine. (B) Endoproteinase Glu-C peptides of CK-II-phosphorylated 32P-labeled FLAG-HOXA9 separated by reverse-phase HPLC generated a single radioactive peak. (C) Edman sequencing of the HPLC radioactive fractions contributing to the peak released the label in cycle 9. (D) Sequence of the FLAG-HOXA9 protein with the predicted endoproteinase Glu-C and trypsin cleavage sites; Endoproteinase Glu-C peptide K167-K176 has S175 at position 9.
HOXA9 protein is phosphorylated at S204 in vivo.
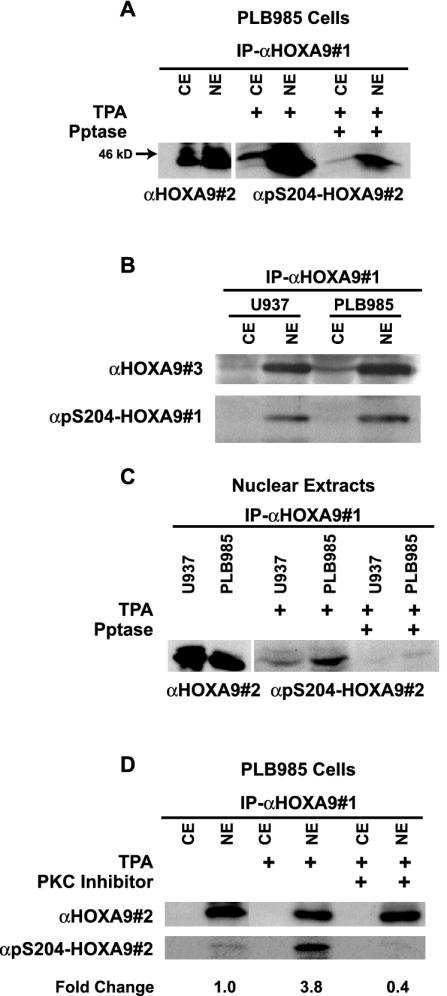
To examine whether S204 within the endogenous HOXA9 protein was phosphorylated in vivo, we generated affinity-purified antibodies specific to the phosphorylated serine residue, α-pS204. Since we have observed that HOX proteins are expressed at quite low levels in most hematopoietic cells, we chose to study PLB985, a myeloid leukemia cell line that we had previously shown expressed relatively high levels of HOXA9 mRNA (30). PLB985 was stimulated with TPA, an activator of PKC (43). Cytoplasmic and nuclear extracts were then immunoprecipitated with a goat α-HOXA9 antibody (antibody 1) and the resulting immunoprecipitates were subjected to Western blot analysis with either affinity-purified chicken HOXA9 antibody (antibody 2) to confirm the presence of HOXA9, or with α-pS204 to measure the degree of phosphorylation at this residue. HOXA9 protein was expressed predominantly in the nuclear extract and was phosphorylated on S204 (Fig. 5A). The endogenous HOXA9 protein, detected with the affinity-purified antibody to the entire HOXA9 protein, migrates on SDS-polyacrylamide gels with an apparent molecular mass of approximately 44 kDa. To confirm that the pS204-specific antibody was detecting the phosphorylated protein, and not residual binding to the HOXA9 peptide backbone, the immunoprecipitates were subjected to phosphatase treatment prior to Western blot analysis with α-pS204 antibody. The signal detected was significantly reduced after treatment with phosphatase, indicating specific phosphorylation of HOXA9 on S204. In a similar manner, HOXA9 immunoprecipitated from TPA-stimulated U937 cells, a second myeloid line known to express HOXA9 mRNA (30), was shown to be predominantly nuclear using a third affinity-purified antibody to HOXA9 (antibody 3) (Fig. 5B). Blotting with the α-pS204 antibody showed that HOXA9 was also phosphorylated on S204 in this myeloid cell line and that this phosphorylation was reduced with phosphatase treatment (Fig. 5C). These results indicated that the endogenous HOXA9 protein in myeloid cells is predominantly in the nuclear fraction. Stimulation with TPA results in the phosphorylation of S204 and that phosphorylation is not associated with change in the subcellular localization of HOXA9.
FIG. 5.
HOXA9 is phosphorylated at S204 in vivo and this phosphorylation is mediated by PKC. (A) Cytoplasmic or nuclear extracts of PLB985 cells were immunoprecipitated with goat α-HOXA9 antibody (αHOXA9#1). Western blot analysis of the immunoprecipitates using affinity-purified chicken α-HOXA9 antibody (αHOXA9#2) showed expression of HOXA9 predominantly in the nuclear extract and to a lesser extent in the cytoplasmic extract. HOXA9 immunoprecipitates from cytoplasmic and nuclear extracts of TPA-stimulated PLB985 cells, treated with or without phosphatase, were probed with α-pS204-HOXA9 antibody (#2). (B) HOXA9 protein from cytoplasmic and nuclear extracts from U937 and PLB985 cells treated with TPA was precipitated with αHOXA9#1 and detected with a rabbit affinity-purified α-HOXA9 antibody (#3) or a different α-pS204-HOXA9 antibody (#1). (C) Immunoprecipitates from nuclear extracts of U937 cells contained HOXA9 protein that was immunoreactive with αHOXA9#2 antibody. HOXA9 immunoprecipitates, from nuclear extracts of TPA-stimulated PLB985 and U937 cells, treated with or without phosphatase, were probed with α-pS204-HOXA9 antibody. (D) PLB985 cells stimulated with either TPA or control were pretreated with or without the PKC inhibitor bisindolylmaleamide1 (5 μM). Cytoplasmic and nuclear extracts were immunoprecipitated with αHOXA9#1, and analyzed by Western blotting with either αHOXA9#2 or α-pS204-HOXA9. Change in HOXA9 phosphorylation level was calculated by NIH ImageQuant analysis.
