Abstract
Protease inhibitor 6 (PI-6/SERPINB6) is a widely expressed nucleocytoplasmic serpin. It inhibits granulocyte cathepsin G and neuronal neuropsin, and it is thought to protect cells from death caused by ectopic release or internalization of protease during stress such as infection or cerebral ischemia. To probe the biological functions of PI-6, we generated mice lacking its ortholog (SPI3/Serpinb6). SPI3-deficient mice developed normally and were fertile, and no abnormal pathology or increased sensitivity to cerebral ischemia was observed. There were no perturbations in leukocyte development or numbers, and recruitment of leukocytes to the peritoneal cavity was normal. SPI3-deficient mice were equally susceptible as wild-type mice to systemic Candida albicans infection, although there was a slight decrease in the ability of neutrophils from SPI3-deficient mice to kill C. albicans in vitro. Increased levels of a related inhibitor Serpinb1 (monocyte/neutrophil elastase inhibitor) in the tissues of targeted mice suggests that compensation by other serpins reduces the impact of SPI3 deficiency in these animals and may explain the lack of a more obvious phenotype.
Serine proteases are involved in numerous physiological processes, including coagulation, cell migration, phagocytosis, fibrinolysis, cell-mediated cytotoxicity, and complement fixation. To avoid nonspecific tissue damage, protease activity is often regulated by inhibitors, such as members of the serpin superfamily. Inhibitory serpins bind their target proteases irreversibly, via a conformational change which deforms the protease and stabilizes the serpin-protease complex (20). Serpin dysfunction or deficiency is the underlying factor in a variety of human diseases, including emphysema, angioedema, thrombosis, and dementia (39, 40).
Serpins are grouped into different phylogenic clades (21). In humans, the largest clade (clade B) contains 13 largely intracellular proteins, among which are serine proteinase inhibitors and cysteine proteinase inhibitors (39). Unlike the plasma or neural serpins, there are no known naturally occurring clade B serpin mutants. This has hindered elucidation of their functions, but proposed physiological roles include the regulation of cell growth and differentiation and cytoprotection (39). For example, maspin is an antiangiogenic tumor suppressor, megsin is a differentiation factor, PAI-2 protects retinoblastoma protein from degradation (13), and several clade B serpins (PAI-2, SCCA-1, SCCA-2, bomapin, and hurpin) protect cells from tumor necrosis factor alpha- or UV-induced cell death. So far, PAI-2-deficient mice represent the only clade B serpin knockouts, and these animals show no obvious defects (15).
A distinct subset of clade B serpins comprises proteinase inhibitor-6 (PI-6), PI-9, and the monocyte/neutrophil elastase inhibitor (MNEI). These inhibitors are encoded by a gene cluster on chromosome 6 (36), and heterozygous deletion of this region is associated with congenital craniofacial deformity, encephalopathy, and cervical carcinoma (14). All three are nucleocytoplasmic proteins (3) that inhibit leukocyte serine proteinases, and it has been suggested that they protect both protease-producing leukocytes and bystanders from adventitious proteolysis during normal function or stress (5). For example, PI-9 inhibits the cytotoxic lymphocyte protease granzyme B. It is present in the nucleocytoplasm of cytotoxic lymphocytes and surrounds granules containing granzyme B, it is up-regulated during degranulation, and it protects cells against granzyme B-mediated apoptosis (4, 18). A similar cytoprotective role can be envisaged for PI-6 and MNEI, which are also nucleocytoplasmic, are coexpressed in leukocytes, and have overlapping inhibitory profiles.
PI-6 is a potent inhibitor of the monocyte/granulocyte protease cathepsin G, which is stored in azurophilic granules and then released into phagolysosomes or secreted during inflammation. PI-6 is present in epithelial cells, endothelial cells, myeloid cells, and platelets (10, 35, 37). In neutrophils, it is nucleocytoplasmic and surrounds cathepsin G-containing granules (C. Bird, unpublished observations). It may therefore protect cells from death induced by ectopic cathepsin G, particularly as this protease can activate procaspase 7 (44). PI-6 also inhibits kallikreins, a family of serine proteinases exemplified by the prostate-specific antigen (PSA). It is coexpressed with kallikreins in the prostate and ovary, and it is found complexed with kallikreins in samples from prostatic tumors (30). It is also is an inhibitor of neuropsin (kallikrein 8), which is present in the brain and skin (24). Neuropsin processes extracellular matrix, is important for neuronal plasticity, and is significantly up-regulated in cases of Alzheimer's disease (17, 38). Coexpression of PI-6 and neuropsin occurs in developing and adult brains (24, 25), and PI-6 is up-regulated following ischemia (31), suggesting that it protects against ectopic neuropsin. As exemplified by PSA (9), kallikreins are stored in cytoplasmic storage vesicles analogous to leukocyte granules, so PI-6 may protect against kallikrein leakage from storage vesicles or from protease that is reinternalized after release.
To probe the physiological functions of PI-6, we generated mice deficient in its ortholog, SPI3. These mice allowed us to examine the effects of the loss of SPI3 in different cells under various types of cellular stress.
MATERIALS AND METHODS
Antibodies.
Rabbit anti-SPI3 polyclonal sera and 1F3, a monoclonal antibody that recognizes the hinge region of many serpins, have been described previously (19, 33). The following rat monoclonal antibodies were used in flow cytometry: anti-mouse CD45R/B220-phycoerythrin (PE), anti-mouse CD4-PE, and anti-mouse CD11b (Mac-1)-PE (BD Pharmingen, San Diego, Calif.); and anti-mouse CD8α-Spectral Red and anti-mouse Ly-6G (Gr-1)-Spectral Red (Southern Biotech, Birmingham, Ala.). The rat anti-green fluorescent protein (GFP) monoclonal antibody was kindly provided by Shinobu Fujita (Mitsubishi Kasei Institute of Life Sciences, Tokyo, Japan). A rabbit anti-human MNEI antibody, which also recognizes mouse serpinb1/elastase inhibitor A (EIA), was kindly provided by E. Remold-O'Donnell (Center for Blood Research, Harvard University, Boston, Mass.). A goat anti-human actin antibody, which cross-reacts with mouse actin, was from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.). A mouse monoclonal antibody to cytochrome c was from Research Diagnostics Inc. (Flanders, N.J.). Anti-rabbit immunoglobulin (Ig)-horseradish peroxidase (HRP) and anti-sheep/goat Ig-HRP conjugates were from Chemicon International (Temecula, Calif.). The anti-rat Ig-HRP conjugate was from Sigma (Sydney, Australia).
SPI3-deficient mice.
