Abstract
We developed a targeted RNA sequencing method that couples microfluidics-based multiplex PCR and deep sequencing (mmPCR-seq) to uniformly and simultaneously amplify up to 960 loci in 48 samples independently of their gene expression levels, and accurately and cost-effectively measure allelic ratios even for low-quantity or low-quality RNA samples. We applied mmPCR-seq to RNA editing and allele-specific expression studies. mmPCR-seq complements RNA-seq and provides a highly desirable solution for future applications.
RNA allelic ratios, including RNA editing and allele-specific expression (ASE), are quantitative traits. Adenosine-to-Inosine (A-to-I) editing, the most common type of RNA editing in metazoans1, is tightly controlled2, 3. The editing level of specific sites is critical as aberrant editing has been linked with various diseases4. ASE is a phenomenon where two alleles of a gene within an individual exhibit unequal expression. It is largely considered to reflect the effects of functional cis acting variants5. The ability to accurately measure RNA allelic ratios is critical to study RNA editing and ASE.
RNA sequencing (RNA-seq) has been used to quantify RNA editing (editotyping) and ASE (allelotyping)6–9. However, the intrinsic limitation of RNA-seq is the dynamic range of RNA expression, which leads to inaccurate quantification of allelic ratios for genes with low to moderate expression levels. This limitation cannot be overcome by the conventional targeted genome resequencing technologies that often capture all desired genes simultaneously in a single reaction10. In targeted RNA-seq, by capturing and sequencing transcripts of interest hybridized with oligonucleotide baits, the dynamic range of RNA is maintained11, 12. The padlock probe-based approach we recently developed for editotyping and allelotyping was unable to evenly amplify different loci, due to the wide range of gene expression and different efficiency among padlock probes3, 13.
To uniformly amplify multiple transcripts and obtain accurate quantification of allelic ratios requires a PCR-based approach that allows individualized and saturated amplification of different loci. Several studies have coupled regular PCR and subsequent deep sequencing to quantify RNA allelic ratios2, 14, 15, however the throughput is very low. To enhance throughput, we developed an assay that couples microfluidics-based multiplex PCR and next generation sequencing (mmPCR-seq) (Fig. 1a; Online Methods). Built on the