Abstract
A major objective of systems biology is to organize molecular interactions as networks and to characterize information-flow within networks. We describe a computational framework to integrate protein-protein interaction (PPI) networks and genetic screens to predict the “signs” of interactions (i.e. activation/inhibition relationships). We constructed a Drosophila melanogaster signed PPI network, consisting of 6,125 signed PPIs connecting 3,352 proteins that can be used to identify positive and negative regulators of signaling pathways and protein complexes. We identified an unexpected role for the metabolic enzymes Enolase and Aldo-keto reductase as positive and negative regulators of proteolysis, respectively. Characterization of the activation/inhibition relationships between physically interacting proteins within signaling pathways will impact our understanding of many biological functions, including signal transduction and mechanisms of disease.
Introduction
Objectives of systems biology include organizing molecular interactions as networks and characterizing their structure, dynamics and controllability. Tremendous progress has been made using “omics” datasets to identify the parts and connections of these networks. For example, protein-protein interactions (PPIs), identified from yeast-two hybrid (Y2H) or affinity purification-mass spectrometry (AP-MS) approaches, have provided information on the biophysical interactions occurring between two or more proteins1–5. Similarly, systematic loss of function analysis such as RNA interference (RNAi) screens have identified sets of genes implicated in specific biological processes6. Integration of omics datasets and inferring information-flow are critical aspects of the reconstruction of signaling networks7. Such reconstructions reveal how proteins communicate and coordinate cellular functions, and allow researchers to explore the emergent properties of networks.
There is a need for systematic approaches to infer causal relationships between interacting proteins, by which we refer to the “direction” (edge direction), “sign” (activation/inhibition) and “mode” (e.g. phosphorylation, ubiquitination) of signal flow in PPI networks. Genome-scale reconstruction of signaling networks remains a challenge8, largely because of the difficulty of predicting such causal relationships, although small scale networks have been successfully reconstructed. Furthermore, databases of signaling pathways are incomplete, and annotations are inconsistent across databases9. Recent studies have attempted to infer direction of information-flow10–14 as well as to reconstruct kinase-substrate networks15 but few attempts have been made to predict activation/inhibition relationships among interacting proteins.
Here, we have developed a computational framework to predict the signs (positive or negative) of physical interactions using RNAi screens. In a positive PPI, proteins A and B interact to form a functional complex in which A activates B (or vice-versa). In a negative PPI, proteins A and B interact to form a protein complex in which A inhibits protein B (or vice-versa), such that one of the proteins is a negative regulator of the complex. We applied this framework to construct a Drosophila melanogaster signed PPI network and thereby identified unexpected roles for the metabolic enzymes Enolase and Aldo-keto reductase as positive and negative regulators, respectively, of proteolysis in Drosophila. Finally, we built a database, the Signed Protein-Protein Interaction network (SignedPPI), to access, build and navigate signed interaction networks (http://www.flyrnai.org/SignedPPI/).
Results
Development of a signed prediction framework
We compiled RNAi screens recording 42 phenotypes from various resources including the Drosophila RNAi Screening Center16, GenomeRNAi17, Neuroblasts Screen online databases18 and Bristle Screen online database19 (Methods and Supplementary Table 1). We also included results from an image-based RNAi screen measuring nucleolus size20 and six other phenotypes (Neumuller et al., unpublished data). With respect to the hits, the screens show an average 14% similarity with each other (Supplementary Fig. 1). Each screen identifies positive and negative regulators of a particular phenotype, allowing us to construct a phenotypic matrix where the rows correspond to genes and columns correspond to 49 different phenotypes (Fig. 1a); positive and negative regulators are color coded differently. Next, we used a simple correlation of phenotypes to predict activation/inhibition relationships, with positive correlations when both genes have the same color, and negative correlation when they have different colors. We compute a sign score (Sscore) when both the interacting proteins in a pair score in two or more screens (Fig. 1a, see methods). The sign score determines if the phenotypes have positive or negative correlations. We predict a positive edge sign (activation) if the Sscore is positive and a negative edge sign (inhibition) if the Sscore is negative.
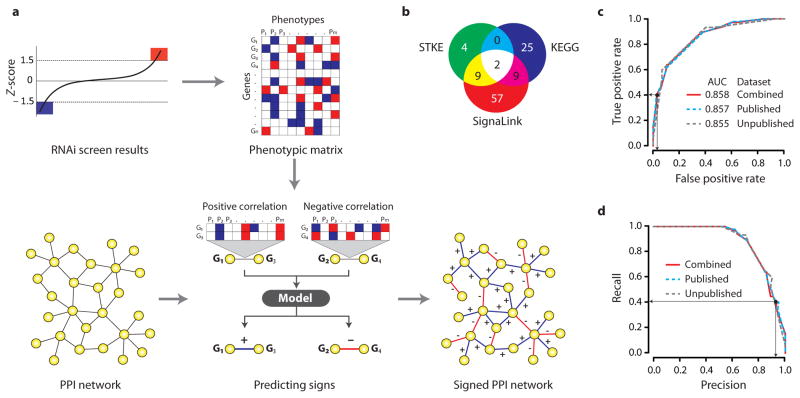
Figure 1.
Framework to predict the signs of protein interactions. (a) Schematic representation of the framework. (b) Sources of signaling PPIs with known edge signs. (c) ROC plot and (d) precision-recall curve shows the performance of the sign prediction model. Black dots and the arrows show the chosen Sscore cutoff (Sscore ≥ 1 or Sscore ≤ −1).