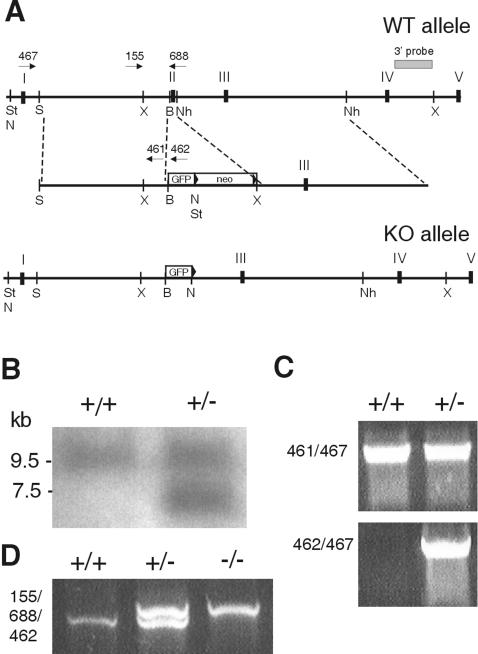
The strategy and targeting vector used to disrupt exon 2 of SPI3 and insert enhanced GFP (EGFP) have been described previously (33). Recombinants were analyzed by Southern blotting with a 32P-labeled 3′ external probe on XbaI-cleaved genomic DNA (Fig. 1). Positive clones were confirmed by using NotI or StuI cleavage with neomycin or EGFP probes (data not shown). PCRs with primers PB467 and PB462 or PB467 and PB461 (Table 1) confirmed that the 5′ end of the targeting construct had integrated correctly (Fig. 1). Chimeric mice were crossed to C57BL/6 Cre-deleter transgenic mice (34) to remove the neomycin cassette from the targeted allele. Screening PCRs on tail DNA from wild-type (+/+), heterozygous (+/−), and SPI3-deficient (−/−) mice used the primers 155 and 688 to amplify a 1.1-kb product from the wild-type allele and the primers 155 and 462 to amplify a 1.2-kb product from the targeted allele.
FIG. 1.
Targeted disruption of the SPI3 gene. (A) Exon 2 of the SPI3 gene was replaced with EGFP fused in frame with the SPI3 initiation codon. The pgk-loxPneo selection cassette was removed by crossing SPI3 heterozygotes with Cre-deleter mice. St, StuI; S, SmaI; X, XbaI; B, BamHI; Nh, NheI; N, NotI; WT, wild type; KO, knockout. (B) XbaI-cleaved DNA from wild-type cells (+/+) and recombinant cells (+/−) was hybridized to the 3′ probe, which distinguishes the wild-type allele (9.5 kb) from the targeted allele (7.5 kb). (C) To confirm correct integration of the 5′ arm, DNA from wild-type (+/+) and heterozygous (+/−) cells was subjected to PCR using primers 461 and 467, which amplify a 4-kb product from the wild-type allele, and primers 462 and 467, which amplify a 4-kb product from the targeted allele. (D) Wild-type (+/+), heterozygous (+/−), and knockout mice were typed by PCR of tail DNA with primers 152, 688, and 462 simultaneously.
TABLE 1.
PCR primers
| Name | Sequence | Target gene (direction) |
|---|---|---|
| PB155 | 5′-GAGATAGGCTCTCATAGTTG-3′ | SPI3 intron A (sense) |
| PB259 | 5′-ACAGCTGGCATGATGACG-3′ | SPI3 exon 7 (sense) |
| PB370 | 5′-GGCAATTGTGCTCAGGGAGAGGAGAACC-3′ | SPI3 3′ UTR (antisense) |
| PB461 | 5′-TGCTTCCTGTAGAGGATCCATGATGG-3′ | SPI3 exon 2 (antisense) |
| PB462 | 5′-ACGCTGAACTTGTGGCCGTTTACGTCG-3′ | EGFP (antisense) |
| PB467 | 5′-GGTTGGGGCCTCCTCATCTCTTCTCTG-3′ | SPI3 exon 1 (sense) |
| PB584 | 5′-GACCCCTTCATTGACCTCAAC-3′ | GAPDH (sense) |
| PB585 | 5′-GATGACCTTGCCCACAGCCTT-3′ | GAPDH (antisense) |
| PB688 | 5′-GGTGCCATTTGCTTCCTGTAGAGG-3′ | SPI3 exon 2 (antisense) |
RT-PCR.
Mice were killed by cervical dislocation, and RNA from various organs was extracted by using the RNAzol B RNA isolation system (Tel-Test). Reverse transcriptase PCR (RT-PCR) analysis was performed as described previously (23) with primers specific for the SPI3 reactive center loop and 3′ untranslated region (PB259 and PB370). Primers specific for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (PB584 and PB585) were used to assess the efficiency of cDNA synthesis. Samples that lacked RT were also subjected to PCR to control for contaminating genomic DNA.
Immunoblotting.
Organs from 8-week-old mice were homogenized in 10 mM Tris-HCl-0.15 mM NaCl (pH 7.6)-1% Triton X-100, containing the protease inhibitors aprotinin (1 μg/ml), phenylmethylsulfonyl fluoride (150 μg/ml), leupeptin (0.5 μM), and pepstatin (1 μM). Approximately 50 μg of lysates was separated on a 12.5% sodium dodecyl sulfate-polyacrylamide gel and transferred to nitrocellulose. Membranes were incubated overnight with the appropriate primary antibody, washed, and incubated for 1 h with HRP-conjugated secondary antibodies that were subsequently detected via chemiluminescence (Western Lightning; Perkin-Elmer Life Sciences Inc., Boston, Mass.).
Flow cytometry.
Blood was collected from mice by retro-orbital bleeding. Cell suspensions from the spleen, thymus, inguinal lymph nodes, and bone marrow from 8- to 10-week-old mice were prepared, and the erythrocytes in the spleen suspensions were lysed by incubation in 84 mM NH4Cl for 5 min at 37°C. Approximately 3 × 106 cells were stained with the various rat monoclonal antibodies described above, collected on a FACSCalibur flow cytometer, and analyzed using Cell Quest software. The numbers and distribution of various cell types in the lymph node, spleen, bone marrow, and thymus were assessed by using specific markers and flow cytometry (CD4+ CD8+ T cells, B cells, macrophages, and granulocytes). Full blood examinations were performed using a Coulter counter on heparinized blood obtained from retro-orbital bleeding. Differential white blood cell counts were performed on blood smears treated with Giemsa-May-Grunwald stain.
Leukocyte recruitment to the peritoneal cavity induced by sterile peritonitis.
Wild-type and SPI3-deficient mice were injected intraperitoneally with 1 ml of 3% sterile thioglycolate broth. Mice were killed 4 or 72 h after injection, and the cells of the peritoneal cavity were harvested with 5 ml of phosphate-buffered saline (PBS) and counted. Flow cytometry with Mac-1 and Gr-1 antibodies to identify elicited cells was performed as described above.
Infection of mice with Candida albicans.