Fluidigm Access Array platform that amplifies 48 PCR products from each of the 48 genomic DNA samples on a single microfluidic chip, we have made several substantial improvements to enable uniform amplification of up to 960 loci from each of the 48 cDNA samples. Resulting PCR amplicons are barcoded for each sample, then subjected to next-generation sequencing to obtain deep coverage allowing accurate measurement of allelic ratios.
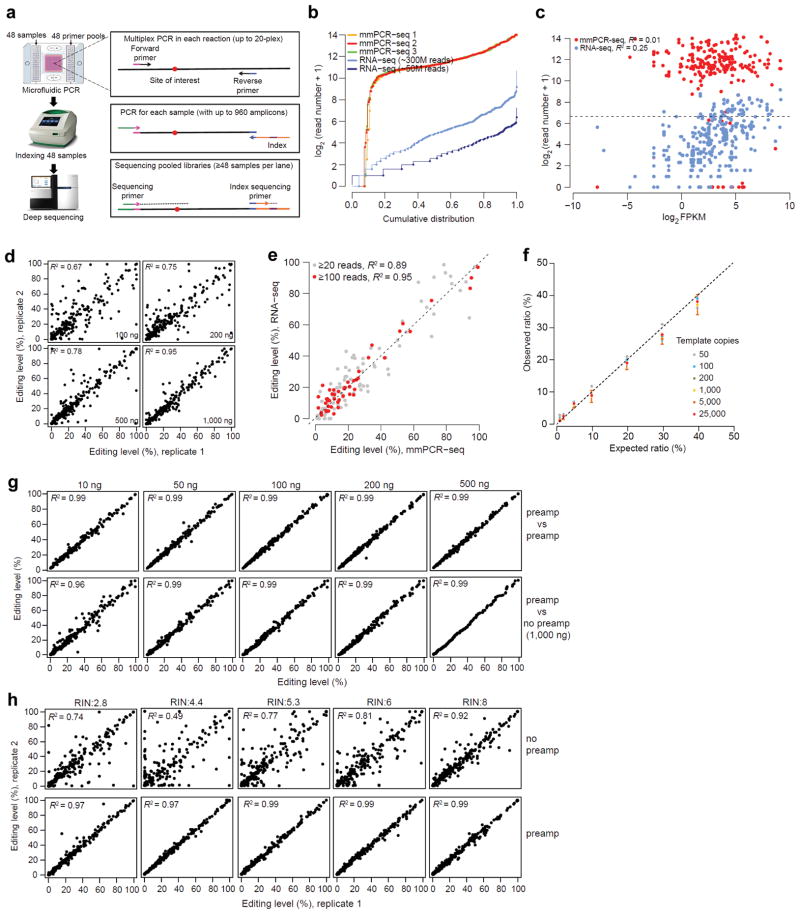
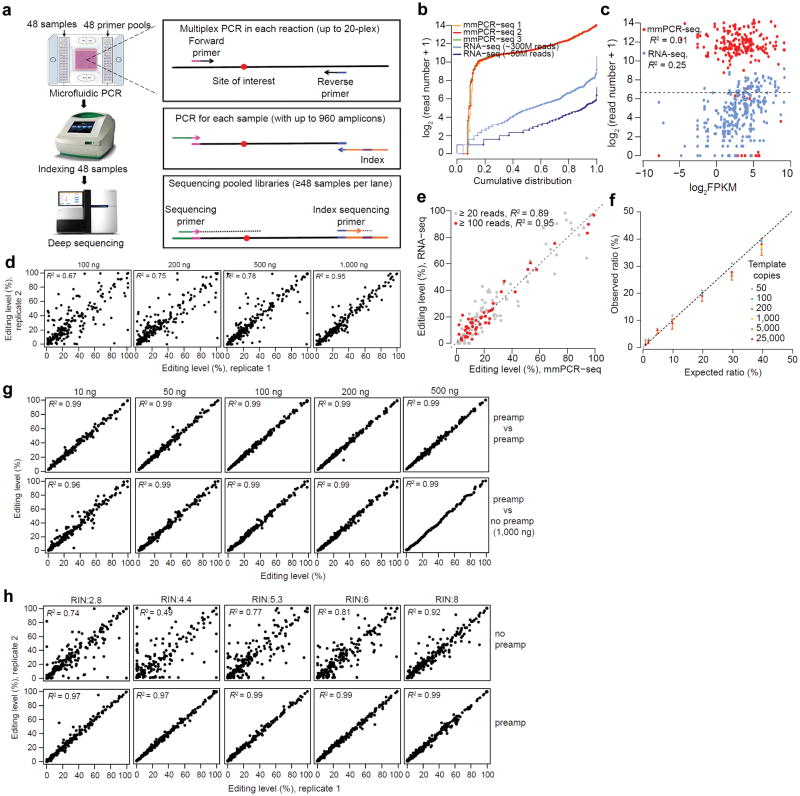
Figure 1.
The development and performance of mmPCR-seq. (a) Schematic diagram of mmPCR-seq. (b) Uniformity of different amplicons. 240 RNA editing loci were amplified using 1 ug of HBRR cDNA sample. Read numbers of three technical replicates were normalized to 0.8 million mapped reads per sample. The coverage of the same sites from RNA-seq data (both the 300 M full set and the 50 M subset) was also shown. (c) Relationship between the coverage of individual sites and the gene expression levels reported in Fragments Per Kilobase of transcript per Million mapped reads (FPKM). For mmPCR-seq, the average depth of three technical replicates was shown. The Pearson correlation coefficient (R2) was indicated. (d) Relationship between the reproducibility of RNA editing measurements in technical replicates and the amount of cDNA input. A full description of comparison among three technical replicates is shown in Supplementary Fig. 3. (e) Comparison of editing levels measured by mmPCR-seq and RNA-seq. Sites with at least 20 or 100 reads in RNA-seq were used, respectively. (f) The comparison between expected allelic ratio and observed ratio measured by mmPCR-seq. The observed allelic ratio is the average of 6 sites. Different amounts of templates (copies of molecules per PCR reaction) were indicated in different colors. Standard deviation for 1,000 copies of template was shown as an example. A full description of comparison is shown in Supplementary Table 2. (g) Reproducibility of editing level measurement using pre-amplified cDNAs. Top row: technical replicates using pre-amplified samples. Bottom row: the pre-amplified cDNA versus the un-amplified 1,000 ng of cDNA. (h) Reproducibility of RNA editing level measurement in technical duplicates using low quality RNA samples, without (top row) and with (bottom row) preamplification. RNA integration number (RIN) was indicated. For un-amplified samples, 1,000 ng of cDNA was used. For pre-amplified samples, 200 ng of cDNA was used.
