Down's syndrome (DS) arises from triplication of genes on chromosome 21. Kurabayashi and Sanada discovered that two genes within human chromosome 21, DYRK1A and DSCR1, are expressed in mouse neural progenitors. Both proteins delay neuronal differentiation through suppressing NFATc. In Ts1Cje mice, a DS mouse model, counteracting this deregulated pathway ameliorates delayed neuronal differentiation. These findings provide a mechanism to explain the neurodevelopmental defects in DS.
Keywords: Down's syndrome, developing neocortex, neurogenesis, neural progenitor
Abstract
Down's syndrome (DS), a major genetic cause of mental retardation, arises from triplication of genes on human chromosome 21. Here we show that DYRK1A (dual-specificity tyrosine-phosphorylated and -regulated kinase 1A) and DSCR1 (DS critical region 1), two genes lying within human chromosome 21 and encoding for a serine/threonine kinase and calcineurin regulator, respectively, are expressed in neural progenitors in the mouse developing neocortex. Increasing the dosage of both proteins in neural progenitors leads to a delay in neuronal differentiation, resulting ultimately in alteration of their laminar fate. This defect is mediated by the cooperative actions of DYRK1A and DSCR1 in suppressing the activity of the transcription factor NFATc. In Ts1Cje mice, a DS mouse model, dysregulation of NFATc in conjunction with increased levels of DYRK1A and DSCR1 was observed. Furthermore, counteracting the dysregulated pathway ameliorates the delayed neuronal differentiation observed in Ts1Cje mice. In sum, our findings suggest that dosage of DYRK1A and DSCR1 is critical for proper neurogenesis through NFATc and provide a potential mechanism to explain the neurodevelopmental defects in DS.
Down's syndrome (DS), which occurs in one in 700–800 live births (Canfield et al. 2006), is caused by the presence of an extra human chromosome 21. DS is characterized by neurological, skeletal, cardiovascular, and immunological defects (Hasle et al. 2000; Roizen and Patterson 2003; Antonarakis et al. 2004). In particular, mental retardation is the most common phenotype in DS (Chapman and Hesketh 2000). DS brains are microcephalic, and structural alterations such as hypoplasia of the forebrain and cerebellum were observed in prenatal and perinatal stages (Schmidt-Sidor et al. 1990; Golden and Hyman 1994; Weitzdoerfer et al. 2001). DS brains also exhibit reduced neuronal density and number (Mann et al. 1987; Tanzi 1996), and neurons in DS brains have shorter dendrites and fewer spines with altered morphology (Takashima et al. 1981; Petit et al. 1984). These observations suggest defects in neurodevelopmental processes in individuals with DS.
Features of DS are generally thought to originate from an increase in the dosage of genes within human chromosome 21. Several studies on humans with partial trisomy 21 targeted a specific region of chromosome 21 called the DS critical region (DSCR). This chromosomal region is hypothesized to be responsible for a subset of DS traits, including neurological phenotypes (McCormick et al. 1989; Korenberg et al. 1990; Delabar et al. 1993). Brain abnormalities similar to DS have been reported in mouse models of DS. In particular, Ts65Dn and Ts1Cje mouse models, which are trisomic for large segments of mouse chromosome 16 (including DSCR), orthologous to human chromosome 21, display abnormal forebrain and cerebellar development (Chakrabarti et al. 2007; Ishihara et al. 2010). In the developing neocortices, proliferation and differentiation of neural progenitors are impaired, suggesting misregulation of neocortical progenitors in DS (Chakrabarti et al. 2007; Ishihara et al. 2010). On the other hand, studies using mouse models that are trisomic for the DSCR alone show that the DSCR is necessary but not sufficient for certain brain phenotypes seen in Ts65Dn mice (Olson et al. 2004, 2007). These studies suggest that phenotypes associated with DS are due to the complex effects of multiple genes in the human chromosome 21. Identification of dosage-sensitive genes within human chromosome 21 and of their mechanisms of actions constitutes an important step in our understanding of DS and toward the better development of therapeutic interventions. Nevertheless, molecular and cellular bases underlying neurodevelopmental defects in DS are incompletely understood.
