Abstract
MAP3K1 is a member of the mitogen-activated protein kinase kinase kinase (MAP3K) family of serine/threonine kinases. MAP3K1 regulates JNK activation and is unique among human kinases in that it also encodes an E3 ligase domain that ubiquitylates c-Jun and ERK1/2. Full length MAP3K1 regulates cell migration and contributes to pro-survival signaling while its caspase 3-mediated cleavage generates a C-terminal kinase domain that promotes apoptosis. The critical function of MAP3K1 in cell fate decisions suggests that it may be a target for deregulation in cancer. Recent large-scale genomic studies have revealed that MAP3K1 copy number loss and somatic missense or nonsense mutations are observed in a significant number of different cancers, being most prominent in luminal breast cancer. The alteration of MAP3K1 in diverse cancer types demonstrates the importance of defining phenotypes for possible therapeutic targeting of tumor cell vulnerabilities created when MAP3K1 function is lost or gained.
Keywords: MAP3K, MEKK, protein kinase, apoptosis
Mitogen-activated protein kinases (MAPKs) are key mediators of evolutionarily conserved signaling networks that play an essential role in multiple aspects of cell physiology.1,2 Activated by diverse stimuli, these signaling networks involve 3 sequential phosphorylation steps from MAPK kinase kinases (MAP3Ks, MEK kinases, or MKKKs) to MAPK kinases (MAP2Ks, MEK, or MKKs) to MAPKs. MAPKs are categorized into 4 subfamilies, extracellular signal response kinase 1/2 (ERK1/2), extracellular signal response kinase 5 (ERK5), c-Jun NH2-terminal kinase (JNK) or stress-activated protein kinase (SAPK), and p38. Twenty MAP3Ks, 7 MAP2Ks, and 11 MAPKs constitute an integrated signaling network responding to diverse stimuli to control critical functions in virtually all cells.3
MAP3K1 or MEKK1 (MEK kinase 1) is a 196-kDa serine-threonine kinase that belongs to the MAP3K family and the STE superfamily.2,4 MAP3K1 was originally identified as the mammalian homolog of the yeast MAP3Ks Ste11 and Byr2 that function in pheromone responsive signaling.5 In addition to the conserved kinase domain, MAP3K1 has several unique structural characteristics that mediate its specific activities compared with other MAP3Ks. Studies have demonstrated that MAP3K1 functions in cell survival, apoptosis, and cell motility/migration in multiple normal and tumor cell types. Genetic alterations in the MAP3K1 gene have been recently found in comprehensive genomic analyses of cancers. As discussed herein, the function of MAP3K1 in apoptosis suggests why it is deleted or mutated in specific cancers.
MAP3K1 Is a Serine/Threonine Protein Kinase, E3 Ubiquitin Ligase, Scaffolding Protein, and Capase-3 Substrate
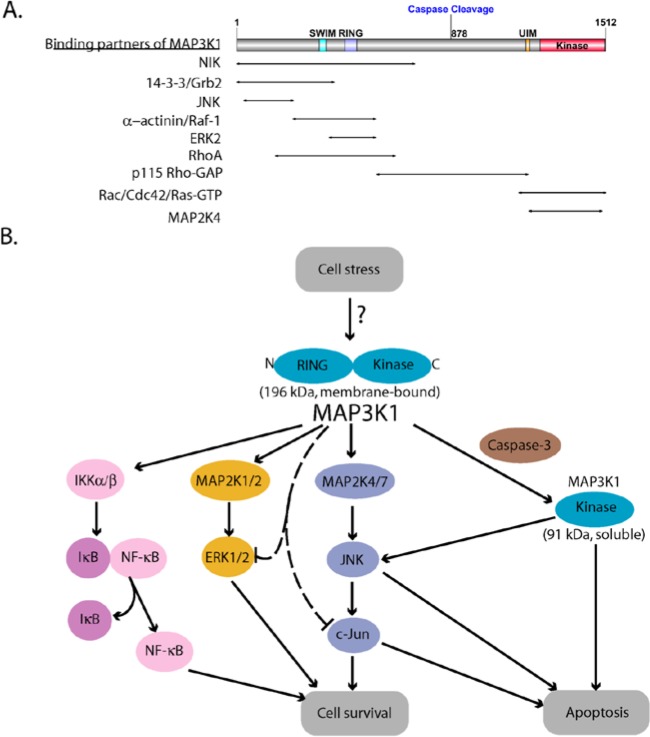
The kinase domain of MAP3K1 is located at the C-terminus (residues 1243-1508) (Fig. 1A). MAP3K1 selectively phosphorylates and activates MAP2K4, which in turn phosphorylates and activates JNK.6,7 MAP3K1 can also phosphorylate MAP2K1/2 and MAP2K7 that phosphorylate and activate ERK1/2 and JNK, respectively.5,8 Following its oligomerization, MAP3K1 can also trans-autophosphorylate, which presumably regulates its activation.9,10 Located upstream of the kinase domain are a conserved caspase-3 cleavage site (875DTLDG879 in the human homolog) and an Ubiquitin Interacting Motif (UIM).11 Although the role of the UIM is unclear, caspase-3 cleavage separating the C-terminal kinase domain from the regulatory N-terminus is required for apoptosis induction by MAP3K1. Unique among MAP3Ks, the N-terminus of MAP3K1 has 2 zinc-finger domains, a SWIM domain (residues 338-366), and a RING domain (residues 443-492), which are characterized by specific spacing of the conserved metal chelating cysteine and histidine residues.12,13 SWIM domains are ancient domains found in several functionally unrelated proteins such as the bacterial SWI2/SNF2 ATPase and the plant MuDR transposases, among others.13 Residues 443 to 492 of MAP3K1 show the typical pattern of the plant homeodomain (PHD) and its closely related RING domain. Like many RING-domain-containing proteins, MAP3K1 exhibits E3 ubiquitin ligase activity. MAP3K1 catalyzes the polyubiqitylation of c-Jun and ERK1/2, resulting in their degradation by proteasomes.12,14 The SWIM domain directly binds c-Jun and is required for MAP3K1-mediated c-Jun ubiquitylation.15 MAP3K1 can also ubiquitylate itself without causing its degradation, and this auto-ubiquitylation leads to the inhibition of downstream signaling.16 MAP3K1 ubiquitylation requires both functional RING and kinase domains.12
Figure 1.