We used interactions with known activation/inhibitory relations from the literature to test our model and find an appropriate cutoff value for the sign score. We compiled such interactions from signaling pathway databases such as SignaLink21, Database of Cell signaling (http://stke.sciencemag.org/cm/) and KEGG22 (Supplementary Table 2 and methods). We selected 106 literature-based interactions where both proteins scored in two or more RNAi screens and define these as the positive reference set (PRS) (Fig. 1b, Supplementary Table 3). By reversing the original signs, we created a negative reference set (NRS; 106 interactions, see methods). Next, we used our model to predict sign and compared the results to the original annotations. We used three different RNAi datasets to assess the robustness of our model: published (42 phenotypes), unpublished (seven phenotypes), and combined (49 phenotypes). The results show that our model has good predictive power (area under ROC curve = 0.858) and is robust to various subsets of RNAi screen data (Fig. 1c–d and Supplementary Table 4). However, the predictive performance of sign score is lost with a randomized phenotype matrix (Supplementary Fig. 2). We found that the performance is comparable between a subset of reference interactions with respect to the source database or considering only positive or negative interaction signs (Supplementary Fig. 3a–b). A minimum of nine RNAi screens is needed to make a reliable prediction, but coverage increases with increasing number of screens (Supplementary Fig. 4a–b). Further, the relationship between the RNAi screens and number of hits in RNAi screens influences coverage but not the predictive performance (Supplementary Fig. 4c–f).
We identified an appropriate sign score cutoff value of Sscore ≥ 1 for positive signs and Sscore ≤ −1 for negative signs (Fig. 1c–d). At this cutoff value, we achieved 90% precision and 41% recall (2.8% false positive rate and 59% false negative rate). Note that we compared the performance of this simple model to various classifiers trained to predict signs using the phenotype matrix as features and found that the simple model performs better (Supplementary Table 5).
Constructing a signed Drosophila PPI network
We collected PPIs from major databases such as BioGrid23, IntAct24, DIP25, MINT26, DroID27 and DPiM1 (Supplementary Table 6), selecting PPIs identified as binary interactions (e.g. Y2H screen), high confidence AP-MS interactions and AP-MS interactions predicted to be direct interactions (see methods). The resulting integrated Drosophila network consists of 47,293 PPIs among 9,107 proteins. We next predicted signs for these Drosophila PPIs based on the 49 phenotypic data sets. The signed network consists of 6,125 PPIs connecting 3,352 proteins, among which 4,135 PPIs are positive interactions and 1,990 PPIs are negative (Fig. 2a and Supplementary Table 7), with the sign score of each interaction indicating the confidence of the predicted sign. Our predicted sign network consists of 13-fold more interactions as compared to the literature-based signed interactions (434 PPIs).
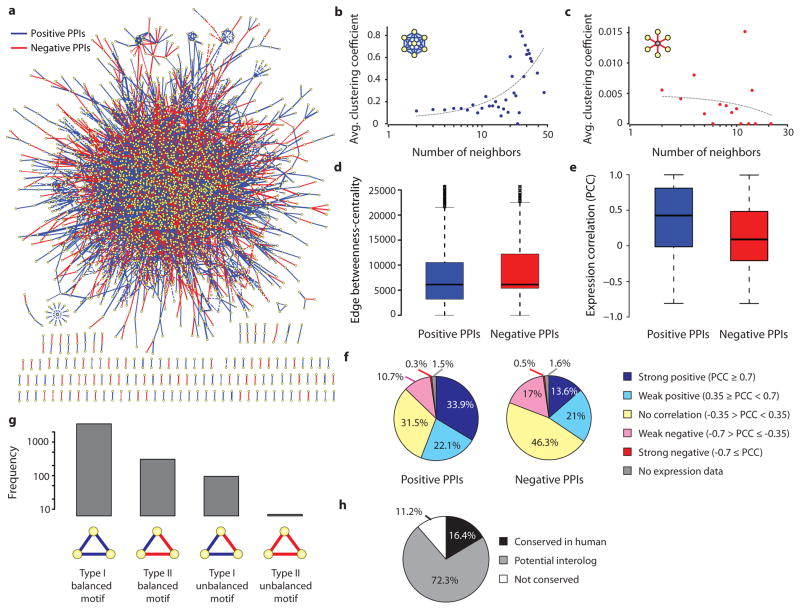
Figure 2.
Drosophila signed PPI network properties. (a) Signed PPI network. (b–f) Comparison of positive and negative interactions with respect to average clustering coefficient (b, c), edge betweenness-centrality (d) and gene expression correlation (e). Grey dotted line in (b, c) corresponds to regression line. (f) Classification of positive and negative interactions with respect to the gene-expression correlation (Pearson correlation coefficient). (g) Frequency of triad motifs in the signed PPI network. (h) Conservation of signed interactions.
We systematically analyzed various properties of the entire signed network and of sub-networks consisting only positive or negative interactions. We observed a positive correlation between the number of phenotypes regulated by a gene and the number of interactions of the gene (Supplementary Fig. 5). Further, sub-networks with positive and negative interactions show a similar degree distribution (Supplementary Fig. 6). Functional enrichment analysis reveals that kinases tend to be hubs with similar proportions of positive and negative interactions, whereas transcription co-regulators tend to be hubs with primarily positive interactions and transcriptional co-repressors are hubs with primarily negative interactions (Supplementary Table 8).
