Abstract
The Saccharomyces cerevisiae Spt16/Cdc68, Pob3, and Nhp6 proteins (SPN or yFACT) bind to and alter nucleosomes in vitro, providing a potential explanation for their importance in both transcription and replication in vivo. We show that nucleosomes bound by either Nhp6 alone or the yFACT complex remain largely intact and immobile but are significantly reorganized, as indicated by changes in the pattern of sensitivity to DNase I and enhanced digestion by some restriction endonucleases. In contrast, yFACT enhanced access to exonuclease III only at very high levels of enzyme, suggesting that the DNA near the entry and exit sites of nucleosomes is largely unperturbed and that the position of the histone octamers relative to the DNA is not altered during reorganization. DNase I sensitivity was enhanced at sites clustered near the center of the nucleosomal DNA, away from the entry and exit points, and the pattern of nuclease sensitivity was only mildly affected by the configuration of linker extensions, further indicating that linkers play only a minor role in the reorganization of nucleosomes by yFACT. The DNA in contact with H2A-H2B dimers is therefore the region whose nuclease sensitivity was the least affected by yFACT reorganization. The most dramatic changes in nucleosome structure occurred when Spt16-Pob3 and the HMG box protein Nhp6 were both present, but Nhp6 alone altered DNase I sensitivity at some specific sites, supporting an independent role for this class of proteins in the general management of chromatin properties. yFACT activity does not require ATP hydrolysis and does not alter the position of nucleosomes, indicating that it acts through a mechanism distinct from chromatin remodeling. The results presented here suggest instead that yFACT promotes polymerase progression by reorganizing nucleosome cores into a less inhibitory conformation in which the properties of DNA sequences near the center of the nucleosomes are altered.
The Saccharomyces cerevisiae Spt16, Pob3, and Nhp6 proteins cooperate in vivo to promote normal DNA replication and RNA transcription (4, 13, 19, 30). They also cooperate in vitro to bind to and alter the properties of nucleosomes (5, 20). Unlike nucleosome remodeling, which involves moving histone octamers relative to DNA (29), this reorganization of nucleosomes does not require hydrolysis of ATP. Spt16, Pob3, and Nhp6 (SPN or yFACT) (4) represent a family of factors that are highly conserved among eukaryotes and includes the human FACT (facilitates chromatin transcription) and the frog DUF (DNA unwinding factor) complexes (15, 17). Overall, the properties of these factors suggest that they are necessary for both DNA replication and transcription because they make nucleosomes less restrictive to the passage of polymerases along chromatin templates (4, 16). This suggests the adjusted definition of “facilitates chromatin transactions” for the FACT family, and we refer to combinations of the yeast proteins Spt16, Pob3, and Nhp6 as yeast FACT (yFACT).
Human FACT allows RNA polymerase II to transcribe through templates incorporated into nucleosomes, which otherwise block progression in vitro (16, 17). We have shown previously that yFACT enhances the sensitivity of specific sites within nucleosomal DNA to DNase I (5), indicating that the structure of nucleosomes is altered by yFACT. Human FACT has been shown to enhance the displacement of one H2A-H2B dimer from a nucleosome (1), and the resulting “hexasomes” have been shown to be less inhibitory than normal nucleosomes to the passage of RNA polymerase II in vitro (9). Genetic evidence suggests that while yFACT may promote partial disruption of nucleosomal components under some conditions, it usually restores nucleosomes to their normal composition afterwards (6). Consistent with this, human FACT has also been shown to be able to promote deposition of core histones onto DNA (1). Current data therefore suggest that FACT family members reorganize nucleosomes into a form that is less inhibitory to polymerases and more susceptible to the loss of H2A-H2B dimers but in which nucleosomal components are usually tethered together and subsequently reassembled. Here, we report further analysis of the nature of the reorganized nucleosome form by examining changes in accessibility of the DNA in yFACT-nucleosome complexes to different types of nucleases.
The human FACT subunit SSRP1 contains a single DNA binding motif found in members of the HMG box family of DNA binding proteins, but this function is supplied in yFACT by the separate subunit Nhp6 (3, 4, 20, 26). Spt16 (also known as Cdc68) and Pob3 form a stable heterodimer called SP or CP (3, 27) that associates only weakly with Nhp6 (2, 5). Nhp6 can bind to nucleosomes and effect some changes in structure, but this requires the simultaneous action of about 10 Nhp6 monomers per nucleosome (20). It is not yet clear how the HMG box motif functions in nucleosome reorganization by FACT, but these results suggest that Nhp6 supports the first step in a two-step mechanism. In this view, binding by multiple Nhp6 molecules, either simultaneously or during a short window of time, induces a change in a nucleosome that converts it to a substrate for Spt16-Pob3 or perhaps for other chromatin-modifying factors.
The two-step reorganization of nucleosomes by yFACT components has been revealed principally by examining changes in sensitivity to DNase I at specific sites in nucleosomes assembled with the positioning sequence derived from the sea urchin 5S ribosomal DNA (rDNA) gene (5, 20, 23). Here, we generate a more comprehensive map of these effects to ask which regions of the nucleosome are most affected by reorganization and whether changes can be promoted by Nhp6 alone or instead require the yFACT complex. We also extended the analysis to a different nucleosome positioning sequence to determine which effects are unique to the 5S rDNA sequence and which are likely to be more general indications of the action of yFACT on all nucleosomes. Finally, we used additional nucleases to refine the picture of the structure of reorganized nucleosomes. The results show that nucleosome cores remain intact and do not move in response to yFACT but attain a degree of disruption near the center of the nucleosomal DNA sequence that is sufficient to allow access by a restriction endonuclease, whereas the DNA at the entry and exit points of the nucleosome remains relatively unaffected. Therefore, while FACT appears to enhance the ability of polymerases to penetrate nucleosomes, reorganization by yFACT seems to principally affect the accessibility of nucleosomal DNA to nucleases at sites distant from the entry and exit points.
MATERIALS AND METHODS
Assembly of nucleosomes.
DNA molecules containing the 146-bp nucleosome positioning sequence from the sea urchin 5S rDNA gene (23) or the very stable 147-bp “601” sequence described by the Widom group (10, 24) flanked by various amounts of “linker” DNA (DNA extending beyond the minimal positioning sequence) were amplified by PCR with appropriate primers (sequences available upon request). To label the top strand of the 5S nucleosomes (see Fig. 1, 5S top), a PCR product was digested with EcoRI, treated with phosphatase, labeled with [γ-32P]ATP and polynucleotide kinase and then digested with EcoRV, producing a 151-nucleotide top strand labeled at the 5′ end and a 147-nucleotide unlabeled bottom strand. The left end extends past the expected 146-bp histone octamer footprint by 1 bp plus the 4-nucleotide single-strand overhang resulting from EcoRI digestion, and the last two bases of the 5S rDNA sequence are changed (CA to AT) to create the EcoRV site. The bottom strand (5S bottom in Fig. 1) was labeled with a similar strategy except that the EcoRI site was placed on the right side of the positioning sequence and the first three bases on the left end were altered (CAA to ATC) to create the EcoRV site. The same restriction enzymes were used for the products with linkers, but their sites were placed farther from the 5S rDNA so that the ends extended 15 bp from the histone octamer footprint on both sides, plus the 4-nucleotide single-strand extension on the EcoRI side (Fig. 1, + linkers). The 147-bp 601 sequence (10, 24) was flanked by EcoRV and PvuII sites and labeled with a similar strategy to produce 153-bp blunt-ended fragments labeled on either strand (601 top and bottom in Fig. 1). Sequences are numbered so that digestion between the first and second nucleotides counting from the left ends of the 146- or 147-bp positioning sequence gives a product of 1 nucleotide. The products of digestion of the top strand are given the prefix T, and those of the bottom strand are given the prefix B (see T1 and B1 in Fig. 1).
