Abstract
Replication initiation and replication fork movement in the subtelomeric and telomeric DNA of native Y′ telomeres of yeast were analyzed using two-dimensional gel electrophoresis techniques. Replication origins (ARSs) at internal Y′ elements were found to fire in early-mid-S phase, while ARSs at the terminal Y′ elements were confirmed to fire late. An unfired Y′ ARS, an inserted foreign (bacterial) sequence, and, as previously reported, telomeric DNA each were shown to impose a replication fork pause, and pausing is relieved by the Rrm3p helicase. The pause at telomeric sequence TG1-3 repeats was stronger at the terminal tract than at the internal TG1-3 sequences located between tandem Y′ elements. We show that the telomeric replication fork pause associated with the terminal TG1-3 tracts begins ∼100 bp upstream of the telomeric repeat tract sequence. Telomeric pause strength was dependent upon telomere length per se and did not require the presence of a variety of factors implicated in telomere metabolism and/or known to cause telomere shortening. The telomeric replication fork pause was specific to yeast telomeric sequence and was independent of the Sir and Rif proteins, major known components of yeast telomeric heterochromatin.
Replication of the linear DNA of a eukaryotic chromosome imposes a problem of end replication, as originally predicted by Watson (40) and Olovnikov (31). While the synthesis of the leading strand can proceed to the very end of the template, the lagging strand is predicted to shorten upon every round of replication in each cell cycle. Most eukaryotes solve the end-replication problem by maintaining specific repetitive DNA sequences at their chromosome ends, called telomeres, by the enzyme telomerase, which elongates the 3′ end of the telomeric DNA in a sequence-specific manner. In those rarer situations in which a eukaryote does not have telomerase, other multiple repeats, such as transposable elements in the fruit fly Drosophila melanogaster, are periodically added to their chromosome ends.
The yeast Saccharomyces cerevisiae has telomeres containing ∼250 to 350 bp of TG1-3 repeats and uses telomerase for their maintenance. About two-thirds of the 32 telomeres in haploid cells carry one or more copies of subtelomeric Y′ elements (see reference 32 for a review). Members of the major class of Y′ elements are 6.7 kb long, and there is a minor 5.2-kb class; they are always arranged in the same orientation, such that multiple Y′ elements form directly repeating arrays separated by short stretches of telomeric TG1-3 DNA.
Replication of eukaryotic chromosomes initiates at autonomously replicating sequence (ARS) elements—origins of replication present at multiple locations on every chromosome. Every Y′ element contains an ARS (4). While many genomic ARSs are fired early in S phase, it has been reported, by using the density transfer method, that Y′ repeats replicate late in S phase (11, 28, 33, 39). Telomeric chromatin has been implicated in determining the timing of activation of subtelomeric ARSs. An ARS placed on a circular plasmid containing telomeric TG1-3 repeats initiates replication early, but if the plasmid is linearized and therefore contains telomeres, the ARS is fired late (11). Furthermore, deletion of Sir3p, one of the components of telomeric heterochromatin, causes the telomeres to replicate in early S phase (39). These results raise the question of whether all the Y′ elements initiate their ARSs synchronously in late S phase or whether the timing of replication is different for the terminal and internal elements, since only the former are positioned next to a terminal telomeric TG1-3 tract.
Replication fork movement does not proceed monotonically. Programmed replication fork pausing is conserved from bacteria to higher plants and animals and can contribute to genomic stability. Polar replication fork blocks can ensure unidirectional replication at a certain region of a genome, thereby playing regulatory roles in different cellular processes. For example, in bacteria, polar replication pausing coordinates termination of bidirectional chromosome replication so that the two replication forks meet at the terminus region where newly synthesized chromosomes are to be decatenated (reviewed in reference 34). On either side of the terminus, three sites (Ter) are bound by the protein Tus, and the resultant DNA-protein complex blocks the progression of replication forks in a polar manner. In the mating type locus of the fission yeast Schizosaccharomyces pombe, replication pausing regulates the orientation of DNA replication, thereby determining whether cells undergo mating type switching (9). Many organisms have been shown to have specific replication blocks near ribosomal DNA (rDNA) genes. In the budding yeast S. cerevisiae, replication fork pausing has been found at rDNA and telomeres, and the Rrm3 helicase has been shown to alleviate it at both loci (19, 20). While the replication pausing at rDNA is caused by the DNA-binding protein Fob1 (21, 22), the key determinants that impose the replication barrier at telomeres are unknown. Toward a fuller understanding of the role of replication in telomere maintenance in vivo, here we have analyzed replication of natural yeast telomeric regions, including the subtelomeric Y′ elements, to address the question of the nature and architecture of replication fork pausing at Y′ telomeres.
MATERIALS AND METHODS
Strains.
All yeast strains used in this work are listed in the Table 1.
TABLE 1.
Strains of S. cerevisiae A364a used in this study
| Strain | Genotype | Parental strain/source/comments |
|---|---|---|
| NK1 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 | From J. Li (YJL310) |
| NK2 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rad9Δ::URA3 | NK1/PCR-based deletion |
| NK3 | MATaura3-52 trp1-289 cdc9-1 | From J. Li (YJL347) |
| NK26 | MATaura3-52 leu2-3,112 bar1::LEU2 cdc9-1 rad9Δ::URA3 | NK2 (MATα) × NK3 |
| NK15 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 sgs1Δ::KAN | NK1/PCR-based deletion |
| NK17 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rrm3Δ::KAN | NK1/PCR-based deletion |
| NK18 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 pif1Δ::KAN | NK1/PCR-based deletion |
| NK23 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 stm1Δ::KAN | NK1/PCR-based deletion |
| NK24 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 cdp1Δ::KAN | NK1/PCR-based deletion |
| NK34 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 tel1Δ::KAN | NK1/PCR-based deletion |
| NK35 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 sir3Δ::KAN | NK1/PCR-based deletion |
| NK60 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 Y′::URA3 | NK1/PCR-based insertion |
| NK78 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rad51Δ::TRP1 | NK1/PCR-based deletion |
| NK80 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rad52Δ::TRP1 | NK1/PCR-based deletion |
| NK88 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 gbp2Δ::TRP1 | NK1/PCR-based deletion |
| NK94 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 yku70Δ::TRP1 | NK1/PCR-based deletion |
| NK96 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 yku80Δ::TRP1 | NK1/PCR-based deletion |
| NK98 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rif1Δ::TRP1 | NK1/PCR-based deletion |
| NK100 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rif2Δ::TRP1 | NK1/PCR-based deletion |
| NK125 | MATaura3-52 leu2-3,112 bar1::LEU2 | NK1/The EcoRI site near ARS1 is removed |
| NK180 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rad50Δ::TRP1 | NK1/PCR-based deletion |
| NK201 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 (pYT30 CEN-ARS URA3 CDC13-EST1) | NK1/transformation |
| NK346 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 Y′::URA3 rrm3Δ::TRP1 | NK60/PCR-based deletion |
| NK350 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rrm3Δ::TRP1 Y′::KAN − R+T+ (Reb1p and Tbf1p binding sites are present)-telomere | NK346/transformation/screen for Ura− |
| NK351 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rrm3Δ::TRP1 Y′::KAN − R−T− (Reb1p and Tbf1p binding sites are absent) - telomere | NK346/transformation/screen for Ura− |
| NK361 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::URA3 | NK1/PCR-based insertion |
| NK370 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::URA3 (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK361/transformation |
| NK380 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rrm3Δ::TRP1 Y′::KAN − R+T+ - telomere sir2::URA3 | NK350/PCR-based deletion |
| NK382 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 rrm3Δ::TRP1 Y′::KAN − R−T− - telomere sir2::URA3 | NK351/PCR-based deletion |
| NK384 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::TG1-3-355 bp (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK370/transformation |
| NK385 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::AC1-3-355 bp (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK370/transformation |
| NK388 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::T2G4-360 bp (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK370/transformation |
| NK389 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::A2C4-360 bp (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK370/transformation |
| NK392 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::TG1-3-355 bp rrm3Δ::KAN (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK384/PCR-based deletion |
| NK393 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::AC1-3-355 bp rrm3Δ::KAN (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK385/PCR-based deletion |
| NK394 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::TG1-3-355 bp rap1ΔC::URA3 (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK384/PCR-based deletion |
| NK395 | MATaura3-52 trp1-289 leu2-3,112 bar1::LEU2 SNQ2::TG1-3-355 bp rrm3Δ::KAN rap1ΔC::URA3 (pYT57 CEN ARS TRP1 RPL4 PSF1 RAD61) | NK392/PCR-based deletion |
Plasmids.