Correlating the “number of neighbors” and “average clustering coefficient” shows that hubs with positive interactions tend to cluster (Fig. 2b–c). Next, we compared positive and negative interactions with respect to the edge betweenness centrality, a measure based on the number of shortest paths that passes through an edge in the network. The inter-modular interactions, bridging different biological processes, tend to have high edge betweenness-centrality scores, whereas intra-modular interactions, such as interactions within a protein complex, tend to have low edge betweenness centrality28. Our analysis revealed that negative interactions tend to have high edge betweenness centrality, meaning that that they are likely to be inter-modular interactions, in contrast to positive interactions which are likely intra-modular interactions (p-value 2.2e-16; Wilcoxon test) (Fig. 2d).
Comparison of edge signs with gene expression from Drosophila developmental time-course data29 reveals that the positive interactions are more likely to show positive expression correlation (Pearson Correlation Coefficient (PCC)) than negative interactions (p-value=2.2e-16; Wilcoxon test) (Fig. 2e and Supplementary Fig. 7).
Although half of the negative interactions show no expression correlation (−0.35 > PCC < 0.35), almost 13% of these interactions have strong positive expression correlations (Fig. 2f), suggesting potential tight negative regulation. Furthermore, half of the positive interactions show positive expression correlation..
Including signs on a PPI network allows the application of “structure balance theory” which is based on the ratios of balanced and unbalanced triad motifs, enabling us to measure the stability of the network in a given condition30. In a triad motif, if the product of the signs is positive, the motif is defined as a balanced motif. Our analysis reveals that similar to social networks, signed PPI networks have more balanced than unbalanced motifs (Fig. 2g, Supplementary Table 9 and methods). Unbalanced motifs are particularly interesting because they are highly dynamic and unstable. For instance, Type-I unbalanced motifs, consisting of two positive and one negative interaction, could potentially function as negative feedback loops or incoherent feed-forward loops, which are both associated with adaptation responses and are crucial for system controllability31. We identified 95 Type-I unbalanced motifs in the signed network (Supplementary Table 10). The resource can also be used to systematically explore larger unbalanced motifs (4 nodes or greater) that could contribute to the network dynamics. Finally, 16% of the signed Drosophila interactions are conserved in human and another 72% are potential human interologs. 35% of the conserved interactions are linked directly to human disease proteins, suggesting the relevance of the signed network to human diseases (Supplementary Table 11).
We constructed sub-networks focusing on major signaling pathways9 and known protein complexes32 to explore novel positive and negative interactions linked to core components (Fig. 3, Supplementary Fig. 8). Compared to other signaling pathways the EGFR/ERK pathway is densely connected because of the availability of both PPI networks and functional datasets for this pathway6, 33. Consistent with Figure 2b, members of the same protein complexes are primarily connected to one another via positive interactions.
Figure 3.
Network representation of signed PPIs connecting known signaling pathways and protein complexes (see methods).
Validating novel regulators of the proteasome
We selected the proteasome complex to further investigate positive and negative interactions inferred using signed PPI networks. The proteasome is essential for regulating proteostasis via degradation of proteins modified by polyubiquitin. Moreover, deregulation of proteasome function is relevant to many human diseases, including neurodegeneration, cancer and cachexia34.
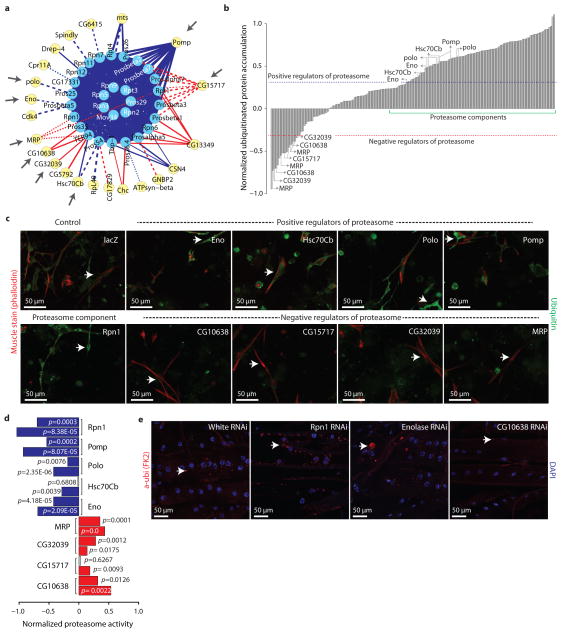
First, we constructed a proteasome sub-network with 51 nodes, including 29 proteins that are part of the proteasome complex and 22 proteins that interact with it (Fig. 4a). We selected these 22 interacting proteins based on high-confidence positive or negative interactions with the proteasome (Sscore ≥ 1.73 and Sscore ≤ −1.73) and for which we have three or more independent RNAi reagents for tissue culture experiments. Next, we knocked down these 51 selected genes (the 29 proteasome components served as controls) in primary embryonic muscle cells and assessed their effect on proteasome activity by measuring the accumulation of ubiquitinated proteins (Fig. 4b and Supplementary Data). Knocking down positive regulators should increase the accumulation of ubiquitinated proteins, whereas knocking down the negative regulators should decrease their accumulation. We identified genes as proteasome regulators if two or more independent RNAi reagents met the cutoff (see methods and Supplementary Table 12). Out of 10 putative hits, tests in primary cell culture showed that eight of them can regulate the proteasome in a manner that is consistent with the predicted edge signs (Figure 4b–c, Supplementary Table 12).
Figure 4.
