Abstract
The conserved protein kinase Chk1 mediates cell cycle progression and consequently the ability of cells to survive when exposed to DNA damaging agents. Cells deficient in Chk1 are hypersensitive to such agents and enter mitosis in the presence of damaged DNA, whereas checkpoint-proficient cells delay mitotic entry to permit time for DNA repair. In a search for proteins that can improve the survival of Chk1-deficient cells exposed to DNA damage, we identified fission yeast Msc1, which is homologous to a mammalian protein that binds to the tumor suppressor Rb (RBP2). Msc1 and RBP2 each possess three PHD fingers, domains commonly found in proteins that influence the structure of chromatin. Msc1 is chromatin associated and coprecipitates a histone deacetylase activity, a property that requires the PHD fingers. Cells lacking Msc1 have a dramatically altered histone acetylation pattern, exhibit a 20-fold increase in global acetylation of histone H3 tails, and are readily killed by trichostatin A, an inhibitor of histone deacetylases. We postulate that Msc1 plays an important role in regulating chromatin structure and that this function modulates the cellular response to DNA damage.
The activation of checkpoint pathways in response to various signals is important for maintaining genome integrity during cell cycle progression. Eukaryotic cells (including those of the fission yeast Schizosaccharomyces pombe) respond to DNA damage by undergoing a transient arrest of the cell cycle. Arrest is dependent on the DNA damage checkpoint pathway, a signal transduction pathway that couples damage detection to cell cycle control (56). In the absence of such a delay, checkpoint-defective cells enter mitosis with damaged DNA and die (2, 75). Inhibiting the checkpoint pathway can cause uncontrolled cell proliferation, which in multicellular eukaryotes can contribute to the onset of cancer (74). Indeed, mutation of the human checkpoint pathway gene ATM leads to the genetic disorder ataxia telangiectasia, which is characterized by a high incidence of neurodegeneration and cancer (66).
Transitions in the cell cycle are regulated by a family of protein complexes called cyclin-dependent kinases, consisting of a cyclin regulatory subunit and a kinase catalytic subunit or cdk (54). Fission yeast Cdc2, the primary cdk in S. pombe, is phosphorylated on several residues in a cell cycle-dependent manner (23). Entry into mitosis in inhibited by phosphorylation of a tyrosine residue at position 15 of Cdc2 and is carried out by kinases in the Wee1 family (45). Dephosphorylation of that same site by the phosphatase Cdc25 activates Cdc2 and promotes entry into mitosis (51). Chk1 inhibits mitotic entry by phosphorylating Wee1 and Cdc25 (59, 60). In response to DNA damage, Chk1 itself is phosphorylated at serine 345 in fission yeast and at the analogous site in Xenopus, mouse, and human cells (10, 29, 41, 42). The phosphorylation of Chk1 is dependent on the protein kinase Rad3 (76) as well as on other established components of the checkpoint pathway, all of which are conserved in mammalian cells (50).
DNA in eukaryotic cells is packaged into chromatin. The main packaging component of chromatin is the nucleosome, which is composed of an octamer of histone proteins (67). Histones are subject to a complex and dynamic set of covalent modifications that are thought to be involved in the modulation of transcription during development, genome stability, and meiotic chromosome dynamics (68). Histone modifications reported to date include acetylation, phosphorylation, methylation, ADP ribosylation, and ubiquitination (5, 25, 32). Multiple residues on each of the four core histones have been identified as potential modification sites. According to the histone code hypothesis, these modifications may be interdependent and provide entry sites for proteins responsible for higher-order chromatin organization and gene activation or inactivation (33).
Acetylation of histones is mediated by two activities, those of histone acetyltransferases (HAT) and histone deacetylases (HDAC). The acetylation of histone within a particular nucleosome can modify chromatin structure locally, leading to the repression or expression of neighboring genes (33). A conserved domain of many transcription activators binds to specifically acetylated lysine residues of histone tails (18, 31). Inactivation of the Rpd3 HDAC complex in yeast leads to hyperacetylation of many genes (39) and disrupts cell cycle-regulated histone acetylation at the HO locus, leading to the suggestion that Rpd3 might act globally to remove acetyl groups from newly replicated chromatin (72).
In mammalian cells the retinoblastoma (Rb) tumor suppressor protein can recruit HDAC to chromatin (8, 46, 47), which represses the transcription of many genes involved in cell cycle regulation that contain sites for the E2F transcription factor in their promoters (19). In tumor cell lines lacking Rb, the failure to recruit HDAC to some E2F-controlled genes results in inappropriate expression (8, 46). In addition to having a role in the control of gene expression, HDAC have also been implicated in DNA replication via association with DNA polymerase ɛ (73) and PCNA (52).
The mammalian CBP/p300 and Gcn5 HAT have been reported to associate with DDB1, a protein involved in recognizing damaged DNA (16, 49). The CBP/p300 acetylase is also found in complex with thymine DNA glycosylase, an enzyme required for base mismatch repair (71). Some recent reports indicate that checkpoints and chromatin may also be intimately connected, as several checkpoint proteins have been found to associate with various chromatin activities. HDAC have been found in complexes or associated directly with ATM (36), ATR (62), the human Hus1 and Rad9 proteins (9), and the breast cancer susceptibility gene product BRCA1 that has been implicated in checkpoint and DNA repair functions in mammalian cells (79).
In this study we isolated msc1 as a multicopy suppressor of a defect in the DNA damage checkpoint pathway in fission yeast. Msc1 shows high-level similarity to a mammalian protein, Rb binding protein 2 (RBP2). RBP2 was identified in a two-hybrid screening for proteins that bind to the tumor suppressor Rb (17, 21). Related sequences were identified subsequently due to homology to RBP2 and were termed RBP2H1 and RBP2H1A. Close analysis of these two cDNA sequences deposited in GenBank suggests that they represent the same gene. The same sequence was also cloned as a gene that is up-regulated in breast cancer cells and named PLU-1 (43). Msc1, RBP2, and PLU-1 each contain jumonji N and jumonji C domains (12) as well as three PHD finger motifs (each being a 50- to 60-amino-acid Zn finger characterized by seven cysteines and a histidine residue that are arranged as Cys4HisCys3) separated by intervening sequences of various lengths and amino acid compositions (1). The PHD motif was originally found in plant homeodomain transcription factors: thus the name PHD (for “plant homeodomain”). Subsequently, the motif has been found in a number of proteins thought to influence chromatin structure either directly (34, 65) or via association with HDAC (64). The results obtained in our studies suggest that Msc1 (encoded by a nonessential gene) associates with chromatin and plays a role in chromatin modification through association with an HDAC.