The Dyrk1a gene (dual-specificity tyrosine-phosphorylated and -regulated kinase 1A), a proline-directed serine/threonine kinase, lies within the DSCR and is overexpressed in fetal and adult DS brains (Guimera et al. 1999). DYRK1A has been implicated as a regulator for CNS development, as Dyrk1a-deficent mice show reduced brain size (Fotaki et al. 2002). Also, Dyrk1a transgenic mice display alteration of brain size and neuronal density in the cerebral cortex (Guedj et al. 2012). Dscr1 also lies within the centromeric border of the DSCR, encodes for an inhibitor of protein phosphatase calcineurin 1, and often is referred to as Rcan1 (regulator of calcineurin 1) (Rothermel et al. 2000). Like DYRK1A, DSCR1 is overexpressed in fetal and adult DS brains (Fuentes et al. 2000), but its role in CNS development is incompletely understood. Furthermore, the contribution of an increased dosage of the two genes to neurodevelopmental defects in DS has not been totally explored in a more pathological context, such as in animal models of DS. Interestingly, in vitro studies have shown that DSCR1 synergizes with DYRK1A to prevent the signaling pathway mediated by NFATc (Arron et al. 2006), a critical regulator for vertebrate development (Tuan and Simone 2008; Müller and Rao 2010). Importantly, Nfatc-deficient mice manifest several features seen in human DS, such as cognitive decline, hypotonia, heart defects, and skeletal abnormalities (Graef et al. 2001; Chang et al. 2004; Arron et al. 2006). However, it has not been examined whether Nfatc deficiency and dysregulation of the NFATc pathway lead to developmental defects in mouse brains.
In this study, we found that DYRK1A and DSCR1 are expressed in neural progenitor cells in the mouse developing neocortex. Furthermore, we discovered that increasing the dosage of DYRK1A and DSCR1 delayed progenitor differentiation and altered their laminar fate via attenuation of NFATc in vivo. In the developing neocortices of Ts1Cje mice, a widely used mouse model of DS, we found dysregulation of NFATc in conjunction with increased levels of both DYRK1A and DSCR1. Furthermore, delayed neuronal differentiation of progenitors observed in the Ts1Cje neocortices was ameliorated by counteracting the dysregulated DYRK1A/DSCR1–NFATc pathway with depletion of DYRK1A/DSCR1 and activation of NFATc. In sum, our work underscores the DYRK1A/DSCR1–NFATc pathway as a critical pathway for proper neuronal differentiation of progenitors, highlighting a potential mechanism for neurodevelopmental defects observed in DS.
Results
Overexpression of DYRK1A and DSCR1 results in decreased neuronal differentiation of cortical progenitors
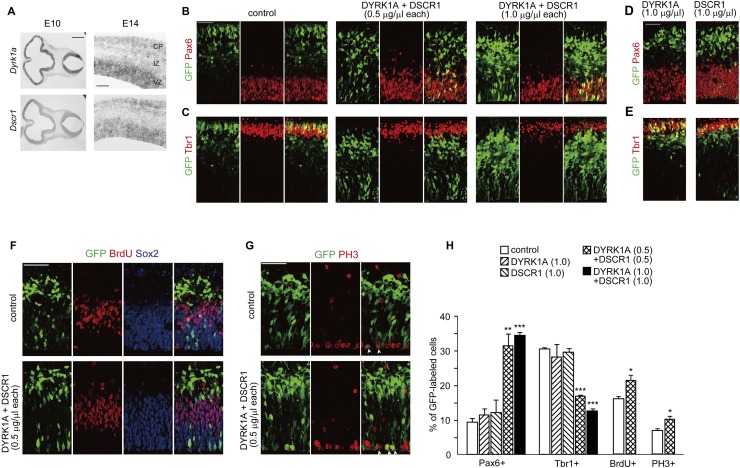
During the development of the mammalian brain, neural progenitor cells located in the ventricular zone (VZ) surrounding the ventricles give rise to neurons. In the developing neocortex, newborn neurons migrate radially from their birthplace in the VZ toward the pia through the intermediate zone (IZ) to form the cortical plate (CP). To examine the expression pattern of Dyrk1a and Dscr1, two genes within human chromosome 21, in developing mouse brains, we performed in situ hybridization analysis using an antisense probe to the mRNAs of the two genes. In agreement with previous studies (Hämmerle et al. 2008), Dyrk1a was expressed at the proliferative regions surrounding the ventricles at embryonic day 10 (E10) (Fig. 1A). In addition, Dscr1 shows an expression pattern similar to Dyrk1a (Fig. 1A). Furthermore, intense expressions of Dyrk1a and Dscr1 were detected in the VZs of E14 mouse neocortices (Fig. 1A). Also, DYRK1A and DSCR1 immunoreactivities were detected in almost all cortical progenitors (Supplemental Fig. S1), suggesting that these genes are expressed in cortical progenitors.