Validation of predicted proteasome regulators. (a) Sub-network of proteasome complex (as in Figure 3). Grey arrows highlights subsequently validated proteins. (b) Results from the image-based RNAi screen measuring the accumulation of ubiquitinated proteins in primary muscle cells in which regulators shown in (a) has been knocked down with RNAi. Blue and red dotted-lines indicated the cut-off values used for positive and negative regulators, respectively. Green line highlights region corresponding to most proteasome core components. (c) Micrographs show muscle cells stained with phalloidin (red) and α-ubiquitin (green), in which the indicated candidate regulator has been knocked down with RNAi. Arrows point to ubiquitinated proteins in cells. (d) Enzymatic activity of the proteasome upon the indicated RNAi treatment in S2R+ cultured cells. Blue and red bars corresponds to significant reduction and increase in proteasome activity, respectively. Independent RNAi reagents are shown for each gene. (e) Micrographs show 3rd instar larval longitudinal muscles expressing the indicated RNAi hairpins under control of the muscle-specific driver line Dmef2-Gal4 (red=phalloidin, blue=DAPI). Arrows point to ubiquitin-labeled aggregates.
Next, we used a luminescent assay that measure the protease activities associated with the proteasome complex in S2R+ cultured cells to further validate our putative regulators. We observed that knockdown of candidate positive regulators (Eno, polo, Hsc70Cb and pomp) with independent RNAi reagents decreased proteasome activity (Fig. 4d, Supplementary Tables 13), while that of the candidate negative regulators (MRP, CG32039, CG15717, CG10638) increased this activity (Fig. 4d). Knockdown efficiency of the RNAi reagents determined by qPCR is shown in Supplementary Figure 9. Our experiments, together with the evidence of direct physical interactions (Supplementary Tables 14), show that the hits we identified using the signed PPI network are bonafide candidate regulators of the proteasome.
To further validate these regulators in vivo, we selected two metabolic enzymes Enolase (Eno) and CG10638, for which two or more transgenic RNAi fly lines were available. Using the muscle specific Dmef2-Gal4 line to drive the expression of the UAS-RNAi hairpins, we assayed the formation of ubiquitinated protein aggregates in the RNAi-treated muscles compared to control Regulatory particle non-ATPase 1 (rpn1, a proteasome component) and white RNAi knockdown. All RNAi constructs targeting Eno (three of three) and CG10638 (two of two) gave consistent phenotypes. In agreement with our predicted signs, knockdown of Eno resulted in an increase in ubiquitin aggregates, whereas knockdown of CG10638 led to a decrease in aggregates (Fig. 4e). Eno is a multifunctional protein with a key role in glycolysis35, and its role in proteasome regulation had not previously been established. CG10638 is an aldo-keto reductase (AKR) family member. AKRs catalyze the NADPH-dependent reduction of aldehydes and ketones to alcohols36. A subset of mammalian AKRs have previously shown to be regulated by drug-induced proteasome inhibition37. However, direct regulation of the proteasome or proteolysis by an AKR has not been previously reported. We note that knockdown of Eno or CG10638 has no effect on ubiquitin gene expression in S2R+ cell lines (Supplementary Fig. 10). Altogether, using the proteasome complex as an example, our results demonstrate the usefulness of predicted edge-signs to discover protein function.
Database to navigate signed PPI network
We created the SignedPPI database (http://www.flyrnai.org/SignedPPI/) to build and navigate signed interaction networks (Supplementary Fig. 11). In addition to PPI network, we also predicted signs for Drosophila functional interaction derived from the STRING database38. We successfully predicted signs for 40,216 functional interactions, including 31,178 positive and 9,038 negative interactions; these data are accessible via the SignedPPI database (Supplementary Table 15). We created prediction tool called SignPredictor that accepts a phenotype matrix and PPIs as input and predicts signed PPIs (Supplementary Software). The tool is implemented as a PERL module that can be downloaded from SignedPPI database and installed locally.
Discussion
Unlike previous studies which use genetic interaction correlation39 or phenotype similarity40 to predict functional interactions, we used phenotype correlation to predict the function of physical interactions (signs). Our method is robust to inherent noise in RNAi screens and has high predictive power. It is limited however to predict context dependent signs such as asymmetric bidirectional signs (e.g. negative-feedback loop between Cdc2 and anaphase-promoting complex (APC), where Cdc2 activates the APC, which in turn inactivates Cdc2).
The signed network we constructed for Drosophila, only covers ~10% of known PPIs, due to the limited number of RNAi data sets available. As more RNAi screens become available, it will become possible to further expand the scope and utility of the constructed network. In addition, the resource will benefit from additional PPI data sets such as ongoing interactome mapping projects41. Finally, the sign prediction approach could be easily applied to other species.
Our analysis of the Drosophila signed network revealed novel insights into the design principles of network organization and identified unexpected roles for two metabolic enzymes, Enolase (Eno) and Aldo-keto reductase in regulating proteasome proteasome function. The signed network opens up new scope for network analysis, such as the application of structure balance theory. Further integration of other information-flow properties such as edge-direction would enable sophisticated flow-based network analysis.
