Abstract
Αn α-helical MA-3 domain appears in several translation initiation factors, including human eukaryotic translation initiation factor 4G (eIF4G) and DAP-5/NAT1/p97, as well as in the tumor suppressor Pdcd4. The function of the MA-3 domain is, however, unknown. C-terminal eIF4G (eIG4Gc) contains an MA-3 domain that is located within the eIF4A-binding region, suggesting a role for eIF4A binding. Interestingly, C-terminal DAP-5/NAT1/p97 contains an MA-3 domain, but it does not bind to eIF4A. Mutation of amino acid residues conserved between Pdcd4 and eIF4Gc but not in DAP-5/NAT1/p97 to the amino acid residues found in the DAP-5/NAT1/p97 indicates that some of these amino acid residues within the MA-3 domain are critical for eIF4A-binding activity. Six Pdcd4 mutants (Pdcd4E249K, Pdcd4D253A, Pdcd4D414K, Pdcd4D418A, Pdcd4E249K,D414K, and Pdcd4D253A,D418A) lost >90% eIF4A-binding activity. Mutation of the corresponding amino acid residues in the eIF4Gc also produced similar results, as seen for Pdcd4. These results demonstrate that the MA-3 domain is important for eIF4A binding and explain the ability of Pdcd4 or eIF4Gc but not DAP-5/NAT1/p97 to bind to eIF4A. Competition experiments indicate that Pdcd4 prevents ca. 60 to 70% of eIF4A binding to eIF4Gc at a Pdcd4/eIF4A ratio of 1:1, but mutants Pdcd4D253A and Pdcd4D253A,D418A do not. Translation of stem-loop structured mRNA is susceptible to inhibition by wild-type Pdcd4 but not by Pdcd4D253A, Pdcd4D418A, or Pdcd4D235A,D418A. Together, these results indicate that not only binding to eIF4A but also prevention of eIF4A binding to the MA-3 domain of eIF4Gc contributes to the mechanism by which Pdcd4 inhibits translation.
Translation initiation of capped mRNA is proposed to occur by a ribosomal scanning mechanism (20). A 43S ribosomal complex comprised of a 40S ribosomal subunit, the initiator Met-tRNAi, and translation initiation factors binds to the cap structure of mRNA at the 5′ end and scans downstream along the 5′-untranslated region (5′UTR) searching for the initiation codon. The rate-limiting step for this process is the binding of the 40S ribosomal subunit to mRNA. Several eukaryotic translation initiation factors (eIFs), including the critical factor eIF4A, participate in this process.
Translation initiation factor eIF4A is a member of the DEAD-box RNA helicase protein family. The DEAD-box family proteins catalyze RNA unwinding and/or rearrangement. eIF4A is an ATP-dependent RNA helicase that functions in translation initiation to catalyze the unwinding of mRNA secondary structure at the 5′UTR (36). Mutations of eIF4A that reduce or inactivate the RNA helicase activity result in inhibition of translation in vitro (28-30). The RNA helicase activity of eIF4A is enhanced by eIF4B or eIF4H binding (1, 35, 37). eIF4A is most active as a helicase when it is a subunit of eIF4F (a complex containing eIF4A, eIF4E, and eIF4G) (38). The eIF4G is a scaffold protein containing two eIF4A-binding sites: one located within the middle one-third (eIF4Gm) (amino acids 635 to 1039) and the other located within the C-terminal one-third (eIF4Gc) (amino acids 1040 to 1560) (24). Binding of eIF4A to eIF4Gm is sufficient for cap-dependent translation (11) and for internal ribosomal entry site-mediated translation (23), but binding of eIF4A to the eIF4Gc and eIF4Gm is required for robust translation (24).
Comparison of the amino acid sequences of several translation initiation factors reveals a conserved α-helical region called an MA-3 domain (or MI domain), which is located in human eIF4G, plant eIF(iso)4F, and eIF4G isoform DAP-5/NAT1/p97 but not in yeast eIF4G (2, 33). This domain is also found in two copies in the programmed cell death 4 (Pdcd4). The MA-3 domain extends over 120 amino acids with 80 to 85% consensus secondary structure. In the human eIF4G, the MA-3 domain is located within an eIF4A-binding region, implying that the MA-3 domain may play a role in binding to eIF4A. Interestingly, the C-terminal half of DAP-5/NAT1/p97 also contains an MA-3 domain, but it does not bind to eIF4A (18). Whether the MA-3 domain is essential for binding to eIF4A is unknown.
Pdcd4 is a ubiquitous protein expressed in all tissues examined (39). Although expression of the pdcd4 gene is upregulated in response to treatment with apoptosis inducers (7, 39) and downregulated by treatment with interleukin-2 or topoisomerase inhibitor (3, 27), no functional significance of Pdcd4 in these responses has been demonstrated. Recently, Pdcd4 was identified as a novel tumor suppressor (43), and loss of Pdcd4 expression is highly correlated to tumor progression (8). Reduction of Pdcd4 protein in JB6 transformation-resistant cells is accompanied by acquisition of a transformation-susceptible phenotype (9). Conversely, elevation of Pdcd4 protein level inhibits tetradecanoyl phorbol acetate (TPA)-induced transformation in JB6 transformation-susceptible cells and suppresses tumor phenotype in JB6 transformed cells (43, 44). Pdcd4 suppresses TPA-induced transformation and expression of tumor phenotype by specifically inhibiting AP-1 but not NF-κB or serum response element-dependent transcription (43, 44). Pdcd4 is a binding partner of eIF4A (14, 42). This was established by yeast two-hybrid and confirmed by glutathione S-transferase pull-down, immunoprecipitation, and immunolocalization (42). Pdcd4 inhibits translation both in the rabbit reticulocyte lysate system and in transfected JB6 cells (42). The inhibition of translation and of AP-1-dependent transcription requires binding to eIF4A because Pdcd4D418A, a mutant inactivated for binding to eIF4A, fails to inhibit cap-dependent translation or AP-1 transactivation (42). An in vitro binding assay indicated that Pdcd4 prevented eIF4A binding to eIF4Gc (42). In addition, Pdcd4 also binds to eIF4Gm independently of eIF4A binding. These findings suggest a possible mechanism in which Pdcd4 inhibits translation by (i) competition with eIF4Gc for binding to eIF4A (ii) inhibition of the helicase activity of eIF4A when Pdcd4 binds to the eIF4Gm (42).
In order to elucidate the mechanism by which Pdcd4 inhibits translation and to determine the function of the MA-3 domain, we performed deletion and mutation analyses of the MA-3 domains of Pdcd4 and eIF4Gc. These analyses demonstrate that the MA-3 domain is essential for binding of both Pdcd4 and eIF4Gc to eIF4A. We also demonstrated that the MA-3 domain in DAP-5/NAT1/p97 is nonfunctional for eIF4A binding. The Pdcd4 mutants that are defective for binding to eIF4A fail to prevent eIF4A binding to eIF4Gc and fail to inhibit translation, indicating that prevention of eIF4A binding to eIF4Gc contributes to the mechanism by which Pdcd4 inhibits translation.
MATERIALS AND METHODS
Construction of plasmids.
The plasmids pCMV-BD-Pdcd4 and pCMV-AD-eIF4A were used for mammalian two-hybrid analysis as previously described (42). To construct pCMV-BD-eIF4Gc, a fragment amplified by Pfu DNA polymerase-dependent PCR, which encodes the eIF4Gc (amino acids 1040 to 1560) was inserted into the EcoRI and ApaI sites of the pCMV-BD vector. The plasmid pCMV-NLS-Pdcd4 used for competition experiments was constructed by deleting the DNA sequence of NF-κB activation domain of pCMV-AD by ClaI and BamHI digestion and ligation with Pdcd4 cDNA. The luciferase cDNA was excised from pGL3-Basic vector (Promega), ligated into the HindIII and BamHI sites of the pcDNA3.1+ vector (Invitrogen), and named pcDNA-LUC. The pcDNA-SL-LUC plasmid was constructed by insertion of an oligonucleotide, 5′-AAGCTTGGGCCCAGATCTACGCGTACGTACGCGTAGATCTGGGCCCAAGCTT-3′ (26), at the HindIII site of pcDNA-LUC plasmid. This plasmid produced a 24-bp stem-loop structure at the 5′UTR of luciferase mRNA with a ΔG of −44.8 kcal/mol (as calculated by the mfold program).
Recombinant protein expression and purification.
His-eIF4A was expressed in Escherichia coli BL21(DE3). The recombinant His-eIF4A expression was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1.0 mM when cells were cultured to mid-log phase (optical density at 600 nm of 0.6 to 1.0). After 4 h, cells were harvested and lysed by using BugBuster reagent (Novagen) including Benzonase (25 U/ml). The His-eIF4A was purified by nickel-agarose beads (Qiagen) as described previously (31).
The recombinant Pdcd4 was expressed in insect Sf9 cells and purified on nickel-agarose resin as described previously (42).
Site-directed mutagenesis.
Site-directed mutagenesis of the MA-3 domains of Pdcd4 or eIF4Gc was performed by using a QuickChange kit (Stratagene), and all mutations were verified by DNA sequence analysis. The mutated Pdcd4 or eIF4Gc cDNAs were excised from the template plasmids and cloned into the EcoRI and ApaI sites of pCMV-BD to produce Gal4 DNA-binding domain-fused Pdcd4 or eIF4Gc constructs. The mutated Pdcd4 cDNAs were also cloned into pcDNA3.1+ or pcDNA4/His/Max B to produce the expression constructs.
Mammalian two-hybrid assay for protein-protein binding.
