Abstract
Expression of carbonic anhydrase 9 (CA9) is associated with poor prognosis and increased tumor aggressiveness and does not always correlate with HIF-1α expression. Presently, we analyzed the regulation of CA9 expression during hypoxia by HIF-1α, Notch3, and the von Hippel-Lindau (VHL) in breast carcinoma cells. Both HIF-1α and Notch3 were absolutely required for the expression of CA9 mRNA, protein, and reporter. Reciprocal co-immunoprecipitation of HIF-1α, Notch3 intracellular domain (NICD3), and pVHL demonstrated their association. The presence of common consensus prolyl hydroxylation and pVHL binding motifs (L(XY)LAP);LLPLAP2191 suggested an oxygen-dependent regulation for NICD3. However, unlike the HIF-1α protein, NICD3 protein levels were not modulated with hypoxia or hypoxia-mimetic agents. Surprisingly, mutations of the common prolyl hydroxylation and pVHL binding domain lead to the loss of CA9 mRNA, protein, and reporter activity. Chromatin immunoprecipitation assay demonstrated the association of NICD3, HIF-1α, and pVHL at the CA9 promoter. Further, the NICD3 mutant defective in prolyl hydroxylation and subsequent pVHL binding caused a reduction in cell proliferation of breast carcinoma cells. We show here for the first time that the interaction of HIF-1α with NICD3 is important for the regulation of CA9 expression. These findings suggest that although CA9 is a hypoxia-responsive gene, its expression is modulated by the interaction of HIF-1α, Notch3, and VHL proteins. Targeting the expression of CA9 by targeting upstream regulators could be useful in cancer/stem cell therapy.
Keywords: hypoxia, Notch3, CA9, PBX1, VHL
Introduction
CAIX, also known as MN or G250, is an isoenzyme belonging to the family of carbonic anhydrases (CAs), whose expression is associated with a large number of human tumors.1-3 CAs promote the acidification of the extracellular environment surrounding cancer cells, which facilitate their survival and growth4 by reversible hydration of carbon dioxide. Both small molecules and antibodies targeting these pH regulators such as CAIX are currently at various stages of clinical development. Presently, compounds based on sulfonamides/sulfamates and coumarins have demonstrated particular promise as potential anticancer agents inhibiting CAIX. Interestingly, these 2 inhibitor classes are mechanistically distinct in their inhibition of CAs. The sulfonamides inhibit CAIX by coordinating with the zinc ion within the active site, while molecules based on coumarin/thiocoumarin act as suicide inhibitors, undergoing hydrolysis to 2-hydroxycinnamic acids and binding irreversibly at the entrance to the active site cavity.5 Girentuximab, a chimeric humanized monoclonal antibody, is currently in phase III clinical trials for kidney carcinoma at Wilex (Munich, Germany). WX-G250 is another CA9-specific chimeric monoclonal antibody that underwent phase III clinical trials as an adjuvant therapy for the treatment of nonmetastasized renal cell carcinoma in patients who are at a high risk of recurrence after resection of the primary tumor.6 Phase III trials are also underway at Wilex for the treatment of metastatic renal cell carcinoma in combination with interleukin-2 or interferon-α.7
These antitumor mechanisms are not exploited by the classic cancer drugs and therefore represent a new anticancer drug discovery strategy. Overexpression of CA9 is consistently seen in a high proportion of carcinomas of the cervix,8 clear cell carcinoma of the kidney,9 and to a lesser extent in breast cancer,10,11 carcinoma of the head and neck,12,13 lung cancer,14 and tumors of the brain.15,16 Elevated expression of CA9 during hypoxia is suggestive of its importance during tumor adaptation to hypoxia.3 Although a significant correlation of hypoxia with CA9 expression was observed in tumors, CA9 expression extended beyond hypoxic regions and was not always specific to hypoxia.17,18
The promoter of CA9 is localized in the region between −173 and +31 bp with respect to the transcription start site.19 Hypoxia activates CA9 expression through hypoxia-inducible factor-1 (HIF-1) binding to the hypoxia-responsive element (HRE) in the CA9 promoter immediately upstream of the transcription start site.17 HIF-1α, Sp1, and CBP/P300 are required for CA9 expression in clear cell renal carcinoma.20 A repressor that negatively regulates the expression of CA9 transcription was recently identified as the MORC2 (micro-orchidia/HDAC2) complex, which causes decreased acetylation of the CA9 promoter.21 Further p66shc/Notch3 interplay was also shown to induce the expression of CA9 and Notch ligand Jagged1 in response to hypoxia in progenitor cells expanded in vitro as multicellular spheroids.22 It was proposed that Notch3 mediates the upregulation of CA9 via the ERK1/2-dependent pathway.22 These findings suggested that RBP-Jκ–independent Notch3 plays an important role in hypoxia-induced expression of CA9. However, the interaction between HIF-1α and Notch3 in regulating the expression of CA9 is not clear. Are these proteins required for hypoxia-induced expression of CA9? Our findings indicate that indeed HIF-1α, von Hippel-Lindau (VHL), and Notch3 intracellular domain (NICD3) interact to regulate the expression of CA9, and its expression is modulated by the interaction of HIF-1α, NICD3, and VHL proteins. Identification of a molecular mechanism as shown in these studies will help in identifying novel targets for therapy.