We developed and optimized mmPCR-seq using RNA editing sites because RNA editing has widely distributed levels, in contrast to ASE whose levels are mostly around 50%. To capture 240 loci containing 287 known RNA editing sites (Supplementary Data 1), we optimized an existing software16 to design multiplex PCR primers (24 pools of 10-plex primers) (Online Methods, Supplementary Data 2).
To achieve uniform amplification of different loci, we first tested different numbers of PCR cycles (30, 35, and 40) using two 10-plex primer pools. We found that 40 cycles led to evenly distributed amplicons and therefore used 40 cycles for subsequent PCR amplifications (Supplementary Note 1, Supplementary Fig. 1). We then carried out mmPCR to amplify 240 loci with 24 pools of 10-plex primers to assess whether our method led to uniform amplification independent of gene expression levels (Supplementary Table 1). We used a cDNA template derived from the Human Brain Reference RNA (HBRR) sample that has deep RNA-seq data available. Additionally, to assess the effect of PCR reaction complexity on uniformity, we carried out 5-plex PCR by splitting each 10-plex reaction into two (Online Methods). After sequencing the pooled amplicons, we observed similar uniformity between 10-plex and 5-plex PCR reactions (Supplementary Fig. 2), suggesting robust design of multiplex primers. Of the 240 primer pairs, 20 (~8.3%) failed, which is consistent with failure rate in conventional single-plex primer designs17 (Fig. 1b). Of the 220 successful amplicons, 201 (91%) were covered with reads within a 16-fold difference (24, from 210 to 214) (Fig. 1b). Importantly, the coverage of amplicons is independent of gene expression levels, in contrast to RNA-seq (Fig. 1c).
We reasoned that the accuracy of allelic ratio quantification using mmPCR-seq may depend on the cDNA input amount due to sampling bottlenecks for sites located in lowly expressed genes (Supplementary Note 2). To assess this, we performed technical replicates using different amounts (100, 200, 500, and 1000 ng) of cDNA. As expected, the reproducibility of measurements increased with more input template (Fig. 1d, Supplementary Figs. 3, 4). We estimate that 1000 ng of input cDNA converts to ~200 cells of RNA materials used in each one of the 48 PCR reactions, thus demonstrating accurate measurement for sites in genes even with few copies per cell (Supplementary Note 2).
To confirm that our reproducible measurements are also accurate, we first compared our results with the editing levels measured by RNA-seq (a combination of 300 million reads of the HBRR sample) (Online Methods). The measurements from two independent methods were highly similar, particularly when sufficient numbers of RNA-seq reads were available (Fig. 1e). Furthermore, we compared allele ratios measured by mmPCR-seq with the known ratios in the prepared mixtures. We mixed two different alleles from the same locus (differing by a single base) at different frequencies (1, 2, 5, 10, 20, 30, and 40%), and measured the allelic frequency using mmPCR-seq. We tested a total of 6 loci and found that the observed allelic frequency is consistent with the expected frequency across a wide range of template concentrations (Fig. 1f, Supplementary Table 2, Online Methods).
To assess the performance of mmPCR-seq on RNA samples with low quantity, we tested how preamplification of low amounts of cDNA would impact the accuracy and uniformity (Online Methods). We estimated that, with preamplification, target regions can be amplified by ~1000 fold (Supplementary Fig. 5). When we applied the preamplification to low amounts (10, 50, 100, 200, and 500 ng) of the HBRR sample in technical replicates, the uniformity is comparable to that achieved without preamplification (Supplementary Fig. 2). More importantly, the measurements were not only reproducible, but also in agreement with the measurements using cDNA without preamplification (Fig. 1g). Additionally, we showed that preamplification enhanced the accuracy of quantifying allelic ratios for low quality RNA samples (Fig. 1h), showcasing the relevance of our method to study tissue-banked samples. We further summarized the measurement variances for different types of input samples (Supplementary Fig. 6). The optimized parameters of mmPCR for different types of input samples were also summarized (Supplementary Table 3).