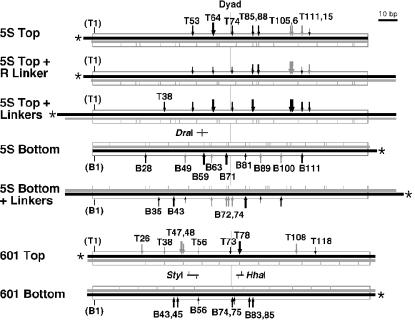
FIG. 1.
Nucleosomal DNAs and the sites digested by DNase I. The DNAs used in these studies are shown schematically, with the 146-bp 5S rDNA (23) and 147-bp 601 (10, 24) sequences represented as boxes, the labeled 5′ end indicated by an asterisk, and the labeled (black) and unlabeled (gray) strands drawn to scale. The sites of preferred DNase I digestion in free nucleosomes are indicated as vertical gray lines. Black arrows indicate sites where DNase I digestion is enhanced by yFACT, and gray arrows indicate sites at least partially enhanced by Nhp6 alone. The size of the arrow is proportional to the strength of the enhancement. Numbers indicate the position of the digestion site in nucleotides, counting from the left end of the nucleosome positioning sequence, with T and B indicating the top and bottom strands, respectively (the presence of linkers therefore does not alter the numbering). Sites common to all of the 5S top types with different linker sizes are labeled only in the 5S top line, and those common to both 5S bottom types are also labeled only in the line without linkers. The position of the DraI site and the sites of digestion in the 5S rDNA sequence (T58 and B58), as well as the sites for StyI (T49 and B55) and HhaI (B77 and T79) in the 601 sequence are indicated.
Labeled DNA was assembled into nucleosomes by slow dialysis from a high-salt solution in the presence of histone octamers, essentially as described previously (5, 12, 28). Nucleosomes were then purified by sedimentation through a 5 to 30% sucrose gradient. Histone octamers prepared from chicken erythrocytes were a generous gift from V. Graziano and V. Ramakrishnan (8). Recombinant histone octamers with yeast histone sequences were prepared as described previously (20, 28). Nucleosomes prepared with histones from chicken erythrocytes may retain some modifications and have been found to have subtle quantitative differences with fully recombinant nucleosomes in binding assays (20), but we have not observed any differences in patterns of DNase I digestion or other assays described here (data not shown).
Binding and DNase I assays.
Spt16-Pob3 was purified as a heterodimer from Saccharomyces cerevisiae cells overexpressing both proteins as described previously (27). Binding reactions were incubated for 10 min at 30°C and typically contained 5 to 10 fmol of DNA or nucleosomes in a volume of 10 μl with final concentrations of 1 mM disodium EDTA, 20 mM HEPES (pH 7.6), 120 mM NaCl, 0.2 mM 2-mercaptoethanol, 0.9 mg of human serum albumin (Sigma) per ml, 12% (wt/vol) sucrose, 2% (wt/vol) glycerol, and various amounts of Spt16-Pob3 and Nhp6 as indicated in each experiment. Spt16-Pob3 displays half-maximal effects in these assays at about 7 nM, and Nhp6 displays half-maximal effects in these assays at about 460 nM (20), and they were typically used at 200 nM for Spt16-Pob3 and 2 to 10 μM for Nhp6. Estimates of the concentrations of these proteins in cells range from 5 to 25 μM for Spt16-Pob3 and 4 to 30 μM for Nhp6 (3, 7, 18, 27).
For DNase I digestions, 2 μl of a solution containing bovine pancreatic DNase I (Worthington; 1× is 100 ng per reaction), 60 mM MgCl2, 0.5 mg of human serum albumin per ml, and 300 mM Tris-HCl (pH 7.5) was added to 10 μl of a binding reaction and incubated for 3 min at 30°C. The reaction was stopped by adding 50 μl of a solution containing 6 μg of salmon testes DNA (Sigma), 3 mM disodium EDTA, 20 mM Tris-HCl (pH 7.5), and 100 mM NaCl at 0°C. For denaturing polyacrylamide gel electrophoresis (PAGE) analysis, samples were extracted with 150 μl of CHCl3 containing 1/25 volume of isoamyl alcohol, the aqueous phase was precipitated with 2.5 volumes of 95% ethanol, and the DNA was recovered by centrifugation. DNA was dissolved in 76% formamide-16 mM disodium EDTA-0.04% each bromophenol blue and xylene cyanol FF dyes. Samples were incubated for several minutes at 65°C and then subjected to electrophoresis through 0.4-mm gels consisting of 8% polyacrylamide (0.4% bisacrylamide), 7 M urea, and 70% Tris-borate-EDTA (21). Gels were fixed in 12% methanol and 10% acetic acid, dried, and then autoradiographed.
Exonuclease III (New England Biolabs) digestions were performed as for DNase I except that various amounts of exonuclease III were added and incubated at 30°C for 10 min before the reaction was terminated and samples were prepared for denaturing PAGE as above. DraI, HhaI, and StyI (New England Biolabs) were used in the same way except the NaCl was omitted from the binding reaction and replaced with 1× reaction buffer supplied with the enzyme, and 20 U of each enzyme was added per 100 fmol of DNA. Reactions were terminated as above, and digestion was assessed by denaturing PAGE followed by phosphorimaging and analysis of the products with ImageQuant software (Molecular Dynamics).
To recover intact nucleosomes, 32 μg of salmon testes DNA (Sigma) was added to each 12-μl sample after the DNase I digestion, and then native electrophoresis was performed with gels containing 3.8% polyacrylamide (0.1% bisacrylamide) and 25% Tris-borate-EDTA (21). Nucleosomes were detected by autoradiography, regions of interest were excised, and the DNA was recovered from the gel fragments by overnight extraction at 37°C with 0.2% sodium dodecyl sulfate in 20 mM Tris-HCl (pH 7.5), followed by precipitation with ethanol and PAGE as described above.
RESULTS
Linkers are not required for reorganization of nucleosomes by yFACT.
We previously showed that while yFACT is similar in mass to a nucleosome and binds with high affinity in a 1:1 complex with nucleosomes, it does not create a footprint in the DNase I digestion pattern that would indicate that it blocks access to specific sites in nucleosomal DNA (5, 20). Instead, yFACT causes enhanced digestion at a subset of sites, indicating that these sites either are more accessible or are in a configuration that makes them better substrates for DNase I. The affected sites were not symmetrical about the dyad axis of the nucleosome; the enhanced digestion sites were instead clustered towards the end of the nucleosome that had a protruding linker sequence (5) (summarized in Fig. 1; 5S Top + R Linker). We therefore considered a model for nucleosome reorganization in which Nhp6 binds to the DNA near the entry and exit points of the nucleosome, creating a bend in the DNA that disturbs local histone-DNA contacts. In this model, linker DNA attracts Nhp6 and initiates destabilization that is subsequently exploited by Spt16-Pob3, but the effects are localized and therefore occur preferentially near the linker. To test this model for linker DNA in the reorganization of nucleosomes by yFACT, we reconstituted nucleosomes without linkers or with 15-bp linkers extending from both ends of the 5S rDNA positioning sequence (Fig. 1).
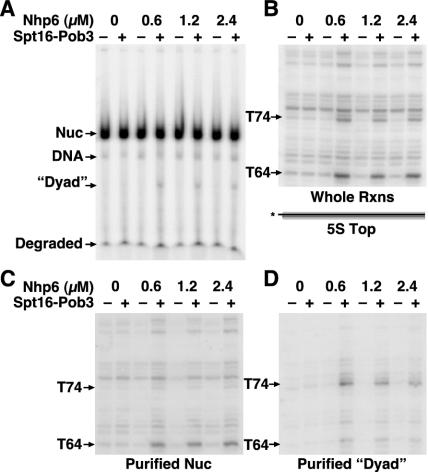
FIG. 5.