Results
HIF-1α and NICD3 Are Both Required for the Expression of CA9
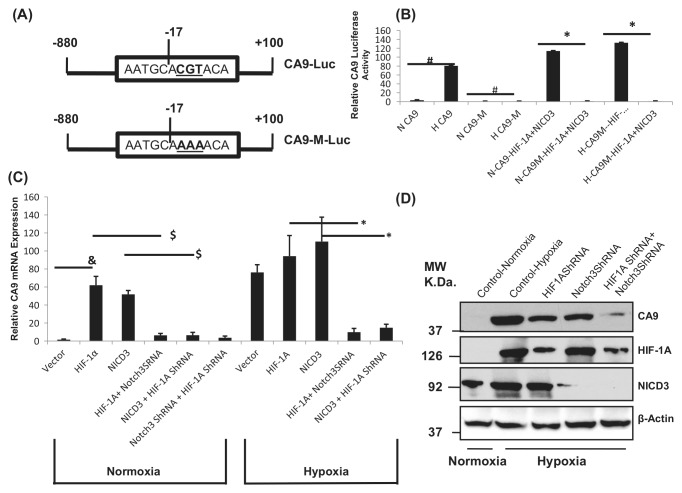
To understand the role of HIF-1 and NICD3 in regulating the expression of CA9 during normoxia and hypoxia, the CA9 luciferase reporter23 was mutated for HIF-1 binding (−17ACGT to −17AAAA)17 (Fig. 1A). The CA9 reporter defective in HIF-1 binding is referred to as CA9M-Luc. The CA9M luciferase reporter did not show any activity or very minimal activity as compared to the wild-type CA9 luciferase reporter, either during normoxia or hypoxia (Fig. 1B). Overexpression of both NICD3 and HIF-1α was able to significantly induce the wild-type CA9 luciferase reporter compared to the CA9 reporter defective in binding to HIF-1 (CA9M-Luc) during normoxia (Fig. 1B), suggesting that HIF-1 binding to the CA9 promoter is required for the induction of CA9 reporter activity during hypoxia or with overexpression of HIF-1α or NICD3. MCF10A cells transfected with shRNA against HIF-1α or NICD3 or both decreased the CA9 mRNA expression studied by real-time RT-PCR (Fig. 1C) or Western blot analysis (Fig. 1D). Overexpression of HIF-1α or NICD3 while concomitantly repressing the expression of Notch3 or HIF-1α, respectively, also does not induce the expression of CA9 mRNA (Fig. 1C). Overexpression of HIF-1α or Notch3 shRNA or both HIF-1α and Notch3 shRNA reduced the expression of the CA9 protein (Fig. 1D). Hypoxia induced about a 2-fold increase in the expression of NICD3 (Fig. 1D) that was also seen in other experiments including immunoprecipitation (Fig. 2A) and hypoxia-mimetic agents (Fig. 3A) as well as expression in MCF10A from different cell lines Western blot analysis (Fig. 4A). About a 50% and 90% reduction in protein levels was seen with HIF-1α and Notch3 shRNA, respectively, as assessed by densitometric analysis. Immunofluorescence for CA9 expression also showed similar results (Suppl. Fig. S1A).
Figure 1.
HIF-1α and NICD3 are both required for the expression of CA9. (A) Schematic representation of the CA9 luciferase and CA9 mutant reporter. The HRE was mutated to create a CA9 promoter defective in HIF-1 binding (CA9M-Luc). (B) CA9 reporter assay: MCF10A cells were transfected with CA9-Luc or CA9M-Luc and HIF-1α and/or NICD3 and exposed to hypoxia. (C) CA9 expression by real-time RT-PCR: MCF10A cells were transfected with HIF-1α and NICD3 shRNA against HIF-1α or Notch3 or both HIF-1α and Notch3 and the relative CA9 mRNA expression as compared to the β-actin expression was analyzed. In addition, the cells were transfected with HIF-1α or Notch3 with Notch3 shRNA or HIF-1α shRNA, respectively, and CA9 mRNA expression was analyzed by real-time RT-PCR. (D) Western blot analysis showing the expression of CA9: MCF10A cells were transfected with vector alone or HIF-1α or Notch3 shRNA or both, and protein lysates were analyzed for the expression of CAIX, HIF-1α, NICD3, and β-actin (loading control). #Significant difference between normoxia- versus hypoxia-induced CA9 reporter activity and loss of CA9M activity during hypoxia at P ≤ 0.0005. *Significant lack of CA9M-Luc induction with overexpression of HIF-1α and NICD3 as compared to the wild-type CA9 promoter with overexpression of HIF-1α and NICD3 at P ≤ 0.05. &Significance between normoxia- versus hypoxia-induced CA9 mRNA expression at P ≤ 0.005. $Significant reduction in CA9 mRNA expression with HIF-1α shRNA or Notch3 shRNA or both HIF-1α and NICD3 shRNA at P ≤ 0.001. *Significant loss of CA9 mRNA expression with overexpression of HIF-1α and NICD3 with Notch3 shRNA and HIF-1 shRNA, respectively, as compared to the overexpression of HIF-1α and NICD3, respectively, at P ≤ 0.01. Each experiment has been repeated at least 3 times. H = hypoxia; N = normoxia.
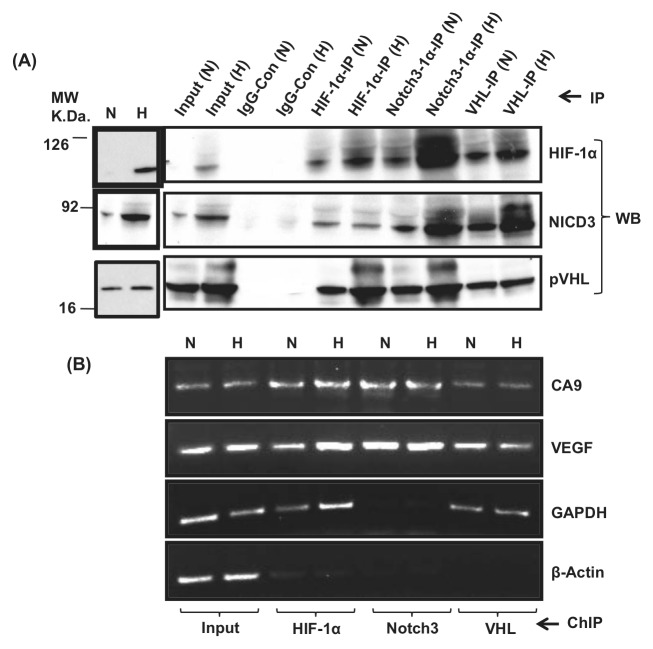
Figure 2.
(A) Reciprocal immunoprecipitation shows the association of HIF-1α, NICD3, and VHL: Normoxic or hypoxic MCF10A cell (80% confluent) lysates in RIPA buffer were immunoprecipitated with antibodies against HIF-1α, NICD3, and pVHL, and the immunoprecipitates were probed with antibodies against HIF-1α, NICD3, and pVHL using Western immunoblot analysis. (B) ChIP shows the localization and association of HIF-1α, NICD3, and VHL at the CA9 promoter: MCF10A extracts were immunoprecipitated with antibodies against HIF-1α, NICD3, and pVHL and nonspecific IgG, and the DNA from immunoprecipitates was PCR amplified for the CA9 promoter, VEGF, GAPDH, and β-actin promoter. These figures are representative of 3 experiments.