Figure 1.
Overexpression of DYRK1A and DSCR1 results in decreased cortical progenitor differentiation. (A) Expression of Dyrk1a and Dscr1 in the developing brain. In situ hybridization analyses with Dyrk1a and Dscr1 probes were carried out on sections of E10 embryos and E14 brains. Images of the horizontal sections of E10 embryos (left panels) and coronal sections of E14 neocortices (right panels) are shown. Bars: in E10 sections, 500 μm; in E14 sections, 200 μm. (B–E) Plasmids encoding DYRK1A and/or DSCR1 were electroporated, together with the GFP-expressing plasmid, in E11 embryos. Thereafter, the E13 brain sections were immunostained with antibodies against Pax6 (B,D) and Tbr1 (C,E). Images of the entire cerebral wall are shown. Plasmids were electroporated, and their concentrations are indicated above the images. Bar, 50 μm. (F,G) Plasmids encoding DYRK1A and DSCR1 were electroporated, together with the GFP-expressing plasmid, in E11 embryos. BrdU was administrated at E12, and brains were fixed 30 min after the BrdU pulse labeling. The brain sections were then immunostained with antibodies against GFP, BrdU, and Sox2 (F) and phospho-Histone H3 (PH3) (G). Images of the entire cerebral wall are shown. Plasmids were electroporated, and their concentration are indicated. Bars, 50 μm. (H) Quantification of a fraction of GFP-positive cells that was also positive for Pax6, Tbr1, BrdU, and PH3. Data are presented as mean ± SEM (n = 3–4 embryos for each group). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 versus control by a two-tailed Student's t-test. The concentration of electroporated plasmids for DYRK1A and DSCR1 (in micrograms per microliter) is presented in brackets.
In the fetal DS brain, DYRK1A and DSCR1 are overexpressed (Guimera et al. 1999; Fuentes et al. 2000). We then sought to determine the potential effect of DYRK1A and DSCR1 overexpression on neurogenesis. For this, plasmids encoding DYRK1A and DSCR1 were electroporated, together with the GFP-expressing plasmid, into the VZ of E11 mouse brains. In the control cortices at E13, the majority of GFP-labeled cells were located in the IZ and CP, and fewer cells remained in the VZ (VZ: 17.6% ± 2.3%; IZ: 50.9% ± 2.0%; CP: 31.5% ± 4.1%; n = 3 embryos) (Fig. 1B,C). When either DYRK1A or DSCR1 was singly electroporated at a concentration of 1.0 μg/μL plasmid (Fig. 1D,E), a normal distribution pattern of GFP-labeled cells was observed. To our surprise, when both DYRK1A and DSCR1 (0.5 μg/μL plasmid each) were introduced, a sizable population of GFP-positive cells remained in the VZ (VZ: 35.3% ± 3.1%, P < 0.05 vs. control by a Student's t-test; IZ: 50.1% ± 2.0%; CP: 14.5% ± 1.2%, P < 0.05 vs. control by a Student's t-test; n = 3 embryos) (Fig. 1B,C). To determine whether the increase in the VZ remaining cell fraction is due to decreased neuronal differentiation of cortical progenitors, GFP-labeled cells were immunostained with Pax6, a marker for apical progenitors (Götz et al. 1998), and Tbr1, a marker for early-born cortical neurons (Bulfone et al. 1995). A Pax6-positive population of GFP-labeled cells in DYRK1A- and DSCR1-introduced neocortices was remarkably increased, as compared with that in control neocortices (control, 9.4% ± 1.0%; DYRK1A/DSCR1, 31.5% ± 3.6%; n = 3 embryos) (Fig. 1B,H). On the other hand, a significant decrease in a proportion of Tbr1-positive cells (control, 30.7% ± 0.4%; 0.5 μg/μL DYRK1A/DSCR1, 17.0% ± 0.4%; n = 3 embryos) was found (Fig. 1C,H). Furthermore, increasing the concentration of the plasmids electroporated (1.0 μg/μL plasmid each) exacerbated the abnormalities (Pax6: 34.5% ± 1.0%; Tbr1: 12.6% ± 0.8%; n = 3 embryos) (Fig. 1B,C,H), suggesting the dose-dependent effect of these proteins on the phenotype. These results suggest that DYRK1A and DSCR1 overexpression results in a decreased propensity for progenitors to differentiate into neurons.
