Abstract
Flap endonuclease 1 (FEN1) is a central component of Okazaki fragment maturation in eukaryotes. Genetic analysis of Saccharomyces cerevisiae FEN1 (RAD27) also reveals its important role in preventing trinucleotide repeat (TNR) expansion. In humans such expansion is associated with neurodegenerative diseases. In vitro, FEN1 can inhibit TNR expansion by employing its endonuclease activity to compete with DNA ligase I. Here we employed two yeast FEN1 nuclease mutants, rad27-G67S and rad27-G240D, to further define the mechanism by which FEN1 prevents TNR expansion. Using a yeast artificial chromosome system that can detect both TNR instability and fragility, we demonstrate that the G240D but not the G67S mutation increases both the expansion and fragility of a CTG tract in vivo. In vitro, the G240D nuclease is proficient in cleaving a fixed nonrepeat double flap; however, it exhibits severely impaired cleavage of both nonrepeat and CTG-containing equilibrating flaps. In contrast, wild-type FEN1 and the G67S mutant exhibit more efficient cleavage on an equilibrating flap than on a fixed CTG flap. The degree of TNR expansion and the amount of chromosome fragility observed in the mutant strains correlate with the severity of defective flap cleavage in vitro. We present a model to explain how flap equilibration and the unique tracking mechanism of FEN1 can collaborate to remove TNR flaps and prevent repeat expansion.
Eukaryotic flap endonuclease 1 (FEN1) is critical for removing initiator RNA primers from Okazaki fragments during DNA lagging-strand synthesis (20, 50, 60). The enzyme belongs to a highly conserved 5′ endo-exonuclease superfamily (53). FEN1 is a structure-specific nuclease in that it specifically removes the 5′ unannealed flap in a branch structure resulting from strand displacement synthesis. This property allows the enzyme to actively participate in both Okazaki fragment processing and DNA long-patch base excision repair (12, 37) since both of these processes involve creation of a 5′ flap by strand displacement synthesis. The removal of initiator RNA primers in Okazaki fragments has been proposed as a two-step mechanism in which an endonuclease, Dna2 protein, removes a part of the flap (2). The remainder of the flap is then cut off by FEN1, leaving a nick to be ligated. Other studies suggest that FEN1 alone is sufficient for removal of most flaps (1). Studies in vivo demonstrate a critical role for FEN1 since the absence of enzyme activity in mice leads to embryonic lethality (30). Knocking out one copy of FEN1 along with one of the adenomatous polyposis coli genes in mice results in an increased number of adenocarcinomas, presumably due to the partial loss of FEN1 function in mammalian DNA replication and repair (30).
In Saccharomyces cerevisiae the FEN1 homologue, RAD27/RTH1 (47) demonstrates a high homology (79% conserved and 60% identical) with human FEN1 (53). A deletion of RAD27/RTH1 leads to conditional lethality phenotypes (54). The mutant cells exhibit temperature sensitivity and cell cycle arrest at G2 with phenotypes resembling those of Polδ and ligase mutants known to have a DNA replication defect (10). In addition, the rad27Δ mutant is highly sensitive to methyl methanesulfonate, a DNA-alkylating agent, but only moderately sensitive to UV and X-ray irradiation (47, 54), consistent with the importance of FEN1 in DNA base excision repair.
FEN1 is critical in maintaining genome stability. The rad27Δ mutant exhibits increased spontaneous mutation, chromosomal loss, and mitotic recombination (47, 54, 58, 61). Tishkoff et al. identified a unique mutator phenotype in the rad27Δ mutant distinct from that of mismatch repair gene mutations by specifically examining the spectrum of Lys+-reverting and Canr mutations (58). Most of the mutations (79%) have a duplicated region flanked by short direct repeats (58). Rad27 was proposed to inhibit recombination between short DNA sequences by endonucleolytically removing the 5′ overhang that mediates the process (44). Furthermore, mutation of Rad27 leads to the instability of repeated sequences including dinucleotide (27, 29, 65) and trinucleotide repeats (TNRs) (15, 52, 56, 62) as well as minisatellites (35, 36). Rad27 also plays a role in maintaining the stability of telomere repeat sequences (45, 46).
TNR expansion is now known to be the cause of at least 15 hereditary human neurodegenerative diseases including Huntington's disease, spinocerebellar ataxia, and myotonic dystrophy (40, 48). The role of FEN1 in maintaining the integrity of repeated sequences implies that the enzyme may participate in preventing these human diseases. The types of genome instability and duplication mutations observed in the absence of FEN1 formed a basis for Gordenin et al. to propose a model in which the inability of FEN1 to resolve a flap with a secondary structure allows sequence expansion (19). The model points out that formation of a bubble intermediate, resulting from misaligned annealing of an unresolved flap to the template, may be an essential step in sequence expansion.
The repeat expansion model has been supported by studies in vivo and in vitro. In the rad27Δ mutant, significantly enhanced triplet repeat expansion is observed (15, 52, 56, 62). Recent data show that mice heterozygous for FEN1 and an expanded CAG tract at the Huntington's disease locus also have increased repeat instability (55). In vitro studies have shown that triplet repeats can form fold-back and bubble structures in the 5′ flap of unprocessed Okazaki fragments that equilibrate with unstructured flaps (17, 56). These fold-back structures are inhibitory to both FEN1 endonucleolytic cleavage and ligation (21, 32, 56), thereby leading to an accumulation of unresolved flaps. Bubbles also inhibit FEN1 cleavage; however, they are favored by ligase and have been demonstrated as an essential step for sequence expansion (34). Although FEN1 cleavage is inhibited by secondary structures in vitro, the length of triplet repeats is maintained successfully in wild-type yeast strains. This suggests that under normal circumstances FEN1 can avoid inhibition in order to cleave CTG flaps. However, the mechanisms employed by the nuclease to protect against expansion remain unclear.
Unresolved flaps are a potential precursor of chromosome breakage. Consistent with this idea is the observation that TNRs are preferential sites of chromosome fragility in both human and yeast cells (see reference 33 for a review). Both CCG and CTG repeats have been shown to induce double-strand breaks in a tract length-specific manner by physical and genetic assays in yeast (3, 8, 15, 25). The level of CTG fragility increases dramatically in a rad27Δ background, suggesting that the absence of efficient flap processing is a source of TNR tract breaks (8). Furthermore, the increasing fragility observed with increasing CTG tract length suggests that a structure inhibitory to ligation is present in the rad27Δ background. The most likely candidate is a self-annealed CTG fold-back structure on the unprocessed flaps.
Previously, we characterized two yeast FEN1 mutants, rad27-G67S and rad27-G240D, that exhibit defective substrate cleavage and a dinucleotide repeat expansion mutator phenotype. Studies of human FEN1 mutants have shown that FEN1 employs endonuclease rather than exonuclease activity to compete with DNA ligase I (34). It is thought that FEN1 endonuclease efficiently removes a TNR flap before it can form an intermediate that leads to expansion upon ligation or chromosome breakage (34). However, it is unknown how this enzyme can employ its endonuclease activity to resolve a triplet repeat flap.
Here we have obtained evidence that flap equilibration plays an important role in the prevention of TNR expansion. Flap equilibration is defined as the ability of a flap to partially or completely reanneal to the template in a way that forms a variety of double-flap intermediates with an adjacent primer. In this study, we used the yeast FEN1 mutants rad27-G67S and rad27-G240D to examine the role of the endonuclease in TNR expansion both in vivo and in vitro. Unexpectedly, we found that the most important activity to prevent TNR expansion was not cleavage efficiency per se but, rather, the ability to capture a cleavable flap structure among equilibrating intermediates. Results suggest that the flap-tracking mechanism of the wild-type FEN1 nuclease interacts effectively with equilibrating TNR intermediates, leading to effective cleavage.
MATERIALS AND METHODS
Yeast strains and YACs.
The rad27-G67S, rad27-G240D, and corresponding rad27Δ and wild-type strains were derived from the isogenic FY strain background (63): FY23 (MATa ura3-52 leu2Δ trp1Δ63), EAY607 (MATα leu2Δ his3Δ200 ura3-52 lys2BglII, trp1Δ63 ade8 rad27-G67S), EAY611 (MATa leu2Δ ura3-52 lys2BglI trp1Δ63 rad27-G240D), and EAY615 (MATα leu2Δ ura3-52 lys2BglI trp1Δ63 his3Δ200 rad27Δ::HIS3) (65). Yeast artificial chromosomes (YACs) with various CTG tract lengths (8) were introduced into wild-type and rad27 mutant backgrounds by a kar cross (14).
CTG repeat stability and fragility assays.
Cells of each strain were plated for single colonies on yeast complete (YC)-Ura-Leu medium and grown at 30°C. The CTG tract length of individual colonies was checked by colony PCR. The CTG tract was amplified from colonies by using a small number of cells as the template, primers that flank the CTG tract (CTG rev2, CCCAGGCCTCCAGTTTGC; T7, TAATACGACTCACTATAGGG), and Clontech Advantage-GC-Genomic kits in 12.5 μl-reaction mixtures. Reactions were run on 2% Metaphor gels (Fisher Scientific) for 2 h at ∼75 V, and CTG tract size was estimated by comparison with DNA Marker VI (Roche). CTG repeat sizes are accurate to within ∼15 bp.
