Abstract
The transcription factor CCAAT/enhancer binding protein β (C/EBPβ) is a key regulator of growth and differentiation in many tissues. C/EBPβ is expressed as several distinct protein isoforms (LAP1, LAP2, and LIP) whose expression is regulated by alternative translational initiation at downstream AUG start sites. The dominant-negative LIP isoform is predominantly expressed during proliferative cellular responses and is associated with aggressive tumors. In this study, we investigated a mechanism by which the LIP isoform is translationally regulated in mammary epithelial cells. We have demonstrated that LIP expression is increased in response to activation of the epidermal growth factor receptor (EGFR) signaling pathway and that the increased expression of LIP is regulated in part by an RNA binding protein referred to as CUG repeat binding protein (CUG-BP1). Our data demonstrate that EGFR signaling results in the phosphorylation of CUG-BP1 and this leads to an increase in the binding of CUG-BP1 to C/EBPβ mRNA and elevated expression of the LIP isoform. Phosphorylation is necessary for the binding activity of CUG-BP1 and the consequent increase in LIP expression, as determined by binding assays and a cell free, transcription-coupled translation system. CUG-BP1 is thus a previously unidentified downstream target of EGFR signaling and represents a new translational regulator of LIP expression in human mammary epithelial cells.
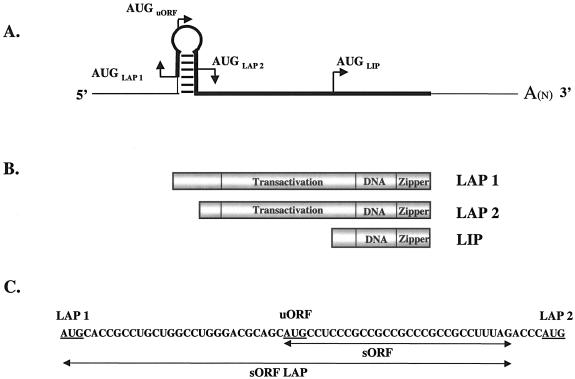
CCAAT/enhancer binding protein β (C/EBPβ), a member of the basic leucine zipper family of transcription factors, is a key regulator of growth and differentiation in many tissues. The gene for C/EBPβ is intronless and is transcribed into a single mRNA (Fig. 1A) that gives rise to multiple protein isoforms, referred to as 38-kDa liver-enriched activating protein (LAP1), 35-kDa LAP2, and a 20-kDa liver-enriched inhibitory protein (LIP) in rodent tissues (Fig. 1B). In human tissue, C/EBPβ is commonly referred to as NF-IL-6 (nuclear factor of interleukin-6) or IL-6DBP, and the LAP isoforms are larger (approximately 46 to 42 kDa) while the size of the LIP isoform remains the same. For clarity, we will use the C/EBPβ, LAP, and LIP nomenclature regardless of the origin of the mammary tissue or cells. All of the C/EBPβ isoforms have the same DNA binding and dimerization domains, but because LIP is translated from the third in-frame AUG start codon, LIP lacks much of the trans-activation domain and has an increased binding affinity for DNA relative to the LAP isoforms (6). Consequently, LIP is unable to activate gene transcription and thereby functions as a dominant-negative factor by antagonizing the transcriptional activity of LAP via a LAP-LIP heterodimer or by competing for DNA binding sites as a LIP homodimer (6).
FIG. 1.
Schematic of C/EBPβ mRNA and protein isoforms. (A) Line drawing of C/EBPβ mRNA. To show the hairpin structure and translation start sites in greater detail, the diagram has not been drawn to scale. The thin line represents the untranslated noncoding sequence, and the thick line represents the translated coding sequence. (B) Schematic of the predominant C/EBPβ protein isoforms. The transactivation, DNA binding (DNA), and leucine zipper (zipper) regions are indicated. (C) Ribonucleotide sequence of the murine C/EBPβ hairpin region from the LAP1 AUG codon to the LAP2 AUG codon to show the sequences spanned by the murine oligomers used in the RNA binding assay. The mouse and human ribonucleotide sequences are 81% identical in this region.
Although the functions of C/EBPβ and its target genes remain to be defined, studies have shown that the C/EBPβ isoforms are overexpressed in multiple types of cancer (42, 31, 23) and can mediate the cdk-independent actions of cyclin D1 in a broad range of human tumors (15). The LIP isoform in particular has been found to be overexpressed and associated with more aggressive tumors. For example, in breast cancer, LIP is overexpressed in 23% of infiltrating ductal carcinomas and is associated with tumors that are estrogen and progesterone receptor negative, aneuploid, highly proliferative, and poorly differentiated (42). In the ovary, LIP expression is detected only in malignant tumors compared to benign tumors (31), and in colorectal cancer, LIP is expressed at higher levels in more-invasive Duke's stage B tumors than in less-invasive Duke's stage A tumors (23). Similarly, full-length LAP1 has also been found to be associated with breast tumors that are estrogen receptor negative and high grade and show nodal involvement (18).
Studies to address the functions of LAP1, LAP2, and LIP have been conducted with mice and cell cultures and support an emerging role for C/EBPβ as an oncogene. For example, forced expression of LIP under the control of the whey acidic promoter in the mouse mammary gland results in the formation of hyperplastic tissue and carcinomas (41). Likewise, chronic expression of LIP in cultured mouse mammary epithelial cells (TM3) or 3T3-L1 adipocytes results in proliferation, focus formation, and loss of contact inhibition (41, 5). LAP1 has been shown to interact with the SWI/SNF ATPase/helicase chromatin remodeling complex (11), while LAP2 may have a role in transformation and epithelial-to-mesenchymal transition (4).
Consequently, in light of the unique roles that each of the C/EBPβ isoforms plays in cell growth, differentiation, and cancer, it is important to understand how these isoforms are translationally regulated. Because our laboratory is particularly interested in the regulation and role of LIP in aggressive breast cancer, this study is focused on understanding the regulation of LIP translation in mammary epithelial cells. It is not well understood how LAP1, LAP2, and LIP isoform expression is regulated, but it has been proposed that the isoforms are differentially translated by a leaky ribosomal scanning mechanism (6, 12, 40). Extracellular stimuli such as cytokines or retinoic acid may affect the alternative translation of the isoforms (10), as well as mRNA sequences in the 5′ untranslated region (40) and sequence-specific mRNA binding proteins that facilitate the interaction of ribosomes with upstream AUG (uAUG) codons (36). The 5′ end of the C/EBPβ mRNA contains a uAUG and a small open reading frame (sORF) that is situated between the translation initiation sites for LAP1 and LAP2 (Fig. 1). This region has been shown to be important for the translational control of the C/EBPβ isoforms (40, 17), and LIP expression appears to be strictly regulated by the uORF (5). Most importantly, the 5′ region of the C/EBPβ mRNA near the uORF contains several binding sites for the RNA binding protein CUG repeat binding protein (CUG-BP1) (36).
CUG-BP1 (hNab50) was first identified in 1996 as a novel RNA binding protein that interacts only with (CUG)8 or (CCG)n triplet repeats and not with single- or double-stranded DNA CTG repeats or RNA triplet repeats of a different sequence such as CGG (34). Later investigations showed that CUG-BP1 binds to GC-rich RNA sequences that potentially might form stable secondary structures (32). Experiments with rat liver after partial hepatectomy, in the acute-phase response to inflammation and the pathology of myotonic dystrophy showed that, in each physiological response, CUG-BP1 can bind near uAUGs in the uORF of C/EBPβ mRNA to regulate the translation of the C/EBPβ LIP isoform (36, 38, 35).
Consequently, we hypothesized that CUG-BP1 may play a role in the translational regulation of LIP in mammary epithelial cells. However, we speculated that this translational process must also be regulated because LIP expression is not constant and changes in relation to epithelial cell growth or mammary gland development. For example, LIP expression increases in cultured epithelial cells that mitotically expand in response to serum and growth factors (41) or proliferate in response to the hormones and growth factors associated with pregnancy and breast cancer (25, 24, 42). Because mammary epithelial cells require epidermal growth factor (EGF) for growth and the EGF signaling pathway is associated with breast cancer, we investigated whether activation of the EGF signaling pathway in mammary epithelial cells can regulate the activity of CUG-BP1 and result in increased translation and expression of LIP.