MAP3K1 domain organization and dual roles in cell survival and apoptosis. (A) Schematic structure of MAP3K1. It contains a SWIM and a RING zinc finger domain near the N-terminus and a serine/threonine kinase domain at the C-terminus. Caspase-3 cleavage occurs at the aspartate 874 of the mouse MAP3K1, which is equivalent to residue 878 of the human homolog. The protein domain structure was created using DOG 1.0 program.17 MAP3K1 also harbors binding sites for multiple upstream and downstream proteins indicated by the arrows. (B) MAP3K1 has a switch-like function that determines cell fate. Activation of MAP2K4/7-JNK-c-Jun, MAP2K1/2-ERK1/2, and NF-κB mediated by full length MAP3K1 promotes cell survival while caspase cleavage, which generates the soluble active kinase domain, induces apoptosis. MAP3K1 also ubiquitylates c-Jun and ERK1/2, leading to their degradation.
As an additional facet of dynamic signaling control, MAP3K1 has a scaffold function, capable of binding multiple signaling proteins. MAP3K1 binds MAP2K4 and JNK via sequences within its kinase domain and N-terminus.18,19 GTP-bound Ras, Raf-1, MAP2K1, and ERK2 were also purified in complexes with MAP3K1.20,21 MAP3K1 binds GTPases of the Rho superfamily controlling the actin cytoskeleton and cell migration. Rac and Cdc42 bind the C-terminal kinase domain of MAP3K1,22 while RhoA and Rho-GAP binding sites are N-terminal of the kinase domain.23 RhoA binding requires the GTP bound state of the GTPase and increases MAP3K1 kinase activity.
MAP3K1 integrates signals from multiple upstream stimuli by interacting with several different kinases, including c-Abl,24 NIK,25 GCK,9 HPK1,26 RIP,27 PKCβ,28 and GSK3β.29 MAP3K1 can also bind several adaptor proteins such as TRAF2,30 Grb2,31 and Axin.32 Interaction of MAP3K1 with different proteins controls the subcellular localization of MAP3K1 signaling complexes. For example, FAK and α-actinin tether MAP3K1 to actin stress fibers entering focal adhesions.33,34 Caspase cleavage of MAP3K1 releases the kinase domain into the cytosol.35 Thus, the specific signal output emanating from MAP3K1 depends on its specific posttranslational modifications, cellular binding partners, and subcellular localization.
MAP3K1 Has Both Anti- and Pro-Apoptotic Functions
MAP3K1 is activated by a variety of stimuli such as growth factors, proinflammatory cytokines, microtubule disruption, cell shape disturbance, cold temperature, mild hyperosmolarity, and other cell stresses.36,37 Activation of full length MAP3K1 stimulates both MAP2K4/7-JNK and MAP2K1/2-ERK1/2 pathways.4,5,38 MAP3K1 has also been reported to activate the p38 pathway possibly due to the ability of MAP2K4 to phosphorylate p38.4,18,39 However, the effect of MAP3K1 on ERK and p38 is modest compared with its ability to activate JNK. MAP3K1 gene knockouts defined the essential role of MAP3K1 for JNK activation in response to several of the stress stimuli listed above.36,40 In response to proinflammatory cytokines and specific stresses, MAP3K1 may phosphorylate and activate IκB kinase α and β (IKKα/β), leading to NF-κB activation.41,42 ERK and NF-κB pathways promote cell survival while JNK, acting through the AP-1 transcription factors (particularly c-Jun), has both pro- and anti-apoptotic effects depending on the cell type and stimulus.4 MAP3K1 deletion further substantiated its pro-survival function, as demonstrated by increased cell death in response to mild hyperosmotic stress, cold temperature, and microtubule disruption in mouse embryonic stem cells and to oxidative stress in stem cell–derived cardiac myocytes.37,43
Confirming its critical role in balancing cell fate decisions, MAP3K1 has been shown to be important in mediating apoptosis in response to multiple cell stresses that include genotoxins,11 anoikis,44 growth factor withdrawal,45 and Fas ligand.46 Following an apoptotic signal, MAP3K1 is cleaved by activated caspase-3 to generate a 91-kDa C-terminal fragment that contains the kinase domain.11 The soluble C-terminal fragment of MAP3K1 is cytosolic, and the kinase activity induces apoptosis. This catalytic fragment exhibits selectivity toward JNK with little activation of ERK1/2.5,35,38,46 The strong activation of JNK promotes stabilization of p53 and phosphorylation of Bcl family proteins.35,46-51 MAP3K1 cleavage also suppresses activation of the pro-survival NF-κB pathway.46 Ectopic expression of MAP3K1 often leads to apoptosis because of its susceptibility to cleavage, leading to the generation of the constitutively active 91-kDa fragment.45,52 Mutation of the caspase cleavage site, expression of kinase-inactive MAP3K1, or tethering the kinase domain to membranes strongly suppresses MAP3K1-induced apoptosis.35,53 Loss of caspase-3 cleavage of MAP3K1 confers resistance to the chemotherapeutic agent cisplastin.54 Functionality of the PHD/RING domain was reported to be involved in MAP3K1-mediated JNK activation and apoptosis in response to cytoskeleton-disrupting drugs, but the mechanism is unclear.55 The cumulative properties determined for MAP3K1 suggests it has an unusual molecular switch that changes its function in regulating cell fate controlled by caspase 3 and subcellular localization (Fig. 1B).