We also assessed a fraction of S-phase cells in GFP-labeled cells after a short-term BrdU pulse. A BrdU-positive fraction of GFP-labeled cells was significantly increased by DYRK1A and DSCR1 overexpression (Fig. 1F,H). Also, a phospho-histone H3-positive, M-phase cell fraction was increased (Fig. 1G,H). Thus, increased dosage of DYRK1A and DSCR1 in progenitors results in decreased neuronal differentiation and concomitant cell cycle re-entry. In addition, we did not observe any increase in cleaved caspase 3-positive cells following DYRK1A and DSCR1 overexpression (Supplemental Fig. S2A,B), indicating that apoptosis does not account for the phenotypes observed. We also found that almost all of the DYRK1A- and DSCR1-introduced cells became located in the CP and differentiated into neurons by E16 (see below). Taken together, we conclude that DYRK1A and DSCR1 overexpression delays neuronal differentiation of neural progenitors.
We next assessed the protein levels of DYRK1A and DSCR1 in cortical progenitors electroporated. For this, a neocortical cell culture was prepared from the E14 brains electroporated at E15 and then immunostained at DIV1 (1 d in vitro) with antibodies against DYRK1A and DSCR1. Consistent with a previous study (Hämmerle et al. 2008), DYRK1A was located in both the cytoplasm and nucleus, whereas DSCR1 was predominantly present in the cytoplasm. DYRK1A immunofluorescence intensities in DYRK1A-introduced progenitor cells were 1.40-fold ± 0.07-fold (0.5 μg/μL plasmid electroporated) and 1.90-fold ± 0.10-fold (1.0 μg/μL plasmid electroporated) above the endogenous levels (Supplemental Fig. S3A,C). On the other hand, DSCR1 immunofluorescence intensities in DSCR1-introduced progenitor cells were 1.59-fold ± 0.10-fold (0.5 μg/μL plasmid electroporated) and 2.01-fold ± 0.13-fold (1.0 μg/μL plasmid electroporated) above the endogenous levels (Supplemental Fig. S3B,D). In addition, coexpression of DYRK1A and DSCR1 had almost no effect on each other's expression and subcellular distribution (Supplemental Fig. S3E–G). These results suggest that an ∼1.5-fold modest increase of both DYRK1A and DSCR1 impairs normal neuronal differentiation.