The Pdcd4 and its site-specific mutated cDNAs were fused to Gal4 DNA-binding domain (BD) of plasmid pCMV-BD and named pCMV-BD-Pdcd4. The eIF4A cDNA was fused to NF-κB activation domain (AD) of plasmid pCMV-AD and named pCMV-AD-eIF4A. A luciferase reporter becomes activated when the BD-Pdcd4 (wild-type or mutant) fusion protein binds to the AD-eIF4A fusion protein. JB6 RT101 cells (104 cells) were seeded in 24-well plates in Eagle minimal essential medium (EMEM) with 4% fetal bovine serum (FBS). Cells were transfected with 50 ng of pCMV-BD-Pdcd4 (wild type or mutants), 50 ng of pCMV-AD-eIF4A, 25 ng of Gal4-luciferase reporter gene, and 10 ng of thymidine kinase-Renilla luciferase gene with 2 μl of Lipofectamine (Invitrogen). For eIF4Gc-eIF4A binding assays, 400 ng of pCMV-AD-eIF4A was used. After 4 to 6 h, cells were incubated with EMEM containing 4% FBS. After 48 h, cells were lysed in 1× passive lysis buffer (Promega), and the luciferase activity was measured as previously described (43). The transfection efficiency was normalized to Renilla luciferase activity.
Pull-down and immunoprecipitation assays.
The pull-down assays were performed as previously described (42). In brief, 5 × 105 JB6 RT101 cells were transfected with 10 μg of wild type or a site-specifically mutated Pdcd4 expression construct. After 48 h, cells were harvested and lysed in lysis buffer (20 mM HEPES-KOH [pH 7.6], 100 mM KCl, 0.5 mM EDTA, 20% glycerol, 0.5% Triton X-100, 50 μg of RNase A/ml, and 1× protease inhibitor cocktail [Boehringer Mannheim]). A total of 50 μg of His-eIF4A recombinant protein was mixed with a 10-μl bed volume of Co-Sepharose resin (Pierce). After a wash with lysis buffer, His-eIF4A bound resin was added to cell lysates, followed by incubation for 2 h at 4°C. For immunocoprecipitation assays, 5 × 105 JB6 RT101 cells were transfected with 10 μg of xpress-tagged wild-type or site-specifically mutated Pdcd4 expression construct. After 48 h, 2 μg of xpress antibody (Invitrogen) was added to the cell lysate, followed by incubation for 2 h at 4°C. Then, 10 μl of protein G-Sepharose beads (Pharmacia) was washed with 250 μl of lysis buffer twice, added to the cell lysates, and rotated overnight at 4°C.
The beads were washed three times with 250 μl of lysis buffer and boiled in sodium dodecyl sulfate (SDS) sample buffer. The bound proteins were resolved on a 10% Tris-Bis NuPage gel (Invitrogen) and detected by using Pdcd4 or eIF4AI antibody (Santa Cruz).
Competition assays.
JB6 RT101 cells (104 cells) were seeded in 24-well plates in EMEM with 4% FBS. Cells were transfected with increasing amounts (0 to 400 ng) of pCMV-NLS-Pdcd4 (wild type or site-specific mutant), 50 ng of pCMV-BD-eIF4Gc, 400 ng of pCMV-AD-eIF4A, 25 ng of Gal4-luciferase reporter gene, and 10 ng of thymidine kinase Renilla luciferase gene with 2 μl of Lipofectamine (Invitrogen). After 4 to 6 h, cells were incubated with EMEM with 4% FBS. After 48 h, cells were lysed in 1× passive lysis buffer (Promega), and the luciferase activity was measured as previously described (43). The transfection efficiency was normalized to the Renilla luciferase activity.
In vitro transcription and in vitro translation.
The luciferase and stem-loop luciferase mRNAs were in vitro transcribed by using a MEGAscript T7 kit (Ambion) with pcDNA-LUC and pcDNA-SL-LUC as templates, respectively. Portions (10 μl) of nuclease-treated rabbit reticulocyte lysate (Promega) were mixed with 1 μl of SUPERase.In (40 U; Ambion), 1 μl of amino acid mixture-Met (1 mM each amino acid), 0.2 μg of luciferase or stem-loop luciferase mRNA, 15 μCi of [35S]methionine, and recombinant Pdcd4 protein (0 to 2.0 μg). The reaction mixture was added to buffer A (20 mM HEPES-KOH [pH 7.6], 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 10% glycerol) to a final volume of 20 μl, followed by incubation at 30°C for 1 h. The products of translation were resolved by SDS-polyacrylamide gel electrophoresis, fixed with 40% methanol-7% acetic acid, and treated with Amplify (Amersham). The intensity of bands was determined by a Storm 850 PhosphorImager (Molecular Dynamics).
Transient-transfection assays of translation.
An in vivo translation assay of the luciferase and stem-loop luciferase reporter system was described previously (42). Briefly, 8 × 104 JB6 RT101 cells were seeded in a six-well plate in EMEM with 4% FBS. After transfection with 0 to 2 μg of pcDNA-Pdcd4 (or Pdcd4 mutants) or 0.2 μg of pcDNA-LUC (non-stem-loop luciferase) or pcDNA-SL-LUC (stem-loop luciferase), cells were allowed to recover for 12 to 18 h in EMEM with 4% FBS. Cells were then serum starved with 0.2% FBS in EMEM medium for 24 h, followed by incubation in EMEM-4% FBS for an additional 24 h. Cells were lysed in 1× passive lysis buffer (Promega), and the luciferase activity was measured as previously described (43).
Northern blot analysis.
RT101 cells (5 × 105 cells) were transiently transfected with 10 μg of pcDNA-Pdcd4 expression plasmid (or Pdcd4 mutants) and 1 μg of pcDNA-SL-LUC as described above. After serum starvation for 24 h, cells were incubated with EMEM-4% FBS medium for an additional 24 h. Cells were then harvested and lysed in TRIzol reagent (Invitrogen). The total RNA was isolated according to the manufacturer's protocol. Then, 10 μg of RNA was separated on a 1% formaldehyde gel and transferred to a nylon membrane (Boehringer Mannheim) as previously described (45). The membrane was prehybridized with NorthernMax Prehybridization/Hybridization buffer (Ambion) for 30 min, hybridized by incubation with 32P-labeled luciferase cDNA in NorthernMax Prehybridization/Hybridization buffer, and then washed according to the manufacturer's instructions.
RESULTS
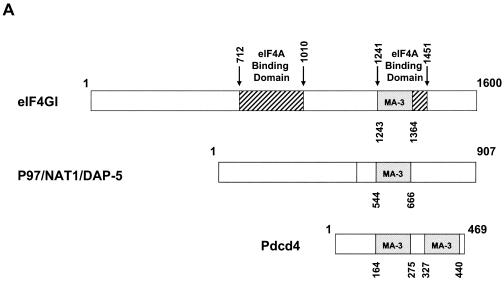
The MA-3 domains of Pdcd4 are required for binding to eIF4A but the MA-3 domain of DAP-5/NAT1/p97 is not functional for eIF4A binding.
Previously, we demonstrated that eIF4A is a binding partner of Pdcd4 (42), but the Pdcd4 domains used for binding to eIF4A are not known. Analysis of the amino acid sequence of Pdcd4 indicates that Pdcd4 contains two MA-3 domains. One MA-3 domain consists of amino acids 164 to 275 (N-terminal MA-3 domain), and the other spans amino acids 327 to 440 (C-terminal MA-3 domain) (Fig. 1A). The eIF4G protein also contains an MA-3 domain located within the second eIF4A-binding domain (amino acids 1241 to 1451) (24) (Fig. 1A). Because MA-3 domains are prominent in two proteins that bind to eIF4A, namely, Pdcd4 and eIF4G, the MA-3 domain may play a role in binding to eIF4A. To determine whether the MA-3 domains in Pdcd4 function as mediators for binding to eIF4A, we performed mammalian two-hybrid assays with the Gal4 DNA-binding domain fused to various lengths of deleted Pdcd4 plasmids and the NF-κB activation domain fused to full-length eIF4A plasmid. The mammalian two-hybrid assay is a sensitive method for detecting protein-protein interaction. The luciferase assay contributes to the high sensitivity, as does the fact that DNA-binding domain-Pdcd4 and activation domain-eIF4A translocate into the nucleus, thereby decreasing the background interference. Although overexpression of Pdcd4 and eIF4A may force binding, the physical interaction between these two proteins has been carefully evaluated previously by independent methods in vitro and in vivo (42). The mammalian two-hybrid assay renders likely the possibility that mutants scored as inactivated are not physically interacting with each other.
FIG. 1.
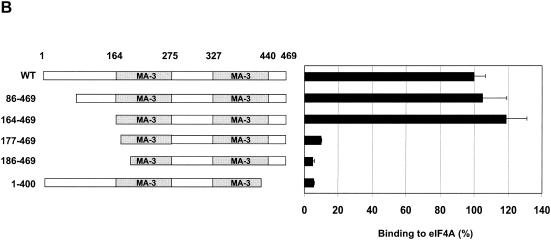
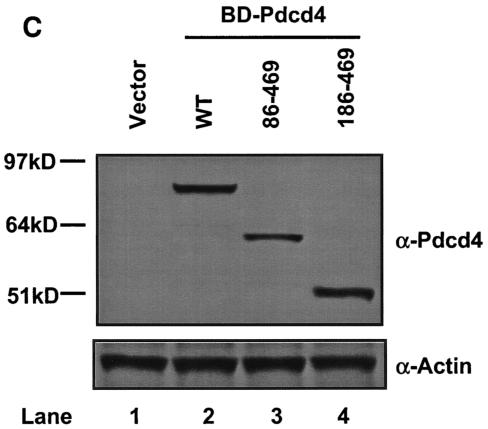
Both MA-3 domains of Pdcd4 are required for eIF4A-binding activity. (A) Structures of eIF4G1, DAP-5/NAT1/p97, and Pdcd4. The numbers refer to the size (in amino acids) of eIF4G1, DAP-5/NAT1/p97, and Pdcd4 and to the location of the eIF4A-binding domain and the MA-3 domain (2, 17, 24). The MA-3 domains (gray box) in eIF4G1, DAP-5/NAT1/p97, and Pdcd4 are indicated schematically. The eIF4A-binding domains (strip box and arrows) in eIF4G1 are indicated schematically. (B) Partial deletion of N- or C-terminal MA-3 domain of Pdcd4 dramatically decreases the eIF4A-binding activity. Plasmid pCMV-BD-Pdcd4 (50 ng) (wild type [WT] or deletion mutants) was transiently transfected with pCMV-AD-eIF4A (50 ng) and Gal4-luciferase reporter DNA (25 ng) into JB6 RT101 cells. After 48 h, cells were lysed and the luciferase activity was measured. The luciferase activity with wild-type Pdcd4 was designated as 100%. These experiments were repeated three times with five independent transfections, and representative data are shown. The results are expressed as the means plus the standard deviation. (C) Protein expression level of wild-type and truncated Gal4 DNA-binding domain-Pdcd4. Cell lysates (15 μg) from transiently transfected empty vector (lane 1), wild-type (WT) Pdcd4 (lane 2), or truncated Pdcd4 (lanes 3 and 4) were separated on 10% Bis-Tris NuPage gels, transferred to nitrocellulose, and subjected to immunoblotting with Pdcd4 or actin antibody with visualization by chemiluminescent detection.
