Abstract
In the adult mammalian brain, neural stem cells in the subventricular zone continuously generate new neurons for the olfactory bulb. Cell fate commitment in these adult neural stem cells is regulated by cell fate-determining proteins. Here, we show that the cell fate-determinant TRIM32 is upregulated during differentiation of adult neural stem cells into olfactory bulb neurons. We further demonstrate that TRIM32 is necessary for the correct induction of neuronal differentiation in these cells. In the absence of TRIM32, neuroblasts differentiate slower and show gene expression profiles that are characteristic of immature cells. Interestingly, TRIM32 deficiency induces more neural progenitor cell proliferation and less cell death. Both effects accumulate in an overproduction of adult-generated olfactory bulb neurons of TRIM32 knockout mice. These results highlight the function of the cell fate-determinant TRIM32 for a balanced activity of the adult neurogenesis process.
Keywords: neurogenesis, cell fate specification, neural stem cells
During embryonic and early postnatal periods of development, the mammalian brain develops through precisely coordinated processes of neural stem cell (NSC) proliferation, fate specification and migration (reviewed in Gotz and Huttner1). In the adult mammalian brain, neurogenesis is restricted to the subventricular zone (SVZ) of the lateral ventricles and the dentate gyrus of the hippocampus.2 In the SVZ, the adult NSCs are glial fibrillary acid protein (GFAP)-expressing astrocytes (type B cells). As a defining feature of all stem cells, these adult NSCs have the ability to simultaneously self-renew and generate more fate-committed daughter cells. The progeny of type B cells are rapidly dividing transient amplifying cells (type C cells). Type C cells give rise to young neuroblasts (type A cells) that eventually will become olfactory bulb (OB) neurons or callosal oligodendrocytes.3 Neuroblasts generated in the SVZ leave their place of birth and migrate via chain migration along the rostral migratory stream (RMS) towards the OB. After reaching the OB, they populate different OB layers where they differentiate and maturate into fully functional neurons.4 How the sequential transition between these cell fates, which represent diverse levels of differentiation, is ensured is largely unknown.
TRIM32 belongs to the family of TRIM-NHL proteins. Previously, this protein family has been implicated in the regulation of proliferation and differentiation of stem cells and progenitor cells in Drosophila,5, 6, 7, 8 Caenorhabditis elegans9 and the developing mammalian brain10, 11 as well as the mammalian skeletal muscle.12
Here, we analyzed the function of TRIM32 during neurogenesis in the adult mammalian brain. Our data indicate that TRIM32 is upregulated during differentiation of SVZ-generated neuroblasts and that it is important for the induction of neuronal differentiation of these cells. Finally, by analyzing neurogenesis with TRIM32 gain-of-function and loss-of-function approaches we show that TRIM32 indeed regulates fate specification in adult NSCs. Interestingly, the TRIM32-deficiency-induced overproduction of adult-generated OB neurons is caused by a combined effect of increased progenitor proliferation and less apoptosis in the OB. These results highlight the function of the cell fate-determinant TRIM32 for a balanced activity of the adult neurogenesis process.
Results
In the adult mammalian brain, NSCs (type B cells) localized in the SVZ give rise to fast-dividing transient amplifying cells (type C cells). These differentiate into the more lineage-restricted neuroblasts (type A cells). Neuroblasts migrate from the SVZ along the RMS towards the OB where they differentiate into neurons and get integrated into the existing network (Figure 1a).
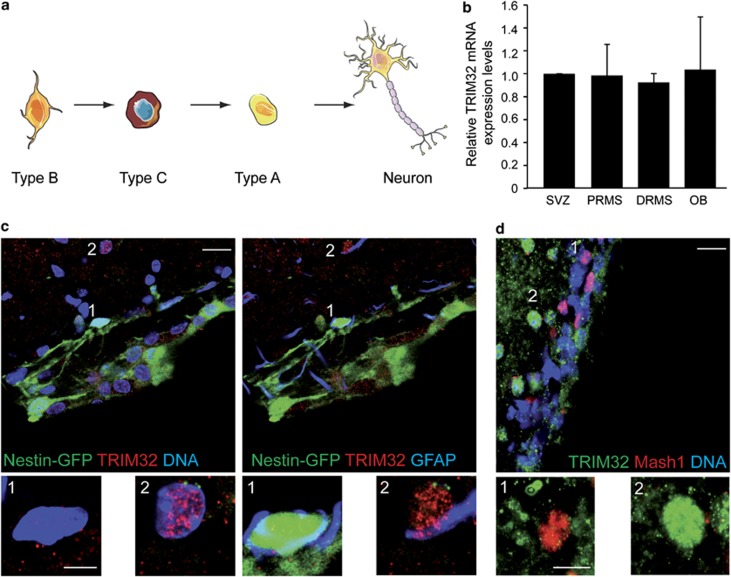
Figure 1.
TRIM32 mRNA is expressed throughout the SVZ–OB system, but TRIM32 protein is virtually absent from the majority of type B and C cells. (a) Schematic drawing of the neuronal lineage: type B cells (stem cell astrocytes) in the SVZ give rise to type C cells (transient amplifying cells), which differentiate into type A cells (neuroblasts). Type A cells further differentiate into mature neurons. (b) RT-qPCR measuring the relative TRIM32 mRNA expression levels within the different indicated adult brain regions derived after micro-dissection and Nestin-GFP-based FACS analysis (N=3; mean±S.E.M.; t-test). (c and d) Immunostainings of mouse brain sections labeled with the indicated antibodies. Images were taken in the SVZ. Bars=20 μm, for high magnifications 5 μm (c) and 10 μm (d)
TRIM32 is expressed in neuroblasts and neurons
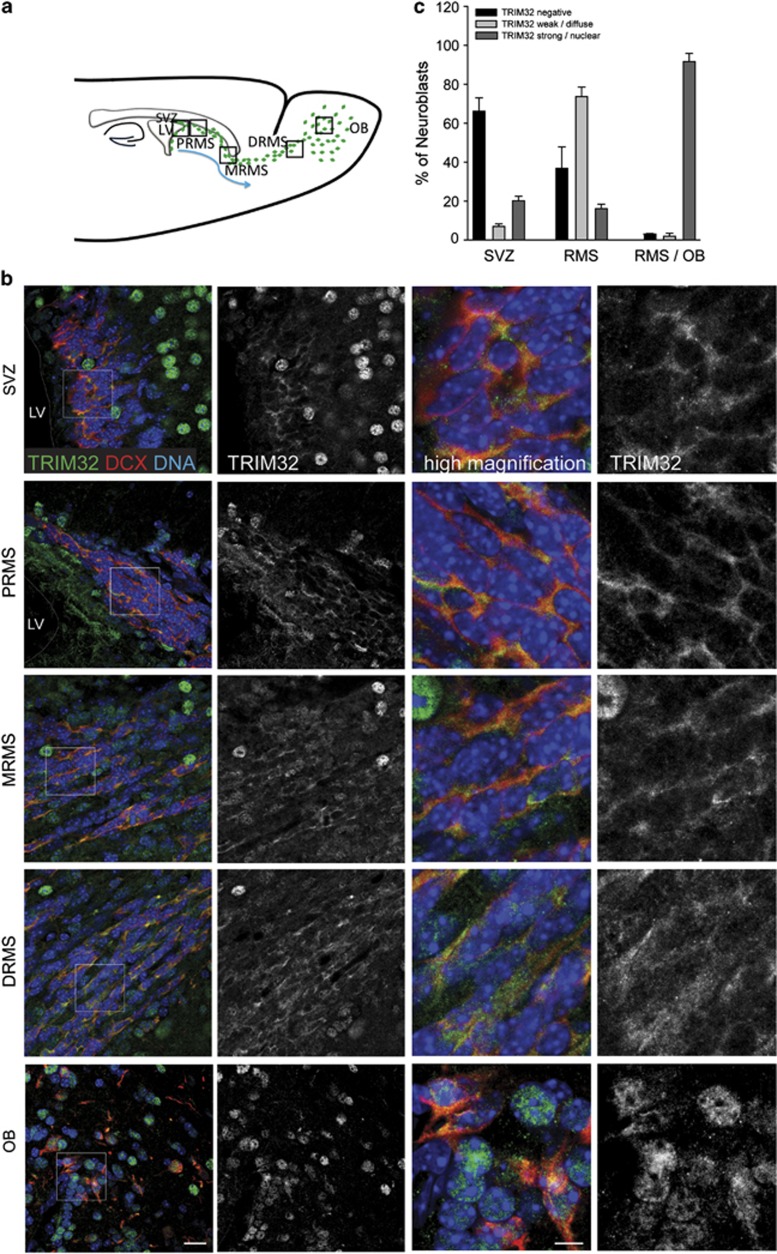