Role of MAP3K1 in Regulation of Cell Migration
In addition to promoting survival or apoptotic decisions, MAP3K1 regulates the motility and migration of various cell types. Newborn MAP3K1 knockout mice fail to close their eyelids because of a defect in epithelial sheet migration.40 Embryonic fibroblasts and stem cells lacking MAP3K1 have impaired motility while overexpression of MAP3K1 induces lamellipodia-like structures.36,40 MAP3K1 regulation of cell motility is, in part, mediated by the protease calpain localized at focal adhesions.34 ERK1/2 activation by MAP3K1 at the site of actin fibers associating with focal adhesions leads to calpain activation. Calpain catalyzes the cleavage of several focal adhesion proteins involved in the control of rear-end detachment during migration. MAP3K1 also regulates the expression of urokinase-type plasminogen activator (uPA) by modulating the expression, activity, and stability of AP-1 transcription factors.56,57 uPA catalyzes the proteolysis of plasminogen to produce active plasmin, leading to extracellular matrix degradation. Loss of MAP3K1 expression reduces cell migration and invasion and delays tumor metastasis.57,58
Additional Functions of MAP3K1
MAP3K1 has been implicated in Wnt signaling. Wnt stimulation of cells was shown to induce association of MAP3K1 and Axin, a negative regulator of Wnt signaling.59 Knockdown of MAP3K1 impairs the expression of Wnt target genes. Interestingly, the integrity of the PHD/RING domain, but not the kinase domain, is essential for MAP3K1 regulation of Wnt signaling. The association of Axin and MAP3K1 also activates JNK via MAP3K1-MAP2K4 when Axin is overexpressed.32
Crosstalk between MAP3K1 and hormone receptor signaling has also been reported. MAP3K1 can activate the transcriptional activity of estrogen receptor (ER) in endometrial and ovarian cancer cells60 as well as androgen receptor in prostate cancer cells,61 possibly through JNK and p38 pathways. In endometrial and ovarian cancer, overexpression of kinase active MAP3K1 shifts 4-hydroxytamoxifen from an antagonist to an agonist of ER signaling, indicating that the agonistic activity of this endocrine therapeutic compound can be modulated by MAP3K1.60 In prostate cancer cells, apoptosis induced by overexpression of the constitutively active MAP3K1 is dependent on the presence of androgen receptor.61 Another transcription factor, Stat3, which is the effector of interferon and cytokine signaling, is phosphorylated and activated by MAP3K1 following EGF stimulation, and MAP3K1 activation of Stat3 requires the known upstream activators of Stat3, Jak, and Src.62
Genetic investigation of familial and sporadic cases of the 46,XY disorder of sex determination revealed that the disease is associated with mutations of MAP3K1 that increase ERK and, to a lesser extent, p38 phosphorylation in lymphoblastoid cell lines isolated from patients.53 The mutations occur in the Grb2, FAK, and RhoA binding regions, causing an enhancement of RhoA binding, which potentially up-regulates MAP3K1 activity. This observation emphasizes a role of MAP3K1 in human development of sexual organs and sex determination.
