Abstract
Purpose
To investigate the relationship between LDH-A expression, lactate concentration, cell metabolism and metastases in murine 4T1 breast tumors.
Experimental Design
Inhibition of LDH-A expression and protein levels were achieved in a metastatic breast cancer cell line (4T1) using shRNA technology. The relationship between tumor LDH-A protein levels and lactate concentration (measured by magnetic resonance spectroscopic imaging-MRSI) and metastases was assessed.
Results
LDH-A knockdown cells (KD9) showed a significant reduction in LDH-A protein and LDH activity, less acid production, decreased transwell migration and invasion, lower proliferation, reduced glucose utilization and glycolysis and increase in oxygen consumption, ROS and cellular ATP levels, compared to control (NC) cells cultured in 25 mM glucose. In vivo studies showed lower lactate levels in KD9, KD5, KD317 tumors than in NC or 4T1 wild-type tumors (p<0.01), and a linear relationship between tumor LDH-A protein expression and lactate concentration. Metastases were delayed and primary tumor growth rate decreased.
Conclusions
We show for the first time that LDH-A knockdown inhibited the formation of metastases, and was accompanied by in vivo changes in tumor cell metabolism. Lactate MRSI can be used as a surrogate to monitor targeted inhibition of LDH-A in a pre-clinical setting and provides a non-invasive imaging strategy to monitor LDH-A targeted therapy. This imaging strategy can be translated to the clinic to identify and monitor patients who are at high risk of developing metastatic disease.
Keywords: Breast cancer, Metastases, Lactate, LDH-A, MRI, MRS
Introduction
Metabolic alterations in tumors impact their evolution, progression, and development of metastases (1-7). Cancer cells modify their energy metabolism as they proliferate and adjust to a changing microenvironment to meet their energy and macromolecular synthetic demands. Many cancer cells have high glucose and glutamine utilization and high rates of aerobic glycolysis (1) which impact the development of the metastatic phenotype (2, 8).
This manuscript focuses on lactate dehydrogenase A (LDH-A). LDH-A provides a link between several metabolic pathways. The product of LDH-A activity (lactate) can be assessed non-invasively and quantitatively using magnetic resonance spectroscopic (MRS) imaging (MRSI). The low sensitivity and overlapping peaks of the lactate and lipid MR spectra present technical difficulties during MR spectroscopy, but can be overcome by the use of spectral editing methods, such as SELective Multiple Quantum Coherence (SEL-MQC) techniques (9). We previously showed that tumor lactate concentration can be measured in murine breast tumor models by in vivo 1H MRSI (10).
The inhibition of LDH-A has an anti-proliferative effect on primary breast tumors (5). However, there have been no studies investigating the effect of LDH-A inhibition on the development and progression of metastases, or to evaluate methods to noninvasively monitor LDH-A inhibition. We focused on the role of lactate and LDH-A in the development of metastases, since the cause of death in breast cancer is almost always due to metastases. We hypothesized that there is a relationship between LDH-A expression, tumor lactate concentration, and the growth and metastatic potential of murine breast tumor models. We employ shRNA knockdown technology to inhibit LDH-A expression, and to study the effects of LDH-A inhibition on tumor cell metabolism, growth, and the development of metastases. We address several new questions: 1) Will shRNA-mediated reduction of LDH-A expression in 4T1 breast cancer cells alter orthotopic 4T1 tumor lactate concentrations?; 2) Does shRNA-mediated reduction of LDH-A expression in 4T1 breast cancer cells alter metabolism and sensitivity to metabolic stress (reactive oxygen species - ROS)?; 3) Does shRNA-mediated reduction of LDH-A expression in 4T1 breast cancer cells alter the development of metastases and tumor growth?; 4) Is there a relationship between tumor LDH-A protein levels and MRSI-measured tumor lactate concentration?
Materials and Methods
Cell culture
4T1 cells derived from a spontaneous breast tumor in a BALB/c mouse and were provided by Fred Miller (Karmanos Cancer Institute)(11). The cell line was not authenticated by the authors since cell authentication testing can be performed only on human cell lines. KD (knockdown) and NC (control) cells, derived from 4T1 murine breast cancer cells, were grown in standard DMEM containing 10% FCS supplemented with either 25 or 5 mM glucose and 6 mM L-glutamine, penicillin/streptomycin and 4 mg/L of puromycin.
Generation of LDH-A knockdown and control cell lines
4T1 cells were transfected with SureSilencing™ shRNA plasmids (SABiosciences) to specifically knockdown expression of the mouse LDH-A gene. The vectors contained shRNA under the control of the U1 promoter and included a puromycin resistance gene. Stably transduced clones (KD cell lines) were developed, along with a control (NC) cell line bearing a scrambled shRNA. LDH-A qRT-PCR RNA and immunoblotting protein assays confirmed successful transduction (see Supplemental Data – Methods).
In Vitro Assays
Cell proliferation and metabolic assays (glucose utilization, glycolysis, LDH activity, lactate production, oxygen consumption rate, oxidative phosphorylation, reactive oxygen species (ROS), cellular mitochondria) and cell migration and invasion assays were performed (see Supplemental Data).
Experimental Animal model
Cells were orthotopically implanted as described previously (10). Primary tumor volume was determined by caliper measurements and tumor doubling times were calculated from the tumor volume vs. time profiles (12).
In vivo lactate detection
MRSI experiments were performed on a 7T Bruker Biospec Spectrometer. The lactate signal was acquired using a selective multiple-quantum coherence transfer (SelMQC) editing sequence in combination with chemical shift imaging (CSI) (9, 10, 13) as detailed in the Supplemental Data.
MR images
Lung metastases were imaged using the Bruker gradient echo fast imaging (GEFI) sequence with TR=300ms, TE=2.5ms, NA=4, Matrix=512×256. Gated respiration was used to reduce respiratory artifacts.
Analysis of Breast Cancer Microarray Datasets
A compendium of four breast cancer microarray datasets were analyzed using the Bioconductor set of tools (www.bioconductor.org) in R statistical language (www.r-project.org). Data was downloaded from GEO. The four breast cancer datasets that were analyzed included: 1. MSKCC-82 GSE-2603 (14), 2. EMC-286 GSE-2034 (15), 3. ECM 192 GSE12276: 204 samples (16), 4. EMC-344 (EMC 286 AND 58 cases of ER- tumors, GSE 5327) (17). Data were normalized using the standard gcrma (18) procedure. Survival analysis was performed using R package survival. Details are provided in Supplemental Methods.
Statistical analysis
Results are presented as mean ± standard deviation. Statistical significance was determined by a two-tailed Student T-test. A p-value of <0.05 was considered significant.
