Mitochondrial dysfunction is implicated in a spectrum of human diseases. In this review, Pagliarini and Rutter recap the discoveries that defined the core functions of mitochondria, the mitochondrial proteome, and the functions of unannotated proteins and pathways. They also offer a view of the challenges facing mitochondrial biochemists in the coming decades.
Keywords: biochemistry, mitochondria, proteomics
Abstract
Stemming from the pioneering studies of bioenergetics in the 1950s, 1960s, and 1970s, mitochondria have become ingrained in the collective psyche of scientists as the “powerhouses” of the cell. While this remains a worthy moniker, more recent efforts have revealed that these organelles are home to a vast array of metabolic and signaling processes and possess a proteomic landscape that is both highly varied and largely uncharted. As mitochondrial dysfunction is increasingly being implicated in a spectrum of human diseases, it is imperative that we construct a more complete framework of these organelles by systematically defining the functions of their component parts. Powerful new approaches in biochemistry and systems biology are helping to fill in the gaps.
Mitochondria are iconic structures in biochemistry and cell biology. Their unmistakable double-membrane architecture rife with tortuous cristae—first revealed to the world in detail by the electron micrographs of Palade and Sjostrand in the early 1950s (Palade 1953; Sjostrand 1953)—are recognizable to anyone who has taken a high school biology course. That same group could also convey the raison d'etre of mitochondria: They are the cellular “powerhouses” that burn fuel from the food we eat to generate energy. However, while it is certainly still true that the 10 million billion mitochondria throughout our bodies produce the bulk of the ATP for nearly all of our cells (Lane 2006), mitochondrial biology has undergone major transformations unforeseen by the bioenergeticists who unlocked the mysteries of “oxidative phosphorylation” (OxPhos) decades ago. The simplistic concept of these organelles as discrete, kidney bean-shaped energy factories has given way to that of a cell-specific and dynamic organellar network that fuses, divides, and directs a vast array of functions central to cellular life, death, and differentiation (McBride et al. 2006; Nunnari and Suomalainen 2012). Importantly, it has also become clear that mitochondrial dysfunction underlies >50 inborn errors of metabolism (Calvo and Mootha 2010; Koopman et al. 2012); strongly contributes to a growing list of common disorders, including type 2 diabetes (Szendroedi et al. 2012), neurodegenerative diseases (Lin and Beal 2006), and cancer (Wallace 2012); and is central to the aging process (Balaban et al. 2005; Bratic and Larsson 2013).
Despite these revelations, there are miles to go to achieve a systems-level understanding of mitochondria. This is clearly exemplified by recent large-scale efforts to define the mitochondrial proteome. These studies have revealed the existence of hundreds of mitochondrial proteins with no known biochemical function while simultaneously providing a framework for resolving long-standing mysteries of mitochondrial biology. Here, we recap the seminal early discoveries that defined the core functions of mitochondria, review the recent systems-level analyses that have established the mitochondrial proteome, highlight a few recent efforts that have leveraged the proteome to systematically elucidate the functions of unannotated proteins and pathways in this organelle, and offer a view of the challenges facing mitochondrial biochemists in the coming decades.
The early years: mitochondria as the centers of cellular respiration
The concept that organisms “burn” fuel for energy can be traced back to Lavoisier, who, in 1790, described respiration as being literally “…a slow combustion of carbon and hydrogen, similar in every way to that which takes place in a lamp or lighted candle.…” However, it was not until 1870 that Eduard Pfluger demonstrated that respiration took place in cells, and it was not until the early 1910s—a full century ago—that Kingsbury presciently postulated that this process occurs in “mitochondria,” a term coined by Benda in 1898 to describe the ubiquitous cellular structures likely first observed in the 1840s (Ernster and Schatz 1981; Lane 2006).
The demonstration that respiration occurred in cells and not in the blood led scientists to search for a “respiratory pigment” with hemoglobin-like properties. In 1925, building on initial observations by Charles MacMunn decades earlier, David Keilin identified such pigments and dubbed them “cytochromes,” a term quite familiar to mitochondrial biologists today. This paved the way for Keilin, Warburg, Hartree, and others to flesh out the concept of a “respiratory chain,” with dehydrogenases on one end and Warburg's oxygen-reducing respiratory enzyme (the Atmungsferment) on the other (Mitchell 1979; Ernster and Schatz 1981). In 1931, in part for describing the nature of the cytochromes, Warburg won the Nobel Prize “for his discovery of the nature and mode of action of the respiratory enzyme” (Box 1).
Box 1. Notable events in the discovery of mitochondria and their proteomes.
1840s—First observations of cellular structures that were likely to be mitochondria.
1884—Charles MacMunn identifies a “respiratory pigment” that could enable respiration to occur inside cells (Keilin 1925).
1890—Altman recognizes the ubiquitous nature of the “bioblasts” structures first seen in the 1840s. He proposes that they are “elementary orgnaisms” (Altman 1890).
1898—Benda coins the term “mitochondrion” (Benda 1898).
1925—David Keilin (Cambridge) rediscovers MacMunn's pigments, further characterizes them, and names them “cytochromes” (Keilin 1925).
1930s—Keilin proposes the idea of a “respiratory chain.” Warburg identifies coenzymes and wins a Nobel Prize for his characterization of the “respiratory enzyme” (Ernster and Schatz 1981) (Nobel Prize 1931).
1937—Hans Krebs publishes a paper on the Krebs cycle (Krebs and Johnson 1937) (Nobel Prize 1953 with Fritz Lipmann).
1946—Albert Claude works out the first differential centrifugation techniques to isolate mitochondria and demonstrates that succinoxidase and cytochrome oxidase are mitochondrial (Claude 1946).
1948–1951—Eugene Kennedy, Albert Lehninger, David Green, and others demonstrate that the enzymes of the citric acid cycle, fatty acid oxidation, and oxidative phosphorylation are mitochondria (Ernster and Schatz 1981).
1949–1956—Using new isolation protocols, various other enzyme systems are localized to mitochondria, including adenylate kinase, glutamate dehydrogenase, transaminases, pyruvate carboxylase, nucleoside diphosphokinase, nicotinamide nucleotide transhydrogenase, enzymes involved in the synthesis of phorphyrin and heme, citrulline, phospholipids, and the substrate-level phosphorylation linked to α-ketoglutarate oxidation.
1952–1953—First high-resolution images of mitochondria are published by George Palade and Fritiof Sjostrand (Palade 1953; Sjostrand 1953).
1950s, early 1960s—Experiments describing the nature of key elements of oxidative phosphorylation, including coenzyme Q, nonheme iron, and metalloflavoproteins.
1961—Peter Mitchell publishes his chemiosmotic coupling hypothesis (Nobel Prize 1978), which was refined, expanded, and generalized in 1966 (Mitchell 1979).
1964—Effraim Racker observes ATP synthase particles via microscopy. He refers to them as the “elementary particles of life” or “fundamental particles of biology” (Lane 2006).
1966—Identification of anion translocators.
1970s—Reconstitution of oxidative phosphorylation in artificial phospholipid vesicles and evidence for electron transport-linked proton pumps.
1981—The sequence of the human mitochondrial genome is determined by scientists at the MRC Laboratory of Molecular Biology in Cambridge (Anderson et al. 1981).
1998—First mitochondrial proteomics study is published.
2003—Albert Sickmann, Eric Lander/Matthias Mann, and Steven Taylor publish papers describing more a comprehensive catalog of the mitochodrial proteome (Mootha et al. 2003; Sickmann et al. 2003; Means et al. 2011).
2008—Vamsi Mootha, Steven Carr, and colleagues publish the MitoCarta compendium, which is the most complete catalog of mitochondrial proteins to date (Pagliarini et al. 2008).
2013—Alice Ting and colleagues perform live-cell proteomic mapping of the mitochondrial matrix (Rhee et al. 2013).
In addition to these seminal advancements in understanding the nature of cellular respiration, the period between the late 1920s and the early 1940s provided significant progress in defining other key aspects of aerobic metabolism. ATP was discovered by Lohmann in 1929 (Lohmann 1929) and shown to be required for muscle contraction by Engelhardt a year later. Engelhardt soon proposed ATP generation to be the central function of oxygen respiration—an idea validated by Kalckar and Lipmann, who later proclaimed ATP to be the universal energy currency of life. Not to be upstaged, 1937 saw the elucidation of the citric acid cycle by Krebs and Johnson (1937), earning them a Nobel Prize in 1953. However, all of these discoveries occurred before mitochondria became a key feature of the story.