In this report, we present evidence that EGF receptor (EGFR) signaling increases the binding activity of CUG-BP1 to C/EBPβ mRNA and leads to increased translation of LIP in MCF10A cells. Phosphorylation of CUG-BP1 appears to be a critical component of CUG-BP1 binding activity, without which translation of LIP will be decreased. CUG-BP1 is thus a previously unidentified downstream target of EGFR signaling and represents a new translational regulator of LIP expression in human mammary epithelial cells.
MATERIALS AND METHODS
Pituitary isografts.
Mice were housed in an Association for Assessment and Accreditation International-accredited facility in the Sidney Kimmel Cancer Center at Johns Hopkins and were provided a standard irradiated mouse diet and acidified water ad libitum on a 12-h light-dark cycle. Prior to surgery, animals were anesthetized with Avertin (1% solution given intraperitoneally at 0.0375 ml/g of body weight). Pituitaries were removed from 12-week-old nontransgenic sibling mice and transplanted into the thoracic mammary glands of host whey acidic protein (WAP)-TGF-α transgenic mice (kindly provided by Eric Sandgren) or nontransgenic littermates with an 18-gauge trocar. Inguinal mammary glands from both transgenic and nontransgenic mice were harvested at 1, 2, 3, 5, 8, and 12 weeks postisografting and processed for histological analysis or protein extraction.
Cell culture.
Cultured MCF10A cells were grown in Dulbecco modified Eagle medium (DMEM)/F12 (Invitrogen, Carlsbad, Calif.) supplemented with 5% donor horse serum (Invitrogen), 20 ng of recombinant human EGF (Invitrogen) per ml, 10 μg of bovine pancreatic insulin (Sigma, St. Louis, Mo.) per ml, 100 ng of cholera toxin (Sigma) per ml, 0.5 μg of hydrocortisone (Sigma) per ml, and 5 μg of gentamicin sulfate (Invitrogen) per ml. Cells were plated at a density of 1.7 × 106/100 mm, and when the monolayers were approximately 75 to 80% confluent, the growth medium was removed and replaced with a serum-free, defined medium containing DMEM/F12, 100 ng of cholera toxin per ml, 0.5 μg of hydrocortisone per ml, and 5 μg of gentamicin sulfate per ml. Cells were maintained in defined medium for 24 h prior to the addition of 10 ng of EGF or TGF-α per ml and harvested at either 5 min or 16 h after the addition of ligand. The antagonists and inhibitors AG1478 (Calbiochem, San Diego, Calif.), OSI-774 (OSI Pharmaceuticals Inc., Melville, N.Y.), UO126 (Calbiochem), and cycloheximide (Sigma) were added 30 min before addition of EGF.
Isolation of poly(A)+ RNA and Northern blot analysis.
Poly(A)-enriched RNA was isolated and purified from MCF10A cells by a modification of previously described methods (2, 14). Cells were lysed in a buffer containing proteinase K (0.2 mg/ml), and mRNA was purified by affinity chromatography with oligo(dT) cellulose (Ambion, Austin, Tex.). mRNA (7 μg) was size fractionated via gel electrophoresis in a 1% agarose-6.6% formaldehyde gel, transferred to a nylon membrane (Hybond-XL, Amersham Pharmacia Biotech, Piscataway, N.J.), and hybridized with an [α-32P]dCTP random primed human C/EBPβ cDNA probe. Hybridization-positive bands were visualized with a phosphorimager (Molecular Dynamics, Sunnyvale, Calif.).
Protein isolation and Western blot analysis.
Tissue and/or cells were homogenized in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.25% deoxycholate, 150 mM NaCl, 10 mM EGTA, 0.2% sodium dodecyl sulfate [SDS]) containing a protease inhibitor cocktail (Sigma) and a phosphatase inhibitor II mixture (Sigma). Aliquots of these lysates containing 100 μg of protein were boiled at 100°C for 10 min, electrophoresed on denaturing SDS-12% polyacrylamide minigels, and then transferred to polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). Blots for C/EBPβ detection were blocked for 2 h in TBST (20 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Tween 20) containing 10% nonfat dry milk (Bio-Rad, Hercules, Calif.) and then incubated for 2 h in TBST-10% nonfat dry milk (NFDM) containing C/EBPβ polyclonal antibodies (C-19; Santa Cruz Biotechnology, Santa Cruz, Calif.) at a 1:500 dilution. Blots were washed with TBST three times for 5 to 10 min each time with agitation and then incubated for 1 h with goat anti-rabbit-horseradish peroxidase (HRP) conjugate (Bio-Rad) in TBST-10% NFDM. Proteins were visualized by enhanced chemiluminescence (Super Signal; Pierce, Rockford, Ill.) with X-Omat-LS film (Kodak). Blots were stripped in Re-blot Plus Mild Solution (Chemicon, Temecula, Calif.) and prepared for β-actin immunodetection via a 2 × 10-min block in 5% NFDM-TBST, followed by a 2-h incubation with a β-actin polyclonal antibody (1:1,000; Santa Cruz) in 5% NFDM-TBST. Membranes were washed, incubated with HRP-conjugated donkey anti-goat antibody (Santa Cruz), and developed as described above. Blots for phosphorylated and total p44/42 MAPK detection were blocked with 5% NFDM-TBST for 1 h, followed by incubation with either phospho-p44/42 polyclonal antibody or total mitogen-activated protein kinase (MAPK) antibody (1:1,000; Cell Signaling, Beverly, Mass.) in 5% NFDM-TBST overnight at 4°C. CUG-BP1 expression was detected with a 5% NFDM-TBST block for 1 h, followed by incubation with a CUG-BP1 monoclonal antibody (1:1,000; Santa Cruz) in 5% NFDM-TBST for 2 h at room temperature (RT). Blots were washed, incubated with HRP-conjugated goat anti-mouse antibody, and visualized as described above.
Pulse-chase.
MCF10A cells were grown to a confluency of approximately 80% in 100-mm-diameter dishes, rinsed three times with Hanks' balanced salt solution, and incubated for 24 h in serum-free DMEM/F12 containing 100 ng of cholera toxin per ml, 0.5 μg of hydrocortisone per ml, and 5 μg of gentamicin sulfate per ml. Cells were then rinsed for 10 min in Met- and Cys-free DMEM (Sigma) at RT and then placed in Met- and Cys-free DMEM containing 0.2 mCi of [35S]Met-Cys (Amersham) and 10 ng of EGF per ml for 3.5 h. Cells were washed and chased twice with DMEM/F12 containing unlabeled Cys and Met and then placed in serum-free defined medium containing EGF (10 ng/ml) for 20.5 h (24 h total of EGF treatment). Harvested cell pellets were solubilized in RIPA buffer (without SDS) for 2 h on ice, and 250 μg of whole-cell protein lysate was then incubated with 1.6 μg of C/EBPβ polyclonal antibody (Santa Cruz) overnight at 4°C. Immunoprecipitates were incubated with 30 μl of protein G-PLUS agarose for 2 h at 4°C, and agarose beads were rinsed three times with TBST and twice with phosphate-buffered saline, followed by addition of sample buffer (0.125 M Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 10% β-mercaptoethanol), and heated at 100°C for 10 min. Immunoprecipitates were analyzed via 12% polyacrylamide gel electrophoresis (PAGE) and standard autoradiography.
32Pi labeling of CUG-BP1.