An overnight culture of C. albicans (ATCC 10231) was grown in Sabouraud's broth, washed three times with PBS, and counted. Mice were injected intravenously in the tail vein with 100 μl of culture containing 2.5 × 106 CFU of C. albicans and monitored and weighed daily for 3 weeks. Surviving mice were killed and the kidneys, lungs, liver, and spleen were removed and weighed. Organs were homogenized in 2 ml of PBS, and various dilutions were plated onto Sabouraud's agar plates. Colonies were counted 2 days later, and yeast load was calculated and expressed as CFU per mg of tissue.
Elicitation of neutrophils with C. albicans culture filtrate.
An overnight culture of C. albicans (ATCC 18804) grown in Sabouraud's broth was washed three times with modified Hanks’ balanced salt solution (16) and counted, and the concentration was adjusted to 5 × 108 CFU/ml in modified Hanks’ balanced salt solution. The yeast was incubated at 37°C in a shaking incubator for a further 2 h, after which time the yeast was centrifuged at 700 × g for 5 min, and the supernatant was removed and filtered. Culture filtrate from approximately 2 × 108 to 3 × 108 CFU of C. albicans was injected intraperitoneally into mice in order to elicit neutrophils. Four hours later, the peritoneal cells were harvested and counted, and the concentration was adjusted to 5 × 106 cells/ml in complete media (CM) (RPMI containing 5% heat-inactivated fetal calf serum, 20 mM HEPES, 1% glutamine, and 1% sodium pyruvate) (41). Approximately 1 × 106 cells were stained with antibodies for Mac-1 and Gr-1 and analyzed by flow cytometry to assess the elicitation of neutrophils.
In vitro C. albicans killing assay. An overnight culture of C. albicans grown in Sabouraud's broth was washed three times in saline and counted. To determine whether there was a difference in the ability of peritoneal neutrophils to kill either the yeast or hyphal form of C. albicans, the C. albicans blastoconidia were converted to hyphae by incubating 1 × 107 CFU of the yeast in RPMI-10% fetal calf serum at 37°C and 5% CO2 for 2 h (6). Approximately 95% of the yeast converted to the hyphal form under these conditions. The hyphae were harvested by gently scraping them off the dish, centrifuged, and resuspended in CM. Yeast and hyphal forms of the C. albicans at 2 × 105 CFU/ml were opsonized by incubating in CM containing 2.5% fresh normal mouse sera at 24°C for 45 to 60 min. Approximately 5 × 105 filtrate-enriched neutrophils from the peritonea of wild-type or SPI3-deficient mice were incubated with 2 × 104 CFU of C. albicans yeast or hyphae in duplicate in a 96-well plate at 37°C for 2 h. The well contents were harvested, each well was washed with 4 × 200 μl of water, and the washes were combined. Various dilutions of the harvested wells were plated onto Sabouraud's agar plates and incubated at 37°C for 48 h, and the colonies were counted. Yeast or hyphae were incubated without peritoneal cells to determine 100% survival. The proportion of surviving C. albicans was calculated as follows: (average number of yeast colonies after exposure to peritoneal cells/average number of yeast colonies alone) × 100.
Induction of focal cerebral ischemia.
Induction of stroke by midcerebral artery occlusion was performed as previously described (11). Briefly, the right proximal common carotid artery was ligated in anesthetized mice, and then a 6-0 nylon monofilament with a heat-blunted tip was introduced into the distal internal carotid artery and advanced 11 to 12 mm distal to the carotid bifurcation. This resulted in the occlusion of the midcerebral artery, where it junctions off the Circle of Willis. Blood flow was monitored via a Perimed PX5010 laser doppler (PERIMED, Järfälla, Sweden). Twenty-four hours after surgery, the mice were killed and the brains were removed into ice-cold PBS and then sectioned coronally into 1-mm slices that were stained with 2% 2,3,5-triphenyltetrazolium chloride monohydrate in order to assess the infarct size. Images of sections were scanned onto an image scanner, and the area of ischemic damage was measured by an analytic imaging system.
RESULTS
SPI3-deficient mice.
To investigate the physiological role of SPI3, we generated mice lacking SPI3 (Fig. 1). In addition to disrupting exon 2 of the SPI3 gene, the targeting vector fused EGFP in frame with the initiation codon of SPI3 so that EGFP could be used to mark cells that normally express SPI3. Preliminary experiments showed that retention of the neomycin selectable marker gene in the mutated SPI3 allele results in dysregulated EGFP expression (33), so chimeric mice were crossed with transgenic Cre-deleter mice to remove the neomycin gene from all cells early in development. Heterozygous mice lacking neomycin were intercrossed to produce mice of all three genotypes.
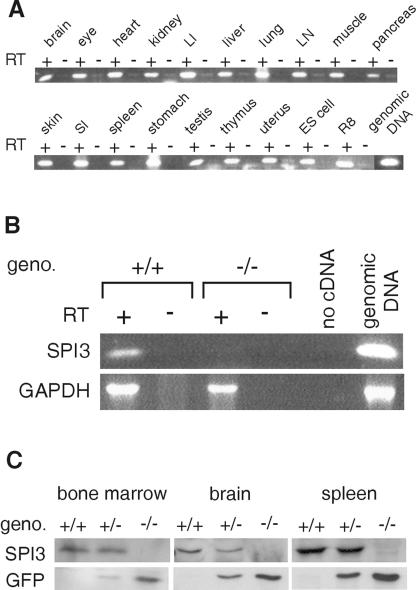
By RT-PCR analysis, SPI3 is found present in a wide range of cells and tissues (Fig. 2A). To demonstrate a SPI3 null mutation, RNA from the brains of wild-type and SPI3-deficient mice was compared. Figure 2B shows a product of the correct size in wild-type samples, whereas no product was amplified from the brains of SPI3-deficient mice. Immunoblotting of protein samples isolated from the tissues of wild-type, heterozygous, and SPI3-deficient mice showed that the 42-kDa SPI3 protein is present in wild-type and heterozygous mice but not in SPI3-deficient mice (Fig. 2C). The 27-kDa EGFP was also evident in these tissues in heterozygous and knockout mice but not in wild-type animals.
FIG. 2.
Distribution of SPI3 mRNA and protein. (A) RT-PCR analysis of SPI3 mRNA in normal C57BL/6 mouse tissues. cDNA synthesized in the presence (+) or absence (−) of RT was amplified with SPI3-specific primers 259 and 370. LI, large intestine; LN, lymph node; SI, small intestine; R8, mouse cytotoxic T-cell line; ES, embryonic stem cells. (B) cDNA synthesized in the presence (+) or absence (−) of RT from wild-type (+/+) and SPI3-deficient (−/−) mouse brains was amplified with SPI3-specific primers 259 and 370 (top panel) or GAPDH-specific primers (bottom panel). (C) Immunoblots of marrow, spleen, and brain protein from wild-type (+/+), heterozygous (+/−), and SPI3-deficient (−/−) mice were probed with either a rabbit anti-SPI3 serum (top panel) or a rat anti-GFP antibody (bottom panel).