For the fragility assay, 10 starting colonies with correct tract lengths were used to inoculate 10 separate 1-ml cultures grown on YC-Leu medium. These cultures were grown to about three doublings at 30°C in YC-Leu medium to maintain selection for the YAC but to allow loss of the right arm of the YAC. A total of 100 μl of each culture was plated on fluoroorotic acid (FOA)-Leu medium to select for breakage events. At the same time, a portion of each culture was combined and then diluted to plate for single colonies on YC-Leu medium for a total cell count. Cells were grown into individual colonies at 30°C. The number of colonies was counted, and rates of FOA resistance were calculated by the method of the median (31). Significance was determined by a pooled variance t test by using Systat software. Each assay was repeated a minimum of three times.
To analyze FOA-resistant (FOAR) YAC structure, FOAR colonies were grown for 24 h in YC-Leu medium, and genomic DNA was purified by the glass bead method. Genomic DNA was digested with BstEII (New England Biolabs) at 65°C for 2 days and separated on a 1% agarose gel. The digested DNA was probed with a digoxigenin-labeled probe of HindIII-digested λ DNA and detected by a colorimetric system (Roche).
For the CTG repeat instability assay, 42 to 100 colonies on the total cell count plates were analyzed by colony PCR to detect changes in CTG tract length. The assay was repeated at least three times for each strain and each tract length; significance was determined by a Fisher's exact test.
Oligonucleotide substrates.
Two classes of oligonucleotides were designed to construct either fixed- or equilibrating-flap substrates. Generally, a substrate was constructed by annealing a downstream primer and an upstream primer to a template primer. A fixed-flap substrate is a double flap containing a downstream 5′ unannealed flap and an upstream 1-nucleotide (nt) 3′ tail. An equilibrating-flap substrate is constructed by annealing an upstream 3′ flap and a downstream 5′ flap that has identical sequences annealed to the same region of the template. This allows upstream- and downstream-flap equilibration. All the oligomer sequences are listed in Table 1.
TABLE 1.
Oligonucleotide sequences
| Primer | Size (mer) | Sequencea |
|---|---|---|
| CTG repeat substrates | ||
| Fixed | ||
| DFCTG10 | 54 | 5′-CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGTTCCAAGTAAAACGACGGCCAGTG-3′ |
| DFCTG20 | 78 | 5′-CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGTGCTGCTGCTGCTGCTGCTGCTGCTGCTGGTAAAACGACGGCCAGTG-3′ |
| TFCAG | 44 | 3′-GCGGTCCCAAAAGGGTCAGTGCTGGCATTTTGCTGCCGGTCACG-5′ |
| UFCTG | 26 | 5′-CGCCAGGGTTTTCCCAGTCACGACCG-3′ |
| Equilibrating | ||
| DECTG10 | 49 | 5′-CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGGTAAAACGACGGCCAGTG-3′ |
| ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪ | ||
| DECTG20 | 78 | 5′-CTGCTGCTGCTGCTGCTGCTGCTGCTGCTGTGCTGCTGCTGCTGCTGCTGCTGCTGCTGGTAAAACGACGGCCAGTG-3′ |
| ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪ | ||
| TECAG10 | 65 | 3′-GCGGGTCAGTGCTGGGACGACGACGACGACGACGACGACGACGACCATTTTGCTGCCGGTCACG-5′ |
| TECAG20 | 95 | 3′-GCGGGTCAGTGCTGGGACGACGACGACGACGACGACGACGACGACGACGACGACGACGACGACGACGACGAGACCCATTTTGCTGCCGGTCACG-5′ |
| UECTG5 | 30 | 5′-CGCCCAGTCACGACCCTGCTGCTGCTGCTG-3′ |
| ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪ | ||
| UECTG10 | 45 | 5′-CGCCCAGTCACGACCCTGCTGCTGCTGCTGCTGCTGCTGCTGCTG-3′ |
| ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪ | ||
| UECTG20 | 75 | 5′-CGCCCAGTCACGACCCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTGCTG-3′ |
| ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪ | ||
| Unique flap substrates | ||
| Fixed | ||
| DF15 | 40 | 5′-AGGTCTCGACTACCACCCGTCCACCCGACGCCACCTCCTG-3′ |
| DF30 | 55 | 5′-AGGTCTCGACTAACTCTAGTCGTTGTTCCACCCGTCCACCCGACGCCACCTCCTG-3′ |
| TF | 51 | 3′-GCTGGCACGGTCGGATTTAAAGTTAGGGCAGGTGGGCTGCGGTGGAGGACG-5′ |
| UF | 26 | 5′-CGACCGTGCCAGCCTAAATTTCAATC-3′ |
| Equilibrating | ||
| DE15 | 58 | 5′-TAGGTCTCGACGACTAACTCTAGTCGTTGTTCCACCCGTCCACCCGACGCCACCTCCTG-3′ |
| ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪ | ||
| TE | 85 | 3′-GCTGGCACGGTCGGATTTAAAGTTATCCAGAGCTGCTGATTGAGATCAGCAACAAGGTGGGCAGGTGGGCTGCGGTGGAGGACG-5′ |
| UE15 | 39 | 5′-CGACCGTGCCAGCCTAAATTTCAATAGGTCTCGACGACT-3′ |
| ▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪▪ |
Annealed regions of the primers are underlined. Unannealed residues are in boldface. The upstream and downstream annealed regions are represented as thin and thick lines, whereas the dotted lines in the equilibrating substrates indicate the annealed regions that allow equilibration to occur between the 5′ end of a downstream primer and the 3′ end of an upstream primer.
Substrates were radiolabeled at the 3′ end of their downstream primers. Details about radiolabeling and purification of downstream primers were described by Xie et al. (65). Briefly, a downstream primer was initially annealed to a template resulting in a 5′ template overhang. Subsequently, the 3′ end of the downstream primer was extended with [α-32P]dCTP (New England Nuclear) by Klenow fragment (Roche Molecular Biochemicals) at 37°C for 2 h. The radiolabeled primer was subjected to urea denaturing gel purification after the removal of unincorporated radionucleotides by a spin column (Bio-Rad).
Substrates were constructed by annealing a 3′ radiolabled downstream primer (D), a template (T), and an upstream primer (U) at a molar ratio of 1:5:20, respectively. At this ratio, annealing of a downstream primer, DFCTG10 or DFCTG20, and the upstream primer UFCTG to template TFCAG results in a fixed flap containing 10 and 20 CTG repeats (CTG-10 and CTG-20), respectively. This further created a double-flap substrate having a 5′ unannealed CTG-10 or CTG-20 flap and an upstream 1-nt 3′ tail. An equilibrating CTG substrate was generated by annealing a downstream primer, DECTG10 or DECTG20 (DECTG10/20), to template TECAG10 or TECAG20 (TECAG10/20) along with annealing an upstream primer, UECTG5 or UECTG10 or UECTG20 (UECTG5/10/20), at the DECTG10/20/TECAG10/20/UECTG5/10/20 ratio of 1:5:20. As the downstream 5′ CTG repeats and the upstream 3′ CTG repeats can compete to anneal to CAG repeats on the template, this allows a CTG-5, CTG-10, and CTG-20 flap equilibration between the downstream and the upstream primer. The nonrepeat fixed-flap substrates were constructed by annealing a downstream primer, DF15 or DF30 (15 or 30 nt), and upstream primer UF to template TF. This allowed the creation of a 15-nt fixed-double-flap substrate. Annealing of a downstream primer, DE15, and an upstream primer, UE15, to a template, TE, resulted in equilibrating-flap structures having unique DNA sequences. All the oligoprimers were synthesized by Integrated DNA Technologies (Coralville, Iowa).
FEN1 protein expression and purification.
S. cerevisiae FEN1 (Rad27p), Rad27-G67Sp, and Rad27-G240Dp proteins were expressed in Escherichia coli strain BL21(DE3) codon plus (Stratagene, Inc.) by utilizing a T7 expression vector, pET24b. This expression vector allows a six-His tag to be attached to the C terminus of the expressed proteins. Protein expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Gibco, Inc., Gaithersburg, Md.), and the expressed protein was subjected to purification through successive chromatographic columns including nickel-agarose, carboxymethyl-Sepharose, Mono-S, hydroxyapatite, and phenyl-Sepharose. The procedure was described in detail by Xie et al. (65) and Kao et al. (28).
Enzymatic assay.
Wild-type and mutant FEN1 proteins were incubated with various flap substrates at the amounts indicated in the figure legends in a final volume of 20 μl of reaction buffer (30 mM HEPES, 0.5% inositol, 0 to 40 mM KCl, 4 mM Mg2+, 0.01% NP-40, 0.1 mg of bovine serum albumin/ml, and 1 mM dithiothreitol; pH 8.0) as described by Liu and Bambara (34). The buffer used for the experiments where both FEN1 and DNA ligase I were present also contained 1 mM ATP (34). The products were resolved in a urea denaturing polyacrylamide gel and detected by a PhosphorImager (Molecular Dynamics). Quantitative analysis of the products was performed by using ImageQuant version 1.2 software from Molecular Dynamics. The percentage of products was calculated as described previously (34).
Enzyme kinetics.
The kinetic study of S. cerevisiae FEN1 (Rad27p), G67Sp, and G240Dp was performed at 30°C under the conditions described in “Enzymatic assay” above. The constant amounts of enzymes (1 fmol) and various amounts (5, 10, 20, and 40 fmol) of a 6-nt double-flap substrate were utilized as described previously (34). The kinetic experiments were initiated by combining the reaction buffer, substrates, and enzymes sequentially. Aliquots (20 μl) were removed from the reaction mixture at various time points as indicated previously (34). The initial velocity was determined by measuring the intensities of substrate and product. The velocity of the reaction was calculated by using an approach demonstrated previously (59). The values of Km and Vmax were obtained by fitting the data to a Michaelis-Menten equation.