PKC-mediated phosphorylation of HOXA9 on S204.
To investigate the role of PKC in the phosphorylation of S204 in HOXA9, PLB985 cells were treated with control, TPA, or bisindolylmaleamide1, a potent and relatively specific PKCα inhibitor, (11) prior to stimulation with TPA. HOXA9 was again detected predominantly in the nuclear extracts, and phosphorylation of the protein on S204 was enhanced 3.8-fold upon stimulation of the cells with TPA (Fig. 5D). HOXA9 S204 phosphorylation was reduced 60% compared to control, in the nuclear extracts of TPA-stimulated cells pretreated with the PKC inhibitor. These results indicated that TPA enhanced the phosphorylation of the HOXA9 protein on S204 in the PLB985 myeloid leukemia cell line and that this phosphorylation was mediated by PKCα. One possible result of phosphorylation might be increased protein degradation. However, Western blotting with a second HOXA9 antibody (antibody 2) showed that the immunoreactive material precipitated with the first HOXA9-specific serum did not change appreciably following TPA treatment (Fig. 5D and below).
Phosphorylation of HOXA9 protein by PKC decreases its DNA binding affinity.
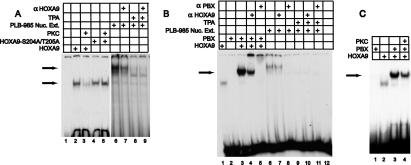
Since the N-terminal region of the HD binds DNA, we next examined the effect of PKC-mediated phosphorylation of this region of HOXA9 on its DNA binding ability. In contrast to the HOX proteins from paralog groups 1 to 8, HOXA9 can bind to DNA efficiently alone as well as with PBX and MEIS cofactor proteins (49, 50). We first explored the binding of HOXA9 alone to a target containing its consensus binding site. Wild-type and a S204A/T205A double-mutant HOXA9 protein, which cannot be phosphorylated by PKC (Fig. 2D), were synthesized in vitro and then subjected to phosphorylation with either PKC or a mock control prior to EMSA analysis. Proteins synthesized in the presence of [35S]methionine were immunoprecipitated with αFLAG serum to demonstrate that phosphorylation did not alter the amount of HOXA9 during a subsequent 30-min incubation in the TNT lysate (data not shown). In the absence of PKC, HOXA9 protein bound to the target DNA (Fig. 6A). Preincubation with PKC substantially reduced the HOXA9 EMSA band. HOXA9 mutant protein lacking the PKC phosphorylation sites was able to bind to the target DNA following PKC treatment as well as in the absence of PKC (Fig. 6A, lanes 4 and 5). These results suggested that PKC-mediated phosphorylation of HOXA9 on S204 and T205 reduced the protein's DNA binding affinity.
FIG. 6.
Phosphorylated HOXA9 exhibits impaired DNA binding ability. (A) HOXA9-containing EMSA complexes are sensitive to PKC and TPA. EMSA was used to assess DNA binding of in vitro-translated FLAG-HOXA9 and FLAG-HOXA9-S204A/T205A proteins. PKC treatment eliminated HOXA9-DNA binding, but did not affect binding by the mutant protein that could not be phosphorylated on S204 and T205 (compare lanes 3 and 5). The nuclear extracts from PLB985 cells stimulated with TPA that are shown in Fig. 5D were also used for EMSA. These extracts showed reduced EMSA bands compared to controls (compare lanes 6 and 8). The presence of HOXA9 in the EMSA complex formed from PLB985 nuclear extracts was demonstrated using α-HOXA9 antibody (compare lanes 6 and 7). (B) PBX-HOXA9 containing EMSA complexes formed from PLB985 nuclear extracts are sensitive to TPA. The arrow denotes the EMSA complexes formed by HOXA9 and PBX1 (lane 3), which migrate between the band for HOXA9-alone-bound DNA (lane 1) and the band formed by PLB nuclear extract (lane 6). The presence of PBX1 in the extract-derived EMSA band was shown using α-PBX1 antibody (lane 8), while α-HOXA9 antibody partially reduced this complex (lane 7). TPA treatment that induces PKC-mediated phosphorylation of HOXA9 abolishes the EMSA (compare lanes 6 and 9). EMSA was performed using an oligonucleotide probe containing a consensus-binding site for HOXA9 and PBX. (C) In vitro-transcribed and -translated HOXA9 and PBX1a proteins were used for EMSA analysis with a consensus PBX1-HOXA9 target. HOXA9 protein was incubated with PKC buffer control (lane 3) or with PKC (lane 4), prior to EMSA analysis. The arrow marks the migration position of the heterodimeric PBX1a-HOXA9 complex established in previous studies (50), which is reduce by pretreatment of the HOXA9 by PKCα.