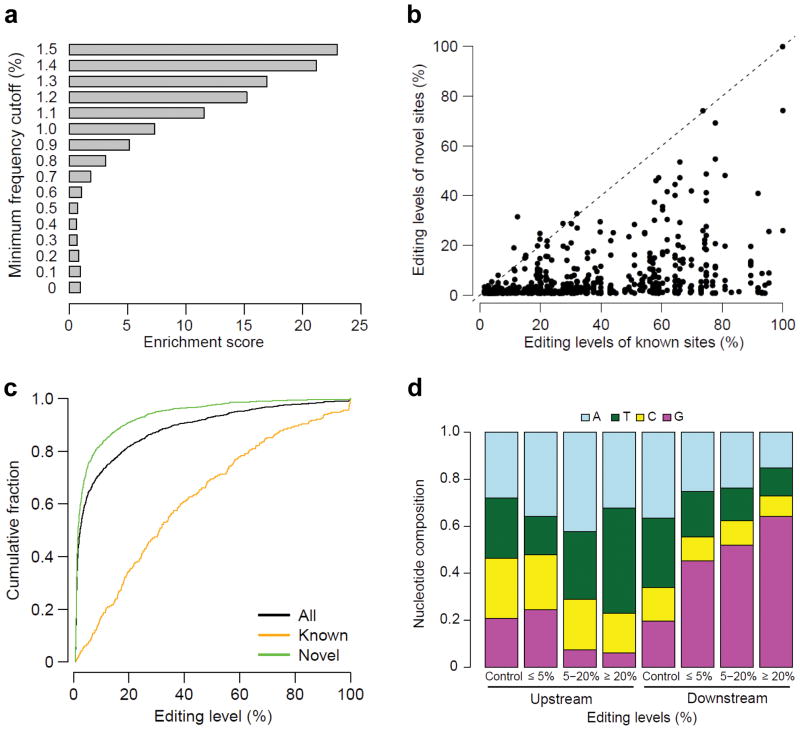
We next took advantage of the ultra-deep coverage of mmPCR-seq to identify novel editing sites surrounding known editing sites. To distinguish RNA editing events from sequencing errors, we determined 1.1% as the cutoff variant frequency, with an estimated false discovery rate of 1.4–8.6% (Fig. 2a, Supplementary Note 3, Supplementary Data 3). In the HBRR sample, we identified 418 novel sites that are usually edited at lower levels as compared to the nearby known sites (Fig. 2b). We further examined 7 post-mortem brain Brodmann area 44 samples. In total, we identified 914 novel sites that are usually edited at extremely low level (Fig. 2c). Nevertheless, 109 novel sites were edited at ≥20% editing level, suggesting that mmPCR-seq complements RNA-seq to identify more moderately and highly edited sites. Of all novel sites, 136 are nonrepetitive nonsynonymous events, greatly expanding the repertoire of recoding targets (Supplementary Data 4). As expected18, increased editing levels are associated with the more obvious trends of the under- and over- representation of guanosines immediately 5′ and 3′ of the edited adenosine (Fig. 2d, Supplementary Fig. 7) as well as the higher TAG and AAG triplet fraction (Supplementary Fig. 8). In contrast to our observation that a large number of lowly edited sites were nearby moderately and highly edited events, there was a deficit of lowly edited sites at loci that lack known RNA editing sites. This is consistent with the coupling hypothesis19, while not fully supporting the continuous probing hypothesis20 (Supplementary Note 4).
Figure 2.
Characterizing novel RNA editing sites identified via mmPCR-seq. (a) Relationship between the enrichment score and the minimum frequency of variant nucleotide. The enrichment score was calculated as the number of detected A-to-G or T-to-C mismatches per 10 kb in RNA samples divided by the counterpart in DNA samples. The average value of two samples was shown. Sites with ≥1,000 reads were used. (b) Pairwise comparison of the editing level of each novel site and the nearest known site. (c) The cumulative distribution of RNA editing levels for different groups of sites. For each site, the highest editing level among 8 samples was shown. (d) Nucleotide composition in positions immediately upstream and downstream of the edited sites. The control is all “A” sites that are covered by mmPCR-seq reads and not edited in any samples tested.
We next reasoned that mmPCR-seq would provide high-resolution ASE measurements. We examined 960 SNPs (Online Methods) in lymphoblastoid cell lines (LCLs) from 16 individuals within a three-generation family (Supplementary Fig. 9), with additional genome and RNA sequencing data available (X. Li et al., unpublished). We designed 48 pools of 20-plex primers; this higher throughput allowed us to examine all 960 SNPs on a single chip.