RESULTS
Mutation of FEN1 endonuclease leads to CTG tract expansions in vivo.
In order to determine how the S. cerevisiae FEN1 protein affects CTG tract instability in a chromosomal context in vivo, we utilized a YAC with various lengths of a CTG repeat tract (8). We tested the effect of three different RAD27 mutations (rad27-G67S, rad27-G240D, and rad27Δ) on CTG repeat stability by replacing the wild-type copy of RAD27 with mutant versions of the gene. Both point mutations have a severe exonuclease defect, but they differ in the amount of residual endonuclease cleavage activity. The rad27-G67S mutant has a moderate endonuclease defect, whereas the rad27-G240D mutant exhibits much less endonuclease cleavage activity in vitro (65). Both mutant strains are slightly sensitive to the DNA-damaging agent methyl methanesulfonate; however, in the absence of the drug, they grow as well as the wild-type strain (65).
Individual yeast colonies of either wild-type or mutant strains containing a CTG tract YAC were grown, and colony PCR was used to identify starting colonies that contained a full-length CTG tract. The CTG repeat lengths chosen were 85 (CTG-85) and 155 CTG (CTG-155). These repeat sizes are well over the threshold of instability at TNR disease loci. Ten starting colonies were used to inoculate 10 liquid cultures that were grown for three doublings to allow expansion or contraction of the repeat tract to occur. The use of multiple cultures maximized the chance that the expansions analyzed arose as independent events. The cultures were mixed, a dilution was plated for single cells that were grown into daughter colonies, and CTG tract length was analyzed by colony PCR.
In the wild-type strain, the frequency of CTG tract expansion was very low (0.5 to 1.4%) (Table 2). Contractions were markedly higher than expansions and increased with increasing tract length (8.8% for CTG-85; 23% for CTG-155) (Table 2). This result is consistent with the known bias toward contractions in yeast cells (16, 38, 39). As has been seen previously, repeat sequence instability was dramatically increased in the rad27Δ strain with a strong bias towards expansion (Table 2) (8, 15, 52, 56).
TABLE 2.
Frequency of CTG/CAG tract expansions and contractions in wild-type and rad27 mutant backgrounds
| Strain background | Total no. of coloniesa
|
% Expansion (fold)b
|
% Contraction (fold)b
|
% No change (fold)b
|
||||
|---|---|---|---|---|---|---|---|---|
| CTG-85 | CTG-155 | CTG-85 | CTG-155 | CTG-85 | CTG-155 | CTG-85 | CTG-155 | |
| Wild type | 192 | 144 | 0.5 (1.0) | 1.4 (1.0) | 8.8 (1.0) | 23 (1.0) | 90 (1.0) | 76 (1.0) |
| rad27-G67S | 144 | 144 | 0 | 3.5 (2.5) | 13 (1.5) | 30 (1.3) | 87 (0.97) | 67 (0.9) |
| rad27-G240D | 144 | 144 | 12 (23)** | 20 (15)** | 18 (2.0)* | 27 (1.2) | 70 (0.8) | 53 (0.7) |
| rad27Δ | 138 | 148 | 40 (78)** | 16 (12)** | 20 (2.3)** | 50 (2.2)** | 40 (0.4) | 34 (0.4) |
Number of colonies of each strain tested by colony PCR is shown.
Frequencies are expressed as percentages; increase over wild type is indicated in parentheses. Statistical significance was determined by a Fisher's exact test. *, P < 0.05; **, P < 0.01.
The rad27-G240D strain showed a significant increase in expansions. The expansion frequency increased with increasing tract length from 12% for CTG-85 to 20% for CTG-155, whereas in the rad27Δ strain the expansion frequency decreased from 41% for CTG-85 to 16% for CTG-155 (Table 2). Thus, the expansion frequency of CTG-155 was even higher in the rad27-G240D strain than in the absence of the FEN1 protein. In contrast, neither expansions nor contractions in the rad27-G67S strain were significantly different from those in the wild type (Table 2). Since the rad27-G67S mutation causes a severe defect in exonuclease activity (65), these results indicate that FEN1 exonuclease activity has little effect on CTG tract instability in vivo.
We also analyzed the mean and the range of the tract length changes seen in wild-type cells and the RAD27 mutant backgrounds (Table 3). For rad27-G67S, repeat length changes were similar to those of the wild type. However, in both the rad27-G240D and rad27Δ strains, we saw some large expansions of the CTG-85 tract that double, or almost double, the size of the tract and are therefore unlikely to be simply due to religation of an unprocessed flap during Okazaki fragment maturation. The frequency of these large expansions differed in the two strains: 2 out of 17 (12%) expanded CTG-85 tracks in the rad27-G240D strain versus 19 out of 56 (34%) expanded CTG-85 tracts in the rad27Δ strain. These results suggest that the mechanism that generates very large expansions is less common in the nuclease-defective rad27-G240D strain than in the absence of the Rad27 protein.
TABLE 3.
Size of CTG/CAG tract length changes observed in stability assaysa
| Strain | CTG-85
|
CTG-155
|
||||||
|---|---|---|---|---|---|---|---|---|
| Expansions
|
Contractions
|
Expansions
|
Contractions
|
|||||
| Range | Mean | Range | Mean | Range | Mean | Range | Mean | |
| Wild type | 15 | 15 | 5-60 | 35 | 5-10 | 5 | 35-130 | 75 |
| rad27-G67S | 0 | 0 | 5-60 | 10 | 5-10 | 10 | 10-155 | 70 |
| rad27-G240D | 5-75 | 35 | 5-65 | 30 | 5-55 | 15 | 30-130 | 75 |
| rad27Δ | 5-80 | 45 | 5-60 | 25 | 5-15 | 10 | 35-130 | 85 |
Changes are expressed as the numbers of CTG/CAG repeats added (for expansions) or subtracted (for contractions) from the original tract length. Sizing is accurate to five repeats (15 bp).
Mutation of FEN1 endonuclease increases CTG repeat fragility.
CTG repeats have been shown to cause chromosomal breakage in yeast in a tract-specific manner by both physical analysis and a genetic assay based on recombination (15). We have also developed a novel genetic assay to monitor chromosomal breakage at CTG tracts based on the aforementioned YACs containing repeat tracts of various lengths (8). This assay has the advantage that it does not depend on recombination. Thus, mutations that increase recombination, as RAD27 mutations do, should not affect the assay.
To study CTG repeat fragility, various repeat lengths were cloned on the right arm of YAC-CF1 (Fig. 1). The URA3 gene on YAC-CF1 is distal to the CTG repeats but is far enough away from the telomere that it is not affected by telomeric silencing (51). Proximal to the repeats there is a 108-bp stretch of C4A4/T4G4 sequence, the backup telomere, which can be recognized by yeast telomerase as a substrate for new telomere addition. Cells containing the original YAC are Ura3+ and 5-FOA sensitive (FOAS) (Fig. 1). If breakage occurs at the CTG tract, the broken end of the tract will be degraded, exposing the backup telomere and allowing telomere addition and, thus, recovery of the broken chromosome. These cells will have lost the URA3 gene and will thus be FOAR (Fig. 1). The rate of generation of FOAR cells can be considered a monitor of the rate of breakage (8).
FIG. 1.
The YAC breakage assay. Cells containing YAC CF1, which contains a CAG/CTG tract and URA3 gene, are FOAS. When breakage occurs inside the CTG tract (heavy black line), the broken YAC can be rescued by the addition of a new telomere to the backup telomere (C4A4 sequence). The resulting cells will be Leu2+ and FOAR. The repeat tract is oriented such that the CAG sequence is on the lagging-strand template and the CTG sequence is on the Okazaki fragments. YAC CF1 is ∼62 kb long and contains 41 kb of lambda sequences between the LEU2 gene and the C4A4 sequence indicated by hatch marks. Drawing is not to scale.
In order to test the effect of the FEN1 functional defects on CTG fragility, we performed the YAC breakage assay in strains containing a YAC with 0 (CTG-0), 85, or 155 CTG repeats. As expected, the wild-type strains displayed an increasing rate of generation of FOAR cells with increasing tract length (Table 4). The rad27Δ strains showed a significant increase in the rate of FOAR cell generation over wild type (3.4-fold increase compared to wild type for CTG-85 and 8-fold compared to wild type for CTG-155) (Table 4), and the rate of FOAR cell generation also increased with increasing tract length. The increased FOAR cell generation rate for the CTG-0 YAC in the rad27Δ strain indicates that the absence of yeast FEN1 increases overall chromosomal breakage. However, subtraction of the rad27Δ CTG-0 value from the CTG-85 and CTG-155 values shows that the presence of a triplet repeat tract significantly exacerbates chromosomal breakage.
TABLE 4.
Fragility of YAC-CF1, containing 0, 85, or 155 CTG/CAG repeats in wild-type and rad27 mutant backgrounds
| Strain background and repeat | Rate of FOAR cell generation (10−6) ± SDa | Increase (n-fold) over wild type |
|---|---|---|
| CTG-0 | ||
| Wild type | 1.7 ± 0.3 | 1.0 |
| rad27-G67S | 0.86 ± 0.1 | 0.5 |
| rad27-G240D | 2.4 ± 0.7 | 1.4 |
| rad27Δ | 3.5 ± 1.1 | 2.0 |
| CTG-85 | ||
| Wild type | 8.2 ± 1.8 | 1.0 |
| rad27-G67S | 3.1 ± 0.7* | 0.4 |
| rad27-G240D | 18 ± 2.9* | 2.2 |
| rad27Δ | 28 ± 6.5* | 3.4 |
| CTG-155 | ||
| Wild type | 14 ± 2.6 | 1.0 |
| rad27-G67S | 9.4 ± 1.0 | 0.7 |
| rad27-G240D | 20 ± 1.6 | 1.4 |
| rad27Δ | 112 ± 30* | 8.0 |
Statistical significance was determined by a pooled variance t test. SD, standard deviation; *, P < 0.05.