As nothing is known about the function of TRIM32 during adult neurogenesis, we investigated the expression of TRIM32 in the adult mouse SVZ–OB system. For studying mRNA expression levels of TRIM32 in neural progenitor cells, we used adult mice expressing green fluorescent protein (GFP) under the control of the promoter of the neural progenitor marker Nestin. In these mice, newly generated neurons, which are generated in the SVZ and migrate rostrally, express GFP until they reach the OB.13 We isolated GFP-positive neural progenitors from microdissected SVZ, proximal part of the RMS (PRMS), distal part of the RMS (DRMS) and OB from these mice via FACS analysis. The RNA was isolated from these fractioned cells, and the TRIM32 mRNA levels were investigated through reverse transcription-quantitative PCR (RTq-PCR). We were able to detect expression of TRIM32 in progenitors from all regions. Additionally, the expression levels of TRIM32 mRNA were comparable in neural progenitor cells from all investigated regions (Figure 1b and Supplementary Figure S1a). In the next steps, we made further use of these Nestin-GFP mice to determine the expression of the TRIM32 protein via immunofluorescence stainings. Adult NSCs (type B cells) are characterized by the expression of the two markers Nestin and GFAP. By using Nestin-GFP mice together with immunostainings against GFAP and TRIM32, we were able to show that the TRIM32 protein is virtually absent from type B cells in the SVZ (Figure 1c). Specificity of the anti-TRIM32 antibody has been shown previously14 and is confirmed by TRIM32 short hairpin RNA (shRNA) experiments (see below). Additionally, staining of sections from wild-type mouse brain with the type C cell marker Mash-1 showed that also the majority of transient amplifying type C cells were not positive for TRIM32 (Figure 1d, Supplementary Table S1). Neuroblasts, which are already committed to the neuronal cell fate, leave the SVZ in an immature state and become more mature during their migration along the RMS towards the OB. These neuroblasts are positive for the microtubule-associated protein doublecortin (DCX). Co-stainings for DCX and TRIM32 revealed that neuroblasts express TRIM32 (Figure 2). This expression of TRIM32 strongly increases when neuroblasts mature and reach the OB (Figure 2b and c). The TRIM32 expression further increases when the cells leave the RMS, populate the OB and start to express the neuronal marker neuronal nuclei (NeuN; Figure 3). Interestingly, early in the proximal RMS the expression of TRIM32 is mainly cytoplasmic, while it becomes more nuclear in neuroblasts at the distal RMS and OB. In NeuN-positive neurons, TRIM32 is strongly expressed and exclusively localized in the nucleus of the cells. Among the NeuN-positive cells in the granular cell layer (GCL), 95% express also TRIM32, while 70% of the NeuN-positive neurons in the glomerular cell layer (GL) show a strong expression of nuclear TRIM32 (Figure 3, Supplementary Figures S1n and o). Together, these expression data suggest that TRIM32 mRNA levels are present through the entire SVZ–RMS–OB system without presenting any significant changes. However, the TRIM32 protein becomes upregulated during neuronal cell fate commitment. Most probably, post-transcriptional events are responsible for a lack of significant translation of the TRIM32 mRNA in type B and C cells.
Figure 2.
Expression of TRIM32 increases in neuroblasts through the RMS into the OB. (a) Schematic representation of the SVZ–RMS–OB system is shown. (b) Immunostainings of mouse brain sections, taken from regions indicated in panel (a), labeled with the indicated antibodies. Bars=20 μm, high magnification 10 μm. (c) Quantification of the percentage of neuroblasts expressing TRIM32 (mean±S.E.M., t-test, n≥735 cells, N=3 mice). The expression of TRIM32 increases from immature cells in the SVZ to mature neurons in the OB
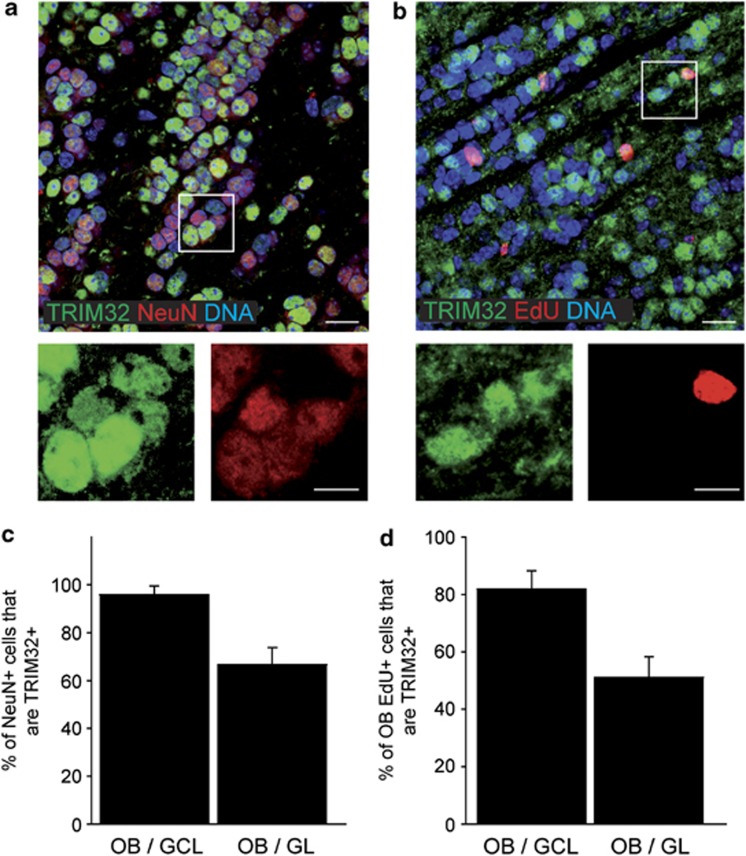
Figure 3.
TRIM32 is strongly expressed in adult-generated neurons of the OB. (a and b) Immunostainings of mouse brain sections, taken in the OB, labeled with the indicated antibodies. Bars=20 μm, for high magnifications 10 μm. (c and d) Diagrams showing the percentage of cells in the GL and GCL of the OB that are double positive for NeuN and TRIM32 (c) and EdU and TRIM32 (d) (mean±S.E.M., n≥3 mice). These results indicate that TRIM32 is expressed in adult-generated neurons of the OB
In good agreement with the protein expression pattern, the majority of mitotically active Ki67-positive cells, as well as dividing PH-3-positive cells, are negative for TRIM32 (Supplementary Figures S1b–e). Additionally, astrocytes and oligodendrocytes show only very low levels of TRIM32 immunostaining (Supplementary Figures S1f–h, Supplementary Table S1).
Next we analyzed whether TRIM32 is indeed expressed in adult-generated neurons. Therefore, we stained mouse brain sections 8 days after injection of the nucleotide analog 5-ethynyl-2'-deoxyuridine (EdU) for EdU and TRIM32 (Figures 3b and d). EdU-positive cells were detectable all along the SVZ–OB. Especially in the middle and DRMS, EdU/TRIM32 double-positive cells were detectable (Supplementary Figures S1i–m.). This fraction of EdU/TRIM32 double-positive cells increased strongly within the OB (Figure 3d and Supplementary Figures S1i–m). In the GCL, 80% of the EdU-positive cells express TRIM32, while in the GL TRIM32 was detectable in 50% of the EdU-labeled cells (Figure 3d).