Results
Selection/characterization of KD9 and NC 4T1 cells
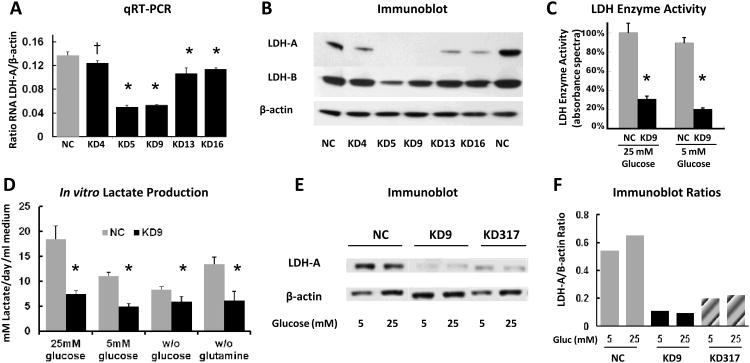
To assess the link between LDH-A expression and the metabolic and metastatic characteristics of an established murine breast cancer model, we transfected 4T1 breast tumor cells with four different SureSilencing shRNAs plasmids specifically targeting mouse LDH-A mRNA (KD), and a non-specific scrambled shRNA (NC), respectively. Several knockdown clones with different levels of LDH-A protein expression were isolated for further experiments. The shRNA knockdown efficiency was evaluated by analyzing LDH-A mRNA expression using qRT-PCR and protein expression by immunoblotting. KD cells have significantly lower levels of LDH-A mRNA (Fig. 1A) and decreased LDH-A protein expression (Fig. 1B) compared to NC cells. Clone #9 (KD9) transduced with shRNA #2 had the lowest LDH-A mRNA and protein levels, and an unchanged LDH-B level (Fig. 1A, B). Another clone, KD317, was developed from cells bearing the plasmid with shRNA#3 (Fig. 1E).
Figure 1. Selection and characterization of LDH-A knockdown cells.
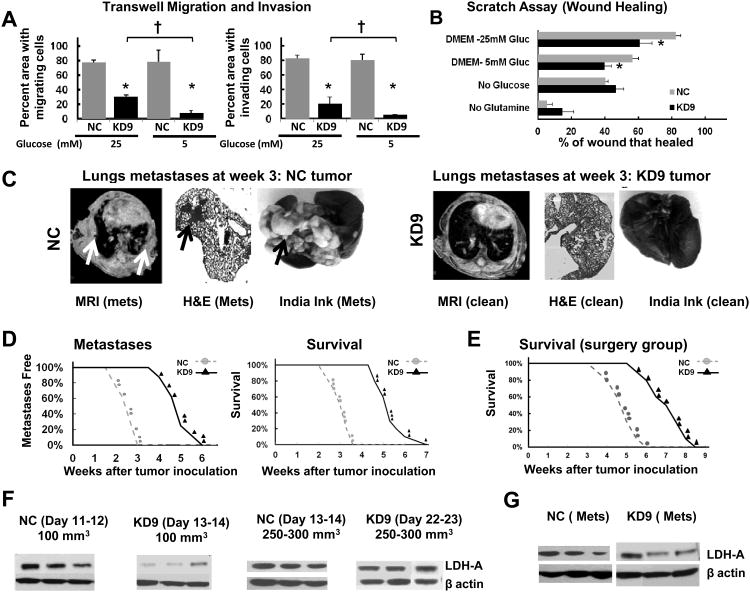
(A) qRT-PCR analysis of LDH-A mRNA expression in the 4T1 cell lines transfected with scrambled shRNA (NC, control) and shRNA to mouse LDH-A mRNA (KD, knockdown) n=3. P values for KD4, KD5, KD9, KD13 and KD16 cells were p=0.03, p<0.0001, p<0.0001, p<0.01 and p<0.01, respectively (*: p<0.01; †: p<0.05). (B) Western blot analyses on whole cell lysates prepared from NC and KD clones. (C) Total LDH enzyme activity in NC and KD9 cells cultured in DMEM with 25 mM or 5 mM glucose, 6 mM L-glutamine and 10% FCS) (* p<0.01). (D) Lactate production: appearance of lactate in different culture medium between NC and KD9 cells (* p<0.01 comparing NC and KD9 cells). (E) Western blot analysis of LDH-A expression: cells were grown in DME media with 5 or 25 mM glucose, and whole cell lysates were analyzed for LDH-A, and β-actin expression. (F) LDH-A/β-actin proteins bands ratio were assessed by ImageJ software.
To validate the correlation between LDH-A expression levels and functional activity of the LDH enzyme complex, we performed an enzymatic assay on viable KD9 and NC tumor cells in growth medium containing 25 or 5 mM glucose. KD9 cells have three-fold lower LDH activity than NC cells when cultured in 25 mM glucose-containing media and more than a four-fold difference in 5 mM glucose (p<0.01; Fig. 1C). KD9 cells also produce significantly less lactate (p<0.01; Fig 1D) than NC cells. We also found that LDH-A expression (Western blot) remains high in the control group (NC) and low in the KD group (KD9, KD317) in both high glucose and low glucose culture medium (Fig. 1E, F).
Metabolic properties of KD9 and NC cells
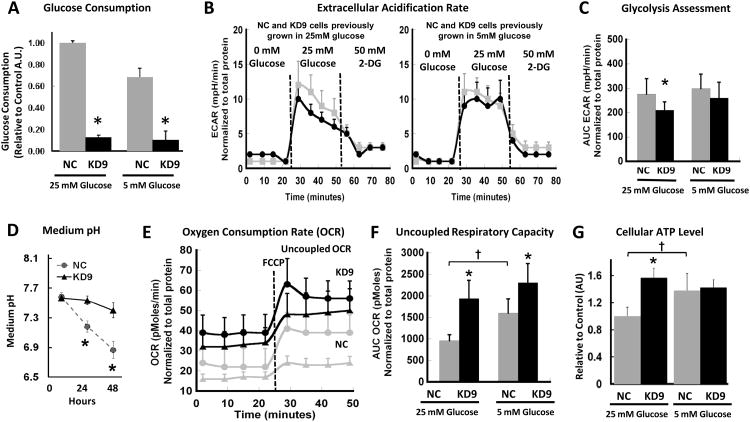
Glucose utilization was significantly less in KD9 cells compared to NC cells, growing in either 25 or 5 mM glucose-containing medium (p<0.01) (Fig. 2A). We used a Seahorse Bioscience XF96 Extracellular Flux Analyzer to measure the extracellular acidification rate (ECAR) and the oxygen consumption rate (OCR) of these cells. We obtained a base-line measure of ECAR using basic glucose-free XF assay medium, then added glucose to assess glycolysis, and then inhibited the process by adding 2-deoxyglucose (2-DG) to the incubation medium. The injection of glucose (final concentration of 25 mM) caused a significant increase in ECAR in both cell lines, with a higher increase in NC cells compared to KD9 cells (Fig. 2B). The subsequent injection of 2-DG (final concentration of 50 mM), decreased ECAR to basal levels. The effects of these treatments are reflected in the integrated areas under the profile measurements (Fig. 2C); the differences between KD9 and NC cells were significant (p<0.01) for cells growing under 25 mM of glucose; a similar trend was noted for cells growing in 5 mM glucose, but the difference was not significant (p=0.147). These results demonstrated a lower glycolytic rate for KD9 cells compared to NC cells (Fig. 2B, 2C) growing under a high concentration of glucose, and a corresponding lower rate of acidification of the incubation medium over 48 hours (Fig. 2D).
Figure 2. Metabolic properties of NC and KD9 cells in vitro.