MAP3K1 and the Cancer Genome
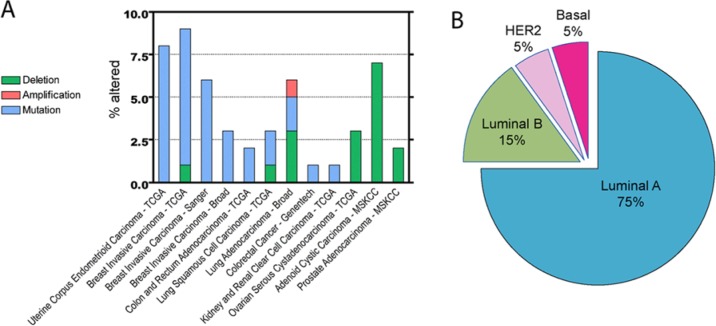
Gene expression, whole exome sequencing, and copy number variation analyses of substantial numbers of multiple human tumor types hold the promise of deciphering the genomic heterogeneity of cancer. While deregulation of the MAPK signaling architecture has long been appreciated as a frequent event in tumorigenesis, recent efforts to systematically categorize the human cancer genome have illuminated the potential contribution of specific gene alterations. In a study aimed at identifying novel somatic mutations, Kan et al. utilized >400 tumor samples from breast, lung, ovarian, pancreatic, and prostate cancers.63 MAP3K1 copy number loss and somatic missense or nonsense mutations were observed in a significant fraction of the tumor data set. MAP3K1 mutations were more frequently observed in hormone receptor positive (HR+) breast cancer as compared with other tumor types and additional breast cancer subtypes, including HER2+ and triple-negative. Subsequent to this study, increasing numbers of cancer genome analyses from a multitude of tumor types have enriched our appreciation of alterations that occur at lower frequencies but may have significant impact on tumor characteristics and patient outcomes. This is certainly true for the deletion or mutation of MAP3K1 found at a low to modest frequency in many tumor types (Fig. 2).
Figure 2.
Alterations in MAP3K1 genes are observed at relatively low frequency in multiple cancers with prevalent occurrence in luminal A breast cancer. (A) Histogram illustrating the percentage of MAP3K1 alterations for each cancer study: deletion (green), amplification (red), mutation (blue). (B) Alterations of MAP3K1 in the TCGA Breast Invasive Carcinoma data set are predominant in the luminal A, followed by luminal B, breast cancer subtypes. The data were obtained and analyzed by cBioPortal (http://www.cbioportal.org/public-portal/).64,65
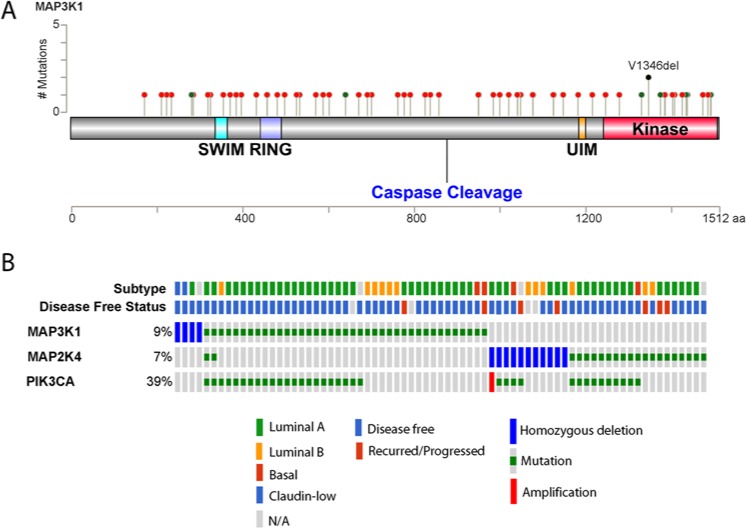
Assessment of MAP3K1 alteration frequency across multiple data sets using the cBio Cancer Genomics Portal64,65 further substantiates the notion that MAP3K1 loss-of-function contributes to specific cancer phenotypes (Fig. 2A). Ovarian, prostate, and adenoid cystic carcinoma exhibit copy number loss of MAP3K1, while uterine and breast carcinomas predominantly harbor somatic mutations. The extensive genomic analyses of breast cancer samples have been augmented by the classification of distinct subtypes (luminal A, luminal B, HER2, and basal) based on their mRNA expression patterns. Estrogen receptor (ER) positive breast cancer includes the luminal A and luminal B subtypes, differentiated generally by higher grade and poorer prognosis in luminal B as compared to luminal A. Amplification of the HER2/ERBB2 gene typifies the HER2 subtype, while the basal subtype (also termed triple-negative breast cancer, TNBC) lacks expression of ER, progesterone receptor, and HER2. Somatic mutations in the Cancer Genome Atlas study of breast invasive carcinoma in the context of mRNA expression subtypes revealed that MAP3K1 alterations were enriched in the luminal A subtype (Fig. 2B).66 Strikingly, the occurrence of mutations in MAP3K1 among luminal A samples was second only to mutations in PIK3CA. While the observation of copy number loss may suggest a putative tumor suppressor function of the gene product, the interpretation of somatic mutation data requires more rigorous analysis. Visualization of the mutations in MAP3K1 from the TCGA breast cancer data set reveals the absence of any hotspots or domain-specific alterations (Fig. 3A). Nonsense mutations were observed (19% of observed mutations) but the majority of mutations result in a frame-shift deletion or insertion (59%) and are predicted to be inactivating. Only 1 specific somatic mutation (an in-frame deletion) occurred in 2 unique samples and 4 of the 6 missense mutations observed occurred in the kinase domain of MAP3K1. Down-regulation, loss of function mutations, and homozygous deletion of the MAP3K1 downstream target MAP2K4 have also been reported in clinical samples from several types of cancer such as prostate, pancreatic, and ovarian cancer.50,66-69 Comparable portraits of MAP3K1 and MAP2K4 alterations were observed in separate large-scale breast cancer data sets.67,70
Figure 3.
