Abstract
The ribosomal protein S19 (RPS19) is located in the small (40S) subunit and is one of 79 ribosomal proteins. The gene encoding RPS19 is mutated in approximately 25% of patients with Diamond-Blackfan anemia, which is a rare congenital erythroblastopenia. Affected individuals present with decreased numbers or the absence of erythroid precursors in the bone marrow, and associated malformations of various organs are common. We produced C57BL/6J mice with a targeted disruption of murine Rps19 to study its role in erythropoiesis and development. Mice homozygous for the disrupted Rps19 were not identified as early as the blastocyst stage, indicating a lethal effect. In contrast, mice heterozygous for the disrupted Rps19 allele have normal growth and organ development, including that of the hematopoietic system. Our findings indicate that zygotes which are Rps19−/− do not form blastocysts, whereas one normal Rps19 allele in C57BL/6J mice is sufficient to maintain normal ribosomal and possibly extraribosomal functions.
The ribosomal proteins constitute a major component of cellular proteins and are known to be mandatory for cellular growth. A reduced level of one ribosomal protein is rate limiting for the assembly of ribosomes and may constitute a bottleneck for protein synthesis in tissues with a high proliferative activity. Recently, Draptchinskaia et al. found that the gene encoding ribosomal protein S19 (RPS19) was mutated in 25% of patients with Diamond-Blackfan anemia (DBA) (5). DBA is a rare, congenital, and chronic anemia that is characterized by the absence or decreased numbers of erythroid precursors in the bone marrow but an otherwise normal cellularity (1, 4). Approximately 30% of affected patients with DBA show one or several dysmorphic features including growth retardation, hand and/or limb malformations, urogenital anomalies, and congenital heart defects (1, 2, 10). Clinical expression in DBA is highly variable (14, 21), and the role of RPS19 in the pathogenesis of the disease is presently unknown.
RPS19 is part of the 40S ribosomal unit and shows a high degree of sequence conservation across mammalian species at the protein level (5). In addition to its implication in erythropoiesis, there are indications that RPS19 has extraribosomal functions.
A previous study has shown that a dimer of RPS19 may mediate chemotaxis (13). Free RPS19 has also been shown to interact with fibroblast growth factor 2 (FGF-2), and a possible role for RPS19 in embryonic development has been suggested (17). We hypothesized that an Rps19 null mutant would be deleterious for development and that mice heterozygous for Rps19 may present with hematological abnormalities. To clarify the role of RPS19 in erythropoiesis, we created a null mutation for murine Rps19 by homologous recombination in embryonic stem (ES) cells. The targeted Rps19 was introduced on a C57BL/6J background. We present here the results from our studies of this mouse model.
MATERIALS AND METHODS
Construction of the Rps19 targeting vector.
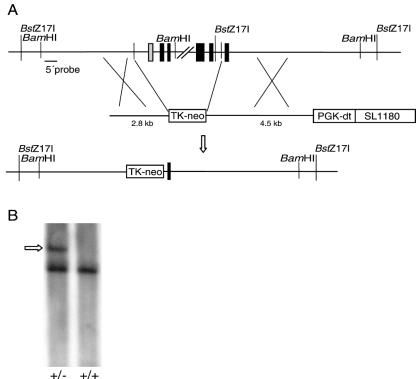
An Rps19 intron probe was used to screen a mouse 129 strain bacterial artificial chromosome (BAC) library (BAC mouse release II; Genome Systems Inc., St Louis, Mo.). Two clones (356H01 and 480F08) were isolated and used for the construction of the Rps19 targeting vector. A 2.8-kb NheI-NheI BAC sequence 5′ of the RPS19 gene was subcloned into a Bluescript II KS vector. A 1.4-kb sequence with a thymidine kinase promoter-driven neomycin resistance gene flanked by loxP sites was used as the replacement cassette for a region including the 5′ untranslated region (UTR) and exons 1 to 4 of the Rps19 gene. A 4.5-kb HindIII-BamHI BAC sequence 3′ of the RPS19 gene was used, followed by a phosphoglycerate kinase promoter-driven diphtheria toxin gene (Fig. 1A).
FIG. 1.
Targeting of Rps19 in ES cells. (A) Structures of the wild-type Rps19 gene, the Rps19 targeting construct, and the resulting targeted Rps19 gene. Black boxes represent protein-coding exons, and the grey box is the 5′ UTR. A linearized construct was used with a phosphoglycerate kinase promoter-driven diphtheria toxin (PGK-dt) gene followed by a SL1180 cloning vector. A thymidine kinase promoter-driven neomycin resistance gene (Tk-neo) was used to replace the 5′ UTR and exons 1 to 4 of Rps19 by homologous recombination together with the 2.8- and 4.5-kb murine wild-type sequences surrounding the Tk-neo cassette. (B) BstZ17I-digested genomic DNA from an Rps19+/− mouse and an Rps19+/+ mouse was analyzed by Southern hybridization using the 5′ probe. The larger fragment (arrow) of 10.1 kb was produced in the absence of the BstZ17I site in intron 5 of the targeted allele.