MATERIALS AND METHODS
Strains and growth conditions.
Standard genetic methods and growth conditions were utilized (53). Cells were grown at 30°C unless otherwise indicated. Survival following UV treatment was determined as described previously (75). For drug sensitivity experiments, cells were grown to mid-log phase and 10-fold serial dilutions were made from 107 cells/ml. Aliquots of 5 μl for each dilution were spotted on agar plates. For trichostatin A (TSA; Sigma) sensitivity assays, serially diluted cells were spotted either on YEA (0.5% yeast extract, 3% glucose, 20 μg of adenine per ml) plates (−TSA) or on YEA plates containing 25 μg of TSA/ml (+TSA). Plates were incubated at 30°C and examined after 3 days. To facilitate detection of Msc1, the method of PCR-based tagging described by Bahler et al. (3) was used to integrate a triple hemagglutinin (HA) tag at the C terminus of the msc1 gene in the chromosome. The msc1:HA-tagged strain was tested for TSA sensitivity. While an msc1 deletion strain is sensitive to TSA and is unable to form colonies on TSA-containing plates, the msc1:HA strain behaves exactly like the wild type (data not shown).
To assess checkpoint proficiency, cells synchronized in G2 were exposed to 100 J of UV light/m2 and the percentage of cells passing through mitosis was determined microscopically using 4′,6′diamidino-2-phenylindole (DAPI)-stained cells as described previously (77). For assessing phosphorylation of Chk1 in the DNA ligase-deficient cdc17-K42 mutant background, chk1::HA cdc17-K42 strains containing either empty vector plasmid or msc1 plasmid were grown to mid-log phase at 25°C in minimal medium and then shifted to 32°C for 6 h to reduce DNA ligase activity. Lysates were prepared, and Western blotting was performed using 12CA5 antibody as described previously (76).
Immunofluorescence studies.
Immunofluorescence studies were performed using exponentially growing cells essentially as described by Hagan and Hyams (30). HA-Msc1 was detected using HA (F-7) antibody (Sc-7392) (Santa Cruz Biotechnology) at a 1:30 dilution, incubated overnight at room temperature with rotation, washed, and then detected with secondary antibody coupled to CY3 at a dilution of 1:100 and incubated at room temperature for 4 h. Cells were washed and suspended in 10 μl of Vecta Shield (Vector Laboratory). The cell suspension (1 μl) was analyzed using a fluorescence microscope (Zeiss Axioplan 2). Images were captured with a Zeiss AxioCam and analyzed with Openlab software. For DAPI staining, 1 μl of cells was mixed with 0.1 μl of 100 μg of DAPI solution/ml.
Chromatin fractionation assay.
The chromatin fractionation assay was performed using log-phase cells (107 cells/ml) as described by others (28). Msc1 was detected with anti-HA antibody (12CA5). Antibody to histone H3 (catalog no. 05-499; Upstate Biotech) was used as a marker for soluble chromatin protein, while antibody to Ded1 was used as a cytosolic marker (40).
Histone purification.
Histones were purified as described by Edmondson and Roth (20) from a 500-ml yeast culture grown to late log phase (1.5 × 107 cells/ml). Acetylation of histones was determined by running 25 μg of histones on a 15% polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane and then immunoblotted with anti-diacetylated (K9 and K14) histone H3 (catalog no. 06-599; Upstate Biotech), anti-tetra-acetylated (K5, K8, K12, and K16) histone H4 (catalog no. 06-598; Upstate Biotech), or anti-histone H3 (catalog no. 05-499; Upstate Biotech).
Chromatin immunoprecipitation of Msc1.
To assess association of HDAC activity with Msc1, a chromatin immunoprecipitation protocol (38) was utilized to isolate chromatin-associated Msc1. Briefly, HA-tagged Msc1 cells were cross-linked with 1% formaldehyde at room temperature for 15 min, washed, and then lysed with glass beads. The lysate was transferred to several separate tubes and sonicated six times at 30 to 40% output (90% duty cycle; 5 s). The lysate was centrifuged at 8,160 × g in a microcentrifuge for 5 min, transferred to another tube, and again centrifuged at 8,160 × g in a microcentrifuge for 15 min. The lysate was collected, and protein levels were estimated by the method of Bradford (Bio-Rad). A total of 35 μl of HA (F-7) antibody (Sc-7392) (Santa Cruz Biotech) was added to 4 mg of protein lysate and rocked overnight at 4°C. Recombinant protein A-Sepharose beads (50 μl) were added to capture the immune complexes and rocked for another 1 to 2 h at 4°C. Immunoprecipitation products were collected by centrifugation, washed three times with lysis buffer, and then equilibrated in HDAC assay buffer.
HDAC activity was measured using an HDAC assay kit from Upstate Biotech (catalog no. 17-320). In summary, biotinylated histone H4 peptide was labeled with [3H]acetyl coenzyme A and collected on streptavidin agarose beads as described by the manufacturer. A total of 3 μl of labeled histone H4 (∼10,000 cpm) was added to 1× HDAC buffer containing immunoprecipitated protein (HA-tagged Msc1) bound on recombinant protein A-Sepharose beads in a total volume of 200 μl. The reaction was incubated on a rotating wheel at room temperature for 24 h. The mix was centrifuged at 16,000 rpm in a microcentrifuge for 4 min, and released counts were assayed by transferring 100 μl of supernatant to a scintillation vial containing scintillation fluid, mixed thoroughly, and counted.
Construction of msc1 deletions.
To express the msc1 gene ectopically, an HA epitope tag was engineered in frame at the 3′ end of the gene immediately upstream of the stop codon. The full-length gene was expressed from its own promoter in the pSP1 plasmid (15). Deletion constructs were made either by restriction digestion followed by religation to generate in-frame deletions or by PCR amplification of fragments that were subsequently ligated into the pSP1/msc1HA plasmid.