Characterization of MAP3K1 alterations in breast cancer. (A) Somatic mutations found in MAP3K1 gene by the TCGA Breast Invasive Carcinoma study are predicted to be loss-of-function. The mutations are represented by circles: nonsense, splicing mutations, frameshift deletion, and insertion (red), missense mutations (green), and inframe deletion (black). (B) MAP3K1 alterations in breast cancer are mutually exclusive with those of MAP2K4 and partially overlap with those of PIK3CA. Shown is the oncoprint of MAP3K1, MAP2K4, and PIK3CA genes, in which individual samples are represented as columns and individual genes are represented as rows. Subtype assignment and disease-free status of the patients are shown in the first 2 rows. The data were obtained and visualized by cBioPortal database using the TCGA Breast Invasive Carcinoma data set with modifications.
Functional Consequences of MAP3K1 Perturbation in Cancer
The significance of specific genetic alterations in cancer genome data sets can be bolstered by the concurrent analysis of additional components that constitute a given pathway or functional complex. Alterations of 2 genes that affect the same pathway are typically not seen in a single individual. For example, samples with MAP3K1 alterations in the TCGA breast invasive carcinoma data set are notably distinct from those with alterations in MAP2K4 (Fig. 3B). Given the interplay of MAP3K1 and MAP2K4 in cell fate decisions (Fig. 1B), it can be hypothesized that the loss of function of either gene product is sufficient to deregulate the pathway in predominantly luminal A breast cancer (Fig. 3B). In contrast, activating mutations in PIK3CA are observed in approximately 50% of samples harboring MAP3K1 or MAP2K4 alterations. While it is known that PI3K-AKT signaling negatively regulates MAP3K1-JNK, the interplay between these 2 pathways in luminal A breast cancer deserves more study.
The majority of breast cancers are luminal, and the luminal A subtype is characterized by better prognosis and lower rate of recurrence than the other subtypes (Fig. 3B).67 The reconciliation of MAP3K1 alterations in cancer, and the luminal A breast cancer subtype specifically, with known MAP3K1 function remains an open question. Mammary gland involution is a complex process of coordinated apoptosis and tissue remodeling that occurs postlactationally and, in a related but distinct process termed lobular involution, occurs with age.71,72 Delay or failure to undergo both types of involution has been identified as a breast cancer risk factor.71 One hypothesized explanation for this correlation is that involution removes the pool of epithelial cells that may represent cancer progenitors. Given the strong association between caspase activation and mammary gland involution, it is reasonable to hypothesize that MAP3K1 might play an important role in the process of involution. Loss of MAP3K1 function causes insensitivity to cell death, potentially promoting the emergence of cancer cells. Moreover, with roles in the degradation of focal adhesion and extracellular matrix as discussed above, MAP3K1 might be essential for the detachment of epithelial cells from alveolar cells, causing the cells to die during anoikis.72 MAP3K1 is also capable of activating NF-κB, Stat3, and calpains, all of which have been reported to induce the lysosomal pathway of cell death during involution.72 Future studies dissecting the role of involution as a determinant of breast cancer subtype and outcome with regard to MAP3K1 functioning will be enlightening.
Perturbation of MAP3K1 signaling affects multiple pathways—IKK-NFκB, ERK1/2, and JNK. Thus, the impact of MAP3K1 mutation and deletion on cancer cell signaling networks demand further investigation. For example, TCGA cases of uterine corpus endometroid carcinoma with observed MAP3K1 alterations trend toward better overall survival (P = 0.071) when compared with unaltered MAP3K1 cases.73 Concomitantly with defective apoptosis, MAP3K1 impairment can result in defects in pro-survival signaling and cell migration, which have the potential to reduce tumor growth and metastasis. Whether this is related to the better prognosis of luminal A breast cancer and uterine corpus endometroid carcinoma with MAP3K1 mutations needs evaluation. Contrary to this hypothesis, MAP3K1 and MAP2K4 were identified as metastasis suppressors in ovarian, prostate, and gastric cancer through both clinical studies and experimental models.50 Therefore, it will be of interest to determine if other cancer types with functional loss of MAP3K1 are similar to the luminal A breast cancer subtype in molecular phenotype and clinical outcome. In a similar manner, elucidation of the specific contribution of MAP3K1 signaling (and possibly MAP2K4) to the luminal A mRNA signature may provide insight into the subtype-specific phenotype.
Recently, an in vivo mutagenesis screen in mice identified MAP3K1 as a driver in melanomagenesis. Ni et al. utilized a piggyBac transposon strategy that identified multiple activating insertions in introns 9 and 10 of the MAP3K1 gene.74 These insertions were shown to result in the production of truncated MAP3K1 products lacking the N-terminal SWIM and RING domains (Fig. 1A). On ectopic expression, these aberrant forms of MAP3K1 activated ERK1/2 that was sustainable even in the presence of a BRAF inhibitor and promoted proliferation in the absence of growth factors in melanocytes. Thus, constitutively active MAP3K1 functioned as a driver of melanoma in their model. To support this observation, the TCGA melanoma data set available on cBioPortal database showed MAP3K1 amplification and mRNA up-regulation in approximately 5% of human melanomas.64,65 Taken together, these data suggest that MAP3K1 may function as a driver in some cancer types (e.g., melanoma) and is inactivated in other types (e.g., luminal A breast cancer). This dichotomy is consistent with MAP3K1 having a molecular switch-like function for cell survival or apoptosis controlled, in part, by caspase 3 and subcellular localization.