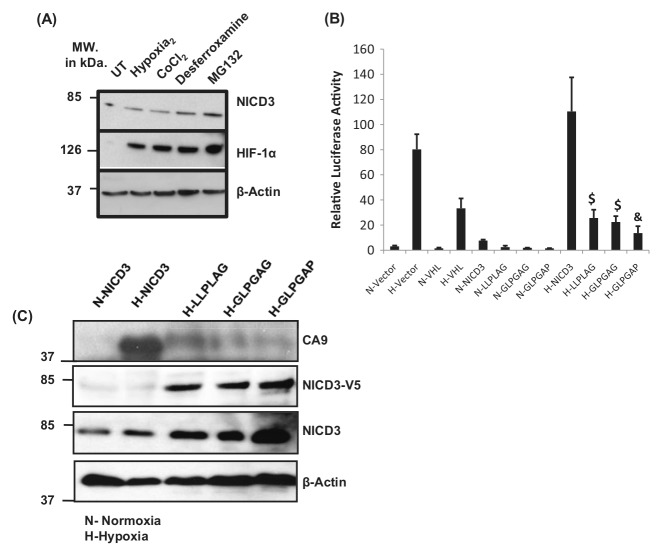
Figure 3.
NICD3 mutants defective in VHL binding repress the expression of CA9. (A) NICD3 is not regulated by prolyl hydroxylation and subsequent VHL binding. MCF10A cells were untreated or exposed to hypoxia, 100 μM CoCl2, 100 μM desferrioxamine, 20 μM MG132, and the protein extracts were probed for the expression of NICD3, HIF-1α, and β-actin by Western immunoblot analysis. (B) NICD3 was mutated for VHL binding by site-directed mutagenesis ((NICD3P2191G [LLPLAG], NICD3L2189G;P2191G [GLPGAG], and NICD3L2186G;L2189G [GLPGAP]). MCF10A cells were transfected with CA9-Luc with or without NICD3, NICD3P2191G (LLPLAG), NICD3L2189G;P2191G (GLPGAG), and NICD3L2186G;L2189G (GLPGAP) in the presence of normoxia or hypoxia, and the CA9 reporter activity was measured. (B) MCF10A cells were transfected with mutants of NICD3 (as shown by the V5 epitope). Expression of CA9, NICD3-V5, NICD3, and β-actin was analyzed. These figures are representative of 3 experiments (A). Each experiment has been repeated at least 3 times (B, C).
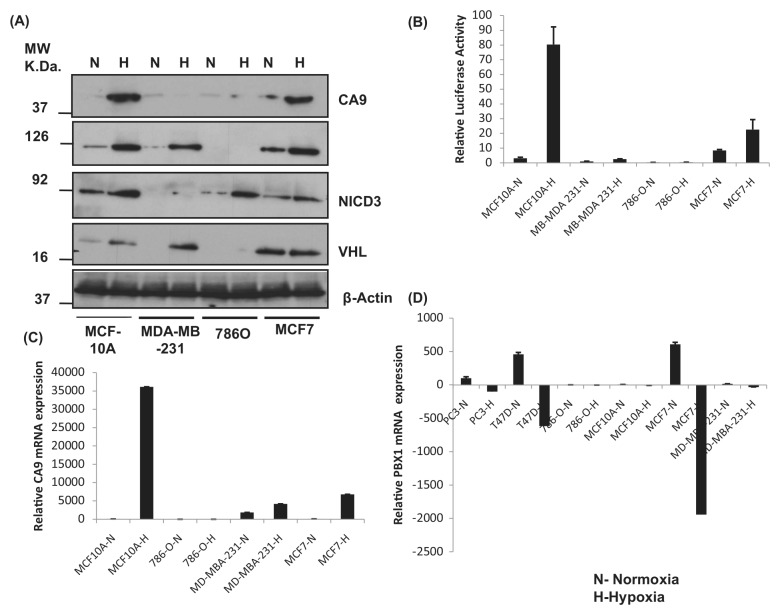
Figure 4.
Expression of CAIX and proteins regulating CA9 expression. (A) By Western blot analysis: Different cells as indicated were grown in normoxia or hypoxia, and protein extracts were subjected to Western blot analysis for CAIX, HIF-1α, NICD3, VHL, and β-actin serving as a loading control. (B) By reporter assay: Different cells were transiently transfected with the CA9 luciferase reporter (CA9M-Luc) and exposed to normoxia or hypoxia and the relative expression of the reporter as compared to Renilla luciferase used as an internal control. (C) Real-time RT-PCR: Different cells as indicated were grown in normoxia or hypoxia, and total RNA extracts were subjected to quantitative RT-PCR for CAIX, HIF-1α, NICD3, VHL, and β-actin serving as a loading control. N = normoxia; H = hypoxia. These figures are representative of 3 experiments (A). Each experiment has been repeated at least 3 times (B, C).
Reciprocal immunoprecipitations show the association of HIF-1α, NICD3, and pVHL
To discover if HIF-1α, NICD3, and VHL interact with each other, a reciprocal immunoprecipitation was carried out. HIF-1α immunoprecipitates showed the presence of HIF-1α along with NICD3 and pVHL with higher levels in hypoxic extracts (except the NICD3 protein). Similarly, Notch3 immunoprecipitates showed the presence of HIF-1α and pVHL along with NICD3. VHL immunoprecipitates also showed the association of VHL with HIF-1α and NICD3. The association between HIF-1α, NICD3, and VHL seems to be enhanced during hypoxia. Furthermore, a negative control, constituting an immunoprecipitation using nonspecific rabbit IgG, lacked HIF-1α, NICD3, or VHL (Fig. 2A). A regular Western blot analysis may not show the presence of HIF-1α (see Fig. 2A; HIF-1α is absent in the normoxic cell lysate), but enrichment using immunoprecipitation shows its presence even during normoxia (Fig. 2A).