To investigate if phosphorylation of the endogenous HOXA9 protein modulated its ability to bind to DNA we performed EMSA analysis using the same TPA- or control-stimulated nuclear extracts from PLB985 cells shown in Fig. 5D. Nuclear extract from control-stimulated PLB985 cells shifted the oligonucleotide probe, and α-HOXA9 antibody reduced this EMSA band (Fig. 6A, lanes 6 and 7). Nuclear extract from TPA-stimulated PLB985 cells exhibited only a very weak or no capacity to shift the oligonucleotide probe (Fig. 6A, lane 8, and Fig. 6B, lane 9). Since TPA treatment enhanced S204 phosphorylation of endogenous HOXA9, without altering the amount of HOXA9 protein (Fig. 5 and below), these results suggested that TPA-stimulated phosphorylation of HOXA9, mediated by PKC, decreased ability of the endogenous protein to bind to a target DNA site.
The EMSA band observed with the nuclear extracts migrated at a higher position than that seen in the EMSA assays with in vitro-translated HOXA9 protein (Fig. 6A, compare lanes 2 and 6). We hypothesized that this could be due to the presence of other proteins such as PBX in the PLB985 nuclear extracts. As previously reported (50), in vitro-translated PBX and HOXA9 formed a heterodimeric DNA binding complex that migrated between the band formed with HOXA9 alone and the endogenous EMSA complex (Fig. 6B, compare lanes 3 and 6), suggesting the presence of multiple proteins in this latter band. When the extracts from control-stimulated cells were preincubated with antibody against PBX1 protein, the EMSA band was completely disrupted indicating the presence of PBX in the complex with HOXA9 (Fig. 6B, lane 8). Since we have previously demonstrated that in nuclear extracts from other myeloid cells HOXA9 can form a DNA binding triple complex with PBX proteins and MEIS1 (51), MEIS1 or other MEIS-like proteins may also be present in this EMSA complex. Although α-HOXA9 antibody blocks only a fraction of the shifted complex, TPA treatment eliminates the entire EMSA band. This observation suggests that the other proteins in the endogenous EMSA complex are also sensitive to the TPA-stimulated phosphorylation event. We next explored whether PKC-mediated phosphorylation of HOXA9 influenced its DNA binding in complexes with its known cofactors. PKC treatment reduced the capacity of HOXA9 to form a cooperative complex with PBX on an oligonucleotide target containing a PBX-HOXA9 site (Fig. 6C). A similar analysis showed that PKC treatment of in vitro-synthesized HOXA9 also reduced its capacity to form a heterodimeric complex with MEIS1 on an oligonucleotide target containing a MEIS1-HOXA9 site (data not shown).
PKC activation is required for TPA-mediated monocytic differentiation of PLB985 cells and HOXA9-immortalized bone marrow cells.
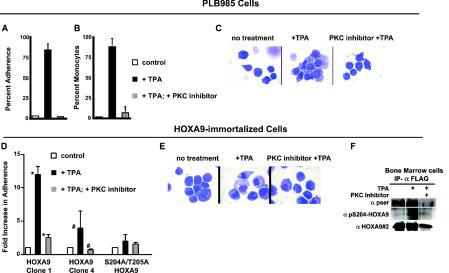
TPA is known to induce monocytic differentiation in PLB985 cells (58). To assess the potential role of the PKC pathway in TPA-induced monocytic differentiation, PLB985 cells were treated with the PKC inhibitor bisindolylmaleamide1 prior to treatment with TPA. Consistent with previously published observations, TPA treatment resulted in an increase in monocytic differentiation as measured by adherence and cellular morphology (Fig. 7A to C). PKC inhibition essentially blocked TPA-induced monocytic differentiation, as measured by adherence (Fig. 7A) and by analysis of morphological features of monocytic differentiation (Fig. 7B), compared to PLB985 cells treated with TPA alone, while cell staining indicated partial differentiation occurred in the presence of the inhibitor (Fig. 7C). Since treatment of PLB985 cells with TPA also causes S204 phosphorylation (Fig. 5), these data are consistent with a model in which PKC-mediated phosphorylation of HOXA9 on S204 attenuates the protein's DNA binding capacity, leading to cellular differentiation.
FIG. 7.