Overexpression of DYRK1A and DSCR1 in progenitors alters laminar fate of their neuronal progeny
In the developing neocortex, the laminar fates of projection neurons are highly correlated with their birthdate; early-born neurons are destined for deeper-layer neurons, whereas later-born neurons become upper-layer neurons. Therefore, it is expected that a delay in neuronal differentiation of progenitors following DYRK1A and DSCR1 overexpression will cause an alteration of the laminar fate of their neuronal progeny. To examine this, E16 cortices electroporated at E11 were immunostained with Cux1 and Tbr1, markers for layer 2–4 neurons and layer 5/6 neurons, respectively (Bulfone et al. 1995; Nieto et al. 2004). Note that during this period of corticogenesis, Cux1 protein is expressed in both progenitors and immature neurons destined for layers 2–4 (Nieto et al. 2004). In the control cortices, 32.2% ± 1.7% and 48.3% ± 0.9% (n = 3 embryos) of GFP-labeled cells were positive for Cux1 and Tbr1, respectively (Fig. 2), which is consistent with previous reports (e.g., Yoshida et al. 2010; Dominguez et al. 2013). In the DYRK1A- and DSCR1-introduced cortices, the location of GFP-labeled cells was relatively more superficial than control cells. Also, a Cux1-positive fraction of GFP-labeled cells was remarkably increased, and instead a significantly smaller fraction of Tbr1-positive cells was detected (Cux1: 32.2% ± 1.7% vs. 41.5% ± 2.2% in control and DYRK1A/DSCR1, respectively, n = 3 embryos, P < 0.05 by a Student's t-test; Tbr1: 48.3% ± 0.9% vs. 40.7% ± 1.6% in control and DYRK1A/DSCR1, respectively, n = 3 embryos, P < 0.05 by a Student's t-test) (Fig. 2). In addition, increasing the concentration of the plasmids electroporated (1.0 μg/μL each) exacerbated the phenotype (Fig. 2). Taken together, these results confirm that overexpression of DYRK1A and DSCR1 in progenitors causes delay in their neuronal differentiation, which ultimately leads to alteration of laminar positioning and the fate of their neuronal progeny.
Figure 2.
Overexpression of DYRK1A and DSCR1 in progenitors causes alteration of their laminar fate. (A–C) Plasmids encoding DYRK1A and DSCR1 were electroporated in E11 embryos, together with the GFP-expressing plasmid. The E16 brain sections were then immunostained with antibodies against Cux1 (A) and Tbr1 (B). Images of the entire cerebral wall are shown. The plasmids were electroporated, and their concentrations are indicated above the images. Bar, 50 μm. (C) Quantification of a fraction of GFP-positive cells that was also positive to Cux1 and Tbr1. Data are presented as mean ± SEM (n = 3 embryos for each group). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 versus control by a two-tailed Student's t-test. The concentration of electroporated plasmid for DYRK1A and DSCR1 (in micrograms per microliter) is presented in brackets.
Overexpression of DYRK1A and DSCR1 inhibits NFATc activity to delay neuronal differentiation in progenitor cells
NFATc is a transcription factor regulated by its phosphorylation/dephosphorylation (Crabtree 1999). It is known that the phosphorylated form of NFATc, which is retained in the cytoplasm, enters the nucleus through its dephosphorylation by Ca2+/calcineurin and activates the transcription (Crabtree 1999). In this activation process, DSCR1 inhibits calcineurin and thereby suppresses dephosphorylation and nuclear entry of NFATc (Rothermel et al. 2000). On the other hand, nuclear export of NFATc is accelerated upon its priming phosphorylation of the serine/proline repeats by DYRK1A (Arron et al. 2006; Gwack et al. 2006) and subsequent phosphorylation by GSK3 (Graef et al. 1999). Of note, in HEK cells, overexpression of DYRK1A and DSCR1 synergistically prevents NFATc-dependent transcription (Arron et al. 2006). Four members of NFATc (NFATc1–4) have been identified in mammals (Müller and Rao 2010), and multiple NFATc family members were found to be expressed in the VZ in the developing neocortex (Supplemental Fig. S4). This raised the possibility that NFATc is a potential candidate mediating the effects of DYRK1A and DSCR1 overexpression in cortical progenitors. We then examined the potential effect of DYRK1A and DSCR1 overexpression on subcellular distribution of NFATc in cortical progenitor cells. For this purpose, the plasmids encoding GFP-NFATc4, DYRK1A, and/or DSCR1 were electroporated into the VZ of E14 brains, and the neocortical cell culture prepared from the E15 brains was immunostained at DIV2 to examine the subcellular distribution of GFP-NFATc4. We found that the nuclear occupancy of GFP-NFATc4 was decreased when both DYRK1A and DSCR1 were introduced, whereas expression of either of these genes had no significant effect on GFP-NFATc4 distribution (Fig. 3A–C). When cortical progenitor cells were treated with ionomycin, which increases the intracellular Ca2+ level, nuclear occupancy of NFATc4 was strongly enhanced (Fig. 3D). Even in this condition, introduction of both DYRK1A and DSCR1 diminished nuclear localization of GFP-NFATc4 (Fig. 3D,E). These results indicate that overexpression of DYRK1A and DSCR1 cooperatively reduces nuclear NFATc in progenitors.