HIF-1α, NICD3, and VHL Are Associated on the CA9 Promoter
It was earlier shown that FIH-1 binds to VHL and that VHL also functions as a transcriptional co-repressor that inhibits the HIF-1α transactivation function by recruiting histone deacetylases.24 To show if HIF-1α, NICD3, and pVHL are engaged on the CA9 promoter as a complex, chromatin immunoprecipitation (ChIP) was carried out using anti–HIF-1α, -Notch3, and -VHL antibodies on nucleoprotein extracts made from normoxic or hypoxic cells. PCR amplification of the CA9 promoter using the DNA immunoprecipitated by these antibodies indicates the enrichment of the CA9 promoter but not the nonspecific β-actin gene, suggestive of a specific interaction of HIF-1α, NICD3, and VHL and localization at the CA9 promoter. The human β-actin gene was used as an internal control for ChIP and did not show any enrichment with HIF-1α or NICD3 or VHL. The presence of HIF-1 at its target gene promoters as seen by us during normoxia was also reported earlier for stem cell factor25 and Glut1.26 Further, the VEGF gene promoter, a known target of HIF-1, was used as a positive control for ChIP and showed enrichment in HIF-1α or NICD3 or VHL ChIP (Fig. 2B). The presence of Notch3 at the VEGF promoter suggests the possibility of a similar mechanism of regulation of VEGF by binding HIF-1α to NICD3.
Hypoxia and Hypoxia-Mimetic Agents Do Not Stabilize the Notch3 (or NICD3) Protein
Hydroxylation of either human HIF-1α Pro402 or Pro564 promotes the interaction with pVHL, leading to the recruitment of ubiquitin complexes and degradation of HIF-1α.27 We analyzed the protein sequence of Notch3 for the presence of a common consensus prolyl hydroxylation and VHL binding site L(XY)LAP as seen in HIF-1α.28 Indeed, one such site (LLPLAP2191) was present in the primary protein sequence of Notch3 (Suppl. Fig. S2). This suggests that the posttranslational levels of Notch3 or NICD3 could be regulated in an oxygen-dependent manner like that of HIF-1α. However, neither hypoxia nor hypoxia-mimetic agents, such as cobalt chloride and desferrioxamine, could stabilize NICD3 as it does in the case of the HIF-1α protein (Fig. 3A). MG132, a proteosomal inhibitor, induced the protein expression of both Notch3 and HIF-1α, suggesting a proteosome-mediated degradation for Notch3 (Fig. 3A) unrelated to prolyl hydroxylases and pVHL.
NICD3 Mutant Defective in Putative VHL Binding Represses the Expression of CA9
pVHL is known to repress the transcriptional activity of HIF-1 by forming a complex with it at the HIF-1 target promoters.29 Hydroxylation of either human HIF-1α Pro402 or Pro564 promotes the interaction with pVHL, leading to the recruitment of ubiquitin complexes and degradation of HIF-1α.12 FIH-1 was initially identified as a protein binding to HIF-1α and VHL and regulating the transcriptional activity of HIF-1.24 Lando et al.30 showed that FIH-1 is an asparaginyl hydroxylase that regulates the transcriptional activity of HIF-1 by hydroxylating Asn803.31 To find out if the mutation of VHL binding in NICD3 (Suppl. Fig. S1B) could therefore lead to the loss of this repression and transcriptional activation of the CA9 target of Notch3 since the repressor VHL cannot bind to it, a reporter assay and a Western immunoblot were carried out. In contrast, overexpression of these Notch3 mutants led to the repression of CA9 reporter luciferase activity and CA9 protein expression (Fig. 3B and 3C), suggesting a requirement of pVHL binding for the transcriptional activity of the HIF-1/NICD3 complex.
PBX1, a Direct Target of Notch3, Could Be a Negative Marker of Hypoxia
We studied the expression of HIF-1α, Notch3 (NICD3), and VHL in different cells (MCF10A, MCF7, and MDA-MB-231.) and one renal carcinoma cell (786-O) using Western blot analysis and RT-PCR to look at its correlation with the expression of CA9. We also studied the modulation of expression of PBX1, a direct target of Notch3.32 PBX1 is a proto-oncogene with a consensus NICD/CSL binding site in its promoter and is a direct target of Notch3.32 Further, the CA9 promoter lacks a consensus NICD/CSL binding site (AG/CCGTGGGAAA/C).33
Expression of CA9 was observed in MCF10A and MCF7 but not MDA-MB-231 cells and 786-O cells. Renal carcinoma 786-O cells lack the expression of HIF-1α, VHL, and CA9. MDA-MB-231 cells show a reduced expression of NICD3 (during normoxia) and VHL (during normoxia) and CA9 (Fig. 4A and Suppl. Fig. S3B-D). Between MCF10A and MCF7 cells, a higher expression of CA9 was seen in MCF10A cells. Expression of Notch3 was low in MCF7 cells, and expression of VHL was high in MCF7 cells compared to that seen in MCF10A cells (Fig. 4A and Suppl. Fig. S2). This was also reflected in reporter expression and mRNA profiles (Fig. 4B and 4C and Suppl. Fig. S3). Interestingly, the expression of PBX1 was always found to be downregulated in all the cell lines studied, including prostate carcinoma PC3 cells in response to hypoxia (Fig. 4D).
shRNA
against Notch3 and NICD3 Mutants Defective in VHL Binding Causes a Decrease in Cell Proliferation
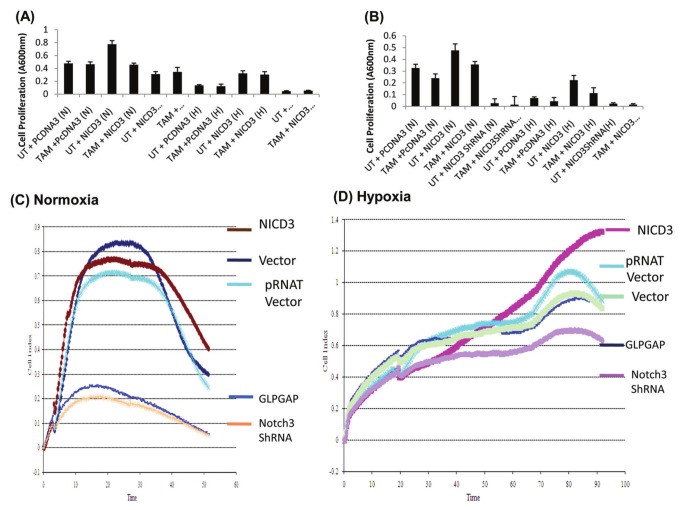
Tamoxifen has been widely used for more than 30 years in breast cancer treatment and prevention.34 To examine if combining the inhibition of Notch3 with tamoxifen improves the treatment outcome, we combined Notch3 inhibition by shRNA with 1 μM of tamoxifen. Maximal reduction in cell proliferation was observed when Notch3 shRNA was combined with 1 μM of tamoxifen as measured by the Cell Titer Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI) in both the MCF7 (Fig. 5A) and MDA-MB-231 cells (Fig. 5B).