(A) Glucose consumption: Cell culture media (standard DMEM with 5 or 25 mM glucose) was assayed for Glucose following 48 hours incubation. Fluorescence intensity was normalized to the number of viable cells and background fluorescence (* p<0.01). (B) Extracellular Acidification Rate (ECAR): measurements were obtained prior to and after injection of glucose (initiate glycolysis) and 2-DG (block glycolysis), sequentially. A typical experiment is shown. (C) Glycolysis Assessment calculated from ECAR results (Panel B) for cells previously growing in 25 and 5 mM glucose-containing media; the mean ECAR area under the curve (AUC; mpH/min) (* p<0.01 comparing NC and KD9 cells) normalized to total protein (μg). (D) Cell culture medium acidification during NC and KD9 cell growth in standard DMEM with 25 mM glucose (* p<0.01). (E) Oxygen consumption rate (OCR) and the maximal mitochondrial capacity of cells cultured in 25 mM (triangles) and 5 mM (circles) glucose-containing media. FCCP (carbonyl cyanide p-trifluoro methoxy phenylhydrazone) was used as a potent uncoupler of oxidative phosphorylation in mitochondria. (F) The uncoupled respiratory capacity was calculated from Panel E results: AUC OCR (pMoles) (* p<0.01 comparing NC and KD9 cells; † p< 0.01 comparing NC cells in 25 vs 5 mM glucose-containing media). (G) ATP levels were measured in cells growing in standard DMEM containing 25 or 5 mM glucose; results were normalized to the number of viable cells, and corrected for background luminescence (* and † p<0.01).
Oxygen consumption is an indicator of mitochondrial respiration. A Seahorse Bioscience XF96 Analyzer was used to measure the real-time oxygen consumption rate (OCR, pMoles/minute) in serum free DMEM with 25 and 5 mM glucose and 6 mM of glutamine (Fig. 2E, 2F). FCCP (carbonyl cyanide p-trifluoromethoxy phenylhydrazone) was added to uncouple oxidative phosphorylation from the electron transport chain in order to measure the maximum respiratory capacity (19). The respiratory rate (OCR) of KD9 cells was significantly higher (2- and 1.5-fold) than that of NC cells cultured in 25 and 5 mM glucose, respectively (p<0.01). OCR increased by a similar level for KD9 and NC cells after FCCP was added (Fig. 2F). Similar results were obtained in separate experiments using trypsinized cells growing in the media with 25 mM of glucose using an Oxylite fiber optic probe; KD9 cells had a 60% higher oxygen consumption rate (p<0.01) than NC cells (Fig. S1A).
The high basal level of OCR is consistent with the 40% higher ATP levels observed in KD9 cells compared to NC cells (p<0.01) (Fig. 2G), and suggests that ATP production in KD9 cells may be associated with greater proton leak and ROS production than in NC cells. However this trend occurs only in cells growing in 25 mM glucose. The ATP level was notably higher in NC cells cultured in 5 mM of glucose (compared with 25 mM glucose), while the difference in ATP levels for KD9 cells growing under 25 mM and 5 mM of glucose was not significant (Fig. 2G). These differences in ATP levels between NC and KD9 cells at 25 mM glucose can be explained by the higher mitochondrial oxidative phosphorylation of KD9 cells in high glucose-containing media. It is interesting to note the higher OCR and ATP results in NC cells growing in 5 mM glucose compared to 25 mM glucose media (Fig. 2F,G). These data support the concept that some cancer cells which use aerobic glycolysis, can also switch from glycolysis to oxidative phosphorylation under glucose-limiting conditions (20). This plasticity reflects the interplay between glycolysis and oxidative phosphorylation and the ability to adapt metabolism and energy production to changes in the microenvironment, and to adapt to differences in tumor energy needs or biosynthetic activity (21).
We measured mitochondrial reactive oxygen species (ROS) in cells growing under 25 mM of glucose (Fig. S1B). Intracellular ROS was significantly higher in KD9 cells compared to NC cells (p<0.01), consistent with prior findings (4). However, the level of mitochondrial mass was similar in these two cell lines (Fig. S1C). These findings are consistent with a higher proton leak and consequent increase in ROS formation in the LDH-A knockdown cells. These results demonstrate that LDH-A shRNA knockdown in 4T1 cells leads to an increase in TCA activity and mitochondrial respiration (in KD9 cells compared to NC cells), by switching from aerobic glycolysis (NC cells) to enhanced mitochondrial respiration. However these cell-response properties are more evident when cells are grown under high concentrations of glucose, and this may reflect their “addiction” to high glucose levels.
In summary, the magnitude of glucose consumption, ECAR and pH changes (Fig. 2A,B,C,D) indicate higher aerobic glycolysis in NC compared to KD9 cells under high concentrations of glucose. In contrast, O2 consumption (Fig. 2E,F and Fig. S1A) and ROS (Fig. S1B) were greater for KD9 than NC cells, and O2 consumption was higher at 5 mM glucose than at 25 mM glucose for both cell lines (Fig. 2E,F).
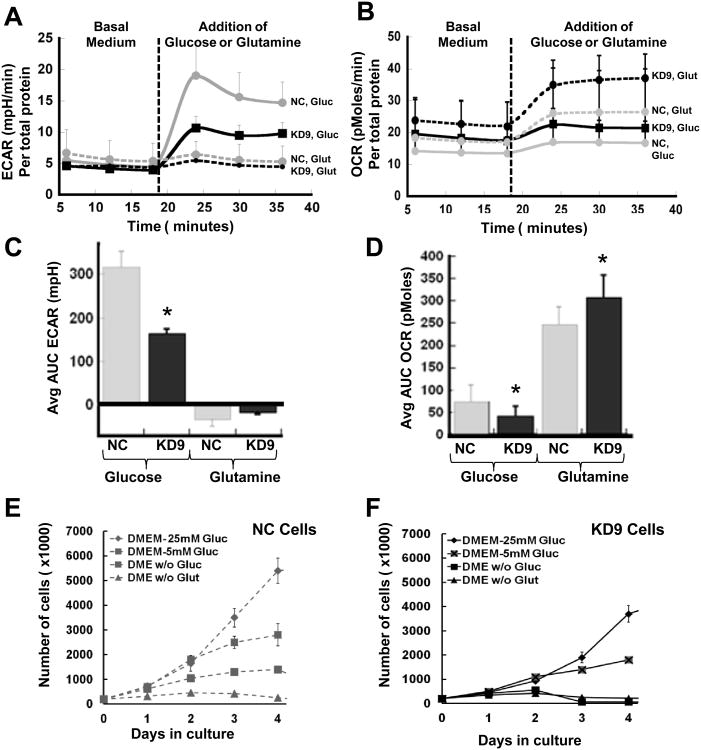
We also studied the effect of excluding either glucose or glutamine from the incubation medium on the two major energy producing pathways of the cell – mitochondrial respiration and glycolysis – using the XF96 Extracellular Flux Analyzer. ECAR in basic glucose-free XF assay medium was low in both cell lines (Figs. 2B, 3A). ECAR (reflecting glycolysis) markedly increased on the addition of 25 mM glucose alone, whereas there was little or no change in glycolysis following the addition of 6 mM glutamine alone (Fig. 3A, 3C). We intentionally used 25 mM of glucose in order to saturate the glycolytic pathway and measure the maximal up-regulation of ECAR. In contrast, OCR increased markedly in both cell lines following the addition of 6 mM glutamine alone, whereas a smaller increase in OCR was observed following the addition of 25 mM glucose alone (Fig. 3B, D). The ECAR and OCR responses of KD9 and NC cells to the addition of glucose and glutamine indicate that glucose metabolism (not glutamine metabolism) is the primary source of extracellular acidification and aerobic glycolysis in both cell lines and that glutamine metabolism is the significant source of oxidative phosphorylation. The results also suggest that inhibition of LDH-A in 4T1 breast cancer cells leads to enhanced mitochondrial respiration through the glutamine pathway, and is associated with an increase in mitochondrial activity (Fig. S1A) and ROS (Fig. S1B), but not in mitochondrial number (Fig. S1C and Fig S2).