We carried out mmPCR-seq for 16 LCLs in technical triplicates. By sequencing all 48 samples in one Illumina HiSeq lane with 101 bp single-end reads we obtained an average of ~1.6 million mapped reads per sample (Supplementary Table 4) and an average of 770 sites (91%) had ≥100 reads per LCL. At 100 reads, this provides an ability to detect allelic effects of 1.56-fold with a binomial probability of 0.05. We confirmed the accuracy of allelic ratio measurements and the more uniform distribution of amplicons independent of gene expression level as in the editotyping assays (Supplementary Note 5, Supplementary Figs. 10–13, Supplementary Table 5). To determine the relative performance of mmPCR-seq using a personal genome sequencer, which can be more amenable to targeted studies, we sequenced the same samples on one llumina MiSeq run with 150bp single-end reads. Although 10 times fewer reads were obtained, an average of 647 sites (80%) had ≥100 reads per LCL (Supplementary Note 5). Additionally, the ASE was highly similar between HiSeq and MiSeq measurements (Supplementary Fig. 14). These data show the feasibility of using MiSeq to quantify ASE of hundreds of SNPs, as well as the possibility of using HiSeq to examine more SNPs and/or samples.
We next assessed the ability of ASE detection from mmPCR-seq. Of all heterozygous SNPs we investigated, 14–22% showed ASE in genes expressed at any level (Fig. 3a). In contrast, RNA-seq detected a substantially smaller fraction of ASE, especially in lowly or moderately expressed genes, when using the matched sites of mmPCR-seq (Fig. 3a). The same conclusion was obtained when combining all heterozygous SNPs in the same gene to call ASE (Supplementary Note 6, Supplementary Figs. 15, 16). As accumulation of regulatory variations, and therefore expectation of ASE discovery, could potentially differ for highly versus lowly expressed genes, we further tested the heritability of the discovered effects across expression levels. We discovered better capture of heritable effects for mmPCR-seq compared to RNA-seq across all expression levels (Supplementary Fig. 17). These results highlight the utility of mmPCR-seq to detect ASE for genes independent of gene expression. We further determined the optimal read depth for capturing heritable ASE. By taking random subsets of reads from mmPCR-seq, we observe that heritability estimates become saturated for sites with 400 or more reads (Fig. 3b), which suggests the requirement of high coverage to capture ASE more accurately.
Figure 3.
ASE analysis of mmPCR-seq data. (a) Proportion of sites with ASE among all heterozygous sites using mmPCR-seq or RNA-seq for genes with various expression levels. The matched sites obtained from mmPCR-seq and RNA-seq data were used. (b) Correlation of ASE of IBD siblings calculated from sub-samplings of mmPCR-seq data. Pearson correlation coefficient R2 reflects degree of correlation between ASE among IBD siblings. Standard deviation was shown. (c) The percentage and number of cis-eQTL genes identified using different permutation p value thresholds. Only genes with mmPCR-seq sites were used in the analysis. Three models were used for the mapping. TReC: Total Read Count, an association model using gene expression information only, TReCASE(RNA-seq): a joint model of Total Read Count and ASE measured by RNA-seq, TReCASE(mmPCR-seq): a joint model of Total Read Count and ASE measured by mmPCR-seq.
By combining genotypic and allelic effects on expression, it is possible to jointly detect cis-eQTL21. However, adopting this approach using RNA-seq data has limited success because of insufficient sequencing coverage for accurate ASE quantification21. To assess the impact of accurate measurement of ASE on cis-eQTL mapping, we compared the number of cis-eQTL genes identified using gene expression information only, gene expression information and ASE measured by RNA-seq, and gene expression information and ASE measured by mmPCR-seq. We found that a combination of RNA-seq and ASE measured by mmPCR-seq nearly doubled the power to detect cis-eQTL (Fig. 3c).
In summary, we demonstrated that mmPCR-seq provides a flexible high-throughput methodology to measure RNA allelic ratios. mmPCR-seq is cost-effective (as low as $24 per sample) and efficient (<8 minutes per sample) (Supplementary Table 6). These features, along with its utility for low-quantity or low-quality RNA samples, will enable future large-scale editotyping and allelotyping studies. At its current throughput, mmPCR-seq enables measuring RNA editing levels for all known nonrepetitive recoding sites, thus providing a one-size-fits-all solution. For ASE studies, by focusing on a customized set of SNPs, mmPCR-seq may help identify regulatory effects linked to GWAS variants22, map causal regulatory variation23 or identify epistatic interactions24, pinpoint genetic interactions that define the variable penetrance of coding variants25, and provide more complete insight into the genetics of gene expression.
