Abstract
The outer membrane porin proteins are the major factors in controlling the permeability of cell membrane. OmpF is an example of porin proteins in Esherichia coli. In normal growth condition a large amount of this protein is synthesised, but under stress condition, such as the presence of antibiotics in environment its expression is decreased inhibiting the entrance of antibiotics into cell. The expression of ompF is inhibited by antisense RNA transcribed from micF. In normal condition the expression of micF is low, but in the presence of antibiotics its expression is increased and causes multiple resistances to irrelevant antibiotics. The aims of this research were to study first, the intactness of micF and then quantify the expression of ompF in ciprofloxacin and tetracycline resistant mutants of E. coli. For this purpose the 5’ end of micF was amplified and then sequenced. None of these mutants except one and its clone has a mutation in this gene. Then the relative expression of ompF in these mutants was quantified by real time PCR. There was no significant difference between ompF transcription of mutants and wild type strain. Based on this study and previous study it is concluded that low to intermediate levels of resistance to ciprofloxacin and tetracycline does not decrease ompF transcription.
Keywords: micF, multiple antibiotic resistance, ompF, real time PCR
Small noncoding RNAs (snRNAs) are present in all organisms. They are antisense RNA that regulate post-transcriptionally gene expression and promote adaptation of cells to various growth conditions[1]. There are more than 80 snRNAs in Esherichia coli (E. coli) genome[2]. Among these snRNAs, some regulate the expression of outer membrane porins, such as micC and micF RNAs. micF RNA regulates negatively the expression of ompF in the presence of antibiotics such as tetracycline[3]. About 25 nucleotide (nt) at the 5′ end of micF RNA base pairs to ompF 5′ untranslated region (5′ UTR) mRNA and forms a duplex (fig. 1). As the formation of duplex is not perfect, the small stem loop structure produces. micF RNA in the duplex structure, covers the Shine-Dalgarno sequence (ribosome binding site) and AUG start codon of ompF mRNA and thereby inhibits its translation[3].
Fig. 1.
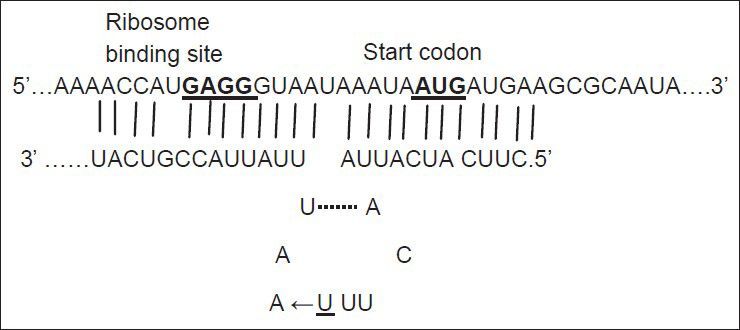
Duplex formation between micF RNA and 5′UTR of ompF mRNA.
The underlined U is the nucleotide changed in one of mutants. Modified and adapted from Vogel and Papenfort, 2006.
In addition to general transcriptional regulator, including H-NS, HU and Lrp, specific transcriptional regulator, such as MarA positively regulates micF transcription[4]. There is a mar box next to and partly overlapping the -35 region of the micF promoter[5]. marA is located in marRAB operon whose up and down regulations are under the control of MarA and MarR, respectively[5]. Binding of different ligands, such as antibiotics to MarR, dissociates this repressor from the operator site of marRAB operon[6]. Then binding of MarA to mar box upstream of -35 region of marRAB operon activates this operon expression[5,7]. Over activity of MarA enhances the transcription of micF and thereby decreases the translation of ompF mRNA[4]. To decrease the translation of ompF mRNA, an intact micF locus is required[8]. Additionally, expression of ompF is regulated at transcription level as well.
Outer membrane proteins (porins) such as ompF and OmpC are abundant proteins and form trimeric β-barrels in the OM[4]. They form channels through the outer membrane (OM) for the entry of different compounds for example nutrients and antibiotics, such as ciprofloxacin and tetracycline. Thus, decreased levels of ompF prevent the entry of above antibiotics to E. coli cells and cause multiple antibiotic resistance phenotype[4].