Figure 3. Effect of LDH-A knockdown on in vitro growth and metabolic properties.
Changes in ECAR (A) and OCR (B) following the separate addition of glucose (final concentration 25 mM) or glutamine (final concentration 6 mM) to a non-buffered glucose-free DMEM culture medium without FCS (the standard medium for the XF assays). The mean AUC ECAR (mpH) (C) and OCR (pMoles) (D) results are compared, respectively. The mean of 12 independent measurements normalized to total amount of protein (μg) was calculated. Significant differences between NC and KD9 cells are indicated (* p<0.01). Growth profiles of NC (E) and KD9 (F) cells in standard DMEM (25 mM glucose, 6 mM glutamine) compared to growth profiles with 5 mM glucose or with the absence of glucose or glutamine.
Growth profiles of KD9 and NC cells
The proliferation of KD9 and NC cells was studied in DMEM medium with different D-glucose (0, 5, 25 mM) and L-glutamine (0, 6 mM) concentrations. KD9 cells have a considerably slower growth rate compared to NC cells (Fig. 3E, 3F; Table 1). The doubling times were calculated and showed no differences between cells growing under high (25 mM) and a more physiological concentration of glucose (5 mM) in the media (Table 1). The growth of the cells in 5 mM glucose are similar to their growth in 25 mM glucose medium for the first 48 hours, but slows after day 2, compared to their growth in 25 mM glucose (p<0.01), as glucose is consumed from the medium. Both cell lines have markedly reduced growth in the absence of glucose or glutamine, and the effect was greater in KD9 cells (Fig. 3E, 3F).
Table 1.
| Cell Line Doubling Time (hours)† | |||
|---|---|---|---|
| Medium Glucose Concentration | KD9 | NC | 4T1 WT |
| 5 mM | 19.6 ± 0.5 | 15.3 ± 0.3 | 14.7 ± 0.3 |
| 25 mM | 20.5 ± 0.6. | 15.4 ± 0.3 | 14.5 ± 0.2 |
Estimated over two days following cell plating and media change.
Growth and metabolic profiles of KD9 and NC tumors
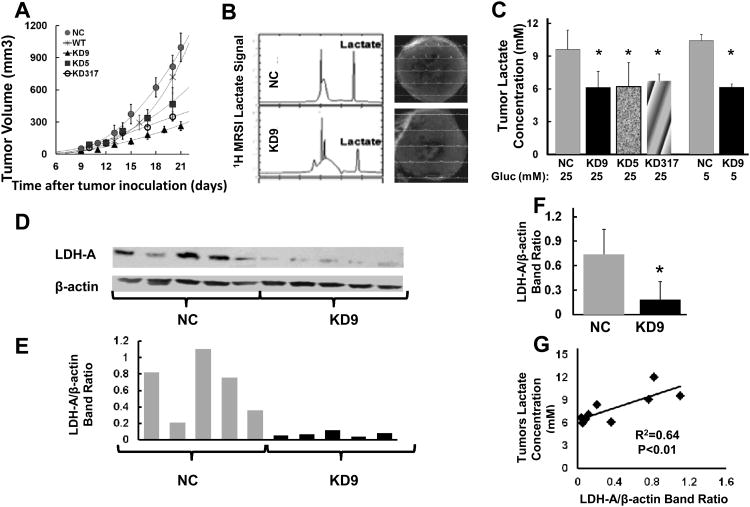
Downregulation of LDH-A expression leads to slower growth, reduced glycolytic flux and increased mitochondrial respiration in vitro. We asked whether enhanced mitochondrial respiration and dependence on glutamine could affect in vivo tumor growth, lactate production and the potential for developing distant metastases. We injected cells into the mammary fat pad and evaluated the effect of LDH-A suppression on the tumorigenicity of the 4T1 breast cancer cell clones. We compared the growth profiles of KD9, KD5 and KD317 with NC and wild-type 4T1 tumors (Fig. 4A). By 2 weeks after injection, a significant tumor volume differences had developed, and LDH-A knockdown KD9, KD5 and KD317 tumors had significantly longer doubling times compared to NC and 4T1 wild-type tumors (Table 2). However, there was no difference in the doubling time of tumors derived from cells cultured in 5 mM compared to 25 mM glucose.
Figure 4. Effect of LDH-A knockdown on tumor growth, lactate and LDH-A protein expression.
(A) Growth profiles of KD9, KD5, KD317 orthotopic breast tumors compared to NC and wild-type 4T1 tumors (n=13 (NC), n=18 (KD9), n=6 (KD5), n=4 (KD317)). (B) Representative lactate spectra from NC and KD9 tumors are shown. (C) Mean lactate concentration of small (∼100mm3) KD9, KD5, KD31 orthotopic breast tumors compared to NC and wild-type 4T1 tumors (* p<0.01). (D) LDH-A expression assessed by Western blotting from 10 small (100 mm3) NC and KD9 tumors. (E) LDH-A/β-actin protein band ratios of Western blots shown in Panel D were analyzed by ImageJ software. (F) The mean LDH-A/β-actin ratio assessed in small (100 mm3) orthotopic tumors (9 NC and 9 KD9; * p<0.01). (G) Tumor lactate concentration assessed by MRS was plotted vs. LDH-A protein level of the corresponding tumor assessed by Western blotting.
Table 2.
| Doubling Time (days)† | ||
|---|---|---|
| Glucose Concentration* | ||
| Tumor cell line | 25 mM | 5 mM |
| KD9 | 3.9 ± 0.5 | 3.5 ± 0.2 |
| KD5 | 3.6 ± 0.3 | - |
| KD317 | 3.4 ± 0.4 | - |
| NC | 2.3 ± 0.2 | 2.0 ± 0.2 |
| 4T1 | 2.2 ± 0.4 | - |
Estimated over first two weeks following implantation
Glucose concentration in pre-implantation culture medium
4T1 tumors are known to undergo necrosis at volumes greater than approximately 100 mm3 (10), and necrosis increases significantly as tumors grow beyond 200-300 mm3 (10). Therefore, we measured in vivo lactate levels using MRSI (Fig. 4B) in tumors that were approximately 100 mm3 in size, in order to avoid the complicating effects of tumor necrosis. We found a significant reduction of lactate in KD9, KD5 and KD317 tumors compared to NC and 4T1 wild-type tumors (Fig. 4C); the mean lactate levels measured in small (100 mm3) KD9 and NC tumors (n=13) were significantly different: 6.1±1.5 and 9.6±1.8 mM (p<0.01), respectively.