Figure 5.
Effects of NICD3, NICD3 mutant defective in VHL binding, and Notch3 shRNA on cell proliferation. (A) Notch3 shRNA chemosensitizes MCF7 (A) and MDA-MB-231 (B) cells during normoxia and hypoxia. MCF10A and MDA-MB-231 cells were transfected with pcDNA3, Notch3 shRNA, or NICD3 and either untreated or exposed to 1 μM of tamoxifen citrate (TAM). NICD3 caused an increased cell proliferation during normoxia. Notch3 shRNA sensitized both the MCF10A and MDA-MB-231 cells to TAM. MCF7 cells transfected with vector, pRNAT vector, NICD3, NICD3 GLPGAP, or Notch3 shRNA exposed to normoxia (C) or hypoxia (D), and cell proliferation was assessed by RT-CES. Time in hours is shown on the x-axis. Each experiment has been repeated at least 3 times.
The RT-CES system monitors the overall size and morphology of the cells noninvasively and in real time. For measuring cell proliferation, MCF7 cells were transfected with vector (pRNAT), NICD3, NICD3 GLPGAP, or Notch 3 shRNA and seeded in the wells of E-Plates (Roche, Basel, Switzerland) at a density of 10,000 cells/well overnight. The cells were continuously monitored for about 60 to 100 hours using the RT-CES system. Continuous monitoring by the RT-CES system clearly shows a significant difference in multiple aspects of cellular behavior, particularly the attachment quality, the period of lag phase, and the onset and rate of the proliferation phase. NICD3 promoted cell proliferation in all phases both during normoxia (Fig. 5C) and hypoxia (Fig. 5D). However, both NICD3 mutants defective in binding to pVHL (NICD3 GLPGAP) as well as shRNA against Notch3 showed a decrease in cell proliferation at all phases during normoxia (Fig. 5A) as well as hypoxia (Fig. 5B).
Discussion
Notch signaling is involved in cell fate determination, morphogenesis, and oncogenesis, which are mediated by unique Notch downstream effectors. Expression of specific Notch downstream genes is tissue and context dependent. Although a repertoire of Notch-controlled genes has been identified, the Notch effectors that are involved in the development of human cancer remain elusive. In this study, we report a novel interaction of Notch3 with HIF-1α and pVHL in breast cancer cells and show that transcriptional regulation of CA9 is a direct consequence of NICD3 interacting with hypoxia-induced HIF-1α. Our studies provide evidence that the expression of CA9, at least in part, is regulated by the interaction of HIF-1α, NICD3, and pVHL. Our results also further suggest that direct targets of Notch3, such as PBX1, are downregulated during hypoxia possibly as a consequence of the increased interaction of HIF-1α and NICD3. Tumor cells alter their metabolism when in hypoxia to survive, proliferate, and metastasize.35 HIF-1 regulates the gene expression of proteins responsible for alterations in hypoxic tumor cell metabolism, one core aspect of which is the shift from aerobic to fermentative glycolysis,35,36 leading to elevated lactic acid production, and a drop in the intracellular pH (pHi) of hypoxic tumor cells is expected unless pH homeostasis is established.3 CA enzymes, specifically isozymes CA IX and CA XII, robustly regulate the pH system by maintaining a pHi range compatible with cell viability and proliferation while contributing to the extracellular acidification of the tumor microenvironment. These enzymes confer a survival advantage to tumor cells growing in a hypoxic and acidic microenvironment.37,38 Thus, regulation of pH by tumors growing in hypoxia could be a good target for cancer therapy, and hence, it is important to understand the molecular mechanisms regulating the expression of enzymes responsible for pH regulation. This report is one such effort in understanding the mechanism of acid regulation as a function of hypoxia. We have identified novel players and interaction in regulating the expression of CA9. Although HIF-1α,17 Notch3,22 and pVHL4 were individually shown to regulate the expression of CA9, our studies, for the first time, show that these proteins interact with each other and regulate the expression of CA9. VHL was earlier shown to repress the expression of CA9. If VHL is not needed for the activation of CA9 expression, renal carcinoma cells null for the VHL gene should show maximal CA9 expression, which is not observed in our findings (Suppl. Fig. S3D) or earlier studies.4 We observed a dramatic decrease in the expression of PBX1, a direct target of NICD3, in response to hypoxia in all cancer cells studied. Thus, PBX1 could be a novel negative biomarker of hypoxia in several different tumors. The decrease in the expression of PBX1 could be explained by the hypothesis that NICD3 regulates the expression of genes like PBX1 through its interaction and recruitment through RBP-Jκ. During hypoxia, an increase in HIF-1α levels could lead to more NICD3/HIF-1 complex formation, leading to the decrease in the NICD3/RBP-Jκ complex and thus dramatic reduction in the expression of PBX1. Further studies utilizing genome-wide approaches could identify novel targets of NICD3 regulated in a manner that is similar to that of CA9 and PBX1.
