Abstract
ROSA3 mice were developed by retroviral insertion of the βGeo gene trap vector. Adult ROSA3 mice exhibit widespread expression of the trap gene in epithelial cells found in most organs. In the central nervous system the highest expression of βGeo is found in CA1 pyramidal cells of the hippocampus, Purkinje cells of the cerebellum, and ganglion cells of the retina. Characterization of the genomic insertion site for βGeo in ROSA3 mice shows that the trap vector is located in the first intron of Fem1c, a gene homologous to the sex-determining gene fem-1 of Caenorhabditis elegans. Transcription of the Rosa3 allele (R3) yields a spliced message that includes the first exon of Fem1c and the βGeo coding region. Although normal processing of the Fem1c transcript is disrupted in homozygous Rosa3 (Fem1cR3/R3) mice, some tissues show low levels of a partially processed transcript containing exons 2 and 3. Since the entire coding region of Fem1c is located in these two exons, Fem1cR3/R3 mice may still be able to express a putative FEM1C protein. To this extent, Fem1cR3/R3 mice show no adverse effects in their sexual development or fertility or in the attenuation of neuronal cell death, another function that has been attributed to both fem-1 and a second mouse homolog, Fem1b. Examination of βGeo expression in ganglion cells after exposure to damaging stimuli indicates that protein levels are rapidly depleted prior to cell death, making the βGeo reporter gene a potentially useful marker to study early molecular events in damaged neurons.
Promoter trap experiments using the reverse orientation splice acceptor (ROSA) βGeo retroviral gene trap vector (9) have been useful in identifying genes involved in mouse development (4, 5). The coding region for the trap vector contains bacterial β-galactosidase fused in frame to neomycin phosphotransferase (Neo) to produce a fusion protein (βGeo) that processes substrates like X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and neomycin. Transcription of βGeo is dependent on the trap vector being inserted into the vicinity of an existing gene in the mouse genome, where the presence of a 5′ splice acceptor facilitates the splicing of the βGeo coding region into the nearest upstream exon. The ROSA3 line of mice was initially part of 31 lines established using retroviral insertion of the βGeo vector into embryonic stem cells (9). The insertion of the βGeo gene caused embryonic recessive lethality in some of the lines. ROSA3 mice showed no overt defects, however, and exhibited a widespread pattern of staining by embryonic day 12.
Here, we report the further characterization of the ROSA3 line of mice. As with the results seen with the embryos, ROSA3 mouse adult tissues show widespread staining (including selective staining of certain populations of neurons in the central nervous system) in β-galactosidase assays. Analysis of the genomic insertion site shows that the βGeo gene has been inserted into the first intron of Fem1c, the third member of a murine gene family that shares homology with the Caenorhabditis elegans fem-1 gene.
MATERIALS AND METHODS
Animals.
Mice used in this study were maintained and handled in accordance with the guidelines established by the Association for Research in Vision and Ophthalmology Statement on animals in research and overseen by the animal use committee at the University of Wisconsin. ROSA3 mice were a gift (to L.H.P.) from Phillip Soriano. These mice had a C57BL/6J background of unknown congenic generation. This strain was backcrossed further into C57BL/6J mice (N14) obtained from the Jackson Laboratory (Bar Harbor, Maine). A second line of ROSA3 mice was also established by backcrossing the Rosa3 (R3) allele into DBA/2J inbred mice, which were also obtained from the Jackson Laboratory. For the experiments shown here, the R3 allele was considered congenic on C57BL/6J mice and was in the sixth generation of the backcrosses onto DBA/2J mice.
Histochemical staining for β-galactosidase activity.
Staining for β-galactosidase activity was conducted on freshly isolated organs dissected from mice euthanized by pentobarbital overdose. Tissues were snap frozen in liquid nitrogen, embedded in OCT compound, and sectioned (10-μm thickness) on a Jung Frigocut 2800E cryomicrotome (Leica, Nussloch, Germany). Frozen sections were collected on charged Plus slides (Fisher Scientific, Chicago, Ill.) and briefly fixed in 4% paraformaldehyde in 0.1 M phosphate-buffered saline (PBS) (pH 7.4) containing 0.15 M NaCl. Slides were washed in PBS containing 2 mM MgCl2 and 2 μm of CaCl2 and stained in this same solution containing 4.5 mM potassium ferrocyanide, 4.5 mM potassium ferricyanide, 0.3% Triton X-100, and 1 mg of X-Gal/ml. In some cases 1 mg of BluoGal/ml was substituted for X-Gal. Slides were stained overnight in a humidified chamber at 37°C, after which they were washed in PBS, counter-stained with eosin or nuclear fast red, and coverslipped for viewing under bright-field microscopy using an Axiphot photomicroscope (Zeiss, Thornwood, N.Y.).
Staining for β-galactosidase was also conducted on retinal whole mounts. Mice were killed by pentobarbital overdose, and the eyes were enucleated. A small incision was made through the globe with a No. 57 surgical blade, and the eyes were fixed for 30 min in 4% paraformaldehyde in PBS. The eyes were washed in PBS, and the anterior chamber and lenses were removed. The resulting eye cups were stained in either X-Gal or BluoGal overnight at 37°C as described above. After staining the eyecups were washed and fixed in 4% paraformaldehyde in PBS, after which the retinas were dissected free and whole mounted on glass slides for viewing and photographing using a Leica dissecting microscope with a 2× objective and 35-mm camera attachment. Higher-magnification images were obtained on a Zeiss Axiophot Fluorescent microscope with a conventional 35-mm camera attachment. In some experiments, the retinas were eluted from the slides and embedded in JB4-Plus (Polysciences, Inc., Warrington, Pa.) for sectioning and cell quantification as described elsewhere (17). Only retinas stained with BluoGal were embedded in JB4-Plus.
ELISAs for β-galactosidase.
Enzyme-linked immunosorbent assay (ELISA) quantification of βGeo was performed using a β-galactosidase ELISA kit available from Roche Diagnostics Corp. (Indianapolis, Ind.). Retinas were harvested from sacrificed animals and stored frozen at −80°C. When all samples were collected, 1 ml of lysis buffer from an ELISA kit was added to each retina and sonicated for 10 pulses at low intensity. Homogenized samples were cleared by centrifugation. The protein concentration for each sample was determined in a 100-μl aliquot with a bicinchoninic acid protocol (30), and βGeo levels were determined (using the ELISA) from triplicate 200-μl aliquots for each retina as described by the manufacturer. ELISA plates were read using an ELx800 universal microplate reader (Bio-Tek Instruments, Inc., Winooski, Vt.). The relative amount of βGeo in each aliquot was determined from β-galactosidase standards (Roche Diagnostics Corp.) included in each ELISA plate. Retinas from wild-type mice were included in the assays to monitor background levels. The level of βGeo in each retina was calculated as follows. The mean of the triplicate measurements was calculated and normalized to the amount of total protein in the sample. This value was corrected for background by subtracting the equivalent values obtained for wild-type retinas. Finally, a change in the βGeo level for each experimental eye was calculated as a percentage of the level in the control eye for each mouse.
Retrograde labeling of retinal ganglion cells.