Future Directions/Concluding Remarks
At present, a signaling network displaying MAP3K1 as the central hub illustrates the complex and intricate nature of its function in vivo. Undoubtedly, cell type-specificity and even tissue environment are critical determinants of the cell fate decisions mediated by MAP3K1. Recently, the comprehensive nature of genomic efforts to categorize human cancer has provided further insight into the molecular contribution of heretofore unappreciated and understudied gene products. Certainly, MAP3K1 is an understudied kinase in relation to its function in cancers. Given the relative frequency of MAP3K1 mutations in many tumor types, ongoing investigation of MAP3K1 function will be important for providing answers to questions surrounding specific cancer phenotypes and their vulnerabilities when MAP3K1 is mutated or deleted.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by US Army Medical Research Grant W81XWH-12-1-0129.
References
- 1. Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807-69 [DOI] [PubMed] [Google Scholar]
- 2. Uhlik MT, Abell AN, Cuevas BD, Nakamura K, Johnson GL. Wiring diagrams of MAPK regulation by MEKK1, 2, and 3. Biochem Cell Biol. 2004;82:658-63 [DOI] [PubMed] [Google Scholar]
- 3. Cuevas BD, Abell AN, Johnson GL. Role of mitogen-activated protein kinase kinase kinases in signal integration. Oncogene. 2007;26:3159-71 [DOI] [PubMed] [Google Scholar]
- 4. Hagemann C, Blank JL. The ups and downs of MEK kinase interactions. Cell Signal. 2001;13:863-75 [DOI] [PubMed] [Google Scholar]
- 5. Lange-Carter CA, Pleiman CM, Gardner AM, Blumer KJ, Johnson GL. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science. 1993;260:315-9 [DOI] [PubMed] [Google Scholar]
- 6. Siow YL, Kalmar GB, Sanghera JS, Tai G, Oh SS, Pelech SL. Identification of two essential phosphorylated threonine residues in the catalytic domain of Mekk1. Indirect activation by Pak3 and protein kinase C. J Biol Chem. 1997;272:7586-94 [DOI] [PubMed] [Google Scholar]
- 7. Yan M, Dai T, Deak JC, et al. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798-800 [DOI] [PubMed] [Google Scholar]
- 8. Hirai S, Noda K, Moriguchi T, et al. Differential activation of two JNK activators, MKK7 and SEK1, by MKN28-derived nonreceptor serine/threonine kinase/mixed lineage kinase 2. J Biol Chem. 1998;273:7406-12 [DOI] [PubMed] [Google Scholar]
- 9. Chadee DN, Yuasa T, Kyriakis JM. Direct activation of mitogen-activated protein kinase kinase kinase MEKK1 by the Ste20p homologue GCK and the adapter protein TRAF2. Mol Cell Biol. 2002;22:737-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deak JC, Templeton DJ. Regulation of the activity of MEK kinase 1 (MEKK1) by autophosphorylation within the kinase activation domain. Biochem J. 1997;322(pt 1):185-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Widmann C, Gerwins P, Johnson NL, Jarpe MB, Johnson GL. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol Cell Biol. 1998;18:2416-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell. 2002;9:945-56 [DOI] [PubMed] [Google Scholar]
- 13. Makarova KS, Aravind L, Koonin EV. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem Sci. 2002;27:384-6 [DOI] [PubMed] [Google Scholar]
- 14. Xia Y, Wang J, Xu S, Johnson GL, Hunter T, Lu Z. MEKK1 mediates the ubiquitination and degradation of c-Jun in response to osmotic stress. Mol Cell Biol. 2007;27:510-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rieger MA, Duellman T, Hooper C, Ameka M, Bakowska JC, Cuevas BD. The MEKK1 SWIM domain is a novel substrate receptor for c-Jun ubiquitylation. Biochem J. 2012;445:431-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Witowsky JA, Johnson GL. Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways. J Biol Chem. 2003;278:1403-6 [DOI] [PubMed] [Google Scholar]
- 17. Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009;19:271-3 [DOI] [PubMed] [Google Scholar]
- 18. Xia Y, Wu Z, Su B, Murray B, Karin M. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 1998;12:3369-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu S, Cobb MH. MEKK1 binds directly to the c-Jun N-terminal kinases/stress-activated protein kinases. J Biol Chem. 1997;272:32056-60 [DOI] [PubMed] [Google Scholar]
- 20. Karandikar M, Xu S, Cobb MH. MEKK1 binds raf-1 and the ERK2 cascade components. J Biol Chem. 2000;275:40120-7 [DOI] [PubMed] [Google Scholar]
- 21. Russell M, Lange-Carter CA, Johnson GL. Direct interaction between Ras and the kinase domain of mitogen-activated protein kinase kinase kinase (MEKK1). J Biol Chem. 1995;270:11757-60 [DOI] [PubMed] [Google Scholar]
- 22. Fanger GR, Johnson NL, Johnson GL. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 1997;16:4961-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gallagher ED, Gutowski S, Sternweis PC, Cobb MH. RhoA binds to the amino terminus of MEKK1 and regulates its kinase activity. J Biol Chem. 2004;279:1872-7 [DOI] [PubMed] [Google Scholar]