Finally, we aimed to analyze the TRIM32-expressing neuronal subtypes in more detail. We found that TRIM32 is not only expressed in NeuN-positive neurons of the GCL and GL (Supplementary Figures S1n and o) but that it is also expressed in the Calbindin-positive neurons of the GL (Supplementary Figure S1p). Additionally, TRIM32 is expressed in GABAergic, glutamatergic and dopaminergic neurons in the OB (Supplementary Figures S1q–s). Interestingly, the expression of TRIM32 in neurons and neuroblasts seems to be conserved from mice to primates (Marmoset and Macaque monkeys) (Supplementary Figures S1t and u).
This expression analysis revealed that in cells with high potential for self-renewal and proliferation (like type B and C cells) TRIM32 is only very weakly expressed. However, during neuronal cell fate specification the levels of TRIM32 increase strongly.
Absence of TRIM32 leads to an overproduction of adult-generated OB neurons
As TRIM32 is upregulated during the critical period of differentiation of neuroblasts into neurons in the adult SVZ–OB system, we analyzed the activity of neurogenesis in TRIM32-deficient mice. Wild-type and TRIM32 knockout mice were injected with bromodeoxyuridine (BrdU) on 3 consecutive days, and brains were fixed 14 days after the last injection (Figure 4a).
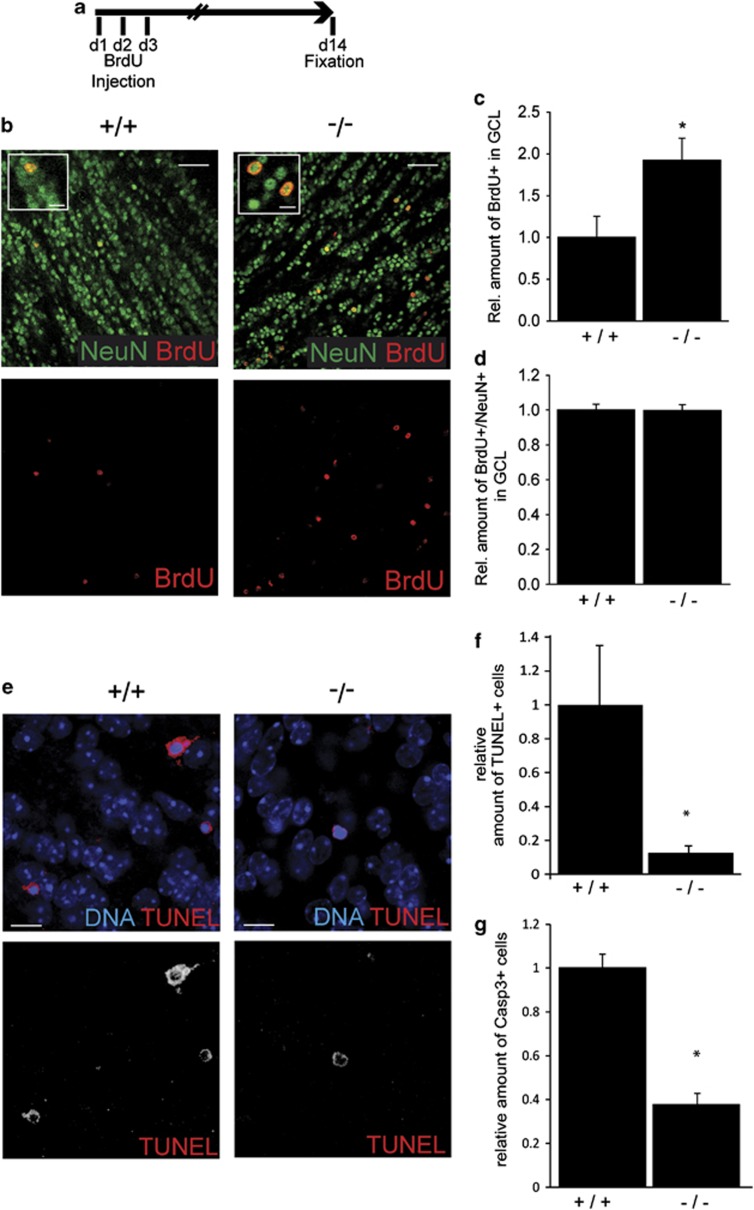
Figure 4.
TRIM32 knockout mice show an increase in the number of newborn neurons and decreased rates of apoptosis. (a) Schematic drawing of the experimental paradigm. For 3 days, mice received a daily injection of BrdU; 14 days later, the brains were fixed and analyzed. (b) Immunostainings of sections from the OB-GCL of wild-type and TRIM32 knockout mice labeled with the indicated antibodies. Bars=50 μm, for high magnifications 15 μm. (c–d) Diagrams showing the relative amount of BrdU-positive cells in the OB-GCL (c) and the amount of cells that are double positive for BrdU and NeuN in the OB-GCL (d). All quantifications were conducted in wild-type and knockout mice (mean±S.E.M., N≥4 mice; t-test, * P<0.05). (e) TUNEL stainings of sections from wild-type and TRIM32 knockout mice OB-GCL. Bars=15 μm. (f and g) Diagrams showing the relative amount of TUNEL+ (f) and Casp3+ (g) cells in the OB-GCL. (Median±S.E.M., N≥4 mice, Mann–Whitney U Test *P<0.05). These results show that in the absence of TRIM32 in the adult OB-GCL more cells are generated and that these cells eventually undergo neuronal differentiation
By comparing wild-type with TRIM32 knockout mice, we found significantly more BrdU+ cells in the GCL of these knockout mice (Figures 4b–d, Supplementary Table S2). Interestingly, although significantly more new cells (BrdU positive) were generated for the OB of TRIM32 knockout mice, the neuronal differentiation of these cells was not defective. Both in wild-type mice as well as in TRIM32 knockout mice, nearly all BrdU-positive cells differentiated into neurons within 14 days (Figure 4d, Supplementary Table S2). Additionally, the amount and differentiation status of BrdU+ cells in the GL was unchanged in TRIM32 knockout mice (Supplementary Figure S2, Supplementary Table S2). Finally, differentiation into glia cells is unchanged in the absence of TRIM32 (data not shown). An overproduction of new neurons for the OB could be caused by the generation of more new cells or a decreased rate in cell death. To first address the latter, apoptotic profiles in the GCL of TRIM32 knockout and WT mice were compared using terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining as well as cleaved caspase-3 (Casp3) staining (Figure 4e). Indeed, we found significantly less apoptotic cells in the GCL of TRIM32 knockout mice (Figures 4e and f and Supplementary Table S2). We conclude that in the absence of TRIM32 in the adult SVZ–OB system more newly generated neurons populate the GCL. The overproduction is caused at least partially by a reduced apoptosis rate in TRIM32 knockout mice (analysis of proliferation: see below). However, within the long time period of 14 days from the birth of these cells to analysis of their fate their ability to undergo neuronal differentiation was not impaired. Clearly, with this assay we would be unable to detect a cell autonomous delay of differentiation at early stages of differentiation that is potentially caused by the absence of TRIM32. Therefore, in the next step we conducted cell autonomous knockdown of TRIM32 and did the analysis 8 days after this knockdown.