Retinal ganglion cells were labeled using retrograde transport of Fluorogold injected stereotactically into the superior colliculus on both sides of the brain. Briefly, the skulls of anesthetized mice were exposed and a 1.5-mm-diameter hole was drilled through the skull at a position 4 to 5 mm posterior to the bregma. A 10-μl Hamilton syringe attached to a micromanipulator was inserted to a depth of 1.5 to 2 mm from the surface of the brain. In each side of the superior colliculus a total of 1 μl of a 1% solution of Fluorogold (Fluorochrome, Inc., Denver, Colo.) in PBS was delivered in two separate injections of 0.5 μl, waiting 1 min between injections and 1 min before withdrawing the needle. After injections were complete, the skin over the skull hole was glued with liquid sutures. Mice were allowed to recover and were sacrificed 2 to 3 days thereafter, and the eyes were enucleated and stained with X-Gal as described above. Whole mounts of these retinas were viewed by combined fluorescent and bright-field microscopy using a Zeiss Axiophot fluorescent microscope.
Acute models of retinal ganglion cell death.
Two acute models of damage were used as described previously (17) to activate retinal ganglion cell death in adult mice. Briefly, mice were anesthetized by intraperitoneal injection of ketamine (6 mg/ml) and xylazine (0.4 mg/ml). For optic nerve crushing, the optic nerve was exposed using an intraorbital approach through the conjunctival layers and then clamped with self-closing N7 microforceps (Fine Science Tools, Vancouver, British Columbia, Canada) for 3 s. This procedure produces synchronous loss of retinal ganglion cells over a 3-week period. Ganglion cell death was also stimulated by exposure to elevated levels of the glutamate analog N-methyl-d-aspartate (NMDA) (Research Biochemicals Int., Natick, Mass.). A small puncture was introduced (using a 30-gauge needle) into one eye of each anesthetized mouse. A glass micropipette was then inserted through this hole into the vitreous, and 2 μl of an 80 mM solution of NMDA (for a total of 160 nmol) was injected. This procedure produces synchronous loss of retinal ganglion cells over a 4-day period.
Cell loss induced by these methods was quantified from sections of retina harvested from both experimental and control fellow eyes of each mouse as described previously (17). At various times after damage, the mice were killed and the eyes were harvested and fixed for 1 h at 22°C in 4% paraformaldehyde in PBS. After an hour, the anterior chambers and lenses were removed and the eyecups were postfixed overnight in 0.4% paraformaldehyde in PBS at 4°C. Retinas were embedded in JB4-Plus, and 1- to 2-μm-thick sections were cut from the superior quadrant. Sections were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Roche) and viewed and photographed using a Zeiss Axiophot fluorescent microscope. Cell counts were made from digitized photographs of 6 to 10 sections of each eye of a mouse. Cell loss was quantified as the percentage of cells lost over a defined length of retina (800 μm) in the experimental eye relative to the results seen with the control eye of the same mouse. Mean percentages were calculated for four to six mice at each time point for comparisons.
Analysis of the Rosa3 insertion site in genomic DNA.
All primer sequences used for PCR-based experiments are shown in Table 1.
TABLE 1.
Sequences of primers used for genomic PCR and RT-PCR experiments
| Primera | Sequence |
|---|---|
| LacZ.1 (a) | 5′ CGTGACTGGGAAAACCCTGGC |
| LacZ.2 (b) | 5′ GAGCGAGTAACAACCCGTCGG |
| βGeoE (c) | 5′ GCGATACCGTAAAGCACGAGG |
| SupF (d) | 5′ GGAGCAGACTCTAAATCTGCC |
| BF4.1 (e) | 5′ GCGGAGTAGGGCATCCCAGG |
| BF4.2 (f) | 5′ GCAGAAGCGGCCCATAAAGG |
| BF4.5 | 5′ AGTTAAAGGCTGGAAAGGCG |
| R3B | 5′ CCAGCCCAGCTTTGTTCTGG |
| BF4.6 (g) | 5′ GAAGTGTCCAACCGCCATGG |
| BF4.7 (h) | 5′ TTGTCTGGGCATGGTGCG |
| β-Actin.1 | 5′ CTCTCCCTCACGCCATCCTG |
| β-Actin.2 | 5′ CCGCCTAGAACTTGCGG |
The letters in parentheses correspond to the locations of select primers relative to the genomic map of this locus shown in Fig. 6.
Inverse PCR was carried out essentially as described by Groden et al. (11) with the following modifications. Total genomic DNA from spleens of R3 heterozygous mice was isolated. DNA (16 μg) was digested to near completion with EcoRI at 37°C, after which the reaction was heat inactivated for 20 min at 65°C. Digested DNA was subsequently ligated at a concentration of 5 to 10 ng/μl overnight at 15°C to allow circularization of the EcoRI fragments. Long-range PCR was carried out using the system provided by Roche Diagnostics Inc. Briefly, 10 to 20 ng of total genomic DNA per reaction was amplified with the primers βGeoE and SupF. Amplification conditions were as follows: 10 cycles of 94°C (10 s), 60°C (30 s), and 68°C (8 min) followed by 15 cycles of 94°C (10 s), 60°C (30 s), and 68°C (8 min) (with an additional 20 s added per cycle).
Once the genomic position of the R3 allele was established, primers were synthesized that amplified both wild-type and R3-containing alleles. The wild-type allele was amplified using primer BF4.5 (which corresponds to a sequence of the first intron of Fem1c upstream of the βGeo trap vector insertion site) and primer R3B (which corresponds to a downstream sequence of the first intron). The R3 allele was amplified using the SupF and R3B primers. The genotyping of mice was carried out using DNA isolated from tail cuttings and the following PCR conditions: 30 cycles of 94°C (30 s), 60°C (30 s), and 72°C (1 min).
Total RNA isolation, first-strand cDNA synthesis, and reverse transcriptase PCR (RT-PCR) were performed as described previously (18, 27). First-strand cDNA was made using 5 μg of total RNA isolated from the brains of R3/R3, R3/+, and +/+ animals. Approximately 2 ng of single-stranded cDNA was used for each RT-PCR. Conditions for PCR amplification were as follows: 30 cycles of 95°C (30 s), 55°C (30 s), and 72°C (60 s). In some cases, PCR was extended to 40 cycles to enhance the amplification of rare transcripts. Reactions were separated on 1% agarose gels containing ethidium bromide. Images of the gels were digitized using a Kodak EDAS 290 digital imaging camera (Eastman Kodak, Rochester, N.Y.) on a UV light box.
Statistical analyses.
The distribution of sex of the offspring of R3/+ parents was analyzed using a 2-by-3 contingency table and χ2 analysis. Expected frequencies were calculated using the assumptions that the R3 allele would segregate according to Mendel's Law of Equivalency and that there would be a 50:50 distribution of males and females in the litters. Cell density in the ganglion cell layers of the retinas of R3/R3 and wild-type mice was assessed using a Student's t test of the mean values accumulated from four mice of each genotype.
RESULTS
Distribution of β-galactosidase activity in adult ROSA3 mice.
To determine the cell types expressing βGeo, we stained frozen sections of tissues harvested from ROSA3 mice. In previous studies (9), the pattern of β-galactosidase activity in ROSA3 mice was not examined beyond E12 embryos. Staining was first observed on embryonic day 12 and was considered widespread, with some cells exhibiting stronger staining than others. Similarly, β-galactosidase activity in frozen sections of selected adult tissues was also widespread (Fig. 1). Staining was most prominent and nearly ubiquitous in the small intestine and spleen. The kidney, skin, lung, and liver exhibited more-selective staining. Both smooth and skeletal muscle cells (including heart muscle) were negative for β-galactosidase activity. In nearly all cases, the most prominently stained cells appeared to be epithelial in origin or function.