The rad27-G240D mutation also increased the rate of FOAR cell generation compared to wild type in a tract-specific manner (Table 4). The rate of FOAR cell generation for CTG-85 approached the rate of FOAR cell generation for the rad27Δ strain. These results indicate that the rad27-G240D mutation causes a significant increase of double-strand breaks in a CTG-85 repeat tract and suggest the presence of unresolved flaps. Interestingly, unlike the rad27Δ strain, the rate of breakage rose only slightly as the tract length increased to CTG-155 repeats, suggesting some differences in the fate of unprocessed flaps in the two strains. Surprisingly, we saw a decrease in the rate of FOAR cell generation in the rad27-G67S strain for all tract lengths, with the decrease being statistically significant for the CTG-85 tract (P = 0.03; Table 4).
To verify that the rates of FOAR cells reflect that of CTG breakage in the RAD27 mutant backgrounds, DNA was purified from at least six FOAR colonies for each tract length for each mutant, and the structure of the YAC was examined by Southern blotting (data not shown). The majority of the time (average, 74%), YACs were healed at the backup telomere, indicating that the assay worked as expected (data not shown). The rad27-G67S and rad27-G240D mutations did not alter the spectrum of healing compared to wild type, and no cases of CTG telomere healing were observed (data not shown).
Mutant enzymes are mainly defective in catalysis.
To understand why the G67S and G240D mutations can lead to triplet repeat expansion and fragility and how the mechanism of FEN1 relates to these biological processes, we performed biochemical analyses with the mutant proteins. Specifically, we analyzed the biochemical kinetics and cleavage specificity of intermediates that could potentially allow sequence expansion. By correlating the in vivo phenotypes of the mutants with the in vitro biochemical properties of the mutant proteins, we expected to establish how FEN1 contributes to the stability of triplet repeats.
A previous biochemical characterization of yeast FEN1 mutants Rad27-G67S and Rad27-G240D has shown that the mutant enzymes are defective in endo- and exonucleolytic cleavage (65). These defects may be a main cause of dinucleotide repeat expansion (65) and TNR expansion (the present study). To further analyze the nature of the mutations, we began with basic biochemical kinetic parameters. Table 5 lists the results of kinetic analyses of wild-type and mutant proteins on a 6-nt double-flap substrate with a 1-nt 3′ tail as previously described (34). As shown in Table 5, both mutant enzymes have a significantly reduced Vmax compared with the wild-type enzyme. The G67S and G240D mutations decreased Vmax of the enzyme by about 5- and 10-fold, respectively. However, they did not alter the Km value significantly, although G67S increased the Km value by about 1.8-fold. This suggests that the mutations mainly create a defect in catalysis rather than substrate binding ability. These results are consistent with the fact that the mutant proteins did not show substrate binding defects (65).
TABLE 5.
Enzymatic kinetics of FEN1 on a double-flap substrate
| FEN1 protein | Km (nM) | Vmax (10−4 nM/s) |
|---|---|---|
| Wild-type Rad27p | 4.8 ± 0.8 | 27.0 ± 4.7 |
| Rad27-G67Sp | 8.6 ± 1.9 | 5.5 ± 1.0 |
| Rad27-G240Dp | 4.5 ± 0.2 | 2.3 ± 0.1 |
A FEN1 endonucleolytic defect allows triplet repeat expansion in vitro.
Unresolved triplet repeat flaps should accumulate as a result of the FEN1 endonucleolytic defects. To examine whether this accumulation leads to sequence expansion, we utilized a substrate system in vitro that simulates dynamic CTG repeat flaps in vivo. In this system, both the 5′ end of a downstream primer and the 3′ end of an upstream primer contain a CTG repeat sequence that competes for base pairing to the same complementary CAG repeat region of the template (21, 34). This results in an equilibration (referred to here as an equilibrating flap) between up- and downstream CTG flaps. By varying the length of a CTG repeat tail at the 3′ end of the upstream primer, CTG flap overlaps ranging from 5 to 20 repeats were created. We examined the accumulation of expanded products when G67Sp- and G240Dp-mutant FEN1 nucleases utilized a CTG-5 equilibrating-flap substrate (Fig. 2). FEN1 converted 80% of the substrate to either a nonexpanded ligation product or cleavage products. Only about 10% of substrate molecules were ligated into expanded products at 5 fmol of FEN1 (Fig. 2, lane 6). Endonucleolytic cleavage of a double flap with a 1-nt 3′ tail is the precursor for nonexpanded ligation, whereas exonucleolytic cleavage products result from progressive cleavage by FEN1 after flap removal. Expanded products result from direct ligation of bubble intermediates formed by misaligned base pairing. The G67S protein allowed about 20% expanded product and less than 5% of nonexpanded product unless the enzyme level was raised to 50 fmol (lanes 10 to 14). At this level, G67Sp produced 10% expanded and 70% nonexpanded product (lane 14). The G240D protein allowed 25% expanded product, but little nonexpanded product was made (lanes 16 to 20) even when a large amount of enzyme (50 fmol) was used.
FIG. 2.
A FEN1 functional defect allows triplet repeat expansion in vitro. Wild-type and mutant FEN1 were incubated with a CTG-5 repeat equilibrating substrate radiolabeled at the 3′ end of the downstream primer. The 5′ end of the downstream primer was also phosphorylated. A 5-fmol substrate containing primers DECTG10, UECTG5, and TECAG10 was incubated with increasing amounts of FEN1 (0.5, 1, 5, 10, and 50 fmol) and 5 fmol of DNA ligase I in the reaction buffer containing 1 mM ATP as described in Materials and Methods. The reaction was performed at 30°C for 10 min in a total volume of 20 μl. The substrate and products were subsequently separated by electrophoresis on a 15% urea denaturing polyacrylamide gel. The reaction mixtures containing FEN1, G67Sp, or G240Dp are shown in lanes 4 to 8, lanes 8 to 14, and lanes 16 to 20, respectively. The reaction mixture in lane 1 contains only substrate. The reaction mixture in lane 2 contains DNA ligase I and the substrate. Lanes 3, 9, and 15 represent the reaction mixture containing either FEN1 or G67Sp or G240Dp. A schematic diagram is shown above the figure. The light gray line in the template represents CAG repeats. The gray textured lines on the up- and downstream primers represent CTG repeats. The product representing triplet repeat expansion is shown as longer (expanded) than its template. Nonexpanded product is shown as having the same length as its template. The bands occurring in between the nonexpanded product and the substrate are deletion products that are described by Liu and Bambara (34).
With an increase of wild-type nuclease, the expanded product was reduced, indicating that the competition between FEN1 and DNA ligase I determines the occurrence of triplet repeat expansion. In contrast, even increased amounts of mutant proteins, particularly G240Dp, failed to deplete the expanded products significantly. Since the proportion of nonexpanded product is an indicator of the efficiency of FEN1 endonucleolytic cleavage, these results suggest that the increase in CTG expansion is a direct result of an endonucleolytic cleavage defect.
G67Sp and G240Dp are defective in cleaving equilibrating flap substrates.
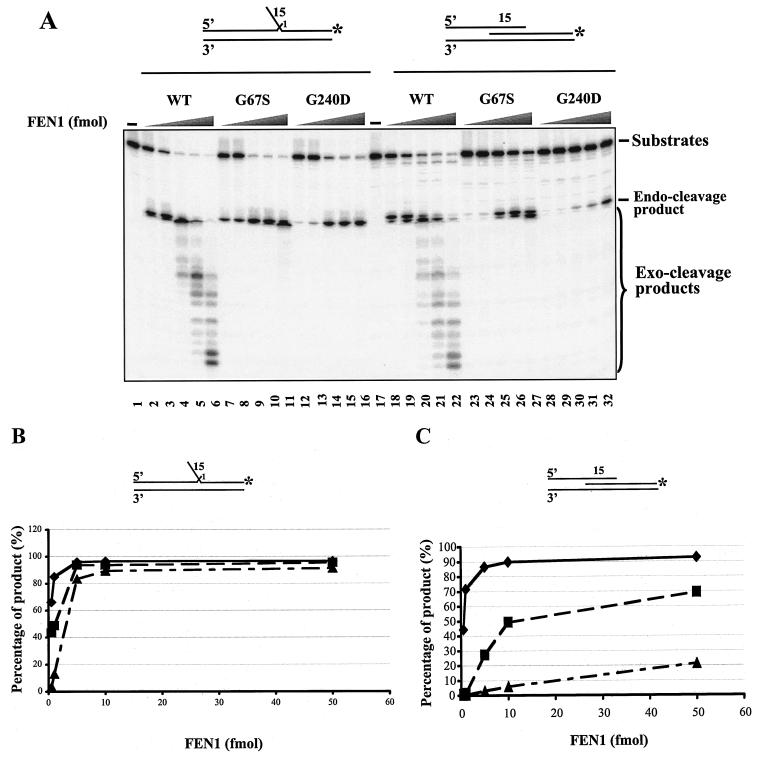
We next assessed whether the mutant nucleases are more defective in cleaving equilibrating flaps than fixed flaps. The equilibrating substrate was constructed by annealing two primers with an overlapping region to a template. The 5′ end of the downstream primer had 15 nt of identical sequences with the 3′ end of the upstream primer, allowing for competitive annealing (Fig. 3A, top right). The corresponding fixed-flap substrate had a 15-nt 5′ flap, not complementary to the template, and a 1-nt 3′ tail (Fig. 3A, top left). Note that the 3′ tails on our fixed substrates are complementary to the template but compete for template binding with a complementary nucleotide on the downstream primer. The mutant proteins (Fig. 3A) did not show a significant difference from wild type in cleaving the fixed-double-flap substrate, except for their lack of exonuclease activity. The G67Sp protein had almost the same enzymatic activity as wild type on the fixed double flap at all enzyme levels tested (Fig. 3A, compare lanes 2 to 6 with lanes 7 to 11). Lowering the level (0.5 fmol and 1 fmol) of G240Dp significantly reduced cleavage on the 5-fmol fixed-double-flap substrate (Fig. 3A, lanes 12 and 13). However, when the enzyme level of G240Dp was raised to 5 fmol, such that the enzyme-substrate ratio reached 1:1, substrate cleavage efficiency was similar to the wild-type level (Fig. 3).