TRIM32 is regulating neuronal differentiation of adult NSCs
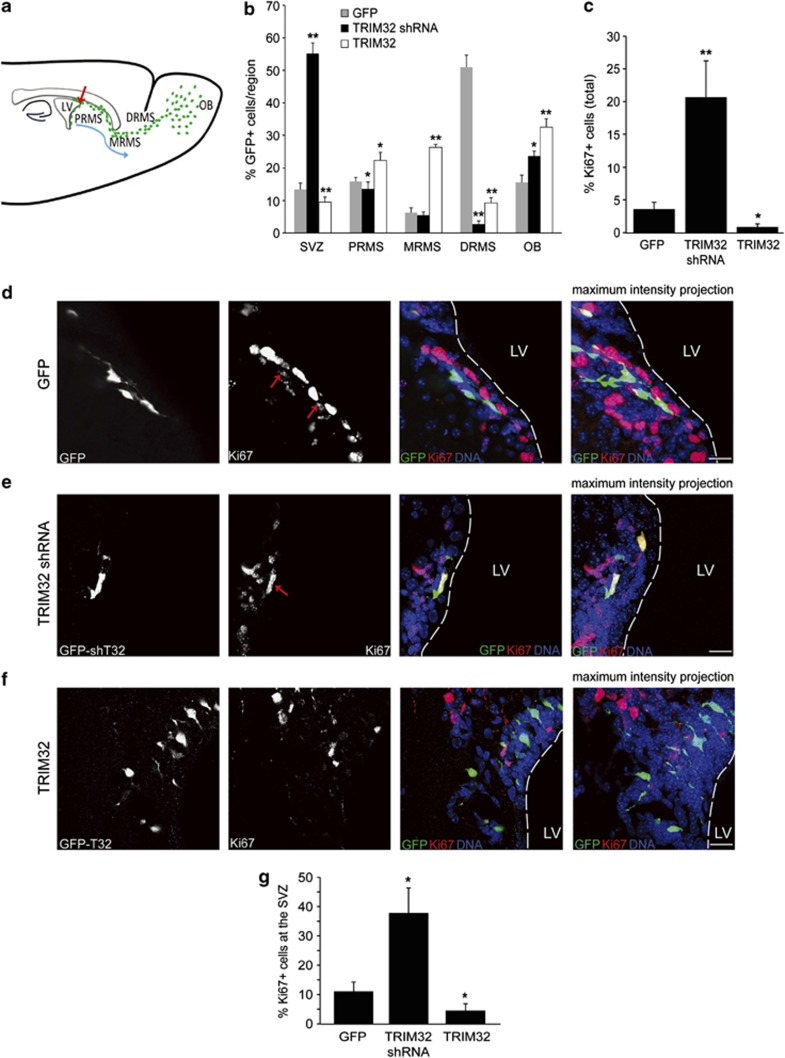
TRIM32 knockout mice have more adult-generated cells in their OB (Figure 4). Fourteen days after birth, neuronal differentiation seems not to be affected in those adult-generated cells. However, there might be an impairment of differentiation at early stages that might be rescued at 14 days after birth and therefore not detected with the assay conducted above. Furthermore, because this is a complete knockout it is impossible to study cell autonomous effects. In order to conduct a detailed analysis of TRIM32 function during fate specification, we overexpressed or knocked down this gene in individual adult NSCs of the SVZ and analyzed manipulated cells at 8 days after infection. Therefore, we stereotactically injected retroviruses for overexpression (TRIM32 with IRES-EGFP (internal ribosome entry site-enhanced GFP)), knockdown (two independent TRIM32 shRNA constructs, EGFP expression from a separate promoter) and control (scrambled shRNA and EGFP expression) into the adult SVZ (Figure 5). Functionality of the shRNA constructs was proven by western blotting and immunofluorescence stainings in vitro and in vivo (Supplementary Figures S3a–d). Furthermore, these results confirm the specificity of our anti-TRIM32 antibodies. The fate of transduced cells was analyzed 8 days after injection. At this time point, the majority of the control cells (expressing only EGFP) were localized in the DRMS (Figure 5b). In contrast, a knockdown of TRIM32 led to a significant increase in the amount of cells that remained in the SVZ, while overexpression of TRIM32 resulted in the accumulation of transduced cells in the PRMS and middle RMS (Figure 5b).
Figure 5.
Knockdown of TRIM32 induces proliferation in neural progenitor cells. (a) Schematic representation depicting the migration process of the neural progenitor cells from the SVZ to the OB. Viruses for expression of a scrambled shRNA sequence (control), TRIM32-shRNA or overexpression of TRIM32 were injected in the SVZ. All constructs express GFP under an independent promoter. Eight days after injection, brain sections were analyzed. (b) Quantification of the position of GFP-positive cells in injected brains (mean±S.E.M., N≥700; N>8 mice, Mann–Whitney U test, *P<0.05; **P<0.01). (c) Ki67 was used as a proliferation marker. A quantification of GFP-positive cells that are also positive for Ki67, summarizing all brain regions, is shown (mean±S.E.M., n≥700; N>8 mice, Mann–Whitney U test, *P<0.05; **P<0.01). (d–f) Immunostainings of mouse brain sections after injection of viruses for GFP, TRIM32-shRNA or TRIM32, taken from the SVZ and labeled with the indicated antibodies. Bars=15 μm. (g) Quantification of GFP-positive cells in the SVZ, which are also positive for Ki67 (mean±S.E.M., n≥500; N≥8 mice, Mann–Whitney U test, *P<0.05; **P<0.01). The results presented here indicate that TRIM32 regulates proliferation of neural progenitor cells
To investigate the effect of TRIM32 on proliferation in transduced cells, brain sections were stained with the cell cycle marker Ki67. A knockdown of TRIM32 increased the amount of proliferative cells strongly, whereas TRIM32 overexpression resulted in a significant reduction of Ki67-positive cells. This effect was detectable irrespectively of the localization of the transduced cells (Figure 5c) and was not limited to cells within the SVZ (Figures 5d–g). Strikingly, upon knockdown of TRIM32 we were even able to detect some mitotic cells (showing characteristically condensed chromatin) within the distal RMS (Supplementary Figure S3d); this is an observation that we never observed for EGFP-expressing control cells.
To investigate neuronal fate commitment, we used the marker neuronal class III β-tubulin (TuJ1). Overexpression of TRIM32 led to a small but significant increase of TuJ1-positive cells. In contrast, a knockdown of TRIM32 strongly reduced the amount of transduced TuJ1-positive cells (Supplementary Figure S4). The effects observed for TuJ1 are probably not so pronounced, because this marker not only labels postmitotic neurons but is also expressed in neuroblasts. However, when we analyzed the TuJ1 stainings after TRIM32 knockdown, we sometimes found clusters of TuJ1 negative, transduced round cells within the RMS that were reminiscent of type C-cell cluster that are usually found in the SVZ (Supplementary Figure S5).