Online methods
Compiling RNAi screens
We compiled Drosophila RNAi screens that cover 49 phenotypes from the following resources: 1) Drososophila RNAi Screening Center (DRSC, www.flyrnai.org/) ; 2) GenomeRNAi (http://genomernai.de/GenomeRNAi/); 3) Neuroblasts Screen online database (http://neuroblasts.imba.oeaw.ac.at/); 4) Bristle Screen online database (http://bristlescreen.imba.oeaw.ac.at); and 5) unpublished data (7 phenotypes). Refer to Supplementary Table 1 for more details. The datasets are preprocessed based on general guidelines provided by DRSC to handle potential off-targets (only for datasets where amplicon level information is available). This includes: 1) filtering out the amplicons that have predicted off-target effects; 2) Averaging values of multiple RNAi reagents targeting same gene to consider the most consistent phenotype; 3) Filtered out the hits that are not shown to be expressed in a given cell line. The screens reported the phenotypic contributions as either Z-Score or categorical data. For screens with Z-scores, we used a standard cutoff of 1.5/−1.5, and define those genes with Z-score ≥ 1.5 as negative regulators and genes with Z-score ≤ −1.5 as positive regulators of the phenotype. For the categorical data, we directly used the annotation, e.g. “Down regulation of Wg Pathway” or ”Upregulation of Wg pathway,” to define positive and negative regulators of the phenotypes. Note that we only selected screens that include both positive and negative regulators of a specific phenotype.
Model for predicting signs
For each RNAi screen, the positive and negative regulators were distinguished with values +1 and −1 respectively. Genes that did not score in a particular screen was assigned value zero. We constructed a phenotype matrix by combining multiple RNA screens where the rows correspond to genes and columns correspond to the RNAi screens. In a given RNAi screen, if both interacting proteins have non-zero values, then the relationship is classified as either positive correlation (both +1 or both −1) or negative correlation (one is +1 and another is −1). For each interacting pair, we computed the total number of positive and negative correlations. Then we used a simple model to calculate a sign score (Sscore) for each interaction as follows
Pc, Nc corresponds to the number of positive and negative correlations, respectively. Tp is the total number of matching phenotypes (Pc + Nc). Note that Tp should be ≥ 2 in order to be considered for sign predictions (to compute correlation minimum two data points are needed). √Tp is the weight factor to assign more confidence for signs predicted based on a larger number of phenotypes. If a score has a positive value then the interaction gets a positive sign (activation); similarly, interactions with negative values are assigned a negative sign (inhibition).
Validation of the model
To validate the model, we compiled known signaling PPIs from SignaLink (http://signalink.org/), KEGG (http://www.genome.jp/kegg/pathway.html) and Database of Cell Signaling (STKE) (http://stke.sciencemag.org/cm/). All the data corresponds to the versions available online at those sites as of September 2012. The three datasets were integrated and we selected signaling PPIs with two or more matching phenotypes in phenotype matrix as the reference sets (Supplementary Table 2 and 3). Next, we manually curated the entire reference set by verifying the literature for the validity of the signs assigned by the databases. The sign annotation from the signaling databases was used as a PRS. To construct NRS, the signs from the signaling databases were reversed. Since our objective is to predict signs of physical interactions both the PRS and the NRS includes physically interacting pairs. However, PRS has true signs and NRS has incorrect signs. Consider example reference interactions, Dsor1-rolled from MAPK signaling pathway and Akt - Foxo from insulin signaling pathway. In the first example, Dsor1 (Drosophila ortholog of MAP2K) activates rolled (Drosophila ortholog of ERK/MAPK) by phosphorylation. In other words, whenever an interaction between these two proteins occur, the Dsor1 only activates rolled and never inhibits it and the interaction sign is always positive. In the next example, Akt inhibits Foxo by phosphorylation, hence the interaction sign is negative. In PRS the Dsor1 - rolled interaction is assigned a positive sign and Akt - Foxo interaction assigned a negative sign, but in NRS, they are assigned inverse signs (negative sign for Dsor1 – rolled and positive sign for Akt - Foxo interaction). Based on the current literature, Dsor1 never inhibits rolled and Akt never activates Foxo, making valid negative reference sets. Using the PRS and NRS, the true positive rate, false positive rate, precision and recall were calculated at various Sscore cutoff values. We then plotted receiver operating characteristic (ROC) and Precision-Recall (PR) curves.
Compiling PPI networks
We compiled experimentally identified PPIs from BioGrid, IntAct, DIP, MINT, DroID and DPiM (Supplementary Table 6). The data corresponds to versions available in September 2012. Next, we grouped the PPIs as direct (e.g. yeast-two hybrid screen) or indirect (e.g. affinity-purification followed by mass-spec) based on the experimental approach used to detect PPIs. We constructed binary interaction network as follows: 1) all interactions identified as direct interactions were selected; 2) high-confidence AP-MS interactions reported by literature were selected; 3) We analyzed the rest of the AP-MS interactions network to look for additional evidences such as domain-domain interactions, kinase-substrate interactions, interologs and genetic interactions as described by 33. We selected the AP-MS interactions that overlap with any of these networks and considered them as direct interactions.
Network analysis and visualization
To analyze properties of the signed network, we used various publicly available tools. Networks were visualized using Cytoscape, an open source platform for network analysis and visualization42. NetworkAnalyzer, a cytoscape plugin for analysis of network properties, was used to analyze the degree distribution, clustering coefficient, and edge betweenness centrality43. Both Cytoscape and NetworkAnalyzer were downloaded from www.cytoscape.org and installed locally. The triad motif enrichment analysis was performed using FANMOD tool44 (Supplementary Table 9). Triad motifs were extracted using perl scripts developed in house and freely available upon request.