ONLINE METHODS
Multiplex PCR primer design
To design multiplex PCR primers, we modified the yamPCR program which designs multiplex PCR primers based on genome sequence information16. For a given group of loci of interest, yamPCR involves four steps to design primers: (i) cutting flanking sequences of given loci and identifying candidate primers using a modified Primer 3 program, (ii) searching all partial and complete matches of the candidate primers on the genome by Blast, (iii) predicting the amplification products of all possible primer combinations based on a thermodynamic model, and (iv) deducing a group of compatible primers that are specific to the loci of interest. We made three modifications on this program to design multiplex primers based on transcriptome information. First, for a given site, we mapped it to a specific transcript based on gene annotation information and cut flanking cDNA sequences from given genome file. Notably, for RNA editing site primer design, since editing occurs before splicing, the editing level of a specific site should be the same for different isoforms of a gene. Therefore the selection of different transcripts in this step does not affect the quantification of editing levels. For ASE site primer design, the incorporation of transcript selection in this step can be used to examine ASE in different isoforms of a selected gene. Second, we made a database that contains cDNA sequences of all human genes and used this database instead of genome sequence for Blast search. For each gene, we merged all its exons to generate a representative transcript. Third, the editing or ASE sites were designed to be within 100 bp from the 5′ end of one primer. To avoid that the designed primers are located in regions with variants, which may potentially affect the allelic ratio quantification, the genome that has been masked using known SNPs and editing sites is used to design primers. A Perl script used to design the primers is available in http://lilab.stanford.edu/mmPCR/.
Editing site collection
We collected a total of 330 exonic A-to-I editing sites from two resources: (i) a subset (88) of 400 nonrepetitive editing sites identified in a genome-wide editing site scanning3. Our previous study identified 400 nonrepetitive editing sites in human genome. To obtain high confidence sites, we selected 88 of them that are edited in the brain RNA-seq datasets we collected (Supplementary Table 7). (ii) 49 newly identified nonrepetitive nonsynonymous editing sites and 176 UTR sites located in nonrepetitive regions8.
We designed 24 10-plex primers which cover 287 editing sites (Supplementary Data 1) within 240 loci. The sizes of amplicons range from 150 to 350 bp. Primer sequences are listed in Supplementary Data 2.
Allele-specific expression site collection
We selected two sets of sites from 16 LCL samples. Group 1 contains 617 sites that are known eQTL genes6. Group 2 contains 755 non-eQTL sites6. All selected SNPs are within expressed genes, and heterozygous in at least three individuals within the family. We designed 48 20-plex primers which cover 960 sites, including 410 and 550 Group 1 and 2 sites respectively (Supplementary Data 1). The sizes of amplicons range from 150 to 350 bp. Primer sequences are listed in Supplementary Data 2.
RNA and cDNA preparation
Total RNAs were extracted with RNeasy Kit (Qiagen) or Trizol (Invitrogen). After DNase I treatment, 2–10 ug of total RNA was used to synthesize the cDNA with iScript™ Advanced cDNA Synthesis Kit (Bio-Rad). cDNA was purified with MinElute PCR Purification Kit (Qiagen) and concentrated using SpeedVac if needed.
Preamplification of cDNA samples
To preamplify cDNA samples before the microfluidic multiplex PCR, 1 ul of cDNA sample was added to 9 ul of pre-sample mix containing 5 ul of KAPA2G Fast Multiplex Mix (2X) and 2.4 ul of primer pool (104 nM per primer). We used the following PCR program for preamplification: 95°C 10 min, and 15 cycles of 95°C 15 sec and 60°C 4 min. Following preamplification, amplified product was purified with MinElute PCR Purification Kit (Qiagen).
Preparation of mixtures with known allelic ratio
We randomly chose 6 SNPs from the set of 960 ASE sites with one requirement: we are able to find an individual from the 16-person family with homozygous AA genotype and another with homozygous BB genotype. For each of the 6 loci, we carried out PCR to obtain amplicons with either A or B allele, and mixed them at different allelic frequency (A/(A+B) = 1, 2, 5, 10, 20, 30, and 40%). In addition to testing different allelic frequency, we also titrated the concentration of different templates in the Fluidigm reactions to test the effect of different amounts of template molecule copies on the accuracy of allelic ratio measurements.