The KpnI-XbaI fragment (which contains the CDC13-EST1 fusion) of pVL1091 (10) was ligated into pRS316 digested with KpnI and XbaI to obtain pYT30. pYT57 includes the region between bp 471061 and 476983 of S. cerevisiae chromosome IV (see http://www.yeastgenome.org), which was cloned between the KpnI and EcoRI sites of the vector pRS314 by the gap rescue technique. First, ∼300-bp fragments encompassing sequences on either side of this genomic region were PCR amplified as MfeI-XhoI (upstream of RPL4B) and XhoI-KpnI (downstream of RAD61) fragments and cloned together in pRS314 digested with EcoRI and KpnI. The resultant plasmid (pYT55) was linearized with XhoI and transformed into trp1 yeast (NK1). The recovered plasmids were purified from yeast and transformed in Escherichia coli K-12 DH5α for amplification and subsequent restriction analysis. pYT58 is a pRS316 derivative that carries the RRM3-containing fragment (region, bp 170335 to 173335 of chromosome VIII) cloned between SalI and BamHI sites by the method described above.
Yeast manipulations.
Yeast cultures were grown in standard rich medium or minimal media (when plasmid maintenance was desired). Knockout strain derivatives were obtained using the PCR-based deletion method (26).
Cell synchronization.
Yeast cells (NK125) were grown in liquid rich medium to an optical density at 600 nm of 0.350 and arrested in G1 with 500 ng of α-factor/ml. After 2.5 h the cells were harvested, washed once with a small volume (1/50 of the original volume) of fresh medium, and resuspended in two volumes of prewarmed fresh medium. Culture aliquots (0.5 liters) were taken at appropriate time intervals after the release and used for DNA purification.
Telomere preelongation.
pYT30 that carried the CDC13-EST1 fusion (10) was introduced into wild-type cells (NK1) and propagated for about 80 generations on selective medium lacking uracil (−URA plates) to allow elongation and resetting of the telomere length equilibrium. The resultant strain (NK201) and wild-type (wt) yeast with normal telomere length (NK1) were used for transformations to knock out EST2, TLC1, YKU70, RAD50, or TEL1. The colonies of the transformants were screened for the correct integration of the knockout cassettes and immediately streaked on 5-fluoro-orotic acid (5-FOA) plates to select against pYT30. Untransformed cells were streaked in parallel along with a wt control. Growing colonies were scraped from the 5-FOA plates, grown in 500 ml of rich medium to log phase, and harvested for DNA purification.
Insertion of yeast or Tetrahymena telomeric sequences in yeast genomic locus.
The URA3 cassette was inserted between SNQ2 and RPL4B first. The resultant yeast (NK361) was transformed with pYT57 (NK370) and then with DNA fragments containing either yeast or Tetrahymena TG repeats flanked by ∼0.5 kb of DNA, homologous to the genomic DNA of the region, and selected for 5-FOA-resistant colonies. The resultant strains (NK384, NK385, NK388, and NK389) preserved all the original genomic DNA sequence interrupted by the insertion at the HpaI site.
DNA purification and analysis by 2D GE.
DNA for two-dimensional (2D) gel electrophoresis (GE) was purified by CsCl gradient centrifugation according to the protocol of Gerbi and Bielinsky (14). 2D GE experiments were performed as described in reference 13 with the following modifications: the first-dimension gels were run at ∼0.4 V/cm for 45 to 50 h, and the second-dimension gels were run at 3 V/cm for 12 to 18 h. The DNA was transferred from the gel onto an Osmonics (Minnetonka, Minn.) MagnaGraph membrane and hybridized to radioactively labeled probes according to the manufacturers' recommendations. Either the PrimeIt-II kit (Stratagene, La Jolla, Calif.) and [α-32P]dCTP (NEN, Boston, Mass.) or T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) and [γ-32P]ATP (NEN) were used for probe labeling. A Molecular Dynamics Storm 800 PhosphorImager and ImageQuant software were used for image scanning and quantification.
Size fractionation of telomeric DNA fragments by agarose GE and alkaline GE.
To resolve the telomeric fragments by molecular mass, i.e., telomere length, the samples were run at a low voltage on 1.5% agarose gel until the marker bands roughly corresponding to the shortest and the longest Y′ telomere fragments (0.7 and 1.2 kb for the KpnI digests and 1.0 and 1.6 kb for the XhoI digests) were separated by about 4 to 4.5 cm. The agarose block of the sample lane between the corresponding marker sizes was excised from the gel and cut to 0.5-cm slices. The DNA from every slice was purified by using a Qiagen gel extraction kit and then run on an alkaline 2% agarose gel to separate newly synthesized and parental strands (35). The DNA was analyzed by Southern blotting using different oligonucleotides as probes as described above.
RESULTS
DNA replication in terminal regions of Y′ class telomeres.
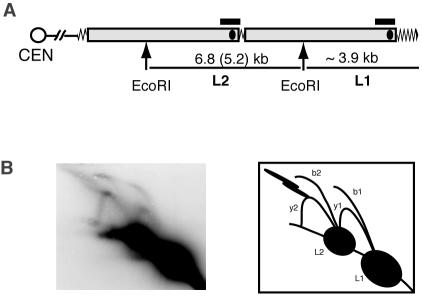
We used nondenaturing 2D GE followed by Southern blotting to detect the intermediates arising during the replication of telomeres and subtelomeric Y′ elements. Genomic DNA from nonsynchronized populations of cells was digested with EcoRI restriction enzyme. Each Y′ element contains a unique EcoRI site, generating two major classes of restriction fragments containing Y′ DNA: ∼3.9-kb terminal fragments of the chromosomes that include telomeric TG1-3 repeat tracts, and internal fragments that are the length of a single Y′ element, resulting from EcoRI digestion of the tandem Y′ elements (Fig. 1A). The majority of the Y′ elements in our strain were the long class, i.e., 6.8 kb rather than 5.2 kb. Therefore, we were able to ignore replication intermediates from the 5.2-kb fragments in our experiments, since they did not yield enough signal to interfere with the signals from the replicating 6.8- and 3.9-kb fragments.
FIG. 1.
Replication intermediates of Y′ telomeres detected by 2D gel electrophoresis. (A) Schematic diagram of an EcoRI digest of a chromosome arm containing two tandem Y′ elements (gray bars). Here and in the following figures, zigzag line represents TG1-3 tracts, black ellipsoids within analyzed DNA fragments show the position of the known ARSs, and black horizontal bars correspond to the fragments that were used as probes for the Southern hybridizations shown. (B) Replication intermediates of Y′ telomeres analyzed by 2D GE (see the text for explanations).
Two sets of replication intermediates could be seen on a 2D gel when probed with a Y′ sequence. One set arose from replication of the 3.9-kb terminal fragments (L1 fragments; Fig. 1A) and consisted of a fork arc and a bubble arc, depicted as y1 and b1, respectively, in Fig. 1B. The other set represented replication intermediates of the 6.8-kb internal Y′ elements (L2 fragments; Fig. 1A) and also consisted of a fork arc, y2, and a bubble arc, b2. The internal EcoRI fragments each contain the previously mapped Y′ ARS, which is centrally located in each 6.8-kb fragment (Fig. 1A). The DNA bubble structures in arc b2 were therefore formed as a result of activation of these ARSs in the internal Y′ elements. When these ARSs are not active, passive replication through the 6.8-kb internal EcoRI segments produce the Y-shaped forked structures seen collectively as arc y2. The replication pattern of the terminal 3.9-kb EcoRI fragments was not as readily explainable and is analyzed and discussed below.
The two streaks that were seen in the top-left corner of many of our gels are high-molecular-weight (HMW) structures that apparently were a result of in vitro interactions of G-strand overhangs of telomeric fragments. This was concluded because these streaks were abolished or alleviated when, after cell lysis, DNA purification and consequent analysis were performed in the continued presence of excess amounts (10-fold and more molar excess over telomeres) of added 80-bp single-stranded oligonucleotides, with the telomeric consensus sequence of either the G- or the C-strand of yeast telomeres (data not shown). The HMW structures remained intact if the oligonucleotides were added only after the EcoRI digestions. Our conclusions on the nature of the HMW structures were also supported by the observations that DNA samples treated with 3′→5′ single-stranded specific nucleases (ExoI and ExoT) prior to the EcoRI digestion did not form these structures, whereas treatments with a 5′→3′ nuclease (RecJ) increased their abundance (data not shown).
Timing of telomere replication during the cell cycle.
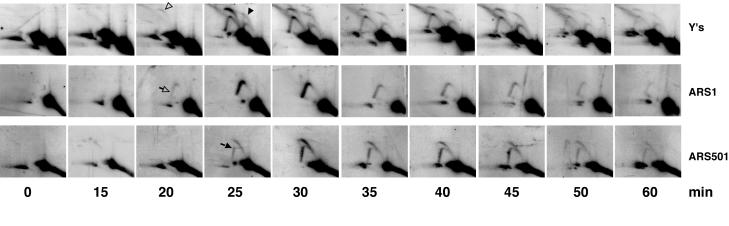
We compared the timing of replication of Y′ telomeres with genomic DNA fragments containing a known early-mid-S-phase origin (ARS1) or a known late-firing origin (ARS501). Cells were synchronized in G1 with α-factor and released into the cell cycle upon removal of the pheromone. Aliquots taken at different time points were monitored for replication by 2D GE as described above.