Computing gene expression correlation
To compute correlation coefficients from gene expression profiles we used Drosophila developmental time course data from the modENCODE Consortium29. A gene expression matrix was generated from RNA-Seq data using the latest fly genome annotation as provided by modENCODE (http://www.modencode.org/). The profiles characterize expression dynamics for 15,998 coding and non-coding genes during 27 distinct stages of development, including 12 embryonic, six larval, six pupal and three sexed adult stages (30 data points in total). The Pearson correlation coefficient was computed for 6125 signed PPIs across 30 data points.
Constructing sub networks around signaling pathways and protein complexes
Annotation of signaling pathways was obtained from the SignaLink database21. The protein complex annotations are used as defined in protein complex enrichment analysis tool (COMPLEAT) resource32. For each pathway/protein complex, direct signed interactions were extracted and integrated with gene-expression correlation. Note that for this analysis we excluded signed interactions from the literature but not those predicted by our approach.
Preparation of dsRNAs
Gene-specific amplicons (<200–500 bp) were amplified by PCR using Choice Taq Mastermix (Denville Scientific, CB4070-8) from genomic DNA, using synthesized oligos with an attached T7 sequence. dsRNA was then synthesized from PCR templates using the T7 Megascript kit (Ambion), and product size was confirmed by gel electrophoresis. Following purification with Millipore Multiscreen PCR plates (#MANU03050), dsRNAs were quantified by measurement of the OD260 (Nano-drop 8000, Fisher Scientific), then stored at −20°C until use.
Primary cell culture RNAi experiments
Embryonic primary cell cultures were isolated from gastrulating Oregon R embryos as described previously45–47 and seeded in 384-well plates at 4 × 104 cells (10μl volume) per well. Each well contained 5μl dsRNA in water (0.25ug dsRNA) targeting a gene from the proteasome network or control dsRNAs targeting lacZ or thread. Following 20 hours incubation in serum-free M3 medium at 18°C, 30μl of serum-containing medium was added to each well for a final fetal calf serum concentration of 10%. Primary cells were then cultured for an additional 5 days at 18°C before fixation for 2hr in 2% formaldehyde. Cells were stained overnight at 4°C with α-ubiquitin mAb FK2 (Enzo Life Sciences, 1:400), then washed and stained for 2hrs with Alexa Fluor 594 goat α-mouse (Molecular probes, 1:1000), phalloidin Alexa Fluor 635 (Molecular Probes; 1:2000), and DAPI (Sigma, 1:5000), then washed again. All antibody incubations and washes were performed in PBT, except for a final rinse in PBS prior to image analysis. This experiment was repeated to give 4 biological replicates.
Image analysis
Acquisition of high quality images of the primary cell culture was performed with the Evotec Opera microscope at the DRSC (www.flyrnai.org/). Using a 20X water immersion lens, 24 microscope fields were obtained per well for both α-ubiquitin and phalloidin stains. Images were analyzed with MetaXpress High Content Image Acquisition & Analysis Software (Molecular Devices). For each microscope field, muscles were identified from the mixed population of cells by positive phalloidin staining. Then, ubiquitin aggregates were identified within these muscles using the MetaXpress granularity application module. Total ubiquitin aggregate area was divided by total muscle area for each field. These values were then combined to give a measure of the total area of aggregates per muscle cell area per well (ubiquitin accumulation). Normalized ubiquitinated protein accumulation was computed by taking log2 fold change of ubiquitin accumulation over lacZ control. For each amplicon (unique dsRNA design) the replicates (four) were combined and median values were obtained. A gene is considered a regulator of proteolysis if two or more amplicons targeting that gene met the threshold (±0.32 log2 fold change). Note, we did not consider a gene as a regulator if different dsRNAs gave inconsistent results, i.e. one scores as positive and another as negative. The dsRNAs used for the screening and the results are shown Supplementary Table 12.
Proteasome Activity Assay
For each experiment, 5 × 103 S2R+ cells in serum-free Schneider’s medium were aliquoted per well (384 well tissue culture plate) and incubated with 0.25μg dsRNA for 30′, before adding serum to a final volume of 30 μl. Following 1 day of incubation, 30 μl of Proteasome-Glo Trypsin-like Cell-Based reagent (Promega G8760) was added to each well, and the plate was incubated for 30′ at room temperature. Luminescence was measured with a SpectraMax Paradigm Plate Reader. Readings from cell-culture medium (no cells) control wells were subtracted from all experimental values. Four replicates were performed for each dsRNA, and readings were normalized to lacZ control RNAi wells. Normalized proteasome activity was computed by taking log2 median values from four replications.