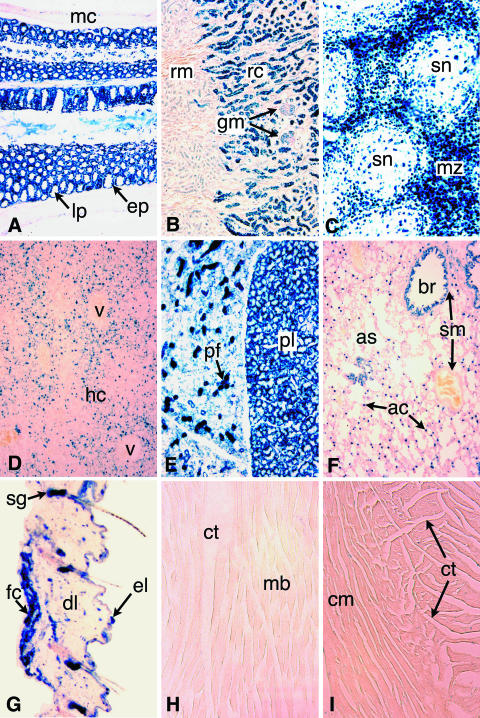
FIG. 1.
β-Galactosidase expression is widespread in the adult ROSA3 mouse. X-Gal staining was used to determine the βGeo expression patterns in frozen sections of various tissues harvested from adult R3/+ mice. Each panel is a micrograph taken at ×100 magnification. (A) Intestine. Epithelial cells (ep) lining the villi, which are seen mostly in cross section, are strongly and uniformly stained. No staining is detected in the lamina propria (lp), which form the cores of the villi, or in the circular and longitudinal muscles forming the muscular coat (mc) surrounding the intestine. (B) Kidney. Staining is present in the convoluted tubules of the renal cortex (rc) and absent from the renal medulla (rm). Glomeruli (gm) are less intensely stained. (C) Spleen. Staining is mostly restricted to cells in the marginal zone (mz) surrounding splenic nodules (sn). (D) Liver. Sparse staining is mainly detected in hepatocytes of the hepatic cords (hc). Vascular endothelial cells forming hepatic blood vessels (v) are unstained. (E) Thyroid. Intense staining is present in follicle cells of the pyramidal lobe (pl). In an adjacent lobe, follicular cells are lightly stained whereas presumptive parafollicular cells (pf) are intensely stained. (F) Lung. Intense staining is present in the columnar epithelium of the bronchioles (br) and in a population of (type II) alveolar cells (ac) lining alveoli and alveolar sacs (as). No staining is detected in smooth muscle (sm) surrounding the bronchioles and blood vessels (arrows). (G) Skin. Intense labeling is present in the sebaceous glands (sg) of each hair follicle. Some label is detected in the cells of the epidermal (el) and dermal layers (dl). Fat cells (fc) underlying the dermis also appear labeled. (H and I) Skeletal muscle (H) and cardiac muscle (I). No staining is detected in muscle cell bundles (mb) or cardiac muscle cells (cm). Cells comprising connective tissue (ct) appear to be lightly or negatively stained. Eosin was used for counterstaining.
Neuronal distribution of βGeo expression.
The brain and retina were also examined for β-galactosidase activity (Fig. 2). Staining in the brain was moderate, and several regions (such as the medulla oblongata, inferior colliculus, and corpus striatum) exhibited numerous positively stained cells scattered throughout. Some regions (including CA1 pyramidal neurons of the hippocampus and Purkinje cells of the cerebellum) of the brain, however, showed distinct populations of neurons with strong activity. In addition, pyramidal neurons in the outer granular layer of the cerebral cortex appeared to be preferentially stained relative to those of cells in adjacent layers.
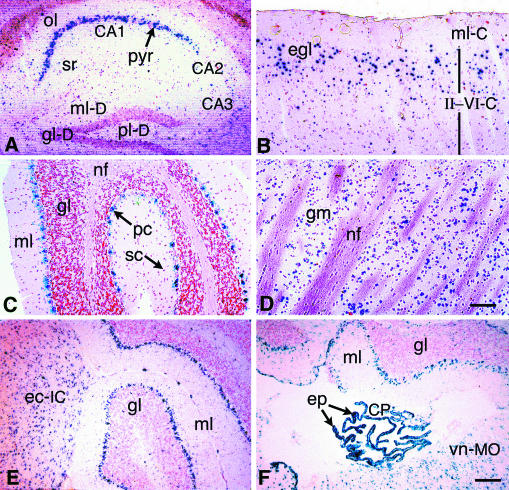
FIG. 2.
Distinct neuronal cell populations express βGeo in the adult Rosa3 brain. Brains were harvested from adult R3/+ mice and processed as described for Fig. 1. (A) Hippocampus. Intense staining is present in the CA1 pyramidal neurons (pyr [arrow]), and light staining is present in the CA3 pyramidal cells. Light staining to no staining is visible in the CA2 neurons, the oriens layer (ol), and the stratum radiatum (sr). Only diffuse staining is present in the dentate gyrus. ml-D, molecular layer; gl-D, granular layer; pl-D, polymorphic layer. (B) Cerebral cortex. Variable staining is present in the cerebrum. The molecular layer (ml-C), which is comprised mainly of nerve fibers, is unstained. Although cortical layers II to VI (II-VI-C) are not clearly discernible, the majority of stained neurons are present in the external granular layer (egl). (C) Cerebellum. Purkinje cells are strongly stained (pc [arrow]), but only sparse staining is present in the granular layer (gl). Stellate cells (sc [arrow]) of the molecular layer (ml) are unstained. nf, nerve fibers. (D) Striatum. Some neurons making up the gray matter (gm) of the striatum are stained. nf, nerve fiber bundles. (E) Inferior colliculus. Staining is present in cells throughout the external cortex of the inferior colliculus (ec-IC) adjacent to the cerebellum. gl, granular layer; ml, molecular layer of the cerebellum. (F) Choroid plexus. Intense staining is evident in the epithelial cells of the choroid plexus (CP) forming the roof of the fourth ventricle. Stained cells are present throughout the vestibular nucleus of the medulla oblongata (vn-MO) region. Nuclear fast red was used for counterstaining. Bar (in panel D) for panels A to D, 75 μm. Bar (in panel F) for panels E and F, 125 μm.
The retina also exhibited a localized staining pattern for β-galactosidase, with the large majority of labeling restricted to the ganglion cell layer (Fig. 3A and B). To determine whether staining in the ganglion cell layer was limited to ganglion cells or displaced amacrines or both, the ganglion cells were first back labeled (using Fluorogold injected into the superior colliculus) by retrograde axonal transport. After 3 days, the retinas were harvested and labeled with X-Gal (Fig. 3C). Quantification of Fluorogold and X-Gal labeling in randomly chosen regions of four retinas showed that 91 ± 1% of the Fluorogold-labeled cells also expressed βGeo. No βGeo-positive cells in the ganglion cell layer were Fluorogold negative. A sparse population of stained cells was also evident in the inner nuclear layer. In section, these cells were found in the inner region of this layer (which is the typical location of amacrine cells and displaced ganglion cells) (Fig. 3A and B). These cells remained labeled 2 weeks after optic nerve crushing, which causes the death of ganglion cells but not of amacrine cells (Fig. 3D and E).