Nucleosomes remain intact after reorganization by yFACT and digestion by DNase I except when digested at site T74. (A) 5S rDNA nucleosomes (5S top in Fig. 1) reconstituted with yeast histone octamers were mixed with the amount of Nhp6 indicated with or without 0.2 μM Spt16-Pob3 as indicated and then treated with 1× DNase I as described in Materials and Methods; 32 μg of genomic competitor DNA was added to bind the Nhp6 (which disrupts all complexes formed with DNA or nucleosomes), and the samples were subjected to native PAGE and autoradiography. The positions of free nucleosomes (Nuc), intact free DNA, and small degraded fragments are indicated. “Dyad” denotes a discrete form observed only in the presence of yFACT that migrated on the native gel at a position consistent with a free double-stranded DNA molecule of about 74 bp. (B) Aliquots of the samples shown in panel A were reserved before addition of competitor and analyzed by denaturing PAGE without first performing native PAGE to purify the nucleosomes. Products resulting from digestion at sites T74 and T64 are indicated. (C) DNA was recovered from the band marked Nuc in panel A and extracted, and the purified nucleosomes were subjected to denaturing PAGE. (D) DNA was recovered from the region marked dyad in panel A and subjected to denaturing PAGE. Panels B to D show the region containing T64 and T74 from the same exposure of the same denaturing gel.
As shown in Fig. 2, nucleosomes without linkers adopted a single rotational phase, leading to a pattern of DNase I digestion products that peaked approximately every 10 bp. In contrast, the same DNA free of histones produced a more random pattern. Addition of yFACT had no effect on the pattern obtained with naked DNA but enhanced DNase I digestion at specific sites within nucleosomes. For example, digestion was more common in the presence of yFACT at T64 (the nomenclature is described in Materials and Methods and in the legend to Fig. 1) and also at other sites indicated with circles in Fig. 2 (also see reference 20). Notably, most of the sites in the 5S rDNA sequence that were affected by yFACT were the same in the nucleosomes without linkers as in the nucleosome with a linker at the right side (Fig. 1) (5). For example, digestion of the nucleosome with a linker was partially enhanced by Nhp6 alone in the region near T105 to T106, and this effect was then strengthened by further addition of Spt16-Pob3. The same pattern was observed in Fig. 2 with a nucleosome lacking linkers. A linker is therefore not necessary for reorganization of nucleosomes by Nhp6 or yFACT. Consistent with this, linkers also did not enhance the affinity of Nhp6 for nucleosomes (20).
FIG. 2.
Products of DNase I digestion of nucleosomes without linkers. DNA or nucleosomes (5S Top as in Fig. 1; presented schematically at the bottom of the figure) reconstituted with histone octamers from chicken erythrocytes were mixed with 10 μM Nhp6 and 0.2 μM Spt16-Pob3 as indicated and then treated with 1×, 2×, 4×, 8×, or 16× DNase I as described in Materials and Methods. Products were separated by denaturing PAGE and then detected by autoradiography. Labels indicate the size of the product as described in Fig. 1. Sizes on the left indicate major cut sites observed with free nucleosomes, and those on the right indicate sites enhanced by addition of Nhp6 or yFACT. These sites are also denoted with circles in the center of the figure; open circles indicate regions partly affected by Nhp6 alone, and solid circles indicate effects that depend on yFACT.
Addition of linkers influences but does not determine the DNase I digestion pattern.
To further assess the effect of linkers on the DNase I sensitivity induced by Nhp6 and yFACT, we constructed nucleosomes with the same 5S rDNA sequence but with 15-bp linkers extending from both ends (Fig. 1, +linkers). Figure 3 shows that many of the effects of yFACT were still observed on the top strand at the same sites within the 5S rDNA sequence when linkers were added (summarized in Fig. 1). For example, sensitivity to DNase I was enhanced at T64, T105, and T106 by yFACT (compare Fig. 3A with Fig. 2). While many elements of the pattern of DNase I sensitivity were unaffected by the addition of linkers, the effects of Nhp6 alone were diminished at sites such as T105 and T106 on the top strand as well as at B89 and B100 on the bottom strand (described below). The linkers protrude into the region near the dyad axis (see Fig. 8) and might therefore diminish access to some sites by steric hindrance. This effect should be minimal farther from the linkers, as observed with the retention of the pattern near T105-T106. We conclude that the linkers can interfere with DNase I digestion but they do not determine the pattern of sites whose sensitivity is enhanced by yFACT. Linkers therefore do not appear to have a significant role in the mechanism of nucleosome reorganization by Nhp6 or yFACT.
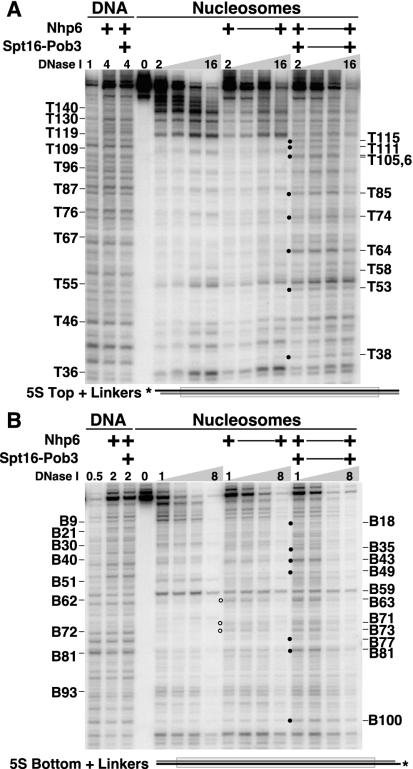
FIG. 3.
Linkers influence but do not dictate the sites of DNase I sensitivity induced by yFACT. Nucleosomes with 15-bp linkers extending from both ends of the nucleosome positioning sequence (Fig. 1, + linkers) were mixed with 10 μM Nhp6 and 0.2 μM Spt16-Pob3 as indicated and then treated with 0.5×, 1×, 2×, 4×, 8×, or 16× DNase I as described in Materials and Methods. Digestion of the top strand (A) and the bottom strand (B) is shown schematically below each panel. Circles indicate enhanced sensitivity induced by Nhp6 (open) or yFACT (solid), and the products are labeled as in Fig. 1, with prominent sites in free nucleosomes labeled on the left and sites whose sensitivity to DNase I is enhanced by Nhp6 or yFACT labeled on the right.
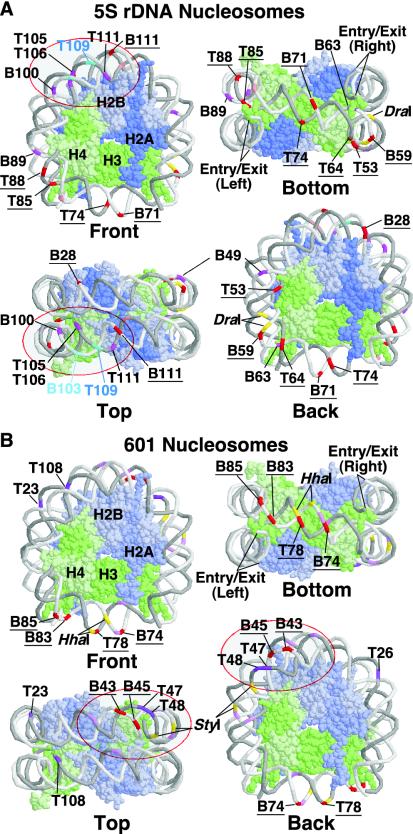
FIG. 8.