In some animals, tumors were removed and underwent histological examination. Tumor histology showed similar features for both the KD9 and NC tumors at small volumes (Fig. S2). Hematoxylin and eosin (H&E) staining of small NC and KD9 tumors showed only minimal necrosis, predominantly in the tumor core. Hoechst perfusion staining showed a heterogeneity of perfusion across the tumor sections. LDH-A enzyme expression levels in small (∼100 mm3) KD9 tumors were compared to small NC tumors by immunoblot analyses of whole tumor lysates. Ten representative immunoblots (5 from each group) and their band intensity ratios are shown in Fig. 4D, 4E. The mean Western blot band intensity for LDH-A from 18 mice, 9 from each group, showed that LDH-A expression was ∼4-fold lower in KD9 tumors compared with NC tumors (p<0.01) (Fig. 4F). We also noted that there was a linear relationship between tumor lactate concentration and the corresponding tumor LDH-A/β-actin protein band ratio (R2 = 0.64; p<0.01) (Fig. 4G), indicating the potential of lactate MRS to monitor the expression of LDH-A and the extent of LDH-A knockdown or inhibition.
Migration potential of KD9 and NC cells
Migration and invasiveness of tumor cells are important aspects of metastasis formation. In transwell migration assays at 25 and 5 mM glucose, fewer KD9 cells migrate through the 8 μm pores of a transwell chamber compared to NC cells (Fig. 5A). Invasion through a Matrigel environment (3 mm thick) also demonstrated a significantly smaller number of invading KD9 cells compared to NC cells. These differences were more profound in 5 mM glucose (p<0.001) (Fig. 5A), but were significant in both glucose environments. In the in vitro scratch assay (Fig. 5B), KD9 cell migration (wound closure) was significantly slower compared to NC cell migration over 6 hours in both 25 and 5 mM glucose (p<0.01). In the absence of glucose or glutamine, wound closure was similar for both cell lines and the reduction in closure was marked in the absence of glutamine (Fig. 5B).
Figure 5. Metastatic profile of LDH-A knockdown (KD9) and control (NC) cells and tumors.
(A) Inhibition of LDH-A in KD9 tumor cells decreases their migration and invasiveness compared to NC tumor cells (* p<0.01 comparing NC and KD9; † p<0.01 comparing 5 and 25 mM glucose). (B) The effect of nutrient absence on wound healing is plotted (* p<0.01). (C) MR, H&E and India Ink imaging of lungs show extensive metastases in the control group (NC) compared to no visible metastases in the LDH-A knockdown group (KD9) at week three. (D) Percent of metastatic free lungs and survival of animals bearing NC and KD9 tumors is plotted vs. time (weeks after orthotopic inoculation). Survival was determined from the day of orthotopic tumor cell implantation until the day of death or day of euthanasia (primary tumor volume reached 1 cm3, or severe stress, weight loss, or immobility were noted). (E) Survival following surgical removal of the primary orthotopic tumor (100 mm3; ∼1-2 weeks after orthotopic inoculation). Animal survival is plotted vs. time (weeks after inoculation). (F) Western blots of LDH-A protein from primary tumors harvested at various times after orthotopic inoculation. All NC tumors had high expression of LDH-A. In contrast, small (∼100 mm3) KD tumors had low LDH-A, whereas larger KD tumors (∼250-1000 mm3) had high LDH-A expression, similar to NC tumors. (G) Western blots of LDH-A protein from distant metastases are shown. The primary tumor was removed surgically (at ∼100 mm3), and distant NC and KD9 lymph node and lung metastases were assayed; all metastases (both NC and KD9) were found to have high LDH-A expression.
Metastatic potential of KD9 and NC tumors
We tested the effects of LDH-A knockdown on the development of metastases by comparing orthotopic KD9 and NC primary tumors. All animals bearing orthotopic NC tumors developed lung metastases 1-3 weeks after tumor inoculation, whereas no animals bearing orthotopic KD9 tumors developed visible metastases over the first three weeks. This MRI observation was confirmed by India ink injection and histology in one set (5 animals per group) of animals that were sacrificed and examined after 21 days of primary tumor growth (Fig. 5C). The mean lung weight for animals bearing primary KD9 tumors was 0.16±0.03g, whereas the corresponding lung weight of NC tumor-bearing animals was 0.27±0.03g (p<0.01) (Fig. S3). In another cohort of animals, MRI-identified lung metastases and survival were monitored. A 2-to-3 week delay in the development of lung metastases and survival were observed between KD9 and NC tumor bearing mice (Fig. 5D).
The growth of primary NC and 4T1 tumors were significantly more rapid than that of primary KD9, KD5 and KD317 tumors (Fig. 4A, Table 2). NC and KD9 cells cultured for 2 weeks in DME medium with 5 mM glucose were also injected orthotopically into the mammary fat pad. These tumor growth profiles and calculated tumor doubling times were similar to that obtained when the glucose concentration in the culture medium was 25 mM (Table 2).
To control for the effect of differing tumor volumes and growth rates, and to more closely simulate the clinical situation where the primary tumor is removed, we studied a separate group of animals (10/group) and surgically removed the primary mammary tumor (tumor volume ∼100 mm3). The NC tumor-resected animals began to die 4 weeks after tumor inoculation, whereas the KD9 tumor-resected animals began to die 6 weeks after tumor inoculation (Fig. 5E). A 2 week increase was observed in the survival of the KD9 tumor-resected animals, compared to NC tumor-resected animals (Fig. 5E). The mean survival time for tumor resected animals was 33.5±6 days for NC and 44±7 days for the KD9 (p<0.01).
LDH-A expression in NC and KD9 tumors and metastases
LDH-A protein (immunoblotting) levels in small (∼100 mm3) KD9 tumors were ∼4 fold less than in comparable, small size NC tumors (p<0.01, Fig. 4D, F; Fig. 5F). However, there was a reappearance of LDH-A protein expression in KD9 tumors as they enlarged (from ∼100 to 250-300 mm3), and LDH-A protein levels in the larger KD9 and NC tumors were similar (Fig. 5F). Metastases to lymph nodes and lungs, as well as recurrent tumor at the primary site (KD9 and NC) had similar high expression of LDH-A (Fig. 5F, G). Thus, the larger KD9 and NC tumors, and metastatic nodules from both tumors, had similar LDH-A protein expression levels.
Discussion
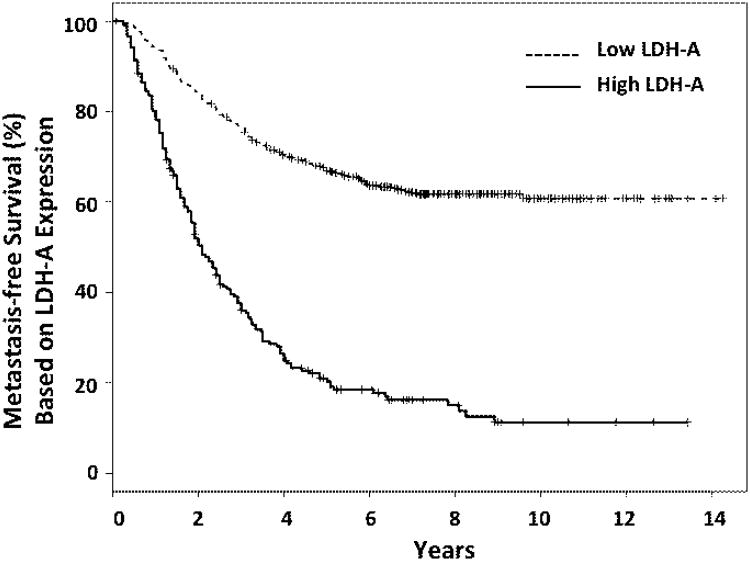
Tumors with high tissue lactate concentrations and high LDH-A expression have been linked to poor prognosis (22-25), and are associated with greater metastatic potential (24, 25). Shifts in metabolism have been shown to have a significant impact on the tumor microenvironment, disease evolution, progression, and development of metastases (1, 3-6, 26-28). In our analysis of 4 clinical data sets (622 breast cancer patients) (14-17), we show that patients with high levels of LDH-A expression have a significantly higher probability (p<10−16) of developing metastases compared to women with low levels of LDH-A (Fig. 6).