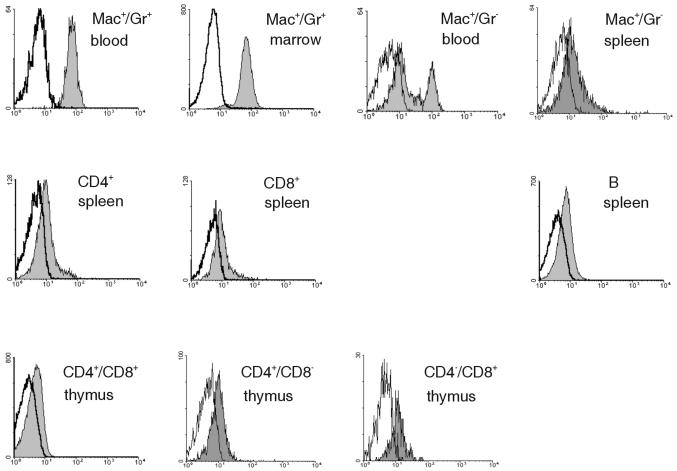
By immunoblotting and immunohistochemical analysis, EGFP expression also correlated with SPI3 expression in the heart, lung, stomach, small and large intestines, kidney, liver, and skin (data not shown). As shown in Fig. 3, flow cytometry demonstrated the highest levels of EGFP in granulocytes (Mac+/Gr+) from blood and bone marrow. Lower levels of EGFP were observed in blood monocytes (Mac+/Gr−) and splenic macrophages. Lower levels of EGFP were also evident in T and B cells in the central and peripheral lymphoid organs of SPI3-deficient mice. Overall, this pattern is consistent with the expression of PI-6 in human blood cells, which is found in low levels in lymphocytes and in higher levels in granulocytes and monocytes (18, 37).
FIG. 3.
Expression of GFP in immune cells. Leukocytes from the spleen, thymus, blood, or bone marrow were stained with the indicated antibodies and analyzed by flow cytometry. The appropriate cell populations were gated and analyzed for GFP expression.
Viability, growth, and fertility of SPI3-deficient mice.
Intercrossing of heterozygous mice generated a 40:78:35 Mendelian ratio of wild-type (+/+) to heterozygous (+/−) to homozygous (−/−) mice. SPI3-deficient mice were physically indistinguishable from their wild-type and heterozygous littermates, and both sexes were fertile and produced normal litter sizes. No increase in morbidity or mortality of the SPI3-deficient mice was observed up to 24 months of age, and histopathological analysis of various organs uncovered no obvious defects. We conclude that SPI3 is not required for the normal growth, development, and fecundity of mice.
Response to cerebral ischemia.
PI-6 and SPI3 are efficient inhibitors of neuropsin/kallikrein 8. SPI3 is coexpressed with neuropsin in various regions of the brain, particularly in neurons of the hippocampus (25), and its expression is up-regulated during ischemia (31). Other serpins also expressed in the rodent brain have been shown to protect neurons from damage induced by stroke (8, 42). To determine whether SPI3 is required to protect neurons from damage during ischemia, stroke was induced in SPI3-deficient mice by midcerebral artery occlusion. Twenty-four hours after surgery, the mice were killed, and the brains were examined to determine the volume of the infarct. There were no differences observed in the infarct volumes in the SPI3-deficient mice and wild-type mice (mean = 43 ± 9 mm3 for knockout mice [n = 6]; mean = 39 ± 4 mm3 for wild-type mice [n = 9]). This suggests that SPI3 does not protect neurons from the effects of ischemia.
Development of cells of the immune system.
Proteases such as elastase, proteinase 3, and cathepsin G are synthesized early in granulocyte maturation and have been implicated in the differentiation and mobilization of these cells (1, 27). Because PI-6 and SPI3 are cathepsin G inhibitors, we postulated that uncontrolled cathepsin G in SPI3-deficient mice may influence myeloid cell (or other leukocyte) differentiation or maturation. Cells in the lymph node, spleen, bone marrow, and thymus from five mice of each genotype were analyzed by flow cytometry. As shown in Table 2, there were no differences in the numbers or proportions of CD4+ T cells, CD8+ T cells, B cells, macrophages, or granulocytes in any organ, comparing those of wild-type mice to those of SPI3-deficient mice. Full blood examinations were performed on six wild-type and six SPI3-deficient mice, and no significant differences were observed in any of the parameters measured (Table 3). Manual differential white blood cell counts on blood smears also did not demonstrate any perturbations in the number and morphology of blood cells or platelets in the SPI3-deficient mice (data not shown).
TABLE 2.
Leukocytes in immune tissue
| Organ | Population | No. of leukocytes (106) for indicated phenotype
|
||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Spleen | Total | 98.2 ± 13 | 92.6 ± 20 | 102 ± 35 |
| CD4+ T cells | 13.9 ± 1.4 | 14 ± 2.8 | 15.3 ± 4.8 | |
| CD8+ T cells | 8.4 ± 0.8 | 8.3 ± 1.7 | 9.4 ± 2.8 | |
| B cells | 65 ± 12 | 60.9 ± 14 | 66.3 ± 24 | |
| Macrophages (Mac+/Gr−) | 5.5 ± 1.5 | 5.8 ± 1.8 | 5.4 ± 1.3 | |
| Neutrophils (Mac+/Gr+) | 1.9 ± 1 | 2.2 ± 0.7 | 1.7 ± 0.7 | |
| Lymph node | Total | 3.7 ± 1.1 | 4.5 ± 2.1 | 4 ± 1.6 |
| CD4+ T cells | 1.0 ± 0.3 | 1.4 ± 0.6 | 1.2 ± 0.5 | |
| CD8+ T cells | 0.8 ± 0.4 | 1.1 ± 0.5 | 1 ± 0.3 | |
| B cells | 1.8 ± 0.7 | 1.9 ± 1 | 1.7 ± 0.8 | |
| Thymus | Total | 90.4 ± 44 | 148 ± 61 | 132 ± 54 |
| CD4+/CD8− | 8 ± 4.1 | 11.7 ± 5.1 | 10 ± 3.2 | |
| CD4−/CD8+ | 2.9 ± 1.8 | 3.7 ± 1.4 | 3.3 ± 1.1 | |
| CD4+/CD8+ | 74.2 ± 36 | 123 ± 52 | 109 ± 47 | |
TABLE 3.
Full blood examinations
| Phenotype | Hemoglobin (g/liter) | Total white blood cells (109/liter) | Neutrophils (109/liter) | Lymphocytes (109/liter) | Monocytes (109/liter) | Eosinophils (109/liter) | Platelets (109/liter) |
|---|---|---|---|---|---|---|---|
| +/+ | 144.4 ± 5.9 | 9.0 ± 1.0 | 1.2 ± 0.4 | 7.2 ± 0.8 | 0.3 ± 0.2 | 0.3 ± 0.2 | 647.8 ± 141.1 |
| −/− | 141.7 ± 5.0 | 11.7 ± 2.1 | 2.0 ± 1.3 | 8.7 ± 1.8 | 0.6 ± 0.5 | 0.5 ± 0.2 | 664.7 ± 96.4 |
Leukocyte recruitment to the peritoneal cavity.