The decade from 1946 to 1956 saw mitochondria come into focus—literally and figuratively—as the centers of aerobic metabolism. This was catalyzed by the work of Claude at the Rockefeller Institute who, in 1946, devised the first reliable cellular fractionation procedures using differential centrifugation, allowing for the first isolation of intact mitochondria (Claude 1946). Using this procedure, Claude and colleagues (Hogeboom et al. 1946) demonstrated that succinoxidase and cytochrome oxidase were localized exclusively to mitochondria. Kennedy and Lehninger (1949) then demonstrated that fatty acid oxidation and the citric acid cycle also occurred exclusively in these organelles and that these processes were linked to the synthesis of ATP via NADH. Demonstration of the mitochondrial localization of myriad other enzymes soon followed, including adenylate kinase, glutamate dehydrogenase, transaminases, pyruvate carboxylase, nucleoside diphosphokinase, nicotinamide nucleotide transhydrogenase, and enzymes involved in the synthesis of heme, citrulline, and phospholipids (Ernster and Schatz 1981). Simultaneously, the pioneering electron microscopy of Palade (1953) and Sjostrand (1953) in the early 1950s revealed the unique morphology of these organelles in striking detail, including the characteristic double-membrane structure and the convoluted invaginations that Palade termed “cristae.” Visually and biochemically, the foundations of mitochondrial biology were laid.
With mitochondria as the newly minted cellular “powerhouses,” mitochondrial biologists became dedicated to working out the details of OxPhos. Indeed, the late 1950s introduced the concept of respiratory control, the development of sensitive methods to study electron transport, the isolation of individual OxPhos complexes, and roles for coenzyme Q, nonheme iron, and metalloflavoproteins as redox carriers (Ernster and Schatz 1981; Tager et al. 1983). Despite these advancements, the field was nonetheless baffled by how the oxidation of substrates could be coupled to the synthesis of ATP from ADP and inorganic phosphate. Using substrate-level phosphorylation as the biochemical model, many of the greatest minds in biochemistry spent years fruitlessly searching for an elusive (and nonexistent) “high-energy intermediate” that could directly link the oxidation and phosphorylation components of OxPhos (Nicholls and Ferguson 2002; Scheffler 2008). Peter Mitchell finally solved the mystery in 1961 with his radical new theory of “chemiosmotic coupling” (Mitchell 1961). Essentially, Mitchell proposed that the energy derived from oxidation of fuels was used to pump protons across the inner mitochondrial membrane from the matrix to the intermembrane space, creating what he called the “proton motive force.” His theory explained that it was the energy captured from those protons returning to the matrix and not energy stored in an undefined high-energy covalent chemical intermediate that was then used to drive ATP synthesis. This Nobel Prize-worthy concept (1978) was so counter to the leading theories of the day that the field spurned it for the better part of a decade. Nonetheless, following seminal experiments by Jagendorf (1967) and others demonstrating that ATP could be synthesized by a proton motive force and ATP synthase in the absence of an electron transport chain (ETC), the field had largely come around to Mitchell's hypothesis (Boyer et al. 1977). The golden age of OxPhos had come to a close.
Mitochondria make a comeback
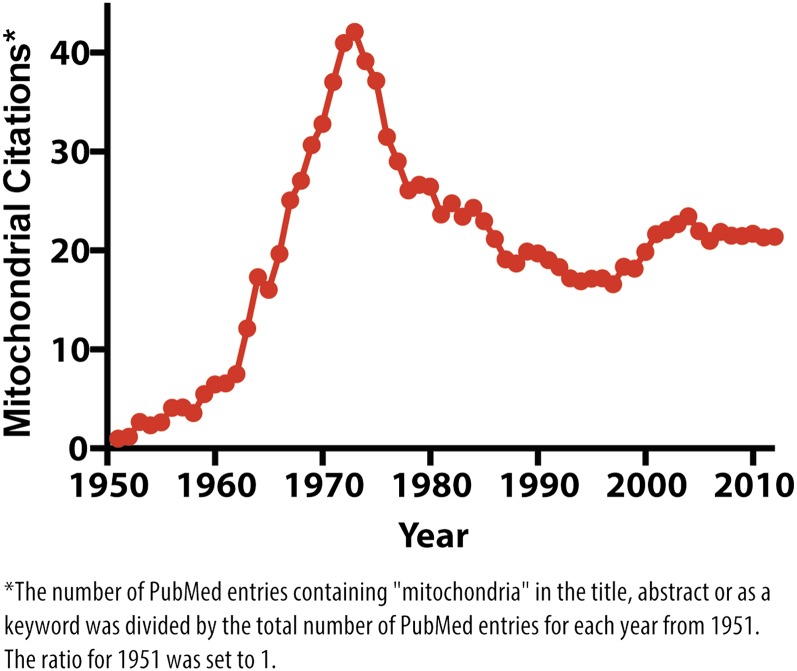
The elucidation of the general mechanistic basis of OxPhos by the mid-1970s brought mitochondrial biology into a new age. Visionary scientists had uncovered most of the secrets underlying how this organelle made and consumed organic substrates and generated ATP—the specifics of which later led to a Nobel Prize for Paul Boyer and John Walker in 1997. With these fundamental questions largely answered, there was a sense that mitochondria were a tired subject, and interest in this organelle steadily decreased over the ensuing two decades (Fig. 1). A telling anecdote, recapped by Hiltunen et al. (2010), describes a young Michael Yaffe—now well known for his pioneering work on mitochondrial dynamics—being admonished at a Gordon Conference in the 1980s for pursuing a subject for which “everything important was already known.” Undeterred, Yaffe and many others forged ahead with important work in areas beyond ATP production and intermediary metabolism. They made key discoveries that helped to shape our understanding of mitochondrial fission and fusion, inheritance, the genetic basis of mitochondrial disease, protein import, and OxPhos complex assembly. However, mitochondrial biology had to wait until the mid-1990s to return to the fore. This renaissance was such that Science magazine published a special mitochondria-focused issue in 1998 introduced by an editorial entitled “Mitochondria Make a Comeback” (Kiberstis 1999).
Figure 1.
The changing tides of mitochondrial research. To estimate the relative attention given to mitochondrial biology in each year from 1951 to 2012, we divided the number of PubMed entries containing “mitochondria” in the title or abstract or as a keyword by the total number of PubMed entries. The ratio for each year was normalized to that of 1951, which was set to 1.
A principal harbinger of this renaissance was the discovery of the integral role of mitochondria in the process of programmed cell death. Pioneering work from the laboratories of Xiaodong Wang, Stan Korsmeyer, Bob Horvitz, John Reed, Doug Green, Guido Kroemer, and many others unequivocally established that mitochondria and mitochondrial proteins underlie the commitment to apoptosis in most situations (Green and Reed 1998). These studies culminated with the profound discovery that cytochrome c, an electron carrier operating within the OxPhos system, was released from mitochondria upon apoptopic insult, whereupon it initiated caspase activation and the execution of apoptosis (Liu et al. 1996; Li et al. 1997). Anti-apoptotic BCL proteins, originally discovered for their oncogenic potential, reside on the mitochondrial outer membrane and antagonize the proapoptotic function of Bax and Bak to promote cytochrome c release (Oltvai et al. 1993; Kiefer et al. 1995; Kluck et al. 1997; Yang et al. 1997; Shimizu et al. 1999). We now know this system to be more complicated, but the fundamental observations of the 1990s form the foundation for our current understanding of mitochondrial cell death pathways.
Work in the 1990s and early 2000s continued to flesh out these processes and solidified the role of mitochondrial dysfunction in many rare and common human diseases (Wallace 1999; DiMauro and Schon 2003). While the examples above are only a few of many, they underscore a very surprising development: Mitochondria might spend their days as the “powerhouses of the cell” but clearly moonlight in an array of other activities.
Expanding the powerhouse
In 1924, commenting on the speculation that mitochondria house the machinery required for cellular respiration, Edmund Cowdry stated, “…it is quite obvious that the investigation of mitochondria will never achieve the usefulness which it deserves as an instrument for advance in biology and medicine until we know much more of their chemical constitution…” (Bechtel 2006). This statement proved to be as appropriate in the 1990s as it was in the 1920s. The expanding roles for mitochondria in cell and molecular biology prompted scientists to return to the fundamental question first addressed following Claude's isolation of these organelles in the late 1940s: What proteins reside in mitochondria? Studies spanning the decade from 1998 to 2008 provided many new answers to that question and, with them, the realization that our understanding of mitochondrial form and function is, surprisingly, still in its infancy.