Homologous recombination, breeding, and genotyping.
The targeting vector was linearized using a unique NotI site, and 45 μg was electroporated using 30 × 106 (129/Sv)/(129/SvJ) F1 hybrid ES cells previously cultured on mouse fibroblast feeder cells. ES cells were cultured in G418-containing medium (300 μg/ml), and DNA from surviving clones was purified. Ten micrograms of genomic ES cell DNA from each G418-resistant clone was digested with BamHI (New England Biolabs), separated on agarose gels, and transferred to Hybond N+ nylon filters (Amersham Pharmacia Biotech) by Southern blotting. Filters were hybridized using a [32P]dCTP-labeled 5′ intron probe (Fig. 1A) at 42°C overnight followed by washing using standard protocols. Filters were exposed to X-ray films (Kodak BioMax MR). Cells from one ES clone positive for homologous recombination were injected into C57BL/6J blastocysts and implanted into the oviducts of C57BL/6J pseudopregnant foster females. Chimeras were continuously bred onto a C57BL/6J background for at least five generations. Mice from consecutive breeding were screened for targeted disruption of Rps19 by Southern blotting using BamHI or BstZ17I (New England Biolabs) and by PCR using three primers that allowed for simultaneous amplification of wild-type and targeted allele in each sample: GCC GGA AAT GGG AAT AGT TAC G (reverse wild-type primer), CTT TCC TCT CCG CGA GCG (forward common primer), and CAA TGG CCG ATC CCA TAT TGG C (reverse neomycin resistance gene primer). Each PCR sample included at least 100 ng of phenol-chloroform-extracted DNA from a tail tip sample together with a 0.2 mM total concentration of deoxynucleoside triphosphates, 3 pmol of each primer, and 0.5 U of Taq polymerase (Promega) with supplied PCR buffer and was cycled at 95°C for 6 min followed by 30 cycles at 95°C for 30 s, 50°C for 1 min, and 72°C for 45 s, with a final extension at 72°C for 10 min.
Embryo genotyping was performed on day 11 and day 12 embryos. Dissected embryo tissues were washed in phosphate-buffered saline (PBS), treated with proteinase K (500 μg/ml) overnight at 55°C, ethanol precipitated, and subjected to PCR genotyping.
Blastocyst genotyping assay.
Blastocysts were harvested on gestation day 3.5 from pregnant Rps19+/− female mice previously mated with Rps19+/− male mice. Individual blastocysts were washed in PBS-1% fetal calf serum (FCS), freeze-thawed in buffer containing 50 mM Tris-HCl (pH 8.0), 0.5% Triton X-100, and 200 μg of proteinase K (Amersham Biosciences)/ml, and incubated at 55°C for 16 h followed by proteinase K deactivation at 95°C for 10 min. The blastocyst samples were amplified as described above using a two-step PCR approach with CTT TCC TCT CCG CGA GCG (forward common primer), ACG TAA ACT CCT CTT CAG ACC (reverse neomycin resistance gene primer), and GCC GGA AAT GGG AAT AGT TAC G (reverse wild-type primer) for the first PCR performed at 95°C for 6 min followed by 44 cycles at 95°C for 30 s, 55°C for 1 min, and 72°C for 45 s, with a final extension at 72°C for 7 min. Primers ATG GAG CTG AAG CAA CTT CC (forward common primer), AGA CTC GAG GAA TTC CGA TC (reverse neomycin resistance gene primer), and TCC AGG GAA AAG GCA CTA TG (reverse wild-type primer) were used for the second amplification performed at 95°C for 6 min followed by 29 cycles at 95°C for 30 s, 62°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min.
Analysis of Rps19 transcript levels.
Total RNA was prepared from individual spleens derived from heterozygous and wild-type sibling mice by using Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples were subjected to cDNA synthesis using Moloney murine leukemia virus reverse transcriptase (USB Corp.) and random hexamers (Amersham Biosciences) according to the manufacturers' recommendations. For real-time PCR (9, 12), FAM/TAMRA-labeled probes (MedProbe) were used with sequences 5′-CCA AGA TCT ATG GAG GAC G-3′ for Rps19 and 5′-CCT GGC CAA GGT CAT CCA TGA CAA CTT T-3′ for murine GAPDH (endogenous reference). The cycling conditions for real-time PCR were 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The primers used for Rps19 cDNA amplification were 5′-CAG CAC GGC ACC TGT ACC T-3′ and 5′-GCT GGG TCT GAC ACC GTT TC-3′, and those for murine GAPDH cDNA amplification were 5′-CTT CAC CAC CAT GGA GAA GGC-3′ and 5′-GGC ATG GAC TGT GGT CAT GAG-3′. Standard curves were created using a serial dilution of PCR products from Rps19 and murine GAPDH cDNA by using the primers described above. cDNA samples from three adult Rps19+/− and two Rps19+/+ mice were analyzed in triplicate on an ABI Prism 7000 sequence detection system (Applied Biosystems).