The Pdcd486-469 and Pdcd4164-469 mutants have the first 85 and 163 amino acid residues of Pdcd4 deleted, respectively, and contain both MA-3 domains intact. Two further N-terminal deletions, Pdcd4177-469 and Pdcd4186-469, are partially deleted in the N-terminal MA-3 domain and retain the intact C-terminal MA-3 domain. The Pdcd41-400 is partially deleted in the C-terminal MA-3 domain and retains the N-terminal intact MA-3 domain (Fig. 1B). As shown in Fig. 1B, the full-length Pdcd4, Pdcd486-469 and Pdcd4164-469 show similar eIF4A-binding abilities. Deletion mutants Pdcd4177-469, Pdcd4186-469 and Pdcd41-400 showed ∼10% of the eIF4A-binding activity seen for full-length Pdcd4, indicating that partial deletion of either the N- or the C-terminal MA-3 domain dramatically decreases binding to eIF4A. These results indicate that both MA-3 domains of Pdcd4 are critical for binding to eIF4A. The decrease in the eIF4A-binding activity is not due to reduced Pdcd4 protein expression since wild-type Pdcd4 and Pdcd4 deletion mutants show similar expression (Fig. 1C).
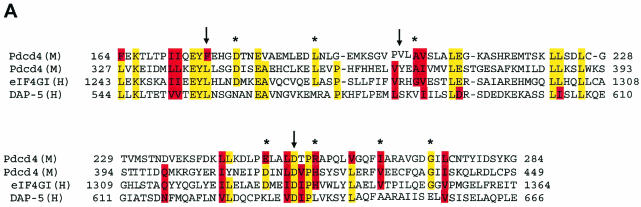
To further define the amino acid residues of the Pdcd4 MA-3 domain that mediate binding to eIF4A, we created site-specific mutations in the MA-3 domains. In the human eIF4G, the MA-3 domain is located within the second eIF4A-binding domain (amino acids 1241 to 1451) (24). However, DAP-5/NAT1/p97, although containing an MA-3 domain in the C-terminal half (2, 33), fails to bind to eIF4A (18). Comparison of the amino acid sequences in Pdcd4, eIF4G, and DAP-5/NAT1/p97 indicates that several amino acid residues are conserved between Pdcd4 and human eIF4G but not in DAP-5/NAT1/p97 (Fig. 2A). Our strategy to generate the Pdcd4 mutants was to change the amino acid residues that are identical between Pdcd4 and eIF4G but differ in DAP-5/NAT1/p97 to the amino residues found in the DAP-5/NAT1/p97 (marked as asterisks in Fig. 2A). In addition, we randomly created three site-specific mutations (Pdcd4L339A, Pdcd4Y365E, and Pdcd4D418A) within the C-terminal MA-3 domain (Fig. 2A, arrows). The wild-type or mutated Pdcd4 cDNA was cloned into the Gal4 DNA-binding domain vector (pCMV-DB), and the eIF4A cDNA was cloned into the NF-κB activation domain vector (pCMV-AD). These two plasmids and Gal4-luciferase reporter plasmid were then cotransfected into JB6 RT101 cells. The binding of eIF4A to various Pdcd4 mutants was determined by mammalian two-hybrid assay.
FIG. 2.
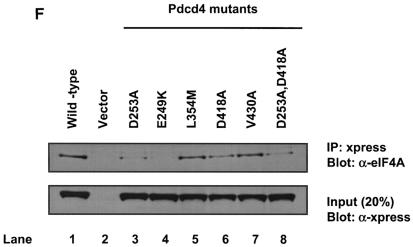
The MA-3 domain of DAP-5/NAT1/p97 is not functional for binding to eIF4A. (A) Alignment of the MA-3 domain of eIF4G1, DAP-5/NAT1/p97, and Pdcd4. Conserved (yellow box) and similar (red box) amino acid residues between eIF4G1, DAP-5/NAT1/p97, and Pdcd4 are indicated. The amino acids that are conserved between eIF4G1 and Pdcd4 but are different from DAP-5/NAT1/p97 are marked with asterisks. (B and C) Point mutation analyses of the C-terminal (B) and N-terminal (C) MA-3 domain of Pdcd4 for eIF4A-binding activity. Plasmid pCMV-BD-Pdcd4 (50 ng) (wild type [WT] or site-specific mutants) was transiently transfected with pCMV-AD-eIF4A (50 ng) and Gal4-luciferase reporter DNA (25 ng) into mouse JB6 RT101 cells. After 48 h, cells were lysed and the luciferase activity was measured. The luciferase activity with wild-type Pdcd4 was designated as 100%. These experiments were repeated three times with five independent transfections, and representative data are shown. The results are expressed as the means plus the standard deviations. In panel C, the corresponding mutant in the C-terminal MA-3 domain is indicated in parentheses. (D) Similar expression of wild-type and mutant Gal4 DNA-binding domain-Pdcd4. RT101 cell lysates (10 μg) from transient transfection with wild-type (lane 1) and mutant (lanes 2 to 8) pCMV-BD-Pdcd4 expression plasmids were separated on 10% Bis-Tris NuPage gels, transferred to nitrocellulose, and subjected to immunoblotting with Gal4 DNA-binding domain antibody, with visualization by chemiluminescent detection. (E) Pull-down assay of Pdcd4 with His-eIF4A. RT101 cell lysates from a transient transfection with Pdcd4 wild-type expression plasmid (lane 1), empty vector (lane 2), or site-specific Pdcd4 mutant expression plasmids (lanes 3 to 6) were pulled down with His-eIF4A. The bound proteins were resolved by using a 10% Bis-Tris NuPage gel, transferred to nitrocellulose, and subjected to immunoblotting with Pdcd4 or eIF4A antibody. (F) Coimmunoprecipitation of eIF4A with xpress-tagged Pdcd4. RT101 cell lysates from transient transfection with xpress-tagged Pdcd4 wild-type expression plasmid (lane 1), empty vector (lane 2), or site-specific xpress-tagged Pdcd4 mutant expression plasmids (lanes 3 to 8) were immunoprecipitated with mouse monoclonal xpress antibody. The bound proteins were resolved by using a 10% Bis-Tris NuPage gel, transferred to nitrocellulose, and subjected to immunoblotting with eIF4A or xpress antibody.
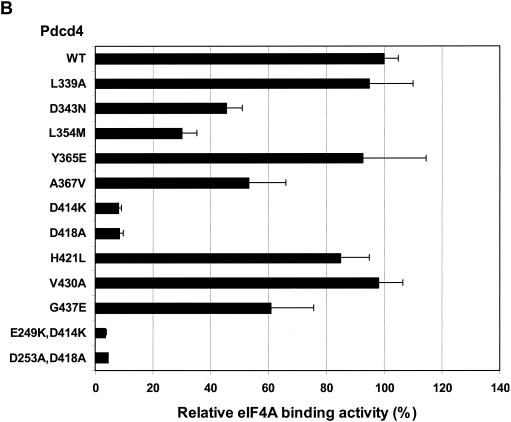
We first assessed site-specific mutations within the C-terminal MA-3 domain of Pdcd4. Pdcd4L339A, Pdcd4Y365E, Pdcd4H421L, and Pdcd4V430A showed no significant difference in eIF4A-binding activity compared to the wild-type Pdcd4 (Fig. 2B). When 343Asp was changed to Asn, 354Leu was changed to Met, 367Ala changed to Val, or 437Gly changed to Glu, these mutants of Pdcd4 showed a partial (40 to 60%) decrease in eIF4A-binding activity (Fig. 2B). In agreement with previous observations (42), the Pdcd4D418A showed a dramatic decrease in binding to eIF4A. In addition to Pdcd4D418A, the Pdcd4D414K, in which Asp was changed to Lys, also showed ca. 10% of wild-type eIF4A-binding activity, indicating that this amino acid residue is also critical for eIF4A-binding activity in the C-terminal MA-3 domain of Pdcd4.
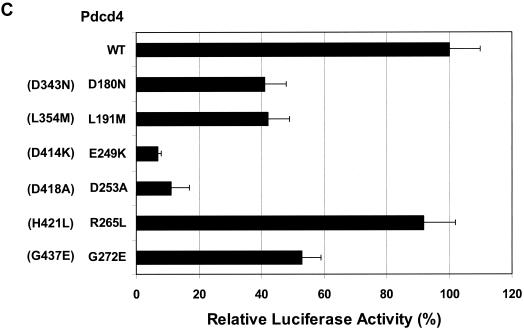
To define N-terminal MA-3 residues of Pdcd4 that contribute to eIF4A-binding activity, we created several mutations within the N-terminal MA-3 domain. As shown in Fig. 2C, amino acid residues mutated to correspond to the mutations made in the C-terminal MA-3 domain have eIF4A-binding activity similar to the C-terminal MA-3 domain mutants. The Pdcd4R265L corresponding to Pdcd4H421L in the C-terminal MA-3 domain, showed-binding activity similar to wild-type Pdcd4. The Pdcd4 D180N, Pdcd4L191M, and Pdcd4G272E corresponding to Pdcd4D343N, Pdcd4L354M, and Pdcd4G437E, respectively, have 40 to 50% of the eIF4A-binding activity seen for wild-type Pdcd4. Mutation of 249Glu to Lys or of 253Asp to Ala in the N-terminal MA-3 domain of Pdcd4, corresponding to Pdcd4D414K and Pdcd4D418A in the C-terminal MA-3 domain, produced ∼10% of wild-type eIF4A-binding activity. Double mutations Pdcd4E249K,D414K and Pdcd4D253A,D418A, in which 249Glu and 414Asp were changed to Lys and 253Asp and 418Asp were changed to Ala, respectively, showed 5 to 8% of wild-type Pdcd4-binding activity (Fig. 2B; see also Fig. 3D). As shown in Fig. 2D, the expression of wild-type Pdcd4 (lane 1), a mutant with high eIF4A-binding activity (Pdcd4V430V) (lane 7), two mutants showing a partial reduction of eIF4A-binding activity (Pdcd4L180M and Pdcd4L354M) (lanes 2 and 4), and mutants with very low eIF4A-binding activity (Pdcd4D235A, Pdcd4D141K, Pdcd4D418A, and Pdcd4D235A,D418A) (lanes 3, 5, 6, and 8) all show similar protein expression levels, indicating that loss of eIF4A-binding activity was not attributable to loss of expression of mutant Pdcd4s.