The hypothesis that cancer is a stem cell disease is widely supported.39 Thus, Comprehension of the basic biology of stem cells will provide an insight into the pathogenesis of hypoxia, microenvironment and cancer.40 Further, there is evidence that hypoxia affects stem cell function and survival.41-43 In cell culture, hypoxia maintains a stem cell/immature phenotype, induces a loss of differentiation markers, and blocks differentiation.44,45 In addition, intact Notch signaling is required for maintaining a hypoxia-induced undifferentiated stem cell state.46 In vivo, stem cells express higher levels of hypoxia-regulated genes than do the more mature progeny emerging from them.47 Thus, stem cells reside in a tissue niche that has poor vasculature, leading to the development of hypoxia.48 Therefore, linking stem cells with hypoxia survival leads to the hypothesis that the control of stem cell survival and the regulation of hypoxia response are intimately coupled and that they may share common control pathways. Thus, the interactions observed in our in vitro studies using breast carcinoma cells suggest that it is likely that these interactions also play a role in stem cell maintenance and phenotype.
Materials and Methods
Cell Lines and Transient Transfections
Breast carcinoma MCF7 and MDA-MB-231 cells and renal carcinoma cells (786-O) (VHL–/–) were grown in RPMI 1640 medium with 1% penicillin/streptomycin and 10% fetal bovine serum. MCF10A (immortalized nontumorigenic breast epithelial) cells were grown in DMEM F12 medium with 1% penicillin/streptomycin, 5% horse serum, EGF (20 ng/mL), hydrocortisone (0.5 μg/mL), cholera toxin (100 ng/mL), and insulin (10 µg/mL). Cells were transiently transfected using Lipofectamine (Life Technologies, Grand Island, NY). For normoxia, cells were exposed to 21% O2 and 5% CO2 at 37°C for 24 hours. For hypoxic conditions, cells were placed in hypoxia chambers with 0.1% O2 and 5% CO2 at 37°C for 24 hours.
Western Blot Analysis
Protein extracts from normoxic and hypoxic breast cells and 786-O cells were differentially transfected and subjected to Western blot analysis,49,50 using anti-CAIX, –HIF-1α, -Notch3, –β-actin, -pVHL, and -V5 antibody. The bound immune complexes were detected using chemiluminescent substrate (Millipore, Billerica, MA). Extracts were made in T-Per reagent (Pierce Chemical, Rockford, IL).
Plasmids and shRNA constructs
CA9 luciferase constructs have been described previously.23
Briefly, the CA9 gene promoter (spanning −885 to +95) was PCR amplified using human genomic DNA and cloned into the Kpn1/Sac1 restriction site in the pGL3 luciferase vector (Promega). Activated Notch3 (NICD3) cloned in pcDNA3.1/V5-His plasmid was a gift from Dr. Bonafe.22 Three different VHL binding mutants (NICD3P2191G [LLPLAG], NICD3L 2189G;P2191G [GLPGAG], and NICD3L2186G;L2189G [GLPGAP]) were made by a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). HIF-1α51 and VHL52 cloned in pcDNA3 and pRC/CMV, respectively, were purchased from Addgene (Cambridge, MA). HIF-1α and Notch3 shRNA were made by annealing the oligos (Suppl. Table S1) and cloning the duplex into the BamHI/HindIII site in the pRNAT vector (GenScript, Piscataway, NJ) and confirming the insertion by restriction digestion and sequencing. About 1,000 ng of the CA9 reporter was used in a dual luciferase reporter assay. About 500 ng of HIF-1α and NICD3 shRNA was used in transfections. Cells were transfected for 24 hours before exposing to normoxia or hypoxia for another 24 hours.
Dual luciferase assay
Breast carcinoma cells (2 × 105) were transfected with 1 μg of the CA9-Luc or CA9M-Luc reporter vector constructs and phRL-TK (10 ng, Promega). Cells were also transfected with different vectors (at 500 ng), such as NICD3, VHL, HIF-1α, and/or shRNA against Notch3 and HIF-1α. The concentration of the total DNA transfected was made for a total of 2 μg per transfection using the parental vector. Cells were cultured for 24 hours after transfection, and cell lysates (made using lysis buffer from Promega) were subjected to 3 freeze-thaw cycles to achieve complete cell lysis. All experiments were carried out in triplicate.
RNA Extraction and Quantitative RT-PCR Analysis
Total RNA was isolated using Trizol reagent (Life Technologies). cDNA was prepared from 1 μg total RNA by the SuperScript II reverse transcriptase system (Invitrogen, Carlsbad, CA) and amplified using Taqman-based detection designed across the exon-exon junction using an Applied Biosystems 7300 instrument (Foster City, CA). For data analysis, threshold cycles (Ct) for β-actin (internal control) and other genes (HIF-1α, Notch3, VHL, and PBX1) were determined in triplicate (arithmetical mean). The quantity of expression of each gene relative to that of β-actin was calculated using the comparative Ct (ΔΔCt) method.53
Reciprocal Co-immunoprecipitations
Normoxic or hypoxic MCF10A cell (80% confluent) lysates in RIPA buffer were incubated with protein A beads to preclear the lysate, which was incubated with anti–HIF-1α or -NICD3 or -VHL or nonspecific rabbit IgG (ratio of 1:500 [protein:protein] of IgG:lysate) followed by incubation with protein A beads for 4 hours at 4°C. The beads were washed with RIPA buffer followed by washes with 62.5 mM Tris-HCl (pH 6.8). Elution was performed by adding SDS-PAGE sample buffer and boiling at 95°C for 1 minute. The bound proteins were analyzed by Western blot analysis.
Chromatin immunoprecipitation
ChIP assay has been described previously.54 Briefly, nucleoprotein extract was prepared using RIPA buffer from 2 to 5 × 109 normoxic or hypoxic MCF10A cells. After immunoprecipitation with specific antibodies (using 5 μg anti–HIF-1α [rabbit polyclonal, Millipore] or -NICD3 [rabbit monoclonal, Cell Signaling Technology, Danvers, MA] or -VHL [rabbit polyclonal, Cell Signaling Technology] or nonspecific rabbit IgG [Sigma, St. Louis, MO] per immunoprecipitation) and extraction of DNA, PCR was performed using specific primers for CA9 and the VEGF promoter (Suppl. Table S1) that yielded ≈500-bp products. The β-actin gene promoter served as a negative control.
Cell proliferation assay
Cell proliferation was measured using the Cell Titer Aqueous One Solution Cell Proliferation Assay (Promega). MCF7 and MDA-MB-231 cells were transfected with NICD3 and its mutant NICD3 GLPGAP, Notch3 shRNA with/without 1 μM of tamoxifen citrate, and exposed to normoxia or hypoxia.