Efforts to systematically define the mitochondrial proteome began when Rabilloud et al. (1998) identified 46 proteins by two-dimensional (2D) gel electrophoresis of purified human placental mitochondria. A year later, in 1999, the MITOP database was established as a central location for information on both nuclear- and mitochondria-encoded genes and their corresponding proteins, which included 311 human entries (Scharfe et al. 1999). The development of the first mitochondrial localization sequence prediction algorithm, TARGETP (Emanuelsson et al. 2000), and the use of density gradient purification and subfractionation approaches gradually enabled further additions to the list of known mitochondrial proteins, which then saw a marked increase in 2003. In that year, three studies leveraged state-of-the art mass spectrometry (MS)-based proteomics approaches to nearly double the number of known yeast (Sickmann et al. 2003), mouse (Mootha et al. 2003), and human (Taylor et al. 2003) mitochondrial proteins. The study by Mootha et al. (2003) also revealed that mitochondria differed quite considerably between mouse tissues, indicating that these organelles are highly customized to serve local cellular physiology and helping to set a foundation for understanding the nature of the confounding tissue-specific pathophysiology seen in many mitochondrial diseases.
Despite these great strides in defining the mitochondrial proteome, by 2006, the MITOP database still listed only ∼600 genes that encoded mitochondrial proteins for both mice and humans. However, comparisons with yeast and Rickettsia prowazekeii—the closest eubacterial relative of mammalian mitochondria—and 2D gel electrophoresis of purified mitochondria suggested that the full complement of mitochondrial proteins would likely be closer to 1200–1500 (Andersson al. 1998; Lopez et al. 2000). Retrospective analysis of the 2003 MS data revealed that one reason for this large disparity was that only the most abundant mitochondrial proteins were being detected. New MS technology soon fixed this sensitivity issue but simultaneously made it difficult to differentiate low-abundance mitochondrial proteins from contaminating impurities. Individual studies that used microscopy-based colocalization of GFP fusion proteins or computational approaches that leveraged other non-MS-based data helped to sort the wheat from the chaff but still suffered from their own significant caveats.
To date, the most successful efforts to catalog the mitochondrial proteome have come from the integration of multiple approaches. The most comprehensive mitochondrial protein compendium was completed in 2008 by combining literature curation, large-scale GFP microscopy, and the integration of extensive MS proteomics data with six other genome-scale data sets of mitochondrial localization using a Bayesian framework (Pagliarini et al. 2008). This resource, termed MitoCarta, includes 1098 mitochondrial protein-encoding genes and their expression patterns across 14 mouse tissues. Even with this extensive effort, the MitoCarta catalog is estimated to be only 85% complete with a 10% false discovery rate and does not include many proteins (such as protein kinase A [PKA]) that dually localize to mitochondria and other cellular compartments.
Setting aside these significant complexities, the MitoCarta compendium dramatically illustrated the degree to which the mitochondrial proteome was still unexplored. Nearly 300 genes—more than one-quarter of the inventory—had no association with a gene ontology (GO) biological process, suggesting that we have no clear indication of the encoded proteins' functions. As mitochondrial dysfunction continues to be implicated in a broad spectrum of human diseases and with modern genomics approaches rapidly identifying causative mutations affecting mitochondrial proteins, the need for elucidating the biochemical functions of these proteins is clear.
New proteins, old functions
In a 1964 Scientific American article, David Green of the University of Wisconsin quipped, “The mitochondrion is often called the powerhouse of the cell. It is a good deal more than that....” (Green 1964). Indeed, as noted above, the mitochondrial proteomics efforts revealed that many mitochondrial functions—likely much more than Green (1964) anticipated—still await discovery. Additionally, these efforts prioritized candidate proteins that could fulfill important missing biochemical functions. In much the same way that scientists spent years searching for the missing pieces of OxPhos, many other such “known unknowns” plagued mitochondrial biologists for decades. Below, we highlight some powerful and creative new approaches that leveraged the expanded compendium of mitochondrial proteins to fill in key gaps.
Complex I (CI) assembly factors
CI of the ETC couples the oxidation of NADH to the pumping of protons into the intermembrane space, making it a key component of Mitchell's chemiosmotic system of ATP generation. Genomics and MS-based analyses have revealed that human CI includes a staggering ∼45 subunits assembling into an ∼1-MDa complex (Janssen et al. 2006). Mutations affecting many of these proteins give rise to CI deficiency, which is the most common type of respiratory chain disorder. However, nearly half of patients who are diagnosed with an isolated CI disorder lack a mutation in any of the subunit-encoding genes, suggesting that unknown genes that aid in the assembly or activity of CI are mutated in the remaining cases. These so-called CI assembly factors (CIAFs) have been difficult to identify for a variety of reasons: They are typically not part of the mature complex, they do not exist in the workhorse model organism Saccharomyces cerevisiae because this organism lacks CI, and the rare incidence of disorders caused by each assembly factor mutation makes classic linkage analysis ineffective.
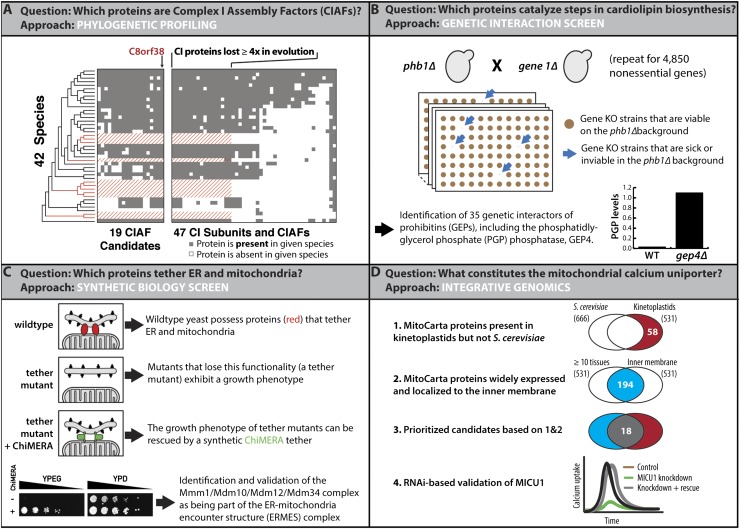
Three recent efforts have accelerated the identification of these missing CIAFs. In each case, the analyses began with the assumption that the CIAFs would themselves exist within mitochondria and thus would be found within the expanded catalog of the mitochondrial proteome. The first example used phylogenetic profiling, which attempts to identify functionally related proteins via shared evolutionary history (Fig. 2A). Building on other recent successes using this approach to identify the CIAF NDUFA12L (Ogilvie et al. 2005), Pagliarini et al. (2008) built a phylogenetic profile of CI subunits across 42 eukaryotic species. This analysis revealed that 15 of the murine CI subunits evolved from ancestral bacterial subunits that were coordinately lost at least four times during evolution. Only 19 of the 1098 proteins in the MitoCarta compendium shared the evolutionary pattern of these COPP (complex one phylogenetic profile) proteins, making them attractive candidates for functional association with CI. Indeed, the top COPP candidate, C8orf38, was validated as a CIAF, and four others now have established connections to CI biogenesis.
Figure 2.
Model studies for identifying new functions for mitochondrial proteins. (A) Phylogenetic profiling leverages shared evolutionary history to highlight functionally related proteins. Pagliarini et al. (2008) used this approach to identify 19 MitoCarta proteins that shared the same evolutionary history as a subset of known CI subunits. These proteins include C8orf38, which harbors causative mutations in a CI deficiency. (B) Genetic interaction screens are another powerful way to associate proteins involved in shared complexes or pathways. Osman et al. (2009) used this approach to identify genes that, when knocked out on a phb1Δ background, caused a synthetic sick/synthetic lethal phenotype. Multiple such genes were found to be involved in lipid metabolism, including GEP4, which catalyzes a key step in CL biosynthesis. The bar graph, adapted by permission from Macmillan Publishers Ltd. from Osman et al. (2010) (© 2010), shows the accumulation of PGP, the GEP4 substrate, in gep4Δ yeast. (C) It has long been known that ER and mitochondria are somehow connected, but the proteins that help form these junctions remained elusive. Kornmann et al. (2009) identified the first components of the so-called ERMES complex using what they called a synthetic biology screen, in which they screened for mutants that could be complemented by a synthetic protein designed to artificially tether the two organelles. From Kornmann et al. (2009). Reprinted with permission from AAAS. (D) Perocchi et al. (2010) used an integrative computational strategy that included comparative physiology, evolutionary genomics, and mitochondrial proteomics information to prioritize candidates for the long-sought-after MCU. Based on decades of literature, the investigators predicted that components of the MCU would be localized to the mitochondrial IM, expressed in the majority of mammalian tissues, and have homologs in kinetoplastids but not in S. cerevisiae. This study validated MICU1 as a component of the MCU.