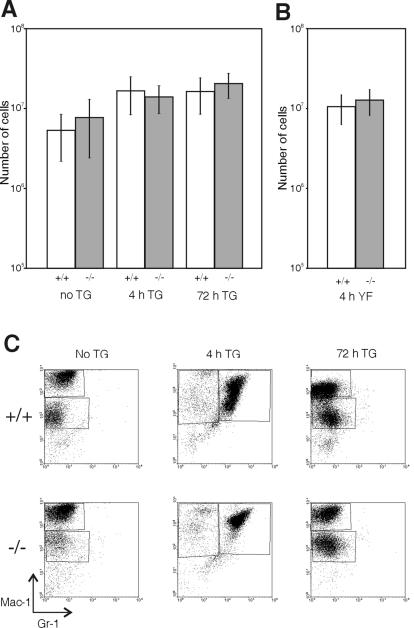
Cathepsin G is a granulocyte chemoattractant (7). To determine whether the lack of SPI3 affects recruitment of these cells into the peritoneal cavity during inflammation, wild-type and SPI3-deficient mice were injected with thioglycolate and killed either after 4 h to examine neutrophils or after 72 h to examine macrophages. As indicated by flow cytometry, there was no difference between wild-type and SPI3-deficient mice in the number (Fig. 4A) or type (Fig. 4C) of cells recruited to the peritoneal cavity or in the number of resident macrophages in noninjected mice.
FIG. 4.
Recruitment of cells to the peritoneal cavity. Wild-type (+/+) and SPI3-deficient (−/−) mice were injected intraperitoneally with either (A) thioglycolate (TG) (n = 11 to 20 for each genotype) or (B) C. albicans culture filtrate (YF) (n = 7 for each genotype) and were killed 4 or 72 h later, and the contents of the peritoneal cavity were harvested and counted. Peritoneal cells from noninjected mice (no thioglycolate) were also evaluated (n = 9 to 16 for each genotype). Results represent the mean ± one standard deviation. (C) Peritoneal cells were stained with antibodies to Mac-1 and Gr-1 and analyzed by flow cytometry to determine the cell types elicited.
To use a more physiological stimulus for leukocyte recruitment, mice were injected with culture filtrate from C. albicans, which has been shown to contain chemoattractants for polymorphonuclear leukocytes (16). The mice were killed 4 h later, and the peritoneal cells were analyzed as described above. There was no difference in the number of cells harvested from the peritoneal cavities of wild-type and SPI3-deficient mice (Fig. 4B) after culture filtrate injection nor was there any difference in the proportion of neutrophils recovered. However, the proportion of neutrophils elicited with yeast culture filtrate was generally lower than when mice were injected with thioglycolate, as 30 to 50% of peritoneal cells were Mac+/Gr+ 4 h after injection of culture filtrate compared to almost 100% of cells recovered 4 h after thioglycolate injection (data not shown).
In vitro killing of C. albicans.
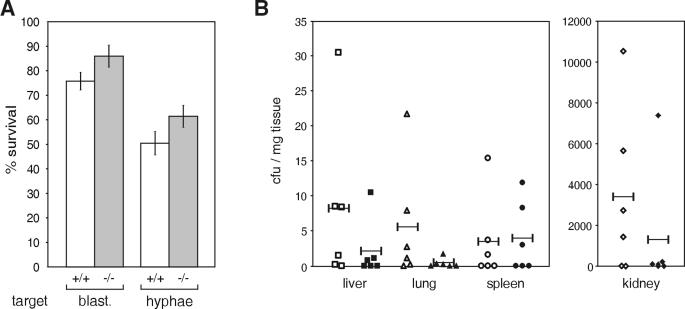
To examine whether lack of SPI3 affects neutrophil function, peritoneal neutrophils elicited with C. albicans culture filtrate from six wild-type and six SPI3-deficient mice were tested for their ability to kill either the blastoconidial or hyphal form of C. albicans in vitro. The hyphal form is the more virulent form of C. albicans (12). As shown in Fig. 5A, there was a consistent trend towards reduced killing of both blastoconidia and hyphae by neutrophils from SPI3-deficient mice (P = 0.11 for blastoconidia; P = 0.13 for hyphae [two-tail t test]). This suggests that neutrophils lacking the cytoprotective serpin SPI3 are not as robust when producing antimicrobial proteases during an immune response.
FIG. 5.
Response to C. albicans in vitro and in vivo. (A) Wild-type (open bar) and SPI3-deficient (grey bar) mice were injected intraperitoneally with culture filtrate from C. albicans. Neutrophils were isolated from the peritoneal cavity 4 h later and tested for the ability to kill either the blastoconidial or hyphal form of C. albicans. Results represent the mean (± one standard error of the mean) (n = 6 for each genotype) proportion of C. albicans surviving after being incubated with peritoneal cells compared to that of C. albicans incubated in the absence of cells. (B) Wild-type (open symbols) and SPI3-deficient (closed symbols) mice were injected intravenously with 2.5 × 106 CFU of C. albicans and killed 3 weeks later, and the yeast load in the liver, lungs, spleen, and kidneys was determined. Yeast load is expressed as CFU/mg of tissue, each symbol represents data from one mouse, and the bar represents the mean for each group (n = 6 for each genotype).
Response to systemic C. albicans infection.
Resolution of C. albicans infection depends mainly on neutrophils and macrophages without generation of an adaptive immune response (22, 26, 29, 32). Lack of control of cathespin G by SPI3 in this context may result in premature phagocyte death and decreased killing potency. To determine whether the lack of SPI3 in neutrophils and macrophages has any affect on the ability of the leukocytes to clear Candida, 10 wild-type and 9 SPI3-deficient mice were injected intravenously with C. albicans and monitored over 3 weeks. There was no difference in the survival rates of the wild-type and knockout mice, with 40% of wild-type mice and 33% of SPI3-deficient mice dying by day 3. The remaining mice survived for 3 weeks after infection, and there was no statistically significant difference (Mann-Whitney test) in the average Candida load for all organs of the wild-type and SPI3-deficient mice (Fig. 5B), suggesting that the lack of SPI3 in neutrophils and macrophages does not impair the clearance of the yeast C. albicans.
Compensation by other serpins.