Histological analysis.
Heart, spleen, kidney, testis, and lung samples were dissected, washed in 1× PBS, fixed in 4% paraformaldehyde for 24 h, and dehydrated using an overnight ethanol gradient from 70 to 100%. Samples were treated with xylene and embedded in paraffin wax. Organ morphology was analyzed using 5-μm-thick sections of each paraffinated sample after staining with Van Giesen, hematoxylin and eosin, and Giemsa dyes according to standard procedures.
Hematological analysis.
Analyses of blood hemoglobin, red and white blood cell counts, hematocrit, mean erythroid corpuscular volume, and mean erythroid corpuscular hemoglobin concentration were done using 300 μl of peripheral blood and were performed at the Clinical Chemistry Center at SVA, Uppsala, Sweden.
Flow cytometry.
Bone marrow cells were flushed from four mouse femurs (two Rps19+/− and two Rps19+/+ mice, respectively) with PBS. One million cells per sample were washed in PBS-1% FCS. Cells were treated with 5 μM H0342 nuclei stain for 30 min at 37°C, chilled on ice for 5 min, and washed in PBS-1% FCS. One microgram each of phycoerythrin-conjugated anti-Ter119 (PharMingen) and fluorescein isothiocyanate-conjugated anti-CD71 (PharMingen) was added per sample followed by incubation at 4°C for 30 min. From each sample, 3 × 105 cells were incubated with Alexa Fluor 647-conjugated annexin V (Molecular Probes) before analysis.
In vitro clonogenic progenitor assay.
For the erythroid progenitor assay, 7.5 × 104 bone marrow cells were plated in duplicate in methylcellulose matrix (Myelocult 3236; Stem Cell Technologies, Inc., Vancouver, Canada) containing 5 U of human erythropoietin (Janssen-Cilag, Sollentuna, Sweden)/ml, 100 ng of human trombopoietin (a gift from Kirin Brewery, Tokyo, Japan)/ml, and mouse stem cell factor (a gift from Amgen, Thousand Oaks, Calif.). For macrophage and granulocyte-macrophage colony formation, 2 × 104 bone marrow cells were plated in duplicate in methylcellulose matrix containing interleukin-3 (10 ng/ml), interleukin-6 (10 ng/ml), and stem cell factor (50 ng/ml) (Myelocult 3534, Stem Cell Technologies, Inc.). After 7 days in culture (37°C and an atmosphere of 5% CO2), colonies were scored under the microscope.
For the CFU-S12 (splenic colony-forming units scored on day 12) assay, bone marrow cells (105 cells) derived from Rps19+/− or control animals were injected into lethally irradiated (975 cGy, 137Cs gamma rays) recipient congenic B6.SJL-PtprcaPepb/BoyJ (CD45.1) mice. Twelve days after injection, animals were sacrificed and the number of macroscopic colonies on the spleen was evaluated.
Experimental animals.
All mice were housed at the Uppsala Biomedical Centre, Uppsala, Sweden, and the construction and analysis of our mouse model was approved by the Uppsala Animal Research Ethics Board (2000-08-25, project number C 159/0).
RESULTS
Generation of mice heterozygous for the disruption of Rps19.
The murine gene encoding RPS19 was isolated from a mouse genomic library (BAC mouse release II; Genome Systems Inc.). The gene consists of a 5′ UTR and five protein-coding exons (Fig. 1A). The construct used for homologous recombination carries the neomycin resistance gene driven by a thymidine kinase promoter that replaces the 5′ UTR and exons 1 to 4 of Rps19 (Fig. 1A). The construct was introduced into murine (129/Sv)/(129/SvJ) F1 hybrid ES cells by electroporation. Screening for homologous recombination of the construct in ES cells by Southern hybridizations revealed one colony positive for the targeted insertion of the neomycin resistance cassette (Fig. 1B). Cells from the positive clone were injected into C57BL/6J blastocysts and transferred into oviducts of pseudopregnant C57BL/6J female mice to generate chimeras. Germ line transmission was obtained in one female chimera. Mice with the disrupted Rps19 were produced and further bred onto a C57BL/6J genetic background.
Analysis of Rps19 transcript levels.