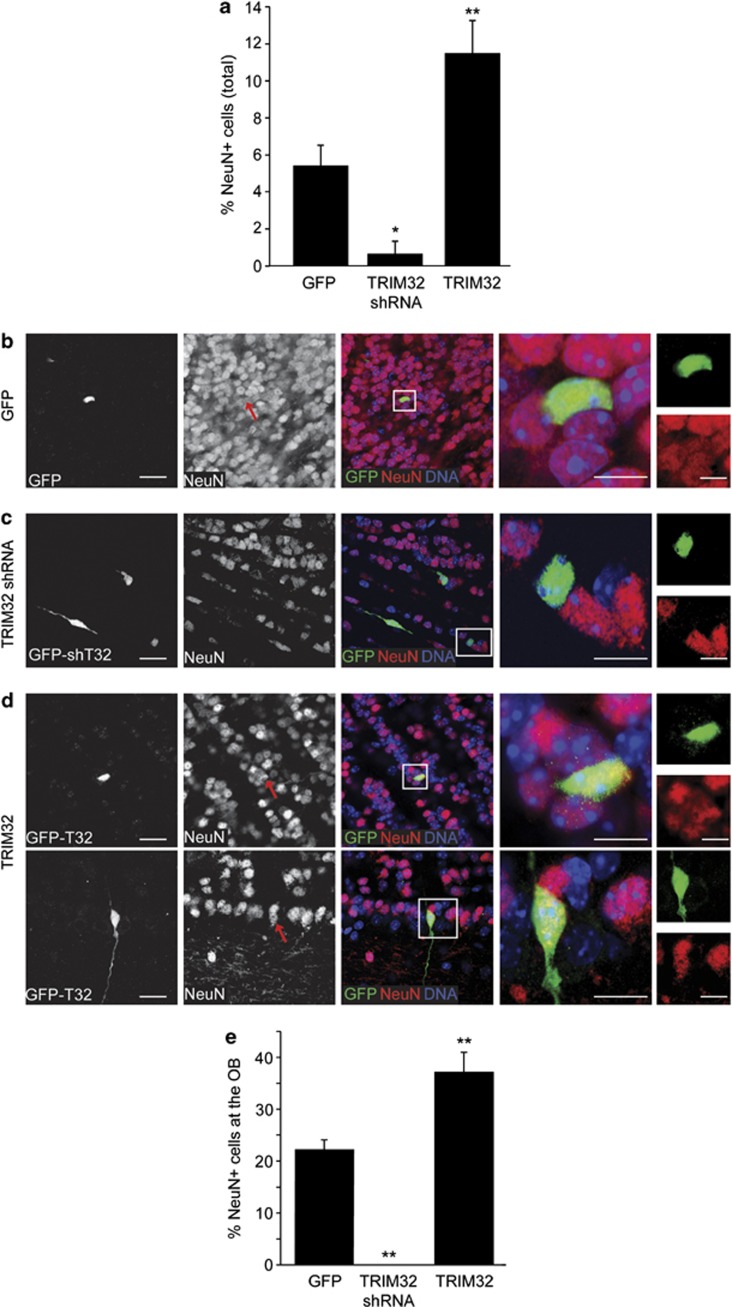
During their differentiation, neuroblasts start to express NeuN and become postmitotic neurons. Therefore, we used NeuN to finally evaluate the effect of TRIM32 on terminal neuronal differentiation. Strikingly, a cell autonomous knockdown of TRIM32 by shRNA 8 days after infection resulted in a strong reduction of NeuN-positive neurons, whereas its overexpression facilitated neuronal differentiation (Figure 6). Altogether, these data indicate that TRIM32 is necessary for the correct induction of neuronal differentiation of adult neural progenitor cells.
Figure 6.
Correct induction of neuronal differentiation depends on the presence of TRIM32. Viruses for expression of a scrambled shRNA sequence (control), TRIM32-shRNA or overexpression of TRIM32 were injected in the SVZ. All constructs express GFP under an independent promoter. Eight days after injection, brain sections were analyzed. (a) NeuN was used as a neuronal differentiation marker. A quantification of GFP-positive cells that are also positive for NeuN, summarizing all brain regions is shown (mean±S.E.M., n≥500; n≥8 mice, Mann–Whitney U test, *P<0.05; **P<0.01). (b–d) Immunostainings of mouse brain sections, taken from the OB and labeled with the indicated antibodies. Boxed areas are shown in higher magnification. Red arrows indicate transduced cells that are positive for NeuN. Bars=15 μm for the first three columns, 10 μm for the fourth and fifth columns. (e) Quantification of GFP-positive cells in the OB, which are also positive for NeuN (mean±S.E.M., n≥500; N≥8 mice, Mann–Whitney U test, *P<0.05; **P<0.01). These results indicate that TRIM32 is necessary for the correct induction of neuronal differentiation of neuroblasts
TRIM32 regulates the gene expression profile of neuroblasts
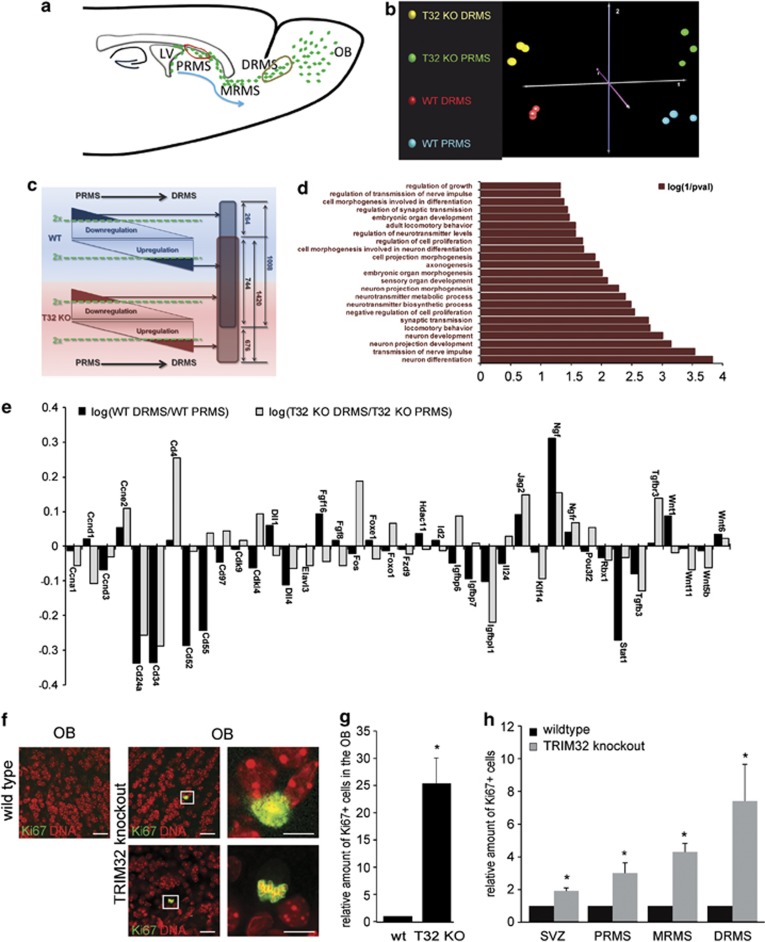
To analyze the molecular changes that occur upon loss of TRIM32, we performed a whole transcriptomic microarray analysis on mRNAs extracted from fresh tissue dissections of the PRMS and DRMS from wild-type and TRIM32 knockout mice (red and brown circles, Figure 7a).
Figure 7.
Molecular profiling demonstrates deregulated neuronal differentiation in TRIM32 knockout mice. (a) Schematic representation reflecting the migration and maturation of neural progenitor cells with regions of isolated tissue samples indicated by red (PRMS) and brown (DRMS) circles. (b) CoA performed on the 5000 transcripts with the highest S.D. throughout the series of 12 microarrays analyzed. (c) Schematic illustration of the expression data filtering process. For wild-type (blue) and TRIM32 knockout (red) mice, gene expression levels were compared between the PRMS and the DRMS. Genes with twofold regulation (dark blue or dark red triangles) were isolated for further data processing. In all, 1008 transcripts were at least twofold regulated when comparing PRMS and DRMS in wild-type mice, whereas 1420 transcripts were twofold regulated in TRIM32 knockout animals. From these genes, 744 transcripts were simultaneously designated as regulated in wild-type and TRIM32 knockout mice and thus are varying independently from the genotype. A total of 676 transcripts were exclusively regulated in TRIM32 knockout mice and are therefore specifically associated with loss of TRIM32. (d) GO term analysis of the 676 transcripts exclusively associated with loss of TRIM32. The bar diagram represents the log(1/P-value) (Fisher's exact test). The entire list of GO terms is shown in Supplementary Figure S6. (e) Bar diagram of the expression changes of genes implicated in NSC fate-regulating mechanisms. Black and grey bars represent the log of the ratio calculated from the gene expression intensity within the DRMS and within the PRMS of wild-type and of TRIM32 knockout mice. (f) Immunostainings of brain sections (wild-type and TRIM32 knockout), taken from the OB and labeled with the indicated antibodies. Boxed areas are shown in higher magnification. Bars=25 μm, for high magnifications 10 μm. (g) Bar diagram of the relative amount of Ki67+ cells in the OB of wild-type and TRIM32 knockout mice (mean±S.E.M., N≥4 mice, Mann–Whitney U test, *P<0.05). (h) Bar diagram of the relative amount of Ki67+ cells in the SVZ and RMS of wild-type and TRIM32 knockout mice (mean±S.E.M., N≥4 mice, Mann–Whitney U test, *P<0.05)
For wild-type and TRIM32 knockout mice, each indicated region was profiled in triplicate. Via a correspondence analysis (CoA), a high degree of similarity between analogous samples was demonstrated (Figure 7b). Additionally, this analysis illustrated that the four triplicate groups strongly differed in terms of anatomic localization (PRMS versus DRMS) and of genotype (wild-type versus TRIM32 knockout).