Figure 6. Metastatic free breast cancer survival: LDH-A gene expression.
Kaplan-Meier estimators for metastasis free survival from a compendium of four breast cancer patient datasets (14-17). Survival data were separated into low-medium and high groups according to the expression level of LDHA: 2/3 of data-points are in the low group (dashed line) and 1/3 are in the high group (solid line). Patients with high levels of LDH-A expression have a significantly higher (p<10−16) probability of developing metastases compared with women with low levels of LDH-A.
We also focused on LDH-A, because it is a bridge between several metabolic pathways, and because the product of LDH-A activity (lactate) can be assessed non-invasively and quantitatively using MRSI. Although it has been shown that the inhibition of LDH-A has an anti-proliferative effect on primary breast tumors (5), human alveolar adenocarcinoma A549 xenografts (29), and human hepatocellular carcinoma HCCLM3 xenografts (30), there have been no studies investigating the effect of LDH-A inhibition on the development and growth of metastases. This is an important clinical issue, since the cause of death in breast cancer patients is almost always due to metastases.
The inhibition of LDH-A by a small molecule inhibitor, FX-11, reduces progression of human lymphoma P493 xenografts (4). The inhibition of pyruvate conversion to lactate by oxamate (pyruvate analog) also re-sensitizes taxol-resistant human MDA-MB-435 breast tumor xenografts (31). The inhibition of LDH-A may enhance oxidative stress, and is linked to tumor cell death (4, 5, 29, 30). We previously measured greater amounts of LDH-A protein and lactate production in 4T1 cells and tumors (metastatic phenotype) compared to isogenic 67NR cells and tumors (non-metastatic phenotype) (10). We selected the murine 4T1 breast cancer metastatic model to test whether LDH-A silencing, using shRNA-knockdown technology, has a significant effect on the development of metastases, as well as on cell/tumor metabolism and cell/tumor growth, and can we measure these changes non-invasively by MRSI using lactate as a surrogate marker of LDH-A.
We show a close association between LDH-A gene and protein expression, LDH enzyme activity, cell proliferation and transwell migration assays between KD9 and NC cells. In vitro experiments also show a reduction in the rate of glucose utilization and glycolysis (ECAR), and a compensatory increase in oxygen consumption (OCR), maximum respiratory capacity, ROS and cellular ATP levels (reflecting increased oxidative phosphorylation) following stable LDH-A shRNA knockdown in 4T1 cells. Glucose and glutamine dependence studies were also performed, and they demonstrated that glucose metabolism (not glutamine metabolism) is the primary source of extracellular acidification and aerobic glycolysis. Glutamine is essential for cell proliferation and wound healing (scratch assay), and may have a significant impact on mitochondrial activity of LDH-A knockdown cells.
4T1 cells and variants reported here (NC, KD9, etc.) were grown in DMEM with high and normal concentrations of glucose (25 and 5 mM, respectively) and glutamine (6 mM). Many tumor cell lines are routinely cultured in media containing high amounts of glucose and glutamine (32-34), ensuring the survival of most cells over longer periods of cell culture, without the need to monitor glucose or glutamine concentration. However, most healthy, non-diabetic adults maintain fasting glucose levels at about 5 mmol/L and glutamine levels at 1 mM/L (35-38), considerably less than that in most cell culture media. Therefore, we performed comparison studies of cells growing in 25 and 5 mM glucose, since it is known that “hyperglycemic” conditions may lead to changes in cellular functions, such as carbohydrate and fatty acid metabolism, proliferation and cell motility (39). In addition, glucose concentrations in tumors can vary considerably in different tumor regions during tumor progression, and can be significantly lower than that in normal tissues (40). Thus, the tumor microenvironment can be variable and characterized as being “glucose starved” to “glucose addicted” in comparison to other tissues, reflecting an imbalance between poor supply and high consumption rate. Despite the differences in glucose and glutamine concentrations between our in vitro and in vivo experiments, we found no significant differences in the tumor growth and metastatic ability when we used cells that had been adapted to 5 mM glucose over 2 weeks. These 5 mM glucose-adapted NC and KD9 cells had similar in vitro and in vivo growth (doubling times) and similar metastatic patterns compared to cells grown in 25 mM glucose.
A close relationship between LDH-A gene and protein expression, tissue lactate levels, and tumor metastatic potential was shown. Significant in vivo biological effects of LDH-A silencing were demonstrated, which included, decreased lactate production in vivo, the slowing of tumor growth and a reduction and delay in the development of metastases of several KD clones (KD9, KD5, KD317). Therefore, we explored the evolution of LDH-A protein expression in primary orthotopic KD9 and NC tumors, and performed immunoblots on enlarging tumors (from ∼100 to 250-300 mm3), and on metastases in the same animals. Most notable were the similar high LDH-A protein levels that were measured in the larger primary KD9 and NC tumors, and in the metastatic lesions as well. These data, along with our previous studies (10), suggest that small (<100 mm3) wild-type 4T1 and NC tumors (with high LDH-A expression and high lactate levels) are already seeding metastatic cells into the circulation. Nevertheless, small KD9 tumors (with lower LDH-A expression and only moderate lactate levels) are capable of forming metastases, but at a delayed rate.
The eventual appearance of delayed metastases in KD tumor bearing mice may be explained in several ways, including the fact that LDH-A silencing was incomplete. We have seen re-expression of LDH-A protein in KD cells, when cells were cultured without antibiotic selection (Fig. S4). We show an outgrowth of non-LDH-A silenced KD9 cells in enlarging primary KD9 tumors (250-300 mm3) that lead to higher lactate levels, and the presence of high LDH-A protein levels in distant KD9 metastases, similar to that measured in distant NC metastases.
Re-appearance of LDH-A in vivo could also explain the moderate levels of lactate in small (100 mm3) primary KD9 tumors, resulting in failure to more effectively suppress tumor growth and development of metastases. Other explanations for the growth and metastatic profile of KD9 tumors include the fact that the KD9 cells use oxidative metabolism more than NC or wild-type 4T1 cells to produce metabolic intermediates, and they are less dependent on glycolysis (greater use of the TCA cycle and oxidative phosphorylation). In addition, enhanced mitochondrial oxidative activity and elevated ROS can induce a more aggressive tumor phenotype through hypoxia inducible factor 1 (HIF-1)(41, 42), as well as increased oxidative stress and cell death (41).