PLB985 and HOXA9-immortalized murine bone marrow cells treated with TPA differentiate into monocytes, and this differentiation is blocked by a PKC inhibitor and appears to require S204/T205 phosphorylation. PLB985 cells (A to C) or HOXA9-immortalized cells (D to F) were treated with dimethyl sulfoxide vehicle or TPA (100 nM) with or without pretreatment with the PKC inhibitor bisindoylmaleamide1 (5 μM) 5 min prior to stimulation with TPA. (A and D) Percent adherence was measured 24 h after addition of TPA. There was a statistically different adherence in the presence of TPA (PLB985 P < 0.001, n = 4) (symbols: *, P < 0.04; #, P < 0.004) but no difference between controls and cells pretreated with inhibitor before TPA treatment. (B) PLB985 differentiation scored on the basis of three monocytic features: nuclear to cytoplasmic ratio, condensed chromatin and cytoplasmic vacuolization. There was a statistically different monocytic differentiation in the presence of TPA (P < 0.001, n = 4) but no difference between controls and cells pretreated with inhibitor before TPA treatment. (C and E) Wright-Giemsa-stained cytospin preparations showing that pretreatment with a PKC inhibitor partially prevented monocytic differentiation of both PLB985 (C) and HOXA9-immortalized cells (E) as reflected by large nuclear size and a smaller cytoplasmic fraction. (F) Endogenous HOXA9 in immortalized marrow line 4 is phosphorylated on S204. Whole-cell lysates were immunoprecipitated with α-FLAG antibody, and the immunoprecipitates were probed with anti-pSer (α-pser), α-pS204-HOXA9, or αHOXA9#2 antibodies. To confirm that S204 and T205 are important for PKC-mediated myeloid cell differentiation, a S204A/T205A HOXA9 protein containing alanines in place of the putative PKC phosphorylation sites was used to immortalize bone marrow cells. The resulting clone showed greatly reduced differentiation response to TPA as measured by cellular adherence (D) or morphology (not shown). Error bars, standard deviations.
PKC-mediated phosphorylation of S204/T205 in HOXA9 is required for myeloid differentiation of HOXA9-immortalized bone marrow cells.
Previous investigators have demonstrated that HOXA9 immortalizes a murine myeloid progenitor with a promyelocytic phenotype capable of differentiating into granulocytes or monocytes under appropriate conditions (6, 7, 47). The role of PKC in the monocytic differentiation of HOXA9-immortalized murine bone marrow-derived myeloid cell lines was explored in a manner similar to PLB985 cells. Stimulation of three independently derived HOXA9-immortalized marrow cell lines with TPA resulted in significant monocytic differentiation while pretreatment with the PKC inhibitor significantly inhibited these effects (Fig. 7D and E and data not shown). Furthermore, following TPA stimulation, there was an increase in phosphorylation of retrovirally expressed FLAG-HOXA9 detected with an antiserum to pSer and a modest increase in signal detected with the specific α-pS204 antibody (Fig. 7F), suggesting that the exogenous HOXA9 in the bone marrow cells is also phosphorylated on this residue under conditions in which differentiation is induced. We note that TPA treatment of HOXA9-immortalized bone marrow cells did not change the levels of endogenous HOXA9 protein detected by antiserum 2 (Fig. 7F and data not shown), confirming that PKC-mediated phosphorylation does not result in rapid degradation of the HOXA9 protein.
To demonstrate a role for phosphorylation of S204 in myeloid differentiation, we tested a mutant HOXA9 protein in which S204 and T205 were changed to alanines. The S204A/T205A protein was equally active as the wild-type HOXA9 protein in immortalizing myeloid bone marrow progenitor cells (not shown). However, in contrast to HOXA9-immortalized cells, cells immortalized with the mutant protein did not differentiate in response to TPA (Fig. 7D). Taken together, these results support a model in which PKC-mediated phosphorylation of HOXA9 at S204 and perhaps T205 plays a central role in monocytic differentiation.
DISCUSSION
In this study we present evidence that HOXA9 is a biological substrate for PKCα and have identified two adjacent PKC phosphorylation sites at S204 and T205 at the extreme N terminus of the HD within an STRK PKC consensus sequence. S204 appears to be the major phosphorylation site in vitro. We used specific antisera to show that this residue is phosphorylated in the endogenous HOXA9 protein in PLB985 and U937 human myeloid cells as well as in HOXA9-immortalized murine myeloid bone marrow cells following TPA stimulation. Phosphorylation of HOXA9 in vitro at this site by PKC decreases it biological activity as measured by its ability to bind DNA. Furthermore, this study demonstrates that PKC-mediated differentiation of the PLB985 hematopoietic cell line as well as HOXA9-immortalized mouse bone marrow cell lines is correlated with phosphorylation of HOXA9 on S204. Finally, a bone marrow-derived cell line, immortalized with a S204A/T205A mutant HOXA9 protein, did not differentiate towards the myeloid lineage following TPA treatment.
Taken together, our data provide support for a model in which HOXA9 plays a role in maintenance of the undifferentiated myeloid state and support for the argument that PKC-mediated phosphorylation of HOXA9 may regulate aspects of terminal myeloid differentiation. Our data are thus consistent with the findings of Calvo et al. that HOXA9 expression blocks the differentiation of primary murine myeloid progenitors in response to GM-CSF, but retaining the capacity to differentiate into neutrophils or macrophages in response to G-CSF or M-CSF, respectively (7). In these studies, the differentiation block required HOXA9 DNA binding activity. Our results show that HOXA9-immortalized murine bone marrow cells are differentiated to a monocytic phenotype in response to TPA. Furthermore, HOXA9 protein is at least partially phosphorylated on S204 under these conditions in both HOXA9-immortalized cells and PLB cells. In the PLB cells phosphorylation is correlated with a loss of biological activity as reflected by the loss of endogenous HOXA9-containing EMSA complexes. Based on these results, we proposed a similar loss of DNA binding-associated biological activity by the endogenous HOXA9 protein in marrow cells. We note that interpretation of our data with regard to the HOXA9-immortalized lines must be tempered by our observation that cell lines that are blocked at slightly different stages of myeloid differentiation appear to arise from the immortalization process. Some lines exhibit more inherent myeloid features and less responsiveness to TPA treatment than others.