Figure 3.
Overexpression of DYRK1A and DSCR1 affects nuclear occupancy of NFATc4 in cortical progenitor cells. (A) Plasmids expressing the indicated proteins were electroporated, together with the GFP-NFATc4-expressing plasmid, in E14 embryos. Neocortical cell cultures were then prepared from the E15 brains. Neocortical cells were fixed at DIV2 and immunostained with antibodies against GFP (green) and Sox2 (blue). Representative images are shown. Bar, 20 μm. (B) Magnified views of the cells indicated by arrowheads a′–f′ in A are shown. Bar, 10 μm. (C) GFP fluorescence intensity of the nucleus and cytoplasm in individual GFP-labeled cells was measured, and the ratio of the intensity values is plotted. (***) P < 0.001 versus control by a two-tailed Welch's t-test. (D,E) An experiment similar to the one in A was performed in the presence of ionomycin. At DIV2, neocortical cells were treated with ionomycin (final concentration, 2 μM) for 1 h. Representative images are shown in D. Bar, 20 μm. (E) GFP fluorescence intensity of the nucleus and cytoplasm in individual GFP-labeled cells was measured, and the ratio of the intensity values is plotted. (***) P < 0.001 versus control by a two-tailed Welch's t-test.
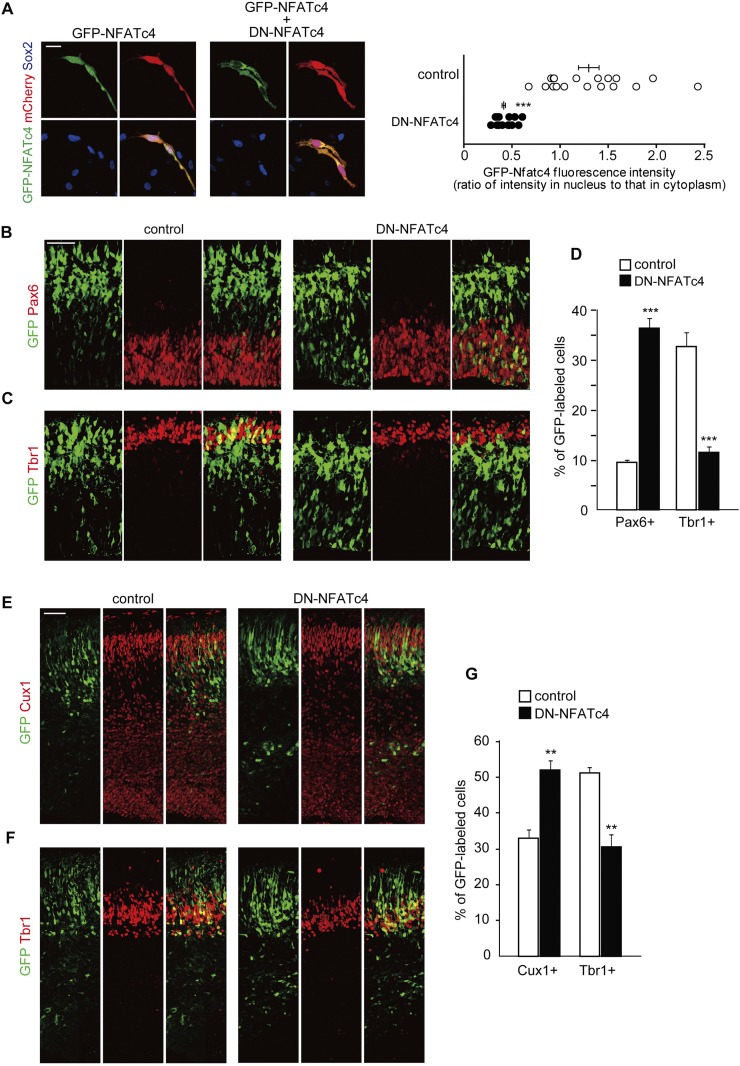
We then asked whether inhibition of NFATc activity in neural progenitors might result in delayed neuronal differentiation, like overexpression of DYRK1A and DSCR1. As multiple NFATc family members are expressed in cortical progenitors, we used a dominant-negative form of NFATc (DN-NFATc4) that blocks nuclear translocation of NFATc members by interfering their interaction with calcineurin (Chow et al. 1999; van Rooij et al. 2002). In fact, when DN-NFATc4 and GFP-tagged wild-type NFATc4 were cointroduced in cortical progenitors, nuclear entry of wild-type NFATc was prevented even when the progenitor cells were treated with ionomycin (Fig. 4A). We then introduced DN-NFATc4 into the VZ of E11 embryos and harvested the embryos at E13 and E16. In the DN-NFATc4-introduced E13 neocortices, a significantly larger fraction of GFP-labeled cells was positive for Pax6 (9.6% ± 0.5% vs. 36.4% ± 2.0% in control and DN-NFATc4, respectively; n = 3–5 embryos; P < 0.001 by a Student's t-test), whereas a smaller fraction of the cells was Tbr1-positive (32.7% ± 2.9% vs. 11.4% ± 1.2% in control and DN-NFATc4, respectively; n = 3–4 embryos; P < 0.001 by a Student's t-test) (Fig. 4B–D). In addition, we did not observe any increase in cleaved caspase 3-positive cells following