FIG. 3.
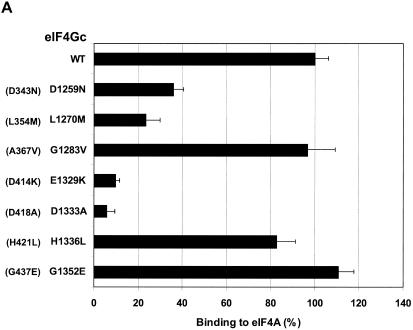
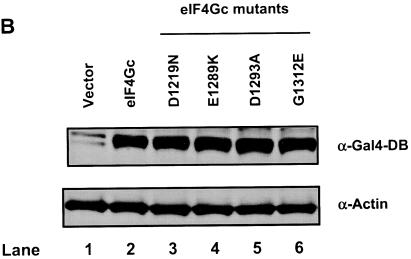
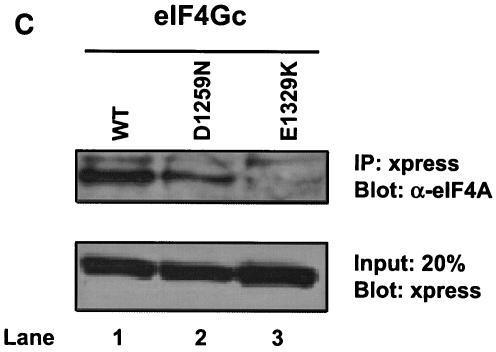
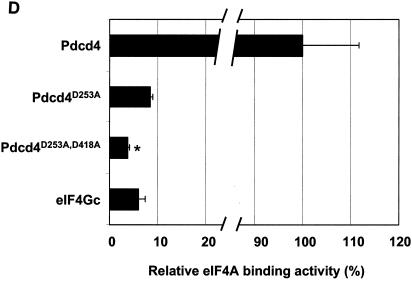
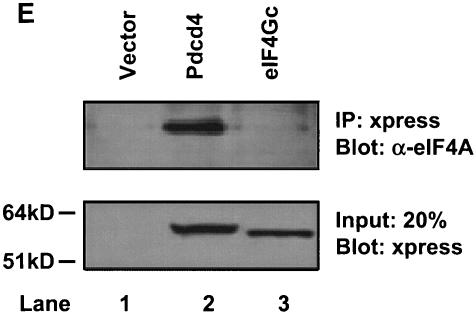
eIF4Gc has an MA-3 domain that is functional for eIF4A binding. (A) Plasmid pCMV-BD-eIF4Gc (50 ng) (wild type [WT] or site-specific mutant) was transiently transfected with pCMV-AD-eIF4A (400 ng) and Gal4-luciferase reporter DNA (25 ng) into JB6 RT101 cells. (B) Similar expression of wild-type and mutant Gal4 DNA-binding domain-eIF4Gc. RT101 cell lysates (10 μg) from a transient transfection with wild type (lane 2) and site-specific mutants (lanes 3 to 6) pCMV-BD-eIF4Gc expression plasmids were separated on 10% Bis-Tris NuPage gels, transferred to nitrocellulose, and subjected to immunoblotting with Gal4 DNA-binding domain antibody, with visualization by chemiluminescent detection. (C) Coimmunoprecipitation of eIF4A with xpress-tagged eIF4Gc. RT101 cell lysates from transient transfection with xpress-tagged eIF4Gc (lane 1), xpress-tagged eIF4GcD1259N (lane 2), or xpress-tagged eIF4GcE1329K (lane 3) expression plasmids were immunoprecipitated with mouse monoclonal xpress antibody. The bound proteins were resolved on a 10% Bis-Tris NuPage gel, transferred to nitrocellulose, and subjected to immunoblotting with eIF4A or xpress antibody. (D) Plasmid pCMV-BD-Pdcd4 (50 ng) (wild type [WT] or point mutation mutants) or pCMV-BD-eIF4Gc (50 ng) was transiently transfected with pCMV-AD-eIF4A (50 ng) and Gal4-luciferase reporter DNA (25 ng) into JB6 RT101 cells. After 48 h, cells were lysed and the luciferase activity was measured. The luciferase activity with wild-type Pdcd4 was designated as 100%. These experiments were repeated three times with five independent transfections, and representative data are shown. The results are expressed as the means plus the standard deviations. An asterisk indicates a significant difference compared to the transfection with pCMV-BD-eIF4Gc, as determined by Student t test (P < 0.005). (E) Relative eIF4A-binding activity of eIF4Gc. RT101 cell lysates from a transient transfection with empty vector (lane 1) xpress-tagged Pdcd4 expression plasmid (lane 2) or xpress-tagged eIF4Gc expression plasmid (lane 3) were immunoprecipitated with mouse monoclonal xpress antibody. The bound proteins were resolved on a 10% Bis-Tris NuPage gel, transferred to nitrocellulose, and subjected to immunoblotting with eIF4A or xpress antibody.
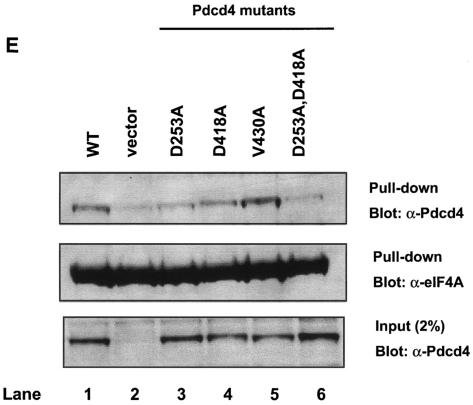
To further confirm the interaction between Pdcd4 mutants and eIF4A, we performed pull-down assays. The recombinant His-tagged eIF4A (His-eIF4A) was expressed and purified from E. coli and added to the RT101 cell lysate after overexpression of wild-type or site specifically mutated Pdcd4. The protein complex was pulled down by Co-Sepharose beads and resolved by SDS-polyacrylamide gel electrophoresis. As shown in Fig. 2E, His-eIF4A, in agreement with the observations seen in the mammalian two-hybrid assay, was able to pull down wild-type Pdcd4 and Pdcd4V430A (lanes 1 and 5). The vector control showed a faint band that corresponded to endogenous Pdcd4 pulled down by His-eIF4A (ca. 5 to 10% of wild-type Pdcd4). His-eIF4A also pulled down Pdcd4D253A and Pdcd4D418A (lanes 3 and 4), but the amount was 30 to 40% of the wild-type Pdcd4 and Pdcd4V430A (lanes 1 and 5), indicating that these Pdcd4 mutants have a weak eIF4A-binding activity. The band intensity of double mutant, Pdcd4D235A, D418A (lane 6), is ca. 10 to 20% of wild-type or Pdcd4V430A, suggesting that this mutant has very weak eIF4A-binding activity that is barely distinguishable from the vector control level. It is noteworthy that the abundant endogenous eIF4A and His-eIF4A compete with each other for binding to Pdcd4 (or Pdcd4V430A), resulting in less Pdcd4 (or Pdcd4V430A) being pulled down by His-eIF4A. This competition may contribute to the observation that the eIF4A-binding activity of Pdcd4D253A or Pdcd4D418A was only reduced two- to threefold in contrast to a 10-fold reduction in the mammalian two-hybrid assay. In addition to pull-down assays, we also performed coimmunoprecipitation assays with xpress antibody. As shown in Fig. 2F, xpress antibody coprecipitated eIF4A from the lysates after transient transfection with xpress-tagged Pdcd4 and Pdcd4V430A (lane 1 and 7). The xpress-tagged Pdcd4L354M coprecipitated ca. 60 to 80% as much eIF4A as Pdcd4 or Pdcd4V430A (lane 5). The xpress-tagged Pdcd4E249K, Pdcd4D253A, Pdcd4D418A, and Pdcd4D253A,D418A also coprecipitated with eIF4A, but the amount was 3 to 15% of the amount for Pdcd4 or Pdcd4V430A (lanes 3, 4, 6, and 8). Interestingly, Pdcd4D418A showed a little higher eIF4A binding than Pdcd4D253A in both pull-down and coimmunoprecipitation assays. It is noteworthy that the RT101 cells express a low level of Pdcd4 (44). The coimmunoprecipitation assays show little interference by endogenous Pdcd4, with the result that the eIF4A-binding activity of Pdcd4D253A or Pdcd4D418A shows a greater fold reduction than that seen in pull-down assays. These coimmunoprecipitation results are in agreement with the results of mammalian two-hybrid (Fig. 2B and C) and pull-down assays (Fig. 2E).
Mutations in either MA-3 domain of Pdcd4 dramatically reduced the eIF4A-binding activity, indicating that both N- and C-terminal MA-3 domains mediate binding to eIF4A. Both MA-3 domains are important and have similar eIF4A-binding activity. It was also demonstrated that the MA-3 domain of DAP-5/NAT1/p97 does not have eIF4A-binding activity because several critical amino acid residues in MA-3 domain are not conserved. For example, negatively charged residues 414Asp of Pdcd4 and 1329Asp of eIF4G are conserved but a positively charged residue 631Lys is found in DAP-5/NAT1/p97.
The MA-3 domain of eIF4Gc is functional for eIF4A binding.