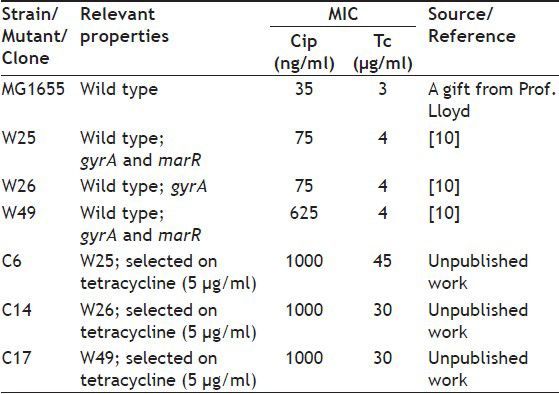
In the previous work gyrA mutants, which are resistant to ciprofloxacin and tetracycline, and with and without a mutation in marR were described[9,10]. These mutants and their increased tetracycline resistant clones could possess low level of ompF. The aims of this research were first, to study the intactness of micF gene and its mar box; and then to study ompF expression in these mutants.
MATERIALS AND METHODS
Tetracycline hydrochloride (Tc) (Sigma) was used to promote resistance in mutants. Stock solution of 4 mg/ml was prepared for the study. Diluted LB broth (Merck, Germany) and LBA containing 1.5% agar (Merck, Germany) were used for cultivation of strain and mutants.
Bacterial strain and mutants:
As defined previously resistance to ciprofloxacin can be divided to three levels, including low levels of resistance (MIC: 0.063 to 1 μg/ml), intermediate levels of resistance (MIC: 1 to 32 μg/ml) and high levels of resistance (MIC: >32 μg/ml)[11]. Additionally, it was described that resistance to tetracycline can also be divided into three levels, including low levels of resistance (MIC: 1 to 10 μg/ml), intermediate levels of resistance (MIC: 10 to 50 μg/ml) and high levels of resistance (MIC: >50 μg/ml)[12]. MG1655 was wild type and control strain. gyrA mutants with and without a mutation in marR gene, and based on above definition were with low to intermediate levels of resistance to ciprofloxacin and tetracycline isolated in previous work[10] are listed in Table 1. Mutants W25, W26 and W49 were isolated from cultivation of wild type strain on LBA plus ciprofloxacin[9]. Clones C6, C14 and C17 were obtained from cultivation of above mutants on LBA agar containing Tc (unpublished work).
TABLE 1.
BACTERIAL STRAIN AND MUTANTS
PCR amplification and DNA sequencing of micF gene:
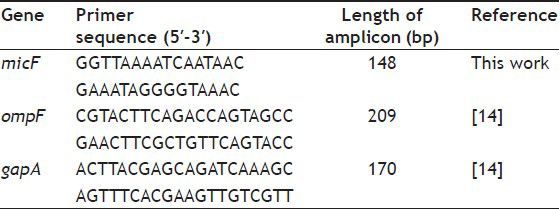
Colony PCR was used to amplify the 5′ end of micF gene and upstream sequences harboring mar box in wild type and mutants[10]. Primers for amplification are listed in Table 2. PCR products (148 bp) were sequenced and compared with MG1655 micF sequence obtained from NCBI.
TABLE 2.
LIST OF PRIMERS
OmpF expression analysis by real time PCR:
After cultivation of bacteria in diluted LB broth and 3 μg/ml Tc (except for wild type) at 37° with shaking at 150 rpm and grown to mid-logarithmic phase (OD600 of 0.6)[11]. Each culture was stabilised by RNA protect bacterial reagent (Qiagen, Germany) and then pelleted by centrifugation (Sigma, Germany). RNA was extracted immediately using an RNeasy Mini Kit (Qiagen, Germany), contaminating genomic DNA was eliminated by RNase-free DNase I treatment according to the manufacturer's instruction (Fermentas, Life science research, Vilnius, Lithuania) and its absence was confirmed by amplification of RNA samples plus a DNA sample as a positive control. Total RNA concentration was estimated at OD260 using spectrophotometer (Ultrospec 1100, Amersham Pharmacia Biothech, UK). Purified total RNA (2 μg) was used as a template in RT-PCR using a RevertAid Reverse Transcriptase kit (Fermentas, Life science research, Vilnius, Lithuania). The cDNAs obtained from reverse transcription were used to quantify the level of ompF and gapA, as an endogenous reference gene by real time PCR in a Rotor Gene 6000 thermocycler (Corbett Research, Australia) using a SYBR Green kit (Takara, Japan). Primers used in this experiment are listed in Table 2. Relative gene expression was calculated using the efficiency corrected calculation method (ratio of ompF expression to gapA expression, which was calculated by following equation, ratio=(EompF)ΔCt/(EgapA)ΔCt…(1)[13]. The efficiency of each gene (E) can be obtained from linear regression plot which is drawn from serial dilution of standard sample and tested samples. The slope of the regression line was used in calculation of PCR efficiency, using equation E=10[-1/slope]…(2). The difference in cycle threshold (Ct) was calculated using equation, ΔCt=Ctwt–Ctmutant…(3).