Model of the sites affected by Nhp6 and yFACT. The sites at which DNase I digestion is enhanced by Nhp6 or yFACT in nucleosomes without linkers (Fig. 1) are shown mapped to the yeast nucleosome structure (25) rendered in RasMol (22). Panel A summarizes the results with the 5S rDNA sequence, and panel B shows results with the 601 sequence. The DNA strands are shown in backbone format, with the top strand in darker gray and the bottom strand in lighter gray. Histones are shown in space-filling format, with H2A in darker blue, H2B in lighter blue, H3 in darker green, and H4 in lighter green, as indicated in the arbitrarily chosen front views. Sites are labeled as in Fig. 1. Strong enhancement of DNase I sensitivity at sites affected at least partially by Nhp6 alone is indicated in purple on the DNA. Strong enhancement of DNase I sensitivity that requires yFACT is indicated in red on the DNA, and the sites are underlined. Weaker enhancement by yFACT is shown in pink, and the sites are not labeled (see Fig. 1). The top cluster of sites, including the T105 to T115 region described in the text in 5S rDNA nucleosomes, is indicated by a red oval. A similar cluster in the 601 nucleosomes is also indicated; this top cluster is in the lower quadrant of the top view in panel A and the upper quadrantof the top view in panel B. This shows that yFACT affects each nucleosome type asymmetrically; the opposing orientations are due to the random assignment of the strands as top and bottom. The sites at which sensitivity is suppressed by yFACT in the 5S rDNA sequence are indicated in shades of light blue, and the sites of digestion by DraI, StyI, and HhaI are indicated in yellow. The entry and exit points from which linkers would extend are labeled in the bottom view.
yFACT effects on nucleosomes are asymmetrical.
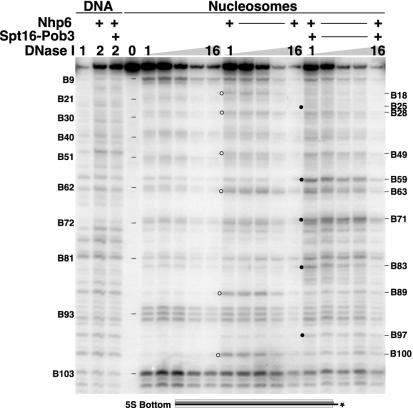
Figure 1 shows that positions on the top strand of the 5S rDNA nucleosomes that are affected by Nhp6 and yFACT are neither randomly distributed nor symmetrical about the dyad axis. This could mean that yFACT reorganizes nucleosomes in an asymmetrical way and affects specific regions of the nucleosome more than others. However, in a perfectly uniform nucleosome structure, the symmetry-related site for a given location on the top strand is on the bottom strand. We therefore examined the bottom strand to see whether the asymmetry noted on the top strand reflected differences that are strand specific or region specific. Figure 4 shows that both Nhp6 and yFACT enhanced DNase I digestion at specific sites on the bottom strand of the 5S rDNA nucleosomes (also see reference 20). On the top strand, 8 of the 11 affected sites displayed enhanced sensitivity only in the presence of yFACT (Fig. 2). Sites such as B59 also required both Spt16-Pob3 and Nhp6, but this pattern was less common on the bottom strand, with 6 of the 11 affected sites being fully enhanced by Nhp6 alone (Fig. 4). Furthermore, the increased sensitivity at two sites (B89 and B100) reverted partially or completely to normal when Spt16-Pob3 was added (Fig. 4) (20). We interpreted this as evidence that Nhp6 at high concentrations promotes the first step of a two-step nucleosome reorganization by yFACT (20). Overall, the major effects on the bottom strand were also asymmetrical relative to the dyad axis and appeared to cluster in the same regions of the nucleosome as the effects on the top strand. This clustering is addressed more directly below (see Fig. 8).
FIG. 4.
DNase I digestion sites on the bottom strand are also enhanced by Nhp6 and yFACT. DNA or nucleosomes labeled on the bottom strand of the 5S rDNA sequence (Fig. 1) reconstituted with yeast histone octamers were mixed with 2 μM Nhp6 and 0.2 μM Spt16-Pob3 as indicated, treated with 1×, 2×, 4×, 8×, or 16× DNase I as described in Materials and Methods, and then processed as for Fig. 2. Circles indicate sites enhanced by Nhp6 alone (open) or by yFACT (solid). Sizes are calculated as in Fig. 1 and are labeled B to indicate the bottom strand. Parallel reactions were performed with 10 μM Nhp6, yielding very similar results (not shown).
As with the top strand, addition of linkers also affected the pattern of DNase I sensitivity on the bottom strand. For example, digestion at B89 was suppressed (see Fig. 3B and 4), and the enhancement at B59 was masked, as this site was yFACT dependent in a nucleosome lacking linkers but prominent even without the addition of yFACT when linkers were present. However, enhanced digestion at B63 was retained with Nhp6 alone and with yFACT. While more complex, the effects of linkers on the bottom strand therefore also indicate that linkers are not a vital component of yFACT reorganization but can influence the strength of DNase I digestion at specific sites.
Reorganized nucleosomes have some features of naked DNA, but only at specific sites.
Incorporation into a nucleosome can either make a site in the 5S rDNA sequence more or less likely to be digested by DNase I (23). For example, digestion at T111 and T115 is common in free DNA but rare in nucleosomes, whereas digestion at T109 is rare in free DNA but common in nucleosomes (Fig. 2). Notably, many of the sites at which DNase I digestion is enhanced by Nhp6 and yFACT are preferred digestion sites in free DNA that are blocked by incorporation of the DNA into nucleosomes (for example, see T85 and B89 in Fig. 2 and 4). Not all preferred sites in free DNA display enhanced sensitivity when yFACT is added; for example, the site at T79 is preferred in free DNA but is not significantly sensitized by yFACT (Fig. 2). yFACT therefore does not appear to convert all regions of nucleosomes to a form that is more like free DNA, but it does have this apparent effect at a subset of sites.
Notably, the change in the pattern of DNase I sensitivity in the T105 to T115 region included a virtually complete loss of sensitivity at T109 (Fig. 2). This effect was remarkably localized, with the pattern being almost unaffected by yFACT at sites T96 and T119, which are adjacent to the T105 to T115 region. The effects in this region were consistent among nucleosomes with various linkers, and nearby sites on the bottom strand (B100 and B111) were also affected (Fig. 4) (20). This region appears to be of special importance in nucleosome reorganization, as discussed further below. Chromatin remodelers tend to convert nucleosomal DNA to the naked DNA configuration at all sites, but yFACT has this effect only in specific regions of the nucleosome, suggesting that reorganization has more localized effects on nucleosome structure.
yFACT alters DNase I digestion within intact nucleosomes.
The ability of human FACT to promote progression of RNA polymerase II through nucleosomes suggests that reorganized nucleosomes might be generally destabilized, particularly at the entry and exit points. However, the pattern of DNase I sensitivity in yFACT-nucleosome complexes is not consistent with complete disruption of the nucleosomes (it is distinct from the pattern obtained with free DNA), nor does the pattern suggest displacement or loosening of the DNA near the entry and exit points, as sensitivity to DNase I was largely unaffected by Nhp6 or yFACT in these regions (Fig. 1 to 4). However, to test directly whether nucleosomes remain intact in the presence of yFACT, we formed yFACT-nucleosome complexes, treated them with DNase I, and then added a large excess of unlabeled genomic DNA to bind the Nhp6 and disrupt all of the complexes. As shown in Fig. 5A, the labeled DNA remained largely in the nucleosomal form after these treatments, indicating that binding by Nhp6 or yFACT and mild DNase I digestion of the complexes resulted in nucleosomes that were unaltered within the resolution of this gel system. This was also true after extended incubations with Nhp6 or yFACT in the presence of 500 mM NaCl, 0.2% NP-40, or 0.2% Triton X-100 (not shown; yFACT-nucleosome complexes remained intact under these conditions, as determined by migration on native gels without the addition of competitor DNA). This assay shows directly that nucleosomal DNA remains largely associated with histones after treatment with Nhp6 or yFACT, demonstrating that nucleosomes are not completely disrupted by the process of reorganization, although we cannot rule out more minor alterations such as partial displacement of H2A-H2B dimers.