- 24. Kharbanda S, Pandey P, Yamauchi T, et al. Activation of MEK kinase 1 by the c-Abl protein tyrosine kinase in response to DNA damage. Mol Cell Biol. 2000;20:4979-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su YC, Han J, Xu S, Cobb M, Skolnik EY. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu MC, Qiu WR, Wang X, Meyer CF, Tan TH. Human HPK1, a novel human hematopoietic progenitor kinase that activates the JNK/SAPK kinase cascade. Genes Dev. 1996;10:2251-64 [DOI] [PubMed] [Google Scholar]
- 27. Kim JW, Joe CO, Choi EJ. Role of receptor-interacting protein in tumor necrosis factor-alpha-dependent MEKK1 activation. J Biol Chem. 2001;276:27064-70 [DOI] [PubMed] [Google Scholar]
- 28. Kaneki M, Kharbanda S, Pandey P, et al. Functional role for protein kinase Cbeta as a regulator of stress-activated protein kinase activation and monocytic differentiation of myeloid leukemia cells. Mol Cell Biol. 1999;19:461-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim JW, Lee JE, Kim MJ, Cho EG, Cho SG, Choi EJ. Glycogen synthase kinase 3 beta is a natural activator of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 1 (MEKK1). J Biol Chem. 2003;278:13995-14001 [DOI] [PubMed] [Google Scholar]
- 30. Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pomerance M, Multon MC, Parker F, et al. Grb2 interaction with MEK-kinase 1 is involved in regulation of Jun-kinase activities in response to epidermal growth factor. J Biol Chem. 1998;273:24301-4 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y, Neo SY, Wang X, Han J, Lin SC. Axin forms a complex with MEKK1 and activates c-Jun NH(2)-terminal kinase/stress-activated protein kinase through domains distinct from Wnt signaling. J Biol Chem. 1999;274:35247-54 [DOI] [PubMed] [Google Scholar]
- 33. Christerson LB, Gallagher E, Vanderbilt CA, et al. p115 Rho GTPase activating protein interacts with MEKK1. J Cell Physiol. 2002;192:200-8 [DOI] [PubMed] [Google Scholar]
- 34. Cuevas BD, Abell AN, Witowsky JA, et al. MEKK1 regulates calpain-dependent proteolysis of focal adhesion proteins for rear-end detachment of migrating fibroblasts. EMBO J. 2003;22:3346-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schlesinger TK, Bonvin C, Jarpe MB, et al. Apoptosis stimulated by the 91-kDa caspase cleavage MEKK1 fragment requires translocation to soluble cellular compartments. J Biol Chem. 2002;277:10283-91 [DOI] [PubMed] [Google Scholar]
- 36. Xia Y, Makris C, Su B, et al. MEK kinase 1 is critically required for c-Jun N-terminal kinase activation by proinflammatory stimuli and growth factor-induced cell migration. Proc Natl Acad Sci U S A. 2000;97:5243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yujiri T, Sather S, Fanger GR, Johnson GL. Role of MEKK1 in cell survival and activation of JNK and ERK pathways defined by targeted gene disruption. Science. 1998;282:1911-4 [DOI] [PubMed] [Google Scholar]
- 38. Minden A, Lin A, McMahon M, et al. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719-23 [DOI] [PubMed] [Google Scholar]
- 39. Guan Z, Buckman SY, Pentland AP, Templeton DJ, Morrison AR. Induction of cyclooxygenase-2 by the activated MEKK1 → SEK1/MKK4 → p38 mitogen-activated protein kinase pathway. J Biol Chem. 1998;273:12901-8 [DOI] [PubMed] [Google Scholar]
- 40. Yujiri T, Ware M, Widmann C, et al. MEK kinase 1 gene disruption alters cell migration and c-Jun NH2-terminal kinase regulation but does not cause a measurable defect in NF-kappa B activation. Proc Natl Acad Sci U S A. 2000;97:7272-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bonvin C, Guillon A, van Bemmelen MX, Gerwins P, Johnson GL, Widmann C. Role of the amino-terminal domains of MEKKs in the activation of NF kappa B and MAPK pathways and in the regulation of cell proliferation and apoptosis. Cell Signal. 2002;14:123-31 [DOI] [PubMed] [Google Scholar]
- 42. Lee FS, Peters RT, Dang LC, Maniatis T. MEKK1 activates both IkappaB kinase alpha and IkappaB kinase beta. Proc Natl Acad Sci U S A. 1998;95:9319-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Minamino T, Yujiri T, Papst PJ, Chan ED, Johnson GL, Terada N. MEKK1 suppresses oxidative stress-induced apoptosis of embryonic stem cell-derived cardiac myocytes. Proc Natl Acad Sci U S A. 1999;96:15127-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cardone MH, Salvesen GS, Widmann C, Johnson G, Frisch SM. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell. 1997;90:315-23 [DOI] [PubMed] [Google Scholar]
- 45. Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326-31 [DOI] [PubMed] [Google Scholar]