Northern analysis.
RNA was isolated (as described by others) (63) from wild-type cells or cells lacking msc1 which had been grown to mid-log phase. DNA sequences of clr3 (6), hda1 (57), clr6 (27), ded1 (40), mst2 (Sanger Centre accession number SPAC17G8.13c), and esa1 (Sanger Centre accession number SPAC637.12c) were amplified by PCR using genomic DNA of wild-type S. pombe as a template. A total of 50 μg of each PCR product was used to make each probe. RNA separation and detection by Northern blot analysis were carried out according to standard methods.
RESULTS
Genetic search to identify suppressors of the DNA damage checkpoint pathway.
To identify additional components of the DNA damage checkpoint pathway, we set up a genetic screening utilizing chk1-dependent loss of viability at 32°C of a DNA ligase-deficient S. pombe strain. As depicted in Fig. 1A, a DNA ligase mutant strain with a temperature-sensitive DNA ligase allele (cdc17-K42) is not able to form colonies at 36.5°C because of complete loss of DNA ligase function but grows well at the permissive temperature of 25°C (55). At 32°C the strain can form colonies, but cells are elongated because of a checkpoint-dependent cell cycle delay due to partial loss of DNA ligase activity (2). Thus, a checkpoint-defective allele of chk1 (or chk1 deletion) makes the DNA ligase-deficient strain inviable at 32°C (75, 77). To identify other proteins involved in the checkpoint pathway we performed a genetic screening for genes that (when present in multiple copies per cell) allowed a cdc17-K42 chk1− strain to form colonies at the restrictive temperature of 32°C. In this screening we found plasmids encoding Cdc17 and Chk1, as well as several novel genes, including msc1 (for “multicopy suppressor of Chk1”) (SPAC343.11c; DDBJ/EMBL/GenBank accession number NP_593431). Transforming msc1 on a multicopy plasmid allows a strain lacking chk1 (chk1::ura4 cdc17K42) to grow at 32°C (Fig. 1B), suggesting that msc1 bypasses the requirement for chk1 function when DNA ligase activity is limiting.
FIG. 1.
Isolation of Msc1 as a multicopy suppressor of loss of checkpoint function. (A) Screening used to isolate the multicopy suppressors of loss of checkpoint function (see text for details). (B) chk1::ura cdc17-K42 strains transformed with either an empty vector or a plasmid with a genomic copy of msc1 (pmsc1) were grown to mid-log phase in liquid culture. Tenfold serial dilutions were made, and aliquots were spotted on plates. Plates were incubated at 25 or 32°C for 3 days. (C) Analysis of Chk1 phosphorylation in strains with defective DNA ligase activity of cdc17-K42 at 32°C. A cdc17-K42 strain with an integrated HA-tagged chk1 allele was transformed with either empty vector (lanes 1 and 2) or with pMsc1 (lanes 3 and 4). Strains were grown at 25°C to mid-log phase and then shifted to 32°C for 6 h. Protein was extracted by glass bead lysis, separated on an sodium dodecyl sulfate-polyacrylamide gel, transferred to nitrocellulose membrane, and blotted with antibody to the HA tag to detect the unphosphorylated and phosphorylated forms of Chk1. (D) A chk1::ura4 deletion strain was transformed with empty vector or plasmids containing genomic copies of msc1 or chk1. Strains were grown in liquid culture to mid-log phase, and 1,000 cells were plated and exposed to the indicated doses of UV light. All plates were incubated at 30°C for 3 days. The percentages of surviving colonies relative to those seen with unirradiated control plates were determined. Values shown are the averages of three independent experiments.
To allow colony formation at 32°C, the msc1 plasmid might either restore function to the defective DNA ligase mutant or compensate for the loss of chk1 function. Msc1 clearly cannot substitute for DNA ligase, as a cdc17-K42 strain with multicopy msc1 fails to grow at 36°C (data not shown), a temperature at which the cdc17-K42 allele is inactive. To test whether Msc1 partially substitutes for or restores function to Cdc17, we also did an assay that monitors DNA damage by way of Chk1 phosphorylation. At 32°C cdc17-K42 cells delay entry into mitosis and Chk1 becomes phosphorylated (Fig. 1C, lane 2). As shown in Fig. 1C, phosphorylation of Chk1 occurs even when msc1 is present in multiple copies (Fig. 1C, lane 4), indicating that damage generated by limiting DNA ligase activity is still present in these cells. Thus, it is likely that msc1 suppresses chk1 rather than cdc17 function.
To find out whether multicopy expression of msc1 can bypass the need for chk1 when cells are exposed to other types of DNA damage, we transformed Msc1 on a multicopy plasmid into a chk1 deletion strain and assayed for UV sensitivity. The chk1::ura4 deletion strain transformed with an empty vector plasmid is UV sensitive, while a wild-type chk1 plasmid confers UV resistance to the chk1::ura4 strain. Transformation with msc1 plasmid makes a chk1 deletion strain less sensitive to UV light (Fig. 1D), indicating that multicopy expression of msc1 can partially compensate for the complete absence of chk1.
Cells lacking Msc1 are viable and checkpoint proficient.
To investigate the function of Msc1 we deleted the coding region of the msc1 gene with a selectable marker (kanR). We were able to obtain viable integrants from a haploid strain, indicating that msc1 is a nonessential gene. Cells lacking msc1 are mildly UV sensitive but in combination with chk1 deletion show a cumulative effect (Fig. 2A), suggesting that the two proteins function in distinct pathways to promote survival after DNA damage. To investigate the role of msc1 in checkpoint function we determined whether cells lacking msc1 delay mitotic entry. While a chk1 deletion strain enters mitosis in the presence of DNA damage, msc1 deletion and wild-type cells delay mitotic entry (Fig. 2B), suggesting that msc1 is not required for the checkpoint that mediates mitotic delay.
FIG. 2.