The expected difference in timing of activation of early versus late ARSs (Fig. 2) was readily detectable. The ARS1 replication intermediates first appeared at the 20-min time point, whereas the ARS501 bubble and forked structures only appeared 5 min later. Unexpectedly, we found that at least some of the ARSs of the internal Y′ elements are activated as early as ARS1 (Fig. 2, 20 min). Moreover, the amount of signal from replication initiated at both ARS1 and the active ARSs located in the internal Y′s (the bubble arc b2) peaked at the same time (Fig. 2, 25 min). As expected, terminal Y′ ARSs were activated at the same time as the late ARS501 (Fig. 2, 25 min). This late activation of the terminal Y′ ARSs is consistent with the previously published data that telomeres replicate late in S phase (11, 28, 33, 39). The observations were highly reproducible in two repeats of the time course experiment. We concluded that during the cell cycle, the ARS in an internal Y′ element fires early but a terminal ARS fires late.
FIG. 2.
Timing of replication of Y′ telomeres during the cell cycle. The DNA samples were digested with EcoRI, separated by native/native 2D gel electrophoresis, blotted, and consecutively rehybridized to three different probes. The Y′-specific probe was the same as in Fig. 1; short PCR products within the corresponding EcoRI fragments were used to monitor replication at the ARS1 and ARS501 regions. The ARS1 replication intermediates appeared as a fork arc (rather than a bubble arc), since ARS1 was close to one of the ends of the EcoRI fragment. The initiation of replication at ARS501 produces bubble structures that, as replication proceeds, break open into forked DNA fragments at the EcoRI site closer to ARS501. The arrowhead symbols point out early (no-fill arrowheads) and late (black arrowhead and arrow) ARS firing during the cell cycle (see the text for explanations).
Pausing of replication forks at telomeres.
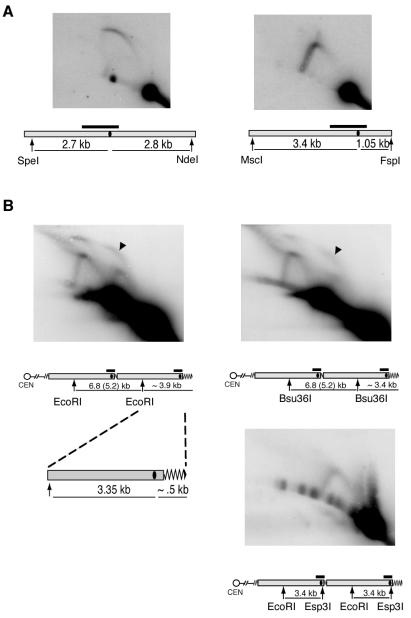
Though an ARS is present in the Y′ element in the terminal 3.9-kb EcoRI fragment (4), we did not expect to see the bubble arc b1 on a 2D gel. This is because the Y′ ARS is so close to the telomeric end of the fragment that if both replication forks move away from the fired ARS at equal rates, the fork moving towards the telomere (rightwards in Fig. 1A) would reach the chromosome end and hence open up well before the fork traveling in the opposite direction approaches the EcoRI site. Therefore, no large bubbles (seen as a bubble arc on a 2D GE) were expected if both forks move at equal rates away from the ARS. As a positive control for our ability to detect bubble arcs under the experimental conditions used here, we analyzed replication intermediates in the ARS1 region (chromosome IV). When the restriction sites were chosen so that the ARS had a central location, we clearly detected the expected replication intermediates which formed a visible bubble arc (Fig. 3A, left panel). Also as expected, when the DNA was digested so that the ARS was positioned close to one of the ends of the fragment, the 2D gel pattern contained only a visible fork arc and no bubble arc (Fig. 3A, right panel).
FIG. 3.
The b1 bubble arc results from replication pausing at telomeres. (A) ARS1 location on the analyzed fragment affects the form of the recovered replication intermediates. ARS1 has a central location at the SpeI-NdeI fragment, and digestion of replicating DNA with these enzymes preserves the bubble structures (left panel). Restriction with MscI and FspI positions ARS1 close to one end of the corresponding restriction fragment and leads to conversion of DNA bubbles into forks. (B) Digestion of the b1 Y′ bubbles (black arrowheads) with EcoRI plus Esp3I, but not Bsu36I, converts them into fork structures, suggesting that replication pausing occurs on the telomeric side (right side) of the Esp3I restriction site. The position of each ARS is presented as the black oval within the gray bar in the map of the chromosome region analyzed. Other symbols are as defined in the legend to Fig. 1.
The presence of the b1 bubble arc in our telomere replication experiments could be explained by two mutually exclusive hypotheses. One is that the previously reported pausing site at telomeres (20) arrests the right-moving fork, preventing it from opening up, thereby leading to the production of large DNA bubbles as replication intermediates as the leftward fork continues to move. Alternatively, there could be an unknown replication start site in the central part of the 3.9-kb fragment. To distinguish between these two possibilities, we cut off ∼0.5 kb from one or the other end of the fragment. If the bubble arc is a result of a replication pausing at telomeres, then cutting off only the telomeric end (thereby cutting off the proposed pause site) would convert bubbles into forks. In contrast, if the replication starts in the middle of the EcoRI fragment and forks proceed at similar rates in both directions, then the loss of 0.5 kb from either side should have equal effect on the 2D GE replication pattern. We chose Esp3I to cut off the telomere and Bsu36I to shorten the telomeric EcoRI fragment from the other (centromere-proximal) side (Fig. 3B). The results were consistent with the first hypothesis: the EcoRI-Esp3I fragment no longer produced a bubble arc, whereas digestion with Bsu36I did not change the overall pattern seen with EcoRI digestion alone (Fig. 3B). Furthermore, because the EcoRI-Esp3I fragment did not form large bubbles, this pausing site was at or located to the right of the Esp3I site (for the diagram, see Fig. 3B, bottom right).
Genetic dependence of replication pausing at telomeres.
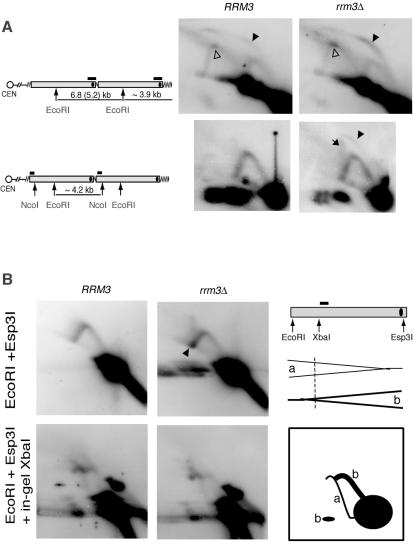
We constructed knockout mutants that lack a variety of proteins that are known, or hypothesized, to play various roles in telomere maintenance and/or replication fork progression at rDNA (see http://www.yeastgenome.org for references). Each strain was assayed for Y′ telomere replication by 2D GE. Deletions of RAD51, RAD52 (homologous recombination and telomere maintenance in the absence of telomerase), GBP2 (telomere clustering), CDP1, STM1 (binding of triplex and quadruplex DNA), RIF1, RIF2, SIR3 (all are components of telomeric chromatin), PIF1 (negative regulator of telomerase, affects replicaton at rDNA locus), SGS1 (telomere maintenance in the absence of telomerase and replication at rDNA), RRM3 (replication fork progression at rDNA and telomeres), and FOB1 (replication pausing at rDNA) have variable effects on telomere length, but none of them greatly shortens telomeres. With one exception, none of these single deletions caused any detectable changes in the 2D replication pattern (data not shown). The exception was RRM3; consistent with previously described experiments (20), deleting RRM3 led to an increase in the b1 bubble arc signal, suggesting accumulation of the large bubbles during replication of the terminal EcoRI fragment (Fig. 4A, upper panels). Also, for the rrm3Δ mutant the accumulation of the signal intensity at the very top of the fork arc y2 was increased (Fig. 4A, upper panels), suggesting a replication pausing site in the middle of the 6.8-kb fragment. The internal Y′ elements are flanked on both sides by short, ∼50- to 100-bp stretches of TG repeats, whereas (in wt cells) each terminal Y′ element is followed by ∼250 to 350 bp of telomeric TG1-3 DNA. Therefore, we also assayed whether a similar kind of replication pausing was present at the homologous regions of the internal repeats. DNA samples from RRM3 and rrm3Δ cells were digested with EcoRI and NcoI. A short region between the internal TG1-3 repeats and the NcoI site differentiates the DNA sequence of the EcoRI-NcoI fragment from the terminal 3.9-kb EcoRI fragment and therefore could be used as a probe specific to internal Y′ elements. No bubble arc was seen at the internal Y′ elements in wt cells (Fig. 4A, lower left), but a weak bubble arc as well as a streak of double-forked 2N structures were clearly present in the rrm3Δ mutant (Fig. 4A, lower right panel).
FIG. 4.