FIG. 3.
Cleavage by wild-type and mutant FEN1 on nonrepeat fixed- and equilibrating-flap substrates. (A) Annealing of primers DF15 and UF to primer TF created a 15-nt fixed-double-flap substrate with a 1-nt 3′ tail, whereas a 15-nt equilibrating-flap substrate was constructed by annealing primers DE15 and UE to TE. Increasing amounts of wild-type and mutant FEN1 (0.5, 1, 5, 10, and 50 fmol) were incubated with either a 15-nt fixed-flap substrate or a 15-nt equilibrating-flap substrate at 30°C for 10 min. Reactions were then subjected to electrophoresis on a 12% denaturing polyacrylamide gel to separate the substrates and the products. Reaction mixtures containing wild-type enzyme and fixed-flap substrate or equilibrating-flap substrate are represented in lanes 2 to 6 and lanes 18 to 22, respectively. The cleavage by G67Sp on fixed- and equilibrating-flap substrates is shown in lanes 7 to 11 (fixed) and lanes 23 to 27 (equilibrating), whereas the cleavage by G240Dp on these substrates is shown in lanes 12 to 16 (fixed) and lanes 28 to 32 (equilibrating). Lanes 1 and 17 contain only substrates. (B) Quantitative analysis of the cleavage of wild-type and mutant proteins on the unique 15-nt fixed flap. (C) The quantitation of cleavage activity of wild type and mutants on a unique 15-nt equilibrating flap substrate. The reactions were performed in the presence of 0.5, 1, 5, 10 and 50 fmol of enzymes, and the total volume of each reaction mixture was 20 μl. A 5-fmol substrate was utilized in each reaction. The substrates are illustrated above the figures. The substrates in panels A and B were radiolabeled at the 3′ end of the downstream primers. An asterisk denotes the position of the radiolabeled nucleotide.
However, both mutant proteins demonstrated significantly reduced activities on the equilibrating-flap substrate, with G240Dp reductions being the most severe (Fig. 3A, lanes 28 to 32, and C). The G67Sp mutant had a cleavage activity between that of the wild type and G240Dp (Fig. 3A and C) on the same substrate. Increasing the amount of G240Dp to 500 fmol did not significantly improve its endonucleolytic cleavage activity (data not shown). We further examined the enzymatic activity of mutants on a 30-nt equilibrating-flap substrate. The results were similar to those with the 15-nt equilibrating-flap substrate (data not shown). Overall, the yeast FEN1 mutant proteins are defective in cleaving a nonrepeat equilibrating flap but effectively cleave a nonrepeat fixed flap.
FEN1 mutants also exhibit a defect in cleaving CTG repeat equilibrating flaps.
To understand how defects in equilibrating-flap cleavage can impair the ability of FEN1 to prevent triplet repeat expansion, we further examined the cleavage by wild-type and mutant enzymes on the CTG-containing equilibrating-flap substrate described in Fig. 2. The quantitative analysis of FEN1 cleavage on the substrate indicated that G67S nuclease generated much less endonucleolytic cleavage product than the wild type from the CTG-5 equilibrating flap (Fig. 4). The G240D protein was even more severely defective in cleaving the CTG-5 substrate (Fig. 4). The mutant nucleases exhibit proficient cleavage on the CTG-5 fixed flap although with a lower efficiency than their cleavage on the nonrepeat 15-nt fixed flap (data not shown). These results indicate that the CTG repeats inhibit endonucleolytic cleavage by the mutant nucleases, particularly on equilibrating CTG flaps. Furthermore, they suggest that the mutants, in particular G240Dp, may have lost their ability to capture the best substrate for cleavage among equilibration intermediates. Alternatively, G240Dp may be defective in either allowing or facilitating the formation of an optimal fixed-flap intermediate for cleavage.
FIG. 4.
Cleavage of wild-type and mutant FEN1 on CTG equilibrating-flap substrates. CTG-5 equilibrating substrate (5 fmol) was incubated with increasing amounts of wild-type and mutant FEN1 (0.5, 1, 5, 10, and 50 fmol) at 30°C for 10 min. The gray textured lines indicate CTG repeats. The light gray line represents 10 CAG repeats within the template. The quantitative data resulting from enzyme cleavage on the equilibrating CTG-5 flap substrate are shown in the graph. The data were processed by the software program ImageQuant, version 1.2. The percentages of endo- and exonucleolytic cleavage products were plotted against the amounts of wild-type and mutant enzymes. ♦, cleavage by wild-type enzyme; ▪, G67Sp cleavage; ▴, G240Dp cleavage.
Flap equilibration facilitates FEN1 resolution of triplet repeats.
Our genetic data have shown that the G67S mutation effectively prevents sequence expansion from CTG-85, only allowing a slight increase of CTG expansion from the CTG-155 tract. The G240D mutation, on the other hand, allows dramatic sequence expansion from both CTG tracts. This finding indicates that in vivo the G240Dp mutant protein is not able to employ a mechanism to bypass an inhibitory effect resulting from CTG repeats.
To explore the possibility of a bypass mechanism, we further examined cleavage by the mutant nucleases on fixed and equilibrating CTG-10 flaps. The CTG fixed-flap substrate contained a CTG-10 flap and a 1-nt 3′ tail. For the equilibrating CTG-10 flap substrate, the downstream CTG-10 competed with upstream CTG-10 repeats to anneal to 10 CAG repeats of the template. Mutant proteins had significantly reduced cleavage on both fixed and equilibrating CTG-10 substrates. The fixed CTG-10 repeat flap was a very poor substrate for both mutant proteins (Fig. 5A and B). Only less than 5 and 1% of the products resulted from endonucleolytic cleavage by the G67Sp and the G240Dp proteins, respectively, throughout the titration ranging from 0.5 fmol to 50 fmol. Cleavage was most likely inhibited by the long self-complementarity of the fixed flap. We note that with nonrepeat flap substrates, cleavage of a fixed flap by G67Sp was much more efficient than the cleavage on an equilibrating flap (Fig. 3). In contrast, G67Sp cleaved the equilibrating CTG-10 flap more efficiently than the fixed CTG flap (Fig. 5A, compare lanes 9 to 11 with lanes 25 to 27; B and C). At 10- and 50-fmol enzyme levels, the percentage of product reached about 10 and 35%, respectively (Fig. 5C). Cleavage by G240Dp on the equilibrating substrate was also improved since 50 fmol of enzyme generated about 8% of the endonucleolytic cleavage products, whereas less than 1% of the products were made with the CTG-10 fixed-flap substrate (Fig. 5B C). Wild-type FEN1 did not exhibit a significant difference in cleaving the fixed and equilibrating CTG-10 substrates (Fig. 5A, compare lanes 2 and 3 with lanes 18 and 19; B and C).
FIG. 5.
Flap equilibration facilitates resolution of triplet repeats by mutant FEN1 proteins. (A) A fixed CTG-10 repeat flap substrate was made by annealing primers DFCTG10 and UFCTG to template TFCaG. Annealing of primers DECTG10 and UECTG10 to template primer TECAG10 constructed a CTG-10 repeat equilibrating substrate. Substrate (5 fmol) was incubated with increasing amounts of either FEN1, G67Sp, or G240Dp (0.5, 1, 5, 10, and 50 fmol) at 30°C for 10 min. Reaction mixtures were subsequently subjected to electrophoresis on a 15% polyacrylamide denaturing gel. Reaction mixtures resulting from wild-type enzyme cleavage are shown in lanes 2 to 6 (fixed) and lanes 18 to 22 (equilibrating). G67Sp cleavage is shown in lanes 7 to 11 (fixed) and lanes 23 to 27 (equilibrating). The enzyme cleavage by G240Dp on the fixed and equilibrating CTG flap is shown in lanes 13 to 16 (fixed) and lanes 28 to 32 (equilibrating). Lanes 1 and 17 show results from reaction mixtures that contained only substrate. (B) The data resulting from enzyme cleavage on the fixed CTG-10 repeat flap substrate shown in panel A were processed by ImageQuant software, version 1.2. The percentage of cleavage products was plotted against the amounts of wild-type and mutant enzymes. (C) The data from wild-type and mutant enzymatic cleavage on a CTG-10 repeat equilibrating substrate were quantitated and plotted as described for panel B. Schematic diagrams of the substrates are presented above the figure. The gray textured lines represent CTG repeats and a light gray line on the template indicates CAG repeats. The substrates were radiolabeled at the 3′ end of the downstream primers; an asterisk denotes the position of the radiolabeled nucleotide. ♦, cleavage by wild-type FEN1; ▪, G67Sp cleavage; ▴, G240Dp cleavage.