Loss of msc1 function compounds the sensitivity to UV light of a chk1-deficient strain. (A) The indicated strains were grown in rich medium to mid-log phase, and 1,000 cells per plate were exposed (or not exposed) to the indicated doses of UV light. Survival after 3 days on plates was determined as described in the legend to Fig. 1D. (B) The UV sensitivity of a strain lacking msc1 is not due to a compromised checkpoint. The indicated strains (each having a cdc25-22 mutant allele) were synchronized in G2 by incubation at 36.5°C, exposed to UV light, and released to permissive temperature to monitor passage through mitosis as described in Materials and Methods.
Msc1 protein contains three PHD fingers and shows high-level similarity to RBP2.
Msc1 contains three PHD fingers, jumonji (jmj) N and C domains, a small Zn finger, and (according to one motif-scanning program) a BRCT domain. Jumonji domains have been found in many transcription factors (4). PHD fingers have a typical structure (C5HC2 or C4HC3) and are found predominantly in proteins associated with chromatin function (1). The BRCT domain originally identified in the tumor suppressor protein BRCA1 has also been found in several proteins involved in DNA damage checkpoint and repair pathways (35, 79). A BLAST search with the Msc1 amino acid sequence identified the human Rb binding protein RBP2 as having 21% identity over a stretch of 900 amino acids. A search using the National Center for Biotechnology Information (NCBI) Conserved Domain Architecture Retrieval Tool (CDART) (http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi?cmd = rps) indicated that the similarity of the domain structures of Msc1 and RBP2 extends over the entire protein (Fig. 3). Apart from its ability to bind Rb in a two-hybrid assay (21, 37), little is known about the function of RBP2. A second protein identified by CDART has been entered into GenBank three times (as PLU-1, RbBP2H1, and RbBP2H1a), but all three entries appear to represent the same protein and to possess similar domain structures. No apparent homologue (with the same arrangement of domains) exists in Saccharomyces cerevisiae.
FIG. 3.
Domain architecture of Msc1. The domain architecture of Msc1 has the same motif arrangement as that of RBP2 (GenBank accession number NP_005047) and PLU-1 (GenBank accession number CAB63108). Msc1 was investigated using the GenBank database and the CDART available at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi?cmd = rps) a.a., amino acids.
Msc1 localizes to the nucleus.
To characterize the Msc1 protein, a triple HA epitope tag was introduced into the genome in frame with the msc1 coding sequence. The HA epitope had no apparent effect on the function of Msc1 (see Materials and Methods). The results of indirect immunofluorescence microscopic studies using antibody against the HA epitope clearly show that Msc1 localizes in the nucleus (Fig. 4A). To determine whether or not Msc1 associates with chromatin, a cell fractionation procedure developed for S. pombe (28) was utilized as depicted in Fig. 4B. As shown in Fig. 4C, Msc1 cofractionates with the chromatin marker histone H4 whereas very little Msc1 is apparent in the cytosolic fraction. Ded1, a putative RNA helicase (40) implicated in translational regulation (11), was used as a marker for the cytosol.
FIG. 4.
Msc1 is a chromatin-associated protein. (A) Nuclear localization of Msc1. Cells with an integrated allele of HA-tagged msc1 were grown to mid-log phase and fixed with glutaraldehyde, and immunofluorescence assays were performed using anti-HA antibody. (B) Schematic representation of chromatin fractionation assay. (C) DNA was isolated (as described in Materials and Methods) from the indicated fractions, run on an agarose gel, and stained with ethidium bromide. (D) Protein samples from the indicated fractions were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Western blot analysis was performed using anti-HA antibody to detect HA-tagged Msc1 (Msc1-HA), anti-Ded1 antibody (Ded1), or antibody to histone H4.
Cells lacking Msc1 are hypersensitive to the HDAC inhibitor TSA.
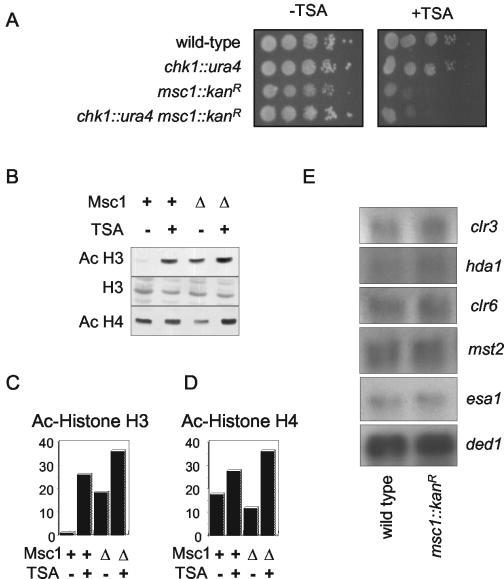
While cells lacking msc1 are mildly sensitive to UV light exposure, the killing caused by UV is not sufficient for genetic screenings to dissect Msc1 function successfully. Therefore, we searched for conditions under which an msc1 mutant could not survive. We tested the sensitivity of the msc1 deletion strain to a variety of drugs: loss of msc1 function had little or no effect on survival upon exposure to the topoisomerase I poison camptothecin, the ribonucleotide reductase inhibitor hydroxyurea, or caffeine (data not shown). Strikingly, msc1::kanR deletion cells were found to be sensitive to TSA, an inhibitor of HDAC (80). As shown in Fig. 5A, wild-type cells and cells lacking Chk1 can tolerate exposure to TSA; however, cells lacking Msc1 function are unable to grow on plates containing the drug.
FIG. 5.
Deletion of msc1 affects cellular sensitivity to TSA and alters the state of histone acetylation in vivo. (A) An msc1::kanR deletion strain is TSA sensitive. Tenfold serial dilutions of the indicated strains were spotted on rich medium (−TSA) or rich medium containing 25 μg of TSA/ml (+TSA) and incubated at 30°C for 3 days. (B) Deletion of Msc1 results in hyperacetylation of histone H3. Histones were isolated from wild-type cells (Msc1 +) or an msc1::kanR deletion strain (Msc1 Δ). Histones (25 μg) were loaded on a 15% polyacrylamide gel and transferred to nylon membrane. Acetylated (Ac) histones were detected using antibody that recognizes histone H3 diacetylated on lysines 9 and 14 (upper panel), total histone H3 (middle panel), or histone H4 tetra-acetylated on lysines 5, 8, 12, and 16 (lower panel). (C and D) The data presented in panel B was quantitated using ImageQuant software normalized with histone H3 values and plotted to convey the relative amounts of acetylation in the different strains. (E) Northern blot analysis of genes affecting histone acetylation. RNA was isolated from wild-type and msc1::kanR cells and probed for the expression level of the indicated genes. The clr3, hda1, and clr6 genes encode HDAC. The mst2 and esa1 genes encode putative HAT, as suggested by sequence similarity to genes in other organisms. The ded1 gene, encoding a DEAD-box helicase involved in translation initiation, was used as a loading control.