Deletion of RRM3 exacerbates replication pausing at both internal and terminal TG1-3 repeats. (A) Accumulation of the bubble replication intermediates (black arrowheads) at Y′ telomeres is stronger than at the homologous internal regions and is increased in the absence of the Rrm3 helicase. No-fill arrowheads point to the replication pausing intermediates in the internal 6.8-kb EcoRI fragment. The black arrow shows double-forked intermediates. (B) Deletion of RRM3 does not affect the frequency of Y′ ARS activation. Replication intermediates of the EcoRI-Esp3I fragment of Y′ repeats form fork arcs both in RRM3 and rrm3 cells (top panels). The fork arcs of replication intermediates arise from right-moving (a, drawn as a thin line in the diagram) and left-moving (b, thick line) forks. The latter arcs result from Y′ARS activation within that Y′ element, whereas the former indicate passive replication through inactive ARSs (see the text for explanation). In-gel digestion of the replication intermediates with XbaI prior to running the second dimension of the gel can distinguish between right- and left-moving forks (lower panels) (3). Bottom right: resolved products of in-gel digestion of the forks approaching the ARS are drawn as a thin line (a), and the ones moving away from the ARS fall into an arc plus a band (b), shown by the thick lines.
The Rrm3p helicase has been shown to promote passage of replication forks through the yeast telomeric TG1-3 repeats (20). Therefore, the accumulation of large bubbles at telomeres in rrm3Δ yeast has been interpreted to be a result of stronger replication pausing in the mutants (20). However, another possibility might account for a stronger b1 bubble arc; namely, that the rrm3Δ mutation might increase the activity, i.e., frequency of firing, of telomeric ARSs. As a result, more replication forks traveling through the Y′ elements would be expected to move from telomere towards centromere. To distinguish between these two possible roles of Rrm3p and also directly address the effect of the rrm3Δ mutation on initiation of replication at telomeric ARSs, we assayed the ratio of right- and left-moving forks through the EcoRI-Esp3I regions of Y′ elements (see Fig. 4B for the diagram). Because the internal Y′ elements replicate earlier than the terminal ones in the cell cycle in both wt and rrm3Δ cells (see Fig. 2; also data not shown), it is unlikely that any left-moving replication fork initiated at an ARS in the more centromere-distal (i.e., more terminal) Y′ elements would often progress into their centromere-proximal neighbors. Therefore, in the EcoRI-Esp3I region of any Y′ element, any replication fork moving towards the EcoRI site (leftwards, i.e., towards the centromere) is very likely to have originated from the ARS within that element (see Fig. 4B for the diagram). In contrast, when the ARS within the Y′ element is not active, a rightward-moving fork due to passive replication travels through this region.
To assay the directionality of fork movements, we used a standard technique of nondenaturing 2D GE with in-gel DNA digestion prior to running the second dimension (3). DNA from RRM3 and rrm3Δ cells was digested with the EcoRI and Esp3I restriction enzymes and run in duplicate on agarose gels. Then, one set of samples was digested in-gel with XbaI and all the four samples were run on the second-dimension gel. Fork arcs that were seen in the “no in-gel digestion” samples were converted to two arcs by XbaI. One of the resulting arcs corresponded to the left-moving forks, and the other corresponded to the right-moving forks, as shown in Fig. 4B (see explanations in the figure legend). The ratio was about the same for wt cells (30% forks moving towards telomeres and 70% towards centromeres) and the rrm3Δ derivative (32% and 68%, respectively). This is consistent with the previously published data for Y′ ARS activity (39). Therefore, the absence of the Rrm3 helicase did not affect the firing activity of the Y′ ARSs. We conclude that the accumulation of the bubbled structures at telomeres in the rrm3Δ cells is caused solely by the replication pausing effect.
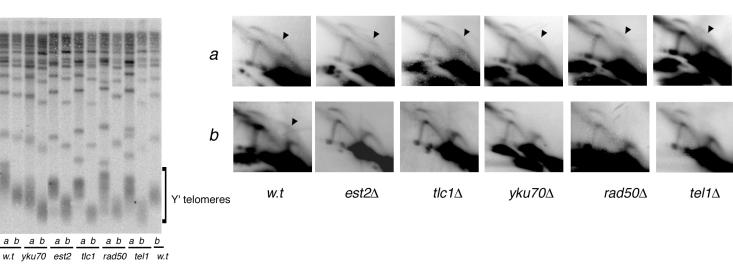
None of the gene deletions tested above led to significant telomere shortening or, as shown above, to loss of the telomeric replication fork pause. In contrast, loss of EST2 or TLC1, which causes short telomeres and eventual senescence (24, 38), or of YKU70, YKU80, RAD50, or TEL1 each resulted in the loss of the bubble arc b1 (Fig. 5, 2D GE panels b; data not shown for yku80). All these mutants have in common a short-telomere phenotype. To determine whether it was the short telomeres per se, or the lack of the product of the disrupted gene, that caused the loss of the bubble arc, we preelongated telomeres with the CDC13-EST1 fusion construct (10) in the plasmid pYT30. Then we knocked out each of the above genes, followed by removal of the CDC13-EST1-carrying plasmid, and immediately assayed telomere replication in the knockouts while their telomeres were still elongated. For all the five mutants (est2, tlc1, yku70, rad50, and tel1), telomere preelongation restored the bubble arc b1 (Fig. 5, 2D GE panels a). Therefore, we conclude that the presence of the persistent large bubbles is dependent on telomere length per se and is independent of functional telomerase, Ku-proteins, Rad50p, or Tel1p.
FIG. 5.
Replication pausing is decreased at short telomeres. The DNA from wt cells and mutants with or without the telomere preelongation step was purified and analyzed for telomere length (left panel; lanes a, DNA from cells with preelongated telomeres; lanes b, DNA from cells with no preelongation step) and replication intermediates by 2D GE (row a, DNA from cells with preelongated telomeres, row b, DNA from cells with no preelongation step). A (GTG1-3T)4 oligonucleotide that specifically hybridizes to telomeric DNA was used as a probe for the teloblot in the left panel. The 2D GE analysis of replication intermediates on the right panel was performed as for Fig. 1.
As discussed above, the formation of large bubbles at Y′ telomeres requires both activation of the ARSs of the terminal Y′ elements and slowed-down progression of the replication fork at the telomeres. Hence, these large bubbles might have been lost because shorter telomeres cause two possible effects: weaker replication pausing and/or a lower efficiency of ARS firing. To test the latter, we carried out an experiment determining the directionality of fork movement similar to the one described above for the rrm3Δ mutant (Fig. 4B). Neither loss of YKU70 nor loss of TEL1 had any effect on the activity of ARSs at the Y′ elements (data not shown). Therefore, we conclude that shorter telomeres cause a loss of large bubbles by alleviating replication pausing at these telomeres.
Rrm3p promotes replication fork movement through inactive ARSs.
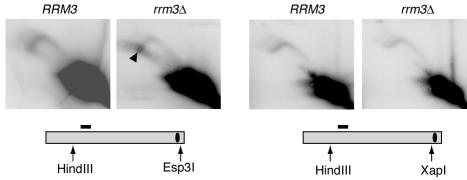
We noticed in the experiment just described (Fig. 4B) that an intense spot was located near one terminus of the fork arc of the rrm3Δ sample: namely, near the 2N DNA content point (Fig. 4B). This suggested that the rrm3Δ mutant accumulated DNA fragments that were almost completely replicated, since their size was nearly twofold higher than that of the linear unreplicated EcoRI-Esp3I fragment and their structure was close to linear. Since the EcoRI-Esp3I fragment did not contain any telomeric sequence, this indicated that there was another replication pausing site not located at the telomeric TG1-3 tracts. We hypothesized that this replication pausing might be caused by inactive Y′ ARSs that were in close proximity to the Esp3I site. Furthermore, we deduced that when the ARSs are active such replication pausing does not occur; otherwise, a bubble arc signal would have been expected for the EcoRI-Esp3I fragments due to delayed opening of the replication bubbles at the Esp3I ends. To test where the replication pause is located in the Y′ ARS region, we digested DNA from RRM3 and rrm3Δ cells with either HindIII and Esp3I or HindIII and XapI. In the former digests, the analyzed fragments contained Y′ ARSs, as in EcoRI-plus-Esp3I restriction digests, whereas in the HindIII-plus-XapI digests the ARSs were cut off (Fig. 6). These experiments showed that the prominent replication pausing site was present within the HindIII-Esp3I fragments (Fig. 6) but not in the HindIII-XapI fragments, and therefore, the pausing site was located between the XapI and Esp3I sites (Fig. 6). These sites are separated by a 189-bp stretch that includes the Y′ ARS. Therefore, we conclude that when a replication fork, moving from a centromere towards the telomere, approaches an inactive Y′ ARS, a pause occurs, and the Rrm3 helicase promotes the fork movement through such an inactive ARS-containing region.
FIG. 6.
Rrm3p promotes replication through inactive ARSs at telomeres. rrm3Δ cells but not RRM3 cells show accumulation of replication forks between Esp3I and XapI restriction sites (black arrowhead). The Esp3I-XapI interval in a Y′ repeat includes the ARS sequence.
What causes replication pausing at telomeres: G-C-rich DNA composition or binding of telomeric protein(s)?