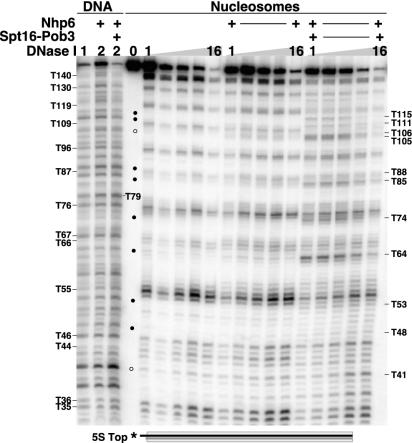
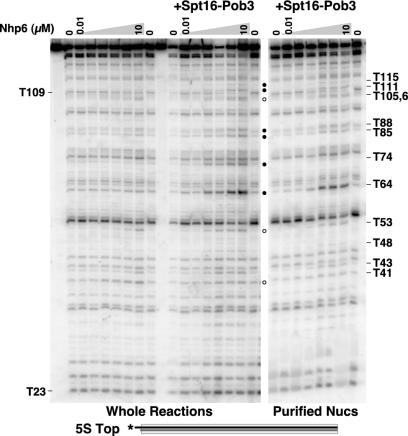
To determine whether the pattern of DNase I digestion observed in the whole reactions described above occurs within intact nucleosomes, we next extracted the DNA from the region of the native gels containing nucleosomes and subjected it to denaturing electrophoresis. Figure 6 shows the pattern obtained with various concentrations of Nhp6 when either whole reactions were analyzed or the nucleosomal form was first purified by native PAGE and then analyzed. The pattern of DNase I sensitivity was generally recapitulated with purified nucleosomes (Fig. 6) (20). For example, the prominent digestion site at T64 induced by yFACT was visible in both the whole reactions and the purified nucleosomes (Fig. 6). The effects of reorganization observed in the region from T105 to T115 described above, including the loss of sensitivity at T109, were also observed in the purified nucleosomes (Fig. 6). We conclude that most of the changes in the DNase I digestion pattern caused by either Nhp6 alone or yFACT reflect events that occur in a nucleosomal context and do not result in disassembly of the nucleosomes.
FIG. 6.
DNase I digestion at most sites occurs in the context of intact nucleosomes. The 5S top nucleosomes (Fig. 1) reconstituted with yeast histone octamers were mixed with 0.2 μM Spt16-Pob3 and increasing amounts of Nhp6 (0, 0.01, 0.1, 0.5, 1, 2, and 10 μM) as indicated; 1× DNase I was then added (except for the first and tenth lanes), and samples were processed as described in Materials and Methods. In the left panel, samples were subjected directly to denaturing PAGE (whole reactions). In the right panel, the same samples were mixed with competitor DNA, and the nucleosomal form was recovered after native PAGE prior to denaturing PAGE as in Fig. 5 (purified nucleosomes). The sizes of the products and enhanced digestion sites are indicated as in Fig. 2.
The notable exception to this conclusion is that the enhanced digestion product at T74 was prominent only in whole reactions and was not found in the purified nucleosomes (Fig. 5 and 6). Examination of the distribution of labeled species after native PAGE in Fig. 5A shows that in reactions in which digestion at T74 was expected (those reactions containing yFACT), an additional band was obtained. This band is marked dyad in the figure to indicate that, compared to size standards, it migrated at about the rate expected for a double-stranded DNA molecule representing digestion at the dyad of the 5S rDNA sequence (not shown). When DNA from this region of the gel was extracted and analyzed by denaturing PAGE, it was found to contain the missing T74 digestion product. Figure 5B shows a portion of the DNase I digestion pattern obtained with whole reactions in this experiment, Fig. 5C shows the pattern obtained with purified nucleosomes in which the T74 product is missing, and Fig. 5D shows the products extracted from the dyad band in which T74 was found to be the dominant product. B71 lies across the minor groove from T74 near the dyad axis (see Fig. 8), and digestion at both B71 and T74 was enhanced by yFACT (Fig. 1, 2, and 4). However, the product of digestion at B71 was found in purified nucleosomes (20). These observations indicate that digestion at T74 in yFACT-nucleosome complexes, unlike digestion at all other sites, leads to dissociation of the nucleosome. The importance of the DNA at the nucleosomal dyad in maintaining the stability of reorganized nucleosomes is addressed further in the Discussion.
A different nucleosome positioning sequence also shows specific yFACT effects.
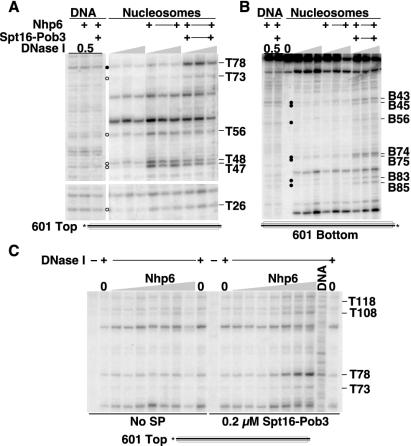
The pattern of digestion by DNase I is influenced by the sequence, the shape, and the accessibility of the DNA. The results discussed above suggest that specific regions of nucleosomes are strongly affected by yFACT, but these results could also indicate that parts of the 5S rDNA sequence are particularly susceptible to yFACT activity in nucleosomes. Furthermore, the 5S rDNA gene is transcribed in vivo by RNA polymerase III, which is not thought to be a target of yFACT activity (9, 14). The role of yFACT in DNA replication suggests that all nucleosomes are likely to be substrates for yFACT and that localized targeting reflects recruitment by other transcription factors, not an inherent affinity for specific nucleosomes. However, to examine the generality of the results obtained with 5S rDNA nucleosomes, we constructed nucleosomes with a different nucleosome positioning DNA sequence, the 147-bp 601 sequence described by the Widom group (10, 24) (Fig. 1). This sequence is unrelated to 5S rDNA and was found to form nucleosomes that were significantly more stable than those formed with 5S rDNA (10, 24). As with 5S rDNA, Nhp6 and yFACT significantly altered the pattern of sensitivity of these 601 nucleosomes to DNase I (Fig. 7), although fewer sites were affected overall, perhaps because of the greater stability of the 601 nucleosomes. For example, digestion at T47 or T48 was enhanced by Nhp6 or yFACT, only yFACT affected digestion at T78, and the region near T108 was partially affected by Nhp6 and this was enhanced by yFACT (the effects were relatively constant with respect to the concentration of DNase I, as shown in Fig. 7A, and were dependent upon the concentration of Nhp6, as shown in Fig. 7C). Effects were also observed on the bottom strand, for example, in the region near B74 (Fig. 7B), but all of these appeared to require yFACT.
FIG. 7.
yFACT and Nhp6 also affect DNase I sensitivity of nucleosomes assembled with a different positioning sequence. Nucleosomes were assembled with yeast histone octamers and the 601 DNA sequence (10, 24) labeled on either the top or bottom strand (Fig. 1), treated with DNase I, and subjected to denaturing PAGE as in Fig. 2. (A) We added 10 μM Nhp6 and 0.2 μM Spt16-Pob3 as indicated to DNA or to nucleosomes labeled on the top strand, and the complexes were treated with 0.5× (DNA), 1×, 2×, or 4× DNase I (nucleosomes) and processed as in Materials and Methods. Circles indicate sites enhanced by Nhp6 alone (open) or by yFACT (solid). (B) As in panel A, except the label was on the bottom strand. (C) DNA or nucleosomes labeled on the top strand were treated as in panel A but with 0, 0.05, 0.2, 0.6, 1.2, 2.0, 10, or 20 μM Nhp6 (triangle) either with or without 0.2 μM Spt16-Pob3.
To analyze the results with the 5S rDNA and 601 nucleosomes, we mapped the affected sites to a three-dimensional model of a nucleosome (Fig. 8). To establish the rotational phase for each DNA, the DNA was threaded through the structure (generated with a symmetrical human α-satellite repeat sequence [11, 25]) until the sites susceptible to DNase I digestion in free nucleosomes occupied positions at the surface of the structure. The models highlight the clustering of affected sites, their asymmetry, and the close correlation of the regions affected in the two different types of nucleosomes. Further analysis of these structural maps is given below in the Discussion.