Cells lacking Msc1 contain hyperacetylated histone H3.
The fact that cells lacking msc1 are hypersensitive to TSA prompted us to check the level of bulk histone acetylation in those cells. Histones were isolated as described previously (20) from wild-type and msc1 deletion cells grown in the presence or absence of TSA. Histones were probed with antibodies that recognize diacetylated histone H3 or tetra-acetylated histone H4. As shown in Fig. 5B, cells lacking Msc1 exhibit a striking (approximately 20-fold) increase in the signal for diacetylated histone H3 compared to wild-type cells (compare lanes 1 and 3). Exposure of wild-type cells to TSA results in hyperacetylation of histone H3 (Fig. 5B, second lane from left) compared to the results seen with untreated wild-type cells (lane 1). Strikingly, a 20-fold increase in diacetylated histone H3 was observed in cells lacking msc1 even in the absence of TSA (Fig. 5B, lane 3, and 5C). TSA treatment of cells lacking msc1 caused a further increase in acetylation (Fig. 5B, lane 4, and 5C). These results suggest that msc1 is required for deacetylation of histone H3 (either through recruitment or activation of a HDAC or through inhibition of a HAT). The effect of msc1 on histone H4 is much less pronounced (Fig. 5B, bottom panel, and 5D).
Given the presence of domains within Msc1 that are found in transcriptional regulators, we considered the possibility that the alteration in histone H3 acetylation might be due to changes in the expression of genes that mediate histone acetylation. Therefore, we generated probes to several known HDAC, clr3 (6), hda1 (57), and clr6 (27), and to mst2 (SPAC17G8.13c) and esa1 (SPAC637.12c), two genes thought to encode HAT by virtue of their homology to such genes in other organisms. RNA was prepared from wild-type cells and cells lacking msc1 and probed by Northern blotting. As shown in Fig. 5E, no change in the level of mRNA for any of these genes was detected.
Msc1 coprecipitates HDAC activity.
Msc1 lacks homology to any known histone-modifying enzymes. Thus, it is possible that the influence of Msc1 on histone acetylation is due to an associated histone-modifying enzyme. We tested whether or not Msc1 associates with a HDAC activity by immunoprecipitating Msc1 and assaying for coprecipitating HDAC activity in the immunoprecipitate. Labeled histone peptides were incubated with either immunoprecipitated Msc1 or a mock immunoprecipitation from a strain lacking the HA tag on Msc1. A threefold increase in released tritiated acetyl coenzyme A levels was observed in the Msc1 coimmunoprecipitated complex compared to the results seen with the mock immunoprecipitated sample (Fig. 6A). The activity was inhibited by sodium butyrate, a known inhibitor of HDAC activity (Fig. 6A).
FIG. 6.
Msc1 associates with a HDAC. Msc1 coprecipitates a HDAC activity. (A) A strain having HA-tagged Msc1 was grown to mid-log phase, and Msc1 was immunoprecipitated (IP) as described in Materials and Methods. Mock or Msc1 immunoprecipitated samples were incubated with labeled histone H4 peptide for 24 h at room temperature. One set of Msc1 immunoprecipitates was incubated in the presence of sodium butyrate (Sod. Butyrate). Released 3H was counted using a scintillation counter. The values shown represent the averages of three assays, and the error bars represent the standard deviations of the data. (B) An msc1::kanr deletion strain was transformed with the indicated plasmids. Transformants were grown to mid-log phase, and 10-fold serial dilutions of the indicated strains were spotted on EMM-leu medium in the absence (−TSA) or presence (+TSA) of 5 μg of TSA/ml. Plates were incubated at 30°C for 4 days. (C) Strains harboring plasmids expressing deletion constructs of HA-tagged Msc1 were grown to mid-log phase, and Msc1 was immunoprecipitated and assayed for HDAC activity as described for panel A.
PHD fingers are necessary for association with HDAC activity and for suppression of TSA sensitivity.
To begin a structure-function analysis of the Msc1 protein, deletion constructs were generated as described in Materials and Methods to express versions of Msc1 in fission yeast that lack particular domains. These plasmids were transformed to an msc1 deletion strain and tested for TSA sensitivity. As shown in Fig. 6B, the wild-type msc1 plasmid confers resistance to TSA whereas the msc1 deletion strain transformed with the empty vector is sensitive to TSA. Transformation of plasmids lacking one or two PHD domains fails to restore TSA resistance, suggesting that these domains are necessary for the activity that is supplied by wild-type Msc1. The jmjN domain does not appear to be important for this activity, as cells expressing the jmjN deletion are TSA resistant. The deletion constructs were also tested for the ability to coprecipitate HDAC activity. As shown in Fig. 6C, the HA-tagged PHD domain deletion proteins encoded by deletion construct 1 (del 1) and del 2 coprecipitated reduced HDAC activity compared to the full-length protein, suggesting that the PHD fingers are important for association with the HDAC protein. A correlation exists between the ability to restore TSA resistance and the ability to coprecipitate HDAC activity, as the jmjN deletion protein encoded by del 4 is competent to do both (Fig. 6B and C). Each of the deletion constructs expresses an HA epitope-tagged protein of the expected molecular weight.
DISCUSSION
We have identified Msc1, a fission yeast protein with motifs reminiscent of an Rb-binding protein, RBP2 (Fig. 3). Msc1 associates with chromatin and coprecipitates HDAC activity. Deletion of Msc1 causes a striking increase in global acetylation of histone H3 and confers cellular sensitivity to the HDAC inhibitor TSA. Cells lacking Msc1 are mildly sensitive to exposure to UV light, and Msc1 (in multiple copies) suppresses the sensitivity of cells lacking the checkpoint kinase Chk1 to DNA damage.