Target amplification on the Fluidigm Access Array microfluidic system
We loaded 4 ul of individual 5-, 10- or 20-plex primer pools (1 uM per primer) into the primer inlets of the 48.48 Access Array IFC (Fluidigm). To prepare the cDNA templates, 2.25 μl of each cDNA sample was added to 2.75 μl of pre-sample mix containing 2.5 ul KAPA2G 2X Fast Multiplex Mix (Kapa Biosystems) and 0.25 ul 20X Access Array sample loading buffer (Fluidigm). After the loading of both samples and primers via IFC Controller AX (Fluidigm), the IFC was subject to thermal cycling using FC1 Cycler (Fluidigm) with the following program: 50°C 2 min, 70°C 20 min, 95°C 10 min, 5, 10, or 15 cycles of 95°C 15 sec, 60°C 30 sec, and 72°C 60 sec, 2 cycles of 95°C 15 sec, 80°C 30 sec, 60°C 30 sec, and 72°C 60 sec, 8 cycles of 95°C 15 sec, 60°C 30 sec, and 72°C 60 sec, 2 cycles of 95°C 15 sec, 80°C 30 sec, 60°C 30 sec, and 72°C 60 sec, 8 cycles of 95°C 15 sec, 60°C 30 sec, and 72°C 60 sec, 5 cycles of 95°C 15 sec, 80°C 30 sec, 60°C 30 sec, and 72°C 60 sec, and 72°C for 3 min.
Sequencing adaptor and barcode addition
For each sample, 0.5 μl of the 100-fold diluted PCR products was added to 9.5 ul of pre-sample mix containing 5 ul of 2X KAPA2G Fast Multiplex Mix, 2 ul of primer mix (an universal forward primer (2 uM) and an reverse primer with different barcode sequence (2 uM)) and 2.5 ul of water. Primer sequences are listed in Supplementary Table 8. We used the following PCR program: 95°C 10 min, 4 cycles of 95°C 30 sec, 55°C 30 sec, and 72°C 1 min, 10 cycles of 95°C 30 sec, and 72°C 1 min, and 72°C 5 min. All pools were then combined at equal volume, and purified via QIAquick PCR purification kit (Qiagen).
Fluidigm library sequencing data analysis
Libraries were pooled and sequenced in Illumina HiSeq with 101 bp single-end reads. We used FASTX Toolkit to demultiplex the raw reads. We used BWA26 to align reads to a combination of the reference genome and exonic sequences surrounding known splicing junctions from gene models annotated in RefSeq and Gencode V12. We chose the length of the splicing junction regions to be slightly shorter than the reads to prevent redundant hits. For allelic ratio count, we took the bases with a minimum quality score of 20. For read depth count, we took the coverage of the representative sites in each amplicon.
To call novel RNA editing sites, we required variants to be supported by at least two mismatch reads with base quality score ≥20 and mapping quality score ≥20. We also removed all known SNPs present in dbSNP (except SNPs of molecular type “cDNA”; database version 135; http://www.ncbi.nlm.nih.gov/SNP/), the 1000 Genomes Project or the University of Washington Exome Sequencing Project (http://evs.gs.washington.edu/EVS/).
RNA-seq data analysis
The Human Brain Reference RNA sample (HBRR, Ambion, Catalog #6050) consists of total RNA extracted from several regions of the brains from 23 adult donors. We obtained three Illumina RNA-seq datasets for this sample from the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra). A list of datasets is shown in Supplementary Table 9. We mapped RNA-seq reads to the human genome as previously described8. To obtain gene expression levels, we used a pipeline comprising Tophat and Cufflinks27, 28.
ASE analysis
Whole genome sequencing data of the family were obtained from Complete Genomics. Samples were sequenced to an average genome-wide coverage of 80X. SNPs are called by Complete Genomics Analysis Pipeline (version 2.0.0).
We performed binomial test to obtain the p values of deviations from 0.5 on the raw allelic counts. q values were then used to estimate the proportion of non-nulls. q values were calculated using qvalue function from R package. A q value <=0.05 was used as the cutoff of statistical significance. We also required that the magnitude of allelic drift be larger than 0.1 (allelic frequency <0.4 or >0.6) to be considered allele-specific. We required ≥5 reads for both reference and alternative alleles with allelic frequency >0.01 and <0.99 to avoid wrongly assigned homozygous SNPs due to potential genotyping or sequencing errors.