Exonuclease III shows that yFACT does not slide nucleosomes and has only a small effect on the stability of the entry and exit points.
Human FACT enables RNA polymerase II to progress through nucleosomal templates (1, 16). This could be accomplished by weakening the association between DNA and histones at the entry and exit points so that the polymerase could more readily disrupt these contacts. However, as shown in Fig. 1 and 8, these regions of the nucleosome are largely silent in the DNase I sensitivity enhancement assay with yFACT with two distinct nucleosome positioning sequences. To more directly probe the properties of these regions, we used exonuclease III. This enzyme degrades duplex DNA from the 3′ end of each strand and pauses at sites of bound proteins. With nucleosomes, exonuclease III produces a characteristic 10-bp stuttering pattern as it pauses at histone-DNA contacts and then rapidly degrades 10-bp regions as each contact is broken. We therefore mixed nucleosomes labeled at the 5′ end of one strand (Fig. 1) with Nhp6 or yFACT, tested an aliquot to ensure that normal complexes were formed by electrophoretic mobility shift assay as previously described (5, 20) (data not shown), and then treated the remainder of the reaction with various concentrations of exonuclease III.
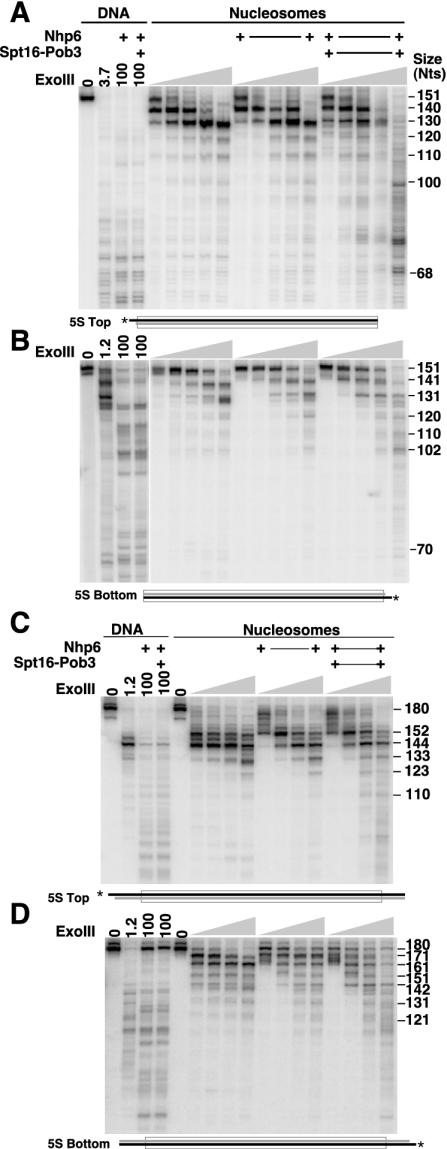
Figures 9A and 9B show that exonuclease III digestion of nucleosomes constructed with the minimal 5S rDNA sequence produced the expected pattern of pausing at the entry and exit points and then about 10 and 20 nucleotides inside the nucleosome. This demonstrates that the nucleosomes have the expected translational phase. Nhp6 alone had a subtle effect on the position of the second pause with the bottom strand labeled (Fig. 9B, 131 nucleotides), but neither Nhp6 nor yFACT had strong effects on the pattern of pausing at low levels of nuclease. At very high levels of exonuclease III, nucleosomes did appear to be significantly destabilized by yFACT. Much of the nucleosomal DNA was either intact or only degraded by 10 or 20 nucleotides at the highest level of nuclease with nucleosomes or nucleosomes with Nhp6. However, the samples with yFACT displayed more severe degradation. As with DNase I, the pattern of products did not match the free DNA pattern, so the mild destabilization does not appear to involve complete disruption of the DNA-histone contacts near the entry and exit points. These results indicate that the DNA near the entry and exit points is largely protected from digestion by exonuclease III even in the presence of yFACT and that the nucleosomal boundaries do not move during reorganization.
FIG. 9.
yFACT affects exonuclease III access to nucleosomes only at high concentrations of nuclease and does not alter the positions of nucleosomal boundaries. DNA or nucleosomes with the 5S rDNA sequence and labeled on the top strand (A, yeast histones; C, chicken histones) or the bottom strand (B and D, chicken histones) were mixed with 10 μM Nhp6 and 0.2 μM Spt16-Pob3 as indicated. Complexes were treated as described in Materials and Methods with 1.2, 3.7, 11, 33, or 100 U (A and B) or 3.7, 11, 33, or 100 U (C and D) of exonuclease III and then subjected to denaturing electrophoresis and autoradiography. The sizes of prominent bands are indicated in nucleotides.
The boundaries of the nucleosomes lacking linkers shown in Fig. 9A and B did not appear to shift when treated with yFACT, but sliding under these conditions would involve a net loss of DNA-histone contacts. To assess whether yFACT induces sliding or causes destabilization when linkers are available, we repeated the exonuclease III assay with nucleosomes with 15-bp linkers at each end (Fig. 9C and D). Examination of the pattern of digestion from each end indicates that these nucleosomes did not have the expected translational phase; instead, the nucleosomes appeared to occupy a position nearer to the left end of the DNA than predicted, leaving a long linker on the right end and a short one on the left. However, this position was not altered by Nhp6 or yFACT, as the locations of the major pauses were not affected. As with nucleosomes lacking linkers, yFACT enhanced the overall digestion at high concentrations of exonuclease III somewhat but did not change the pattern of pause sites. This suggests again that nucleosomes are mildly destabilized by yFACT but the histone cores maintain their position relative to the DNA.
yFACT enhances access to some restriction endonucleases.
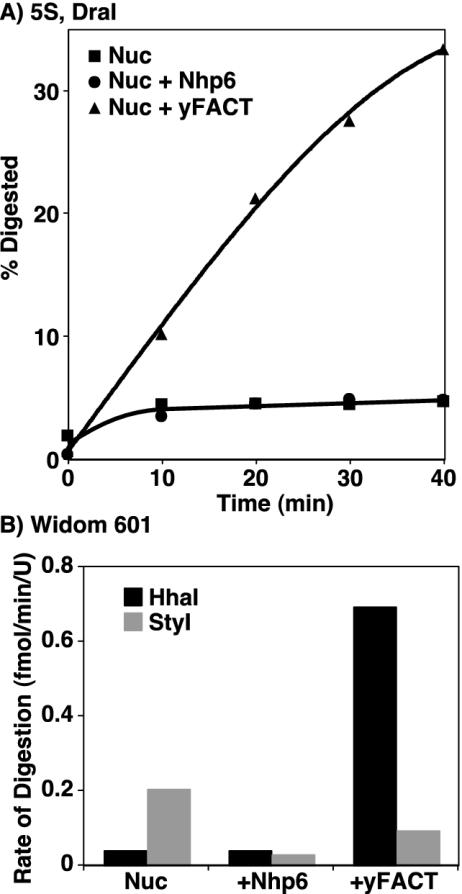
Nucleosomes generally inhibit the access of restriction endonucleases to their recognition sites. ATP-dependent nucleosome remodeling factors overcome this inhibition by moving the histone octamer relative to the DNA, making the recognition site more accessible as it moves to a position outside the nucleosomal core (29). We have demonstrated that nucleosomes remain intact in the presence of yFACT (Fig. 5) and that they maintain their translational phase (Fig. 9). However, as shown in Fig. 10, yFACT enhanced access of DraI to its site near the dyad in nucleosomes assembled with a 5S rDNA sequence (Fig. 1 and 8). Nucleosomes alone or in complexes with either Nhp6 or yFACT were treated with high concentrations of DraI, and samples were withdrawn at various times and examined for digestion as described in Materials and Methods. The nucleosome preparations contained a small amount of free DNA, which was rapidly degraded, followed by slow subsequent digestion of nucleosomes either with or without Nhp6 added (Fig. 10A). In contrast, in the presence of yFACT, nucleosomes were digested at a significant and roughly constant rate, leading to digestion of about one-third of the nucleosomes in a 40-min incubation. This rate of digestion is lower than the rate observed in parallel reactions with free DNA or DNA with Nhp6 or yFACT (not shown; Nhp6 and yFACT inhibited digestion of nonnucleosomal DNA) but was enhanced about 40-fold over the rate observed with nucleosomes with or without Nhp6. These features indicate that yFACT promotes limited but significant access of DraI to its substrate rather than producing a large population of readily digested targets which would instead have caused a single rapid burst of digestion.