We considered two possibilities for why replication forks stall at telomeres: high G-C DNA content that might result in difficulty in DNA unwinding, or telomeric chromatin, i.e., proteins that bind yeast telomeres and may have to be dislodged from the DNA during replication. To distinguish between these two possibilities, we compared the effects of a tract of yeast or Tetrahymena telomeric DNA repeats on replication fork progression in a yeast chromosome. Tetrahymena telomeric repeats G4T2 are as G-C rich as yeast G1-3T telomeric DNA but do not bind yeast Rap1 protein (1) (data not shown). Rap1p is a major sequence-specific DNA binding component at yeast telomeres, and Rif1, Rif2, and Sir2/3/4 localize to telomeres, presumably via interactions with Rap1p, thereby forming telomeric chromatin (2).
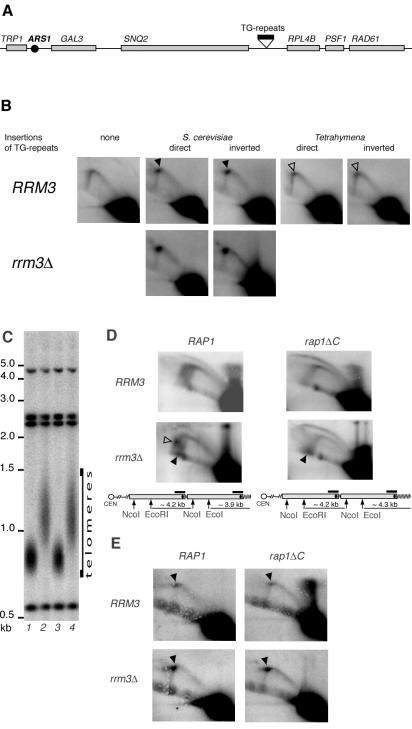
We inserted either 355 bp of yeast or 360 bp of Tetrahymena telomeric DNA in each orientation between the SNQ2 and RPL4B genes on chromosome IV in order to analyze replication intermediates arising from the activation of ARS1, which is located about 8 kb away from the insertion site (Fig. 7A). Though both insertions were designed to minimize any gene disruption, the TG1-3 insertions resulted in poor cell growth, presumably due to silencing of one or more of nearby genes. Since the neighboring RPL4B encodes a ribosomal protein and therefore is likely to be essential, we cloned the RPL4B-RAD61 region in a CEN-ARS vector, pRS314, and transformed it into yeast prior to the introduction of the G1-3T and G4T2 insertions. The presence of this plasmid completely alleviated the growth defect.
FIG. 7.
Replication fork pausing is specific to yeast telomeric sequences and does not depend on the C terminus of Rap1p. (A) Schematic diagram of the region at chromosome IV that was used to insert yeast or Tetrahymena telomeric DNA. (B) DNA samples from nonsynchronized wt yeast (NK1) and the insertion-carrying derivatives (NK384, NK385, NK388, NK389, NK392, and NK393) were digested with KpnI and analyzed by 2D GE. (C) Telomere length in NK384 RRM3 RAP1 (lane 1), NK394 RRM3 rap1ΔC (lane 2), NK392 rrm3Δ RAP1 (lane 3), and NK395 rrm3Δ rap1ΔC (lane 4) cells used for the replication assays in panels D and E. The DNA samples were digested with KpnI, and the blot was probed with the same probe as in panel D. (D) Replication through subterminal and terminal Y′ elements. Black arrowheads point to the accumulation of replication intermediates at telomeres. No-fill arrowhead shows the accumulation of replication intermediates at the inactive Y′ ARSs. Other symbols are as defined in the legend to Fig. 1. (E) Replication pausing at the internally located 355-bp tracts of TG1-3 repeats (see panel A for diagram). The repeats are in direct orientation, i.e., when replication starts at ARS1 the replication fork approaches the insertion from the same side from which a replication fork originating at a Y′ element approaches terminal TG1-3 tracts.
Consistent with previously reported results (20), in our strain the yeast TG1-3 repeats, whether oriented relative to approaching replication forks in the same way as at telomeres or inverted, caused replication pausing. This pausing resulted in a twofold increase in the intensity of the spot on the fork arc (Fig. 7B) compared with the same position on the arc with no insertion at that genome locus (Fig. 7B, “none” panel). In both cases, the Rrm3p helicase alleviated the pausing, since the rrm3Δ derivatives accumulated even more stalled fork structures than the RRM3 cells (Fig. 7B, lower panels). In contrast, insertion of the Tetrahymena telomeric DNA in either orientation caused very little pausing (a 15% increase in the signal on the fork arc at the position of the pause compared with the same position on the arc with no insertion at that genome locus) (Fig. 7B). Therefore, the DNA G-C richness per se does not cause the majority of replication fork pausing at TG1-3 repeats; rather, we infer that proteins that bind yeast telomeric DNA are a likely primary cause of the replication barrier.
Rap1ΔCp bound to DNA is sufficient to cause replication pausing.
Rap1p and Cdc13p bind, respectively, double-stranded and single-stranded yeast telomeric TG1-3 tracts in a sequence-specific manner (1, 6, 27, 30). The finding that internally located TG1-3 repeats can cause replication pausing suggests that Rap1p or its binding partners could be the barriers for replication fork progression. Biochemical and genetic evidence implicate the C terminus of Rap1p in binding of the Sir and Rif proteins (16, 17, 29, 41), which, along with Rap1p, are believed to be the major known components of telomeric heterochromatin (15). Rap1p with the C terminus deleted (Rap1ΔCp) retains the ability to bind TG1-3 tracts sequence specifically; however, rap1ΔC mutants are defective in telomere length regulation as well as in silencing of telomere-positioned genes (23, 25). The former is due to abolition of interactions with the Rif proteins (16, 41), whereas the latter is a result of loss of the Sir proteins from telomeres (29).
To understand whether the C terminus of Rap1p is required for replication pausing, we assayed replication in rap1ΔC mutants both through a tract of internally located TG1-3 repeats and through telomeric regions. A rap1ΔC mutation similar to a well-characterized rap1-17 allele, which lacks the C-terminal 165 amino acids, was introduced into RRM3 and rrm3Δ strains carrying the internally inserted 355-bp TG1-3 repeat tract described above (Fig. 7A). The transformants were screened for the desired deletions and, along with the parental strains, immediately grown for replication assays to minimize the telomere elongation effect caused by the rap1ΔC mutation. By the time of cell harvesting for DNA isolation, the telomeres had elongated by ∼400 bp in these rap1ΔC derivatives of both RRM3 and rrm3Δ cells (Fig. 7C).
To assay replication through the Y′ telomeric regions, the DNA samples were digested with EcoRI plus NcoI so that both subterminal and terminal Y′ elements produced fragments of a similar length and could be combined in the 2D GE assays. As expected, the sample from the RAP1 rrm3Δ cells revealed replication pausing at two positions (Fig. 7D, lower left panel). One was some distance away from the TG1-3 repeats (Fig. 7D) and was likely caused by inactive Y′ ARSs as shown above (Fig. 6). This pausing was greatly alleviated by the rap1ΔC mutation, presumably because in rap1ΔC mutants Sir3p is no longer localized to telomeres, resulting in the increase of the firing frequency of subtelomeric ARSs close to 100% (39), thereby minimizing the portion of inactive ARSs that cause replication pausing. The other pausing site was close to the diagonal line of linear molecules, almost at the 2N DNA content position (Fig. 7D, lower left), and therefore corresponded to the replication pausing at TG1-3 telomeric repeat tracts. These pausing intermediates were generated at the telomeric EcoRI fragments, rather than at the internal EcoRI-NcoI fragments, since no such spot was present when a similar 2D gel was probed selectively for the internal EcoRI-NcoI fragments (Fig. 4A, lower right). This pausing was not relieved by the rap1ΔC mutation, since the 2N spot on the diagonal line of linear molecules was clearly seen in the rap1ΔC rrm3Δ sample (Fig. 7D, lower right). Furthermore, both rap1ΔC RRM3 and rap1ΔC rrm3Δ cells accumulated large bubbles as replication intermediates (Fig. 7D, upper and lower right). We noted that in rap1ΔC cells the Y′ ARS at the terminal EcoRI fragment is located more centrally than in RAP1 yeast, since the rap1ΔC mutation confers the long-telomere phenotype (see the diagrams in Fig. 7D), and theoretically this could naturally delay replication bubble opening on the telomeric side of the analyzed fragment. However, this was not the case. In our experiment the telomere length of the rap1ΔC cells was increased by only ∼400 bp (Fig. 7C), thus positioning the ARS ∼1 kb away from the telomeric end and 3.35 kb away from the EcoRI site. In the control experiment (shown in Fig. 3A), where ARS1 was positioned similarly on an assayed restriction fragment, no bubble arc could be seen. Therefore, in rap1ΔC cells, replication still proceeds more slowly through the telomeric TG1-3 tracts, resulting in the accumulation of large bubble replication intermediates.