To perform cis-eQTL mapping using both genetic data and RNA-seq or by also including ASE information, we employed asSeq R package (http://www.bios.unc.edu/~weisun/software/asSeq.htm)21. We examined all SNPs within the gene body or outside the gene body (within 200 kb of the transcription start or end sites) to identify cis-eQTLs. All 16 individuals were treated as unrelated for mapping. We considered the most significant cis-eQTL for each gene and calculated a permutation p value for each gene using the trecaseP function in the asSeq package with the following parameters: min.AS.reads=20, min.AS.sample=5, min.n.het=5, local.only = TRUE, local.distance = 200000, np.max=500, np=c(20, 100), aim.p=c(0.5, 0.2). For a fair comparison between mmPCR-seq and RNA-seq, we used the same set of ASE sites (selected in the mmPCR-seq assay) for analysis.
We investigated the heritability of ASE within the family by comparing the ASE between Identical-By-Descent (IBD) siblings. IBD sharing block are defined by recombination positions in the family. Haplotype blocks are inferred by method PedIBD29. Pearson correlation coefficient R2 was used to reflect degree of correlation between ASE among IBD siblings.
Statistical analysis
All statistical analyses were performed with either R packages or Matlab.
Supplementary Material
Acknowledgments
We thank M. Snyder (Stanford) for access to the Fluidigm Access Array system, and W. Sun (UNC Chapel Hill) for advice on TReCASE analysis. R.Z. was partially supported by a Dean’s fellowship from Stanford University School of Medicine. G.R. was supported by Stanford Graduate Fellowship. This work was supported by US National Institutes of Health (GM102484), Ellison Medical Foundation, and United States - Israel Binational Science Foundation (to J.B.L.), and Edward Mallinckrodt Jr Foundation (to S.B.M.).
Footnotes
Accession codes US National Center for Biotechnology Information Sequence Read Archive: SRP029341.
AUTHOR CONTRIBUTIONS
R.Z. developed and optimized the mmPCR-seq method with the help from G.R., K.S.S., S.B.M, and J.B.L. R.Z. and X.L. performed computational analyses with help from S.B.M, and J.B.L. G.T. provided the brain samples. R.Z., X. L., S.B.M., and J.B.L. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Nishikura K. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahlstedt H, Daniel C, Enstero M, Ohman M. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JB, et al. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 4.Maas S, Kawahara Y, Tamburro KM, Nishikura K. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastinen T, Hudson TJ. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 6.Montgomery SB, et al. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickrell JK, et al. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramaswami G, et al. Nat Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaswami G, et al. Nat Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng SB, et al. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin JZ, et al. Genome biology. 2009;10:R115. doi: 10.1186/gb-2009-10-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer TR, et al. Nature biotechnology. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K, et al. Nat Methods. 2009;6:613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eran A, et al. Mol Psychiatry. 2012 [Google Scholar]
- 15.Main B, et al. BMC Genomics. 2009;10:422. doi: 10.1186/1471-2164-10-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang K, et al. Nature genetics. 2006;38:382–387. doi: 10.1038/ng1741. [DOI] [PubMed] [Google Scholar]
- 17.Andreson R, Mols T, Remm M. Nucleic acids research. 2008;36:e66. doi: 10.1093/nar/gkn290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polson AG, Bass BL. Embo J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enstero M, Daniel C, Wahlstedt H, Major F, Ohman M. Nucleic acids research. 2009;37:6916–6926. doi: 10.1093/nar/gkp731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gommans WM, Mullen SP, Maas S. Bioessays. 2009;31:1137–1145. doi: 10.1002/bies.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun W. Biometrics. 2012;68:1–11. doi: 10.1111/j.1541-0420.2011.01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindorff LA, et al. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery SB, Lappalainen T, Gutierrez-Arcelus M, Dermitzakis ET. PLoS Genet. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lappalainen T, Montgomery SB, Nica AC, Dermitzakis ET. The American Journal of Human Genetics. 2011;89:459–463. doi: 10.1016/j.ajhg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacArthur DG, et al. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, Pachter L, Salzberg SL. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, et al. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Yin X, Li J. Bioinformatics. 2010;26:i191–198. doi: 10.1093/bioinformatics/btq222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.