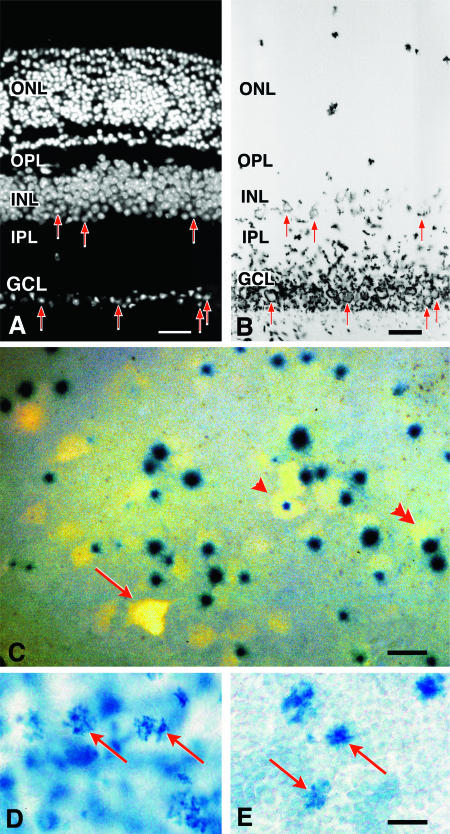
FIG. 3.
β-Galactosidase is primarily expressed in retinal ganglion cells and in a subset of cells in the inner nuclear layer. (A and B) Histological analysis of BluoGal-stained retinas from R3/+ mice. (A) Fluorescent image of a transverse section through a mouse retina (stained with DAPI to highlight nuclei); (B) bright-field image of the same section. BluoGal staining is localized around cells in the ganglion cell layer (GCL). Stain is also deposited in the inner plexiform layer (IPL), which contains the majority of ganglion cell dendritic processes. Some cells in the inner region of the inner nuclear layer (INL) are also labeled. Examples of labeled cells in both the GCL and INL are highlighted with arrows in both panels. No staining is evident in the photoreceptors of the outer nuclear layer (ONL). OPL, outer plexiform layer. Bar, 25 μm. (C) Photomicrograph (taken with both fluorescent and bright-field illumination) of a flat-mounted retina from a R3/+ mouse, showing colocalization of Fluorogold and βGeo activity (as revealed by X-Gal staining) in cells of the ganglion cell layer. Fluorogold labeled the cytoplasm, while X-Gal concentrated in the nucleus. Some ganglion cells have central nuclei (arrowhead), while most cells have nuclei positioned eccentrically (double arrowhead). In the ganglion cell layer, 91% ± 1% of Fluorogold-labeled cells (n = 584) express βGeo. Incomplete penetration of the X-Gal stain could account for the small percentage of Fluorogold-labeled cells that are not associated with βGeo expression (arrow); alternatively, these cells may represent a subset of retinal ganglion cells that do not express the transgene. Bar, 10 μm. (D and E) Bright-field images of whole-mounted retinas stained with BluoGal. The focal plane is centered on the inner nuclear layer of each retina, showing scattered labeled cells (arrows). The control retina has an extensive blue background due tolabeled ganglion cells (D). A retina taken from the contralateral eye of the same mouse 2 weeks after optic nerve crushing shows reduced staining of the ganglion cell layer but no change in labeling in the inner nuclear layer (E). Bar, 5 μm.
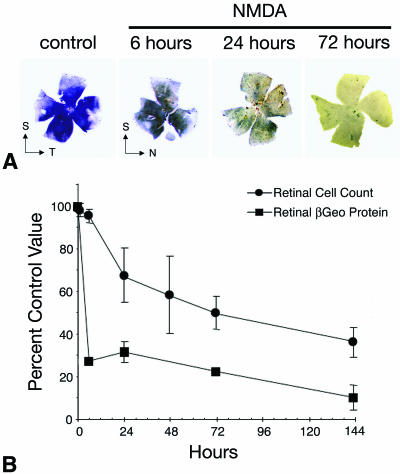
We examined further the change in βGeo expression under conditions that stimulate progressive ganglion cell apoptosis. Single eyes of R3/+ mice were subjected to either optic nerve crushing or an intravitreal injection of 160 nmol of NMDA. Animals were killed at time points consistent with previous studies showing the kinetics of cell loss after these treatments (17, 26). Harvested eyes were first stained with BluoGal, photographed as whole mounts, and then embedded and sectioned for cell counting. In parallel experiments, instead of being stained for β-galactosidase activity the eyes were homogenized and used for ELISAs to quantify βGeo protein levels. Figure 4 shows the results of the optic nerve crush experiments. Ganglion cell layer staining is diminished by 3 and 7 days after crushing and nearly eliminated by 14 days. Microscopic evaluation of the deeper inner nuclear layer showed that staining of putative amacrine cells was unaffected (Fig. 3D and E). ELISA quantification of βGeo showed a rapid 75% drop in protein levels by 7 days after crushing. Figure 5 shows the results of NMDA injection. In similarity to the results seen with crushed optic nerve retinas, staining is dramatically decreased by 24 h after injection of the excitotoxin. ELISA quantification shows a rapid loss of approximately 70% of the βGeo protein by 6 h and a further loss to 90% by 144 h. The greater loss (relative to the results seen with optic nerve crushing) of βGeo protein after NMDA injection may be due to the loss of amacrine cells, which are also sensitive to NMDA (17, 29). In both treatments, the loss of βGeo preceded cell loss in the ganglion cell layer. This loss is consistent with early decreases in gene expression observed for other ganglion cell-specific genes (26).
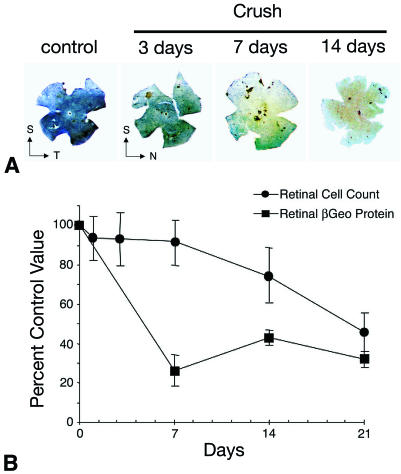
FIG. 4.
A decrease in βGeo protein levels precedes cell loss stimulated by optic nerve crushing. (A) BluoGal-stained retinal whole mounts harvested from mice at selected time points after optic nerve crushing (Crush). BluoGal staining progressively decreases with time after optic nerve crushing. T, temporal; N, nasal; S, superior. (B) Graph comparing the rate of cell loss in the ganglion cell layer (filled circles) to the rate of decrease in βGeo protein (filled squares) after optic nerve crushing. Cell loss was quantified from histological sections, and protein was quantified using a β-galactosidase ELISA. The decrease in βGeo protein levels precedes cell loss in the Rosa3 retina after optic nerve crushing, with similar kinetics displayed by other ganglion cell genes (26).
FIG. 5.
A decrease in βGeo protein levels precedes cell death stimulated by excitotoxins. (A) BluoGal-stained retinal whole mounts harvested from mice at selected time points after intravitreal injection of 160 nmol of the glutamate analog NMDA. BluoGal staining rapidly decreases in the ganglion cell layer after exposure to NMDA. T, temporal; N, nasal; S, superior. (B) Graph comparing the rate of cell loss in the ganglion cell layer (filled circles) to the rate of decrease in βGeo protein (filled squares) after NMDA injection. Cell loss was quantified from histological sections, and protein was quantified using a β-galactosidase ELISA. As seen under the conditions described for optic nerve crushing, the decrease in βGeo transgene expression precedes cell loss in the Rosa3 retina after exposure to NMDA. NMDA injection results in cell loss and reduced βGeo transgene expression more rapidly than optic nerve crushing, but the relationship between protein loss and cell loss is similar.