MCF10A cells were grown to 75% confluency in 100-mm-diameter dishes, serum starved (as previously described) for 24 h, and treated with 10 ng of EGF per ml for 12 h. Cells were then rinsed in phosphate-free DMEM (Invitrogen) for 3 × 1 min and subsequently incubated for 30 min in phosphate-free DMEM plus 10 ng of EGF per ml. Medium was removed, and 6 ml of phosphate-free DMEM containing 0.5 mCi of 32Pi (NEN Dupont, Wilmington, Del.) and 10 ng of EGF per ml was added. Cells were incubated at 37°C for an additional 3.5 h (16 h of EGF treatment). Cells were rinsed three times with cold Hanks' balanced salt solution and harvested. Lysates were prepared by addition of RIPA buffer without SDS to each pellet, incubation for 2 h on ice, and pelleting of unsolubilized debris. Antibody to CUG-BP1 (1.6 μg/dish; Santa Cruz) was added to each sample, and the mixture was incubated overnight at 4°C. Protein G-PLUS agarose (Santa Cruz) was added, and the mixture was incubated for 2 h at 4°C. The complex was rinsed three times with TBST and twice with Tris-buffered saline. Denaturing sample buffer was added, and protein was eluted from the beads by heating at 100°C for 5 min. Proteins were resolved by SDS-12% PAGE, and autoradiography was performed.
RNA-protein UV cross-linking assay.
RNA oligomers sORF-LAP (5′-AUGCACCGCCUGCUGGCCUGGGACGCAGCAUGCCUCCCGCCGCCGCCCGCCGCCUUUAG-3′), corresponding to the C/EBPβ region from the first AUG codon (LAP1) to the third AUG codon (LAP2), including the ORF AUG codon; sORF (5′-AUGCCUCCCCGCCGCCGCCCGCCGCCUUAG-3′) (Fig. 1C); and AU-rich (5′-UAAAUUAAAAUUAAAAAUUUAAAAUUU-3′) were labeled with [γ-32P]ATP by using T4 kinase. Total proteins from whole-cell extracts or immunoprecipitated CUG-BP1 from treated and nontreated MCF10A cells were incubated with an RNA probe for 30 min at RT, exposed to UV light (Stratalinker; Stratagene) for 30 min, and analyzed via electrophoresis on an 8 to 16% gradient denaturing polyacrylamide gel. Proteins were transferred onto nitrocellulose membranes and subjected to autoradiography. After UV cross-linking analysis, the membranes were stained with Coomassie blue to verify the equal loading of protein in each lane.
In vitro TnT of LIP and cleavage assay.
MCF10A cells were grown to 75% confluency in 100-mm-diameter dishes and serum starved (as previously described) for 24 h prior to EGF induction. Cells were treated with AG1478 or vehicle alone for 30 min prior to induction with human EGF. Cells were incubated for 16 h at 37°C, harvested, and incubated on ice with RIPA buffer for 2 h. CUGBP1 was immunoprecipitated from the lysates with a monoclonal antibody to CUG-BP1 for 2 h at 4°C and protein G-PLUS agarose for 2 h at 4°C, followed by three rinses with ice-cold TBST, two rinses with ice-cold Tris-buffered saline (low NaCl), and resuspension in 10 mM Tris (pH 7.4). The immunoprecipitated CUG-BP1 was added to the rabbit reticulocyte lysate transcription-translation (TnT; Promega) reaction mixture in the presence of either a full-length wild-type (WT) C/EBPβ cDNA construct or a mutated construct containing an ATG-to-TTG mutation at the LIP translational initiation AUG codon (mutAUG; provided by J. Papaconstantinou). Immunoprecipitates were incubated with calf intestinal phosphatase (CIP) at 37°C for 30 min prior to the TnT reaction. Translation products were resolved by SDS-12% PAGE and transferred to Immobilon P. C-EBPβ LIP was detected with anti-C/EBPβ antibody and HRP-conjugated goat anti-rabbit antibody, followed by incubation with PICO detection reagents (Pierce). For some reactions, [35S]Met was added to the TnT reaction mixture. These translated proteins were immunoprecipitated with a C/EBPβ polyclonal antibody, separated by SDS-12% PAGE, and autoradiographed.
CUG-BP1 was originally isolated from the cytoplasm of HeLa cells that express high levels of C/EBPβ (33). These studies demonstrated that interaction of CUG-BP1 with the sORF is observed for both bacterially expressed and purified CUG-BP1 and endogenous CUG binding proteins from HeLa cytoplasmic extracts (33). Thus, HeLa whole-cell extracts were used to activate bacterially expressed CUG-BP1 (see Fig. 9) because HeLa cytoplasm contains a kinase that is able to activate CUG-BP1. After activation, recombinant protein was further purified via immunoprecipitation.
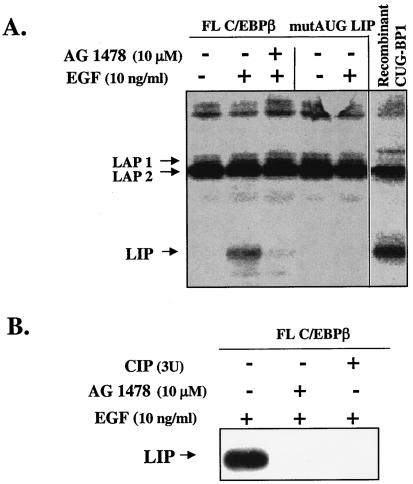
FIG. 9.
CUG-BP1 immunoprecipitated from EGF-treated MCF10A cells induces the translation of LIP in a cell-free system. (A) CUG-BP1 was immunoprecipitated from whole-cell extracts of MCF10A cells that were treated for 16 h with EGF, EGF plus AG1478, or vehicle only. The immunoprecipitated CUG-BP1 protein was added to the rabbit RL in the presence of [35S]methionine and either a full-length WT C/EBPβ construct (FL) or a mutated construct containing an ATG-to-TTG mutation at the LIP AUG codon (mutAUG). The lane designated recombinant CUG-BP1 (see Materials and Methods) is a positive control containing bacterially expressed CUG-BP1 that has been activated with HeLa cell extracts, immunoprecipitated, and added to the 35S-labeled RL programmed with the FL C/EBPβ construct. After translation, the C/EBPβ isoforms were immunoprecipitated, separated by SDS-PAGE, and visualized by autoradiography. (B) CUG-BP1 was immunoprecipitated from whole-cell extracts as described in panel A. The immunoprecipitate from the EGF-treated cells was treated with CIP, washed, and transferred to the TnT reaction mixture, which contained a full-length, WT C/EBPβ cDNA construct. Translated proteins were subjected to Western blot analysis with a polyclonal antibody to C/EBPβ.
For in vitro cleavage assays, the full-length WT C/EBPβ protein was immunoprecipitated and incubated with 20 to 30 μg of whole-cell protein extract from MCF10A cells that were first treated with vehicle, EGF, or EGF and AG1478. Mixtures were incubated for 1 h at 37°C, subjected to SDS-12% PAGE, transferred to nitrocellulose membranes, and visualized by autoradiography.
RESULTS
TGF-α transgenic mouse model.
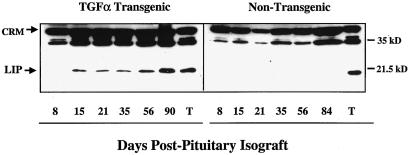
Only three ligands bind exclusively to the EGFR, transforming growth factor α (TGF-α), EGF, and amphiregulin. To test whether EGFR signaling regulates LIP expression, we analyzed both animal and cell culture models in which the EGFR was activated with either TGF-α or EGF. Transgenic mice expressing TGF-α under the control of the WAP promoter were chosen as our in vivo model for analysis of LIP expression. TGF-α overexpression, in this transgenic model, leads to increased EGFR signaling in the mammary gland and the development of a cystic, proliferative, and tumorigenic mammary phenotype (28, 27). We induced TGF-α expression in these mice by implantation of pituitary isografts, which leads to activation of the WAP promoter. The WAP promoter is responsive to hormonal stimulation (16) and is exquisitely sensitive to hormones that are associated with pituitary isografts or pregnancy (7). Western blot analysis of mammary glands from the transgenic mice showed that LIP expression was induced before tumor formation, at 1 to 2 weeks postimplantation of the pituitary isograft, and remained elevated throughout the 12-week regimen (Fig. 2, left panel). All of the mammary tumors that we examined from WAP-TGF-α transgenic mice expressed high levels of LIP (Fig. 2, lane T). LIP expression was not elevated in the pituitary isografted nontransgenic littermates compared to that in their transgenic counterparts (Fig. 2, right panel), but low levels were detectable with longer exposures (data not shown). These data lend support to our hypothesis that LIP expression is strongly regulated by EGFR signaling in the mouse mammary gland.