Phosphorylation of several HD transcription factors has been shown to modulate differentiation, DNA binding, or activity. The DNA binding capacity and transcriptional activity of HOXA10 is reduced by SHP1-mediated tyrosine phosphorylation within the HD (14). A number of HD proteins have been reported to be regulated by CK-II phosphorylation. CK-II modification of HOXB7 has been shown to regulate granulocytic differentiation of 32D cells (62). In a similar manner, both the DNA binding and biological activity of the Drosophila ANTP, which is the homologue of the mammalian HOX6 proteins, was reduced following CK II-mediated phosphorylation (21). CK II phosphorylation of Cut, a non-HOX HD protein, also represses DNA binding (10) and its associated biological activity (9). In the present study, we find that HOXA9 is phosphorylated by CK-II on a noncanonical SE sequence but have not explored the possible regulatory consequences of this modification. PKA-mediated phosphorylation of the FTZ protein on a threonine residue within the N-terminal region of the HD was reported to be required for biological activity (12). The Drosophila SCR protein, which is homologous to HOXA5, is both activated and repressed by different kinases (2). Conversely, phosphorylation of the Engrailed HD protein increases DNA binding (4).
The 39 mammalian HOX genes are arrayed in four parallel chromosomal loci, such that the so-called paralogous proteins display high homology within their respective HDs and variable degrees of sequence conservation outside the HD. Sequence comparison of the HOX proteins reveals that the STRK sequence is conserved within all four HOX9 paralog proteins (in HOXB9, T205 is replaced with S205). Furthermore, 21 of the 39 HOX proteins contain one or both of the possible PKC phosphorylation sites at the start of the HD, plus or minus one amino acid residue, and 16 have serine at the position equivalent to S204 within HOXA9. The STRK site is also conserved among HOXA9 protein species, including human, mouse, chicken, pig, and horn shark, and is partially conserved in the Drosophila homologue, Abd-B. Conservation of this PKC phosphorylation site within the HOX family and in HOXA9 proteins throughout mammalian evolution suggests the importance of this site for the regulation of HOX proteins, and HOXA9 in particular. Crystal structure analysis indicates that HD proteins bind DNA such that the N-terminal region of the HD inserts into the minor groove of the DNA and contributes to DNA binding specificity (16, 23, 61). While this work was in preparation, LaRonde-LeBlanc et al. elucidated the crystal structure of the HOXA9 HD with extended arms and PBX bound to DNA (28). These authors report that S204 and T205 form part of a three-amino-acid linker region which spans the minor groove and tethers the PBX interacting tryptophan within the conserved ANW binding motif, and the HD N-terminal amino acids binding in the DNA minor groove in a highly ordered structure. Although their analysis indicates that S204 and T205 point away from the DNA, this structure was performed on a protein lacking almost the entire 200-amino-acid N terminus of HOXA9. We hypothesize that the full-length protein adopts a structure in which phosphorylation of S204 and/or T205 is incompatible with the presumed stringent requirements within the linker region for efficient DNA binding.
In our studies, PKC-mediated phosphorylation of HOXA9 impaired its ability to bind an oligonucleotide containing a consensus PBX-HOXA9 site. The DNA binding complexes observed using nuclear extracts from PLB985 cells migrated more slowly than a complex formed with in vitro-synthesized PBX and HOXA9. This is presumably due to HOXA9 complex formation with other endogenous proteins such as MEIS as well as PBX (51). PBX1 appeared to be a component in all of the complexes formed by nuclear extracts from PLB985 cells, since the in vivo EMSA band was almost completely shifted by PBX1 antiserum. In contrast, antiserum to HOXA9 shifted only a portion of the endogenous EMSA band, suggesting the presence of other HOX proteins that can partner with PBX1 to recognize the oligonucleotide target in the PLB985 cell extract. However, when the cells were stimulated with TPA the DNA binding of the endogenous complex was completely ablated indicating that at least one component of each of the different complexes in the EMSA band is sensitive to TPA.
The HOXA9 gene plays a role in adult myelopoiesis and has been increasingly implicated in the pathogenesis of human AML. HOXA9 gene expression in human patient bone marrow samples is associated with morbidity in AML (18) and enforced expression of the HoxA9 gene in murine marrow causes AML following a prolonged incubation period (26, 56). This leukemic transformation was shown to depend on HOXA9 DNA binding capacity, as well as being potentiated by the MEIS1 HD protein. Based on our present data, we propose that PKC-mediated phosphorylation of HOXA9 in immortalized myeloid cells leads to a loss of DNA binding activity and the capacity to maintain the undifferentiated myeloid state. In such a model, PKC-mediated phosphorylation of HOXA9 might be predicted to lead to a loss of the HOXA9-associated leukemic phenotype. Future studies will be directed towards the development of model systems to investigate the possible treatment of myeloid leukemias by modulating the phosphorylation status of the HOXA9 protein.