Localization of the βGeo promoter trap vector to the Fem1c gene.
The chromosomal position of the βGeo gene was identified by inverse PCR. Genomic DNA isolated from Rosa3 mice was digested with EcoRI and ligated at low density. Primers specific for the 3′ long terminal repeat (SupF) and β-galactosidase (βGeoE) adjacent to an internal EcoRI site were used to amplify a 1.8-kb fragment that was sequenced with the SupF primer. The resulting sequence was used to design genomic DNA-specific primers. PCR fragments generated using these primers confirmed the location of βGeo inserts. No product was obtained using wild-type or ROSA26 genomic DNA as a template (data not shown).
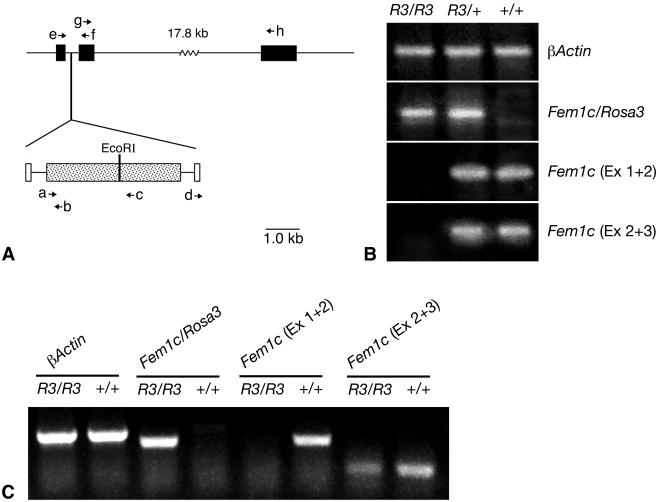
Database (Entrez Genome [http://www.ncbi.nlm.nih.gov/]) searches using the genomic sequence obtained from the inverse PCR fragment yielded a match to region 46788K to 46789K of mouse chromosome 18, which contained an expressed sequence tag (EST) (BF465068) isolated from a subtracted mouse retinal cDNA library (NIH_BMAP_Ret4_S2). The βGeo trap vector was localized to an intron of the EST. Subsequently a full-length cDNA that incorporated EST BF465068 was mapped to the same region (12, 28). This gene encodes a putative protein homologous to mouse Fem1a and was designated Fem1c. The gene structure consists of three exons, with the entire coding region present in exons 2 and 3. The βGeo trap vector is inserted into the first intron (Fig. 6). To determine whether βGeo was spliced to the first exon of Fem1c, a primer corresponding to exon 1 (BF4.1) was synthesized and used with LacZ.2 to amplify a fragment with RT-PCR. As a control, RT-PCR was also conducted using BF4.1 and a second primer corresponding to exon 2 of Fem1c (BF4.2). RT-PCR was carried out on oligo(dT)-primed cDNA made from total RNA isolated from the brains of R3/R3, R3/+, and +/+ mice (Fig. 6). A single fragment was obtained from R3 homozygous and heterozygous mice, but not from wild-type mice, with the BF4.1/LacZ.2 primers. Sequence analysis of this fragment showed correct splicing between exon 1 of Fem1c and βGeo. A spliced RT-PCR fragment of exons 1 and 2 was detected using the BF4.1-BF4.2 primers in R3/+ and wild-type animals but not in R3 homozygous mice. To determine whether Fem1c was alternatively spliced in R3/R3 animals (and therefore potentially translated normally into a protein), we also generated BF4.6 and BF4.7, primers specific for exons 2 and 3 of the Fem1c sequence. RT-PCR using a 30-cycle program on mouse brain samples indicated that a spliced transcript was present only in R3/+ and wild-type mice. When we extended the PCR program to 40 cycles, however, we were able to detect low levels of product corresponding to a transcript containing exons 2 and 3 in R3/R3 mice. This product was present in some tissues such as testes (Fig. 3C) and brain but was not detected in eyes, kidneys, or cardiac muscle (data not shown).
FIG. 6.
βGeo insertion disrupts normal Fem1c expression. (A) Map of Fem1c on chromosome 18 showing the insertion site of the βGeo trap vector. Exons of Fem1c are shown as filled boxes. The βGeo coding region is shown as a stippled box, and the flanking retroviral long terminal repeats are shown as open boxes. βGeo contains an internal EcoRI restriction site that was used for inverse PCR experiments. The relative positions of various primers (a to h) used for PCR experiments are shown with arrows. The sequences and names of these primers are detailed in Table 1. βGeo is inserted into the first intron of the Fem1c gene. (B) Ethidium bromide-stained agarose gels showing the results of RT-PCR analysis of transcripts from the Fem1c alleles. Total RNA was isolated from Rosa3 mouse brains and used to make first-strand cDNA. The genotypes of the mice are indicated above each lane. β-Actin amplification was used as a control for equivalent amounts of template cDNA (top panel). The results of RT-PCR with primers e and b (BF4.1 and LacZ.2, respectively) show that the βGeo coding region is spliced onto exon 1 of Fem1c only in R3/R3 and R3/+ mice (second panel from top). Sequence analysis of this fragment shows correct splicing at the end of exon 1. The results of RT-PCR with primers e and f (BF4.1 and BF4.2, respectively) indicate appropriate splicing of exons 1 and 2 (Ex 1 + 2) of Fem1c in mice that carry one or both wild-type alleles. No splice was evident in R3/R3 mice (third panel). The results of RT-PCR with primers g and h (BF4.6 and BF4.7, respectively) indicate appropriate splicing of exons 2 and 3 of Fem1c. No splice that bypasses exon 1 is detected in R3/R3 mouse brain under these PCR conditions (30 cycles) (fourth panel). These data show that Fem1c expression is knocked out in R3/R3 mice. Ex 2 + 3, exons 2 and 3. (C) Ethidium bromide-stained agarose gel of RT-PCR products generated from extended (40-cycle) PCR. All products were generated from testes RNA isolated from R3/R3 or wild-type mice. Samples from R3/R3 mice exhibit a faint band indicative of a transcript containing correctly spliced exons 2 and 3 of Fem1c. No transcript containing exons 1 and 2 of Fem1c in the same mice was detected. Primers used for these products were the same as those used as describe for panel B, and β-actin was used as a control.
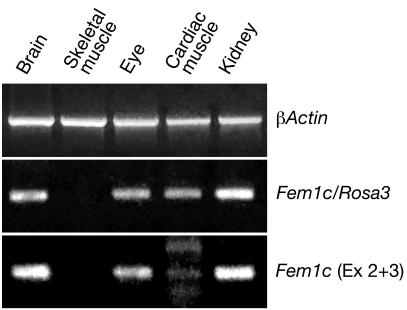
To confirm that the R3 expression pattern matched the expression pattern of Fem1c, we used RT-PCR to survey different tissues harvested from R3/+ mice for transcripts of each allele. Figure 7 shows the results obtained with some of the tissues examined. Transcripts for both the R3 and wild-type alleles were present in brain, eye, and kidney but not in skeletal muscle (consistent with the observations made using histochemical staining for β-galactosidase activity) (Fig. 1, 2, and 3). Cardiac muscle exhibited a band for the R3 transcript but only a very weak band for Fem1c, even though this experiment was conducted using 10 additional cycles (for a total of 40 cycles) for PCR. The presence of the R3 transcript was consistent with the low level of β-galactosidase activity observed in connective tissue cells of this organ (Fig. 1). Fem1c and R3 transcripts were also detected in skin, intestine, lung, liver, spleen, thyroid, and testes harvested from R3/+ animals (data not shown).