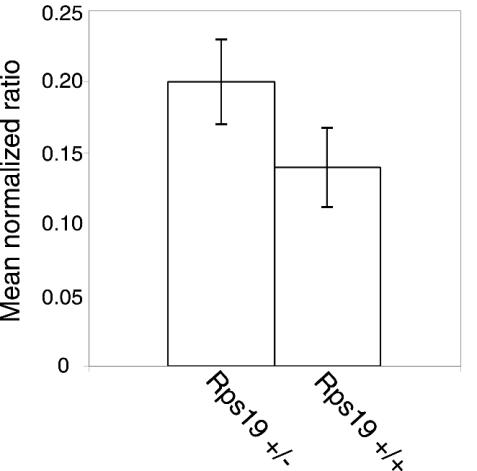
Rps19 mRNA levels were indirectly analyzed in cells from individual spleens taken from adult mice. Total RNA extracted from spleens was used as a substrate for cDNA synthesis using reverse transcriptase. Real-time PCR using GAPDH as the endogenous control revealed similar levels of Rps19 cDNA in samples from mice heterozygous for Rps19 compared to wild-type controls (Fig. 2). The normalized ratios of Rps19 versus GAPDH were 0.20, 0.17, and 0.23 for Rps19+/− mice and 0.12 and 0.16 for Rps19+/+ mice. The difference between Rps19+/− mice and Rps19+/+ mice in real-time PCR products was not significant (P = 0.13, Student's t test). This suggests similar levels of Rps19 mRNA in spleens from Rps19+/− and Rps19+/+ mice.
FIG. 2.
Levels of wild-type Rps19 transcript in spleen cells from adult Rps19+/− mice. PCR products using Rps19 and murine GAPDH cDNA (endogenous control) were quantified by real-time PCR. The mean normalized ratios represent Rps19 transcript levels divided with murine GAPDH from three Rps19+/− mice and two Rps19+/+ mice. Bars indicate standard deviations of the mean normalized ratios.
Absence of blastocysts with homozygous loss of Rps19.
Female and male mice heterozygous for the Rps19 null allele were bred in order to produce offspring homozygous for the targeted Rps19 gene. No homozygous offspring was identified among 52 individuals. The genotyping revealed that 63% of the offspring were heterozygous and 37% were wild type. This distribution of alleles is consistent with a lethal phenotype for the Rps19−/− mice. To further investigate the possibility of embryonic lethality in Rps19−/− mice, embryos from heterozygous male/female breeding were genotyped. A total of 33 embryos from day 11 and day 12 were analyzed. No embryo was homozygous for the disrupted Rps19, whereas 19 embryos were heterozygous and 14 were wild type.
Female and male mice heterozygous for the disrupted Rps19 allele were then bred to screen for Rps19−/− in day 3.5 blastocysts (Fig. 3). Of the 17 blastocysts harvested and analyzed, none carried the Rps19−/− genotype whereas 12 (71%) were heterozygous and five were homozygous for the wild-type allele. The observed genotype distribution of Rps19 in blastocysts differs significantly from the expected distribution (1:2:1) of Rps19+/+, Rps19+/−, and Rps19−/− (P = 0.017, two-sample chi-square test). The combined results indicate early lethality in the Rps19−/− mice and suggest that Rps19−/− zygotes do not develop to normal blastocysts.
FIG. 3.
Blastocyst PCR genotyping. PCR products (1 to 10 of 17) from day 3.5 blastocysts are shown with the 100-bp fragment from a DNA ladder (L), negative control (H2O), wild-type control (+/+), and Rps19 targeting vector as a positive control (C). Amplification of the wild-type allele and the targeted allele resulted in 123- and 109-bp PCR products, respectively.
Organ morphology in Rps19+/− mice.
Mice heterozygous for the disrupted Rps19 showed a normal growth rate and weight compared to what was seen with wild-type littermates. The morphology and histological structure of organ sections of heart, spleen, kidney, testis, and lung were analyzed by confocal microscopy after Van Giesen, hematoxylin and eosin, and Giemsa staining. No gross abnormalities in organ morphology were observed in adult heterozygous animals (data not shown).
Hematological analysis of Rps19+/− mice.