To determine whether the MA-3 domain in the eIF4Gc is also functional for eIF4A binding, point mutations in the MA-3 domain of eIF4Gc were created by changing to the amino acid residues found in the DAP-5/NAT1/p97, which are identical between Pdcd4 and eIF4G but differ in DAP-5/NAT1/p97 (marked as asterisks in Fig. 2A). These mutants were assayed for eIF4A-binding activity by mammalian two-hybrid assays. In order to increase the sensitivity for detecting the binding of eIF4Gc and eIF4A, the ratio of eIF4Gc to eIF4A was increased to 1:8 instead of the 1:1 ratio that was used previously with Pdcd4. As shown in Fig. 3A, eIF4GcG1283V, eIF4GcH1336L, and eIF4GG1352E have an eIF4A-binding activity similar to wild-type eIF4Gc. Two point mutants (eIF4GcD1259N and eIF4GcL1270M) showed ca. 30 to 40% of wild-type binding activity, whereas the eIF4GcE1329K and eIF4GcD1333A expressed ca. 10% of wild-type eIF4A-binding activity. The eIF4A binding activities of eIF4Gc mutants are in agreement with the corresponding mutants created in the N- or C-terminal MA-3 domain of Pdcd4 with the exception of eIF4GcG1352E. The eIF4GcG1352E retained 94% of wild-type eIF4Gc binding to eIF4A, whereas the corresponding mutant in the C-terminal MA-3 domain, Pdcd4G437E, had 60% of wild-type eIF4A-binding activity, suggesting some divergence of the MA-3 domain in Pdcd4 and eIF4Gc. Figure 3B shows that eIF4Gc wild type and mutants have similar protein expression levels, indicating that loss of eIF4A-binding activity was not attributable to the loss of expression of mutant Pdcd4s. Since the specificity of the Gal4 DNA-binding domain antibody is low, it may cross-react with other proteins to generate nonspecific bands (Fig. 3B, lane 1).
To further confirm these interactions, we performed immunoprecipitation assays. In these experiments, we used xpress-tagged wild-type eIF4Gc and two representative xpress-tagged eIF4Gc mutants: eIF4GcD1259N (40% of eIF4A-binding activity) and eIF4GcE1329K (10% of eIF4A-binding activity). Xpress-eIFGc was able to coimmunoprecipitate with eIF4A from the lysates transiently transfected with xpress-tagged eIF4Gc (Fig. 3C, lane 1). The amount of eIF4A coimmunoprecipitated by xpress-eIF4Gc is approximately two- to threefold higher than that by xpress-eIF4GcD1259N (Fig. 3C, lanes 1 and 2). Xpress-eIF4GcE1329K shows little or no coimmunoprecipitation with eIF4A (Fig. 3C, lane 3). These results are in agreement with the results of mammalian two-hybrid assays (Fig. 3A).
What is the relative eIF4A-binding activity for eIF4Gc that has only one intact MA-3 domain? As shown in Fig. 3D, the C-terminal MA-3 mutated Pdcd4D253A and the eIF4Gc each showed ca. 7 to 9% of the eIF4A-binding activity seen with wild-type Pdcd4, and each has a single intact MA-3 domain, whereas a double mutant, Pdcd4D235A,D418A, showed less eIF4A-binding activity than did eIF4Gc. The comparison of eIF4A binding to eIF4Gc and Pdcd4 was further confirmed by a coimmunoprecipitation assay (Fig. 3E). xpress-eIF4Gc showed ca. 5 to 7% of the binding to eIF4A seen with Pdcd4. These findings indicate that eIF4Gc and Pdcd4D253A, each containing only one intact MA-3 domain, have similar eIF4A-binding activities. In addition, these findings also suggest that eIF4A must bind to two MA-3 domains in order to achieve maximal eIF4A-binding activity, while interaction with a single MA-3 domain produces weak binding.
Pdcd4 prevents eIF4A binding to eIF4Gc.
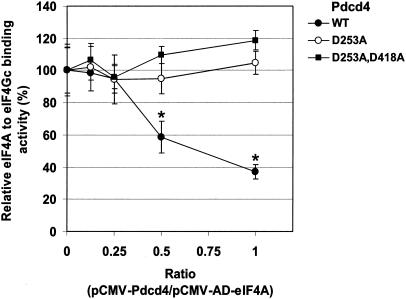
The finding that mutations in the MA-3 domain of Pdcd4 or eIF4Gc inactivate the eIF4A-binding activity (Fig. 2 and 3) suggests that Pdcd4 may compete with eIF4Gc and inhibit its binding to eIF4A. To test this hypothesis, we performed in vivo competition assays. Although Pdcd4 may contain two functional nuclear export signals, it is unclear whether Pdcd4 is able to translocate to the nucleus (4). In order to ensure that Pdcd4 is able to translocate to nuclei, as needed for the two-hybrid assay of binding, a DNA sequence encoding the simian virus 40 nuclear localization signal was ligated in front of Pdcd4 cDNA by deletion of the NF-κB activation domain sequence of pCMV-AD and termed pCMV-NLS-Pdcd4. Plasmids pCMV-BD-eIF4Gc and pCMV-AD-eIF4A were cotransfected with competition plasmid pCMV-NLS-Pdcd4 (or pCMV-NLS-Pdcd4D253A or pCMV-NLS-Pdcd4D253A,D418A) into RT101 cells. After 48 h, cells were lysed, and the luciferase activity was assayed to measure eIF4Gc-eIF4A binding as a function of cotransfected Pdcd4 concentration. As shown in Fig. 4, the binding of eIF4A to eIF4Gc was decreased in a concentration-dependent manner when the amount of wild-type Pdcd4 was increased. In contrast, expression of Pdcd4 mutants Pdcd4D253A and Pdcd4D253A,D418A did not affect eIF4A binding to eIF4Gc. The binding of eIF4A to eIF4Gc was ca. 60 to 70% inhibited by wild-type Pdcd4 at a ratio of pCMV-NLS-Pdcd4 to pCMV-AD-eIF4A of 1:1. The observation that wild type but not Pdcd4 mutant Pdcd4D253A or Pdcd4D253A,D418A (Fig. 2) inhibits eIF4A binding to eIF4Gc in transfected cells supports the hypothesis that Pdcd4 competes with eIF4Gc for binding to eIF4A in vivo.
FIG. 4.
Pdcd4 prevents eIF4A binding to eIF4Gc. Increasing amounts (0, 50, 100, 200, and 400 ng) of pCMV-NLS-Pdcd4 (or pCMV-NLS-Pdcd4D235A or pCMV-NLS-Pdcd4D253A,D418A) were transiently transfected with pCMV-AD-eIF4A (400 ng), pCMV-BD-eIF4Gc (50 ng), and Gal4-luciferase reporter DNA (25 ng) into JB6 RT101 cells. After 48 h, cells were lysed and the luciferase activity was measured. The luciferase activity from 0 ng of pCMV-BD-Pdcd4, pCMV-BD-Pdcd4D253A, or pCMV-BD-Pdcd4D253A,D418A was designated as 100%. These experiments were repeated two times with six independent transfections, and representative data are shown. The results are expressed as means ± the standard deviations. Asterisks indicate significant differences compared to the transfection with 200 or 400 ng of pCMV-BD-Pdcd4D235A, as determined by using the Student t test (P < 0.001).
A stable mRNA secondary structure in the 5′UTR confers susceptibility to inhibition of in vitro and in vivo translation by Pdcd4.
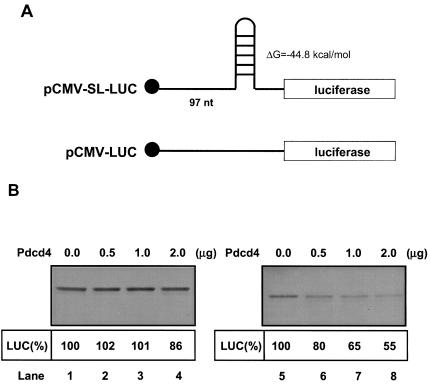
Previous studies (30, 40) suggested that the helicase activity of eIF4A is required for translation, especially translation of mRNAs having secondary structure in the 5′UTR. Because Pdcd4 specifically inhibited the helicase activity of eIF4A (42), we suspected that translation of mRNA with a stable secondary structure at the 5′UTR would be sensitive to suppression by Pdcd4. To test this hypothesis, we created a stable secondary structure introduced in the 5′ leader sequence of a luciferase reporter. The palindromic oligonucleotide sequence (AAGCTTGGGCCCAGATCTACGCGTACGTACGCGTAGATCTGGGCCCAAGCTT) was inserted at the 5′ end of luciferase cDNA, allowing the formation of a 24-bp stem-loop structure positioned 97 nucleotides downstream of the cap of the mRNA. In vitro studies have shown that stem-loop structures with free energies of between −30 and −70 kcal/mol are sufficient to inhibit translation (15). The ΔG value of the 24-bp stem-loop structure used in the present study is −44.8 kcal/mol (as calculated by the mfold program). This free energy of the 24-bp stem-loop is close to that of the TAR (+111) RNA of human immunodeficiency virus, with a stem-loop structure of free energy −49.9 kcal/mol (40).
The effects of Pdcd4 on translation of this stem-loop structured mRNA (or non-stem-loop mRNA) in vitro were measured by adding capped stem-loop luciferase mRNA (or non-stem-loop luciferase mRNA) in a rabbit reticulocyte lysate to which recombinant Pdcd4 was added. As expected, translation of the stem-loop structured mRNA (in the absence of Pdcd4) is relatively inefficient, showing ca. 2% of the translation rate of non-stem-loop structured mRNA (data not shown). Addition of recombinant Pdcd4 (0 to 2.0 μg) to the reticulocyte lysate inhibited stem-loop structured luciferase mRNA translation in a dose-dependent manner (Fig. 5B, lanes 5 to 8). The addition of 0.5 and 1.0 μg of Pdcd4 to the reticulocyte lysate inhibited ca. 20 and 35% of stem-loop structured luciferase mRNA translation, respectively, but did not inhibit non-stem-loop luciferase mRNA translation (Fig. 5B, lanes 2, 3, 6, and 7). A 2-μg portion of recombinant Pdcd4 inhibited translation of stem-loop structured luciferase mRNA by ca. 45% but inhibited translation of non-stem-loop structured luciferase mRNA by ca. 14% (Fig. 5B, lanes 4 and 8). The addition of recombinant Pdcd4 to the rabbit reticulocyte lysate up to 4 μg also showed only 50% inhibition of stem-loop luciferase translation (data not shown). These results indicate that adding secondary structure to the 5′UTR of an mRNA enhances its susceptibility to inhibition of translation by Pdcd4 in vitro.