We also probed for the replication intermediates at the DNA fragment containing the internally located tract of TG1-3 repeats (Fig. 7A) in the rap1ΔC derivatives of RRM3 and rrm3Δ cells. Consistent with our results for the replication of the telomeric regions, the rap1ΔC mutant derivatives retained the replication pausing caused by the insertion of the 355 bp of yeast telomeric TG1-3 tracts (Fig. 7E). We conclude that Rap1ΔCp is sufficient for replication pausing at telomeres as well as at TG1-3 repeat tracts in internal chromosomal regions.
Loss of telomeric Tbf1p and Reb1p binding sites does not affect replication fork pausing at the subtelomeric regions.
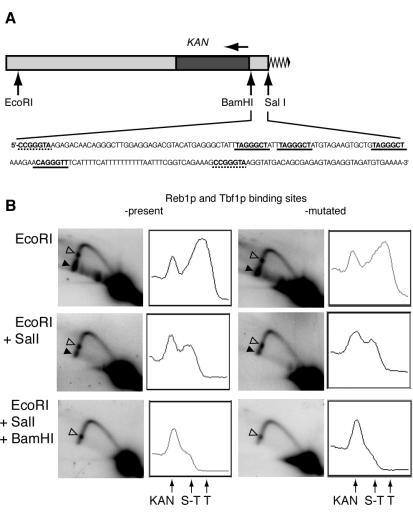
All S. cerevisiae telomeres contain subtelomeric antisilencing region (STAR) elements that include binding sites for Reb1p and Tbf1p (12). Binding of these two proteins was suggested to prevent spreading of heterochromatin seeded at telomeres, thereby counteracting the silencing of genes in subtelomeric regions (12). We tested whether binding of Reb1p and Tbf1p per se or spreading of heterochromatin in the absence of the antisilencers could affect replication of subtelomeric DNA. First, to test the effect of Reb1p and Tbf1p binding, we constructed two rrm3Δ strains that had different Y′ subtelomeric regions substituted in the same arm of the same chromosome. We replaced the Y′ ARS with KAN in both strains and, in one strain, left the STAR element intact, and in the other strain, we mutated all the Reb1 and Tbf1 binding sites by deleting two out of the three essential Gs in each of the binding sequences (Fig. 8A).
FIG. 8.
Replication pausing in the subtelomeric regions. (A) Schematic diagram of telomere constructs used for analysis. Y′ sequences are shown in light gray, the KAN gene is in dark gray, and the telomeric tract is a zigzag line. The direction of KAN transcription is shown by the horizontal arrow above the gene. The sequence of the Y′ STAR element (between the BamHI and SalI sites) is as in wt cells. The binding sites for Reb1p (dashed line) and Tbf1p (solid line) are underlined and in bold. (B) Replication intermediates at telomeres of rrm3Δ mutant yeast with and without Reb1p and Tbf1p binding sites. The black arrowheads point to the telomeric and subtelomeric pauses, and the no-fill arrowheads show the pause within KAN. The graphs to the right of each gel represent PhosphorImager signal intensity analysis scanned along the left arms of the fork arcs (from upper position to lower left). The arrows under the graphs point to the peaks of signal intensity caused by replication pausing inside the KAN cassette (KAN), in the subtelomeric region (S-T), and at the telomeres (T).
DNA samples from nonsynchronized cultures of each strain were digested with EcoRI, EcoRI and SalI, or EcoRI, SalI, and BamHI. Replication intermediates were analyzed by 2D GE, followed by Southern hybridization to a KAN-specific probe. Telomeric TG1-3 repeats were present in the analyzed EcoRI fragments but absent in fragments resulting from the double and triple digests. The EcoRI and EcoRI-SalI fragments but not EcoRI-BamHI fragments contained either the native or the mutated STAR element. Because the analyzed fragments did not contain an ARS, all the replication forks were expected to move in one direction, towards the telomeres. Therefore, Y-like structures with paused replication forks in the analyzed subtelomeric or telomeric regions were expected at the termini of the arcs, close to the 2N points on the diagonal line of linear molecules.
As expected, digestion with EcoRI alone revealed replication pausing close to the telomeric ends of the EcoRI fragments (Fig. 8B, top panels). The visible accumulated paused replication intermediates were partly but not completely lost when the telomeric TG1-3 tracts were removed from the analyzed fragment by EcoRI plus SalI double digestion (Fig. 8B, middle panels). The residual pausing was independent of the presence of the wt STAR element. This pausing was located in the STAR element because EcoRI-BamHI fragments no longer showed any pause intermediates (Fig. 8B, bottom panels). There was also an additional replication pausing site, seen as a spot in the middle of the left arm of the arcs at the EcoRI gels and migrating to the bottom of the arcs as the analyzed fragments were shortened by the combined restriction digestions (Fig. 8B). This site was in the KAN gene, since it was still present after EcoRI-SalI-BamHI digestion. All three replication pauses were comparably alleviated in RRM3 yeast, but they were not suppressed by deletion of SIR2 in the rrm3Δ mutants (data not shown). These results, together with the finding that sir2Δ cells still had the telomeric pause, suggest that neither the antisilencers Reb1p and Tbf1p nor the Sir proteins impose any barrier on replication fork movement at telomeres.
Analysis of newly synthesized DNA in ligase-deficient cells.
To verify the location of replication pauses in subtelomeric regions at Y′ telomeres by an independent method, we used a modified version (see Materials and Methods) of neutral/alkaline 2D GE that allows analysis of newly replicated DNA strands (18). During replication, DNA ligase (encoded by CDC9 in budding yeast) ligates the discontinuities that occur on the daughter strands as a result of discontinuous DNA synthesis following RNA primer removal and strand filling-in. When the nascent DNA strands are not ligated, their length and sequence allow one to distinguish between leading strands and Okazaki fragments. We reasoned that such analysis could provide information on the position of initiation of replication as well as directionality of fork movement and perhaps fork pausing. Loss of Cdc9p, predicted to prevent nascent strands from being ligated to one another, is lethal. Mutants carrying the temperature-sensitive allele cdc9-1, when shifted to nonpermissive temperature, arrest in S phase in a RAD9 dependent manner (36); cdc9-1 rad9Δ cells shifted to the restrictive temperature overcome the S-phase arrest and divide once (36). We therefore grew cdc9-1 rad9Δ cells at room temperature until early log phase and then shifted the culture to 37°C for one cell doubling and analyzed the nonligated newly synthesized strands by separating them from the parental DNA under alkaline gel conditions. Such fragments were clearly visible as described below (Fig. 9A). Under similar experimental conditions, no such DNA products were detected in either CDC9 RAD9 cells or CDC9 rad9Δ cells, while cdc9-1 RAD9 yeast produced some intermediates after prolonged incubations at the restrictive temperature (data not shown). As expected, inactivation of polymerase α with the cdc17-1 temperature-sensitive allele suppressed the production of the replication intermediates in cdc9-1 rad9Δ cells and cdc9-1 RAD9 cells (data not shown), confirming that replication was required to generate these observed DNA products.
FIG. 9.
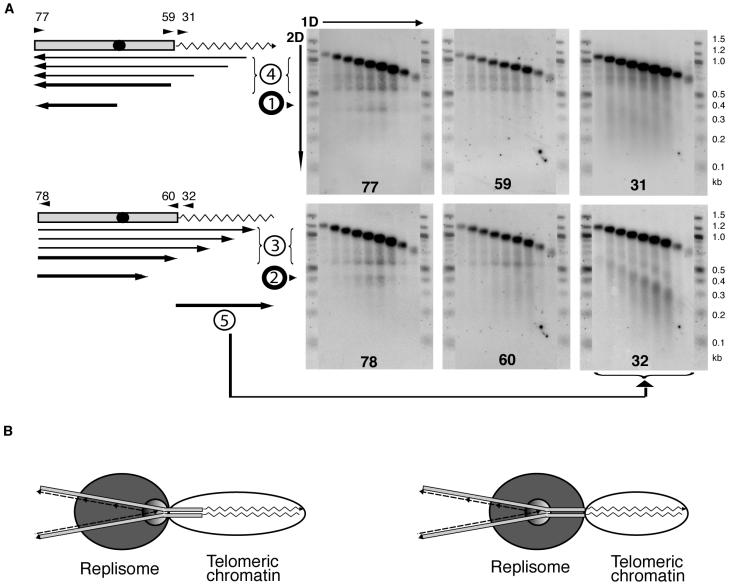
Nonligated replication and recombination intermediates at Y′ telomeres. (A) The telomeric KpnI fragments of Y′ telomeres consist of 517 bp of subtelomeric DNA (gray bar) that contains an ARS (black circle) and telomeric TG1-3 repeats (zigzag line). The upper diagram shows parental G-strand and replication intermediates detected in Southern blots with probes 77, 59, and 31 that consist of G-strand sequences and therefore hybridize to the parental C-strand and C-strand replication intermediates (upper row of panels). The bottom diagram shows parental C-strand and intermediates detected with the C-strand-resembling probes 78, 60, and 32, i.e., parental G-strand and G-strand replication intermediates (bottom row of panels). The arrows below each map represent the replication intermediate fragments detected. The arrows point toward the 3′ end of the single-stranded DNA fragments identified in the blots on the right. Probes 77 and 78 are complementary to each other, and their sequences correspond to the 50 bp of Y′ on the telomere side of the KpnI restriction site. Probes 59 and 60 cover the 50 bp of the immediate telomere-proximal sequence within the subtelomeric region. Probes 31 and 32 hybridize to telomeric DNA TG1-3 repeats. Numbers at the bottom of the gels correspond to the probes used for hybridization. All the detected intermediates are grouped in five different classes (shown as circled numbers) that are discussed in the text (products 1 and 2) and supplementary data (products 3 to 5). A 100-bp ladder (New England Biolabs) was run on both sides of every gel and used to determine the approximate size of the DNA. (B) Two models explaining why replication slows down ∼100 bp away from the telomeric repeats. Left, subtelomeric DNA is included in the telomeric chromatin that inhibits the replisome progression; right, replication slows down in the subtelomeric region because the replication center of the replisome is some distance away from its front surface that encounters the barrier. In this model, the parental DNA strands could be unwound and held as single-stranded DNA at the moment of pausing, but they would reanneal during the DNA purification procedures.