FIG. 2.
LIP expression is increased in WAP-TGF-α mice after transgene stimulation. Western blot analysis of whole-cell extracts (100 μg) prepared from mammary glands removed (8 to 90 days) after implantation of pituitary isografts into the thoracic glands of TGF-α transgenic mice (left panel) and nontransgenic siblings (right panel). The days post pituitary isografting are indicated. Whole-cell extracts from a non-pituitary-isografted TGF-α tumor (T) serves as a positive control for LIP overexpression. The blots were overexposed in an effort to detect LIP in the nontransgenic blot. Cross-reactive material (CRM) serves as an estimate for the total protein loaded and transferred per well.
MCF10A cell model.
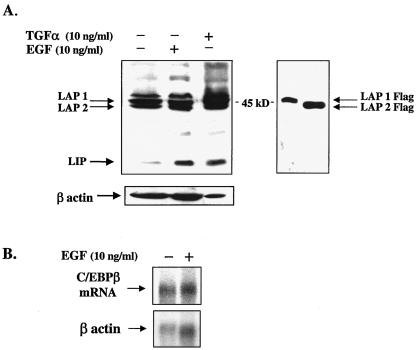
To more thoroughly analyze the mechanisms by which the EGFR signaling pathway regulates LIP expression and to characterize the proteins involved in this regulation requires the use of a mammary epithelial cell line that contains a responsive EGFR signaling pathway. Consequently, MCF10A cells were selected because they are a spontaneously immortalized, normal human breast epithelial line that expresses each of the four ErbB receptor subtypes (30). MCF10A cells have previously been used as a sensitive indicator for EGFR activity in conditioned-medium experiments (8). MCF10A cells were cultured in serum-free, defined medium (as described in Materials and Methods) with either EGF (10 ng/ml) or TGF-α (10 ng/ml) for 16 h. As determined by Western blot analysis, LIP expression was elevated in response to EGF and TGF-α treatment compared to that in nontreated control cells (Fig. 3A). In contrast, the larger LAP1 and LAP2 isoforms showed little or no change in expression after stimulation of the EGFR (Fig. 3A). Likewise, an analysis of C/EBPβ mRNA demonstrated that transcript levels do not change in response to treatment of MCF10A cells with EGF (Fig. 3B). These findings indicate that EGFR signaling selectively controls LIP expression at the posttranscriptional level in mammary epithelial cells. The observed regulation of LIP expression by EGFR signaling is not restricted to the nonmalignant MCF10A cell line but has also been observed in the malignant breast cell lines MDA-MB-231, MDA-MB-468, MCF7, SUM 229 PE, SUM 149 PT, and SUM 102 PT (data not shown).
FIG. 3.
Protein expression of LIP is induced in MCF10A cells after treatment with EGF or TGF-α, but transcriptional regulation of C/EBPβ is relatively unaffected. (A, left side) Western blot analysis of LIP in MCF10A cells treated with EGF or TGF-α. Cells were serum starved for 24 h and then treated with human EGF (10 ng/ml) or TGF-α (10 ng/ml). Cells were harvested 16 h after addition of ligand, and 100 μg of protein from whole-cell extracts was subjected to SDS-PAGE and Western blot analysis. Rehybridization of the blot with a β-actin polyclonal antibody served as an estimate of the total amount of protein loaded and transferred per well. (A, right side) Western blot analysis of 293 cells transfected with LAP1 or LAP2 constructs that were Flag tagged at the amino terminus. LAP isoforms were immunodetected with Flag antibody (Sigma) and serve as controls to aid in the identification of the endogenous LAP isoforms that often migrate with cross-reactive material and are often difficult to resolve on Western blots. (B) Northern blot analysis of poly(A)+ mRNA (7 μg) isolated from EGF-treated and nontreated MCF10A cells hybridized with a cDNA probe to human C/EBPβ. Blots were stripped and rehybridized with a cDNA for β-actin to estimate RNA loading per well.
De novo protein synthesis.
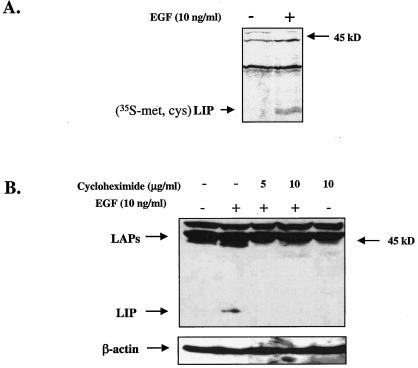
To investigate whether the changes in LIP expression were the result of de novo protein synthesis, MCF10A cells were metabolically labeled (pulsed) with [35S]Met-Cys for 3.5 h. 35S-labeled LIP was undetectable in non-EGF-treated cells but was detectable in protein extracts from cells that were treated with EGF for 24 h (Fig. 4A). To address the possibility that LIP is generated as a proteolytic cleavage product of the larger LAP precursors, EGF-treated cells were treated with cycloheximide to halt protein synthesis and LIP levels were then examined. Western blot analysis of these protein extracts demonstrated that a proteolytically cleaved LIP product was not detectable in cells that were treated with cycloheximide for 16 h (Fig. 4B). Taken together, the data in Fig. 3 and 4 suggest that the increase observed in LIP expression is primarily the result of EGF-induced protein synthesis and not likely the result of transcriptional regulation or proteolytic cleavage.
FIG. 4.
EGF-induced expression of LIP is the result of nascent protein synthesis. (A) MCF10A cells were serum starved for 24 h and then pulsed with [35S]Met-Cys for 3.5 h (10 ng/ml). Cells were chased with unlabeled Met-Cys medium and then harvested 24 h after addition of EGF. 35S-labeled proteins were immunoprecipitated with a C/EBPβ polyclonal antibody and analyzed via SDS-PAGE and autoradiography. (B) Western blot analysis of whole-cell protein extracts (100 μg) from MCF10A cells that were treated with cycloheximide (5 to 10 μg/ml) for 30 min prior to the 16-h EGF treatment (10 μg/ml). The blot was stripped and reprobed with a β-actin polyclonal antibody to estimate loading and transfer of proteins (bottom).
EGFR signaling.
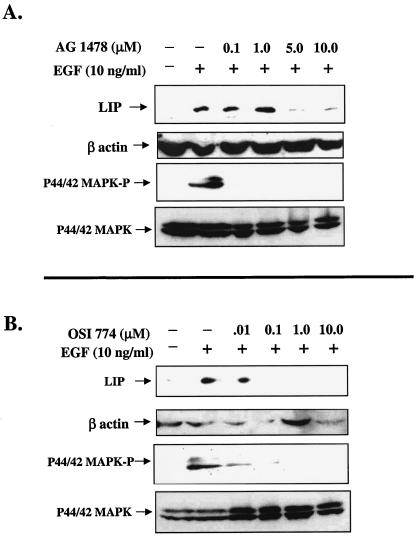
Although it is well established that EGF and TGF-α bind exclusively to EGFR/ErbB1 (29), we wanted to confirm that the increase observed in LIP expression was mediated by the EGFR. Consequently, cells were treated with two well-characterized, low-molecular-weight antagonists of the EGFR tyrosine kinase. AG1478 is a quinazoline that inhibits EGFR kinase activity by competition with ATP binding, whereas CP-358,774 (OSI-774) is a more directly acting and reversible ATP-competitive inhibitor of EGFR tyrosine phosphorylation (20). Treatment of MCF10A cells for 16 h with both antagonists decreased the EGF-induced expression of LIP (Fig. 5A and B, upper panels). These data demonstrate that LIP expression is regulated through EGFR signaling.
FIG. 5.