To examine whether wild-type enzyme exhibits a propensity for cleavage of the equilibrating substrate if its endonucleolytic cleavage activity is inhibited by a longer CTG flap, cleavage activity on a fixed and an equilibrating CTG-20 flap was examined. The fixed CTG-20 flap had a sufficiently long self-complementarity region to cause substantial inhibition (Fig. 6A, lanes 2 to 6; product percentage ranges from 0.5 to 3.5%). In this case, cleavage of the equilibrating CTG-20 flap was significantly improved over cleavage of the fixed CTG-20 flap (Fig. 6A, lanes 8 to 12; product percentage ranges from 1 to 10%), indicating that wild-type FEN1 behaves in a similar manner to the G67S mutant if the CTG flap is sufficiently long. These results suggest that flap equilibration helps FEN1 to resolve a CTG repeat flap by allowing the nuclease to bypass the effects of complementarity.
FIG. 6.
FEN1 employs a flap equilibration to resolve a CTG-20 repeat. (A) A CTG-20 fixed-flap substrate was made by annealing primers DFCTG20 and UFCTG to a template, TFCTG. Annealing of primers DECTG20 and UECTG20 to template primer TECAG20 constructed a CTG-20 repeat equilibrating substrate. Substrates (5 fmol) were incubated with increasing amounts of FEN1 at 30°C for 10 min. The reaction was performed in a mixture containing 0 mM KCl. Cleavage of fixed and equilibrating CTG flaps by FEN1 is indicated in lanes 2 to 6 (fixed) and lanes 8 to 12 (equilibrating). Lanes 1 and 7 represent the reaction mixtures that contain only substrate. (B). A time course study was performed by incubating 10 fmol of FEN1p with 5 fmol of CTG-20 repeat equilibrating substrate at 30°C. Aliquots (20 μl) were removed from the reaction mixture at 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 min. Schematic diagrams of the substrates are presented above the figures. The gray textured lines represent CTG repeats, and a light gray line on the template indicates CAG repeats. The substrates were radiolabeled at the 3′ end of the downstream primers; an asterisk denotes the position of the radiolabeled nucleotide.
We note that the relative rates of cleavage of fixed versus equilibrating flaps are likely to be influenced not only by flap length but also by reaction conditions, nucleotides flanking the repeat region, and type of repeat. Such influences will be examined in future work.
FEN1 resolves an equilibrating CTG flap mainly as a double-flap structure with a 1-nt 3′ tail.
Employing flap equilibration, FEN1 might utilize two possible mechanisms to remove CTG repeats. One would be that FEN1 can capture and bind a short flap intermediate formed during a flap equilibration. The short flap would have little secondary structure, so the binding would be efficient. The nuclease would then cleave a segment of several nucleotides from the 5′ end of the flap. The flap would then reequilibrate to create another short flap, onto which FEN1 could load and cleave again. The enzyme would progressively cleave off the CTG flap through a repeated process of binding, cleavage, and dissociation steps. In this scenario, the enzyme employs a distributive mechanism to trim down the flap.
Another possibility would be that the flap equilibration allows the enzyme to capture a double-flap intermediate. The enzyme subsequently stays on the intermediate, either allowing or facilitating the flap to equilibrate into a double flap with a 1-nt 3′ tail and without secondary structure, the optimal substrate for FEN1 cleavage. To distinguish these two mechanisms, the cleavage of a CTG-20 equilibrating-flap substrate was measured over time (Fig. 6B). Some progressive, distributive cleavage was observed. However, the major product appears to result from direct cleavage on the optimal FEN1 substrate, immediately producing a nicked product.
Minor products corresponding to apparent progressive cleavage were mostly observed at higher FEN1 levels (10 and 50 fmol) (Fig. 6B, lanes 11 and 12). However, these did not appear to be effective precursors to the final nicked product. This is also consistent with our previous findings that double flaps with a 2- or 3-nt 3′ flap are poor substrates for both yeast and human FEN1 (28 and data not shown).
DISCUSSION
In the present study we have examined the ability of yeast FEN1 mutants rad27-G67S and rad27-G240D to allow TNR expansion in vivo and in vitro and found that they are defective in their ability to cleave equilibrating flaps. The degree of the endonuclease cleavage defect of the mutants on equilibrating flaps approximately correlates with their mutator phenotypes, suggesting that FEN1 uses flap equilibration to resolve a TNR flap before it can lead to double-stranded DNA breaks or sequence expansion. Moreover, with an appropriate length substrate, the mutant and wild-type FEN1 enzymes could actually be more effective at cleaving equilibrating flaps than nonequilibrating TNR flaps. This observation further suggested a role for flap equilibration as part of the normal cleavage mechanism.
The endonucleolytic cleavage defect of FEN1 on an equilibrating flap leads to CTG repeat expansion.
The defect in FEN1 endonuclease activity caused by the rad27-G240D mutation increased the percentage of expansion for both CTG-85 and CTG-155 tracts in vivo, whereas the rad27-G67S mutation had little effect on repeat instability. We considered that TNR expansion could directly result from a defect of the nuclease in utilizing a structure resembling the fixed CTG flap. However, this interpretation is inconsistent with the fact that G67Sp did not significantly increase CTG fragility and expansion in vivo, even though the mutant nuclease is severely defective in cleavage of fixed CTG flaps. The cleavage activity of both mutants on the CTG-5 and CTG-10 equilibrating flaps approximately agrees with the frequency of triplet repeat expansion in vivo. This suggests that the critical defect of the rad27-G240D mutant protein is its inability to interact with an equilibrating flap in a way that leads to cleavage. Further support for this hypothesis comes from the result that both the G67Sp and the wild-type FEN1 could cleave equilibrating CTG repeat flaps more efficiently than fixed ones in vitro and handle TNR flaps well in vivo. This finding suggests that both the rad27-G67S mutant and wild-type nucleases utilize CTG flap equilibration to create a cleavable substrate, bypassing the inhibitory effect of flap self-complementarity. In contrast, the G240Dp mutant is both ineffective in cleaving equilibrating flaps and susceptible to TNR expansion. Therefore, we conclude that FEN1 works with flap equilibration to resolve a triplet repeat flap.
If the G240Dp endonuclease defect increases expansions by inefficient capture of the flap structure, the equilibrium would be driven towards fold-back and bubble structures. An increase in fold-back structures, which are poor structures for ligation, could account for the increase in fragility observed. The ligation of bubble structures is predicted to generate expansions (21). FEN1 has been shown to increase the rate of nick ligation in the presence of Polδ (1). Therefore, binding of the mutant enzyme could facilitate ligation of a bubble intermediate, thus leading to the observed increase in expansions in the rad27-G240D strain. An increase of tract length from CTG-85 to CTG-155 further increased the frequency of expansions in the rad27-G240D strain, which suggests that as the tract length gets longer, the likelihood of bubble intermediate formation increases or that there are more CTG-containing flaps per replicating chromosome for CTG-155 versus CTG-85. In contrast, there was a decrease in expansion frequency in the rad27Δ cells when the tract increased from CTG-85 to CTG-155 but an increase in fragility and contractions, suggesting that in the absence of FEN1 the equilibrium shifts to unligatable fold-back structures. Unligatable nicks could be healed by a repair process, possibly leading to contractions, or be converted into double-strand breaks and healed by telomere addition, which would be detected in the YAC breakage assay. Interestingly, a further increase in tract length from CTG-85 to CTG-155 did not significantly exacerbate chromosomal breakage in the rad27-G240D strain. If G240Dp stabilizes bubbles as suggested above, the increased formation of fold-back structures leading to breaks would be prevented.
A flap equilibration is employed by FEN1 to resolve triplet repeat flaps by a single endonuclease cleavage event.
We suspected that flap equilibration facilitates FEN1 endonucleolytic cleavage by helping the nuclease to load onto the substrate. This led us to propose and test two potential mechanisms for equilibration-facilitated FEN1 loading (Fig. 7).
FIG. 7.
A proposed model by which FEN1 removes triplet repeats by a unique tracking mechanism. In mechanism A, the fold-back triplet flap is a poor substrate for FEN1, drawn encircling the DNA. However, it equilibrates into a structure with a bubble and a short 5′ flap that FEN1 can access. FEN1 removes the 5′ flap by using its endonucleolytic cleavage activity. The enzyme then dissociates from the substrate until the continued flap equilibration creates another 5′ flap. Then the enzyme rebinds to the substrate and initiates another round of cleavage. The bubble allows the 3′ flap to be short, producing a series of ideal FEN1 substrates. In this manner, FEN1 employs a distributive mechanism to progressively shorten a triplet repeat flap, eventually removing it. In mechanism B, a triplet repeat flap anneals to the template, creating a double-flap intermediate with a short 5′ flap but long 3′ flap. FEN1 can bind the 5′ flap, but the configuration is not ideal for cleavage. FEN1 tracks along the 5′ flap while allowing or facilitating further equilibration into a double-flap structure having a 1-nt 3′ tail. Subsequently, FEN1 efficiently removes the 5′ triplet repeat flap by using its endonucleolytic activity.
In model A, substrate intermediates are formed that have flaps, bubbles, and fold-backs. A subset of these intermediates has 5′ flaps too short to self-anneal in a way that would inhibit FEN1. This group also has a 1-nt 3′ flap that favors FEN1 cleavage. The nuclease could then load, immediately trim away several nucleotides, and then dissociate. Subsequent reorientation of the remaining 5′ strand would again create a short flap that would allow FEN1 loading and cleavage. The process would be repeated until the flap was so short that trimming produced a ligatable nick.