FIG. 7.
Fem1c and R3 alleles exhibit similar patterns of expression. Ethidium bromide-stained agarose gels showing the results of RT-PCR for β-actin (control) and transcripts from the R3 (Fem1c/Rosa3) and Fem1c (exons 2 and 3 [Ex 2 + 3]) alleles in different tissues of R3/+ mice. Both alleles are expressed in brain, eye, and kidney but not in skeletal muscle (consistent with the X-Gal staining pattern shown in Fig. 1). The R3 allele is expressed in cardiac muscle, but only low levels of the Fem1c product were detected in this tissue. The primers used for these products were as described for Fig. 6.
Examination of phenotype in Fem1cR3/R3.
The results of RT-PCR experiments indicate that a transcript containing the coding region of Fem1c either is not expressed or is expressed at very low levels in R3/R3 mice. We investigated whether this reduced or absent expression affected two major functions attributed to Fem1 genes, namely, sex determination and cell death.
fem-1, the C. elegans homolog to mouse Fem1 genes, plays a critical role in sex determination in nematodes. To examine whether Fem1c has important functions in development and sex determination in mice, we established breeding pairs of R3/+ animals in both the C57BL/6J and DBA/2J genetic backgrounds. Table 2 shows the results of these crosses. In C57BL/6J mice, the R3 allele was found to segregate in a normal Mendelian pattern of inheritance (28% R3/R3, 46.5% R3/+, and 25.5% +/+; n = 86 mice). The sex distribution of R3/R3 mice showed that females were slightly more likely to be produced (11.6% R3/R3 male to 16.3% R3/R3 female), but this result was not statistically significant (P > 0.25 [χ2 test]), indicating that reduced Fem1c expression has minimal effect on the development of mice of one sex in preference to that of the other. Since fem-1 null worms are sterile, R3/R3 male or female mice were mated with wild-type animals to determine whether the lack of Fem1c caused sterility. R3/R3 animals of both sex were able to parent offspring (data not shown). Results were also obtained from crosses of R3/+ DBA/2J mice. The initial litters of these animals yielded no R3/R3 offspring (0/29 mice), suggesting that development in mice with this genetic background might have a greater dependence on normal Fem1c function. Statistical analysis of all the DBA/2J offspring indicated that this effect was significant (P = 0.01). Separate analysis of the subsequent litters, however, revealed a Mendelian-like distribution of the R3 allele (26% R3/R3, 44% R3/+, and 30% +/+; n = 27 mice), again with a modest bias toward the production of females (7.4% male and 18.5% female for R3/R3 animals; n = 7). Statistical analysis of the sex distribution of these subsequent litters showed no significant effect (P > 0.25). Additionally, no effect was found when the results for DBA/2J and C57BL/6J mice were combined (P = 0.25).
TABLE 2.
Segregation of the Rosa3 (R3) allele in offspring of R3/+ parents.
| Sex of mouse | Result for straina:
|
||
|---|---|---|---|
| R3/R3 | R3/+ | +/+ | |
| C57BL/6Jb | |||
| Male | 10 | 20 | 12 |
| Female | 14 | 20 | 10 |
| Subtotal | 24 | 40 | 22 |
| DBA/2Jc | |||
| Male | 2 | 14 | 7 |
| Female | 5 | 15 | 13 |
| Subtotal | 7 | 29 | 20 |
| Total (both strains)d | 31 | 69 | 42 |
The distribution of sex among these offspring was determined by χ2 analysis.
C57BL/6J mice showed no significant effect of sex distribution (P > 0.25).
DBA/2J mice did show a significant effect when the results for animals from all the litters were combined (P = 0.01), but no effect was seen when mice from the last four litters were analyzed separately (P > 0.25).
No significant effect was observed when both data sets were combined and analyzed (P = 0.25).
Fem1 genes have also been reported to play a role in apoptosis in mammalian cells (2, 3). We examined whether Fem1c plays a role in retinal ganglion cell death during normal development of the retina and after acute lesion to the optic nerve. During the first 3 weeks of life, approximately 50% of the retinal ganglion cells undergo programmed cell death during the organization of the retina. Eyes were enucleated from killed 3-month-old R3/R3 and wild-type littermates, and the retinas were removed, flat mounted, and stained with cresyl violet (Nissl stain) to identify cells in the ganglion cell layer. Quantification of cells in retinas from mice of both genotypes showed no significant change in cell density (8,075.4 ± 1,116 [mean ± standard deviation] cells/mm2 for R3/R3 mice versus 8,020.8 ± 1,068 cells/mm2 for +/+ mice; P = 0.9 [t test]), indicating no difference in the rates of programmed cell death in this layer.
Ganglion cell death after acute lesion (crushing) of the optic nerve was also assayed in adult R3/R3 and wild-type mice. There was no significant difference in cell loss levels in the ganglion cell layer between R3/R3 and wild-type mice (20.31 ± 5.37% [mean ± standard deviation] cell loss for R3/R3 mice versus 25.47 ± 5.06% cell loss for +/+ mice; n = 8, P = 0.11 [t test]) at 2 weeks after optic nerve crushing.
DISCUSSION
The βGeo trap vector is inserted into the Fem1c of ROSA3 mice.
Characterization of the genomic insertion site of the βGeo trap vector shows that it is located in the first intron of the Fem1c gene of mice. RT-PCR experiments confirm that the βGeo coding region is spliced onto exon 1 of Fem1c and that normal splicing of Fem1c is disrupted in R3/R3 animals. Some tissues from R3/R3 mice were found to have low levels of transcripts containing exons 2 and 3, which encompass the entire coding region for the putative FEM1C protein. Consequently, it is possible that a low level of a functional protein is present in these mice.
Fem1c was initially identified in the RIKEN sequencing project of mouse cDNA libraries (12, 28), but this gene product has not been characterized further. Fem1c is so named because the amino acid identity (similarity) is 69% (83%) and 41% (60%) compared with the amino acids of the mouse Fem1a and Fem1b genes, respectively. Together, the members of this family of genes share significant homology to the sex-determining fem-1 gene of C. elegans, particularly in the presence of a series of seven ankyrin motifs located near the N terminus and the sequence PXXLXXFXXXH at the C terminus (14, 31, 32). In C. elegans, fem-1 is essential for the development of male worms (both in masculinizing somatic cells and in the production of sperm in males and hermaphrodites) (7, 13). Worms mutant for fem-1 become hermaphrodites (even when they have an XO genotype) (7). The expression of fem-1 is not sex type specific, however, and both males and hermaphrodites exhibit widespread expression of this gene in somatic tissues (10). This has led to the speculation that the function of FEM-1 is regulated posttranscriptionally and possibly posttranslationally. We crossed R3/+ animals to test whether reduced Fem1c expression affects sex determination in mice. R3/R3 pups from these crosses were slightly more likely to be females, but this trend was not significant when large numbers of mice were evaluated. Similarly, both male and female R3/R3 mice were able to parent pups when bred to wild-type animals, indicating that reduction of Fem1c did not affect either the male or female germ lines.