qPCR
For each experiment, 2 × 106 S2R+ cells in serum-free Schneider’s medium were aliquoted per well (6 well tissue culture plate) and incubated with 20μg dsRNA for 30′, before adding serum. Two days later, cells were harvested into Trizol (Invitrogen), followed by phenol-chloroform extraction and purification with the RNeasy kit (Qiagen). Complementary DNA (cDNA) was synthesized with the iScript cDNA Synthesis kit (Bio-Rad), and quantitative RT-PCR was performed with the iQ SYBR Green Supermix (Bio-Rad). Rp49 was used as normalization reference. Relative quantitation of mRNA expression was calculated using the comparative CT method. The primers used were: Rp49, 5′-ATCGGTTACGGATCGAACAA-3′ (forward) and 5′-GACAATCTCCTTGCGCTTCT-3′ (reverse); Eno, 5′-CCGAGAACAAGAGCAAGTTCG-3′ (forward) and 5′-CATGGCCTCTGTGAAGCTGG-3′ (reverse); CG10638, 5′-AGCTCGCTCCGACTGTTAAG-3′ (forward) and 5′-AGGCCCAGAATTGGCATCTC-3′ (reverse); Rpn1, 5′-CCGACGCTGGAGAGTATGG-3′ (forward) and 5′-GCATGAACTTCAAAGGCTTGG-3′ (reverse); Hsc70Cb, 5′-CGTGGCCGCTAAGAACCAG-3′ (forward) and 5′-ATGCTCGTGAGTTCGTGTTGT-3′ (reverse); CG32039, 5′-GAGCCTGTCTGTCCTGCTG-3′ (forward) and 5′-AGGCGATGGCATCAGGTT-3′ (reverse); pomp, 5′-TATCAGCCATCACTGAAAGTCCA-3′ (forward) and 5′-GTTGCGGTTGTACTGGTGC-3′ (reverse); MRP, 5′-CGCCTTCTACTGGGCGTTC-3′ (forward) and 5′-ACCAGAGCTTTGCTCACGTTC-3′ (reverse); mts, 5′-ACGGTCAGTTTCACGACCTC-3′ (forward) and 5′-CTCCACGGAGTAGTATCCACG-3′ (reverse); polo, 5′-TCACCGCAGCCTTAACCATC-3′ (forward) and 5′-ACAGCTCCAGCACAATGTAGAT-3′ (reverse); GNBP2, 5′-CCGCCCAAACGATAGTGAG-3′ (forward) and 5′-GATGTCATGCTTCCAGGTGGT-3′ (reverse); CSN4, 5′-AAGTTGCCTGACGATCTGTCC-3′ (forward) and 5′-TATGCCAGCCACTTGCTCTTC-3′ (reverse); CG15717, 5′-AGTCCCTGCAGAATCCCTTT-3′ (forward) and 5′-GGCTTTCGCCTTGTACTGTC-3′ (reverse); Ubi-p5E, 5′-TCTTCACTTGGTCCTGCGTC-3′ (forward) and 5′-ATGGCTCGACCTCCAAAGTG-3′ (reverse); Ubi-p63E, 5′-ACGCACCCTGTCCGATTAC-3′ (forward) and 5′-TGGTCTTTCCGGTCAAAGTCTT-3′ (reverse).
Larval muscle histology
Wandering third instar larvae were dissected in ice-cold PBS and fixed for 20 min in 4% formaldehyde in PBS. After being washed, body wall muscles were incubated overnight at 4°C with α-ubiquitin mAb FK2 (Enzo Life Sciences, 1:250), then washed and stained for 2hrs with Alexa Fluor 594 goat a-mouse (Molecular probes, 1:1000), and DAPI (Sigma, 1:5000), then washed again and mounted in 50% glycerol/PBS. All antibody incubations and washes were performed in PBT, except for a final rinse in PBS prior to mounting. The ventral longitudinal muscles from segment 3 or 4 were imaged with a Leica TCS SP2 confocal laser-scanning microscope.
Fly stocks
Dmef2-Gal448 was used to drive transgene expression specifically in the larval muscles via the Gal4/UAS system49. The following RNAi hairpin lines targeting proteasome network components were obtained from the NIG Japan: enolase (17655R-1 and 17654R-2) and CG10638 (32101R-2 and 32101R-3). An additional line for enolase (JF02070) was obtained from the DRSC/TRiP (www.flyrnai.org/) at Harvard medical school. All RNAi experiments were performed at 25°C and all lines gave phenotype.
Implementation of SignedPPI
The SignedPPI user interface was implemented as a collection of Java servlets, JavaScript, and Adobe Flash components. SignedPPI integrates existing tools, including Cytoscape Web for complex visualization (http://cytoscapeweb.cytoscape.org/), Lucene (http://lucene.apache.org/) for searches, JSON (http://www.json.org/) for communication between Java servlets and JavaScript, and WebFX for parameter adjustment sliders (http://webfx.eae.net/). The application is hosted on the Orchestra cluster supported by the Research IT Group (RITG) at Harvard Medical School. PPIs are maintained as flat files and indexed by Lucene. The Java Servlets and Java Server Pages run within an instance of Tomcat 6.0.18 on the Orchestra cluster. The Entrez Gene IDs, gene symbols, and alias names for Drosophila genes were retrieved from NCBI (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/). FlyBase IDs, CG numbers, gene symbols, and synonyms were retrieved from FlyBase (ftp://ftp.flybase.net/releases/current/). Protein IDs were retrieved from UniProt (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/). A program developed in-house automatically FTPs and processes these files on a monthly basis.
Supplementary Material
Acknowledgments
We thank B. Housden, Y. Kwon, I.T. Flockhart and S. Rajagopal for helpful suggestions for tool development and manuscript preparation. Funding: This work was supported by P01-CA120964, R01-GM067761 and R01DK088718. S.E.M. is also supported in part by the Dana Farber/Harvard Cancer Center (P30- CA06516). N.P. is supported by Howard Hughes Medical Institute.
Footnotes
Author contributions: A.V. and N.P. conceived and designed the project. A.V. developed the computational method, built and analyzed the signed network. J.Z. designed and performed experimental validation. C.R and B.Y. developed the SignedPPI database. A.V and S.E.M. coordinated the development of SignedPPI database. A.V. and Y.H. compiled RNAi screens. A.S. analyzed modENCODE expression data. R.N. contributed unpublished RNAi screen. All authors contributed to manuscript preparation. Competing interests: The authors declare that they have no competing interests.