All data on ompF expression are the average of triplicate analyses. The data was recorded as mean±SD. Statistical analysis of relative expression was done by SPSS version 16 and T-test was used for comparison of relative gene expression data. A P-value of less than 0.05 is considered significant.
RESULTS
Mutations listed in Table 1 with different MIC for ciprofloxacin and tetracycline were analysed for the presence of possible mutation in mar box and 5′ end of micF gene involved in duplex formation with ompF mRNA. Fig. 2 shows the result of gel electrophoresis of the micF PCR products of MG1655 and mutants. The comparison of nucleotide sequence of PCR products with published sequence of micF showed that W26 and its derived clone, C14 had a nucleotide change (T18→A) in micF gene (fig. 3). This change is located in a region which participates in duplex formation with ompF mRNA (fig. 1). As can be seen from fig. 1, this alteration is located in stem loop structure of micF RNA following its base pairing with ompF mRNA. Other mutants showed the same sequence as MG1655. Moreover, the mar box of all mutants was intact (fig. 3).
Fig. 2.
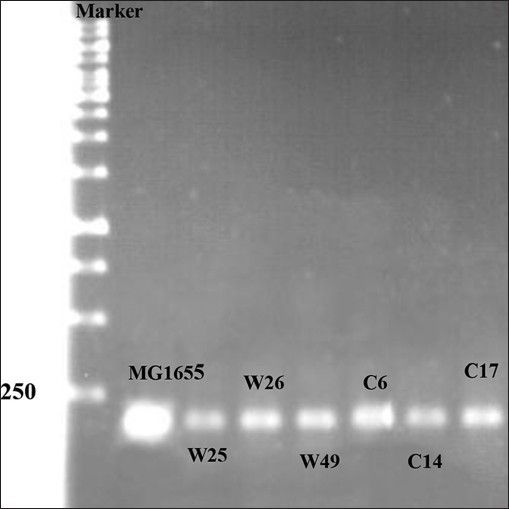
Gel electrophoresis of PCR product.
First lane contains 1Kb DNA ladder and other lanes contain PCR products of wild type and mutants.
Fig. 3.
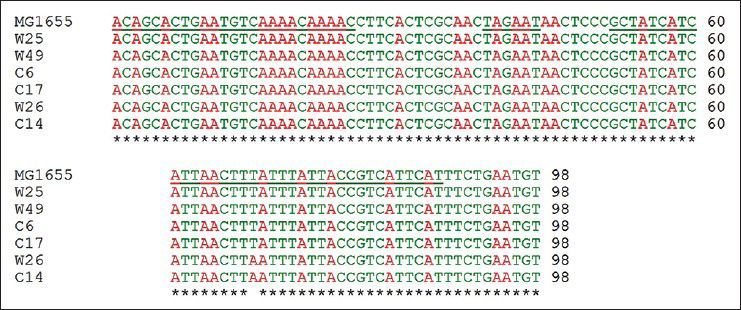
Multiple sequence alignment of 5′ end of micF gene and its upstream region in MG1655 (wild type) and mutants.
The underlined nucleotide sequences shown in order from left side are the mar box, RNP-10 signal and 5′ end of micF gene, respectively.
As mutants used in this study were with or without marR mutation, it was possible that they reduce ompF expression. Purified RNAs were used for real time analysis. Results reveal that the slope of regression line was −3.1 and −3.5 for gapA and ompF, respectively. Thus, the efficiency of ompF and gapA were 1.94 and 2.1, respectively. The melting curve of two genes showed just one major peak which indicates the purity of samples. Fig. 4 shows the melting curve of ompF in wild type and mutants. The melting point of ompF and gapA were 87 and 86°C. Fig. 5 shows the amplification curve of ompF in wild type and mutants. Ct values ranged from 14 to 20. Table 3 shows the relative expression of ompF in these mutants. The t-test analysis showed no significant difference between wild type and mutants for expression of ompF (P<0.05). The reason for this result may be due to low to intermediate level of resistance to ciprofloxacine and Tc (Table 1).
Fig. 4.