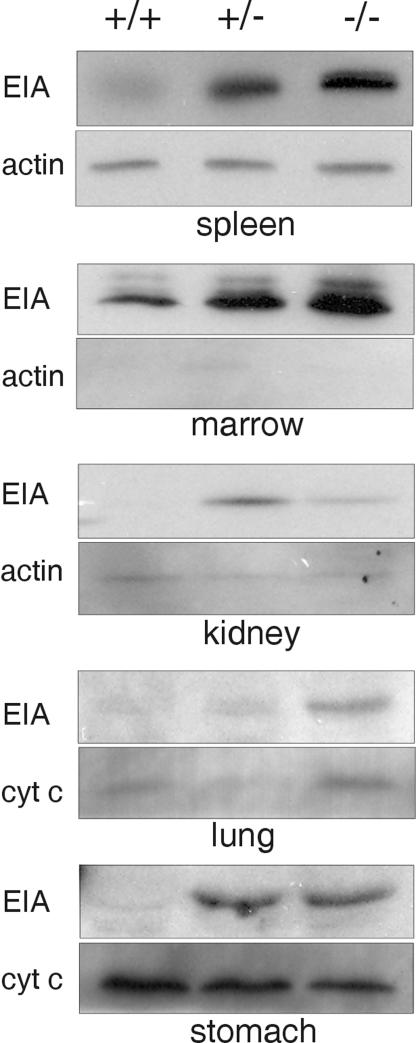
One explanation for the lack of susceptibility of SPI3-deficient mice to stroke or Candida infection is that other protease inhibitors can functionally substitute for SPI3 to protect against excess proteolysis during stress. Probing tissue blots with the pan-serpin monoclonal antibody 1F3 indicated that another serpin is up-regulated in the tissues of SPI3-deficient mice (data not shown). EIA is the ortholog of human monocyte/neutrophil elastase inhibitor and probably has protease targets similar to those of SPI3 (2). To determine whether EIA is up-regulated in SPI3-deficient mice, tissue from wild-type, heterozygous, and SPI3-deficient mice was probed with a rabbit serum specific for EIA. As shown in Fig. 6, there was a significant increase in EIA in the spleen, marrow, stomach, lungs, and kidneys of heterozygous and SPI3-deficient mice compared to that in wild-type mice. By densitometry, the largest increases were observed in the spleen (∼10-fold) and marrow (∼4-fold). EIA was not up-regulated in the liver and was not evident in the thymus, brain, heart, and skin of either wild-type or knockout mice (data not shown). These results indicate that EIA levels are increased in SPI3-deficient mice to compensate for the loss of SPI3.
FIG. 6.
Up-regulation of EIA in SPI3-deficient mice. Immunoblots of protein from tissues of wild type (+/+), heterozygous (+/−), and SPI3-deficient (−/−) mice were probed with a rabbit anti-elastase inhibitor serum (EIA) followed by either a goat anti-human actin antibody or a mouse anti-cytochrome c antibody.
DISCUSSION
Intracellular serpins are believed to protect cells from damage caused by proteases inadvertently entering the cytoplasm, either by leaking from storage compartments or by endocytosis. For example, PI-9 protects cells from apoptosis induced by the cytotoxic protease granzyme B (4). PI-6 probably performs a similar function in granulocytes and macrophages to protect against cathepsin G (37) and in neurons to protect against neuropsin (24).
As shown here, SPI3-deficient mice display no overt phenotype, possibly due to compensation by the related serpin, EIA (Serpinb1). EIA is the ortholog of human MNEI, which has an inhibitory and expression profile that overlaps with that of PI-6. Both serpins are found in myeloid cells, platelets, and epithelia, and both inhibit cathepsin G. Furthermore, it has recently been shown that EIA can interact with the neutrophil granule proteases elastase, cathepsin G, and proteinase 3 (2). The phenotype of the SPI3-deficient mice is therefore consistent with human PI-6 and MNEI forming part of an intracellular protease-regulating system in myeloid (and possibly other) cells. Functional redundancy of PI-6 and MNEI implies that the system is physiologically important and may also explain why naturally occurring PI-6 and MNEI mutants have not been discovered.
The mechanism underlying the increase in EIA levels is currently unclear. Given the large numbers and short life span of granulocytes, and assuming that (i) loss of SPI3 reduces cell viability, (ii) above-average levels of EIA can compensate for loss of SPI3, and (iii) EIA expression in these cells follows a normal distribution, it is possible that a subpopulation of granulocytes at the upper end of EIA expression is selected in SPI3+/− and SPI3−/− mice. This mechanism might also operate in other leukocytes and epithelia that normally coexpress EIA and SPI3.
Alternatively, it might be argued that loss of SPI3 stresses the cell and that transcription of EIA increases in response to stress. In support of this idea, the 5′ flanking region of the human MNEI gene contains an NF-κB binding site important for up-regulating MNEI expression in response to inflammatory stimuli (43), and recent studies have shown that NF-κB also functions as a central regulator of stress responses (28). Examination of the sequence of the 5′ flanking region of the EIA gene with the Alibaba2 program (http://www.gene-regulation.com/pub/programs/alibaba2/intro.html) reveals the presence of at least two potential NF-κB binding sites (D. Kaiserman and P. Bird, unpublished observations). However, it remains to be experimentally verified that these sites participate in the regulation of the mouse EIA.
Although SPI3 is expressed in mouse brain (25), is up-regulated during transient focal ischemia (31), and is known to regulate the neuronal protease neuropsin (24), SPI3-deficient mice do not exhibit increased infarct size in response to stroke. This suggests that SPI3 is not involved in regulating proteases activated during ischemic injury and that other serpins, such as neuroserpin, may be more important for this function. Indeed, it has been shown that intracerebral administration of neuroserpin after induction of stroke in rats significantly reduces infarct volume (42), and transgenic mice overexpressing neuroserpin in the central nervous system also displayed smaller infarcts after midcerebral artery occlusion (8).
In conclusion, the lack of an overt phenotype in SPI3-deficient mice is probably due to compensation by EIA, and it will be necessary to create animals that lack both SPI3 and EIA to further define the physiological role(s) of these serpins. It remains formally possible that one or more of the mouse-specific serpin genes that cluster with SPI3 and EIA on chromosome 13 (23) also compensates for SPI3 deficiency. However, these inhibitors have different reactive center loop sequences and expression profiles than SPI3 and almost certainly control different proteases. So far we have not detected up-regulation of these serpins in SPI3-deficient mice (unpublished data).
Acknowledgments
We thank E. Remold O'Donnell (Center for Blood Research, Harvard University, Boston, Mass.) for MNEI antiserum. We are also grateful to R. Gianello and G. Brownlee for advice and assistance with the Candida infection model and to D. Lane for technical assistance.