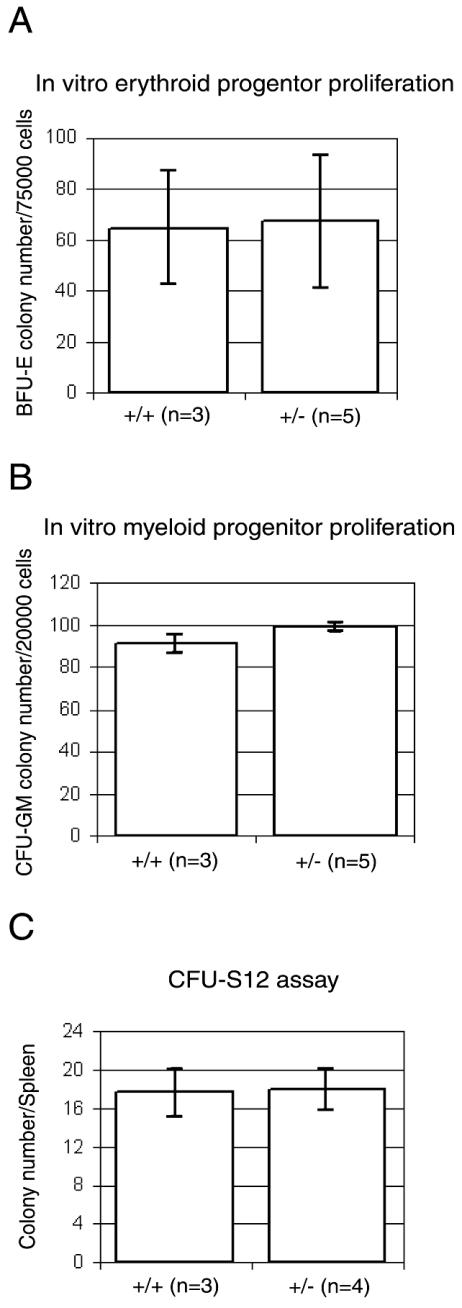
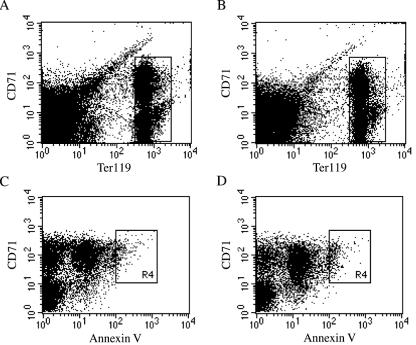
The peripheral blood analysis of wild-type and heterozygous mice included examination of hemoglobin levels, hematocrit, red and white blood cell counts, mean erythrocyte corpuscular volume, and mean erythrocyte corpuscular hemoglobin concentration. The analysis was done on a total of 41 mice who were 3 and 7 weeks of age. The Rps19+/− mice showed no statistically significant differences with respect to these parameters when compared to age-matched wild-type controls (Table 1). The female-to-male distribution was 4:7 for 3-week-old Rps19+/− mice, 5:4 for 3-week-old Rps19+/+ mice, 6:5 for 7-week-old Rps19+/− mice, and 5:5 for 7-week-old Rps19+/+ mice. The total numbers of bone marrow cells from femurs were similar between heterozygous mice and wild-type controls. Colony formation of erythroid and myeloid progenitors was studied in vitro (Fig. 4). Bone marrow cells from heterozygous mice did not differ in their hematopoietic colony-forming abilities (size and number of colonies) when compared to bone marrow cells from littermate controls. This suggests that proliferation and differentiation of bone marrow progenitor cells from Rps19+/− mice is normal. The number and distribution of red-cell progenitors in bone marrow were also analyzed by fluorescence-activated cell sorting. Bone marrow cells from four mice (two Rps19+/− and two Rps19+/+) were analyzed using the cell surface markers Ter119 (associated with glycophorin A) and CD71 (transferrin receptor) to select for proliferating cells of the erythroid lineage. The distribution of CD71+/Ter119high precursor cells in Rps19+/− mice was similar to that in the wild-type mice (Fig. 5A and B). Similarly, cells from heterozygous mice did not show signs of increased apoptosis in the CD71+/Ter119high fraction of bone marrow cells (Fig. 5C and D). The numbers of early hematopoietic cells in bone marrow identified as CD34+/CD45R(B220)− cells were not statistically different between Rps19+/− mice and Rps19+/+ controls (data not shown).
TABLE 1.
Measurements of peripheral-blood parameters in Rps19+/− and Rps19+/+ mice at 3 and 7 weeks of agea
| Age (wk) | Genotype (no. of mice) | B-Hb (g/liter) | RBC (1012/liter) | WBC (109/liter) | Hematocrit (%) | MCV (fl) | MCHC (g/liter) |
|---|---|---|---|---|---|---|---|
| 3 | +/− (11) | 118.7 ± 9.5 | 8.0 ± 0.8 | 8.5 ± 1.9 | 41.9 ± 3.3 | 52.8 ± 2.7 | 283.6 ± 5.6 |
| +/+ (9) | 114.0 ± 9.4 | 7.4 ± 0.6 | 10.4 ± 2.8 | 40.4 ± 3.1 | 54.4 ± 3.2 | 282.2 ± 5.7 | |
| 7 | +/− (11) | 152.3 ± 12.6 | 9.7 ± 0.8 | 6.9 ± 3.6 | 50.5 ± 3.5 | 52.0 ± 1.6 | 302.3 ± 9.0 |
| +/+ (10) | 152.2 ± 4.9 | 9.8 ± 0.3 | 8.9 ± 3.2 | 50.8 ± 1.2 | 51.9 ± 0.7 | 300.0 ± 8.8 |
B-Hb, blood hemoglobin; RBC, red blood cell count; WBC, white blood cell count; MCV, mean corpuscular volume; MCHC, mean corpuscular hemoglobin concentration. Values are means ± standard deviations.