FIG. 5.
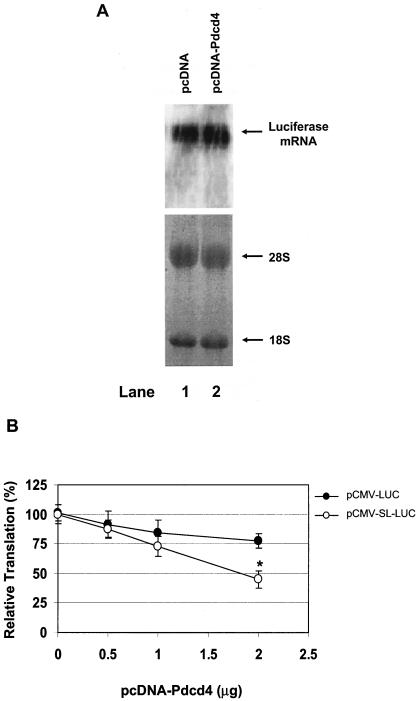
Pdcd4 preferentially inhibits translation of 5′UTR structured mRNA in vitro. (A) Structures of 5′UTR stem-loop structured luciferase (pCMV-SL-LUC) and nonstructured luciferase (pCMV-LUC); (B) in vitro translation of 5′UTR structured and nonstructured luciferase. Rabbit reticulocyte lysate was preincubated with increasing amounts of Pdcd4 (0 to 2 μg) for 5 min at 30°C prior to the addition of the nonstructured luciferase mRNA (0.2 μg) (lanes 1 to 4) or 5′UTR structured luciferase mRNA (0.2 μg) (lanes 5 to 8). The translation was performed in a total volume of 20 μl as described in Materials and Methods. The band intensity was determined with a PhosphorImager. The value obtained for nonstructured or structured luciferase mRNA with 0 μg of Pdcd4 was designated as 100%.
To test whether translation of mRNA with stem-loop structure in the 5′UTR is susceptible to inhibition by Pdcd4 in vivo, the pcDNA-Pdcd4 expression plasmid and the stem-loop structured luciferase reporter, pCMV-SL-LUC (or non-stem-loop structured luciferase reporter, pCMV-LUC), were transiently transfected into JB6 RT101 cells. The effect of Pdcd4 on translation of stem-loop and non-stem-loop luciferase was measured by determining the luciferase activity. As shown in Fig. 6A, the level of luciferase mRNA from cells transfected with Pdcd4 expression plasmid (pcDNA-Pdcd4) is similar to that in the cells transfected with empty vector (pcDNA), indicating that any inhibition of luciferase enzymatic activity cannot be attributed to a decrease in synthesis or stability of the luciferase mRNA but must reflect translational inhibition. Transfection with increasing concentrations of pcDNA-Pdcd4 produced a concentration-dependent decrease in luciferase activity in both stem-loop and non-stem-loop luciferase, but the decrease for stem-loop structured luciferase is greater than that for non-stem-loop structured luciferase (Fig. 6B). Transfection of 2 μg of pcDNA-Pdcd4 inhibited stem-loop structured luciferase expression by 50 to 60%. In contrast, transfection of the same amount of pcDNA-Pdcd4 inhibited non-stem-loop luciferase expression by only 20 to 25%. These results indicate that a structured 5′UTR confers on an mRNA enhanced susceptibility to inhibition by Pdcd4 both in vitro and in transfected cells. It is noteworthy that inhibiting protein translation by 50 to 60% is sufficient to alter a cell's physiological function (41). In addition, Pdcd4 inhibits tumor promoter-induced AP-1-dependent transcriptional activity by only 50% under conditions in which transformation is inhibited (43).
FIG. 6.
Pdcd4 preferentially inhibits translation of 5′UTR structured mRNA in transfected cells. (A) Northern blotting of luciferase mRNA. Empty vector (10 μg) (lane 1) or Pdcd4 expression plasmid (10 μg) (lane 2) was transiently transfected with pCMV-SL-LUC (1 μg) into RT101 cells. After 48 h, the total RNA was isolated, separated on a 1% formaldehyde agarose gel, transferred to a nylon membrane, and visualized by hybridization with 32P-labeled luciferase cDNA. (B) Translation of structured and nonstructured mRNA in transfected cells. The plasmid pcDNA-Pdcd4 (0 to 2 μg) was transiently transfected with pCMV-LUC (0.2.μg) (•) or pCMV-SL-LUC (0.2 μg) (○) into RT101 cells. The total DNA was maintained at 2.2 μg by adding pcDNA3.1+ vector DNA. After transfection, cells were serum starved (0.2% FBS) for 24 h and then incubated with complete medium (4% FBS) for an additional 24 h. The luciferase activity from the cells with 0 μg of pCMV-LUC or pCMV-SL-LUC was designated as 100%. These experiments were repeated three times with five independent transfections, and representative data are shown. The results are expressed as means ± standard deviations. An asterisk indicates significant differences compared to the transfection with 2 μg of pCMV-LUC plasmid, as determined by using the Student t test (P < 0.001).
Pdcd4 mutants that are defective for binding to eIF4A fail to inhibit translation in transfected cells.
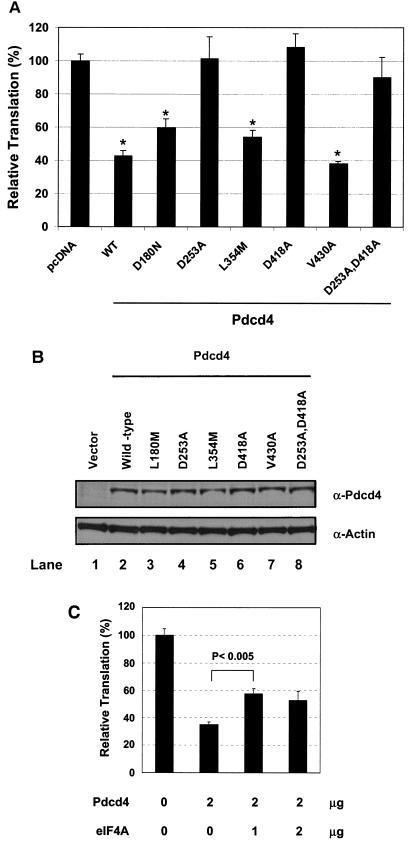
To determine whether the Pdcd4 mutants that inactivate binding to eIF4A lose the ability to inhibit translation in vivo, the Pdcd4 (wild type or mutant) expression plasmids and pCMV-SL-LUC plasmid were transiently transfected into RT101 cells. After serum starvation for 24 h, cells were returned to complete medium for an additional 24 h and then lysed and assayed for luciferase activity. Since Pdcd4 inhibits translation of stem-loop structured luciferase mRNA to a greater extent than that of non-stem-loop structured luciferase mRNA (inhibition of 50 to 60% versus 20 to 30%, Fig. 6), the stem-loop structured luciferase was used for testing the ability of Pdcd4 mutants to inhibit translation in a cultured cell system. As shown in Fig. 7, Pdcd4V430A, has an eIF4A-binding activity similar to that of wild-type Pdcd4, inhibited luciferase translation by 60%, results also similar to those obtained for wild-type Pdcd4. Two Pdcd4 mutants, Pdcd4D180N and Pdcd4L354M, that showed partial decrease in eIF4A-binding activity inhibited ca. 40% of translation. Pdcd4D235A and Pdcd4D418A, two Pdcd4 mutants with ca. 10% of the wild-type eIF4A-binding activity, did not inhibit luciferase activity in the transfected cell system. A double mutant, Pdcd4D235A,D418A, with ca. 5 to 8% of wild-type eIF4A-binding activity also showed no significant inhibition of luciferase translation. These results indicate that transfection of 2 μg of Pdcd4 mutants Pdcd4D235A, Pdcd4D418A, and Pdcd4D235A,D418A into RT101 cells is not sufficient to inhibit stem-loop structured luciferase translation. This inactivation is not attributable to reduced expression of these mutant proteins since all of the Pdcd4 constructs, both wild type and mutant, showed a similar level of expression (Fig. 7B). It is noteworthy that Pdcd4D235A, Pdcd4D418, and Pdcd4D235A,D418A mutants have weak eIF4A-binding activity (Fig. 2), but this weak eIF4A-binding activity is not sufficient for these Pdcd4 mutants to compete with eIF4Gc for binding to eIF4A (Fig. 4). In addition, the finding that the eIF4A-binding activity of Pdcd4 is highly correlated to the ability to inhibit translation of stem-loop structured luciferase (Table 1) suggests that Pdcd4 binding to eIF4A and prevention of eIF4A binding to eIF4G are required for inhibition of translation.
FIG. 7.
Pdcd4 mutants that are defective for binding to eIF4A fail to inhibit translation. (A) Portions (2 μg) of wild-type Pdcd4 expression plasmid (WT), empty vector (pcDNA), or site-specific Pdcd4 mutant expression plasmids (as indicated) were transiently transfected with pCMV-SL-LUC (0.2 μg) into RT101 cells. The total DNA was maintained at 2.2 μg by adding pcDNA3.1+ vector DNA. (B) Similar expression level of wild-type and mutant Pdcd4. RT101 cell lysates (10 μg) from transient transfection with empty vector (lane 1), wild-type Pdcd4 (lane 2), and Pdcd4 mutant expression plasmids (lanes 3 to 8) were separated on 10% Bis-Tris NuPage gels, transferred to nitrocellulose, and subjected to immunoblotting with Pdcd4 antibody, with visualization by chemiluminescent detection (C) Partial reversal by eIF4A of Pdcd4 inhibition of translation. Portions (2 μg) of Pdcd4 expression plasmid and eIF4A expression plasmid (1 or 2 μg) were cotransfected with pCMV-SL-LUC (0.2 μg) into RT101 cells. The total DNA was maintained at 4.2 μg by adding pcDNA3.1+ vector DNA. After transfection, cells were serum starved (0.2% fetal bovine serum) for 24 h and then incubated with complete medium (4% FBS) for an additional 24 h. The luciferase activity from the cells transfected with empty vector was designated as 100%. These experiments were repeated three times with five independent transfections, and representative data are shown. The results are expressed as means plus the standard deviations.