Cell proliferation was also examined using real-time monitoring of the cell proliferation of MCF7 cells (transfected with vector, NICD3, NICD3 GLPGAP, pRNAT vector [parent vector for shRNA]) and Notch3 shRNA and exposed to either normoxia or hypoxia) using the xCELLigence System (ACEA Biosciences, San Diego, CA). Briefly, background measurements were taken after adding 50 μL of the appropriate medium to the wells of the E-Plate (Roche). Then, 5 × 104 cells were seeded in 16-well microtiter plate devices (E-Plate, Roche) for monitoring cell proliferation in real time. Cell proliferation was continuously monitored every hour for a period of 72 hours as indicated using the RT-CES system. Cell sensor impedance is expressed as an arbitrary unit called the Cell Index. The Cell Index at each time point is defined as (Rn – Rb)/15, where Rn is the cell-electrode impedance of the well when it contains cells and Rb is the background impedance of the well with the media alone. Start and end times were selected during the log growth phase and used to calculate doubling time with RTCA Software version 1.2 (ACEA Biosciences) from independent triplicate wells per group. The growth curves with standard deviation were plotted.
Statistics
Statistics were performed using a 2-tailed Student t test. Differences were considered statistically significant at P ≤ 0.05.
Acknowledgments
The authors thank Dr. Alan Pollack, chairman of the Department of Radiation Oncology, for his support. They also thank Dr. Subbarayan Pochi for his help with real-time RT-PCR. They thank Dr. Mansoor M. Ahmed for the use of his facilities.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no conflicts of interest with respect to research, authorship, and/or publication of this article.
Funding: The author(s) received the following financial support for the research, authorship, and/or publication of this article: This work was supported by a grant from the Department of Defense Breast Cancer Research Program to M.M.S. (W81XWH-07-1- 0706).
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
References
- 1. Pastorek J, Pastorekova S, Callebaut I, et al. Cloning and characterization of MN, a human tumor-associated protein with a domain homologous to carbonic anhydrase and a putative helix-loop-helix DNA binding segment. Oncogene. 1994;9:2877-88 [PubMed] [Google Scholar]
- 2. Zavada J, Zavadova Z, Pastorekova S, Ciampor F, Pastorek J, Zelnik V. Expression of MaTu-MN protein in human tumor cultures and in clinical specimens. Int J Cancer. 1993;54:268-74 [DOI] [PubMed] [Google Scholar]
- 3. Ivanov S, Liao SY, Ivanova A, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ivanov SV, Kuzmin I, Wei MH, et al. Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc Natl Acad Sci U S A. 1998;95:12596-601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McDonald PC, Winum JY, Supuran CT, Dedhar S. Recent developments in targeting carbonic anhydrase IX for cancer therapeutics. Oncotarget. 2012;3:84-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bleumer I, Oosterwijk E, Oosterwijk-Wakka JC, et al. A clinical trial with chimeric monoclonal antibody WX-G250 and low dose interleukin-2 pulsing scheme for advanced renal cell carcinoma. J Urol. 2006;175:57-62 [DOI] [PubMed] [Google Scholar]
- 7. Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov. 2011;10:767-77 [DOI] [PubMed] [Google Scholar]
- 8. Airley RE, Loncaster J, Raleigh JA, et al. GLUT-1 and CAIX as intrinsic markers of hypoxia in carcinoma of the cervix: relationship to pimonidazole binding. Int J Cancer. 2003;104:85-91 [DOI] [PubMed] [Google Scholar]
- 9. Liao SY, Aurelio ON, Jan K, Zavada J, Stanbridge EJ. Identification of the MN/CA9 protein as a reliable diagnostic biomarker of clear cell carcinoma of the kidney. Cancer Res. 1997;57:2827-31 [PubMed] [Google Scholar]
- 10. Chia SK, Wykoff CC, Watson PH, et al. Prognostic significance of a novel hypoxia-regulated marker, carbonic anhydrase IX, in invasive breast carcinoma. J Clin Oncol. 2001;19:3660-8 [DOI] [PubMed] [Google Scholar]
- 11. Tan EY, Yan M, Campo L, et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br J Cancer. 2009;100:405-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmitt A, Barth TF, Beyer E, et al. The tumor antigens RHAMM and G250/CAIX are expressed in head and neck squamous cell carcinomas and elicit specific CD8+ T cell responses. Int J Oncol. 2009;34:629-39 [DOI] [PubMed] [Google Scholar]
- 13. Beasley NJ, Wykoff CC, Watson PH, et al. Carbonic anhydrase IX, an endogenous hypoxia marker, expression in head and neck squamous cell carcinoma and its relationship to hypoxia, necrosis, and microvessel density. Cancer Res. 2001;61:5262-7 [PubMed] [Google Scholar]
- 14. Swinson DE, Jones JL, Richardson D, et al. Carbonic anhydrase IX expression, a novel surrogate marker of tumor hypoxia, is associated with a poor prognosis in non-small-cell lung cancer. J Clin Oncol. 2003;21:473-82 [DOI] [PubMed] [Google Scholar]
- 15. Proescholdt MA, Mayer C, Kubitza M, et al. Expression of hypoxia-inducible carbonic anhydrases in brain tumors. Neuro Oncol. 2005;7:465-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaluz S, Kaluzova M, Liao SY, Lerman M, Stanbridge EJ. Transcriptional control of the tumor- and hypoxia-marker carbonic anhydrase 9: a one transcription factor (HIF-1) show? Biochim Biophys Acta. 2009;1795:162-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wykoff CC, Beasley NJ, Watson PH, et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60:7075-83 [PubMed] [Google Scholar]
- 18. Olive PL, Aquino-Parsons C, MacPhail SH, et al. Carbonic anhydrase 9 as an endogenous marker for hypoxic cells in cervical cancer. Cancer Res. 2001;61:8924-9 [PubMed] [Google Scholar]