FIG. 4.
Bone marrow cell culture assays using mice heterozygous for Rps19 (+/−) and control littermates (+/+). (A) Erythroid progenitor proliferation assay. Erythroid burst forming unit (BFU-E) colonies were scored after 7 days of culture on methylcellulose. (B) Macrophage and granulocyte-macrophage colony formation (CFU-GM) after 7 days of culture of bone marrow cells on methylcellulose. (C) Splenic colony-forming units scored 12 days (CFU-S12) after injection of Rps19+/− and wild-type bone marrow cells into irradiated recipient SJL mice.
FIG. 5.
Erythroid cell distribution and apoptosis in bone marrow of Rps19+/− mice. The fluorescence-activated cell sorting analysis data show cells of the erythroid lineage from adult mouse bone marrow cells of one Rps19+/− male (A and C) and one Rps19+/+ male control (B and D). CD71+/Ter119high cell populations (boxed areas in panels A and B) were gated and analyzed for apoptosis by using annexin V as the marker (C and D). An area (R4 boxes) of increased annexin V binding was defined as containing apoptotic cells and included 1.9% of the total CD71+/Ter119high cells for the Rps19+/− mouse and 3.1% for the wild-type control.
DISCUSSION
We have previously shown that mutations in the gene encoding RPS19 in humans cause erythroblastopenia and DBA. In this report, we discuss the targeted disruption of mouse Rps19 and the phenotypic characterization of this mouse model. The ribosomal proteins are thought to facilitate an optimal configuration of the rRNA, thereby promoting the speed and accuracy of the translation of mRNAs into proteins (24). The ribosomal proteins, including RPS19, are highly conserved, with strong homologies between diverse organisms. RPS19 is presently the only ribosomal protein known to be associated with a human disease (5, 23). The finding of a mutant ribosomal protein as a cause of DBA is unexpected in view of the fact that clinical symptoms in the majority of patients are confined to erythropoiesis and in some cases to organs during embryogenesis. There is now accumulating evidence that some ribosomal proteins have a second, extraribosomal function (22, 23). In Drosophila melanogaster, evidence for extraribosomal functions of ribosomal proteins has emerged from the identification of mutations in the genes encoding RPS2, RPS6, RPL19, and RPS21. The RPS2 (string of pearls) mutations result in an arrest of oogenesis at stage 5 of development (3). Mutations in RPS6 result in hypertrophied hematopoietic organs (19), RPL19 mutants display abnormal wing blade development (11), and down-regulation of RPS21 results in small body size and hypertrophied hematopoietic organs with a reduced number of circulating hemocytes (18). Analogous to what is seen with Drosophila RPS2, RPS6, RPL19, and RPS21 mutants, the clinical features of DBA could suggest extraribosomal and tissue-specific functions for RPS19. In humans, the RPS4X and RPS4Y genes encoding the RPS4 isoforms S4X and S4Y were considered to be candidate genes for the extragonadal phenotypes in Turner syndrome (6), but this hypothesis has been rejected in more recent studies (8, 20). Interestingly, the existence of two different isoforms of S4 led to the formation of “male” and “female” ribosomes, with RPS4X being 10 to 15% as abundant as RPS4Y in male ribosomes (25). RPS4X and RPS4Y are interchangeable in the ribosome, and the amino acid sequence difference between the isoforms suggests that RPS4Y may play a part in determining the sex-specific characteristics of males. NIH 3T3 cells with an enhanced expression of RPS3a can undergo apoptosis if the RPS3a levels are suppressed (15). It is suggested that cells with increased expression of RPS3a are primed for apoptosis.
The reason for the extraribosomal functions of ribosomal proteins is unknown. It has been speculated that proteins that existed prior to incorporation into the ribosome may have retained an original and extraribosomal function. In such a case, the ribosomal proteins would have at least one function apart from their specific function in protein synthesis (23). Recent data suggest the existence of extraribosomal functions for RPS19 as well. In vitro studies have shown that RPS19 may form a homodimer that can act as a chemotactic factor in HL-60 cells derived from human promyelocytic leukemia (13, 16). The homodimerization is mediated in the cells by transglutaminase type II and the dimer complex is presented to the extracellular environment, thereby generating the recruitment of macrophages to areas of apoptotic cells. In addition, free intracellular RPS19 can interact with FGF-2 in NIH 3T3 cells (17). FGF-2 is involved in the differentiation process of different cell types, and an interaction between FGF-2 and RPS19 may suggest a link for RPS19 in embryogenesis (5, 17).