This work was supported by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Aprikyan, A. A., and D. C. Dale. 2001. Mutations in the neutrophil elastase gene in cyclic and congenital neutropenia. Curr. Opin. Immunol. 13:535-538. [DOI] [PubMed] [Google Scholar]
- 2.Benarafa, C., J. Cooley, W. Zeng, P. I. Bird, and E. Remold-O'Donnell. 2002. Characterization of four murine homologs of the human ov-serpin monocyte neutrophil elastase inhibitor MNEI (SERPINB1). J. Biol. Chem. 277:42028-42033. [DOI] [PubMed] [Google Scholar]
- 3.Bird, C. H., E. J. Blink, C. E. Hirst, M. S. Buzza, P. M. Steele, J. Sun, D. A. Jans, and P. I. Bird. 2001. Nucleocytoplasmic distribution of the ovalbumin serpin PI-9 requires a nonconventional nuclear import pathway and the export factor Crm1. Mol. Cell. Biol. 21:5396-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird, C. H., V. R. Sutton, J. Sun, C. E. Hirst, A. Novak, J. A. Trapani, and P. I. Bird. 1998. Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol. Cell. Biol. 18:6387-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, P. I. 1999. Regulation of pro-apoptotic leucocyte granule serine proteinases by intracellular serpins. Immunol. Cell Biol. 77:47-57. [DOI] [PubMed] [Google Scholar]
- 6.Blasi, E., L. Pitzurra, A. R. Chimienti, R. Mazzolla, M. Puliti, R. Barluzzi, and F. Bistoni. 1995. Differential susceptibility of yeast and hyphal forms of Candida albicans to proteolytic activity of macrophages. Infect. Immun. 63:1253-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chertov, O., H. Ueda, L. L. Xu, K. Tani, W. J. Murphy, J. M. Wang, O. M. Z. Howard, T. J. Sayers, and J. J. Oppenheim. 1997. Identification of human neutrophil-derived cathepsin G and azurocidin/CAP37 as chemoattractants for mononuclear cells and neutrophils. J. Exp. Med. 186:739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cinelli, P., R. Madani, N. Tsuzuki, P. Vallet, M. Arras, C. N. Zhao, T. Osterwalder, T. Rulicke, and P. Sonderegger. 2001. Neuroserpin, a neuroprotective factor in focal ischemic stroke. Mol. Cell. Neurosci. 18:443-457. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, R. J., J. E. McNeal, S. G. Edgar, T. Robertson, and H. J. Dawkins. 1998. Characterization of cytoplasmic secretory granules (PSG), in prostatic epithelium and their transformation-induced loss in dysplasia and adenocarcinoma. Hum. Pathol. 29:1488-1494. [DOI] [PubMed] [Google Scholar]
- 10.Coughlin, P., J. Sun, L. Cerruti, H. H. Salem, and P. Bird. 1993. Cloning and molecular characterization of a human intracellular proteinase inhibitor. Proc. Natl. Acad. Sci. USA 90:9417-9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crack, P. J., J. M. Taylor, N. J. Flentjar, J. de Haan, P. Hertzog, R. C. Ianello, and I. Kola. 2001. Increased infarct size and exacerbated apoptosis in the glutathione peroxidase-1 (gpx-1) knockout mouse brain in response to ischemia/reperfusion injury. J. Neurochem. 78:1389-1399. [DOI] [PubMed] [Google Scholar]
- 12.Cutler, J. E. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187-218. [DOI] [PubMed] [Google Scholar]
- 13.Darnell, G. A., T. M. Antalis, R. W. Johnstone, B. W. Stringer, S. M. Ogbourne, D. Harrich, and A. Suhrbier. 2003. Inhibition of retinoblastoma protein degradation by interaction with the serpin plasminogen activator inhibitor 2 via a novel consensus motif. Mol. Cell. Biol. 23:6520-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, A. F., G. Mirza, G. Sekhon, P. Turnpenny, F. Leroy, F. Speleman, C. Law, N. van Regemorter, E. Vamos, F. Flinter, and J. Ragoussis. 1999. Delineation of two distinct 6p deletion syndromes. Hum. Genet. 104:64-72. [DOI] [PubMed] [Google Scholar]
- 15.Dougherty, K. M., J. M. Pearson, A. Y. Yang, R. J. Westrick, M. S. Baker, and D. Ginsburg. 1999. The plasminogen activator inhibitor-2 gene is not required for normal murine development or survival. Proc. Natl. Acad. Sci. USA 96:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edens, H. A., C. A. Parkos, T. W. Liang, A. J. Jesaitis, J. E. Cutler, and H. M. Miettinen. 1999. Non-serum-dependent chemotactic factors produced by Candida albicans stimulate chemotaxis by binding to the formyl peptide receptor on neutrophils and to an unknown receptor on macrophages. Infect. Immun. 67:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirata, A., S. Yoshida, N. Inoue, K. Matsumoto-Miyai, A. Ninomiya, M. Taniguchi, T. Matsuyama, K. Kato, H. Iizasa, Y. Kataoka, N. Yoshida, and S. Shiosaka. 2001. Abnormalities of synapses and neurons in the hippocampus of neuropsin-deficient mice. Mol. Cell. Neurosci. 17:600-610. [DOI] [PubMed] [Google Scholar]
- 18.Hirst, C. E., M. S. Buzza, C. H. Bird, H. S. Warren, P. U. Cameron, M. Zhang, P. G. Ashton-Rickhardt, and P. I. Bird. 2003. The intracellular granzyme B inhibitor, proteinase inhibitor 9, is up-regulated during accessory cell maturation and effector cell degranulation, and its overexpression enhances CTL potency. J. Immunol. 170:805-815. [DOI] [PubMed] [Google Scholar]
- 19.Hirst, C. E., M. S. Buzza, V. R. Sutton, J. A. Trapani, K. L. Loveland, and P. I. Bird. 2001. Perforin-independent expression of granzyme B and proteinase inhibitor 9 in human testis and placenta suggests a role for granzyme B-mediated proteolysis in reproduction. Mol. Hum. Reprod. 7:1133-1142. [DOI] [PubMed] [Google Scholar]
- 20.Huntington, J. A., R. A. Read, and R. W. Carrell. 2000. Structure of a serpin-protease complex shows inhibition by deformation. Nature 407:923-926. [DOI] [PubMed] [Google Scholar]
- 21.Irving, J. A., R. N. Pike, A. M. Lesk, and J. C. Whisstock. 2000. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 10:1845-1864. [DOI] [PubMed] [Google Scholar]
- 22.Jensen, J., T. Warner, and E. Balish. 1993. Resistance of SCID mice to Candida albicans administered intravenously or colonizing the gut: role of polymorphonuclear leukocytes and macrophages. J. Infect. Dis. 167:912-919. [DOI] [PubMed] [Google Scholar]