Expression of LIP is decreased in MCF10A cells after treatment with EGFR antagonists. (A) MCF10A cells were serum starved for 24 h and then treated with increasing concentrations of AG1478 for 30 min prior to addition of EGF (10 ng/ml). Cells were then harvested at either 5 min (for p44/42 MAPK detection; lower panels) or 16 h (for LIP detection; upper panel) after EGF stimulation. Whole-cell extracts (100 μg) were analyzed via SDS-PAGE and Western blotting. C/EBPβ blots were stripped and reprobed with a polyclonal antibody to β-actin (middle panel) to estimate loading and transfer of proteins. (B) Dose response of MCF10A cells to treatment with OSI-774. Experiment was conducted as described for panel A.
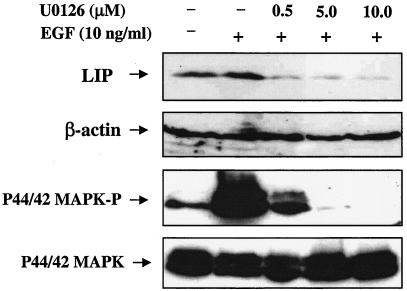
Activation of the EGFR stimulates a number of cytoplasmic signal transduction cascades. However, we limited our analysis to the MAPK (p44/42 MAPK) pathway, which has previously been described as a regulator of C/EBPβ activity (21). To determine whether LIP expression correlates with the signaling activity of the p44/42 MAPK pathway, the phosphorylation status of p44/42 MAPK was analyzed in cells pretreated for 30 min with AG1478 or OSI-774 and then stimulated with EGF for 5 to 10 min (Fig. 5A and B, lower panels). Both AG1478 and OSI-774 decreased p44/42 MAPK phosphorylation (MAPK-P) (Fig. 5A and B, lower panels), with no concomitant change in total p44/42 MAPK levels. Because AG1478 and OSI-774 inhibited both LIP expression and phosphorylation of p44/42 MAPK, we hypothesized that LIP expression may be regulated in part via p44/42 MAPK signaling. To test this hypothesis, an experiment was conducted in which MCF10A cells were stimulated with EGF and treated with increasing doses of the MEK1/2 inhibitor U0126. Both LIP expression and p44/42 MAPK-P activity were decreased by treatment of the cells with a 0.5 μM dose of U0126 (Fig. 6). Comparison of Fig. 5 and 6 demonstrates that basal (nonstimulated) levels of LIP are sometimes elevated and may correlate with the phosphorylation status of p44/42 MAPK. These data suggest that EGFR-mediated p44/42 MAPK activation may play a role in the regulation of LIP expression.
FIG. 6.
Effect of inhibition of p44/42 MAPK signaling on the EGF-induced expression of LIP. Inhibition of p44/42 MAPK activation with U0126 reduces expression of LIP. MCF10A cells were serum starved for 24 h and then treated with increasing concentrations of U0126, a potent MEK1/2 inhibitor, for 30 min prior to addition of EGF (10 ng/ml). Cells were then harvested at either 5 min (for p44/42 MAPK-P and total p44/42 MAPK detection; lower panels) or 16 h (for LIP detection; upper panel) after addition of EGF. Whole-cell extracts (100 μg) were analyzed via SDS-PAGE and Western blotting. The C/EBPβ blot was stripped and incubated with a polyclonal antibody to β-actin to estimate loading and transfer of proteins.
Phosphorylation of CUG-BP1.
EGF stimulation of MCF10A cells results in a posttranslational modification of CUG-BP1 that is detected as a doublet by Western blot analysis (Fig. 7A). Treatment with the EGFR antagonist OSI-774 (Fig. 7A) or phosphatase (Fig. 7B) reduces the CUG-BP1 doublet to a single band of slower mobility, which is suggestive of dephosphorylation. To investigate whether the posttranslational modifications observed for CUG-BP1 are a result of changes in the phosphorylation status of CUG-BP1, MCF10A cells were metabolically labeled with 32Pi and CUG-BP1 was immunoprecipitated and analyzed via SDS-PAGE. Stimulation of cells for 16 h with EGF resulted in increased incorporation of 32P into CUG-BP1 (Fig. 7C). The 32Pi labeling experiment also demonstrated that CUG-BP1 contains a basal level of phosphorylation in the non-EGF-stimulated cells that may be due to low levels of growth factor production (Fig. 7C). Taken together, these data suggest that EGFR signaling can regulate the phosphorylation status of CUG-BP1 in mammary epithelial cells.
FIG. 7.
EGFR signaling regulates the phosphorylation status of CUG-BP1. (A and B) Western blot analysis of whole-cell protein extracts demonstrates that treatment with EGF leads to the appearance of a CUG-BP1 doublet (A and B, lane 2) that is reduced to a single band by treatment with either OSI-774 (A, lanes 4 and 5) or CIP (B, lanes 3 and 4). (C) EGF stimulation leads to increased incorporation of 32P into CUG-BP1. EGF-treated MCF10A cells were metabolically labeled with 32Pi, and CUG-BP1 was immunoprecipitated and analyzed by PAGE and autoradiography. A basal level of phosphorylation of CUG-BP1 is observed in the nontreated cells (−), which is increased upon stimulation with EGF (+).
Binding activity of CUG-BP1 is induced by EGFR signaling.
Because the gene for C/EBPβ is intronless, the LIP isoform can only be generated via translational mechanisms or limited proteolysis (3, 6, 39). Previously, it was found that CUG-BP1 interacts with the 5′ region of C/EBPβ mRNA and increases the translation of LIP (36). We hypothesized that the induction of LIP by EGF in mammary epithelial cells might be mediated in part through CUG-BP1. Consequently, the RNA binding activity of CUG-BP1 in extracts from MCF10A cells treated with EGF, EGF plus inhibitors, EGF plus phosphatase, or vehicle alone was analyzed. As determined by UV cross-linking analysis, CUG-BP1 binding to C/EBPβ RNA is increased in response to treatment of cells for 16 h with 10 to 50 ng of EGF per ml (Fig. 8A, lanes 2 and 5, and B to D, lane 2). Treatment of cells with an inhibitor of the EGFR, AG1478 (Fig. 8A, lanes 3 and 6) or OSI 774 (Fig. 8B, lanes 4 and 5), or a MEK1/2 inhibitor, U0126 (Fig. 8C, left side), reduced the binding activity of CUG-BP1 to near-basal levels. Additionally, treatment of EGF-activated CUG-BP1 with phosphatase abolished the binding of CUG-BP1 to the C/EBPβ RNA (Fig. 8D, lanes 3 and 4). These findings demonstrate that EGFR signaling, specifically, EGF-induced phosphorylation, is important for CUG-BP1 binding activity. These data also suggest that p44/42 MAPK may play a role in the regulation of CUG-BP1 binding activity.
FIG. 8.
The binding activity of CUG-BP1 is regulated by EGFR-induced phosphorylation. (A and B) Binding of CUG-BP1 to C/EBPβ mRNA is increased by stimulation of cells with EGF and inhibited by the EGF antagonists AG1478 and OSI-774 and the MEK1/2 inhibitor U0126. MCF10A cells were serum starved for 24 h and treated with either AG1478 (A), OSI-774 (B), or U0126 (C, left part) for 30 min prior to addition of EGF. Cells were harvested at 16 h, and whole-cell protein extracts were incubated with [γ-32P]ATP-labeled C/EBPβ RNA probes (Fig. 1C) and cross-linked with UV light. RNA-bound proteins were analyzed via electrophoresis on 8 to 16% denaturing polyacrylamide gradient gels, transferred onto nitrocellulose membranes, and visualized by autoradiography. (C, right part) CUG-BP1 interacts specifically with GCN repeats located in the 5′ region of C/EBPβ mRNA. Addition of an excess of an unlabeled RNA oligonucleotide (sORF), but not that of an RNA oligonucleotide (AU-rich), abolished the binding of CUG-BP1 to the labeled (sORF-LAP) probe, as detected by PAGE and autoradiography. (D) Phosphatase treatment of CUG-BP1 abolishes binding to C/EBPβ mRNA, as determined by UV cross-link-immunoprecipitation assay. As described above, MCF10A cells were stimulated with EGF and whole-cell extracts were treated with phosphatase. CUG-BP1 was then immunoprecipitated from both EGF and non-EGF-stimulated cells (lanes 1 and 2) and from CIP-treated, EGF-activated extracts (lanes 3 and 4). The immunoprecipitate was incubated with a [γ-32P]ATP-labeled C/EBPβ sORF probe and analyzed via SDS-PAGE and autoradiography.