Fem1 genes may have a broader function by regulating cell death. Recent reports have found that the C. elegans FEM-1 protein is a substrate for the caspase homolog CED-3 and can stimulate apoptosis in mammalian cells (3). Similarly, the protein product of Fem1b can also stimulate apoptosis (2). FEM1B associates with the cytoplasmic domains of Fas and tumor necrosis factor 1 and is a specific substrate for caspase 3 (2). Deletion analysis of the protein indicates that the core region containing the ankyrin repeats is essential for its death-inducing activity and that these motifs might be necessary for it to bind other proteins (2). To date, no cell death activity has been attributed to the other mouse homologues of fem-1. Cell death is an important phenomenon in neuronal development, however, and it is possible that Fem1c may play a role in this process. Retinal ganglion cells, for example, undergo widespread programmed cell death during the first 3 weeks of life in mice. Some genes play a critical role in this process. Knockout of the Bax gene, or overexpression of Bcl2 in transgenic animals, significantly reduces the loss of ganglion cells during this period (1, 19, 21). Our initial experiments to test for a role of Fem1c in programmed cell death found no difference between the densities of cells in the ganglion cell layers of adult R3/R3 and the densities of cells in ganglion cell layers of wild-type littermates. In addition, stimulation of ganglion cell death by optic nerve lesions showed no attenuation of cell death in adult R3/R3 mice. Together, these data do not support a role for Fem1c in regulating ganglion cell death.
βGeo as a molecular marker for retinal ganglion cells.
Identification of ganglion cells by retrograde labeling with Fluorogold followed by X-Gal staining showed that the majority (91%) of retinal ganglion cells express the βGeo transgene. The 9% of Fluorogold-positive-β-galactosidase-negative cells may represent a subset of ganglion cells that do not express Fem1c. Alternatively, this lack of staining may have been due to incomplete penetration of X-Gal to some ganglion cells in the whole-mount preparations.
Some cells in the inner nuclear layer also exhibited β-galactosidase activity. Their position in the retina (and the lack of effect after crushing) suggests that they are amacrine cells and not displaced ganglion cells. We estimate (on the basis of quantitative histology and comparisons between the amounts of βGeo protein lost in NMDA-treated eyes [where amacrine cells are affected] versus crushed eyes) that 15% of the β-galactosidase activity in the retina is contributed by these cells.
The presence of a versatile reporter gene in retinal ganglion cells may provide an important molecular tool for monitoring the effects of damaging stimuli. There has been significant investigation of the mechanisms of retinal ganglion cell loss, primarily because these cells are directly affected in the common blinding disease glaucoma (23, 24). The effects of experimental manipulations on retinal ganglion cells are typically monitored by assessing differences in cell counts (22). The βGeo reporter gene could be useful to rapidly quantify ganglion cell number as a function of β-galactosidase activity. To test this, we assessed both β-galactosidase activity and βGeo protein levels in R3/+ retinas after optic nerve crushing or intravitreal injection of NMDA. Both treatments stimulate a loss of activity and protein; when the results were compared to those seen with respect to the loss of cells in these retinas, however, we found that protein levels were depleted before detectable cell death had occurred. This result is consistent with previous observations that damaged ganglion cells rapidly degrade some mRNAs (16, 26). The kinetics of protein loss was nearly identical to the kinetics observed for the loss of Thy1 mRNA, a ganglion cell-specific gene (26), suggesting that degradative pathways for mRNA and protein work simultaneously in damaged cells. When βGeo expression is used as a biochemical marker for ganglion cell effects or loss, consequently, the kinetics of protein loss and the expression by putative amacrine cells should be taken into account.
Tissue distribution of βGeo expression.
Examination of β-galactosidase activity in adult tissues shows that βGeo is expressed in a wide variety of cell types. RT-PCR analysis of these tissues shows that Fem1c is also expressed in the same tissues, but at this point it is not known whether the cell specificity characteristics are identical. Cells that most strongly express βGeo appear to be related, being either epithelia (such as the columnar epithelium of bronchioles in the lung) or derived from an epithelium (such as cells in the central nervous system). There is no distinction with respect to what embryological layer (ectoderm, mesoderm, or endoderm) βGeo-expressing cells originated from. Epithelial cells that are specialized to have a secretory or absorptive function are more likely to be labeled than other specialized epithelia, such as endothelia of blood vessels or mesothelia of the gut integument.
βGeo is also expressed in the central nervous system. With some regions, such as the inferior colliculus, it was not possible to determine whether staining was restricted to neurons or glial cells or both. In other regions (such as the retina, optic nerve, and cerebellum) of the central nervous system, however, no glial cells were observed to be X-Gal positive, making it likely that the majority of expressing cells in this tissue were neuronal. The strong expression in subtypes of neurons (particularly the CA1 pyramidal cells of the hippocampus and the Purkinje cells of the cerebellum) is particularly interesting. Like retinal ganglion cells in glaucoma, these cells share the feature of being highly susceptible in a variety of neurodegenerative diseases. The CA1 region is one of the most vulnerable regions affected in Alzheimer's disease and stroke (15, 20, 25), while Purkinje cells exhibit early damage in spinocerebellar ataxia and Creutzfeldt-Jakob disease (8) and may be involved in the cerebellar pathology of multiple sclerosis (6). Ganglion cells and CA1 pyramidal cells share a common pathological link in that they are highly susceptibility to glutamate toxicity. Purkinje cells, however, have not been reported to be similarly sensitive. Further study is required to establish whether Fem1c plays a significant role in the selective sensitivity of these cells to neurodegeneration.
Acknowledgments
This work was funded by grants from the National Eye Institute (R29 EY 12223 and R03 EY13790 to R.W.N.) and by the American Health Assistance Foundation, the Retina Research Foundation, the Glaucoma Foundation, and Research to Prevent Blindness, Inc.
We thank Phillip Soriano for generously providing the initial ROSA3 mice, Gretchen Poulsen for her expertise in processing brain tissue for enzyme histochemistry, and Heather Rae Pelzel for her assistance in conducing RT-PCR experiments.