FIG. 10.
yFACT enhances digestion of a nucleosome by some restriction endonucleases. (A) Nucleosomes alone (Nuc; 5S bottom in Fig. 1, assembled with yeast histone octamers) or mixed with either 2 μM Nhp6 (Nuc + Nhp6) or both 2 μM Nhp6 and 0.2 μM Spt16-Pob3 (Nuc + yFACT) were treated with DraI for various times, and digestion was assessed after denaturing PAGE by phosphorimaging, as described in Materials and Methods. The percentage of the total DNA digested at each time is shown. (B) Similar experiments were performed with 601 nucleosomes assembled with yeast histone octamers with HhaI or StyI and 10 μM Nhp6. The rate of digestion after the initial burst caused by free DNA in the nucleosome preparations was determined from the slope of the linear portion of curves generated as in panel A. In each case, similar experiments were performed with free DNA which showed some inhibition by Nhp6 and yFACT but always resulted in higher rates of digestion than the reactions with nucleosomes (not shown).
To test the generality of this result, we examined the 601 nucleosome sequence, which has sites for the HhaI and StyI endonucleases (Fig. 1). As with the DraI site in 5S rDNA, the HhaI site in the 601 sequence is near the dyad and in a region whose sensitivity to DNase I is strongly enhanced by yFACT. As shown in Fig. 10B, the constant rate of digestion of 601 nucleosomes by HhaI was also enhanced by yFACT, in this case about 26-fold. The StyI site is also in a region whose DNase I sensitivity is somewhat enhanced by yFACT but nearer to the top cluster of affected sites (Fig. 1 and 8). Sensitivity to StyI digestion was mildly inhibited by Nhp6 or yFACT (Fig. 10B). A more careful examination of the effect of yFACT on restriction endonuclease activity will require the use of substrates containing the same recognition site in various contexts so that differences between enzymes and rotational phases of the sites can be determined. However, these initial data suggest that some regions of nucleosomes are dramatically affected by yFACT-mediated reorganization so that they become accessible to endonucleases, including both DNase I and restriction enzymes, whereas other regions are less dramatically affected.
DISCUSSION
Spt16-Pob3 and Nhp6 represent the sequences found in the human FACT complex, which enables RNA polymerase II to progress through nucleosomal templates (1, 4, 16). The ability to reorganize nucleosomes is likely to explain the importance of FACT family members in chromatin-mediated processes such as transcription and replication. Here we used a variety of probes to learn about the nature of the DNA in nucleosomes that have been reorganized by Nhp6 alone or by yFACT. The results suggest that HMG box family members initiate modest changes in nucleosome structure that are then exploited and enhanced by FACT and that these structural changes are focused on the core of the nucleosomal DNA sequence rather than on the ends near the entry and exit points of the nucleosome. This reorganization of the nucleosome is distinct from the activity of chromatin remodeling factors in that it does not require ATP hydrolysis and does not involve sliding of the histone core relative to the DNA.
The importance of Nhp6 in initiating the reorganization of nucleosomes by yFACT suggested a model in which this DNA-binding protein bent the DNA near the entry and exit points of nucleosomes, thereby weakening the association of these sites with the histone octamer. Consistent with previous results showing that linkers do not enhance the affinity of Nhp6 or Spt16-Pob3 for nucleosomes (20), we show here that linkers are not the principal determinants of the changes in DNase I sensitivity induced by Nhp6 and yFACT. Instead, the presence of linkers subtly alters a pattern of changes that appears to be intrinsic to the sequence used to assemble the nucleosomes. Furthermore, the sites whose DNase I sensitivity was affected were largely within the core of the nucleosome, with few changes observed near the entry and exit regions. With the label placed at the 5′ ends of the DNA strands in nucleosomes without linkers, we were only able to examine the pattern of digestion by DNase I at the end of the nucleosome distal to the label, as the proximal products were too short to resolve. The addition of larger linkers allowed both ends to be examined, but this did not reveal changes in the pattern of DNase I sensitivity in the region proximal to the label (data not shown).
As a more direct test of the stability of the association of the DNA with the histone octamer at the edges of the nucleosome, we examined the ability of an exonuclease to penetrate a nucleosome in the presence of yFACT. Exonuclease III produced similar patterns of pausing whether or not Nhp6 or yFACT was present, and the sensitivity to digestion at low levels of nuclease was not significantly enhanced by these factors. An effect was seen at very high levels of exonuclease, which we interpret as an indication that reorganized nucleosomes are somewhat less stable than usual. However, the principal result is that neither Nhp6 nor yFACT dramatically enhance the ability of an exonuclease to degrade DNA near the entry and exit points of nucleosomes, and yFACT does not cause a shift in the position of this boundary. Both DNase I and exonuclease III therefore indicate that yFACT reorganization does not dramatically expose the DNA near nucleosomal entry and exit points to nuclease digestion.
We also show that nucleosomes can be recovered by native PAGE after treatment with yFACT and that the enhanced digestion observed at most sites occurs within apparently intact nucleosomes. The only notable exception is that the prominent digestion of the top strand of a 5S rDNA nucleosome at site T74 near the dyad axis either occurs in or rapidly leads to disrupted nucleosomes, as this product is not recovered efficiently in the nucleosomal fraction. This could mean that the region near the dyad always plays an important role in maintaining nucleosomal stability but DNase I only cuts this region efficiently in yFACT-reorganized nucleosomes. Alternatively, the region near the dyad could become both more exposed and more important than usual for keeping the nucleosome intact in the yFACT-reorganized form.
Consistent with increased exposure of the dyad region in reorganized nucleosomes, we found that the restriction endonucleases DraI and HhaI were also able to access sites in this region of two distinct types of nucleosome in the presence of yFACT. This effect was not universal, as StyI digestion at a site farther from the dyad was not enhanced by yFACT (Fig. 10B). This suggests that restriction endonucleases will be useful probes of the structural changes induced by yFACT in different regions of nucleosomes. The enhanced digestion by DraI and HhaI occurred at a constant rate over a long time period rather than in a fast initial burst. At one extreme, this could mean that reorganization by yFACT switches the nucleosome frequently between at least two states, one refractory to digestion and the other highly accessible. At the other extreme, yFACT could produce a single reorganized state in which the DraI site is only infrequently susceptible to cleavage, leading to slow digestion. Both possibilities are attractive for energetic reasons: unlike nucleosome remodeling, yFACT reorganization does not require ATP hydrolysis and therefore is likely either to be rapidly reversible or to involve the formation of an alternative but energetically similar nucleosomal form. The slow but constant rate of digestion by restriction endonucleases may therefore be a useful tool both for examining the structure of reorganized nucleosomes and for investigating the energetics of their formation.