- 23.Kaiserman, D., S. Knaggs, K. L. Scarff, A. Gillard, G. Mirza, M. Cadman, R. McKeone, P. Denny, J. Cooley, C. Benarafa, E. Remold-O'Donnell, J. Ragoussis, and P. I. Bird. 2002. Comparison of human chromosome 6p25 with murine chromosome 13 reveals a greatly expanded ov-serpin gene repertoire in the mouse. Genomics 79:349-362. [DOI] [PubMed] [Google Scholar]
- 24.Kato, K., T. Kishi, T. Kamachi, M. Akisada, T. Oka, R. Midorikawa, K. Takio, N. Dohmae, P. Bird, J. Sun, F. Scott, Y. Miyake, K. Yamamoto, A. Machida, T. Tanaka, K. Matsumoto, M. Shibata, and S. Shiosaka. 2001. Serine proteinase inhibitor 3 and murinoglobulin I are potent inhibitors of neuropsin in adult mouse brain. J. Biol. Chem. 276:14562-14571. [DOI] [PubMed] [Google Scholar]
- 25.Kishi, T., P. Matsuhashi, P. I. Bird, and K. Kato. 2002. Distribution of serine proteinase inhibitor, clade B, member 6 (Serpinb6) in the adult mouse brain. Gene Expr. Patterns 1:175-180. [DOI] [PubMed] [Google Scholar]
- 26.Kullberg, B. J., M. G. Netea, A. G. Vonk, and J. W. van der Meer. 1999. Modulation of neutrophil function in host defense against disseminated Candida albicans infection in mice. FEMS Immunol. Med. Microbiol. 26:299-307. [DOI] [PubMed] [Google Scholar]
- 27.Levesque, J. P., J. Hendy, Y. Takamatsu, B. Williams, I. G. Wingler, and P. J. Simmons. 2002. Mobilization by either cyclophosphamide or granulocyte colony-stimulating factor transforms the bone marrow into a highly proteolytic environment. Exp. Hematol. 30:440-449. [DOI] [PubMed] [Google Scholar]
- 28.Li, X., and G. R. Stark. 2002. NFkappaB-dependent signaling pathways. Exp. Hematol. 30:285-296. [DOI] [PubMed] [Google Scholar]
- 29.Loyola, W., D. A. Gaziri, L. C. Gaziri, and I. Felipe. 2002. Concanavalin A enhances phagocytosis and killing of Candida albicans by mice peritoneal neutrophils and macrophages. FEMS Immunol. Med. Microbiol. 33:201-208. [DOI] [PubMed] [Google Scholar]
- 30.Mikolajczyk, S. D., L. S. Millar, K. M. Marker, H. G. Rittenhouse, R. L. Wolfert, L. S. Marks, M. C. Charlesworth, and D. J. Tindall. 1999. Identification of a novel complex between human kallikrein 2 and proteinase inhibitor-6 in prostate cancer tissue. Cancer Res. 59:3927-3930. [PubMed] [Google Scholar]
- 31.Nakaya, N., M. Nishibori, Z. Wang, J. Sakiyama, and K. Saeki. 1998. The expression and localization of serine proteinase inhibitor PI-6 mRNA in developmental and ischemic mouse brain. Neurosci. Res. 32:221-230. [DOI] [PubMed] [Google Scholar]
- 32.Qian, Q., M. A. Jutila, N. van Rooijen, and J. E. Cutler. 1994. Elimination of mouse splenic macrophages correlates with increased susceptibility to experimental disseminated candidiasis. J. Immunol. 152:5000-5008. [PubMed] [Google Scholar]
- 33.Scarff, K. L., K. S. Ung, J. Sun, and P. I. Bird. 2003. A retained selection cassette increases reporter gene expression without affecting tissue distribution in SPI3 knockout/GFP knock-in mice. Genesis 36:149-157. [DOI] [PubMed] [Google Scholar]
- 34.Schwenk, F., U. Baron, and K. Rajewsky. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, F. L., P. B. Coughlin, C. Bird, L. Cerruti, J. A. Hayman, and P. Bird. 1996. Proteinase inhibitor 6 cannot be secreted, which suggests it is a new type of cellular serpin. J. Biol. Chem. 271:1605-1612. [DOI] [PubMed] [Google Scholar]
- 36.Scott, F. L., H. J. Eyre, M. Lioumi, J. Ragoussis, J. A. Irving, G. A. Sutherland, and P. I. Bird. 1999. Human ovalbumin serpin evolution: phylogenic analysis, gene organization, and identification of new PI-8-related genes suggest that two interchromosomal and several intrachromosomal duplications generated the gene clusters at 18q21-q23 and 6p25. Genomics 62:490-499. [DOI] [PubMed] [Google Scholar]
- 37.Scott, F. L., C. E. Hirst, J. Sun, S. P. Bottomley, C. H. Bird, and P. I. Bird. 1999. The intracellular serpin proteinase inhibitor 6 (PI-6) is expressed in monocytes and granulocytes and is a potent inhibitor of the azurophilic granule proteinase, cathepsin G. Blood 93:2089-2097. [PubMed] [Google Scholar]
- 38.Shimizu-Okabe, C., G. M. Yousef, E. P. Diamandis, S. Yoshida, S. Shiosaka, and M. Fahnestock. 2001. Expression of the kallikrein gene family in normal and Alzheimer's disease brain. Neuroreport 12:2747-2751. [DOI] [PubMed] [Google Scholar]
- 39.Silverman, G. A., P. I. Bird, R. W. Carrell, F. C. Church, P. B. Coughlin, P. G. W. Gettins, J. A. Irving, D. A. Lomas, C. J. Luke, R. W. Moyer, P. A. Pemberton, E. Remold-O'Donnell, G. S. Salvesen, J. Travis, and J. C. Whisstock. 2001. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. J. Biol. Chem. 276:33293-33296. [DOI] [PubMed] [Google Scholar]
- 40.Stein, P. E., and R. W. Carrell. 1995. What do dysfunctional serpins tell us about molecular mobility and disease. Struct. Biol. 2:96-113. [DOI] [PubMed] [Google Scholar]
- 41.Vonk, A. G., C. W. Wieland, M. G. Netea, and B. J. Kullberg. 2002. Phagocytosis and intracellular killing of Candida albicans blastoconidia by neutrophils and macrophages: a comparison of different microbiological test systems. J. Microbiol. Methods 49:55-62. [DOI] [PubMed] [Google Scholar]
- 42.Yepes, M., M. Sandkvist, M. K. K. Wong, T. A. Coleman, S. Smith, S. L. Cohan, and D. A. Lawrence. 2001. Neuroserpin reduces cerebral infarct volume and protects neurons from ischemia-induced apoptosis. Blood 96:569-576. [PubMed] [Google Scholar]
- 43.Zeng, W., and E. Remold-O'Donnell. 2000. Human monocyte/neutrophil elastase inhibitor (MNEI) is regulated by PU.1/Spi-1, Sp1, and NF-kappaB. J. Cell. Biochem. 78:519-532. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, Q., and G. S. Salvesen. 1997. Activation of pro-caspase-7 by serine proteases includes a non-canonical specificity. Biochem. J. 324:361-364. [DOI] [PMC free article] [PubMed] [Google Scholar]