The DNA from cdc9-1 rad9Δ cells incubated for one cell doubling at the nonpermissive temperature was purified and digested with KpnI or XhoI (data not shown). The telomeric fragments were fractionated by size (see Materials and Methods), and the fractions were run on an alkaline 2% agarose gel to separate newly synthesized and parental strands. The DNA was analyzed by Southern blotting, using different oligonucleotides as probes (Fig. 9A).
The strong upper band in every sample represents intact parental DNA strands and nascent strands that were synthesized as leading strands that extend throughout the analyzed fragment. Several species of replication intermediates were observed. We detected a single-stranded discrete DNA molecule of ∼350 nucleotides that was inferred to originate at the Y′ ARSs and be the product of leading-strand DNA synthesis moving toward centromeres (replication product 1) (Fig. 9A). We also detected replication products slightly over 400 bases long, whose 3′ends terminated in the subtelomeric region containing the binding sites for Reb1p and Tbf1p (replication product 2). Such an intermediate is exactly that predicted to result from replication pausing at telomeres mapped as described above. The presence of this intermediate (product 2) thus independently confirmed our data indicating that when the replisome stalls at the telomeric TG1-3 tract DNA, replication itself actually stops in the subtelomeric region, a short distance away from the actual boundary of the TG1-3 tract sequence. A possible model to explain the nature of the other single-stranded DNA species (products 3 to 5) is described in the supplemental material.
DISCUSSION
We have studied DNA replication in the major class of natural yeast telomeres, those that contain Y′ elements. With the eventual goal of a complete understanding of the replication process for telomeres, we analyzed telomere replication by focusing on Y′ ARS firing activity during the cell cycle and replication fork progression through subtelomeric and telomeric DNA. Previous work on these questions has in many cases used artificial yeast telomeres in vivo. We also analyzed the previously reported replication pausing at telomeres (20) for its dependence on telomeric DNA sequence as well as a variety of proteins involved in telomere maintenance. Our detailed analysis of replication intermediates at telomeres maps the major replication pausing site to a position ∼100 bp in from the telomeric DNA tract itself.
Timing of telomere replication in the cell cycle is controlled by differential activity of Y′ ARSs.
The isotope density transfer technique has previously been used to analyze ARS firing and replication timing at different loci of the yeast genome, including telomeres (7, 11, 28, 33, 39). With this method, Y′ element sequences were reported to replicate overall late in S phase (7, 28, 39). Here, by using neutral/neutral 2D GE to distinguish between internal and terminal Y′s, we have independently confirmed that the ARSs of the terminal Y′ elements fire at the same time as a canonical late ARS and therefore, by inference, that the terminal repeats are replicated late in S phase. However, a new finding was that the internal Y′ ARSs were activated earlier, at a time indistinguishable from the known early-firing origin ARS1. These findings indicate that the internal and terminal Y′ elements are under different replication timing controls. Since the terminal and internal repeats are completely identical in their DNA sequences yet their ARSs have different activation times, this system can be very useful for studying the regulation of ARS firing in yeast. At present, we can suggest two explanations for our results. One is based on the earlier data that the proximity of an ARS to a telomere, rather than simply to a stretch of TG1-3 repeats, causes its late activation (11). By this model, the ARSs of the internal Y′s are fired earlier primarily due to their remoteness from the telomeres. The other possibility is that the timing of ARS firing is defined by the length of the TG1-3 tract next to the internal and terminal Y′s. Sufficient telomere length is required for the spreading of telomeric heterochromatin, via the Sir2/3/4 proteins, into the subtelomeric DNA, causing gene silencing and late ARS activation in those regions (39). Short telomeres, as well as sir mutations, alleviate silencing, and lack of Sir3p has been shown to result in early activation of the Y′ ARSs (39). However, it is unknown whether the terminal ARSs at short telomeres are activated early. Such a possibility is indirectly supported by the finding that inactivation of the YKU genes, which causes a short-telomere phenotype, leads to early activation of subtelomeric ARSs (7). Therefore, the length of TG1-3 stretches may be the determinant influencing the timing of ARS activation at the neighboring Y′s, through its effect on the seeding and spreading of heterochromatin.
What causes replication pausing at telomeres?
A replication fork pause at telomeres, which is exacerbated in the absence of the Rrm3 helicase, has been previously reported (20). Using a gene candidate approach, we further analyzed this pausing in order to explore its primary cause(s). First, we tested whether the pausing is a result of some (hypothetical) telomere-specific DNA structure that could arise from telomere-telomere or telomere-internal locus interactions. Such structures could include strand invasion of the 3′ end of telomeric DNA in cis or in trans (expected to be dependent on RAD52 in combination with RAD50 or RAD51), triplex or quadruplex DNA formation (possibly dependent on Cdp1p and Stm1p, which each bind these DNA structures in vitro), or other hypothetical interactions that result in telomere clustering (which is abolished in gbp2 mutants). However, inactivation of each of these genes named here failed to cause any detectable change in the replication pattern or pausing as assayed by 2D GE (data not shown).
We next tested whether the telomeric pausing results solely from the high G-C DNA content of the TG1-3 tracts at telomeres, which might cause difficulties in strand separation during replication. However, when we compared replication pausing at the internally located yeast TG1-3 or a Tetrahymena T2G4 telomeric repeat tract, only the yeast repeats, but not the comparably G-C-rich Tetrahymena repeats, caused replication pausing. Therefore, the replication barrier is specific to the DNA sequence of yeast telomeres rather than being caused by G-C richness per se. This pausing is not polar, because at least when the TG1-3 repeats are in an internal location, they pause progression of a replication fork approaching from either direction (Fig. 7). Two known proteins, Rap1 and Cdc13, bind, respectively, double-stranded and single-stranded yeast telomeric DNA in a sequence-specific manner. Since internally located TG1-3 repeats, which are not a substrate for Cdc13p binding, can cause replication pausing (20; also our results [Fig. 7]), then the binding of Rap1p (or its associated proteins) on telomeric DNA is the most likely cause of replication pausing.
Rif1p, Rif2p, and the Sir proteins are the known Rap1-interacting molecules at telomeres (2, 16, 29, 41). However, deletion of RIF1, RIF2, SIR2, or SIR3 had no effect on the replication pausing pattern. Furthermore, the replication pausing was not alleviated, either at telomeres or at an internally located tract of yeast telomeric TG1-3 repeat sequence, by deleting the C terminus of Rap1p. This domain of Rap1p is required for recruiting of both Rif and Sir proteins by Rap1 to telomeres and other loci in the genome. Together these data indicate that neither Sir nor Rif proteins are the cause of replication pausing at telomeres. Furthermore, the removal of binding sites for Reb1p and Tbf1p, which is predicted to increase the amount of the Sir proteins at the modified telomere (12), did not affect the replication pausing. Therefore, neither the silencers Sir2/3/4 nor the antisilencers Reb1 and Tbf1 are necessary to impose the barrier on replication fork progression at telomeres; rather, DNA-bound Rap1p itself, or potentially an interacting partner(s) that can bind Rap1p without the C terminus, imposes a barrier for replication fork movement through a DNA locus.
Deleting a variety of genes is known to lead to a short-telomere phenotype. Each of these deletions alleviated the telomeric replication pause. By preelongating telomeres, we have shown that these deleted proteins are not required for the pausing. These results are consistent with our data on replication through the short internal tracts of TG1-3 located between tandem Y′ elements, where replication pausing was undetectable in wt cells and even in rrm3Δ mutants was much less prominent than at the telomeres (Fig. 4A). Hence, we propose that the strength of the replication barrier caused by yeast telomeric repeats is proportional to the length of the TG1-3 tract and likely related to the number of Rap1 or other protein molecules bound to that DNA. An interesting predicted consequence of the lack of pausing at a short telomere is that the replication fork reaches the short-telomere tip sooner in the cell cycle than it does for a longer telomere. This might allow telomerase activity to start preferentially early on that telomere, thereby ensuring that it has time in that cell cycle to elongate the shortened telomere. The recent finding that DNA polymerase α in S. pombe complexes with telomerase (8) is consistent with this proposal.