Other possible factors include the use of other metabolic fuels, such as glutamine, which could feed in through the TCA cycle and provide metabolic intermediates (43). Stromal tissue surrounding the tumor cells can also play an important role in tumor progression and metastasis (44). This mechanism could account for KD9 tumor growth and metastases, where the production of either lactate or pyruvate by the tumor stroma could be used to “feed” the malignant tumor cells and to alter the microenvironment (45). The production of lactate by tumor stromal cells (requiring LDH-A) would not be affected by LDH-A silencing in tumor KD9 cells, since the stromal cells originate from the host animal. Thus, several possible explanations exist that could account for the modest and increasing levels of LDH-A expression and lactate concentration in small and larger KD9 tumors. Such changes would be consistent with the slower growth profile of primary orthotopic KD9 breast tumors and their delayed ability to form metastases.
Imaging paradigms developed in this study can be translated to the clinic for selecting patients at high risk for developing subsequent metastases and who need closer surveillance, or patients appropriate for treatment with metabolic inhibitors. The non-invasive monitoring of LDH-A targeted therapy using lactate MRSI is novel, even though lactate 1H MRSI has been performed in a limited number of oncology studies previously (9, 10, 13, 46, 47). In contrast to hyperpolarized 13C NMR studies, lactate 1H MRSI measurements do not require special instrumentation and injection of hyperpolarized molecules. Pulse sequences for lactate detection have been implemented on clinical scanners (46-49). Therefore, the barrier to translation is modest and these studies can be implemented with currently available technology. These pre-clinical studies support the wider clinical application of lactate 1H MRSI, including the identification and monitoring of women with breast cancer who are at high risk of developing metastatic disease, and to specifically monitor LDH-A targeted drug treatment.
Conclusions
Since elevated LDH-A is a component of many aggressive tumors (4-6, 22, 28), it is a potential drug target for cancer therapy (4, 31). Our results show for the first time that LDH-A inhibition reduces and delays the development of metastases, effects tumor cell metabolism, and confirms the reduction of primary tumor growth in an established murine model of breast cancer (4T1). Our LDH-A shRNA silencing experiments show a strong association between LDH-A gene and protein expression, tumor lactate levels and development of metastases. LDH-A targeted drug therapy may avoid the loss of shRNA-based LDH-A silencing, that we observed, and it will also affect both tumor and stromal components.
These results suggest that LDH-A drug-targeted therapy is likely to be effective in aggressive breast cancer, and that lactate MRSI could serve as a surrogate for measuring LDH-A expression and target inhibition in the development of LDH-A targeted therapies. Inhibition of LDH-A in patients is expected to be tolerable and associated with low toxicity. LDH-A deficiency is a rare, but well-characterized human disease that can lead to exercise intolerance, cramps, myoglobinuria, but is not associated with severe dysfunction of major organs (50, 51).
Supplementary Material
Translational Relevance.
Elevated LDH-A expression is characteristic of many aggressive tumors and is associated with development of metastases. We show that decreased lactate concentration alters tumor cell metabolism, delays metastases and reduces tumor growth rate, and that LDH-A breakthrough is associated with disease progression. Our focus on tumor LDH-A and non-invasive monitoring of LDH-A targeted therapy using lactate MRSI, where lactate concentration is a potential surrogate marker for LDH-A expression, is novel and extends our prior studies. We suggest that this imaging, treatment, and monitoring strategy can be translated to the clinic, where lactate MRSI can be used to identify and monitor women at high risk of developing metastatic disease, and to monitor LDH-A targeted drug treatment.
Acknowledgments
Grant Support: This work was supported in part by grants entitled Breast Cancer Molecular Imaging Fund (JA Koutcher), P01-CA94060, (JA Koutcher); P30 CA08748 (JA Koutcher); DOD BCRP W81XWH-09-1-0042 (S Thakur) and MSKCC Center for Molecular Imaging in Cancer 2P50CA086438 (RG Blasberg).
Research funding: Breast Cancer Molecular Imaging Fund, P01-CA94060, P30 CA08748, DOD BCRP W81XWH-09-1-0042 and MSKCC Center for Molecular Imaging in Cancer (2P50CA086438).
Footnotes
Conflict of Interest: None.
- Conception/design: Asif Rizwan, Inna Serganova, Ronald Blasberg, Jason Koutcher
- Development of methodology: Asif Rizwan, Inna Serganova
- Acquisition of data: Asif Rizwan, Inna Serganova, Raya Khanin, Xiaohui Ni, Hazem Karabeber
- Analysis and interpretation of data: Asif Rizwan, Inna Serganova, Raya Khanin, Xiaohui Ni, Sunitha Thakur, Hazem Karabeber, Kristen L. Zakian, Ronald Blasberg, and Jason A. Koutcher
- Writing, review, and/or revision of the manuscript: Asif Rizwan, Inna Serganova, Ronald Blasberg, and Jason A. Koutcher
- Administrative, technical, or material support: Ronald Blasberg, and Jason A. Koutcher
- Study supervision: Ronald Blasberg, and Jason A. Koutcher
- Pathological diagnosis, analyzation, and interpretation of the immunohistochemical data: Asif Rizwan
- Carrying out experiments and analyzing data: Asif Rizwan, Inna Serganova
References
- 1.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 2.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nature reviews Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 3.Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2008;49(2):24S–42S. doi: 10.2967/jnumed.107.047258. [DOI] [PubMed] [Google Scholar]
- 4.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107:2037–42. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–44. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.He Q, Shungu DC, van Zijl PC, Bhujwalla ZM, Glickson JD. Single-scan in vivo lactate editing with complete lipid and water suppression by selective multiple-quantum-coherence transfer (Sel-MQC) with application to tumors. J Magn Reson B. 1995;106:203–11. doi: 10.1006/jmrb.1995.1035. [DOI] [PubMed] [Google Scholar]
- 10.Serganova I, Rizwan A, Ni X, Thakur SB, Vider J, Russell J, et al. Metabolic imaging: a link between lactate dehydrogenase A, lactate, and tumor phenotype. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:6250–61. doi: 10.1158/1078-0432.CCR-11-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–405. [PubMed] [Google Scholar]
- 12.Mehrara E, Forssell-Aronsson E, Ahlman H, Bernhardt P. Specific growth rate versus doubling time for quantitative characterization of tumor growth rate. Cancer research. 2007;67:3970–5. doi: 10.1158/0008-5472.CAN-06-3822. [DOI] [PubMed] [Google Scholar]
- 13.Yaligar J, Thakur SB, Bokacheva L, Carlin S, Thaler HT, Rizwan A, et al. Lactate MRSI and DCE MRI as surrogate markers of prostate tumor aggressiveness. NMR Biomed. 2012;25:113–22. doi: 10.1002/nbm.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 16.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A. 2007;104:6740–5. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–17. [Google Scholar]
- 19.Moran M, Rivera H, Sanchez-Arago M, Blazquez A, Merinero B, Ugalde C, et al. Mitochondrial bioenergetics and dynamics interplay in complex I-deficient fibroblasts. Biochimica et biophysica acta. 2010;1802:443–53. doi: 10.1016/j.bbadis.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Griguer CE, Oliva CR, Gillespie GY. Glucose metabolism heterogeneity in human and mouse malignant glioma cell lines. J Neurooncol. 2005;74:123–33. doi: 10.1007/s11060-004-6404-6. [DOI] [PubMed] [Google Scholar]