To investigate a possible role for RPS19 in organ development and erythropoiesis, we produced mice with a targeted disruption of the murine homologue Rps19. The absence of the Rps19−/− genotype in mice, embryos, and blastocysts in our study indicates an early lethal effect in Rps19 null mice. Ribosome assembly and function require a stoichiometric balance of ribosomal proteins. Reduced levels of one or several ribosomal proteins may reduce the number of ribosomes and the maximal protein synthesis rate. This may increase apoptosis as a result of reduced levels of antiapoptotic factors (22). Accordingly, the complete absence of one ribosomal component may prevent ribosome formation and a normal development of the Rps19−/− zygote to a blastocyst.
Heterozygous animals present with a normal growth as well as a normal organ morphology and histology. No abnormalities of the hematopoietic system were detected in Rps19+/− mice. A statistically significant reduction in the number of white blood cells was observed in 3-week-old male Rps19+/− mice compared to Rps19+/+ male controls (P = 0.004). The absence of a detectable phenotype in the heterozygous (Rps19+/−) mice may have several explanations. The control of ribosomal protein synthesis is, at least in part, regulated at the translational level. Our results from real-time PCR suggest that the regulation of Rps19 occurs also at the transcriptional level. As a consequence, a normalization of Rps19 may occur. Another possibility for the normal phenotype in Rps19+/− mice is that the postulated role of RPS19 in erythropoiesis is confined to certain mammals or that mice have compensatory mechanisms for haploinsufficiency of Rps19. An alternative explanation for the lack of an Rps19+/− phenotype is the genetic background. The mutant allele is bred on a C57BL/6J background, which is a strongly inbred strain. In humans, the DBA phenotype is highly variable and sometimes even subclinical. The large clinical variation in DBA is not related to the nature of RPS19 mutations, and it has been suggested that other modifying genetic factors play an important role in the expression of the disease (7). Accordingly, the specific genetic background in the C57BL/6J strain may compensate for an Rps19+/− phenotype.
We present the first example of a targeted disruption of a gene encoding a ribosomal protein. Our results show that the Rps19−/− genotype in C57BL/6J mice is lethal prior to implantation whereas mice with disruption of one mutant Rps19 allele have a normal phenotype, including the hematopoietic system. Studies of the mutant allele on different genetic backgrounds are needed to clarify the possible effect of different mouse strains on the phenotypic expression of Rps19+/−. Further studies of Rps19+/− mice may also include stress induction of the hematopoietic system. The repopulation of blood cells in Rps19+/− mice can then be compared to that in Rps19+/+ mice. An understanding of normal Rps19 regulation at the transcriptional and translational levels is also required in order to clarify possible mechanisms compensating for the loss of one Rps19 allele in mice.
Acknowledgments
We thank Erik Larsson for advice and pathological examinations of mouse organs. We acknowledge the Transgenic Mouse Facility at the Uppsala Biomedical Centre for the ES cell injections.
This work was supported by grants from the Children's Cancer Foundation of Sweden, the Swedish Medical Research Council, the Swedish Cancer Society, the DBA Foundation Inc., the Ronald McDonald Fund, the Torsten and Ragnar Söderberg Fund, and Uppsala University.
REFERENCES
- 1.Alter, B. P. 1980. Childhood red cell aplasia. Am. J. Pediatr. Hematol. Oncol. 2:121-139. [Google Scholar]
- 2.Ball, S. E., C. P. McGuckin, G. Jenkins, and E. C. Gordon-Smith. 1996. Diamond-Blackfan anaemia in the U.K.: analysis of 80 cases from a 20-year birth cohort. Br. J. Haematol. 94:645-653. [DOI] [PubMed] [Google Scholar]
- 3.Cramton, S. E., and F. A. Laski. 1994. string of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics 137:1039-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond, L. K., W. C. Wang, and B. P. Alter. 1976. Congenital hypoplastic anemia. Adv. Pediatr. 22:349-378. [PubMed] [Google Scholar]
- 5.Draptchinskaia, N., P. Gustavsson, B. Andersson, M. Pettersson, T. N. Willig, I. Dianzani, S. Ball, G. Tchernia, J. Klar, H. Matsson, D. Tentler, N. Mohandas, B. Carlsson, and N. Dahl. 1999. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21:169-175. [DOI] [PubMed] [Google Scholar]