To specifically isolate genes with expression profiles differing between wild-type and TRIM32 knockout samples during the neuronal maturation, that is, when cells migrate and differentiate from PRMS to DRMS, we used the following filtering steps (Figure 7c): In the first step, genes that were two times upregulated or downregulated in wild-type or in TRIM32 KO animals were isolated. Through superimposing both groups (1008 transcripts regulated in wild-type and 1420 transcripts regulated in the TRIM32 knockout), we were able to distinguish between those genes that are regulated according to the anatomic localization (744 transcripts) and those genes that are exclusively regulated because of loss of TRIM32 (676 transcripts). In order to determine the function of these 676 TRIM32 regulated genes, we used a Gene Ontology (GO) analysis approach. Interestingly, this analysis significantly related the TRIM32-regulated genes to terms, including ‘neuron differentiation', ‘neuron development', ‘morphogenesis' and multiple terms associated with neuronal maturation, such as ‘transmission of nerve impulse' and ‘neuron projection development' (Figure 7 and Supplementary Figure S6a). In the next step, we focused the analysis on mechanisms that are known to be involved in stem cell fate decisions. By using the gene expression data, we investigated how the expression levels of key genes being involved in stem cell fate decisions change during neuronal maturation (comparing PRMS with DRMS) in the absence and presence of TRIM32. Indeed, a series of genes that are differentially expressed when comparing wild-type and TRIM32 knockout animals were detectable (Figure 7e). Together, this gene expression analysis supports the notion that TRIM32 is an important regulator of neuronal differentiation and that loss of TRIM32 severely impairs and decelerates neuronal commitment.
To finally address whether the loss of TRIM32 indeed impairs neuronal differentiation, we made use of the fact that neuronal differentiation is accompanied by cell cycle exit. Additionally, the GO analysis (Figure 7d), as well as the analysis of selected deregulated genes (Figure 7e, Supplementary Table S3), strongly suggests that the cell cycle might be a valid readout for TRIM32 activity. Indeed, when we analyzed the amount of Ki67-positive cells in the OB of wild-type and TRIM32 knockout mice, we detected a strong increase for cell cycle active (Ki67 positive) cells in the absence of TRIM32 (Figures 7f and g, Supplementary Figure S6b and Supplementary Table S4). The same effect was detectable from SVZ throughout the RMS (Figure 7h). Strikingly, this increase in the amount of Ki67-positive cells correlates with the continuous increase of TRIM32 expression from SVZ to OB (compare with Figure 2c). Additionally, these results indicate that the increased amount of adult-generated cells in the OB of TRIM32 knockout mice is not only the result of decreased apoptosis but also that increased proliferation has a role.
Thus this analysis suggests that TRIM32 is important for neuronal differentiation and in the absence of TRIM32 numerous genes associated to this process are deregulated leading to increasing amounts of cells remaining in the cell cycle.
Discussion
Although mechanisms regulating the generation and specification of neurons are well described, only little is known about factors that initiate exit from an immature neuronal precursor state and induce terminal neuronal differentiation during adult mammalian neurogenesis. Data presented in this study indicate that the expression of the cell fate determinant TRIM32 increases gradually from young neuroblasts to postmitotic neurons in the SVZ–OB system. Cell autonomous TRIM32 gain-of-function and loss-of-function approaches revealed that TRIM32 is able to induce cell cycle exit of mitotic progenitor cells and is necessary for accurate timing of terminal neuronal differentiation in adult NSCs. Finally, investigating the molecular mechanism, microarray data indicate that the differentiation-associated translocation of TRIM32 into the nucleus has extensive consequences for the transcriptome of differentiating neuroblasts.
The expression of the TRIM32 protein increases continuously from low levels in just specified neuroblasts in the SVZ to high levels in neuroblasts at the DRMS where neuroblasts differentiate into postmitotic neurons of the OB. These increasing levels of TRIM32 suggest that neuroblasts maturate during their migration from the SVZ to the OB. What might be the contribution of TRIM32 to this differentiation/maturation process? Our data indicate that neuroblast maturation is not only associated with increasing levels of TRIM32 but also with a translocation of TRIM32 into the nucleus. Previously, we have shown that TRIM32 on the one hand binds to Argonaute proteins and activates micro-RNAs,10 while on the other hand it induces degradation of c-Myc after translocation into the nucleus.11 Therefore, it is tempting to speculate that cytoplasmic TRIM32 in neuroblasts of the PRMS initiates differentiation/maturation via micro-RNA activation, while the differentiation process is finalized in neuroblasts of the distal RMS and young neurons of the OB through TRIM32 translocation to the nucleus. In line with this, it has been recently shown that during neuronal differentiation processes TRIM32 is involved in retinoic acid receptor-mediated transcription.14, 15 Furthermore, the involvement of retinoic acid signaling in adult neurogenesis has been described intensively.16, 17, 18 Additionally, human TRIM32 (HT2A) has been reported to undergo nuclear translocation and to interact with the activation domain of lentiviral Tat proteins.19 Interestingly, a similar nuclear translocation to induce differentiation has been shown for other tumor-suppressor proteins, such as PTEN.20 In good agreement with the hypothesis that nuclear translocation of TRIM32 induces terminal neuronal differentiation, a huge fraction of the genes with functions in cell cycle regulation, neuronal differentiation, maturation and related processes are deregulated in the absence of TRIM32. However, because eventually neuronal differentiation is also occurring in the absence of TRIM32, its function seems to be transient, mainly to induce differentiation and thereby to regulate the exact timing of neuronal specification.
A remaining question is how these functions on the molecular level actually affect cellular features like proliferation, differentiation and survival. With a BrdU-incorporation protocol designed to evaluate the amount of newborn neurons, knockout mice lacking TRIM32 had nearly 90% more new adult-generated granule neurons than their WT counterparts. As TRIM32 is absent from the majority of type B and C cells and its expression is upregulated in neuroblasts (type A cells) along the RMS together with the results that we obtained by acute overexpression or knockdown of TRIM32, it seems likely that this increased amount of newly generated cells is the result of increased proliferation of neuroblasts along the RMS. Strikingly, this inverse correlation between TRIM32 expression and proliferation becomes obvious in TRIM32 knockout mice. In regions where TRIM32 is usually only expressed at low levels (e.g., the SVZ) TRIM32 knockout mice only show a moderate increase in proliferative cells. However, in regions with increasing TRIM32 levels (RMS to the OB), TRIM32 knockout mice show comparable increasing levels of proliferation (compare with Figures 2c, 7g and h). Additionally, to this effect on proliferation we found reduced levels of apoptosis in the OB of TRIM32 knockout mice. This is in good agreement with the recently reported pro-apoptotic function of TRIM32.21 Interestingly, other reports describe TRIM32 as an inhibitor of apoptosis.22 Seemingly the pro- or anti-apoptotic function of TRIM32 depends on the cell type and probably even on other circumstances, such as stress level or differentiation status.
Adult neurogenesis holds great regenerative potential for restoring affected brain regions of patients suffering from neurodegenerative diseases. Experimental manipulation of TRIM32 in transplanted or endogenous NSCs could be an interesting new therapeutic approach for cell replacement therapies.