- 21.Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor's dilemma? Biochim Biophys Acta. 2011;1807:552–61. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Koukourakis MI, Giatromanolaki A, Simopoulos C, Polychronidis A, Sivridis E. Lactate dehydrogenase 5 (LDH5) relates to up-regulated hypoxia inducible factor pathway and metastasis in colorectal cancer. Clin Exp Metastasis. 2005;22:25–30. doi: 10.1007/s10585-005-2343-7. [DOI] [PubMed] [Google Scholar]
- 23.Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer research. 2006;66:632–7. doi: 10.1158/0008-5472.CAN-05-3260. [DOI] [PubMed] [Google Scholar]
- 24.Ryberg M, Nielsen D, Osterlind K, Andersen PK, Skovsgaard T, Dombernowsky P. Predictors of central nervous system metastasis in patients with metastatic breast cancer. A competing risk analysis of 579 patients treated with epirubicin-based chemotherapy. Breast Cancer Res Treat. 2005;91:217–25. doi: 10.1007/s10549-005-0323-x. [DOI] [PubMed] [Google Scholar]
- 25.Koukourakis MI, Giatromanolaki A, Sivridis E, Bougioukas G, Didilis V, Gatter KC, et al. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br J Cancer. 2003;89:877–85. doi: 10.1038/sj.bjc.6601205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brizel DM, Schroeder T, Scher RL, Walenta S, Clough RW, Dewhirst MW, et al. Elevated tumor lactate concentrations predict for an increased risk of metastases in head-and-neck cancer. International journal of radiation oncology, biology, physics. 2001;51:349–53. doi: 10.1016/s0360-3016(01)01630-3. [DOI] [PubMed] [Google Scholar]
- 27.Walenta S, Chau TV, Schroeder T, Lehr HA, Kunz-Schughart LA, Fuerst A, et al. Metabolic classification of human rectal adenocarcinomas: a novel guideline for clinical oncologists? J Cancer Res Clin Oncol. 2003;129:321–6. doi: 10.1007/s00432-003-0450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walenta S, Wetterling M, Lehrke M, Schwickert G, Sundfor K, Rofstad EK, et al. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60:916–21. [PubMed] [Google Scholar]
- 29.Seth P, Grant A, Tang J, Vinogradov E, Wang X, Lenkinski R, et al. On-target inhibition of tumor fermentative glycolysis as visualized by hyperpolarized pyruvate. Neoplasia. 2011;13:60–71. doi: 10.1593/neo.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng SL, Liu JJ, Dai YH, Sun XG, Xiong XP, Huang G. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. The FEBS journal. 2012;279:3898–910. doi: 10.1111/j.1742-4658.2012.08748.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhou M, Zhao Y, Ding Y, Liu H, Liu Z, Fodstad O, et al. Warburg effect in chemosensitivity: targeting lactate dehydrogenase-A re-sensitizes taxol-resistant cancer cells to taxol. Molecular cancer. 2010;9:33. doi: 10.1186/1476-4598-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–62. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nature cell biology. 2012;14:1295–304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang X, Zhuang X, Ju S, Zhang S, Jiang H, Mu J, et al. miR-155 promotes macroscopic tumor formation yet inhibits tumor dissemination from mammary fat pads to the lung by preventing EMT. Oncogene. 2011;30:3440–53. doi: 10.1038/onc.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tannock IF, Steele D, Roberts J. Influence of reduced concentration of L-glutamine on growth and viability of cells in monolayer, in spheroids, and in experimental tumours. British journal of cancer. 1986;54:733–41. doi: 10.1038/bjc.1986.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coutinho M, Gerstein HC, Wang Y, Yusuf S. The relationship between glucose and incident cardiovascular events. A metaregression analysis of published data from 20 studies of 95,783 individuals followed for 12.4 years. Diabetes care. 1999;22:233–40. doi: 10.2337/diacare.22.2.233. [DOI] [PubMed] [Google Scholar]
- 37.Elia M, Neale G, Livesey G. Alanine and glutamine release from the human forearm: effects of glucose administration. Clinical science. 1985;69:123–33. doi: 10.1042/cs0690123. [DOI] [PubMed] [Google Scholar]
- 38.Minokoshi Y, Alquier T, Furukawa N, Kim YB, Lee A, Xue B, et al. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature. 2004;428:569–74. doi: 10.1038/nature02440. [DOI] [PubMed] [Google Scholar]
- 39.Sedoris KC, Thomas SD, Miller DM. c-myc promoter binding protein regulates the cellular response to an altered glucose concentration. Biochemistry. 2007;46:8659–68. doi: 10.1021/bi7003558. [DOI] [PubMed] [Google Scholar]
- 40.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer research. 2009;69:4918–25. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 41.Klimova T, Chandel NS. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ. 2008;15:660–6. doi: 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- 42.Patten DA, Lafleur VN, Robitaille GA, Chan DA, Giaccia AJ, Richard DE. Hypoxia-inducible factor-1 activation in nonhypoxic conditions: the essential role of mitochondrial-derived reactive oxygen species. Mol Biol Cell. 2010;21:3247–57. doi: 10.1091/mbc.E10-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–24. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Kruijf EM, van Nes JG, van de Velde CJ, Putter H, Smit VT, Liefers GJ, et al. Tumor-stroma ratio in the primary tumor is a prognostic factor in early breast cancer patients, especially in triple-negative carcinoma patients. Breast cancer research and treatment. 2011;125:687–96. doi: 10.1007/s10549-010-0855-6. [DOI] [PubMed] [Google Scholar]
- 45.Rattigan YI, Patel BB, Ackerstaff E, Sukenick G, Koutcher JA, Glod JW, et al. Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and glycolytic tumor cells in the tumor microenvironment. Experimental cell research. 2012;318:326–35. doi: 10.1016/j.yexcr.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mellon EA, Lee SC, Pickup S, Kim S, Goldstein SC, Floyd TF, et al. Detection of lactate with a hadamard slice selected, selective multiple quantum coherence, chemical shift imaging sequence (HDMD-SelMQC-CSI) on a clinical MRI scanner: Application to tumors and muscle ischemia. Magn Reson Med. 2009;62:1404–13. doi: 10.1002/mrm.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adalsteinsson E, Spielman DM, Pauly JM, Terris DJ, Sommer G, Macovski A. Feasibility study of lactate imaging of head and neck tumors. NMR Biomed. 1998;11:360–9. doi: 10.1002/(sici)1099-1492(1998110)11:7<360::aid-nbm518>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 48.Pan JW, Hamm JR, Hetherington HP, Rothman DL, Shulman RG. Correlation of lactate and pH in human skeletal muscle after exercise by 1H NMR. Magn Reson Med. 1991;20:57–65. doi: 10.1002/mrm.1910200107. [DOI] [PubMed] [Google Scholar]
- 49.Smith MA, Koutcher JA, Zakian KL. J-difference lactate editing at 3.0 Tesla in the presence of strong lipids. J Magn Reson Imaging. 2008;28:1492–8. doi: 10.1002/jmri.21584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spriet LL, Howlett RA, Heigenhauser GJ. An enzymatic approach to lactate production in human skeletal muscle during exercise. Med Sci Sports Exerc. 2000;32:756–63. doi: 10.1097/00005768-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 51.Maekawa M, Sudo K, Li SS, Kanno T. Genotypic analysis of families with lactate dehydrogenase A (M) deficiency by selective DNA amplification. Hum Genet. 1991;88:34–8. doi: 10.1007/BF00204925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.