DN-NFATc4 electroporation, excluding apoptosis as an explanation for the observed phenotypes (Supplemental Fig. S2C). Furthermore, in the E16 cortices, a fraction of GFP-labeled cells that were also positive for Cux1 was significantly increased following DN-NFATc4 electroporation (33.1% ± 2.3% vs. 52.0% ± 2.8% in control and DN-NFATc4; n = 3–4 embryos; P < 0.01 by a Student's t-test) (Fig. 4E,G). On the other hand, a Tbr1-positive fraction was significantly decreased (51.2% ± 1.7% vs. 30.4% ± 3.6% in control and DN-NFATc4, respectively; n = 3 embryos; P < 0.01 by a Student's t-test) (Fig. 4F,G). These observations indicate that blockage of NFATc causes a delay in neuronal differentiation of progenitors, like DYRK1A and DSCR1 overexpression.
Figure 4.
Interfering with NFATc activity impairs normal neuronal differentiation. (A) Plasmids expressing DN-NFATc4, together with plasmids encoding GFP-NFATc4 and mCherry, were electroporated in E14 embryos. Neocortical cell cultures were then prepared from the E15 brains. At DIV2, neocortical cells were treated with ionomycin (final concentration, 2 μM) for 1 h, fixed, and immunostained with antibodies against GFP (green) and Sox2 (blue). Representative images are shown. Bar, 20 μm. GFP fluorescence intensity of the nucleus and cytoplasm in individual GFP-labeled cells was measured, and the ratio of the intensity values is plotted on the right. (***) P < 0.001 versus control by a two-tailed Welch's t-test. (B–G) A plasmid expressing either control or DN-NFATc4 was electroporated, together with the GFP-expression plasmid, in E11 embryos, and the E13 (B–D) and E16 (E–G) brain sections were immunostained with the various antibodies indicated. (B,C) E13 brain sections immunostained with antibodies against Pax6 (B) and Tbr1 (C). Images of the entire cerebral wall electroporated with control (left panels) or DN-NFATc4 (right panels) are shown. Bar, 50 μm. (D) Quantification of a fraction of GFP-positive cells that was also positive for Pax6 and Tbr1. (***) P < 0.001 versus control by a two-tailed Student's t-test. (E,F) E16 brain sections immunostained with antibodies against Cux1 (E) and Tbr1 (F). Images of the entire cerebral wall electroporated with control (left panels) or DN-NFATc4 (right panels) are shown. Bar, 50 μm. (G) Quantification of a fraction of GFP-positive cells that was also positive for Cux1 and Tbr1. (**) P < 0.01 versus control by a two-tailed Student's t-test. In the graphs, data are presented as mean ± SEM (n = 3–5 embryos for each group).