TABLE 1.
Comparison of eIF4A-binding activity and inhibition of translation between wild-type and mutant Pdcd4s
| Mutant | Binding or inhibitiona
|
||||||
|---|---|---|---|---|---|---|---|
| WT | D180N | D253A | L354M | D418A | V430A | D235A, D418A | |
| eIF4A binding | +++ | ++ | + | ++ | + | +++ | + |
| Inhibition of translation | ++ | + | − | + | − | ++ | − |
+++, strong binding; ++, medium binding or inhibition; +, weak binding or inhibition; −, no inhibition.
To test whether overexpression of eIF4A is able to reverse the inhibition of translation by Pdcd4, we cotransfected Pdcd4 expression plasmid and eIF4A expression plasmid with stem-loop luciferase reporter into RT101 cells and then carried out an assay for luciferase activity. Cotransfection of 1 μg of eIF4A expression plasmid partially reversed the inhibition of translation by Pdcd4 (Fig. 7C). Cotransfection of 2 μg of eIF4A expression, however, did not produce further reversal (Fig. 7C), suggesting that other factors are also limited by Pdcd4 expression. Together, our findings suggest that binding to eIF4A is a primary requirement for Pdcd4 to inhibit translation since Pdcd4 mutants defective in binding to eIF4A fail to inhibit translation (Fig. 7A). In addition to binding to eIF4A, other events, such as inhibition of eIF4A's helicase activity, also play a role in the inhibition of translation by Pdcd4.
DISCUSSION
Computational analysis of protein sequences from the sequence database reveals an MA-3 domain with an α-helical secondary structure extended by approximately 120 amino acids (2, 33). This MA-3 domain appears in several translation initiation factors, including human eIF4G, mouse eIF4G, plant eIF(iso)4F, and DAP-5/NAT1/p97 (2). Although several amino acid residues of MA-3 domain are conserved between the eIF4G and Pdcd4 (Fig. 2A), no consensus sequence has been identified. The present study demonstrates for the first time a function of the α-helical MA-3 domains in Pdcd4 and in the human eIF4G, which is the scaffold for binding to eIF4A (Fig. 2 and 3). In contrast, DAP-5/NAT1/p97 contains an MA-3 domain in the C-terminal half, but it is defective for eIF4A-binding activity. This discrepancy is now explained by the demonstration that several amino acid residues in the MA-3 domain of DAP-5/NAT1/p97 are different from those in the Pdcd4 and eIF4Gc (Fig. 2A). In particular, the negatively charged amino acid residues 414Asp of the C-terminal MA-3 domain of Pdcd4 (or 249Glu of N-terminal MA-3 domain of Pdcd4) and 1329Asp of eIF4Gc are conserved, but positively charged amino acid residue 631Lys is found in the DAP5/NAT1/p97. The Pdcd4 mutants that are defective in binding to eIF4A fail to inhibit translation in vivo (Fig. 7), suggesting that binding to the eIF4A is required for inhibition of translation by Pdcd4. The results also establish that translation of mRNA with a stem-loop structure at the 5′UTR is more sensitive to inhibition by Pdcd4 than translation of nonstructured mRNA in vitro and in transfected cells (Fig. 5 and 6), supporting the hypothesis that Pdcd4 preferentially inhibits translation of mRNAs with structured 5′UTRs. Although it is true that the 5′UTR of stem-loop luciferase is 48 nucleotides longer than that of non-stem-loop luciferase (Fig. 5A), the length of the 5′UTR per se is not expected to lower translation efficiency. Indeed, a long nonstructured 5′UTR enhances translation (19, 21). Therefore, the preferential inhibition by Pdcd4 of the translation of 5′UTR-structured luciferase (Fig. 5) is likely to be caused by its high stability.
The finding that partial deletion of N- or C-terminal MA-3 domain of Pdcd4 decreased by approximately 90%, the binding to eIF4A (Fig. 1) suggests that both intact MA-3 domains are important for eIF4A-binding activity. Mutation analyses revealed that several amino acids in the MA-3 domains of Pdcd4 are critical for binding to eIF4A, including 249Glu, 253Asp, 414Asp, and 418Asp (Fig. 2B). These Pdcd4 mutants that are inactivated for eIF4A-binding activity fail to inhibit translation (Fig. 7) and fail to compete with eIF4Gc for binding to eIF4A (Fig. 4), indicating that the inhibition of translation by Pdcd4 occurs, at least in part, through binding to the eIF4A and preventing eIF4A binding to eIF4Gc. The eIF4Gc has been shown to have a regulatory function for translation initiation. Mutations in eIF4Gc that decrease binding to eIF4A result in decreased ribosome binding (24). In addition, by using the 2Apro-treated rabbit reticulocyte lysate, this eIF4Gc mutant shows a lower capacity to stimulate cap-dependent translation than the wild-type eIF4G (24). Therefore, inhibition of the eIF4A binding to the eIF4Gc by Pdcd4 is expected to decrease the rate of translation. Our findings (Fig. 4 and 7) further support our previous model that inhibition of translation by Pdcd4 occurs in part through prevention of eIF4A binding to eIF4Gc (42). Moreover, the residues in the eIF4G MA-3 domain (namely, 1329Glu and 1333Asp) that are here defined as important for binding to eIF4A have been found by Bellsolell et al. to exist on the surface of eIF4Gc (L. Bellsolell, P. F. Cho-Park, F. Poulin, N. Sonenberg, and S. K. Burley, unpublished data).
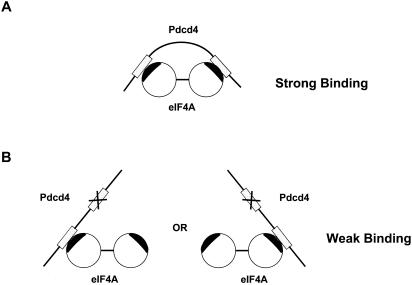
Translation initiation factor eIF4A is one of the more highly conserved proteins among different species. The amino acid sequences of the yeast eIF4A and the human eIF4A are 68% identical and 81% similar (22, 25). Recent resolution of the crystallographic structure of full-length yeast eIF4A reveals a dumbbell structure consisting of two compact domains connected by an extended 11-amino-acid linker (6). Mutations on either compact domain of the mouse eIF4AII result in dramatic decrease in Pdcd4-binding activity (H. Zakowicz et al., unpublished data). These findings, in combination with the results of the present study that deletions or mutations on either MA-3 domain of Pdcd4 dramatically lose the eIF4A-binding activity (Fig. 1 and 2), suggest that Pdcd4 may bind to the two compact domains of eIF4A through the two MA-3 domains (Fig. 8), i.e., one MA-3 domain binds to one compact domain of eIF4A. The fact that Pdcd4 mutants Pdcd4E249K, Pdcd4D253A, Pdcd4D414K, and PdcdD418A mutated on either the N- or the C-terminal MA-3 domain of Pdcd4 lose ca. 90% of eIF4A-binding activity indicates that both intact MA-3 domains are required for maximal eIF4A binding. This finding also suggests a cooperative interaction between the two MA-3 domains of Pdcd4 and both domains of eIF4A. However, our data do not indicate the orientation of the interaction between Pdcd4 and eIF4A. It is unknown whether the N-terminal MA-3 domain of Pdcd4 binds to the N-terminal compacted domain of eIF4A or the C-terminal compacted domain of eIF4A. It is noteworthy that the regions around amino acid residues 249 to 253 of the N-terminal MA-3 domain of Pdcd4 or amino acid residues 414 to 418 of the C-terminal MA-3 domain of Pdcd4 appear to play a critical role in binding to eIF4A since Pdcd4E249K, Pdcd4D253A, Pdcd4D414K, and Pdcd4D418A showed a dramatic decrease in eIF4A binding activity (Fig. 2). Further mutations of this region will be important.
FIG. 8.
Model for Pdcd4 binding to eIF4A. (A) Two MA-3 domains (gray box) of Pdcd4 bind to two compacted domains of eIF4A within the Pdcd4-binding region (black) to achieve a maximal binding activity. (B) When N- or C-terminal MA-3 domain of Pdcd4 is mutated and is defective in binding to the compacted domain of eIF4A, the mutated Pdcd4 only contains one intact MA-3 domain, which is able to bind to the compacted domain of eIF4A. In this case, the mutated Pdcd4 has a very weak eIF4A-binding activity.