REFERENCES
- 1.Bonfanti, L., E. Strettoi, S. Chierzi, M. C. Cenni, X.-H. Liu, J. C. Martinou, L. Maffei, and S. A. Rabacchi. 1996. Protection of retinal ganglion cells from natural and axotomy-induced cell death in neonatal transgenic mice overexpressing bcl-2. J. Neurosci. 16:4186-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, S. L., K. O. Tan, L. Zhang, K. S. Y. Yee, F. Ronca, M. Y. Chan, and V. C. Yu. 1999. F1Aα, a death receptor-binding protein homologous to the Caenorhabditis elegans sex-determining protein FEM-1, is a caspase substrate that mediates apoptosis. J. Biol. Chem. 274:32461-32468. [DOI] [PubMed] [Google Scholar]
- 3.Chan, S. L., K. S. Y. Yee, K. M. L. Tan, and V. C. Yu. 2000. The Caenorhabditis elegans sex determination protein FEM-1 is a CED-3 substrate that associates with CED-4 and mediates apoptosis in mammalian cells. J. Biol. Chem. 275:17925-17928. [DOI] [PubMed] [Google Scholar]
- 4.Chandross, K. J., R. I. Cohen, P. Paras, Jr., M. Gravel, P. E. Braun, and L. D. Hudson. 1999. Identification and characterization of early glial progenitors using a transgenic selection strategy. J. Neurosci. 19:759-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Z., G. A. Friedrich, and P. Soriano. 1994. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 8:2293-2301. [DOI] [PubMed] [Google Scholar]
- 6.Craner, M. J., A. C. Lo, J. A. Black, D. Baker, J. Newcombe, M. L. Cuzner, and S. G. Waxman. 2003. Annexin II/p11 is up-regulated in Purkinje cells in EAE and MS. Neuroreport 14:555-558. [DOI] [PubMed] [Google Scholar]
- 7.Doniach, T., and J. Hodgkin. 1984. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev. Biol. 106:223-235. [DOI] [PubMed] [Google Scholar]
- 8.Ferrer, I. 2002. Synaptic pathology and cell death in the cerebellum in Creutzfeldt-Jacob disease. Cerebellum 1:213-222. [DOI] [PubMed] [Google Scholar]
- 9.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513-1523. [DOI] [PubMed] [Google Scholar]
- 10.Gaudet, J., I. VanderElst, and A. M. Spence. 1996. Post-transcriptional regulation of sex determination in Caenorhabditis elegans: widespread expression of the sex-determining gene fem-1 in both sexes. Mol. Biol. Cell 7:1107-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groden, J., A. Thliveris, W. Samowitz, M. Carlson, L. Gelbert, H. Albertsen, G. Joslyn, J. Stevens, L. Spirio, M. Robertson, et al. 1991. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66:589-600. [DOI] [PubMed] [Google Scholar]
- 12.Kawai, J., A. Shinagawa, K. Shibata, M. Yoshino, M. Itoh, Y. Ishii, T. Arakawa, A. Hara, Y. Fukunishi, H. Konno, J. Adachi, S. Fukuda, and K. Aizawa. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420:563-573. [DOI] [PubMed] [Google Scholar]
- 13.Kimble, J., L. Edgar, and D. Hirsh. 1984. Specification of male development in Caenorhabditis elegans: the fem genes. Dev. Biol. 105:234-239. [DOI] [PubMed] [Google Scholar]
- 14.Krakow, D., E. Sebald, L. M. King, and D. H. Cohn. 2001. Identification of human FEM1A, the ortholog of a C. elegans sex-differentiation gene. Gene 279:213-219. [DOI] [PubMed] [Google Scholar]
- 15.Leifer, D., and N. W. Kowall. 1993. Immunohistochemical patterns of selective cellular vulnerability in human cerebral ischemia. J. Neurol. Sci. 119:217-228. [DOI] [PubMed] [Google Scholar]
- 16.Levin, L. A., C. L. Schlamp, R. L. Spieldoch, K. M. Geszvain, and R. W. Nickells. 1997. Identification of bcl-2 family genes in the rat retina. Investig. Ophthalmol. Vis. Sci. 38:2545-2553. [PubMed] [Google Scholar]
- 17.Li, Y., C. L. Schlamp, and R. W. Nickells. 1999. Experimental induction of retinal ganglion cell death in adult mice. Investig. Ophthalmol. Vis. Sci. 40:1004-1008. [PubMed] [Google Scholar]
- 18.Li, Y., C. L. Schlamp, G. Poulsen, M. Jackson, A. Griep, and R. W. Nickells. 15 September 2002, posting date. p53 regulates apoptotic ganglion cell death induced by N-methyl-D-aspartate. Mol. Vis. 8:341-350. [Online.] http://www.molvis.org/molvis/v8/a41. [PubMed] [Google Scholar]
- 19.Li, Y., C. L. Schlamp, K. P. Poulsen, and R. W. Nickells. 2000. Bax-dependent and independent pathways of retinal ganglion cell death induced by different damaging stimuli. Exp. Eye Res. 71:209-213. [DOI] [PubMed] [Google Scholar]
- 20.Mattson, M. P., P. B. Guthrie, and S. B. Kater. 1989. Intrinsic factors in the selective vulnerability of hippocampal pyramidal neurons. Prog. Clin. Biol. Res. 317:333-351. [PubMed] [Google Scholar]
- 21.Mosinger Ogilvie, J., T. L. Deckwerth, C. M. Knudson, and S. J. Korsmeyer. 1998. Suppression of developmental retinal cell death but not of photoreceptor degeneration in Bax-deficient mice. Investig. Ophthalmol. Vis. Sci. 39:1713-1720. [PubMed] [Google Scholar]
- 22.Neufeld, A. H., A. Sawada, and B. Becker. 1999. Inhibition of nitric-oxide synthase 2 by aminoguanidine provides neuroprotection of retinal ganglion cells in a rat model of chronic glaucoma. Proc. Natl. Acad. Sci. USA 96:9944-9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickells, R. W. 1999. Apoptosis of retinal ganglion cells in glaucoma: an update of the molecular pathways involved in cell death. Surv. Ophthalmol. 43(Suppl.):S151-S161. [DOI] [PubMed] [Google Scholar]
- 24.Quigley, H. A., R. W. Nickells, L. A. Kerrigan, M. E. Pease, D. J. Thibault, and D. J. Zack. 1995. Retinal ganglion cell death in experimental glaucoma and after axotomy occurs by apoptosis. Investig. Ophthalmol. Vis. Sci. 36:774-786. [PubMed] [Google Scholar]
- 25.Rissman, R. A., A. J. Mishizen-Eberz, T. L. Carter, B. B. Wolfe, A. L. De Blas, C. P. Miralles, M. D. Ikonomovic, and D. M. Armstrong. 2003. Biochemical analysis of GABA(A) receptor subunits α1, α5, β1, β2 in the hippocampus of patients with Alzheimer's disease neuropathology. Neuroscience 120:695-704. [DOI] [PubMed] [Google Scholar]
- 26.Schlamp, C. L., E. C. Johnson, Y. Li, J. C. Morrison, and R. W. Nickells. 15 August 2001, posting date. Changes in Thy1 gene expression associated with damaged retinal ganglion cells. Mol. Vis. 7:192-201. [Online.] http://www.molvis.org/molvis/v7/a27. [PubMed] [Google Scholar]
- 27.Schlamp, C. L., G. L. Poulsen, T. M. Nork, and R. W. Nickells. 1997. Nuclear exclusion of wild-type p53 in immortalized human retinoblastoma cells. J. Natl. Cancer Inst. 89:1530-1536. [DOI] [PubMed] [Google Scholar]
- 28.Shibata, K., M. Itoh, K. Aizawa, S. Nagaoka, N. Sasaki, P. Carninci, et al. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409:685-690. [DOI] [PubMed] [Google Scholar]
- 29.Siliprandi, R., R. Canella, G. Carmignoto, N. Schiavo, A. Zanellato, R. Zanoni, and G. Vantini. 1992. N-methyl-D-aspartate-induced neurotoxicity in the adult rat retina. Vis. Neurosci. 8:567-573. [DOI] [PubMed] [Google Scholar]
- 30.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 31.Ventura-Holman, T., and J. F. Maher. 2000. Sequence, organization, and expression of the human FEM1B gene. Biochem. Biophys. Res. Commun. 267:317-320. [DOI] [PubMed] [Google Scholar]
- 32.Ventura-Holman, T., M. F. Seldin, W. Li, and J. F. Maher. 1998. The murine Fem1 gene family: homologs of the Caenorhabditis elegans sex-determination protein FEM-1. Genomics 54:221-230. [DOI] [PubMed] [Google Scholar]