Materials and Methods
Material and plasmids
For immunolabeling, the following antibodies were used: anti-TRIM32 (Gramsch Laboratories, Schwabhausen, Germany), anti-Nestin (BD Biosciences, Franklin Lakes, NJ, USA), anti-GFAP, anti-GFP (Abcam, Cambridge, UK), anti-polysialylated-neural cell adhesion molecule (Millipore, Billerica, MA, USA), anti-DCX (Millipore), anti-Mash-1 (BD Biosciences), anti-NeuN (Millipore), anti-Ki67 (BD Pharmingen, San Diego, CA, USA and Vector Labs, Burlingame, CA, USA), glutathione S-transferase π (anti-GST π; Abcam), anti-gamma-amino butyric acid (anti-GABA; Abcam), anti-vesicular glutamate transporter 2 (Abcam), anti-Tuj1 (Covance, Princeton, NJ, USA), anti-tyrosin hydroxylase (Chemicon Millipore, Temecula, CA, USA), anti-BrdU (AbDSerotec, Kidlington, UK) and anti-Casp3 (Asp175, Cell Signaling Technology, Danvers, MA, USA). DNA was stained with Hoechst (Abcam). For virus production, the retrovirus packaging vector PCL-ECO23 and the following transfer plasmids were used: pMX-GFP23 for control experiments, the coding sequence of TRIM32 was cloned into the pMSCV-CITE-EGFP-PGK-Puro vector (kindly provided by Dr. Manuel Grez, Georg Speyer Haus, Biomedical Research Institute, Frankfurt, Germany), and retroviral TRIM32 shRNA constructs were obtained commercially (Genescript, Hong Kong).
Cell culture, transfection and western blotting
N2A cells were grown in DMEM supplemented with 10% heat-inactivated FCS, 2 mM L-Glutamine, 100 U/ml Penicillin and 100 μg/ml Streptomycin. The cells were transfected using Turbofect (Fermentas, Burlington, ON, Canada) according to the manufacturer's instructions and were lysed 48 h after transfection with lysis buffer 1 (2% Triton X-100, Complete protease inhibitor cocktail (Roche, Basel, Switzerland), 20 mM NaF and 20 mM Na3VO4 in phosphate-buffered saline (PBS)) for 30 min at 4 °C. The cell lysates were centrifuged for 30 min at 13 000 r.p.m. at 4 °C, the supernatant was mixed with sample buffer, boiled at 95 °C for 5 min and used for western blotting for further analysis. To ensure loading of equal amounts of protein for SDS-PAGE and western blotting, the total concentration of proteins was determined with a BCA protein assay kit (Thermo Scientific, Waltham, MA, USA) according to the manufacture's instruction. Furthermore, equal amounts of blotted protein were verified by Ponceau S (Sigma, St Louis, MO, USA) staining on the membrane. Analysis of the western blots was done using the Image J software (National Institute of Mental Health; http://rsb.info.nih.gov/ij).
RTq-PCR
For isolation of neural progenitors, adult mice expressing GFP under the control of the promoter of the neural progenitor marker Nestin were used. Newly generated neurons, which are generated in the SVZ and migrate rostrally, express GFP until they reach the OB.13 Tissue from the SVZ, pRMS, dRMS and the OB of four 5–month-old Nestin-GFP mice was dissected under a binocular and collected in cold DMEM. For dissociation of the tissue into a single-cell suspension, cells were incubated with papain at 37 °C and flushed through a pipette tip followed by a cell strainer (BD Biosciences). Sorting was done using a FACS Aria II cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA).
Total RNA was extracted from Nestin-GFP-positive cells by the miRNAeasy kit (Qiagen, Venlo, Netherlands) following the manufacturer's recommendations. TRIM32 expression levels were evaluated by the KAPA SYBR Fast Master Mix (Peqlab, Erlangen, Germany) following the manufacturer's recommendations. TRIM32-related intensity levels were evaluated upon normalization with GAPDH levels.
Used primers are listed below:
TRIM32-F: 5′-GCATCCAGGAAGAGCTAG-3′
TRIM32-R: 5′-CTCTACCACTTGACTGTTG-3′
GAPDH-F: 5′-ACAATGAATACGGCTACAG-3′ and
GAPDH-R: 5′-GGTCCAGGGTTTCTTACT-3′.
Immunohistochemistry of free-floating sections
All animals used in this study were maintained and treated according to the approved protocols and in accordance with the institutional and national guidelines and regulations. Mice were deeply anaesthetized by intraperitoneal injection of 0.017 ml of 2.5% Avertin (100% stock solution: 10 g 2, 2, 2-Tribromoethanolin 10 ml tert-Amylalcohol) per gram of body weight and killed by perfusion. Brains were fixed overnight at 4 °C in 4% paraformaldehyde in PBS. Later, sections of 40 μm were prepared using a vibratome (Leica, Wetzlar, Germany) and blocked for at least 1 h in TBS (0.1 M Tris,150 mM NaCl, pH 7.4) containing 0.5% Triton X 100, 0.1% Na-Azide, 0.1% Na-Citrate, and 5% normal goat serum. Immunostainings were performed by incubation of the sections with primary antibodies diluted in the blocking solution for 48 h at 4 °C on a shaker, followed by incubation with the secondary antibody diluted in the blocking solution for 2 h at room temperature. Finally, sections were mounted in AquaMount (DAKO, Glostrup, Denmark). Images were collected by confocal microscopy using the ZEN software (Zeiss, Jena, Germany); image analysis was performed with the ZEN software (Zeiss), Adobe Photoshop (San Jose, CA, USA), Image J software and Imaris software (Zurich, Switzerland).
Immunohistochemistry of free-floating sections (Callithrix jacchus and Macaca)
Marmoset (Callithrix jacchus) and macaque (Macaca) monkeys were raised in the institutional breeding facility of the Centre of Reproductive Medicine and Andrology. Breeding, maintenance and experimental procedures were performed in accordance with the German Federal law on the Care and Use of laboratory animals. The animals were deeply sedated using ketamine and killed by exsanguination. Brains from adult marmoset and macaque monkeys were obtained from local colonies at the Institute of Reproductive Medicine, University of Münster, Münster, Germany. Those brains were processed for immunohistochemistry as described above for mouse brains.
TUNEL and Casp3 staining
TUNEL staining was used to detect DNA fragmentation in situ and performed with the In Situ Cell Death Detection Kit, TMR red (Roche, Cat. No. 12156792910) according to the manufacturer's instructions. In brief, 40-μm brain sections of mouse brains were obtained as described above and blocked for 1 h at room temperature in TBS containing 0.5% Triton X-100, 0.1% Na-Azide, 0.1% Na-Citrate and 5% normal goat serum. Sections were washed in PBS twice for 5 min each in PBS and incubated with the TUNEL labeling solution. Therefore, two brain sections were simultaneously incubated with 250 μl of TUNEL labeling solution in one well of a 24-well plate for 1 h at 37 °C covered with aluminium foil. Sections were once washed with PBS containing Hoechst for 10 min at room temperature to stain nuclei. Before mounting sections in AquaMount (DAKO), they were once washed in PBS for 10 min at room temperature. TUNEL-positive (TUNEL+) cells were counted and by normalizing the quantified values to the area of the granule cell layer of the according OB values were expressed as densities (cells/mm2).
Apoptotic cells were additionally labeled with a Casp3 antibody (Asp175, rabbit) from Cell Signaling Technology. Casp3+ cells were counted and by normalizing the quantified values to the area of the granule cell layer of the according OB values were expressed as densities (cells/mm2). P-values are calculated by the Mann–Whitney U test.
EdU and BrdU injections
Cell proliferation in vivo was analyzed by using the Click-iT EdU Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions or injection of BrdU followed by antibody staining with an anti-BrdU antibody (AbDSerotec). For both the procedures, animals were injected with 50 mg/kg EdU or BrdU on 3 consecutive days and killed after the indicated time. For BrdU staining, sections were incubated with 2 M HCl in PBST (PBS+0.3% Triton) for 25 min at 37 °C for denaturation of the DNA, neutralized with 0.1 M sodium tetraborate (pH 8.5) for 7 min at room temperature and stained with an anti-BrdU antibody.