A second effort, by Calvo et al. (2010), combined the COPP genes with other prioritized genes from the MitoCarta compendium to generate 103 candidate genes for human CI deficiency. They then performed high-throughput sequencing approaches to sequence all 103 genes in 103 individuals with CI deficiency of unknown origin along with 42 healthy controls. This so-called “Mito10K” project revealed that mutations in NUBPL and FOXRED1 can cause CI deficiency, making them likely CIAFs. A third study, by Heide et al. (2012), leveraged MS analyses of intact complexes separated by blue-native electrophoresis. This “complexome” analysis showed that TMEM126B, an uncharacterized mitochondrial transmembrane protein, was found in a complex with three other known CIAFs, forming what the investigators termed the mitochondrial CI assembly factor (MCIA) complex. They further demonstrated that TMEM126B is required for the formation of CI, making it another prime candidate gene for mutation in patients with CI deficiency. Of note, TMEM126B was not part of the original MitoCarta compendium, emphasizing the need to identify and annotate the missing ∼15% of the mitochondrial proteome.
Cardiolipin (CL) biosynthesis
The newfound ability to isolate mitochondria in the late 1940s also enabled the analysis of this organelle's lipids (Marinetti et al. 1958). Mitochondrial membranes, like all other cellular membranes, have their own distinct lipid composition. The signature mitochondrial lipid is CL, a lipid dimer consisting of two phosphatidyl groups connected by a glycerol backbone. CL was first isolated in 1945 (Pangborn 1945) and shown to reside nearly exclusively in mitochondria in 1968 (Getz et al. 1968), where it constitutes ∼20% of the total lipid mass of the inner membrane (IM). CL is important for a range of mitochondrial processes, including fission/fusion, OxPhos complex and supercomplex assembly, and cristae structure.
In eukaryotes, CL is synthesized in mitochondria, beginning with phosphatidic acid (PA) derived from the endoplasmic reticulum (ER). Once in mitochondria, PA is processed into CDP-diacylglycerol (CDP-DAG) and then to phosphatidylglycerolphosphate (PGP) by PGP synthase (Claypool and Koehler 2012). PGP must then be dephosphorylated before condensing with another CDP-DAG molecule via CL synthase to form CL. However, until recently, it was not known how PA was delivered to the IM, how it was processed into CDP-DAG, or how PGP was dephosphorylated.
Recently, through a combination of rigorous and clever methodologies, orphan mitochondrial proteins have been assigned to each of these missing functions. To begin, new candidates for proteins involved in CL were identified through a yeast genetic interaction screen (Fig. 2B). In this approach, yeast strains lacking nonessential genes are systematically crossed to form a series of double knockouts, which are then analyzed for growth phenotypes. The missing genes are considered to have a genetic interaction when the resulting growth of the double knockout is either significantly better (a positive interaction) or significantly worse (a negative interaction) than would be expected from the combined effects of the single knockouts. A survey of ∼4600 such strains by Osman et al. (2009) identified 35 genes that were essential for growth in the absence of prohibitins—proteins that, among other functions, are key to maintaining mitochondrial CL and phosphatidylethanolamine (PE) levels. Using MS-based lipidomics and yeast genetics, the investigators then demonstrated that one of these genes, GEP4, encoded the missing yeast PGP phosphatase (Osman et al. 2010). Simultaneously, Zhang et al. (2011) showed that the unrelated mitochondrial phosphatase PTPMT1 (Pagliarini et al. 2005) performed this same reaction in mice, which lack a GEP4 ortholog.
A second gene from the Osman et al. (2009) screen, UPS1, is a member of the conserved PRELI family of proteins that localize to the mitochondrial intermembrane space. Through a series of elegant lipidomic and lipid transfer assays, the Langer group (Connerth et al. 2012) revealed that Ups1 serves as a lipid transfer protein that can shuttle PA between mitochondrial membranes, thereby providing a route for this lipid from the ER to the CL biosynthetic machinery in the IM and matrix. Completing a final piece of the CL biosynthesis puzzle, Tamura et al. (2013) showed that Tam41 is the mitochondrial CDP-DAG synthase. Disruption of Tam41 had previously been shown to affect early steps in CL synthesis, but its specific role remained unknown. Using careful biochemistry and fractionation techniques, the investigators revealed that Cds1, previously thought to serve this function in mitochondria, was found only in the ER and that Tam41 directly catalyzes the formation of CDP-DAG from PA in the mitochondrial IM.
The ER–mitochondria encounter structure (ERMES) complex
There has long been an appreciation of the fact that mitochondria have some type of intimate relationship with the ER. More than five decades ago, electron micrographs showed physical connections between these two organelles (Robertson 1960). This seemingly unfortunate fact has also been encountered repeatedly by researchers trying (and failing) to purify one of these organelles away from the other. An understanding of the molecular nature of at least one of the important systems that maintain this physical connection came from an artful genetic screen (Kornmann et al. 2009). The screen, which the investigators describe as a “synthetic biology” screen, used yeast genetics to identify mutant strains that were unusually dependent on an artificial protein that ectopically tethered mitochondria and the ER (Fig. 2C). The foundational logic was that mutations that disrupted mitochondria/ER connections should have severe phenotypic consequences, but these might be alleviated by expression of an alternative tethering system. This primary screen identified two mutations in MDM12, and subsequent follow-up work showed that Mdm12 and three other proteins form a complex, now named the ERMES complex. The ERMES complex, which serves to tether the ER and mitochondria, spans the ER membrane and mitochondrial outer membrane and forms about two to 10 foci per yeast cell (Kornmann et al. 2009).
The discovery of ERMES was of great importance, as, for the first time, it brought ER–mitochondrial connections into the realm of genetics. It raised a number of questions, however, some of which remain unanswered these 4 years later. Most importantly, what are the specific roles of the ERMES complex, and how do these relate to all of the disparate functions that have been proposed for ER/mitochondrial connections? The function that featured the most prominently in the original description of ERMES was that of phospholipid synthesis (Kornmann et al. 2009). As described above, the metabolic pathways that generate phospholipids are comprised of both ER and mitochondrial enzymes, and therefore substrates and products must transit between the two organelles for efficient phospholipid biosynthesis. Indeed, mutations disrupting the ERMES complex slowed, but did not abrogate, the conversion of phosphatidylserine to phosphatidylcholine, which requires such transitions. This hypothesis is particularly attractive, as Mdm12, Mdm34, and Mmm1 all contain an SMP domain, which is predicted to adopt a fold similar to the tubular lipid-binding (TULIP) domain, which directly binds lipid moieties (Kopec et al. 2010). The ERMES deletion phenotype does not seem to be due to a defect in phosphatidylserine transfer from the ER to mitochondria, as this process is unaffected in ERMES mutants (Nguyen et al. 2012). This one function appears to be the tip of the iceberg, however, as ERMES and its components have been implicated in a striking array of additional functions (Michel and Kornmann 2012), including as mediators in mitochondrial fission at sites of ER/mitochondrial contact (Friedman et al. 2011; Murley et al. 2013).
The calcium uniporter
It is difficult to overstate the significance of Ca2+ signaling in the regulation of eukaryotic cell biology, as it plays a role in almost all aspects of cell function. Rapid, transient, and high-amplitude alterations in Ca2+ concentration are critical in hormone release, transcriptional regulation, cell death initiation, and many other processes. It has been appreciated for decades that mitochondria play a role in the cellular system of Ca2+ handling. Isolated mitochondria were shown to be capable of specific Ca2+ uptake in the early 1960s (Deluca and Engstrom 1961; Vasington and Murphy 1962; Lehninger et al. 1963). The biophysical properties of this current were studied in great detail over the ensuing decades, and the significance of mitochondrial calcium uptake was emphasized in the 1990s when it was demonstrated that increased cytoplasmic Ca2+ leads to high-amplitude Ca2+ accumulation in the mitochondria (Rizzuto et al. 1993). This raised the possibility that, in addition to Ca2+ playing important roles within the mitochondria, Ca2+ uptake by mitochondria might also serve to buffer changes in the cytosol. Both of these roles were subsequently verified: Mitochondrial Ca2+ stimulates respiration and ATP production (McCormack et al. 1990; Brini et al. 1999), and mitochondrial Ca2+ uptake reduces the amplitude of Ca2+ increases in specific situations and locations in the cytoplasm (Rizzuto et al. 2012).