References for main text
- 1.Guruharsha KG, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 3.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 4.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 5.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 6.Friedman A, Perrimon N. Genetic screening for signal transduction in the era of network biology. Cell. 2007;128:225–231. doi: 10.1016/j.cell.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Kholodenko B, Yaffe MB, Kolch W. Computational approaches for analyzing information flow in biological networks. Science signaling. 2012;5:re1. doi: 10.1126/scisignal.2002961. [DOI] [PubMed] [Google Scholar]
- 8.Hyduke DR, Palsson BO. Towards genome-scale signalling network reconstructions. Nat Rev Genet. 2010;11:297–307. doi: 10.1038/nrg2750. [DOI] [PubMed] [Google Scholar]
- 9.Kirouac DC, et al. Creating and analyzing pathway and protein interaction compendia for modelling signal transduction networks. BMC Syst Biol. 2012;6:29. doi: 10.1186/1752-0509-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinayagam A, et al. A directed protein interaction network for investigating intracellular signal transduction. Science signaling. 2011;4:rs8. doi: 10.1126/scisignal.2001699. [DOI] [PubMed] [Google Scholar]
- 11.Liu W, et al. Proteome-wide prediction of signal flow direction in protein interaction networks based on interacting domains. Mol Cell Proteomics. 2009;8:2063–2070. doi: 10.1074/mcp.M800354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeger-Lotem E, et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat Genet. 2009;41:316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitter A, Carmi M, Barkai N, Bar-Joseph Z. Linking the signaling cascades and dynamic regulatory networks controlling stress responses. Genome Res. 2013;23:365–376. doi: 10.1101/gr.138628.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh R. PhD thesis. Chapter 6. MIT; 2012. pp. 91–102. [Google Scholar]
- 15.Linding R, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flockhart IT, et al. FlyRNAi.org--the database of the Drosophila RNAi screening center: 2012 update. Nucleic Acids Res. 2012;40:D715–719. doi: 10.1093/nar/gkr953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilsdorf M, et al. GenomeRNAi: a database for cell-based RNAi phenotypes. 2009 update. Nucleic Acids Res. 2010;38:D448–452. doi: 10.1093/nar/gkp1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumuller RA, et al. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 2011;8:580–593. doi: 10.1016/j.stem.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mummery-Widmer JL, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neumuller RA, et al. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Science signaling. 2013;6:ra70. doi: 10.1126/scisignal.2004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korcsmaros T, et al. Uniformly curated signaling pathways reveal tissue-specific crosstalks and support drug target discovery. Bioinformatics. 2010;26:2042–2050. doi: 10.1093/bioinformatics/btq310. [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark C, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerrien S, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salwinski L, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Licata L, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murali T, et al. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39:D736–743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girvan M, Newman ME. Community structure in social and biological networks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Facchetti G, Iacono G, Altafini C. Computing global structural balance in large-scale signed social networks. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20953–20958. doi: 10.1073/pnas.1109521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinayagam A, et al. Protein complex-based analysis framework for high-throughput data sets. Science signaling. 2013;6:rs5. doi: 10.1126/scisignal.2003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman AA, et al. Proteomic and functional genomic landscape of receptor tyrosine kinase and ras to extracellular signal-regulated kinase signaling. Science signaling. 2011;4:rs10. doi: 10.1126/scisignal.2002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahlmann B. Role of proteasomes in disease. BMC Biochem. 2007;8 (Suppl 1):S3. doi: 10.1186/1471-2091-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz-Ramos A, Roig-Borrellas A, Garcia-Melero A, Lopez-Alemany R. alpha-Enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol. 2012;2012:156795. doi: 10.1155/2012/156795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mindnich RD, Penning TM. Aldo-keto reductase (AKR) superfamily: genomics and annotation. Hum Genomics. 2009;3:362–370. doi: 10.1186/1479-7364-3-4-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebert B, Kisiela M, Wsol V, Maser E. Proteasome inhibitors MG-132 and bortezomib induce AKR1C1, AKR1C3, AKR1B1, and AKR1B10 in human colon cancer cell lines SW-480 and HT-29. Chem Biol Interact. 2011;191:239–249. doi: 10.1016/j.cbi.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 38.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 40.Fuchs F, et al. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol Syst Biol. 2010;6:370. doi: 10.1038/msb.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doncheva NT, Assenov Y, Domingues FS, Albrecht M. Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc. 2012;7:670–685. doi: 10.1038/nprot.2012.004. [DOI] [PubMed] [Google Scholar]
- 44.Wernicke S, Rasche F. FANMOD: a tool for fast network motif detection. Bioinformatics. 2006;22:1152–1153. doi: 10.1093/bioinformatics/btl038. [DOI] [PubMed] [Google Scholar]
- 45.Bai J, Sepp KJ, Perrimon N. Culture of Drosophila primary cells dissociated from gastrula embryos and their use in RNAi screening. Nat Protoc. 2009;4:1502–1512. doi: 10.1038/nprot.2009.147. [DOI] [PubMed] [Google Scholar]
- 46.Bai J, et al. RNA interference screening in Drosophila primary cells for genes involved in muscle assembly and maintenance. Development. 2008;135:1439–1449. doi: 10.1242/dev.012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sepp KJ, et al. Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet. 2008;4:e1000111. doi: 10.1371/journal.pgen.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ranganayakulu G, Schulz RA, Olson EN. Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev Biol. 1996;176:143–148. doi: 10.1006/dbio.1996.9987. [DOI] [PubMed] [Google Scholar]
- 49.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.