Acknowledgments
This study was supported by the Department of Veterans Affairs (C.L. and H.J.L.), NIH grant RO1CA80029 (C.L.), and NIH grant RO1 GM55814001A2 (C.L.).
We thank Cynthia Wolberger for helpful conversations, Alan Pollock for advice on use of the retroviral packaging line, Steve Fong and Sophia Rosenfeld for technical assistance, and the UCSF Cancer Center cell sorting facility for performing FACS.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology, p. 12.1.1-12.1.8. John Wiley & Sons, Hoboken, N.J.
- 2.Berry, M., and W. Gehring. 2000. Phosphorylation status of the SCR homeodomain determines its functional activity: essential role for protein phosphatase 2A,B′. EMBO J. 19:2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow, J., A. M. Shearman, V. P. J. Stanton, R. Becher, T. Collins, A. J. Williams, I. Dube, F. Katz, Y. L. Kwong, C. Morris, K. Ohyashiki, K. Toyama, J. Rowley, and D. E. Housman. 1996. The t(7;11)(p15;p15) translocation in acute myeloid leukemia fuses the genes for nucleoporin NU98 and class I homeoprotein HOXA9. Nat. Genet. 12:159-167. [DOI] [PubMed] [Google Scholar]
- 4.Bourbon, H. M., E. Martin-Blanco, D. Rosen, and T. B. Kornberg. 1995. Phosphorylation of the Drosophila engrailed protein at a site outside its homeodomain enhances DNA binding. J. Biol. Chem. 270:11130-11139. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-143. [DOI] [PubMed] [Google Scholar]
- 6.Calvo, K. R., P. S. Knoepfler, D. B. Sykes, M. P. Pasillas, and M. P. Kamps. 2001. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc. Natl. Acad. Sci. USA 98:13120-13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo, K. R., D. B. Sykes, M. Pasillas, and M. P. Kamps. 2000. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced Meis expression. Mol. Cell. Biol. 20:3274-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo, K. R., D. B. Sykes, M. P. Pasillas, and M. P. Kamps. 2002. Nup98-HoxA9 immortalizes myeloid progenitors, enforces expression of Hoxa9, Hoxa7 and Meis1, and alters cytokine-specific responses in a manner similar to that induced by retroviral co-expression of Hoxa9 and Meis1. Oncogene 21:4247-4256. [DOI] [PubMed] [Google Scholar]
- 9.Coqueret, O., G. Berube, and A. Nepveu. 1998. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 17:4680-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coqueret, O., N. Martin, G. Berube, M. Rabbat, D. W. Litchfield, and A. Nepveu. 1998. DNA binding by cut homeodomain proteins is down-modulated by casein kinase II. J. Biol. Chem. 273:2561-2566. [DOI] [PubMed] [Google Scholar]
- 11.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, J., L. H. Hung, R. Strome, and H. M. Krause. 1998. A phosphorylation site in the ftz homeodomain is required for activity. EMBO J. 17:2308-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorsam, T. S., M. C. Ferrell, P. G. Dorsam, U. Vijapurkar, D. Khodabakhsh, B. Pau, H. Bernstein, M. C. Haqq, C. Largman, and H. J. Lawrence. 2004. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood 103:1676-1684. [DOI] [PubMed] [Google Scholar]
- 14.Eklund, E. A., A. Jalava, and R. Kakar. 2000. Tyrosine phosphorylation of HoxA10 decreases DNA binding and transcriptional repression during interferon gamma-induced differentiation of myeloid leukemia cell lines. J. Biol. Chem. 275:20117-20126. [DOI] [PubMed] [Google Scholar]
- 15.Gehring, W. J. 1987. Homeo boxes in the study of development. Science 236:1245-1252. [DOI] [PubMed] [Google Scholar]
- 16.Gehring, W. J., Y. Q. Qian, M. Billeter, K. Furukubo-Tokunaga, M. Affolter, G. Otting, and K. Wuthrich. 1994. Homeodomain-DNA recognition. Cell 69:211-223. [DOI] [PubMed] [Google Scholar]
- 17.Ghaffari, M., X. Zeng, J. A. Whitsett, and C. Yan. 1997. Nuclear localization domain of thyroid transcription factor-1 in respiratory epithelial cells. Biochem. J. 328:757-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golub, T. R., D. K. Slonim, P. Tamayo, C. Huard, M. Gaasenbeek, J. P. Mesirov, H. Coller, M. L. Loh, J. R. Downing, M. A. Caligiuri, C. D. Bloomfield, and E. S. Lander. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531-537. [DOI] [PubMed] [Google Scholar]
- 19.Hall, M. N., C. Craik, and Y. Hiraoka. 1990. Homeodomain of yeast repressor alpha 2 contains a nuclear localization signal. Proc. Natl. Acad. Sci. USA 87:6954-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izon, D. J., L. G. Komuves, S. Rozenfeld, S. Fong, C. Largman, and H. J. Lawrence. 1998. Loss of function of the homeobox gene HoxA-9 retards T-cell development and induces apoptosis. Blood 92:382-389. [PubMed] [Google Scholar]