Previous studies have established the specificity of the interaction between CUG-BP1 and C/EBPβ RNA (32, 36, 38). Addition of CUG-BP1 antibody to the binding reaction mixture leads to complete neutralization of the 51-kDa protein-RNA complex (26). Likewise, previous studies confirmed that CUG-BP1 specifically binds to the 5′ region of C/EBPβ mRNA because when antibodies were added to the binding reaction mixtures a supershifted complex was observed (36). As shown in Fig. 8C (right side), incorporation of specific competitors into the binding reaction mixture demonstrates that CUG-BP1 interacts specifically with GCN repeats located in the 5′ region of the C/EBPβ mRNA. Addition of excess unlabeled oligomer (sORF), but not that of an RNA oligomer (AU-rich), abolished the binding of CUG-BP1 to the labeled (sORF-LAP) probe (Fig. 8C, right side).
CUG-BP1 immunoprecipitated from EGF-treated cells induces translation of LIP in a cell-free system.
The rabbit RL in vitro TnT system was used to directly test whether CUG-BP1, when activated by EGFR signaling, can increase the translation of LIP. We also examined whether phosphorylation is important for CUG-BP1 activity by testing whether the dephosphorylation of CUG-BP1 affects the in vitro translation of LIP. Translated products were detected in one of two ways. Either [35S]methionine was added to the TnT reaction mixture and proteins were immunoprecipitated with antibody to C/EBPβ and resolved via PAGE, or [35S]methionine was not added to the TnT reaction mixture and total proteins were resolved and analyzed by Western blot analysis with antibody to C/EBPβ. Figure 9A is a representative [35S]methionine-based TnT reaction in which immunoprecipitated CUG-BP1 was added to the RL along with a full-length C/EBPβ cDNA construct (FL C/EBPβ) or a mutated full-length construct in which the LIP translation start site was changed from ATG to TTG (mutAUG). Addition of CUG-BP1 from EGF-activated cells resulted in increased translation of LIP from the FL C/EBPβ construct. In comparison, addition of CUG-BP1 immunoprecipitated from non-EGF-stimulated cells or cells treated with the EGFR antagonist AG1478 reduced the translation of LIP to low or nondetectable levels (Fig. 9A, lanes 1 to 3 [starting from the left]). Similar results were observed with OSI-774 (data not shown). Expression of the LAP isoforms was unaffected by EGFR signaling. Both murine and human FL C/EBPβ cDNA constructs were tested in this in vitro system, and LIP was translated from either construct in the presence of EGF-activated human CUG-BP1. As expected, LIP was not translated from the mutAUG construct containing the mutated LIP translation start site (Fig. 9A, lanes 4 and 5). These data demonstrate that the LIP translational initiation AUG codon is necessary for the production of LIP in EGF-stimulated MCF10A cells. As a positive control, we activated a recombinant CUG-BP1 with extracts from HeLa cells (which contain the kinases necessary to phosphorylate and activate CUG-BP1), immunoprecipitated the protein, and added it to the TnT reaction mixture. Our results demonstrated that recombinant, HeLa cell-activated CUG-BP1 or mammary epithelial cell EGF-activated CUG-BP1 (Fig. 9A, lane 6) can increase the translation of LIP when added to a TnT reaction mixture.
The phosphorylation status of CUG-BP1 is important for the regulation of LIP translation in vitro. Figure 9B is a representative Western blot-based TnT reaction that demonstrates that treatment of CUG-BP1 with CIP results in a dramatic reduction in the translation of the LIP isoform (Fig. 9B, left side, lane 3). This decrease was similar to the reduction in LIP expression observed after addition of CUG-BP1 immunoprecipitated from EGF-stimulated cells treated with AG1478 (Fig. 9B, left side, lane 2). These data suggest that the RNA binding activity and ability of CUG-BP1 to regulate translation of LIP is regulated by EGFR activity and phosphorylation.
There have been reports that the LIP isoform is a product of proteolytic cleavage of the LAP isoforms (17, 39). Consequently, to address the possibility that LIP is generated by proteolysis rather than translation in MCF10A cells, we used the transcription-coupled translation system and a C/EBPβ construct with a mutated LIP AUG codon (mutAUG) (as described in Fig. 9A). Mutation of the LIP ATG codon does not affect the proteolytic cleavage site, and cleaved LIP can still be generated from the mutAUG construct (39). As discussed above, LIP was not translated in vitro from this construct (Fig. 9A, lanes 4 and 5); however, the appearance of LIP would have suggested that LIP could be generated via nontranslational mechanisms such as proteolytic cleavage. Further evidence that proteolytic cleavage is not responsible for LIP expression in MCF10A cells was demonstrated by in vitro cleavage assays. Immunoprecipitated C/EBPβ was incubated with RL or protein extracts from treated MCF10A cells for 1 h at 37°C. SDS-PAGE analysis showed no change in the LAP/LIP ratio under the various experimental conditions (data not shown). Taken together, these results suggest that LIP expression is translationally regulated in MCF10A cells and that LIP is not the product of a proteolytic cleavage event.
DISCUSSION
Although EGFR is a target of small-molecule inhibitors in cancer treatment therapies, much is unknown about the downstream molecular members of this signaling pathway in breast cancer. We have identified two novel downstream targets of EGFR signaling in mammary epithelial cells: the transcription factor C/EBPβ-LIP and the RNA binding protein CUG-BP1. Our study has demonstrated in breast epithelial cells that activation of the EGFR signaling cascade results in an increase in the expression levels of the LIP isoform. Our data show that C/EBPβ mRNA levels are unaffected by EGF stimulation and suggest that the increase in LIP expression is controlled posttranscriptionally and is due in part to the interaction of CUG-BP1 with C/EBPβ mRNA. The lack of C/EBPβ transcriptional regulation observed in our study is consistent with other gene expression studies that have not found changes in C/EBPβ mRNA in response to ErbB receptor activation or breast cancer (1, 37). We have also determined that regulation of the EGFR pathway leads to changes in the phosphorylation status and activity of CUG-BP1, which might be mediated in part by p44/42 MAPK. Our data suggest that phosphorylation of CUG-BP1 is critical for its activity. Dephosphorylation of CUG-BP1 abolishes the binding to C/EBPβ mRNA and results in decreased translation of LIP.
Translational control of mRNA is a crucial step in gene expression that permits cells to quickly regulate levels of specific proteins during cell fate decisions. EGF stimulation often results in small changes in global protein synthesis, but specific mRNAs can show a dramatic increase in translation. Although many of these mRNAs remain to be identified, they usually contain specific cis-regulatory elements such as uAUGs and associated uORFs that facilitate translational control (19). While these cis elements are found in a small percentage of all vertebrate mRNAs, they are conspicuous in two-thirds of oncogenes and growth-controlling genes (such as that which encodes C/EBPβ), reflecting the importance of translational control in cell growth and cancer (19).
Translational control of LIP expression.
C/EBPβ is translated into several distinct protein isoforms (LAP1, LAP2, and LIP) whose expression is regulated by the alternative use of several in-frame translation start sites. Leaky ribosome scanning was first proposed, more than a decade ago, as a mechanism to account for the differential translational initiation from the multiple C/EBPβ AUG start codons (6). The 5′ end of the C/EBPβ mRNA contains a 5′ untranslated region of 298 bases with a GC content of 73% and thus has the potential to form complex, stable secondary structures that can interfere with scanning ribosomes (13, 24). In addition, the 5′ end of the C/EBPβ mRNA contains a uAUG and an sORF that is situated between the translation initiation sites for LAP1 and LAP2 (Fig. 1). This region has been shown to be important for translational control of the C/EBPβ isoforms (17, 40), and LIP expression appears to be strictly regulated by the sORF (5). LAP1 has been shown to be translated by initiation of the ribosomes at the LAP1 AUG codon, whereas LAP2 is produced by leaky ribosome scanning through the sORF AUG, followed by initiation at the LAP2 AUG site (40, 5). Initiation at the uAUG and translation of the sORF may prevent reinitiation at the LAP2 AUG owing to the close proximity of the sORF uAUG to the LAP2 AUG. However, the possibility cannot be discounted that immediate reinitiation after translation of the sORF may occur, and this has also been proposed as a potential mechanism (5). LIP has been shown to be translated predominantly by leaky ribosome scanning over LAP1, initiation of the uORF AUG, and reinitiation at the LIP AUG (5, 40).