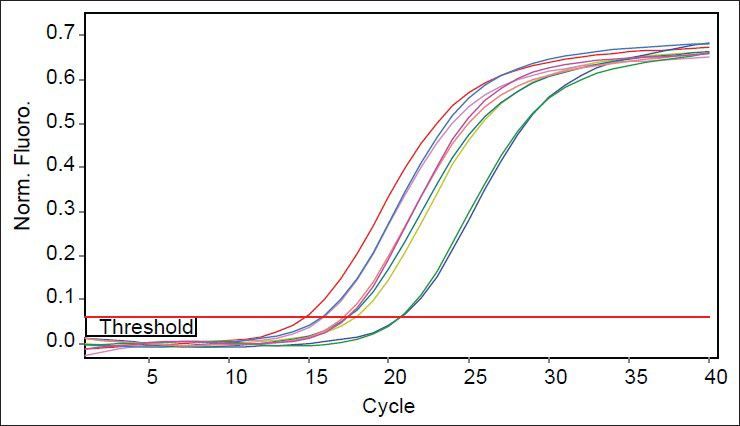
Melting curves of ompF in wild type and mutants.
The yellow color curve belongs to wild type and other colored curves belong to mutants.
Fig. 5.
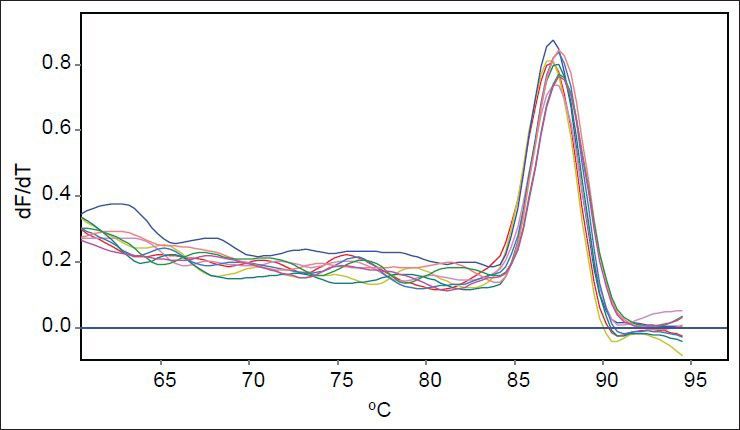
Amplification curves of ompF in wild type and mutants.
The pale blue curve belongs to wild type and other colored curves belong to mutants.
TABLE 3.
RELATIVE EXPRESSION OF OMPF IN WILD TYPE (MG1655) AND MUTANTS

DISCUSION
OmpF is an outer membrane porin found in gram negative bacteria, such as E. coli[3]. This porin is used for entrance of drugs, including quinolones, tetracycline and β-lactams[11]. It was said that down regulation of ompF causes resistance to multiple antibiotics, for example quinolones and tetracycline[11]. Low expression of ompF was frequently found in clinical isolates with high to intermediate levels of resistance to ciprofloxacine. These isolates contain alterations in genes encoding topoisomerase II (gyrase) and topoisomerase IV subunits, including gyrA and parC[11]. The expression of ompF is regulated at both transcriptional and translational levels. At translational level the expression of ompF is negatively controlled by micF RNA, a small antisense RNA that base pairs with ompF RNA[3]. The expression of micF is increased by transcriptional activator called MarA in the presence of ciprofloxacine or tetracycline. Thus, it is expected that the synthesis of OmpF is decreased[14]. However, it was demonstrated that the down regulation of OmpF is not completely dependent on up regulation of MarA activity[15].
In addition, it was shown that salicylate also reduces the translation of ompF RNA mainly by the mechanism similar to antibiotics[16]. On the other hand, at transcriptional level the two component regulatory system, EnvZ-OmpR located in cell membrane is responsible. However, this regulation is dependent on external osmolarity not the presence of antibiotics; in the way that low osmolarity causes ompF up regulation[17]. To see whether the transcription of ompF is changed in the presence of tetracycline, the level of ompF mRNA was quantified by real time PCR in mutants with low to intermediate levels of resistance to ciprofloxacin and tetracycline. Also the intactness of micF was checked in these mutants by PCR amplification of 5′ end and upstream region of this gene and sequencing the PCR products to ensure that they maintain the ability to base pair with ompF RNA following the up regulation of MarA in the presence of antibiotics.