In spite of the clear significance of this mitochondrial Ca2+ uptake system, named the mitochondrial calcium uniporter (MCU), the molecular identify of the transporter remained unknown until recently. The Mootha laboratory (Perocchi et al. 2010), via an integrative strategy that combined information from comparative physiology, evolutionary genomics, and the MitoCarta compendium (Fig. 2D), identified a protein that was required for Ca2+ uptake by mitochondria and that shared many properties with a putative component of the MCU. This protein, which they named MICU1, is essential for mitochondrial Ca2+ uptake; however, it was deemed unlikely to be the pore-forming subunit of the MCU, as it had at most one transmembrane domain. It is now known to be a peripheral membrane protein. Building on this discovery and continued leveraging of MitoCarta, the Mootha and Rizzuto laboratories (Baughman et al. 2011; De Stefani et al. 2011) subsequently identified MCU, which both groups showed to be the likely pore-forming subunit of the MCU. It appears that the search is over for the molecular identity of the MCU. As one might expect, this discovery has enabled a flurry of activity, as now gene-based tools are available for the study of mitochondrial calcium handling. Roles for the MCU have already been firmly established in insulin secretion, neuronal excitation, cell death, and other systems (Alam et al. 2012; Mallilankaraman et al. 2012; Shutov et al. 2013; Tarasov et al. 2013).
The mitochondrial pyruvate carrier (MPC)
The history of the discovery, characterization, and finally molecular identification of the MPC is eerily similar to that of the MCU. The fate of pyruvate, the product of cytosolic glycolysis, is one of the critical metabolic decisions that a cell makes. Pyruvate transport into mitochondria, where it is oxidized to enable efficient ATP production, is the most common fate in the majority of differentiated cells in the human body. Surprisingly, the molecular identity of the transporter that enables pyruvate to traverse the mitochondrial IM remained unknown until 2012. Prior to this time, however, the biochemistry of the MPC was studied extensively, and many of the salient features were already known. Halestrap and Denton (1974) proved in the mid-1970s that pyruvate required a specific transporter to transit into the mitochondrial matrix, and they and others defined the kinetics, substrate specificity, and even some molecular information over the ensuing 35 years.
In 2012, two groups (Bricker et al. 2012; Herzig et al. 2012) simultaneously published reports describing the molecular identification of a two-protein complex that is either the MPC or an obligate component of it. The entrée for both groups into this fruitful endeavor came with the identification of mitochondrial proteins—including the utilization of MitoCarta and work by Da Cruz et al. (2003)—for which no function had previously been described in the literature. Due to the high degree of conservation and the power of the model system, both groups began investigations into the role of the MPC family using the yeast S. cerevisiae. Both groups found that loss of either subunit conferred a defect in mitochondrial pyruvate metabolism that was manifested both in defects of carbohydrate metabolism and in branched chain amino acid and lipoic acid biosynthesis. A series of steady-state metabolomics measurements and biochemical assays isolated this defect to an impairment of mitochondrial pyruvate uptake. Bricker et al. (2012) went on to show that loss of the MPC genes also impaired pyruvate metabolism in flies and mammalian cells, which led to the discovery of human mutations in MPC1 that cause severe defects in carbohydrate metabolism and premature lethality. Herzig et al. (2012) demonstrated that ectopic expression of MPC1 and MPC2 was sufficient to enable pyruvate uptake in bacteria.
The true impact of the MPC on human disease remains to be determined. Several diseases are characterized by decreased pyruvate oxidation, including most cancers and heart disease, in addition to classical inborn errors of metabolism. It remains to be seen whether loss of MPC expression or activity underlies some of the pathology associated with these diseases. Additionally, a provocative set of studies has been recently published that shows that the thiazolidenediones, a billion dollar class of anti-diabetic drugs believed to act through the PPARγ nuclear hormone receptor, bind to the MPC and might function through modulation of mitochondrial pyruvate metabolism (Colca et al. 2013; Divakaruni et al. 2013).
Mitochondrial post-translational modifications (PTMs)
Another clear trend from the recent proteomics efforts is the increase in complexity in the mitochondrial proteome throughout evolution. For example, only 56% of the MitoCarta compendium have direct orthologs in S. cerevisiae (Pagliarini et al. 2008). A case in point is CI; as noted above, Escherichia coli CI is comprised of 14 subunits, whereas yeast CI, when present, has ∼25, and human CI has 45 (Gabaldon et al. 2005; Janssen et al. 2006). For certain, many of these hundreds of “new” mitochondrial proteins enable new biochemical functions; however, it is also anticipated that many of these proteins will confer additional regulatory mechanisms.
One such class of regulatory mechanisms gaining attention among mitochondrial biologists is the use of PTMs, such as phosphorylation (Pagliarini and Dixon 2006) and acetylation (Anderson and Hirschey 2012), to manipulate protein function. The idea that phosphorylation can regulate the activity of mitochondrial proteins dates back to 1968, when Lester Reed's group (Linn et al. 1969) identified the E1 subunit of the pyruvate dehydrogenase complex (PDC) as the first mitochondrial phosphoprotein. Later work in the mid-2000s by the Balaban group (Hopper et al. 2006) and others revealed the presence of many other mitochondrial phosphoproteins. More recent efforts have extensively cataloged and quantified the phosphoproteome in this organelle (Zhao et al. 2011; Grimsrud et al. 2012) and have demonstrated the importance of phosphorylation in regulating such mitochondrial processes as ketogenesis, protein import (Schmidt et al. 2011), and proteolytic processing (Lu et al. 2013). Likewise, lysine acetylation has emerged as a prominent mitochondrial PTM. More than 2000 unique acetylation sites have now been identified, a number of which have been shown to participate in the regulation of pathways, including the urea cycle, fatty acid oxidation, ketogenesis, and the TCA cycle, among others (Kim et al. 2006; Still et al. 2013).
Despite the abundance of these PTMs and their documented ability to regulate a number of mitochondrial proteins, there is a striking lack of understanding regarding the enzymes that manage these modifications. For instance, the best-characterized mitochondrial kinases and phosphatases are those associated with the PDC and branched chain ketoacid dehydrogenase (BCKDH) complex. However, these proteins are physically associated with their corresponding enzyme complexes and are not believed to regulate the phosphorylation of many other mitochondrial substrates. MitoCarta includes five predicted atypical protein kinases (ADCK1–5); however, these kinases also appear to be related to the choline and phosphoinositide kinases (Lagier-Tourenne et al. 2008), and there is currently no direct evidence that they possess protein phosphorylation activity. PKA is clearly anchored to the mitochondrial outer membrane (Means et al. 2011), and the Manfredi group (Acin-Perez et al. 2009) has reported the existence of a complete PKA signaling system in the mitochondrial matrix; however, a widespread role for the latter system remains to be established. Likewise, PINK1, the Parkinson's disease-associated kinase (Valente et al. 2004), is clearly involved in regulating mitochondrial protein and organellar turnover but has few documented substrates. Phosphatases, such as PPTC7 (Martin-Montalvo et al. 2013), DUSP18, and DUSP21 (Rardin et al. 2008), likely have a wide range of substrates but await further characterization.
For acetylation, there is currently only a single well-established, mitochondria-localized protein deacetylase, SIRT3, which appears to regulate only a portion of the mitochondrial acetylome (Hirschey et al. 2011; Feldman et al. 2012; Hebert et al. 2013). Even more strikingly, there remains no known mitochondrial acetyltransferase, although recent work from the Sack group (Scott et al. 2012) suggests that GCN5-L1 is a key player in what may be a protein acetyltransferase complex. Two other sirtuins, SIRT4 and SIRT5, reside in mitochondria but are now known to serve primarily as a demalonylase and desuccinylase, respectively (Laurent et al. 2013; Park et al. 2013). These examples, along with a growing list of additional mitochondrial modifications, including ubiquitylation, methylation, palmitoylation, carbonylation, etc., reveal that much is yet to be determined about the regulation of PTMs in this organelle. Many of the techniques described above (Fig. 2) will likely prove fruitful in identifying orphan mitochondrial proteins involved in these post-translational regulatory processes.
Uncharted territory in a new golden age
The examples above highlight how a well-curated mitochondrial compendium can help match orphan proteins with missing functions; that is, they can accelerate the resolution of the “known unknowns.” However, many other seminal discoveries—from Mitchell's chemiosmotic coupling to the paradigm-shifting apoptosis observations noted above—were quite unexpected. Other such “unknown unknowns” include the observations by the Craig and Lill groups (Kispal et al. 1999; Schilke et al. 1999) in the late 1990s that mitochondria house the protein machinery to generate iron sulfur clusters—key cofactors for a diverse range of cellular processes. Likewise, a surprising recent finding in mitochondrial biology was the identification of a complete type II fatty acid synthase system (FAS II) in this organelle (Hiltunen et al. 2010). Unlike the typical mammalian FAS I system, which is comprised of a single homodimeric complex and produces a single end product (palmitate), the FAS II system consists of a series of dissociated and freely diffusible enzymes that are capable of generating diverse end products. The implications for this pathway in mitochondria are still being worked out.