Several interesting questions are raised by this set of observations. First of all, how might a chromatin-associated protein that clearly affects global histone acetylation patterns influence survival of a checkpoint-defective cell? One possibility is that the ability of multiple copies of Msc1 to improve the survival of cells lacking Chk1 is an indirect consequence of the alteration of chromatin structure. We have demonstrated that Msc1 associates with a HDAC activity. Perhaps when the level of Msc1 is increased, additional HDAC activities are recruited to chromatin. Perhaps alterations in the normal state of histone acetylation alter the sensitivity of chromatin to the effects of DNA damaging agents, possibly limiting the amount of damage done and thereby increasing cell survival. Alternatively, it is possible that multicopy Msc1 brings about changes in chromatin structure that are more favorable for DNA repair, resulting in increased survival of the strain despite the absence of checkpoint function. Finally, altering the copy number of Msc1 may have consequences for gene expression, leading to (for example) the increased expression of DNA repair enzymes. Indeed, a likely Ustilago maydis homologue of Msc1 known as Rum1 (which also displays domain structure similarity to RBP2) has been shown to affect the expression of a number of genes in that organism (58). While cells lacking Msc1 do not exhibit altered expression of genes that mediate histone acetylation (Fig. 5E), further experiments will be needed to evaluate whether Msc1 affects the expression of genes that influence DNA repair.
Msc1 is related to RBP2 and associates with chromatin.
While Msc1 has 21% amino acid identity over 900 amino acids to the human protein RBP2, the domain architecture conservation of Msc1 with the human RBP2 and PLU-1 proteins is particularly striking (Fig. 3). RBP2 was originally identified in a screening for cDNAs encoding proteins capable of interacting with the tumor suppressor protein Rb (17). In a recent study RBP2 was found to be a binding partner for rhombotin-2, a LIM domain protein involved in erythropoiesis and T-cell leukemogenesis (48). RBP2 possesses a motif characterized by the sequence LXCXE, which is typical of Rb binding proteins. Msc1 does not have the LXCXE motif, and S. pombe does not possess any obvious homologue of Rb. RBP2 also binds to the Rb-related protein p107 through the LXCXE motif (37). The interaction between RBP2 and Rb, however, can be accomplished through a distinct motif (37).
Msc1, RBP2, and PLU-1 each contain three PHD fingers and two jumonji (jmj) domains. The jumonji domain was first identified in the jumonji family of transcription factors and subsequently in SMCX, RBP2, and several other proteins (43, 70). Several proteins containing jumonji domains also contain a dead ringer domain and one or more PHD fingers (26). The PHD type of zinc finger, also called leukemia-associated protein finger or trithorax consensus finger (61), is found predominantly in proteins that function at the chromatin level (1).
Although more than 300 (mainly nuclear) proteins containing one or more PHD fingers have been identified, relatively little is known about the function of this domain. Since many PHD finger-containing proteins reside in large multiprotein complexes, these zinc fingers have been proposed to be involved in protein-protein interactions (1). The similarity between PHD fingers and Ring fingers, which possess E3 ubiquitin ligase activity, has prompted tests of PHD domains as E3 ubiquitin ligases. Indeed, several recent studies demonstrated that isolated PHD domains can function in vitro as E3 ligases (7, 13, 14, 44). Thus far, these observations have been made using domains from non-nuclear PHD-containing proteins. If the PHD domains of Msc1 indeed function as E3 ubiquitin ligases, it is tempting to speculate that the target of ubiquitination might be a chromatin-associated protein. This possibility is particularly tantalizing given recent results indicating that ubiquitination of one histone tail is a necessary prerequisite for the methylation of another histone tail (69, 78). The utility of multiple E3 ligase domains in a single protein, such as would be the case for Msc1, might be that of modifying multiple targets simultaneously. Recently, a new function for PHD domains has been suggested from the demonstration that a PHD domain from the chromatin-associated ING2 protein is capable of binding to phosphoinositide, suggesting a possible role for PHD domains as signaling receptors that can regulate nuclear responses (24).
Msc1 is required for global deacetylation of histone H3.
Immunofluorescence studies suggest that Msc1 localizes to the nucleus. Association of Msc1 to chromatin suggests that it is functioning at the level of chromatin and could have a role in transcription regulation through chromatin modifications, as has been shown for other PHD-containing proteins (1). The msc1::kanR strain exhibits sensitivity to the HDAC inhibitor TSA. We isolated histones from wild-type and msc1::kanR cells to evaluate whether Msc1 affects the level of histone acetylation in vivo. As shown in Fig. 5, acetylation of histone H3 in particular is clearly dramatically increased in cells lacking Msc1. This hyperacetylation is further increased upon treatment with TSA. The lethality observed upon incubation of msc1::kanR cells with TSA could result from dramatic changes in chromatin structure due to the compounded effects on acetylation of histone H3 caused by deletion of Msc1 and by TSA treatment. Alternatively, it is possible that simultaneous treatment with TSA of cells lacking Msc1 causes critical changes in gene expression that cannot be tolerated by the cells. The acetylation level of histone H4 (probed with an antibody for tetra-acetylated histone H4) did not reveal dramatic changes in acetylation of this protein in cells lacking Msc1. Nonetheless, treatment of cells with TSA increased the level of acetylated-histone H4 in msc1::kanR cells more than in wild-type cells.
PHD domains have been found in proteins that encode HAT and in proteins that associate with HDAC. Given the fact that Msc1 lacks homology to known histone-modifying enzymes, we theorized that Msc1 might act as a regulator of acetylation rather than as an enzyme that acts directly on histones. Msc1 coprecipitates HDAC activity, and the PHD domains of Msc1 seem to be important for this ability. Furthermore, the deletion mutants with reduced ability to coprecipitate HDAC activity failed to restore resistance to TSA, suggesting that these two properties are linked. There are several known HDAC in S. pombe, including Clr6, Clr3, and Hda1 (27, 57). It is possible that the PHD fingers of Msc1 are required for recruitment of one or more of these enzymes to chromatin. The tumor suppressor Rb has been shown to recruit HDAC to chromatin in mammalian cells, resulting in localized repression of gene expression (22, 81). Thus, whereas Rb itself is not found in yeast it is possible that a protein with which it interacts in mammalian cells and which does have a counterpart in fission yeast might perform a similar function.