In addition to the telomeric tract-induced pause, Ivessa and colleagues (20) observed extra replication pausing sites in the subtelomeric regions and hypothesized that one of them could be caused by inactive ARSs. We extended this observation and mapped the pausing site to a 189-bp locus that contains the Y′ element ARS. When the ARS was active the pausing did not occur, strongly arguing that only an inactive ARS at this location causes replication pausing. We also observed an RRM3-dependent replication pausing site within the KAN cassette. Therefore, the action of Rrm3p in alleviation of replication pausing might not be limited to telomeres and rDNA but occur at many sites throughout the yeast genome. Lack of Rrm3p could therefore prolong S phase, thereby potentially increasing the window of opportunity for telomerase to act in S phase. This may account for the slight increase in telomere length in rrm3Δ mutants (20).
All the replication pauses observed in yeast, as described above and reported by Ivessa et al. (19, 20), are RRM3 dependent, suggesting a general role for this helicase in promoting replication through pausing sites. Rrm3p interacts with PCNA (37) and therefore is likely to function at the replication fork. We therefore also tested the hypothesis that Rrm3p might affect the firing frequency of Y′ ARSs (Fig. 5); however, we showed that the changes in the replication pattern on 2D GE caused by inactivation of RRM3 are, rather, a result of enhanced replication pauses that lead to accumulation of the observed large bubbles and double-fork replication intermediates.
Architecture of replication pausing at telomeres.
All the results presented here and reported previously (20) indicate that replication pausing at telomeres is primarily caused by TG1-3 repeats. However, cutting off solely the telomeric repeat sequence from the analyzed fragment did not completely eliminate the accumulation of forked intermediates at the ∼2N spot (Fig. 8). Thus, the position of the pause indicated that DNA synthesis slowed down or arrested before the replication forks actually reach the TG1-3 regions. This conclusion was independently confirmed by analysis of replication intermediates that accumulate in ligase-deficient cells, which revealed a discrete size class of DNA strands with the 3′ end terminated ∼100 bases away from the telomeric TG1-3 tract (Fig. 9A, see intermediate 2). Although the 200-bp subtelomeric sequence in every yeast telomere (Y′ and X) contains STAR elements characterized by the binding of the two antisilencer proteins, Reb1p and Tbf1p, removal of these Reb1p and Tbf1p binding sites in a Y′ telomere did not eliminate replication pausing in that locus.
We propose two models for why the replication pause maps to the subtelomeric region. First, subtelomeric DNA might be bound by some other, yet-unknown protein(s) or might be a part of a large DNA-protein complex that forms a high-order telomere structure (for example, the previously proposed fold-back loop), and this structure is the cause of replication pausing (Fig. 9B, left). This idea is consistent with the finding that the Tetrahymena nonnucleosomal protected complex extends more than ∼50 bp (but <100 bp) in from the terminal T2G4 repeat tracts themselves (5). Alternatively, we suggest that the 100-bp spacing between the terminal TG1-3 tract and the DNA fork pause represents the distance between the front surface of the replisome as it encounters the barrier and its actual site of DNA polymerization at the fork. In this model, if the replisome starts progressing more slowly once it reaches the TG1-3 repeats, then the rate of replication would begin decreasing some distance behind (Fig. 9B, right).
Supplementary Material
Acknowledgments
We thank members of the Blackburn and Herskowitz labs for support and encouraging discussions. We acknowledge Jennifer Fung, Jue Lin, Tanya Williams, and anonymous referees for critical reading of the manuscript and helpful suggestions.
This work was supported by NIH grants GM59466 (to I.H.) and GM26259 (to E.H.B.)
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
This paper is dedicated to the memory of Ira Herskowitz (deceased on 28 April 2003).
REFERENCES
- 1.Berman, J., C. Y. Tachibana, and B. K. Tye. 1986. Identification of a telomere-binding activity from yeast. Proc. Natl. Acad. Sci. USA 83:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourns, B. D., M. K. Alexander, A. M. Smith, and V. A. Zakian. 1998. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol. 18:5600-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer, B. J., D. Lockshon, and W. L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71:267-276. [DOI] [PubMed] [Google Scholar]
- 4.Chan, C. S., and B. K. Tye. 1983. Organization of DNA sequences and replication origins at yeast telomeres. Cell 33:563-573. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, P., and E. H. Blackburn. 1998. Two types of telomeric chromatin in Tetrahymena thermophila. J. Mol. Biol. 280:327-344. [DOI] [PubMed] [Google Scholar]
- 6.Conrad, M. N., J. H. Wright, A. J. Wolf, and V. A. Zakian. 1990. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63:739-750. [DOI] [PubMed] [Google Scholar]
- 7.Cosgrove, A. J., C. A. Nieduszynski, and A. D. Donaldson. 2002. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 16:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlen, M., P. Sunnerhagen, and T. S. Wang. 2003. Replication proteins influence the maintenance of telomere length and telomerase protein stability. Mol. Cell. Biol. 23:3031-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgaard, J. Z., and A. J. Klar. 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400:181-184. [DOI] [PubMed] [Google Scholar]
- 10.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson, B. M., and W. L. Fangman. 1992. A position effect on the time of replication origin activation in yeast. Cell 68:333-339. [DOI] [PubMed] [Google Scholar]
- 12.Fourel, G., E. Revardel, C. E. Koering, and E. Gilson. 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18:2522-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, K. L., and B. J. Brewer. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262:613-627. [DOI] [PubMed] [Google Scholar]
- 14.Gerbi, S. A., and A. K. Bielinsky. 1997. Replication initiation point mapping. Methods 13:271-280. [DOI] [PubMed] [Google Scholar]
- 15.Grunstein, M. 1998. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell 93:325-328. [DOI] [PubMed] [Google Scholar]
- 16.Hardy, C. F., L. Sussel, and D. Shore. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6:801-814. [DOI] [PubMed] [Google Scholar]
- 17.Hecht, A., S. Strahl-Bolsinger, and M. Grunstein. 1996. Spreading of transcriptional repressor Sir3 from telomeric heterochromatin. Nature 383:92-95. [DOI] [PubMed] [Google Scholar]
- 18.Huberman, J. A., L. D. Spotila, K. A. Nawotka, S. M. el-Assouli, and L. R. Davis. 1987. The in vivo replication origin of the yeast 2 microns plasmid. Cell 51:473-481. [DOI] [PubMed] [Google Scholar]
- 19.Ivessa, A. S., J. Q. Zhou, and V. A. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100:479-489. [DOI] [PubMed] [Google Scholar]
- 20.Ivessa, A. S., J. Q. Zhou, V. P. Schulz, E. K. Monson, and V. A. Zakian. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16:1383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi, T. 2003. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 23:9178-9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, T., and T. Horiuchi. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1:465-474. [DOI] [PubMed] [Google Scholar]
- 23.Kyrion, G., K. Liu, C. Liu, and A. J. Lustig. 1993. Rap1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 7:1146-1159. [DOI] [PubMed] [Google Scholar]
- 24.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 25.Liu, C., X. Mao, and A. J. Lustig. 1994. Mutational analysis defines a C-terminal tail domain of Rap1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics 138:1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 27.Longtine, M. S., N. M. Wilson, M. E. Petracek, and J. Berman. 1989. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from Rap1. Curr. Genet. 16:225-239. [DOI] [PubMed] [Google Scholar]
- 28.McCarroll, R. M., and W. L. Fangman. 1988. Time of replication of yeast centromeres and telomeres. Cell 54:505-513. [DOI] [PubMed] [Google Scholar]
- 29.Moretti, P., K. Freeman, L. Coodly, and D. Shore. 1994. Evidence that a complex of Sir proteins interacts with the silencer and telomere-binding protein Rap1. Genes Dev. 8:2257-2269. [DOI] [PubMed] [Google Scholar]
- 30.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 31.Olovnikov, A. M. 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41:181-190. [DOI] [PubMed] [Google Scholar]
- 32.Pryde, F. E., H. C. Gorham, and E. J. Louis. 1997. Chromosome ends: all the same under their caps. Curr. Opin. Genet. Dev. 7:822-828. [DOI] [PubMed] [Google Scholar]
- 33.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 34.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. 5.36-5.39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schiestl, R. H., P. Reynolds, S. Prakash, and L. Prakash. 1989. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol. Cell. Biol. 9:1882-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt, K. H., K. L. Derry, and R. D. Kolodner. 2002. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 277:45331-45337. [DOI] [PubMed] [Google Scholar]
- 38.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson, J. B., and D. E. Gottschling. 1999. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 13:146-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson, J. D. 1972. Origin of concatemeric T7 DNA. Nature 239:197-201. [DOI] [PubMed] [Google Scholar]
- 41.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748-760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.