- 21.Jaffe, L., H.-D. Ryoo, and R. S. Mann. 1997. A role for phosphorylation by casein kinase II in modulating Antennapedia activity in Drosophila. Genes Dev. 11:1327-1340. [DOI] [PubMed] [Google Scholar]
- 22.Kasahara, H., and S. Izumo. 1999. Identification of the in vivo casein kinase II phosphorylation site within the homeodomain of the cardiac tissue-specifying homeobox gene product Csx/Nkx2.5. Mol. Cell. Biol. 19:526-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kissinger, C. R., B. Liu, E. Martin-Blanco, T. B. Kornberg, and C. O. Pabo. 1990. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution. A framework for understanding homeodomain-DNA interactions. Cell 63:579-590. [DOI] [PubMed] [Google Scholar]
- 24.Kmita, M., and D. Duboule. 2003. Organizing axes in time and space; 25 years of colinear tinkering. Science 301:331-333. [DOI] [PubMed] [Google Scholar]
- 25.Koeffler, H. P., M. Bar-Eli, and M. C. Territo. 1981. Phorbol ester effect on differentiation of human myeloid leukemia cell lines blocked at different stages of maturation. Cancer Res. 41:919-926. [PubMed] [Google Scholar]
- 26.Kroon, E., J. Krosl, U. Thorsteinsdottir, S. Baban, A. M. Buchberg, and G. Sauvageau. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17:3714-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroon, E., U. Thorsteinsdottir, N. Mayotte, T. Nakamura, and G. Sauvageau. 2001. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 20:350-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaRonde-LeBlanc, N. A., and C. Wolberger. 2003. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 17:2060-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence, H. J., C. D. Helgason, G. Sauvageau, S. Fong, D. J. Izon, R. K. Humphries, and C. Largman. 1997. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 89:1922-1930. [PubMed] [Google Scholar]
- 30.Lawrence, H. J., S. Rozenfeld, C. Cruz, K. Matsukuma, A. Kwong, L. Komuves, A. M. Buchberg, and C. Largman. 1999. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia 13:1993-1999. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence, H. J., G. Sauvageau, R. K. Humphries, and C. Largman. 1996. The role of HOX genes in normal and leukemic hematopoiesis. Stem Cells 14:281-290. [DOI] [PubMed] [Google Scholar]
- 32.Look, T. A. 1997. Oncogenic transcription factors in the human acute leukemias. Science 278:1059-1064. [DOI] [PubMed] [Google Scholar]
- 33.Lotem, J., and L. Sachs. 1979. Regulation of normal differentiation in mouse and human myeloid leukemic cells by phorbol esters and the mechanism of tumor promotion. Proc. Natl. Acad. Sci. USA 76:5158-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macfarlane, D. E., and L. Manzel. 1994. Activation of beta-isozyme of protein kinase C (PKC beta) is necessary and sufficient for phorbol ester-induced differentiation of HL-60 promyelocytes. Studies with PKC beta-defective PET mutant. J. Biol. Chem. 269:4327-4331. [PubMed] [Google Scholar]
- 35.Magli, M. C., C. Largman, and H. J. Lawrence. 1997. Effects of HOX homeobox genes in blood cell differentiation. J. Cell Physiol. 173:168-177. [DOI] [PubMed] [Google Scholar]
- 36.McGinnis, W., and R. Krumlauf. 1992. Homeobox genes and axial patterning. Cell 68:283-302. [DOI] [PubMed] [Google Scholar]
- 37.Meisner, H., and M. P. Czech. 1991. Phosphorylation of transcriptional factors and cell-cycle-dependent proteins by casein kinase II. Curr. Opin. Cell Biol. 3:474-483. [DOI] [PubMed] [Google Scholar]
- 38.Mischak, H., J. H. Pierce, J. Goodnight, M. G. Kazanietz, P. M. Blumberg, and J. F. Mushinski. 1993. Phorbol ester-induced myeloid differentiation is mediated by protein kinase C-alpha and -delta and not by protein kinase C-beta II, -epsilon, -zeta, and -eta. J. Biol. Chem. 268:20110-20115. [PubMed] [Google Scholar]
- 39.Moskow, J. J., F. Bullrich, K. Huebner, I. O. Daar, and A. M. Buchberg. 1995. Meis, a PBX1-related homeobox gene involved in myeloid leukemias in BXH-2 mice. Mol. Cell. Biol. 15:5434-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller, M., M. Affolter, W. Leupin, G. Otting, K. Wuthrich, and W. J. Gehring. 1988. Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J. 7:4299-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura, T., D. A. Largaespada, M. P. Lee, L. A. Johnson, K. Ohyashiki, K. Toyama, S. J. Chen, C. L. Willman, I.-M. Chen, A. P. Feinberg, N. A. Jenkins, N. G. Copeland, and J. D. J. Shaughnessy. 1996. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;15) in human myeloid leukemia. Nat. Genet. 12:154-158. [DOI] [PubMed] [Google Scholar]