- 6.Fisher, E. M., P. Beer-Romero, L. G. Brown, A. Ridley, J. A. McNeil, J. B. Lawrence, H. F. Willard, F. R. Bieber, and D. C. Page. 1990. Homologous ribosomal protein genes on the human X and Y chromosomes: escape from X inactivation and possible implications for Turner syndrome. Cell 63:1205-1218. [DOI] [PubMed] [Google Scholar]
- 7.Gazda, H., J. M. Lipton, T. N. Willig, S. Ball, C. M. Niemeyer, G. Tchernia, N. Mohandas, M. J. Daly, A. Ploszynska, K. A. Orfali, A. Vlachos, B. E. Glader, R. Rokicka-Milewska, A. Ohara, D. Baker, D. Pospisilova, A. Webber, D. H. Viskochil, D. G. Nathan, A. H. Beggs, and C. A. Sieff. 2001. Evidence for linkage of familial Diamond-Blackfan anemia to chromosome 8p23.3-p22 and for non-19q non-8p disease. Blood 97:2145-2150. [DOI] [PubMed] [Google Scholar]
- 8.Geerkens, C., W. Just, K. R. Held, and W. Vogel. 1996. Ullrich-Turner syndrome is not caused by haploinsufficiency of RPS4X. Hum. Genet. 97:39-44. [DOI] [PubMed] [Google Scholar]
- 9.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 10.Halperin, D. S., and M. H. Freedman. 1989. Diamond-Blackfan anemia: etiology, pathophysiology, and treatment. Am. J. Pediatr. Hematol. Oncol. 11:380-394. [PubMed] [Google Scholar]
- 11.Hart, K., T. Klein, and M. Wilcox. 1993. A Minute encoding a ribosomal protein enhances wing morphogenesis mutants. Mech. Dev. 43:101-110. [DOI] [PubMed] [Google Scholar]
- 12.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 13.Horino, K., H. Nishiura, T. Ohsako, Y. Shibuya, T. Hiraoka, N. Kitamura, and T. Yamamoto. 1998. A monocyte chemotactic factor, S19 ribosomal protein dimer, in phagocytic clearance of apoptotic cells. Lab. Investig. 78:603-617. [PubMed] [Google Scholar]
- 14.Matsson, H., J. Klar, N. Draptchinskaia, P. Gustavsson, B. Carlsson, D. Bowers, E. de Bont, and N. Dahl. 1999. Truncating ribosomal protein S19 mutations and variable clinical expression in Diamond-Blackfan anemia. Hum. Genet. 105:496-500. [DOI] [PubMed] [Google Scholar]
- 15.Naora, H., I. Takai, and M. Adachi. 1998. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J. Cell Biol. 141:741-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibuya, Y., M. Shiokawa, H. Nishiura, T. Nishimura, N. Nishino, H. Okabe, K. Takagi, and T. Yamamoto. 2001. Identification of receptor-binding sites of monocyte chemotactic S19 ribosomal protein dimer. Am. J. Pathol. 159:2293-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soulet, F., T. Al Saati, S. Roga, F. Amalric, and G. Bouche. 2001. Fibroblast growth factor-2 interacts with free ribosomal protein S19. Biochem. Biophys. Res. Commun. 289:591-596. [DOI] [PubMed] [Google Scholar]
- 18.Torok, I., D. Herrmann-Horle, I. Kiss, G. Tick, G. Speer, R. Schmitt, and B. M. Mechler. 1999. Down-regulation of RpS21, a putative translation initiation factor interacting with P40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol. Cell. Biol. 19:2308-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson, K. L., K. D. Konrad, D. F. Woods, and P. J. Bryant. 1992. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. USA 89:11302-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weil, D., M. F. Portnoi, J. Levilliers, I. Wang, M. Mathieu, J. L. Taillemite, M. Meier, B. Boudailliez, and C. Petit. 1993. A 45,X male with an X;Y translocation: implications for the mapping of the genes responsible for Turner syndrome and X-linked chondrodysplasia punctata. Hum. Mol. Genet. 2:1853-1856. [DOI] [PubMed] [Google Scholar]
- 21.Willig, T. N., N. Draptchinskaia, I. Dianzani, S. Ball, C. Niemeyer, U. Ramenghi, K. Orfali, P. Gustavsson, E. Garelli, A. Brusco, C. Tiemann, J. L. Perignon, C. Bouchier, L. Cicchiello, N. Dahl, N. Mohandas, and G. Tchernia. 1999. Mutations in ribosomal protein S19 gene and Diamond Blackfan anemia: wide variations in phenotypic expression. Blood 94:4294-4306. [PubMed] [Google Scholar]
- 22.Wool, I. G. 1996. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 21:164-165. [PubMed] [Google Scholar]
- 23.Wool, I. G. 1996. Extraribosomal functions of ribosomal proteins, p. 153-178. In R. Green and R. Schroeder (ed.), Ribosomal RNA and group I introns. R. G. Landes Co., New York, N.Y.
- 24.Wool, I. G., Y. Endo, Y. L. Chan, and A. Glück. 1990. Structure, function, and evolution of mammalian ribosomes, p. 203-214. In W. E. Hill, A. Dahlberg, R. A. Garrett, P. B. Moore, D. Schlessinger, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 25.Zinn, A. R., R. K. Alagappan, L. G. Brown, I. Wool, and D. C. Page. 1994. Structure and function of ribosomal protein S4 genes on the human and mouse sex chromosomes. Mol. Cell. Biol. 14:2485-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]