These discoveries and others too numerous to cover in detail here demonstrate that mitochondria are far more complex in form and function than anticipated decades ago. Moreover, as the known mitochondrial proteome remains chock-full of uncharacterized proteins and as more of the missing ∼15% are now being identified (Kazak et al. 2013; Rhee et al. 2013), we can anticipate the coming decades to be as rich in new discoveries as the last.
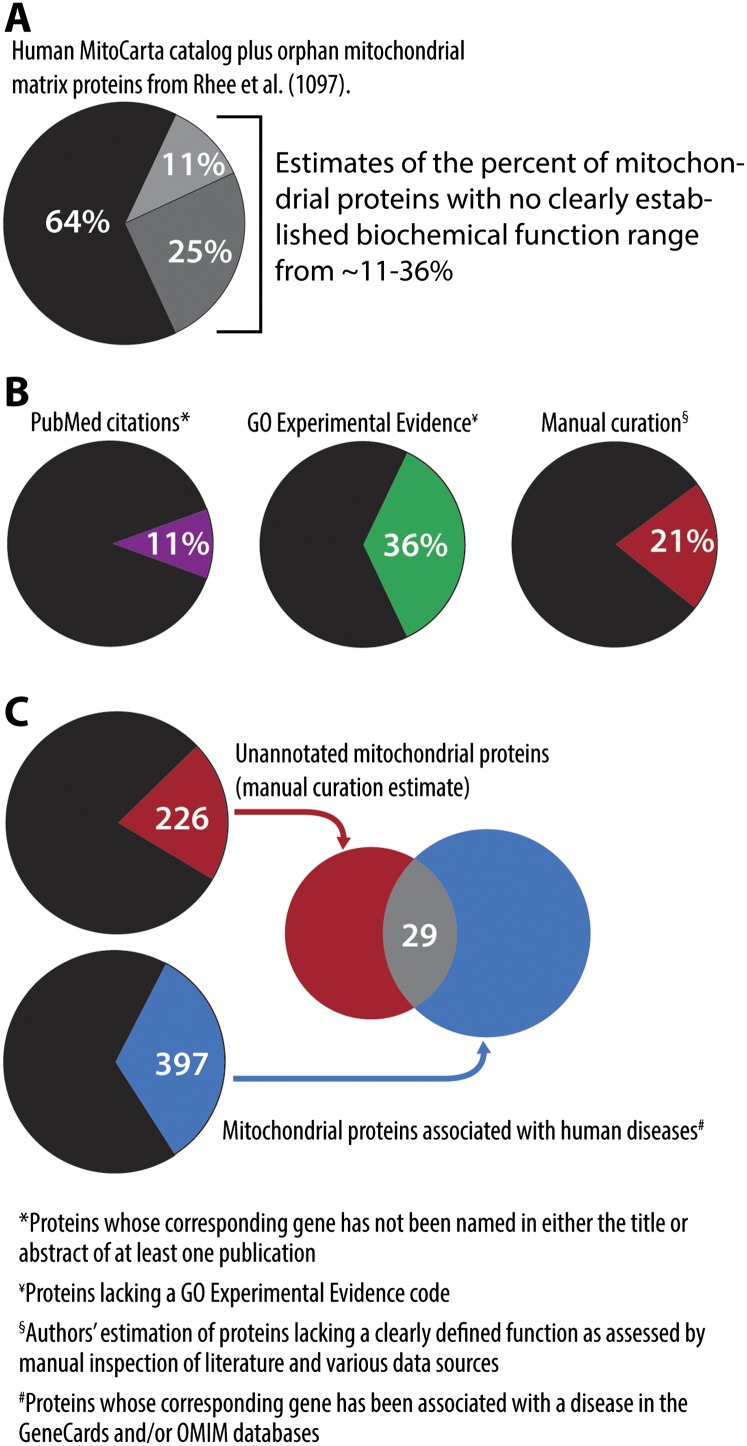
It is difficult to accurately assess the number of mitochondrial proteins that lack a known function, but we estimate that ∼11%–36% of the proteome falls into this category (Fig. 3). Many of these proteins are somehow connected to proteins of known function but are themselves uncharacterized biochemically. Two prime examples are the CIAFs mentioned above and members of the coenzyme Q biosynthetic machinery. Although there are now at least nine CIAFs identified, little to no biochemical function has been attributed to any of them (Mimaki et al. 2012). Likewise, complementation studies in yeast have identified 10 genes (coq1–10) involved in Q production, nearly all of which encode proteins that localize to mitochondria (Tran and Clarke 2007). Intriguingly, this list includes a predicted protein kinase, Coq8 (the ortholog of human ADCK3 noted above), which has been shown to be essential for proper Q production in both yeast and humans, and at least three others (Coq4, Coq9, and Coq10) that likewise have little or no established function.
Figure 3.
Estimating the unannotated mammalian mitochondrial proteome. (A) Collectively, the human MitoCarta catalog and the orphan matrix mitochondrial proteins from Rhee et al (2013). comprise 1097 proteins. We estimate that 11%–36% of these proteins do not have clearly defined functions/biochemical activities. (B) The estimates in A were based on three approaches: (1) appearance of gene names in the title or abstract of PubMed citations, (2) association with an experimental evidence code in the GO database, and (3) manual inspection of the literature and various additional data sources. (C) Three-hundred-ninety-seven genes encoding mitochondrial proteins are associated with a human disease in either the GenCards or Online Mendelian Inheritance in Man (OMIM) databases. Twenty-nine of these genes also encode proteins of unknown function based on our manual inspection criteria.
Many uncharacterized mitochondrial proteins are now known to be mutated in human disease. Another enabling feature of the expanded mitochondrial proteome has been the prioritization of candidate disease-related genes that could harbor causative mutations. Indeed, state-of-the-art high-throughput sequencing technologies are rapidly expanding the list of genes encoding mitochondrial proteins that are linked to disease in humans (Koopman et al. 2012; Vafai and Mootha 2012). Although this represents great progress in the field of mitochondrial medicine, elucidation of the biochemical functions of these disease-related proteins has become a bottleneck in understanding mitochondrial pathophysiology. This list of uncharacterized, disease-related mitochondrial proteins, which is sure to grow in the coming years, represents particularly high-priority targets for investigation.
Finally, there are many proteins that currently have no connection to any human disease or known biochemical process, offering few clues to their roles in mitochondrial biology. As such, moving forward, we must blend targeted analyses with approaches that are agnostic to protein function. Fortunately, as featured above, the large-scale technologies that do and will enable these types of scientific approaches are becoming increasingly powerful and available. These include nuclear magnetic resonance (NMR)- and MS-based protein–protein and protein–metabolite interaction analyses, genome-scale mapping of genetic interactions, phylogenetic profiling and other computational approaches, metabolic profiling, and high-throughput structural genomics efforts. In the end, however, these are tools toward the generation of testable hypotheses. As it has been since Lavoisier and through the days of Warburg, Krebs, and Mitchell, new insights into mitochondrial form and function will also require dedicated scientists toiling away at the bench with open minds as to what they might discover.
In 1967, in a New England Journal of Medicine article entitled simply “The Mitochondrion,” Tapley et al. (1967) stated, “During the past few decades the mitochondrion, as a self-contained unit within the cell, has been a magnificent tool in the hands of biochemists, physiologists and anatomists in their attempts to unravel the complexities of cellular function. It appears that it will continue to be.” Nearly 50 years later, those words still ring true.
Acknowledgments
We thank members of the Pagliarini laboratory, Dennis Winge, and Richard Amasino for critical reading of the manuscript, and Brendan Floyd for assistance with data generation for Figure 3. Support for our mitochondrial research is provided by a Searle Scholar Award, a Shaw Scientist Award, a U.S. Department of Agriculture grant (WIS01671), and National Institutes of Health grants U01GM09462 and R01DK098672 to D.J.P. and R01GM094232 to J.R.
Note added in proof
The Mootha group (Sancak et al. 2013) has recently identified a fourth subunit of the MCU—the essential MCU regulator (EMRE).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.229724.113.