Virus production and stereotactic injections
For virus production, TLA-HEK293T (Open Biosystems, Huntsville, AL, USA) cells were transfected with the retroviral packaging vector PCL-ECO and the according transfer plasmid using Turbofect (Fermentas) following the manufacturer's instructions. The culture media were collected 24, 48 and 56 h after transfection, pooled and cleared from remaining cells by centrifuging 10 min at 1200 r.p.m. at 4 °C. To concentrate the virus, the supernatant was centrifuged at 20 000 r.p.m. at 4 °C for 90 min, and the pellet was resolved in DMEM without any additives.
For transduction of neural progenitors in vivo, mice were deeply anaesthetized by intraperitoneal injection of 0.017 ml of 2.5% Avertin (100% stock solution: 10 g 2, 2, 2-Tribromoethanolin 10 ml tert-Amylalcohol) per gram of body weight and waited until the mice did not respond to pinching with tweezers. In all, 2.5 μl of retrovirus were manually injected (0.5 μl per minute) in the SVZ (stereotactic coordinates: AP+ 0.6, ML+/−0.84, DV—2.5) of each hemisphere of the brain using a stereotactic frame (David Kopf Instruments, Tujunga, CA, USA). The injection of the virus vectors was executed as described previously.24 Mice were killed 8 days after surgery by perfusion, and the dissected brains were used for immunohistochemistry.
Microarray analysis
For comparing gene expression patterns of the PRMS and DRMS, freshly dissected brains were cut with a vibratome (Leica, Wetzlar, Germany) in 100 μm sections, and the corresponding brain regions were isolated using a binocular microscope and shock frozen immediately after isolation. For the PRMS and the DRMS of respectively wild-type and Trim32 KO mice, the tissue extracted from three individual mice was pooled. RNA was isolated using the RNeasy Kit (Qiagen) according to the manufacturer's instructions. mRNA quantity and purity were determined by using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Additional quality check was performed by the Agilent Bioanalyzer (Agilent, Santa Clara, CA, USA). Gene expression profiles were generated at the Genomics Division of the Integrated Functional Genomics (Core Unit of the Interdisciplinary Center for Clinical Research (IZKF) at the Medical Faculty of the University of Münster) using the MouseGene 1.0ST arrays according to manufacturer's recommendations (Affymetrix, Santa Clara, CA, USA). Digitalized images were processed using the Affymetrix Expression Console (PLIER summarization method, Sketch—quantile normalization method and PM-GCBG background analysis method) to obtain intensity files.
Microarray data analysis
For the CoA, we used the appropriate software implemented in the Tigr MEV platform.25 Data analysis were performed on the 5000 transcripts with the highest S.D. The number for neighbors for KNN imputation was equal to 10. GO analysis was performed using the appropriate software implemented in the Database for Annotation, Visualization and Integrated Discovery (DAVID).26 P-values are calculated by the Fisher's exact test.
Acknowledgments
We thank Dr. Manuel Grez (Frankfurt) for plasmids, Kensaku Mori (RIKEN Brain Science Institute, Japan) for providing the Nestin-GFP mouse as well as Anna-Lena Benker, Inga Werthschulte and Thea van Wüllen for excellent technical assistance and Martin Stehling for performing the FACS analysis. A-LH, MASP and LB are supported by the Münster Graduate Program for Cell Dynamics and Disease (CEDAD), TP is supported by a Marie Curie Fellowship. JCS's lab is supported by the German Research Foundation (DFG: Emmy Noether Program, SCHW1392/2-1; SFB629 and SPP1356, SCHW1392/4-1), Kompetenznetzwerk Stammzellforschung NRW, German-Israeli Foundation (GIF) for Scientific Research and Development (G-2226-2034.1/2009), Schram-Stiftung (T287/21795/2011) and Else Kröner-Fresenius-Stiftung (2011_A94). Furthermore, this work was supported by the fund ‘Innovative Medical Research' of the University of Münster Medical School (SC120901 and SC411003) and the Interdisciplinary Center for Clinical Research (IZKF) Münster (SchwJ3/001/11).
Glossary
- BrdU
bromodeoxyuridine
- CoA
correspondence analysis
- Casp3
cleaved caspase-3
- DCX
doublecortin
- DRMS
distal migratory stream
- EdU
5-ethynyl-2'-deoxyuridine
- EGFP
enhanced green fluorescent protein
- GABA
gamma-amino butyric acid
- GFAP
glial fibrillary acid protein
- GFP
green fluorescent protein
- GCL
granular cell layer
- GL
glomerular cell layer
- GO
Gene Ontology
- GST π
glutathione S-transferase π
- IRES
internal ribosome entry site
- NeuN
neuronal nuclei
- mRNA
messenger ribonucleic acid
- NSC
neural stem cell
- PRMS
proximal rostral migratory stream
- OB
olfactory bulb
- RMS
rostral migratory stream
- RTq-PCR
reverse transcription-quantitative PCR
- shRNA
short hairpin RNA
- SVZ
subventricular zone
- Tuj1
neuronal class III β-tubulin
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labelling
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Author contributions
A-LH, MASP and EB: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of the manuscript; MMAW and LB: conception and design, collection and assembly of data, data analysis and interpretation, approval of manuscript; LL: conception and design, collection and assembly of data, data analysis and interpretation; TP: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; JCS: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of the manuscript.
Edited by A Verkhratsky
Supplementary Material
References
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillje AL, Worlitzer MM, Palm T, Schwamborn JC. Neural stem cells maintain their stemness through protein kinase C zeta-mediated inhibition of TRIM32. Stem Cells. 2011;29:1437–1447. doi: 10.1002/stem.687. [DOI] [PubMed] [Google Scholar]
- Nicklas S, et al. TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS One. 2012;7:e30445. doi: 10.1371/journal.pone.0030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Saito H, Suzuki M, Mori K. Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport. 2000;11:1991–1996. doi: 10.1097/00001756-200006260-00037. [DOI] [PubMed] [Google Scholar]
- Sato T, Okumura F, Iguchi A, Ariga T, Hatakeyama S. TRIM32 promotes retinoic acid receptor alpha-mediated differentiation in human promyelogenous leukemic cell line HL60. Biochem Biophys Res Commun. 2012;417:594–600. doi: 10.1016/j.bbrc.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Sato T, Okumura F, Kano S, Kondo T, Ariga T, Hatakeyama S. TRIM32 promotes neural differentiation through retinoic acid receptor-mediated transcription. J Cell Sci. 2011;124:3492–3502. doi: 10.1242/jcs.088799. [DOI] [PubMed] [Google Scholar]
- Jacobs S, Lie DC, DeCicco KL, Shi Y, DeLuca LM, Gage FH, et al. Retinoic acid is required early during adult neurogenesis in the dentate gyrus. Proc Natl Acad Sci USA. 2006;103:3902–3907. doi: 10.1073/pnas.0511294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa N, Cedile O, Pollet-Villard X, Bagnis C, Durbec P, Boucraut J. RAE-1 is expressed in the adult subventricular zone and controls cell proliferation of neurospheres. Glia. 2011;59:35–44. doi: 10.1002/glia.21074. [DOI] [PubMed] [Google Scholar]
- McCaffery P, Zhang J, Crandall JE. Retinoic acid signaling and function in the adult hippocampus. J Neurobiol. 2006;66:780–791. doi: 10.1002/neu.20237. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Harding LS, Bogerd HP, Cullen BR. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu YS, Lee Y, Lee KW, Hwang CY, Maeng JS, Kim JH, et al. TRIM32 protein sensitizes cells to tumor necrosis factor (TNFalpha)-induced apoptosis via its RING domain-dependent E3 ligase activity against X-linked inhibitor of apoptosis (XIAP) J Biol Chem. 2011;286:25729–25738. doi: 10.1074/jbc.M111.241893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano S, Miyajima N, Fukuda S, Hatakeyama S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008;68:5572–5580. doi: 10.1158/0008-5472.CAN-07-6231. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Zhao C, Gage FH. Retrovirus-mediated single-cell gene knockout technique in adult newborn neurons in vivo. Nat Protoc. 2006;1:3049–3055. doi: 10.1038/nprot.2006.473. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.