The mechanism by which CUG-BP1 regulates translation of the LIP isoform is not well understood. In general, RNA binding proteins have been reported to regulate translation by modulation of ribosomal subunit entry and migration and/or stabilization of secondary structure that favors translational initiation at upstream codons such as the uAUG (9). Consequently, increased translation of the sORF uAUG may thus lead to increased scanning over the LAP2 AUG and increased initiation on the LIP AUG.
In addition to CUG-BP1, other RNA binding proteins may be involved in the translational regulation of C/EBPβ. For example, calreticulin has also been shown to interact with GCN repeats of the C/EBPβ mRNA. However, instead of facilitating translation, calreticulin inhibits translation of the protein isoforms (32). These data support the relevance and importance of RNA binding proteins as translational regulators of C/EBPβ; however, the translational regulation of C/EBPβ is most likely complex. In addition to the RNA binding proteins discussed above, the translation initiation factors eIF-2α and eIF-4E have also been shown to play a role in uORF-mediated translation of the LIP isoform (5). Taken together, this information suggests that the uORF and surrounding 5′ region of C/EBPβ is a critical regulatory point for CUG-BP1, as well as several other translation factors (e.g., eIF-2α, eIF-4E, and 4E-BP1) that may play a role in the translational initiation of the C/EBPβ mRNA.
Regulation of CUG-BP1 activity.
Our study shows that CUG-BP1 is expressed in mammary epithelial cells and is phosphorylated by EGFR signaling. Although an analysis of the protein sequence for CUG-BP1 predicts the presence of multiple phosphorylated residues, little is known about the identity of the phosphorylated residues or the kinases responsible for phosphorylating CUG-BP1. Previous studies, however, have demonstrated that myotonin protein kinase can phosphorylate CUG-BP1 in some tissues (26). The concentration of CUG-BP1, as examined in whole-cell extracts from mammary epithelial cells, did not appear to be altered in response to increased EGFR signaling. This is in agreement with a previous study that showed that CUG-BP1 expression in the liver did not change in response to partial hepatectomy (36). However, we cannot exclude the possibility that the intracellular distribution of CUG-BP1 is altered in response to changes in its phosphorylation status. For example, it has previously been shown in cultured cells from patients with muscular dystrophy that the hypophosphorylated isoform of CUG-BP1 translocates and accumulates in the nucleus (26).
EGFR signaling, LIP, and cell growth.
Our study is the first to correlate EGFR signaling with activation of CUG-BP1 and increased expression of LIP. Our data also support the hypothesis that these proteins may cooperate in a signaling pathway to initiate growth, differentiation, and tumorigenesis in the mammary gland. Mammary epithelial cells are highly dependent on EGF for growth, and the EGFR signaling pathway plays an important role in the developing mammary gland, as well as in breast cancer. Similarly, C/EBPβ is necessary for proper mammary development and overexpression can lead to proliferation and mammary tumorigenesis. It is tempting to speculate that the EGFR may exert some of its mitogenic effects through the translational regulation of C/EBPβ expression and reduction of the LAP/LIP ratio. An increase in the expression of the dominant-negative isoform, LIP, might then alter cell fate by preventing the transcription of genes that control differentiation. As suggested in a recent study of Ras/Akt activation (22), signaling pathways associated with growth may first activate translational programs by altering polysome loading, which then results in the increased translation and expression of transcription factors that are necessary for the later transcriptional phase. Several members of the C/EBP family were among the transcription factors that they found were actively recruited to ribosomes upon Ras/Akt activation (22). Likewise, previous studies have demonstrated that both CUG-BP1 and LIP are present in the polysomal fraction isolated from rat livers 3 h after partial hepatectomy and the binding activity of CUG-BP1 to C/EBPβ mRNA is increased when CUG-BP1 is associated with these polysomes (36).
The transcription factor C/EBPβ thereby fits this paradigm because it is translationally regulated by EGFR signaling and has important roles in growth, differentiation, and tumorigenesis in the mammary gland, as well as in other tissues. Taken together, these data lead us to suggest a model in which EGFR stimulation activates the Ras and p44/42 MAPK pathways, leading to the phosphorylation and activation of CUG-BP1, recruitment to the polysome fraction, and consequent translation of the LIP isoform. CUG-BP1 may thus play a crucial role in the translational control of proteins involved in the determination of cell fate in the mammary gland. It will be important to determine how phosphorylation regulates CUG-BP1 binding activity and to identify additional downstream targets of CUG-BP1 translational activity.
Acknowledgments
We thank Mark Ewen, Barry Nelkin, Jeffrey Rosen, Nancy Shaper, and Joel Shaper for helpful discussions or critical review of the manuscript; Eric Sandgren for providing the WAP-TGF-α mice; and Honglin Chen and Jian Huang for providing the flagged C/EBPβ constructs and cell extracts. We acknowledge OSI Pharmaceuticals, Melville, N.Y., for providing erlotinib (OSI-774) for this study.
This work was supported by grants from the Department of Defense Breast Cancer Program (DAMD17-01-1-0287) and the Breast Spore at Johns Hopkins (P50 CA88843). C.A.Z. is a Barbara B. Rubenstein Scholar.