- 42.Naviaux, R. K., E. Costanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niedel, J. E., L. J. Kuhn, and G. R. Vandenbark. 1983. Phorbol diester receptor copurifies with protein kinase C. Proc. Natl. Acad. Sci. USA 80:36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oshevski, S., M. C. Le Bousse-Kerdiles, D. Clay, Z. Levashova, N. Debili, N. Vitral, C. Jasmin, and M. Castagna. 1999. Differential expression of protein kinase C isoform transcripts in human hematopoietic progenitors undergoing differentiation. Biochem. Biophys. Res. Commun. 263:603-609. [DOI] [PubMed] [Google Scholar]
- 45.Pinna, L. A. 1990. Casein kinase 2: an 'eminence grise' in cellular regulation? Biochim. Biophys. Acta 1054:267-284. [DOI] [PubMed] [Google Scholar]
- 46.Rovera, G., D. Ferrero, G. L. Pagliardi, J. Vartikar, S. Pessano, L. Bottero, S. Abraham, and D. Lebman. 1982. Induction of differentiation of human myeloid leukemias by phorbol diesters: phenotypic changes and mode of action. Ann. N. Y. Acad. Sci. 397:211-220. [DOI] [PubMed] [Google Scholar]
- 47.Schnabel, C. A., Y. Jacobs, and M. L. Cleary. 2000. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 19:608-616. [DOI] [PubMed] [Google Scholar]
- 48.Senderowicz, L., J. X. Wang, L. Y. Wang, S. Yoshizawa, W. M. Kavanaugh, and C. W. Turck. 1997. 3-Phosphohistidine cannot replace phosphotyrosine in high-affinity binding to phosphotyrosine binding or Src homology 2 domains. Biochemistry 36:10538-10544. [DOI] [PubMed] [Google Scholar]
- 49.Shen, W.-F., J. C. Montgomery, S. Rozenfeld, H. J. Lawrence, A. Buchberg, and C. Largman. 1997. The Abd-B-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol. Cell. Biol. 17:6448-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen, W.-F., S. Rozenfeld, H. J. Lawrence, and C. Largman. 1997. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J. Biol. Chem. 272:8198-8206. [DOI] [PubMed] [Google Scholar]
- 51.Shen, W. F., S. Rozenfeld, A. Kwong, L. Komuves, H. J. Lawrence, and C. Largman. 1999. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 19:3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sock, E., J. Enderich, M. G. Rosenfeld, and M. Wegner. 1996. Identification of the nuclear localization signal of the POU domain protein Tst-1/Oct6. J. Biol. Chem. 271:17512-17518. [DOI] [PubMed] [Google Scholar]
- 53.Steube, K. G., and H. G. Drexler. 1993. Differentiation and growth modulation of myeloid leukemia cells by the protein kinase C activating agent bryostatin-1. Leuk Lymphoma 9:141-148. [DOI] [PubMed] [Google Scholar]
- 54.Thorsteinsdottir, U., E. Kroon, L. Jerome, F. Blasi, and G. Sauvageau. 2001. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 21:224-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorsteinsdottir, U., J. Krosl, E. Kroon, A. Haman, T. Hoang, and G. Sauvageau. 1999. The oncoprotein E2A-Pbx1a collaborates with Hoxa9 to acutely transform primary bone marrow cells. Mol. Cell. Biol. 19:6355-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorsteinsdottir, U., A. Mamo, E. Kroon, L. Jerome, J. Bijl, H. J. Lawrence, K. Humphries, and G. Sauvageau. 2002. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 99:121-129. [DOI] [PubMed] [Google Scholar]
- 57.Tuazon, P. T., and J. A. Traugh. 1991. Casein kinase I and II—multipotential serine protein kinases: structure, function, and regulation. Adv. Second Messenger Phosphoprotein Res. 23:123-164. [PubMed] [Google Scholar]
- 58.Tucker, K. A., M. B. Lilly, L. Heck, Jr., and T. A. Rado. 1987. Characterization of a new human diploid myeloid leukemia cell line (PLB-985) with granulocytic and monocytic differentiating capacity. Blood 70:372-378. [PubMed] [Google Scholar]
- 59.Vandenbark, G. R., L. J. Kuhn, and J. E. Niedel. 1984. Possible mechanism of phorbol diester-induced maturation of human promyelocytic leukemia cells. J. Clin. Investig. 73:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ways, D. K., B. R. Messer, T. O. Garris, W. Qin, P. P. Cook, and P. J. Parker. 1992. Modulation of protein kinase C-epsilon by phorbol esters in the monoblastoid U937 cell. Cancer Res. 52:5604-5609. [PubMed] [Google Scholar]
- 61.Wolberger, C., A. K. Vershon, B. Liu, A. D. Johnson, and C. O. Pabo. 1991. Crystal structure of a MATα2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell 67:517-528. [DOI] [PubMed] [Google Scholar]
- 62.Yaron, Y., J. K. McAdara, M. Lynch, E. Hughes, and J. C. Gasson. 2001. Identification of novel functional regions important for the activity of HOXB7 in mammalian cells. J. Immunol. 166:5058-5067. [DOI] [PubMed] [Google Scholar]