- 46. Deak JC, Cross JV, Lewis M, et al. Fas-induced proteolytic activation and intracellular redistribution of the stress-signaling kinase MEKK1. Proc Natl Acad Sci U S A. 1998;95:5595-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fuchs SY, Adler V, Pincus MR, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci U S A. 1998;95:10541-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gibson S, Widmann C, Johnson GL. Differential involvement of MEK kinase 1 (MEKK1) in the induction of apoptosis in response to microtubule-targeted drugs versus DNA damaging agents. J Biol Chem. 1999;274:10916-22 [DOI] [PubMed] [Google Scholar]
- 49. Mandic A, Viktorsson K, Molin M, et al. Cisplatin induces the proapoptotic conformation of Bak in a deltaMEKK1-dependent manner. Mol Cell Biol. 2001;21:3684-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Taylor JL, Szmulewitz RZ, Lotan T, et al. New paradigms for the function of JNKK1/MKK4 in controlling growth of disseminated cancer cells. Cancer Lett. 2008;272:12-22 [DOI] [PubMed] [Google Scholar]
- 51. Zebrowski DC, Alcendor RR, Kirshenbaum LA, Sadoshima J. Caspase-3 mediated cleavage of MEKK1 promotes p53 transcriptional activity. J Mol Cell Cardiol. 2006;40:605-18 [DOI] [PubMed] [Google Scholar]
- 52. Johnson NL, Gardner AM, Diener KM, et al. Signal transduction pathways regulated by mitogen-activated/extracellular response kinase kinase kinase induce cell death. J Biol Chem. 1996;271:3229-37 [DOI] [PubMed] [Google Scholar]
- 53. Warr N, Bogani D, Siggers P, et al. Minor abnormalities of testis development in mice lacking the gene encoding the MAPK signalling component, MAP3K1. PloS One. 2011;6:e19572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gebauer G, Mirakhur B, Nguyen Q, Shore SK, Simpkins H, Dhanasekaran N. Cisplatin-resistance involves the defective processing of MEKK1 in human ovarian adenocarcinoma 2008/C13 cells. Int J Oncol. 2000;16:321-5 [DOI] [PubMed] [Google Scholar]
- 55. Tricker E, Arvand A, Kwan R, Chen GY, Gallagher E, Cheng G. Apoptosis induced by cytoskeletal disruption requires distinct domains of MEKK1. PloS One. 2011;6:e17310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cuevas BD, Uhlik MT, Garrington TP, Johnson GL. MEKK1 regulates the AP-1 dimer repertoire via control of JunB transcription and Fra-2 protein stability. Oncogene. 2005;24:801-9 [DOI] [PubMed] [Google Scholar]
- 57. Cuevas BD, Winter-Vann AM, Johnson NL, Johnson GL. MEKK1 controls matrix degradation and tumor cell dissemination during metastasis of polyoma middle-T driven mammary cancer. Oncogene. 2006;25:4998-5010 [DOI] [PubMed] [Google Scholar]
- 58. Su F, Li H, Yan C, Jia B, Zhang Y, Chen X. Depleting MEKK1 expression inhibits the ability of invasion and migration of human pancreatic cancer cells. J Cancer Res Clin Oncol. 2009;135:1655-63 [DOI] [PubMed] [Google Scholar]
- 59. Sue Ng S, Mahmoudi T, Li VS, et al. MAP3K1 functionally interacts with Axin1 in the canonical Wnt signalling pathway. Biol Chem. 2010;391:171-80 [DOI] [PubMed] [Google Scholar]
- 60. Lee H, Jiang F, Wang Q, et al. MEKK1 activation of human estrogen receptor alpha and stimulation of the agonistic activity of 4-hydroxytamoxifen in endometrial and ovarian cancer cells. Mol Endocrinol. 2000;14:1882-96 [DOI] [PubMed] [Google Scholar]
- 61. Abreu-Martin MT, Chari A, Palladino AA, Craft NA, Sawyers CL. Mitogen-activated protein kinase kinase kinase 1 activates androgen receptor-dependent transcription and apoptosis in prostate cancer. Mol Cell Biol. 1999;19:5143-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lim CP, Cao X. Regulation of Stat3 activation by MEK kinase 1. J Biol Chem. 2001;276:21004-11 [DOI] [PubMed] [Google Scholar]
- 63. Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869-73 [DOI] [PubMed] [Google Scholar]
- 64. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Spillman MA, Lacy J, Murphy SK, et al. Regulation of the metastasis suppressor gene MKK4 in ovarian cancer. Gynecol Oncol. 2007;105:312-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yeasmin S, Nakayama K, Rahman MT, et al. Loss of MKK4 expression in ovarian cancer: a potential role for the epithelial to mesenchymal transition. Int J Cancer. 2011;128:94-104 [DOI] [PubMed] [Google Scholar]
- 70. Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Radisky DC, Hartmann LC. Mammary involution and breast cancer risk: transgenic models and clinical studies. J Mammary Gland Biol Neoplasia. 2009;14:181-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Watson CJ, Kreuzaler PA. Remodeling mechanisms of the mammary gland during involution. Int J Dev Biol. 2011;55:757-62 [DOI] [PubMed] [Google Scholar]
- 73. Cancer Genome Atlas Research Network; Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ni TK, Landrette SF, Bjornson RD, Bosenberg MW, Xu T. Low-copy piggyBac transposon mutagenesis in mice identifies genes driving melanoma. Proc Natl Acad Sci U S A. 2013;110:E3640-E3649 [DOI] [PMC free article] [PubMed] [Google Scholar]