Previously, we demonstrated that Pdcd4 specifically inhibited the helicase activity of eIF4A but not of RNA helicase, Ded1p (42). The helicase activity of eIF4A is thought to catalyze the unwinding of the secondary structure of mRNA at 5′UTR, thus allowing the 40S ribosomal subunit to scan the mRNA (36). Recent studies (32) using reconstitution of translation initiation factors showed that a ternary complex comprised of ribosomal 40S subunit, eIF3, eIF2, and eIF1 is able to scan from the 5′-to-3′ direction of an unstructured mRNA and locate the initiation codon. This complex, however, is not sufficient to scan an mRNA containing a stable secondary structure at the 5′UTR. Scanning on such a structured mRNA requires eIF4A helicase activity (32). Mutations of eIF4A that reduce the RNA helicase activity result in inhibition of translation in vitro (28-30). Svitkin et al. (40) showed that dominant-negative eIF4A mutants that are defective in RNA helicase activity, RNA-binding ability, or ATPase activity inhibit translation of stable secondary structured mRNAs more efficiently than less-structured mRNAs. Therefore, translation of 5′UTR structured mRNA is very inefficient and highly dependent on the helicase activity of eIF4A. Our finding that translation of 5′UTR structured mRNA is susceptible to inhibition by Pdcd4 (Fig. 6) provides evidence that the helicase activity of eIF4A is important for translation of 5′UTR structured mRNA in vivo. The finding that translation of 5′ structured mRNA is susceptible to suppression by Pdcd4 (Fig. 5 and 6) suggests that Pdcd4 may preferentially inhibit translation of a set of 5′UTR structured mRNAs. So-called “translationally repressed” mRNAs have been described which contain G- or C-rich regions in the 5′UTR with the potential to form stable secondary structure(s) at the 5′UTR (5, 34). Usually, the products of such translationally repressed mRNAs are involved in cell proliferation; these products include growth factors, growth promotion genes, and proto-oncogenes (10). Inhibiting or decreasing eIF4A helicase activity would be expected to limit translation of this group of mRNAs, resulting in the suppression of cell growth or transformation. Previously, we demonstrated that overexpression of Pdcd4 in JB6 transformation-susceptible cells and transformed cells suppressed tumor promoter-induced transformation (43) and tumor phenotype (44), respectively. This suppression is attributable, at least in part, to inhibiting AP-1-dependent transcription, a required event for tumor promoter-induced transformation (16) and for maintenance of tumor phenotype (12, 13). Pdcd4 does not, however, inhibit NF-κB or serum response element-dependent transcription (43, 44). Inhibition of AP-1-dependent transcription in turn occurs through repressing transactivation of c-Jun and c-Fos (44). A Pdcd4 mutant, Pdcd4D418A, inactivated for binding to eIF4A fails to inhibit AP-1-dependent transcription, suggesting that Pdcd4 binding to eIF4A is required for suppression of tumor promoter-induced transformation and tumor phenotype (42). Overexpression of Pdcd4 does not alter the protein expression level of c-Jun or c-Fos, indicating that Pdcd4 does not directly regulate translation of c-Jun and c-Fos (44). Therefore, Pdcd4, by binding to eIF4A and inactivating the helicase activity of eIF4A, may inhibit translation of activators that activate c-Jun or c-Fos. The target gene(s) whose mRNA translation is regulated by Pdcd4 are currently under investigation.
Acknowledgments
We thank Alyson Baker for technical assistance and the members of Gene Regulation Section of NCI—Frederick for helpful discussions.
REFERENCES
- 1.Abramson, R. D., T. E. Dever, T. G. Lawson, B. K. Ray, R. E. Thach, and W. C. Merrick. 1987. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 262:3826-3832. [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 2000. Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res. 10:1172-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azzoni, L., O. Zatsepina, B. Abede, I. M. Bennett, P. Kanakaraj, and B. Perussia. 1998. Differential transcriptional regulation of CD161 and a novel gene, 197/15a, by IL-2, IL-15, and IL-12 in NK and T cells. J. Immunology 161:3493-3500. [PubMed] [Google Scholar]
- 4.Bohm, M., K. Sawicka, J. P. Siebrasse, A. Brehmer-Fastnacht, R. Peters, and K. H. Klempnauer. 2003. The transformation suppressor protein Pdcd4 shuttles between nucleus and cytoplasm and binds RNA. Oncogene 22:4905-4910. [DOI] [PubMed] [Google Scholar]
- 5.Brown, E. J., and S. L. Schreiber. 1996. A signaling pathway to translational control. Cell 86:517-520. [DOI] [PubMed] [Google Scholar]
- 6.Caruthers, J. M., E. R. Johnson, and D. B. McKay. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. USA 97:13080-13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauhan, D., D. Auclair, E. K. Robinson, T. Hideshima, G. Li, K. Podar, D. Gupta, P. Richardson, R. L. Schlossman, N. Krett, L. B. Chen, N. C. Munshi, and K. C. Anderson. 2002. Identification of genes regulated by dexamethasone in multiple myeloma cells using oligonucleotide arrays. Oncogene 21:1346-1358. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y. H., T. Knosel, G. Kristiansen, A. Pietas, M. E. Garber, S. Matsuhashi, I. Ozaki, and I. Petersen. 2003. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J. Pathol. 200:640-646. [DOI] [PubMed] [Google Scholar]
- 9.Cmarik, J. L., H. Min, G. Hegamyer, S. Zhan, M. Kulesz-Martin, H. Yoshinaga, S. Matsuhashi, and N. H. Colburn. 1999. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc. Natl. Acad. Sci. USA 96:14037-14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Benedetti, A., and A. L. Harris. 1999. eIF4E expression in tumors: its possible role in progression of malignancies. Int. J. Biochem. Cell. Biol. 31:59-72. [DOI] [PubMed] [Google Scholar]
- 11.De Gregorio, E., T. Preiss, and M. W. Hentze. 1998. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA 4:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, Z., V. Lavrovsky, and N. H. Colburn. 1995. Transformation reversion induced in JB6 RT101 cells by AP-1 inhibitors. Carcinogenesis 16:749-756. [DOI] [PubMed] [Google Scholar]
- 13.Dong, Z., R. G. Watts, Y. Sun, S.-N. Zhan, and N. Colburn. 1995. Progressive elevation of AP-1 activity during preneoplastic-to-neoplastic progression as modeled in mouse JB6 cell variants. Int. J. Oncol. 7:359-364. [DOI] [PubMed] [Google Scholar]
- 14.Goke, A., R. Goke, A. Knolle, H. Trusheim, H. Schmidt, A. Wilmen, R. Carmody, B. Goke, and Y. H. Chen. 2002. DUG is a novel homologue of translation initiation factor 4G that binds eIF4A. Biochem. Biophys. Res. Commun. 297:78-82. [DOI] [PubMed] [Google Scholar]
- 15.Gray, N. K., and M. W. Hentze. 1994. Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep. 19:195-200. [DOI] [PubMed] [Google Scholar]
- 16.Hsu, T. C., M. R. Young, J. Cmarik, and N. H. Colburn. 2000. Activator protein 1 (AP-1)- and nuclear factor κB (NF-κB)-dependent transcriptional events in carcinogenesis. Free Radic. Biol. Med. 28:1338-1348. [DOI] [PubMed] [Google Scholar]
- 17.Imataka, H., H. S. Olsen, and N. Sonenberg. 1997. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 16:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imataka, H., and N. Sonenberg. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17:6940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak, M. 1978. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell 15:1109-1123. [DOI] [PubMed] [Google Scholar]
- 20.Kozak, M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 9:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak, M. 1991. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr. 1:117-125. [PMC free article] [PubMed] [Google Scholar]
- 22.Linder, P., and P. P. Slonimski. 1988. Sequence of the genes TIF1 and TIF2 from Saccharomyces cerevisiae coding for a translation initiation factor. Nucleic Acids Res. 16:10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomakin, I. B., C. U. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen, P. J., and H. Trachsel. 1988. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 7:2097-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niepel, M., L. Ling, and D. R. Gallie. 1999. Secondary structure in the 5′-leader or 3′-untranslated region reduces protein yield but does not affect the functional interaction between the 5′-cap and the poly(A) tail. FEBS Lett. 26:79-84. [DOI] [PubMed] [Google Scholar]
- 27.Onishi, Y., and H. Kizaki. 1996. Molecular cloning of the genes suppressed in RVC lymphoma cells by topoisomerase inhibitors. Biochem. Biophys. Res. Commun. 228:7-13. [DOI] [PubMed] [Google Scholar]
- 28.Pause, A., N. Methot, and N. Sonenberg. 1993. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 13:6789-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pause, A., N. Methot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pause, A., and N. Sonenberg. 1992. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 11:2643-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova, T. V., and V. G. Kolupaeva. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes. Dev. 16:2906-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponting, C. P. 2000. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci. 25:423-426. [DOI] [PubMed] [Google Scholar]
- 34.Proud, C. G. 1994. Translation: turned on by insulin. Nature 371:747-748. [DOI] [PubMed] [Google Scholar]
- 35.Richter-Cook, N. J., T. E. Dever, J. O. Hensold, and W. C. Merrick. 1998. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J. Biol. Chem. 273:7579-7587. [DOI] [PubMed] [Google Scholar]
- 36.Rogers, G. W., Jr., A. A. Komar, and W. C. Merrick. 2002. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acids Res. Mol. Biol. 72:307-331. [DOI] [PubMed] [Google Scholar]
- 37.Rogers, G. W., Jr., N. J. Richter, W. F. Lima, and W. C. Merrick. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276:30914-30922. [DOI] [PubMed] [Google Scholar]
- 38.Rozen, F., I. Edery, K. Meerovitch, T. E. Dever, W. C. Merrick, and N. Sonenberg. 1990. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10:1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibahara, K., M. Asano, Y. Ishida, T. Aoki, T. Koike, and T. Honjo. 1995. Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene 166:297-301. [DOI] [PubMed] [Google Scholar]
- 40.Svitkin, Y. V., A. Pause, A. Haghighat, S. Pyronnet, G. Witherell, G. J. Belsham, and N. Sonenberg. 2001. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watkins, S. J., and C. J. Norbury. 2002. Translation initiation and its deregulation during tumorigenesis. Br. J. Cancer 86:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, H. S., A. P. Jansen, A. A. Komar, X. Zheng, W. C. Merrick, S. Costes, S. J. Lockett, N. Sonenberg, and N. H. Colburn. 2003. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol. 23:26-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, H. S., A. P. Jansen, R. Nair, K. Shibahara, A. K. Verma, J. L. Cmarik, and N. H. Colburn. 2001. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-κB or ODC transactivation. Oncogene 20:669-676. [DOI] [PubMed] [Google Scholar]
- 44.Yang, H. S., J. L. Knues, C. Stark, and N. H. Colburn. 2003. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene 22:3712-3720. [DOI] [PubMed] [Google Scholar]
- 45.Yang, H. S., J. I. Morris, Q. Wang, L. G. Korotchkina, M. Kwon, and M. S. Patel. 2001. Human dihydrolipoamide dehydrogenase gene transcription is mediated by cAMP-response element-like site and TACGAC direct repeat. Int. J. Biochem. Cell. Biol. 33:902-913. [DOI] [PubMed] [Google Scholar]