Model B is based on the proposed tracking mechanism of FEN1. A number of studies indicate that the 5′ end of the flap is required to be free before FEN1 can cleave at the flap base. If the 5′ end of a long flap was blocked with streptavidin (59) or a primer (42) or if it could anneal to the template to form a bubble (22), FEN1 could not cleave. This observation led to the proposal that FEN1 must load onto the substrate at the 5′ end of the flap and then track to the site of cleavage (42). In model B, FEN1 loads onto the 5′ end of a short flap in a substrate intermediate that is equivalent to the one envisioned in model A. However, cleavage is delayed because the 3′ flap is too long to induce catalysis. Instead, the nuclease takes advantage of the fluidity of the equilibration process to track further down the flap. When the nuclease has passed enough of the flap, the 5′ end region can self-anneal. This would have the effect of trapping the nuclease on the flap and inducing more movement toward the cleavage site. The nuclease would eventually arrive at a point where the 3′ flap is 1 nt long. This creates the ideal double-flap substrate for cleavage. The yeast FEN1 would then remove the flap as an intact segment and dissociate from a product with a ligatable nick.
Our results suggest that most cleavage occurs by model B for the following reasons. First, FEN1 can only effectively remove a double-flap structure with a 1-nt 3′ tail (28). Other double flaps with ≥2-nt 3′ tails are poor substrates for cleavage (28). This argues that the numerous cleavages required in model A would be suppressed. Second, the major cleavage product obtained with a long CTG equilibrating flap was produced by FEN1 endonucleolytic cleavage on a double flap with a 1-nt 3′ tail (Fig. 6A). Formation of this product does not rely on a FEN1-directed stepwise cleavage process that progressively shortens the flap.
The G240D mutation impairs the ability of FEN1 to employ a flap equilibration.
G67S and G240 are highly conserved amino acids in FEN1 homologues within seven different species including bacteriophage, archeabacteria, yeast, mouse, and human (53). These mutations are located within different domains of the enzyme, with G67 in the N terminus and G240 in the intermediate domain. The crystal structures of FEN1 homologues have been obtained from T5 exonuclease (9), T4 RNase H (41), Methanococcus jannaschii FEN1 (24), and Pyrococcus furiosus FEN1 (23). Based on the P. furious FEN1 structure, which has highest homology with yeast and human FEN1, G67 is located on an α-helix involved in constructing one side of the catalytic groove, whereas G240 is located on a helix-three turn-helix (H3TH) motif which has been proposed to bind to double-strand DNA (23). Our enzyme kinetics data showed that both mutations mainly cause a catalytic but not binding defect on a double-flap substrate.
The H3TH motif in FEN1 homologues is proposed to be involved in enzyme binding onto the downstream double-strand DNA region of a flap substrate (13). Formation of a double flap with a 1-nt 3′ tail requires a flap equilibration that creates a branch migration in the downstream direction. Thus, any change in protein structure that affects enzyme binding on the downstream double-strand DNA could impair branch migration and the accompanying enzyme tracking. Were this hypothesis true, the H3TH motif would be critical for enzyme tracking along a CTG flap and for flap equilibration. The rad27-G240D mutation, located on this motif, leads to a severe endonucleolytic-cleavage defect specifically on equilibrating flaps but not fixed flaps, suggesting that the mutation introduces a blockage in enzyme tracking and DNA branch migration. Such blockage would serve to allow time for the flap substrate to form a bubble intermediate leading to sequence expansion.
The G67S mutation, on the other hand, located on an α-helix proposed as a part of a catalytic groove (23), exhibited a significantly reduced Vmax but a small increase in Km (1.8-fold) on a fixed flap. This result indicates primarily a defect in catalytic steps. This mutation would be predicted to cause a moderate inhibition of the enzyme to either cleave the optimal double flap or allow a flap equilibration or both. The mutation may not directly interfere with enzyme tracking. In essence, the structure-function correlation of these mutations suggests that they employ different mechanisms to impair enzyme cleavage on an equilibrating flap.
A flap equilibration may be important for FEN1 to resolve triplet repeats but not a nonrepeat Okazaki fragment.
Our work suggests that a flap equilibration is not critical for processing nonrepeat Okazaki fragments. First, the cleavage efficiency by mutant FEN1 nucleases on a fixed nonrepeat double flap does not exhibit a significant difference from that of the wild type. This finding is consistent with the fact that the mutations do not produce significant growth defects in vivo (65). Another implication is that during normal Okazaki fragment processing, substrates are mainly fixed double flaps and not equilibrating flaps. This would be the case if FEN1 cleavage usually occurs while the polymerase is generating the flap. FEN1 is an abundant cellular protein (43), and a nonrepeat flap cannot form into a stable secondary structure to inhibit FEN1 endonucleolytic cleavage. Thus, even though the mutants have compromised cleavage activity, they appear to be sufficiently active for prompt removal of most flaps. This possibility is currently being investigated.
In vivo, a FEN1 cleavage defect may be bypassed by repair pathways or interactions with other proteins.
Since a deletion of FEN1 is not lethal to yeast cells, alternative pathways must exist to repair unligated Okazaki fragments. A rad27Δ strain is lethal in combination with proteins involved in homologous recombination (11, 57), implying that the unsealed nick can be repaired by a gap or double-strand break repair pathway. In addition, human cells expressing a nuclease-defective FEN1 protein (D181A) have increased recruitment of ERCC1, a protein involved in both nucleotide excision repair and strand break repair, to repair foci (55). A repair could occur with fidelity or lead to expansion or contraction. In both the rad27-G240D and rad27Δ strains, some large expansions that doubled or almost doubled the size of the CTG-85 tract were observed. It is unlikely that changes larger than 45 to 50 repeats are generated by a single round of flap ligation since this size is as big as an entire Okazaki fragment (∼150 bp, or 50 repeats), whereas the average strand displacement distance is thought to be 30 nt (10 repeats) or less (26). Multiple rounds of replication without correct flap processing could possibly generate the large-size expansions. Alternatively, double-strand break repair, for example, by synthesis-dependent strand annealing (49), could generate large expansions, a hypothesis supported by the fact that the rad27Δ strain showed a large increase in CTG tract fragility. However, since the rad27-G240D strain showed threefold less double-sized expansion and also significantly less fragility than the rad27Δ strain, it is likely that most of the expansions observed in the rad27-G240D background are generated by replication-associated flap ligation rather than by a break repair pathway.
In vivo, the rad27-G240D strain allows substantially more sequence expansion than the wild type, whereas rad27-G67S has only a slightly increased expansion frequency compared with wild type. However, the rad27-G67S strain exhibited a marked defect when attempting to cleave CTG flaps in vitro. An additional factor that could contribute to the discrepancy between in vivo and in vitro observations is the influence of a protein-protein interaction in vivo. Several proteins have been shown to interact with FEN1 to modulate its flap cleavage activity, including proliferating cell nuclear antigen (59, 64), Dna2 (6, 7), the Sgs1 helicase (18), and Werner protein (4, 5).
In summary, CTG repeat expansion phenotypes of yeast FEN1 mutants correlate with cleavage defects of the purified nucleases on equilibrating-flap substrates. FEN1 wild-type endonucleolytic cleavage is also inhibited by secondary structures resulting from triplet repeat flaps. However, our results suggest that wild-type FEN1 employs its unique tracking mechanism in concert with flap equilibration to resolve triplet repeat flaps before they produce intermediates that could expand DNA.
Acknowledgments
We thank the members of the Freudenreich and Bambara laboratories for helpful discussions and our anonymous reviewers for suggestions. We also thank Eric Alani at Cornell University for providing yeast mutant strains and Alison Lauer for technical assistance.
R.A.B., J.V., and Y.L. were supported by National Institutes of Health grant GM24441. C.H.F. and H.Z. were supported by National Institutes of Health grant GM63066.