References
- Acin-Perez R, Salazar E, Kamenetsky M, Buck J, Levin LR, Manfredi G 2009. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab 9: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MR, Groschner LN, Parichatikanond W, Kuo L, Bondarenko AI, Rost R, Waldeck-Weiermair M, Malli R, Graier WF 2012. Mitochondrial Ca2+ uptake 1 (MICU1) and mitochondrial Ca2+ uniporter (MCU) contribute to metabolism–secretion coupling in clonal pancreatic β-cells. J Biol Chem 287: 34445–34454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann R. 1890. Die Elementarorganismen und ihre beziehungen zu den zellen. Veit, Leipzig, Germany. [Google Scholar]
- Anderson KA, Hirschey MD 2012. Mitochondrial protein acetylation regulates metabolism. Essays Biochem 52: 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 [DOI] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133–140 [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T 2005. Mitochondria, oxidants, and aging. Cell 120: 483–495 [DOI] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, et al. 2011. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel W. 2006. Discovering cell mechanisms: The creation of modern cell biology. Cambridge University Press, New York. [Google Scholar]
- Benda C 1898. Ueber die spermatogenese der vertebraten und höherer evertebraten, II. Theil: Die histiogenese der spermien. Arch Anat Physiol 73: 393–398 [Google Scholar]
- Boyer PD, Chance B, Ernster L, Mitchell P, Racker E, Slater EC 1977. Oxidative phosphorylation and photophosphorylation. Annu Rev Biochem 46: 955–966 [DOI] [PubMed] [Google Scholar]
- Bratic A, Larsson NG 2013. The role of mitochondria in aging. J Clin Invest 123: 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, et al. 2012. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini M, Pinton P, King MP, Davidson M, Schon EA, Rizzuto R 1999. A calcium signaling defect in the pathogenesis of a mitochondrial DNA inherited oxidative phosphorylation deficiency. Nat Med 5: 951–954 [DOI] [PubMed] [Google Scholar]
- Calvo SE, Mootha VK 2010. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet 11: 25–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, Rivas M, Guiducci C, Bruno DL, Goldberger OA, et al. 2010. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet 42: 851–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude A 1946. Fractionation of mammalian liver cells by differential centrifugation; problems, methods, and preparation of extract. J Exp Med 84: 51–59 [PubMed] [Google Scholar]
- Claypool SM, Koehler CM 2012. The complexity of cardiolipin in health and disease. Trends Biochem Sci 37: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colca JR, McDonald WG, Cavey GS, Cole SL, Holewa DD, Brightwell-Conrad AS, Wolfe CL, Wheeler JS, Coulter KR, Kilkuskie PM, et al. 2013. Identification of a mitochondrial target of thiazolidinedione insulin sensitizers (mTOT)—relationship to newly identified mitochondrial pyruvate carrier proteins. PLoS ONE 8: e61551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338: 815–818 [DOI] [PubMed] [Google Scholar]
- Da Cruz S, Xenarios I, Langridge J, Vilbois F, Parone PA, Martinou JC 2003. Proteomic analysis of the mouse liver mitochondrial inner membrane. J Biol Chem 278: 41566–41571 [DOI] [PubMed] [Google Scholar]
- Deluca HF, Engstrom GW 1961. Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci 47: 1744–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R 2011. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Schon EA 2003. Mitochondrial respiratory-chain diseases. N Engl J Med 348: 2656–2668 [DOI] [PubMed] [Google Scholar]
- Divakaruni AS, Wiley SE, Rogers GW, Andreyev AY, Petrosyan S, Loviscach M, Wall EA, Yadava N, Heuck AP, Ferrick DA, et al. 2013. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proc Natl Acad Sci 110: 5422–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Ernster L, Schatz G 1981. Mitochondria: A historical review. J Cell Biol 91: 227s–255s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Dittenhafer-Reed KE, Denu JM 2012. Sirtuin catalysis and regulation. J Biol Chem 287: 42419–42427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK 2011. ER tubules mark sites of mitochondrial division. Science 334: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Rainey D, Huynen MA 2005. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I). J Mol Biol 348: 857–870 [DOI] [PubMed] [Google Scholar]
- Getz GS, Bartley W, Lurie D, Notton BM 1968. The phospholipids of various sheep organs, rat liver and of their subcellular fractions. Biochim Biophys Acta 152: 325–339 [DOI] [PubMed] [Google Scholar]
- Green DE 1964. The mitochondrion. Sci Am 210: 67–74 [PubMed] [Google Scholar]
- Green DR, Reed JC 1998. Mitochondria and apoptosis. Science 281: 1309–1312 [DOI] [PubMed] [Google Scholar]
- Grimsrud PA, Carson JJ, Hebert AS, Hubler SL, Niemi NM, Bailey DJ, Jochem A, Stapleton DS, Keller MP, Westphall MS, et al. 2012. A quantitative map of the liver mitochondrial phosphoproteome reveals posttranslational control of ketogenesis. Cell Metab 16: 672–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap AP, Denton RM 1974. Specific inhibition of pyruvate transport in rat liver mitochondria and human erythrocytes by α-cyano-4-hydroxycinnamate. Biochem J 138: 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, et al. 2013. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49: 186–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide H, Bleier L, Steger M, Ackermann J, Drose S, Schwamb B, Zornig M, Reichert AS, Koch I, Wittig I, et al. 2012. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab 16: 538–549 [DOI] [PubMed] [Google Scholar]
- Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, Kunji ER, Martinou JC 2012. Identification and functional expression of the mitochondrial pyruvate carrier. Science 337: 93–96 [DOI] [PubMed] [Google Scholar]
- Hiltunen JK, Chen Z, Haapalainen AM, Wierenga RK, Kastaniotis AJ 2010. Mitochondrial fatty acid synthesis—an adopted set of enzymes making a pathway of major importance for the cellular metabolism. Prog Lipid Res 49: 27–45 [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Huang JY, Schwer B, Verdin E 2011. SIRT3 regulates mitochondrial protein acetylation and intermediary metabolism. Cold Spring Harb Symp Quant Biol 76: 267–277 [DOI] [PubMed] [Google Scholar]
- Hogeboom GH, Claude A, Hotch-Kiss RD 1946. The distribution of cytochrome oxidase and succinoxidase in the cytoplasm of the mammalian liver cell. J Biol Chem 165: 615–629 [PubMed] [Google Scholar]
- Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS 2006. Mitochondrial matrix phosphoproteome: Effect of extra mitochondrial calcium. Biochemistry 45: 2524–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagendorf AT 1967. Acid–base transitions and phosphorylation by chloroplasts. Fed Proc 26: 1361–1369 [PubMed] [Google Scholar]
- Janssen RJ, Nijtmans LG, van den Heuvel LP, Smeitink JA 2006. Mitochondrial complex I: Structure, function and pathology. J Inherit Metab Dis 29: 499–515 [DOI] [PubMed] [Google Scholar]
- Kazak L, Reyes A, Duncan AL, Rorbach J, Wood SR, Brea-Calvo G, Gammage PA, Robinson AJ, Minczuk M, Holt IJ 2013. Alternative translation initiation augments the human mitochondrial proteome. Nucleic Acids Res 41: 2354–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilin D 1925. On cytochrome, a respiratory pigment, common to animals, yeast, and higher plants. Proc Roy Soc B 98: 312–339 [Google Scholar]
- Kennedy EP, Lehninger AL 1949. Oxidation of fatty acids and tricarboxylic acid cycle intermediates by isolated rat liver mitochondria. J Biol Chem 179: 957–972 [PubMed] [Google Scholar]
- Kiberstis P 1999. Mitochondria make a comeback. Science 283: 1475 [Google Scholar]
- Kiefer MC, Brauer MJ, Powers VC, Wu JJ, Umansky SR, Tomei LD, Barr PJ 1995. Modulation of apoptosis by the widely distributed Bcl-2 homologue Bak. Nature 374: 736–739 [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607–618 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD 1997. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 275: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Koopman WJ, Willems PH, Smeitink JA 2012. Monogenic mitochondrial disorders. N Engl J Med 366: 1132–1141 [DOI] [PubMed] [Google Scholar]
- Kopec KO, Alva V, Lupas AN 2010. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics 26: 1927–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P 2009. An ER–mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA, Johnson WA 1937. Metabolism of ketonic acids in animal tissues. Biochem J 31: 645–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Tazir M, Lopez LC, Quinzii CM, Assoum M, Drouot N, Busso C, Makri S, Ali-Pacha L, Benhassine T, et al. 2008. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am J Hum Genet 82: 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. 2006. Power, sex, suicide: Mitochondria and the meaning of life. Oxford University Press, New York. [Google Scholar]
- Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, et al. 2013. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell 50: 686–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehninger AL, Rossi CS, Greenawalt JW 1963. Respiration-dependent accumulation of inorganic phosphate and Ca ions by rat liver mitochondria. Biochem Biophys Res Commun 10: 444–448 [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X 1997. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91: 479–489 [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443: 787–795 [DOI] [PubMed] [Google Scholar]
- Linn TC, Pettit FH, Reed LJ 1969. α-Keto acid dehydrogenase complexes. X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc Natl Acad Sci 62: 234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X 1996. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 86: 147–157 [DOI] [PubMed] [Google Scholar]
- Lohmann K 1929. Über die Pyrophosphatfraktion im Muskel. Naturwissenschaften 17: 624–625 [Google Scholar]
- Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, Robinson M 2000. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis 21: 3427–3440 [DOI] [PubMed] [Google Scholar]
- Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK 2013. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell 49: 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Doonan P, Cárdenas C, Chandramoorthy HC, Müller M, Miller R, Hoffman NE, Gandhirajan RK, Molgó J, Birnbaum MJ, et al. 2012. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell 151: 630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinetti GV, Erbland J, Stotz E 1958. Phosphatides of pig heart cell fractions. J Biol Chem 233: 562–565 [PubMed] [Google Scholar]
- Martin-Montalvo A, Gonzalez-Mariscal I, Pomares-Viciana T, Padilla-Lopez S, Ballesteros M, Vazquez-Fonseca L, Gandolfo P, Brautigan DL, Navas P, Santos-Ocana C 2013. The phosphatase ptc7 induces coenzyme q biosynthesis by activating the hydroxylase coq7 in yeast. J Biol Chem 288: 28126–28137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S 2006. Mitochondria: More than just a powerhouse. Curr Biol 16: R551–R560 [DOI] [PubMed] [Google Scholar]
- McCormack JG, Halestrap AP, Denton RM 1990. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev 70: 391–425 [DOI] [PubMed] [Google Scholar]
- Means CK, Lygren B, Langeberg LK, Jain A, Dixon RE, Vega AL, Gold MG, Petrosyan S, Taylor SS, Murphy AN, et al. 2011. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc Natl Acad Sci 108: E1227–E1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AH, Kornmann B 2012. The ERMES complex and ER–mitochondria connections. Biochem Soc Trans 40: 445–450 [DOI] [PubMed] [Google Scholar]
- Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT 2012. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta 1817: 851–862 [DOI] [PubMed] [Google Scholar]
- Mitchell P 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191: 144–148 [DOI] [PubMed] [Google Scholar]
- Mitchell P 1979. Keilin's respiratory chain concept and its chemiosmotic consequences. Science 206: 1148–1159 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, et al. 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115: 629–640 [DOI] [PubMed] [Google Scholar]
- Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife 2: e00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Lewandowska A, Choi JY, Markgraf DF, Junker M, Bilgin M, Ejsing CS, Voelker DR, Rapoport TA, Shaw JM 2012. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic 13: 880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls DG, Ferguson SJ 2002. Bioenergetics 3. Academic Press, San Diego [Google Scholar]
- Nunnari J, Suomalainen A 2012. Mitochondria: In sickness and in health. Cell 148: 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie I, Kennaway NG, Shoubridge EA 2005. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest 115: 2784–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltvai ZN, Milliman CL, Korsmeyer SJ 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74: 609–619 [DOI] [PubMed] [Google Scholar]
- Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brugger B, Westermann B, Langer T 2009. The genetic interactome of prohibitins: Coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol 184: 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Haag M, Wieland FT, Brugger B, Langer T 2010. A mitochondrial phosphatase required for cardiolipin biosynthesis: The PGP phosphatase Gep4. EMBO J 29: 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Dixon JE 2006. Mitochondrial modulation: Reversible phosphorylation takes center stage? Trends Biochem Sci 31: 26–34 [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Wiley SE, Kimple ME, Dixon JR, Kelly P, Worby CA, Casey PJ, Dixon JE 2005. Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic β cells. Mol Cell 19: 197–207 [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade GE 1953. An electron microscope study of the mitochondrial structure. J Histochem Cytochem 1: 188–211 [DOI] [PubMed] [Google Scholar]
- Pangborn MC 1945. A simplified preparation of cardiolipin, with note on purification of lecithin for serologic use. J Biol Chem 161: 71–82 [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, et al. 2013. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 50: 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK 2010. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature 467: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabilloud T, Kieffer S, Procaccio V, Louwagie M, Courchesne PL, Patterson SD, Martinez P, Garin J, Lunardi J 1998. Two-dimensional electrophoresis of human placental mitochondria and protein identification by mass spectrometry: Toward a human mitochondrial proteome. Electrophoresis 19: 1006–1014 [DOI] [PubMed] [Google Scholar]
- Rardin MJ, Wiley SE, Murphy AN, Pagliarini DJ, Dixon JE 2008. Dual specificity phosphatases 18 and 21 target to opposing sides of the mitochondrial inner membrane. J Biol Chem 283: 15440–15450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339: 1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Murgia M, Pozzan T 1993. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science 262: 744–747 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, De Stefani D, Raffaello A, Mammucari C 2012. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol 13: 566–578 [DOI] [PubMed] [Google Scholar]
- Robertson JD 1960. The molecular structure and contact relationships of cell membranes. Prog Biophys Mol Biol 10: 343–418 [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovács-Bogdán E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, et al. 2013. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science doi: 10.1126/science.1242993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfe C, Zaccaria P, Hoertnagel K, Jaksch M, Klopstock T, Lill R, Prokisch H, Gerbitz KD, Mewes HW, Meitinger T 1999. MITOP: Database for mitochondria-related proteins, genes and diseases. Nucleic Acids Res 27: 153–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler IE. 2008. Mitochondria. Wiley-Liss, New York. [Google Scholar]
- Schilke B, Voisine C, Beinert H, Craig E 1999. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci 96: 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schonfisch B, Guiard B, Sickmann A, Pfanner N, et al. 2011. Regulation of mitochondrial protein import by cytosolic kinases. Cell 144: 227–239 [DOI] [PubMed] [Google Scholar]
- Scott I, Webster BR, Li JH, Sack MN 2012. Identification of a molecular component of the mitochondrial acetyltransferase programme: A novel role for GCN5L1. Biochem J 443: 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Narita M, Tsujimoto Y 1999. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature 399: 483–487 [DOI] [PubMed] [Google Scholar]
- Shutov LP, Kim MS, Houlihan PR, Medvedeva YV, Usachev YM 2013. Mitochondria and plasma membrane Ca2+-ATPase control presynaptic Ca2+ clearance in capsaicin-sensitive rat sensory neurons. J Physiol 591: 2443–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostrand FS 1953. Electron microscopy of mitochondria and cytoplasmic double membranes. Nature 171: 30–32 [DOI] [PubMed] [Google Scholar]
- Still AJ, Floyd BJ, Hebert AS, Bingman CA, Carson JJ, Gunderson DR, Dolan BK, Grimsrud PA, Dittenhafer-Reed KE, Stapleton DS, et al. 2013. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J Biol Chem 288: 26209–26219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szendroedi J, Phielix E, Roden M 2012. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat Rev Endocrinol 8: 92–103 [DOI] [PubMed] [Google Scholar]
- Tager JM, Groen AK, Wanders RJ, Duszynski J, Westerhoff HV, Vervoorn RC 1983. Control of mitochondrial respiration. Biochem Soc Trans 11: 40–43 [DOI] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K, et al. 2013. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab 17: 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley DF, Kimberg DV, Buchanan JL 1967. The mitochondrion. N Engl J Med 276: 1182–1191 [DOI] [PubMed] [Google Scholar]
- Tarasov AI, Semplici F, Li D, Rizzuto R, Ravier MA, Gilon P, Rutter GA 2013. Frequency-dependent mitochondrial Ca2+ accumulation regulates ATP synthesis in pancreatic β cells. Pflugers Arch 465: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, et al. 2003. Characterization of the human heart mitochondrial proteome. Nat Biotechnol 21: 281–286 [DOI] [PubMed] [Google Scholar]
- Tran UC, Clarke CF 2007. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7: S62–S71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK 2012. Mitochondrial disorders as windows into an ancient organelle. Nature 491: 374–383 [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. 2004. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304: 1158–1160 [DOI] [PubMed] [Google Scholar]
- Vasington FD, Murphy JV 1962. Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem 237: 2670–2677 [PubMed] [Google Scholar]
- Wallace DC 1999. Mitochondrial diseases in man and mouse. Science 283: 1482–1488 [DOI] [PubMed] [Google Scholar]
- Wallace DC 2012. Mitochondria and cancer. Nat Rev Cancer 12: 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng TI, Jones DP, Wang X 1997. Prevention of apoptosis by Bcl-2: Release of cytochrome c from mitochondria blocked. Science 275: 1129–1132 [DOI] [PubMed] [Google Scholar]
- Zhang J, Guan Z, Murphy AN, Wiley SE, Perkins GA, Worby CA, Engel JL, Heacock P, Nguyen OK, Wang JH, et al. 2011. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab 13: 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Leon IR, Bak S, Mogensen M, Wrzesinski K, Hojlund K, Jensen ON 2011. Phosphoproteome analysis of functional mitochondria isolated from resting human muscle reveals extensive phosphorylation of inner membrane protein complexes and enzymes. Mol Cell Proteomics 10: M110.000299. [DOI] [PMC free article] [PubMed] [Google Scholar]