Acknowledgments
We thank Marc Gartenberg for helpful discussions and comments on the manuscript.
This work was supported by a grant from the National Institutes of Health (RO1GM53194).
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.al-Khodairy, F., and A. M. Carr. 1992. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 4.Balciunas, D., and H. Ronne. 2000. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem. Sci. 25:274-276. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 6.Bjerling, P., R. A. Silverstein, G. Thon, A. Caudy, S. Grewal, and K. Ekwall. 2002. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 22:2170-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 8.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 9.Cai, R. L., Y. Yan-Neale, M. A. Cueto, H. Xu, and D. Cohen. 2000. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem. 275:27909-27916. [DOI] [PubMed] [Google Scholar]
- 10.Capasso, H., C. Palermo, S. Wan, H. Rao, U. P. John, M. J. O'Connell, and N. C. Walworth. 2002. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J. Cell Sci. 115:4555-4564. [DOI] [PubMed] [Google Scholar]
- 11.Chuang, R. Y., P. L. Weaver, Z. Liu, and T. H. Chang. 1997. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275:1468-1471. [DOI] [PubMed] [Google Scholar]
- 12.Clissold, P. M., and C. P. Ponting. 2001. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem. Sci. 26:7-9. [DOI] [PubMed] [Google Scholar]
- 13.Coscoy, L., and D. Ganem. 2003. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 13:7-12. [DOI] [PubMed] [Google Scholar]
- 14.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cottarel, G., D. Beach, and U. Deuschle. 1993. Two new multi-purpose multicopy Schizosaccharomyces pombe shuttle vectors, pSP1 and pSP2. Curr. Genet. 23:547-548. [DOI] [PubMed] [Google Scholar]
- 16.Datta, A., S. Bagchi, A. Nag, P. Shiyanov, G. R. Adami, T. Yoon, and P. Raychaudhuri. 2001. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone acetyltransferase. Mutat. Res. 486:89-97. [DOI] [PubMed] [Google Scholar]
- 17.Defeo-Jones, D., P. S. Huang, R. E. Jones, K. M. Haskell, G. A. Vuocolo, M. G. Hanobik, H. E. Huber, and A. Oliff. 1991. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature 352:251-254. [DOI] [PubMed] [Google Scholar]
- 18.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 19.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 20.Edmondson, D. G., and S. Y. Roth. 1998. Interactions of transcriptional regulators with histones. Methods 15:355-364. [DOI] [PubMed] [Google Scholar]
- 21.Fattaey, A. R., K. Helin, M. S. Dembski, N. Dyson, E. Harlow, G. A. Vuocolo, M. G. Hanobik, Haskell, K. M., A. Oliff, D. Defeo-Jones, and R. E. Jones. 1993. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene 8:3149-3156. [PubMed] [Google Scholar]
- 22.Ferreira, R., I. Naguibneva, M. Mathieu, S. Ait-Si-Ali, P. Robin, L. L. Pritchard, and A. Harel-Bellan. 2001. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2:794-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gould, K. L., and P. Nurse. 1989. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342:39-45. [DOI] [PubMed] [Google Scholar]
- 24.Gozani, O., P. Karuman, D. R. Jones, D. Ivanov, J. Cha, A. A. Lugovskoy, C. L. Baird, H. Zhu, S. J. Field, S. L. Lessnick, J. Villasenor, B. Mehrotra, J. Chen, V. R. Rao, J. S. Brugge, C. G. Ferguson, B. Payrastre, D. G. Myszka, L. C. Cantley, G. Wagner, N. Divecha, G. D. Prestwich, and J. Yuan. 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114:99-111. [DOI] [PubMed] [Google Scholar]
- 25.Grant, P. A. 2001. A tale of histone modifications. Genome Biol. 2:REVIEWS0003. [Online.] http://genomebiology.com/2001/2/4/reviews/0003. [DOI] [PMC free article] [PubMed]
- 26.Gregory, S. L., R. D. Kortschak, B. Kalionis, and R. Saint. 1996. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 16:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grewal, S. I., M. J. Bonaduce, and A. J. Klar. 1998. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150:562-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths, D., M. Uchiyama, P. Nurse, and T. S. Wang. 2000. A novel mutant allele of the chromatin-bound fission yeast checkpoint protein Rad17 separates the DNA structure checkpoints. J. Cell Sci. 113:1075-1088. [DOI] [PubMed] [Google Scholar]
- 29.Guo, Z., A. Kumagai, S. X. Wang, and W. G. Dunphy. 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagan, I. M., and J. S. Hyams. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:343-357. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 32.Jason, L. J., S. C. Moore, J. D. Lewis, G. Lindsey, and J. Ausio. 2002. Histone ubiquitination: a tagging tail unfolds? Bioessays 24:166-174. [DOI] [PubMed] [Google Scholar]
- 33.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 34.Kalkhoven, E., H. Teunissen, A. Houweling, C. P. Verrijzer, and A. Zantema. 2002. The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Mol. Cell. Biol. 22:1961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerr, P., and A. Ashworth. 2001. New complexities for BRCA1 and BRCA2. Curr. Biol. 11:R668-R676. [DOI] [PubMed] [Google Scholar]
- 36.Kim, G. D., Y. H. Choi, A. Dimtchev, S. J. Jeong, A. Dritschilo, and M. Jung. 1999. Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J. Biol. Chem. 274:31127-31130. [DOI] [PubMed] [Google Scholar]
- 37.Kim, Y. W., G. A. Otterson, R. A. Kratzke, A. B. Coxon, and F. J. Kaye. 1994. Differential specificity for binding of retinoblastoma binding protein 2 to Rb, p107 and TATA-binding protein. Mol. Cell. Biol. 14:7256-7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 39.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 40.Liu, H.-Y., B. S. Nefsky, and N. C. Walworth. 2002. The Ded1 DEAD box helicase interacts with Chk1 and Cdc2. J. Biol. Chem. 277:2637-2643. [DOI] [PubMed] [Google Scholar]