REFERENCES
- 1.Ayyagari, R., X. V. Gomes, D. A. Gordenin, and P. M. Burgers. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278:1618-1625. [DOI] [PubMed] [Google Scholar]
- 2.Bae, S. H., D. W. Kim, J. Kim, J. H. Kim, D. H. Kim, H. D. Kim, H. Y. Kang, and Y. S. Seo. 2002. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 277:26632-26641. [DOI] [PubMed] [Google Scholar]
- 3.Balakumaran, B. S., C. H. Freudenreich, and V. A. Zakian. 2000. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 9:93-100. [DOI] [PubMed] [Google Scholar]
- 4.Brosh, R. J., H. Driscoll, G. Dianov, and J. Sommers. 2002. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry 41:12204-12216. [DOI] [PubMed] [Google Scholar]
- 5.Brosh, R. J., C. von Kobbe, J. Sommers, P. Karmakar, P. Opresko, J. Piotrowski, I. Dianova, G. Dianov, and V. Bohr. 2001. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 20:5791-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budd, M. E., and J. L. Campbell. 2000. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. 459:173-186. [DOI] [PubMed] [Google Scholar]
- 7.Budd, M. E., and J. L. Campbell. 1995. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 92:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callahan, J. L., K. J. Andrews, V. A. Zakian, and C. H. Freudenreich. 2003. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell. Biol. 23:7849-7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceska, T. A., J. R. Sayers, G. Stier, and D. Suck. 1996. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature 382:90-93. [DOI] [PubMed] [Google Scholar]
- 10.Culotti, J., and L. H. Hartwell. 1971. Genetic control of the cell division cycle in yeast. 3. Seven genes controlling nuclear division. Exp. Cell Res. 67:389-401. [DOI] [PubMed] [Google Scholar]
- 11.Debrauwere, H., S. Loeillet, W. Lin, J. Lopes, and A. Nicolas. 2001. Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMott, M. S., B. Shen, M. S. Park, R. A. Bambara, and S. Zigman. 1996. Human RAD2 homolog 1 5′- to 3′-exo/endonuclease can efficiently excise a displaced DNA fragment containing a 5′-terminal abasic lesion by endonuclease activity. J. Biol. Chem. 271:30068-30076. [DOI] [PubMed] [Google Scholar]
- 13.Dervan, J., M. Feng, D. Patel, J. Grasby, P. Artymiuk, T. Ceska, and J. Sayers. 2002. Interactions of mutant and wild-type flap endonucleases with oligonucleotide substrates suggest an alternative model of DNA binding. Proc. Natl. Acad. Sci. USA 99:8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutcher, S. K. 1981. Internuclear transfer of genetic information in kar1-1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freudenreich, C. H., S. M. Kantrow, and V. A. Zakian. 1998. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279:853-856. [DOI] [PubMed] [Google Scholar]
- 16.Freudenreich, C. H., J. B. Stavenhagen, and V. A. Zakian. 1997. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 17:2090-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gacy, A. M., G. Goellner, N. Juranic, S. Macura, and C. T. McMurray. 1995. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81:533-540. [DOI] [PubMed] [Google Scholar]
- 18.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordenin, D. A., T. A. Kunkel, and M. A. Resnick. 1997. Repeat expansion-all in a flap? Nat. Genet. 16:116-118. [DOI] [PubMed] [Google Scholar]
- 20.Harrington, J. J., and M. R. Lieber. 1994. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 13:1235-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henricksen, L., J. Veeraraghavan, D. R. Chafin, and R. A. Bambara. 2002. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J. Biol. Chem. 277:22361-22369. [DOI] [PubMed] [Google Scholar]
- 22.Henricksen, L. A., S. Tom, Y. Liu, and R. A. Bambara. 2000. Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem. 275:16420-16427. [DOI] [PubMed] [Google Scholar]
- 23.Hosfield, D. J., C. D. Mol, B. Shen, and J. A. Tainer. 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell 95:135-146. [DOI] [PubMed] [Google Scholar]
- 24.Hwang, K. Y., K. Baek, H. Y. Kim, and Y. Cho. 1998. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol. 5:707-713. [DOI] [PubMed] [Google Scholar]
- 25.Jankowski, C., F. Nasar, and D. K. Nag. 2000. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl. Acad. Sci. USA 97:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, Y. H., R. Ayyagari, M. A. Resnick, D. A. Gordenin, and P. M. Burgers. 2003. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 278:1626-1633. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, R. E., G. K. Kovvali, L. Prakash, and S. Prakash. 1995. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science 269:238-240. [DOI] [PubMed] [Google Scholar]
- 28.Kao, H. I., L. A. Henricksen, Y. Liu, and R. A. Bambara. 2002. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 277:14379-14389. [DOI] [PubMed] [Google Scholar]
- 29.Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin, and T. D. Petes. 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t). Mol. Cell. Biol. 18:2779-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kucherlapati, M., K. Yang, M. Kuraguchi, J. Zhao, M. Lia, J. Heyer, M. Kane, K. Fan, R. Russell, A. Brown, B. Kneitz, W. Edelmann, R. Kolodner, M. Lipkin, and R. Kucherlapati. 2002. Haploinsufficiency of flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. USA 99:9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 32.Lee, S., and M. Park. 2002. Human FEN-1 can process the 5′-flap DNA of CTG/CAG triplet repeat derived from human genetic diseases by length and sequence dependent manner. Exp. Mol. Med. 34:313-317. [DOI] [PubMed] [Google Scholar]
- 33.Lenzmeier, B. A., and C. H. Freudenreich. 2003. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res. 100:7-24. [DOI] [PubMed] [Google Scholar]
- 34.Liu, Y., and R. Bambara. 2003. Analysis of human flap endonuclease 1 mutants reveals a mechanism to prevent triplet repeat expansion. J. Biol. Chem. 278:13728-13739. [DOI] [PubMed] [Google Scholar]
- 35.Lopes, J., H. Debrauwere, J. Buard, and A. Nicolas. 2002. Instability of the human minisatellite CEB1 in rad27Δ and dna2-1 replication-deficient yeast cells. EMBO J. 21:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maleki, S., H. Cederberg, and U. Rannug. 2002. The human minisatellites MS1, MS32, MS205 and CEB1 integrated into the yeast genome exhibit different degrees of mitotic instability but are all stabilised by RAD27. Curr. Genet. 41:333-341. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto, Y., K. Kim, J. Hurwitz, R. Gary, D. S. Levin, A. E. Tomkinson, and M. S. Park. 1999. Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J. Biol. Chem. 274:33703-33708. [DOI] [PubMed] [Google Scholar]
- 38.Maurer, D. J., B. L. O'Callaghan, and D. M. Livingston. 1998. Mapping the polarity of changes that occur in interrupted CAG repeat tracts in yeast. Mol. Cell. Biol. 18:4597-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miret, J., L. Pessoa-Brandao, and R. Lahue. 1997. Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:3382-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirkin, S., and E. Smirnova. 2002. Positioned to expand. Nat. Genet. 31:5-6. [DOI] [PubMed] [Google Scholar]
- 41.Mueser, T. C., N. G. Nossal, and C. C. Hyde. 1996. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell 85:1101-1112. [DOI] [PubMed] [Google Scholar]
- 42.Murante, R. S., J. A. Rumbaugh, C. J. Barnes, J. R. Norton, and R. A. Bambara. 1996. Calf RTH1 nuclease can remove the initiator RNAs of Okazaki fragments by endonuclease activity. J. Biol. Chem. 271:25888-25897. [DOI] [PubMed] [Google Scholar]
- 43.Murray, J. M., M. Tavassoli, R. al-Harithy, K. S. Sheldrick, A. R. Lehmann, A. M. Carr, and F. Z. Watts. 1994. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol. Cell. Biol. 14:4878-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Negritto, M. C., J. Qiu, D. O. Ratay, B. Shen, and A. M. Bailis. 2001. Novel function of Rad27 (FEN-1) in restricting short-sequence recombination. Mol. Cell. Biol. 21:2349-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parenteau, J., and R. Wellinger. 2002. Differential processing of leading- and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics 162:1583-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parenteau, J., and R. J. Wellinger. 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reagan, M. S., C. Pittenger, W. Siede, and E. C. Friedberg. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177:364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richard, G., C. Hennequin, A. Thierry, and B. Dujon. 1999. Trinucleotide repeats and other microsatellites in yeasts. Res. Microbiol. 150:589-602. [DOI] [PubMed] [Google Scholar]
- 49.Richard, G. F., G. M. Goellner, C. T. McMurray, and J. E. Haber. 2000. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 19:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robins, P., D. J. Pappin, R. D. Wood, and T. Lindahl. 1994. Structural and functional homology between mammalian DNase IV and the 5′-nuclease domain of Escherichia coli DNA polymerase I. J. Biol. Chem. 269:28535-28538. [PubMed] [Google Scholar]
- 51.Schulz, V. P., and V. A. Zakian. 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76:145-155. [DOI] [PubMed] [Google Scholar]
- 52.Schweitzer, J. K., and D. M. Livingston. 1998. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 7:69-74. [DOI] [PubMed] [Google Scholar]
- 53.Shen, B., J. Qiu, D. Hosfield, and J. A. Tainer. 1998. Flap endonuclease homologs in archaebacteria exist as independent proteins. Trends Biochem. Sci. 23:171-173. [DOI] [PubMed] [Google Scholar]
- 54.Sommers, C. H., E. J. Miller, B. Dujon, S. Prakash, and L. Prakash. 1995. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J. Biol. Chem. 270:4193-4196. [DOI] [PubMed] [Google Scholar]
- 55.Spiro, C., and C. T. McMurray. 2003. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol. Cell. Biol. 23:6063-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spiro, C., R. Pelletier, M. L. Rolfsmeier, M. J. Dixon, R. S. Lahue, G. Gupta, M. S. Park, X. Chen, S. V. Mariappan, and C. T. McMurray. 1999. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell 4:1079-1085. [DOI] [PubMed] [Google Scholar]
- 57.Symington, L. S. 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26:5589-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tishkoff, D. X., N. Filosi, G. M. Gaida, and R. D. Kolodner. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88:253-263. [DOI] [PubMed] [Google Scholar]
- 59.Tom, S., L. A. Henricksen, and R. A. Bambara. 2000. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem. 275:10498-10505. [DOI] [PubMed] [Google Scholar]
- 60.Turchi, J. J., L. Huang, R. S. Murante, Y. Kim, and R. A. Bambara. 1994. Enzymatic completion of mammalian lagging-strand DNA replication. Proc. Natl. Acad. Sci. USA 91:9803-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vallen, E. A., and F. R. Cross. 1995. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol. Cell. Biol. 15:4291-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, P. J., R. H. Borts, and M. C. Hirst. 1999. Stability of the human fragile X (CGG)n triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol. Cell. Biol. 19:5675-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 64.Wu, X., J. Li, X. Li, C. L. Hsieh, P. M. Burgers, and M. R. Lieber. 1996. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 24:2036-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie, Y., Y. Liu, J. L. Argueso, L. A. Henricksen, H. I. Kao, R. A. Bambara, and E. Alani. 2001. Identification of rad27 mutations that confer differential defects in mutation avoidance, repeat tract instability, and flap cleavage. Mol. Cell. Biol. 21:4889-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]