REFERENCES
- 1.Alaoui-Jamali, M. A., D. J. Song, N. Benlimame, L. Yen, X. Deng, M. Hernandez-Perez, and T. Wang. 2003. Regulation of multiple tumor microenvironment markers by overexpression of single or paired combinations of ErbB receptors. Cancer Res. 63:3764-3774. [PubMed] [Google Scholar]
- 2.Badley, J. E., G. A. Bishop, T. St John, and J. A. Frelinger. 1988. A simple, rapid method for the purification of poly A+ RNA. BioTechniques 6:114-116. [PubMed] [Google Scholar]
- 3.Baer, M., and P. F. Johnson. 2000. Generation of truncated C/EBPβ isoforms by in vitro proteolysis. J. Biol. Chem. 275:26582-26590. [DOI] [PubMed] [Google Scholar]
- 4.Bundy, L. M., and L. Sealy. 2003. CCAAT/enhancer binding protein beta (C/EBPβ)-2 transforms normal mammary epithelial cells and induces epithelial to mesenchymal transition in culture. Oncogene 22:869-883. [DOI] [PubMed] [Google Scholar]
- 5.Calkhoven, C. F., C. Muller, and A. Leutz. 2000. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 14:1920-1932. [PMC free article] [PubMed] [Google Scholar]
- 6.Descombes, P., and U. Schibler. 1991. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67:569-579. [DOI] [PubMed] [Google Scholar]
- 7.Dux, A., and O. Muhlbock. 1969. Mouse mammary carcinoma induced by pituitary isografts in mammary fat pads. Proc. Soc. Exp. Biol. Med. 130:355-359. [DOI] [PubMed] [Google Scholar]
- 8.Ethier, S. P., R. Moorthy, and C. A. Dilts. 1991. Secretion of an epidermal growth factor-like growth factor by epidermal growth factor-independent rat mammary carcinoma cells. Cell Growth Differ. 2:593-602. [PubMed] [Google Scholar]
- 9.Gray, N. K., and M. Wickens. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14:399-458. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, W., and S. Chen-Kiang. 1993. Convergent regulation of NF-IL6 and Oct-1 synthesis by interleukin-6 and retinoic acid signaling in embryonal carcinoma cells. Mol. Cell. Biol. 13:2515-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 12.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak, M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai, C. C., T. H. Chiu, H. C. Rosenberg, and W. H. Huang. 1993. Improved proteinase K digestion for the rapid isolation of mRNA from mammalian tissues. BioTechniques 15:620-626. [PubMed] [Google Scholar]
- 15.Lamb, J., S. Ramaswamy, H. L. Ford, B. Contreras, R. V. Martinez, F. S. Kittrell, C. A. Zahnow, N. Patterson, T. R. Golub, and M. E. Ewen. 2003. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell 114:323-334. [DOI] [PubMed] [Google Scholar]
- 16.Li, S., and J. M. Rosen. 1994. Glucocorticoid regulation of rat whey acidic protein gene expression involves hormone-induced alterations of chromatin structure in the distal promoter region. Mol. Endocrinol. 8:1328-1335. [DOI] [PubMed] [Google Scholar]
- 17.Lincoln, A. J., Y. Monczak, S. C. Williams, and P. F. Johnson. 1998. Inhibition of CCAAT/enhancer-binding protein alpha and beta translation by upstream open reading frames. J. Biol. Chem. 273:9552-9560. [DOI] [PubMed] [Google Scholar]
- 18.Milde-Langosch, K., T. Loning, and A. M. Bamberger. 2003. Expression of the CCAAT/enhancer-binding proteins C/EBPα, C/EBPβ and C/EBPδ in breast cancer: correlations with clinicopathologic parameters and cell-cycle regulatory proteins. Breast Cancer Res. Treat. 79:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Morris, D. R., and A. P. Geballe. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20:8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moyer, J. D., E. G. Barbacci, K. K. Iwata, L. Arnold, B. Boman, A. Cunningham, C. DiOrio, J. Doty, M. J. Morin, M. P. Moyer, M. Neveu, V. A. Pollack, L. R. Pustilnik, M. M. Reynolds, D. Sloan, A. Theleman, and P. Miller. 1997. Induction of apoptosis and cell cycle arrest by CP-358,774, an inhibitor of epidermal growth factor receptor tyrosine kinase. Cancer Res. 57:4838-4848. [PubMed] [Google Scholar]
- 21.Nakajima, T., S. Kinoshita, T. Sasagawa, K. Sasaki, M. Naruto, T. Kishimoto, and S. Akira. 1993. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 90:2207-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajasekhar, V. K., A. Viale, N. D. Socci, M. Wiedmann, X. Hu, and E. C. Holland. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12:889-901. [DOI] [PubMed] [Google Scholar]
- 23.Rask, K., M. Thorn, F. Ponten, W. Kraaz, K. Sundfeldt, L. Hedin, and S. Enerback. 2000. Increased expression of the transcription factors CCAAT-enhancer binding protein-beta (C/EBβ) and C/EBζ (CHOP) correlate with invasiveness of human colorectal cancer. Int. J. Cancer 86:337-343. [DOI] [PubMed] [Google Scholar]
- 24.Raught, B., A. C. Gingras, A. James, D. Medina, N. Sonenberg, and J. M. Rosen. 1996. Expression of a translationally regulated, dominant-negative CCAAT/enhancer-binding protein beta isoform and up-regulation of the eukaryotic translation initiation factor 2α are correlated with neoplastic transformation of mammary epithelial cells. Cancer Res. 56:4382-4386. [PubMed] [Google Scholar]
- 25.Raught, B., W. S. Liao, and J. M. Rosen. 1995. Developmentally and hormonally regulated CCAAT/enhancer-binding protein isoforms influence beta-casein gene expression. Mol. Endocrinol. 9:1223-1232. [DOI] [PubMed] [Google Scholar]
- 26.Roberts, R., N. A. Timchenko, J. W. Miller, S. Reddy, C. T. Caskey, M. S. Swanson, and L. T. Timchenko. 1997. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc. Natl. Acad. Sci. USA 94:13221-13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose-Hellekant, T. A., K. Gilchrist, and E. P. Sandgren. 2002. Strain background alters mammary gland lesion phenotype in transforming growth factor-alpha transgenic mice. Am. J. Pathol. 161:1439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandgren, E. P., J. A. Schroeder, T. H. Qui, R. D. Palmiter, R. L. Brinster, and D. C. Lee. 1995. Inhibition of mammary gland involution is associated with transforming growth factor alpha but not c-myc-induced tumorigenesis in transgenic mice. Cancer Res. 55:3915-3927. [PubMed] [Google Scholar]
- 29.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 30.Soule, H. D., T. M. Maloney, S. R. Wolman, W. D. Peterson, Jr., R. Brenz, C. M. McGrath, J. Russo, R. J. Pauley, R. F. Jones, and S. C. Brooks. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075-6086. [PubMed] [Google Scholar]
- 31.Sundfeldt, K., K. Ivarsson, M. Carlsson, S. Enerback, P. O. Janson, M. Brannstrom, and L. Hedin. 1999. The expression of CCAAT/enhancer binding protein (C/EBP) in the human ovary in vivo: specific increase in C/EBPβ during epithelial tumour progression. Br. J. Cancer 79:1240-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timchenko, L. T., P. Iakova, A. L. Welm, Z. J. Cai, and N. A. Timchenko. 2002. Calreticulin interacts with C/EBPα and C/EBPβ mRNAs and represses translation of C/EBP proteins. Mol. Cell. Biol. 22:7242-7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timchenko, L. T., J. W. Miller, N. A. Timchenko, D. R. DeVore, K. V. Datar, L. Lin, R. Roberts, C. T. Caskey, and M. S. Swanson. 1996. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timchenko, L. T., N. A. Timchenko, C. T. Caskey, and R. Roberts. 1996. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum. Mol. Genet. 5:115-121. [DOI] [PubMed] [Google Scholar]
- 35.Timchenko, N. A., Z. J. Cai, A. L. Welm, S. Reddy, T. Ashizawa, and L. T. Timchenko. 2001. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 276:7820-7826. [DOI] [PubMed] [Google Scholar]
- 36.Timchenko, N. A., A. L. Welm, X. Lu, and L. T. Timchenko. 1999. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucleic Acids Res. 27:4517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van't Veer, L. J., H. Dai, M. J. van de Vijver, Y. D. He, A. A. Hart, M. Mao, H. L. Peterse, K. van der Kooy, M. J. Marton, A. T. Witteveen, G. J. Schreiber, R. M. Kerkhoven, C. Roberts, P. S. Linsley, R. Bernards, and S. H. Friend. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530-536. [DOI] [PubMed] [Google Scholar]
- 38.Welm, A. L., S. L. Mackey, L. T. Timchenko, G. J. Darlington, and N. A. Timchenko. 2000. Translational induction of liver-enriched transcriptional inhibitory protein during acute phase response leads to repression of CCAAT/enhancer binding protein alpha mRNA. J. Biol. Chem. 275:27406-27413. [DOI] [PubMed] [Google Scholar]
- 39.Welm, A. L., N. A. Timchenko, and G. J. Darlington. 1999. C/EBPα regulates generation of C/EBPβ isoforms through activation of specific proteolytic cleavage. Mol. Cell. Biol. 19:1695-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong, W., C. C. Hsieh, A. J. Kurtz, J. P. Rabek, and J. Papaconstantinou. 2001. Regulation of CCAAT/enhancer-binding protein-beta isoform synthesis by alternative translational initiation at multiple AUG start sites. Nucleic Acids Res. 29:3087-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahnow, C. A., R. D. Cardiff, R. Laucirica, D. Medina, and J. M. Rosen. 2001. A role for CCAAT/enhancer binding protein beta-liver-enriched inhibitory protein in mammary epithelial cell proliferation. Cancer Res. 61:261-269. [PubMed] [Google Scholar]
- 42.Zahnow, C. A., P. Younes, R. Laucirica, and J. M. Rosen. 1997. Overexpression of C/EBPβ-LIP, a naturally occurring, dominant-negative transcription factor, in human breast cancer. J. Natl. Cancer Inst. 89:1887-1891. [DOI] [PubMed] [Google Scholar]