Moreover, as mentioned before for marA dependent reduction of OmpF synthesis, an intact micF locus is required[8]. This includes and intact mar box and 5′ end of micF gene[3,5]. It was found that in all mutants except one (W26) and its clone (C14) the sequence of 5′ end of micF is intact. In W26 and its derived clone (C14) the nucleotide change in micF sequence would be in small loop structure following its attachment to ompF RNA. Possibly, it may not affect on base pairing ability. However, further study is needed to prove it.
Furthermore, statistical analysis did not reveal any significant difference between ompF expression in mutants and wild type strain (P<0.05). This is consistent with previous results on multidrug resistant mutants isolated from calves[18]. Reduced expression of ompF was seen in E. coli mutants with high levels of resistant to ciprofloxacin[11]. However, reduction in ompF expression was not seen in all mutants with low to intermediate levels of resistance to ciprofloxacin[11]. Kishii et al.[11] suggest that different genetic backgrounds are the cause of low or normal expression of ompF in mutants with low and intermediate levels of resistant to ciprofloxacin. Also, it is possible that a change in unknown factor is also necessary for down regulation of ompF[15].
ACKNOWLEDGEMENTS
This work was financially supported and partly conducted in the Institute of Biotechnology, University of Shahrekord.
Footnotes
Jaktaji and Heidari: Study the Expression of OmpF in Mutants
REFERENCES
- 1.Windbichler N, von Pelchrzim F, Mayer O, Csaszar E, Schroeder R. Isolation of small RNA-binding proteins from E. coli. RNA Biol. 2008;5:1–11. doi: 10.4161/rna.5.1.5694. [DOI] [PubMed] [Google Scholar]
- 2.Valentin-Hansen P, Johansen J, Rasmussen AA. Small RNAs controlling outer membrane porins. Curr Opin Microbiol. 2007;10:152–5. doi: 10.1016/j.mib.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006;9:605–11. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Delihas N, Forst S. MicF: An antisense RNA gene involved in response of Escherichia coli to global stress factors. J Mol Biol. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- 5.Martin RG, Gillette WK, Rhee S, Rosner JL. Structure requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–41. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 6.Perera IC, Grove A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol. 2010;2:243–54. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 7.Martin RG, Kam-Wing J, Wolf RE, Rosner JL. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J Bacteriol. 1996;178:2216–23. doi: 10.1128/jb.178.8.2216-2223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen SP, McMurry LM, Levy SB. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistance (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–22. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pourahmad Jaktaji R, Mohiti E. Study of mutations in the DNA gyrase gyrA gene of Escherichia coli. Iran J Pharm Res. 2010;9:43–5. [PMC free article] [PubMed] [Google Scholar]
- 10.Pourahmad Jaktaji R, Ebadi R, Karimi M. Study of organic solvent tolerance and increased antibiotic resistance properties in E. coli gyrA mutants. Iran J Pharm Res. 2011;11:595–600. [PMC free article] [PubMed] [Google Scholar]
- 11.Kishii R, Takei M. Relationship between the expression of OmpF and quinolone resitance in Escherichia coli. J Infect Chemother. 2009;15:361–6. doi: 10.1007/s10156-009-0716-6. [DOI] [PubMed] [Google Scholar]
- 12.George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol and other antibiotics in Escherichia coli: Involvement of a non-plasmid determined efflux of tetracycline. J Bacteriol. 1983;155:531–40. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST© ) for group wise comparison and statistical analysis of relative expression results in real time PCR. Nucl Acids Res. 2002;30:1–10. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, et al. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS One. 2007;4:e365. doi: 10.1371/journal.pone.0000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karczmarczyk M, Martin M, Quinn T, Leonard N, Fanning S. Mechanisms of fluoroquinolone resistance in Escherichia coli isolates from food producing animals. Appl Environ Microbiol. 2011;77:7113–20. doi: 10.1128/AEM.00600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosner JL, Chai TJ, Foulds J. Regulation of OmpF porin expression by salicylate in Escherichia coli. J Bacteriol. 1991;173:5631–8. doi: 10.1128/jb.173.18.5631-5638.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forst S, Delgado J, Ramakrishnan G, Inouye M. Regulation of OmpC and OmpF expression in Escherichia coli in the absence of envZ. J Bacteriol. 1998;170:5080–5. doi: 10.1128/jb.170.11.5080-5085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vinson HM, Gautam A, Olet S, Gibbs PS, Barigye R. Molecular analysis of transcription in heterogenotypic multidrug resistant Escherichia coli isolates from scouring calves. J Antimicrob Chemother. 2010;65:1926–35. doi: 10.1093/jac/dkq246. [DOI] [PubMed] [Google Scholar]