- 19. Kaluz S, Kaluzova M, Opavsky R, et al. Transcriptional regulation of the MN/CA 9 gene coding for the tumor-associated carbonic anhydrase IX: identification and characterization of a proximal silencer element. J Biol Chem. 1999;274:32588-95 [DOI] [PubMed] [Google Scholar]
- 20. Grabmaier K, A de Weijert MC, Verhaegh GW, Schalken JA, Oosterwijk E. Strict regulation of CAIX(G250/MN) by HIF-1alpha in clear cell renal cell carcinoma. Oncogene. 2004;23:5624-31 [DOI] [PubMed] [Google Scholar]
- 21. Shao Y, Li Y, Zhang J, et al. Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX. Nucleic Acids Res. 2010;38:2813-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sansone P, Storci G, Giovannini C, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807-15 [DOI] [PubMed] [Google Scholar]
- 23. Shareef MM, Dancea HC, Gross JL, et al. A noncommercial polymerase chain reaction-based method to approach one hundred percent recombinant clone selection efficiency. Anal Biochem. 2008;382: 75-6 [DOI] [PubMed] [Google Scholar]
- 24. Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han ZB, Ren H, Zhao H, et al. Hypoxia-inducible factor (HIF)-1 alpha directly enhances the transcriptional activity of stem cell factor (SCF) in response to hypoxia and epidermal growth factor (EGF). Carcinogenesis. 2008;29:1853-61 [DOI] [PubMed] [Google Scholar]
- 26. Barliya T, Mandel M, Livnat T, Weinberger D, Lavie G. Degradation of HIF-1alpha under hypoxia combined with induction of Hsp90 polyubiquitination in cancer cells by hypericin: a unique cancer therapy. PLoS One. 2011;6:e22849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Metzen E, Ratcliffe PJ. HIF hydroxylation and cellular oxygen sensing. Biol Chem. 2004;385:223-30 [DOI] [PubMed] [Google Scholar]
- 28. Huang Y, Du KM, Xue ZH, et al. Cobalt chloride and low oxygen tension trigger differentiation of acute myeloid leukemic cells: possible mediation of hypoxia-inducible factor-1alpha. Leukemia. 2003;17:2065-73 [DOI] [PubMed] [Google Scholar]
- 29. Stolze IP, Tian YM, Appelhoff RJ, et al. Genetic analysis of the role of the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (FIH) in regulating hypoxia-inducible factor (HIF) transcriptional target genes [corrected]. J Biol Chem. 2004;279:42719-25 [DOI] [PubMed] [Google Scholar]
- 30. Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McNeill LA, Hewitson KS, Claridge TD, Seibel JF, Horsfall LE, Schofield CJ. Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the beta-carbon of asparagine-803. Biochem J. 2002;367:571-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park JT, Shih Ie M, Wang TL. Identification of Pbx1, a potential oncogene, as a Notch3 target gene in ovarian cancer. Cancer Res. 2008;68:8852-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rivera-Guevara C, Camacho J. Tamoxifen and its new derivatives in cancer research. Recent Pat Anticancer Drug Discov. 2011;6:237-45 [DOI] [PubMed] [Google Scholar]
- 35. Brahimi-Horn MC, Bellot G, Pouyssegur J. Hypoxia and energetic tumour metabolism. Curr Opin Genet Dev. 2011;21:67-72 [DOI] [PubMed] [Google Scholar]
- 36. Brahimi-Horn MC, Pouyssegur J. HIF at a glance. J Cell Sci. 2009;122:1055-7 [DOI] [PubMed] [Google Scholar]
- 37. Chiche J, Brahimi-Horn MC, Pouyssegur J. Tumour hypoxia induces a metabolic shift causing acidosis: a common feature in cancer. J Cell Mol Med. 2010;14:771-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chiche J, Ilc K, Brahimi-Horn MC, Pouyssegur J. Membrane-bound carbonic anhydrases are key pH regulators controlling tumor growth and cell migration. Adv Enzyme Regul. 2010;50:20-33 [DOI] [PubMed] [Google Scholar]
- 39. Polyak K, Hahn WC. Roots and stems: stem cells in cancer. Nat Med. 2006;12:296-300 [DOI] [PubMed] [Google Scholar]
- 40. Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea. A paradigm shift. Cancer Res. 2006;66:1883-90, discussion 1895-6. [DOI] [PubMed] [Google Scholar]
- 41. Cejudo-Martin P, Johnson RS. A new notch in the HIF belt: how hypoxia impacts differentiation. Dev Cell. 2005;9:575-6 [DOI] [PubMed] [Google Scholar]
- 42. Ramirez-Bergeron DL, Simon MC. Hypoxia-inducible factor and the development of stem cells of the cardiovascular system. Stem Cells. 2001;19:279-86 [DOI] [PubMed] [Google Scholar]
- 43. Covello KL, Kehler J, Yu H, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Axelson H, Fredlund E, Ovenberger M, Landberg G, Pahlman S. Hypoxia-induced dedifferentiation of tumor cells: a mechanism behind heterogeneity and aggressiveness of solid tumors. Semin Cell Dev Biol. 2005;16:554-63 [DOI] [PubMed] [Google Scholar]
- 45. Danet GH, Pan Y, Luongo JL, Bonnet DA, Simon MC. Expansion of human SCID-repopulating cells under hypoxic conditions. J Clin Invest. 2003;112:126-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gustafsson MV, Zheng X, Pereira T, et al. Hypoxia requires Notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617-28 [DOI] [PubMed] [Google Scholar]
- 47. Unwin RD, Smith DL, Blinco D, et al. Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood. 2006;107:4687-94 [DOI] [PubMed] [Google Scholar]
- 48. Nilsson GE. Surviving anoxia with the brain turned on. News Physiol Sci. 2001;16:217-21 [DOI] [PubMed] [Google Scholar]
- 49. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680-5 [DOI] [PubMed] [Google Scholar]
- 50. Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237-46 [DOI] [PubMed] [Google Scholar]
- 52. Iliopoulos O, Kibel A, Gray S, Kaelin WG. Tumour suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822-6 [DOI] [PubMed] [Google Scholar]
- 53. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-8 [DOI] [PubMed] [Google Scholar]
- 54. Shareef MM, Cui N, Burikhanov R, et al. Role of tumor necrosis factor-alpha and TRAIL in high-dose radiation-induced bystander signaling in lung adenocarcinoma. Cancer Res. 2007;67:11811-20 [DOI] [PubMed] [Google Scholar]