- 41.Liu, Q., S. Guntuku, X.-S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14:1448-1459. [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Girona, A., K. Tanaka, X. B. Chen, B. A. Baber, C. H. McGowan, and P. Russell. 2001. Serine-345 is required for Rad3-dependent phosphorylation and function of checkpoint kinase Chk1 in fission yeast. Proc. Natl. Acad. Sci. USA 98:11289-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu, F. J., K. Sundquist, D. Baeckstrom, R. Poulsom, A. Hanby, S. Meier-Ewert, T. Jones, M. Mitchell, P. Pitha-Rowe, P. Freemont, and J. Taylor-Papadimitriou. 1999. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 274:15633-15645. [DOI] [PubMed] [Google Scholar]
- 44.Lu, Z., S. Xu, C. Joazeiro, M. H. Cobb, and T. Hunter. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9:945-956. [DOI] [PubMed] [Google Scholar]
- 45.Lundgren, K., N. Walworth, R. Booher, M. Dembski, M. Kirschner, and D. Beach. 1991. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64:1111-1122. [DOI] [PubMed] [Google Scholar]
- 46.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 47.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 48.Mao, S., G. A. Neale, and R. M. Goorha. 1997. T-cell oncogene rhombotin-2 interacts with retinoblastoma-binding protein 2. Oncogene 14:1531-1539. [DOI] [PubMed] [Google Scholar]
- 49.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 51.Millar, J. B. A., C. H. McGowan, G. Lenaers, R. Jones, and P. Russell. 1991. p80cdc25 mitotic inducer is the tyrosine phosphatase that activates p34cdc2 kinase in fission yeast. EMBO J. 10:4301-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milutinovic, S., Q. Zhuang, and M. Szyf. 2002. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J. Biol. Chem. 277:20974-20978. [DOI] [PubMed] [Google Scholar]
- 53.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 54.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 55.Nasmyth, K. A. 1977. Temperature-sensitive lethal mutants in the structural gene for DNA ligase in the yeast Schizosaccharomyces pombe. Cell 12:1109-1120. [DOI] [PubMed] [Google Scholar]
- 56.O'Connell, M. J., N. C. Walworth, and A. M. Carr. 2000. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10:296-303. [DOI] [PubMed] [Google Scholar]
- 57.Olsson, T. G., K. Ekwall, R. C. Allshire, P. Sunnerhagen, J. F. Partridge, and W. A. Richardson. 1998. Genetic characterisation of hda1+, a putative fission yeast histone deacetylase gene. Nucleic Acids Res. 26:3247-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quadbeck-Seeger, C., G. Wanner, S. Huber, R. Kahmann, and J. Kamper. 2000. A protein with similarity to the human retinoblastoma binding protein 2 acts specifically as a repressor for genes regulated by the b mating type locus in Ustilago maydis. Mol. Microbiol. 38:154-166. [DOI] [PubMed] [Google Scholar]
- 59.Raleigh, J. M., and M. J. O'Connell. 2000. The G2 DNA damage checkpoint targets both Wee1 and Cdc25. J. Cell Sci. 113:1727-1736. [DOI] [PubMed] [Google Scholar]
- 60.Rhind, N., B. Furnari, and P. Russell. 1997. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 11:504-511. [DOI] [PubMed] [Google Scholar]
- 61.Saha, V., T. Chaplin, A. Gregorini, P. Ayton, and B. D. Young. 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc. Natl. Acad. Sci. USA 92:9737-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt, D. R., and S. L. Schreiber. 1999. Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry 38:14711-14717. [DOI] [PubMed] [Google Scholar]
- 63.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schultz, D. C., J. R. Friedman, and F. J. Rauscher III. 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15:428-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shamay, M., O. Barak, and Y. Shaul. 2002. HBXAP, a novel PHD-finger protein, possesses transcription repression activity. Genomics 79:523-529. [DOI] [PubMed] [Google Scholar]
- 66.Shiloh, Y., and M. B. Kastan. 2001. ATM: genome stability, neuronal development, and cancer cross paths. Adv. Cancer Res. 83:209-254. [DOI] [PubMed] [Google Scholar]
- 67.Smith, M. M. 1991. Histone structure and function. Curr. Opin. Cell Biol. 3:429-437. [DOI] [PubMed] [Google Scholar]
- 68.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 69.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 70.Takeuchi, T., Y. Yamazaki, Y. Katoh-Fukui, R. Tsuchiya, S. Kondo, J. Motoyama, and T. Higashinakagawa. 1995. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 9:1211-1222. [DOI] [PubMed] [Google Scholar]
- 71.Tini, M., A. Benecke, S. J. Um, J. Torchia, R. M. Evans, and P. Chambon. 2002. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell 9:265-277. [DOI] [PubMed] [Google Scholar]
- 72.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 73.Wada, M., H. Miyazawa, R. S. Wang, T. Mizuno, A. Sato, M. Asashima, and F. Hanaoka. 2002. The second largest subunit of mouse DNA polymerase epsilon, DPE2, interacts with SAP18 and recruits the Sin3 co-repressor protein to DNA. J. Biochem. (Tokyo) 131:307-311. [DOI] [PubMed] [Google Scholar]
- 74.Wahl, G. M., S. P. Linke, T. G. Paulson, and L.-C. Huang. 1997. Maintaining genetic stability through TP53 mediated checkpoint control, p. 183-219. In M. B. Kastan (ed.), Cancer surveys, volume 29: checkpoint controls and cancer. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 75.Walworth, N., S. Davey, and D. Beach. 1993. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature 363:368-371. [DOI] [PubMed] [Google Scholar]
- 76.Walworth, N. C., and R. Bernards. 1996. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271:353-356. [DOI] [PubMed] [Google Scholar]
- 77.Wan, S., and N. C. Walworth. 2001. A novel genetic screen identifies checkpoint-defective alleles of Schizosaccharomyces pombe chk1. Curr. Genet. 38:299-306. [DOI] [PubMed] [Google Scholar]
- 78.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 79.Yarden, R. I., and L. C. Brody. 1999. BRCA1 interacts with components of the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:4983-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 81.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]