Mapping of the sites whose nuclease sensitivity was most significantly affected by Nhp6 and yFACT reveals clustering that suggests two regions of nucleosomes that are altered by reorganization. The first region is near the top of the nucleosome, opposite from the entry and exit points (red ovals in Fig. 8). This group includes many of the sites that were partially affected by Nhp6 alone (marked in purple in Fig. 8), and the only site that was dramatically protected by yFACT (T109 in 5S rDNA nucleosomes, although the adjacent B103 was also somewhat protected; Fig. 3 and not shown). The similarity of the patterns with two different nucleosomes suggests that this region reveals a general feature of reorganized nucleosomes. Notably, each type of nucleosome displayed asymmetry, as one side of each nucleosome was more strongly affected than the region opposing it on the other side of the dyad axis (see the top views in Fig. 8 and the accompanying legend). Human FACT appeared to displace only one H2A-H2B dimer (1); perhaps the choice of site of attack by yFACT is not random but instead reflects a preference for one orientation in each type of nucleosome influenced in some way by the asymmetrical DNA sequence.
As noted above, in the presence of yFACT, the top cluster region of 5S rDNA nucleosomes displayed DNase I sensitivity more similar to free DNA than to nucleosomes, but the effect was very localized. If this region is displaced from the histone octamer, the DNA must be sharply bent, as little disturbance was detected at flanking sites. Alternatively, the changes are consistent with a local change in the rotational phase of the DNA that brings the enhanced sites to the surface and the occluded sites away from the surface. Once again, this effect would have to be quite localized and would be expected to be associated with significant kinking of the DNA at the transition points between this region and the unaffected flanks. It is not clear what feature of this region makes it more likely to be impacted by Nhp6 or yFACT, nor is it clear why the effect is so asymmetrical. However, we note that the effect of occlusion at T109 is almost complete, suggesting that virtually all nucleosomes in the population are affected at least in this region by yFACT-mediated reorganization.
The second region in which DNase I sensitivity was enhanced by yFACT in both 5S rDNA and 601 nucleosomes is the swath along the bottom of the nucleosomes that includes the dyad and 10 to 20 bp on either side of it (Fig. 8, bottom views). The degree of symmetry and the length of the affected regions differ between the two nucleosomes, but as with destabilization of the nucleosomes associated with digestion at T74 in the 5S rDNA nucleosomes (Fig. 5), this again points to particularly significant changes near the dyad in reorganized nucleosomes. Effects in the dyad region require yFACT (underlined sites and sites in red in Fig. 8), indicating that they represent the second step in the two-step reorganization of nucleosomes.
If changes in DNase I sensitivity were caused principally by displacement of an H2A-H2B dimer (1, 17), the region underlying that dimer should be significantly exposed to DNase I digestion, and no pauses in this area should be observed with exonuclease III. As shown in Fig. 8, the sequences whose DNase I sensitivity was affected near the dyad were associated with the (H3-H4)2 tetramer, and the sequences affected at the top of the nucleosome were associated with the interface between this tetramer and an H2A-H2B dimer. H2A-H2B dimers underlie about 40 bp of DNA extending into the nucleosome from each of the entry and exit points, regions whose sensitivity to DNase I and exonuclease III was largely unaffected by yFACT. We cannot conclude that reorganization does not involve displacement of H2A-H2B dimers, but if so, the contacts are either replaced with motifs within yFACT or access to the DNA in these regions is occluded by yFACT in the reorganized form, as neither DNase I nor exonuclease III sensitivity was significantly enhanced. Either substitution or occlusion models are consistent with recent evidence that human FACT can bind to (H3-H4)2 (1). Overall, however, the data presented here suggest that the DNA in contact with the (H3-H4)2 tetramer is the primary site of changes in nuclease sensitivity in response to yFACT binding.
We propose that the Nhp6-induced changes shown in Fig. 8 correspond to the first step in the reorganization of the nucleosome, and this step may involve a localized change in the rotational phase of the DNA in the T100 to T115 region. In the second step, Spt16-Pob3 reinforces this change and also alters the accessibility and importance of the region near the dyad without moving the histone core with respect to the DNA. This model leaves several important questions unresolved. For example, how could Nhp6 rotate DNA, and how does Nhp6 action promote the binding and function of Spt16-Pob3? What is the configuration of the histones, particularly H2A-H2B dimers, in the yFACT-nucleosome complex? How is binding energy alone utilized to effect the reorganization of a complex as stable as a nucleosome? Most significantly, how do these changes in the core of a nucleosome promote progression of polymerases? The results reported here provide clues concerning the structure of the DNA in reorganized nucleosomes and the mechanism of FACT activity which will promote the search for answers to these questions.
Acknowledgments
This work was supported by grant GM-64649 from the National Institutes of Health to T.F.
We thank V. Ramakrishnan and V. Graziano for supplying chicken erythrocyte histone octamers, R. Johnson for Nhp6 expression plasmids, J. Widom for supplying the 601 positioning sequence, J. Wittmeyer and B. Cairns for plasmids expressing yeast histones, and J. Wittmeyer, B. Cairns, and D. Stillman for discussions, advice, and comments on these experiments and the manuscript.
REFERENCES
- 1.Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090-1093. [DOI] [PubMed] [Google Scholar]
- 2.Brewster, N. K., G. C. Johnston, and R. A. Singer. 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21:3491-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewster, N. K., G. C. Johnston, and R. A. Singer. 1998. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem. 273:21972-21979. [DOI] [PubMed] [Google Scholar]
- 4.Formosa, T. 2002. Changing the DNA landscape: putting a SPN on chromatin. Curr. Top. Microbiol. Immunol. 274:171-201. [DOI] [PubMed]
- 5.Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu, and D. J. Stillman. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Formosa, T., S. Ruone, M. D. Adams, A. E. Olsen, P. Eriksson, Y. Yu, A. R. Rhoades, P. D. Kaufman, and D. J. Stillman. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway. Polymerase passage may degrade chromatin structure. Genetics 162:1557-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 8.Graziano, V., S. E. Gerchman, and V. Ramakrishnan. 1988. Reconstitution of chromatin higher-order structure from histone H5 and depleted chromatin. J. Mol. Biol. 203:997-1007. [DOI] [PubMed] [Google Scholar]
- 9.Kireeva, M. L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V. M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9:541-552. [DOI] [PubMed] [Google Scholar]
- 10.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 11.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 12.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304:3-19. [DOI] [PubMed] [Google Scholar]
- 13.Malone, E. A., C. D. Clark, A. Chiang, and F. Winston. 1991. Mutation in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5710-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuhara, K., K. Ohta, H. Seo, M. Shioda, T. Yamada, Y. Tanaka, N. Dohmae, Y. Seyama, T. Shibata, and H. Murofushi. 1999. A DNA unwinding factor involved in DNA replication in cell-free extracts of xenopus eggs. Curr. Biol. 9:341-350. [DOI] [PubMed] [Google Scholar]
- 16.Orphanides, G., G. LeRoy, C.-H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105-116. [DOI] [PubMed] [Google Scholar]
- 17.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises the human SPT16/CDC68 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 18.Paull, T. T., M. Carey, and R. C. Johnson. 1996. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 10:2769-2781. [DOI] [PubMed] [Google Scholar]
- 19.Rowley, A., R. A. Singer, and G. Johnston. 1991. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol. 11:5718-5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruone, S., A. R. Rhoades, and T. Formosa. 2003. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and reorganize nucleosomes. J. Biol. Chem. 278:45288-45295. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374-376. [DOI] [PubMed] [Google Scholar]
- 23.Simpson, R. T., and D. W. Stafford. 1983. Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. USA 80:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thastrom, A., P. T. Lowary, H. R. Widlund, H. Cao, M. Kubista, and J. Widom. 1999. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 288:213-229. [DOI] [PubMed] [Google Scholar]
- 25.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20:5207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittmeyer, J., and T. Formosa. 1997. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17:4178-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wittmeyer, J., L. Joss, and T. Formosa. 1999. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38:8961-8971. [DOI] [PubMed] [Google Scholar]
- 28.Wittmeyer, J., A. Saha, and B. Cairns. 2003. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 377:322-343. [DOI] [PubMed]
- 29.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 30.Yu, Y., P. Eriksson, and D. J. Stillman. 2000. Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol. 20:2350-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]