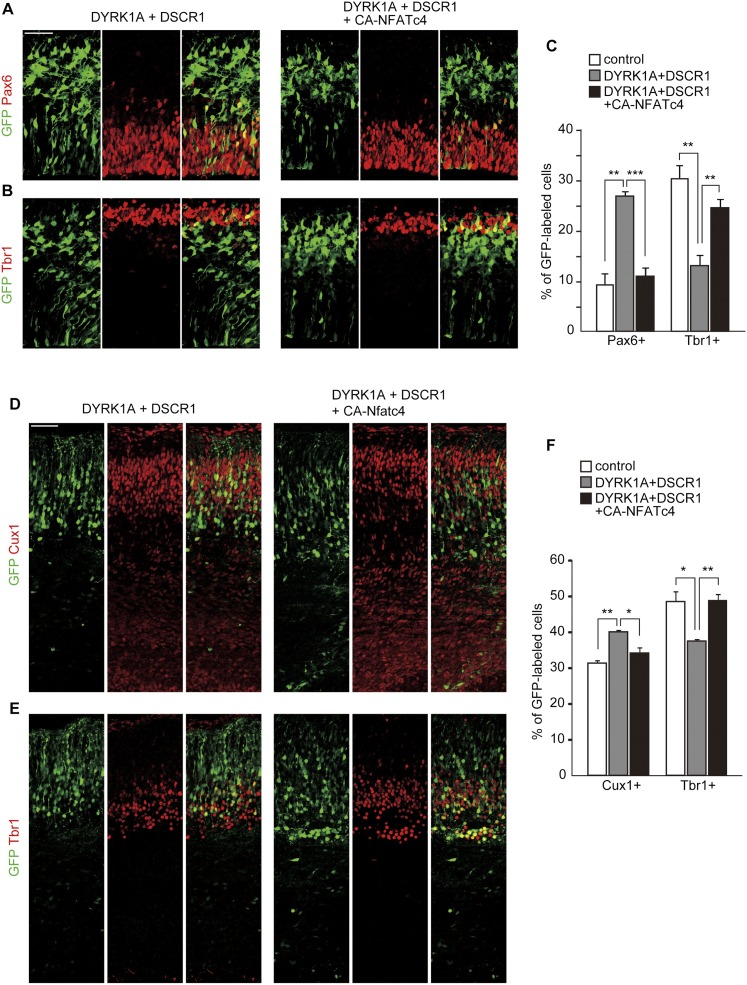
To further inspect whether delay in neuronal differentiation following DYRK1A and DSCR1 overexpression is caused by dysregulation of NFATc, we attempted to rescue the phenotype of DYRK1A and DSCR1 overexpression by gain of function of NFATc. For this purpose, we used a constitutively active form of NFATc4 (CA-NFATc4) that lacks most of its regulatory domain and no longer requires Ca2+/calcineurin to become active (Molkentin et al. 1998). When CA-NFATc4 was expressed in cortical progenitors, CA-NFATc4 was predominantly found in the nucleus even in the condition where DYRK1A and DSCR1 were overexpressed (Supplemental Fig. S5A). We then introduced CA-NFATc4 together with DYRK1A and DSCR1 into the VZ of E11 embryos and harvested the embryos at E13 or E16. In the E13 neocortices, coexpression of CA-NFATc4 almost completely reversed the abnormalities seen with the introduction of DYRK1A and DSCR1; the proportion of GFP-labeled cells positive for Pax6 was reduced, and instead the fraction of Tbr1-positive cells was increased to levels comparable with control (Fig. 5A–C). In addition, we did not observe any increase in cleaved caspase 3-positive cells following CA-NFATc4 overexpression (Supplemental Fig. S2D). Moreover, in the E16 cortices, we found that cointroduction of CA-NFATc4 almost completely reversed the abnormalities (i.e., increased the Cux1-positive fraction and decreased the Tbr1-positive fraction) seen with introduction of DYRK1A and DSCR1 (Fig. 5D–F). Of note, when the expression plasmid encoding CA-NFATc4 was solely electroporated into the VZ of E11 embryos, we observed no significant effect on fractions of GFP-labeled cells that were also positive for Pax6/Tbr1 at E13 and Cux1/Tbr1 at E16 (Supplemental Fig. S5B–G). Taken together, these results suggest that delayed neuronal differentiation induced by an increased dosage of DYRK1A and DSCR1 in progenitors is mediated by attenuation of NFATc.
Figure 5.
Delay in neuronal differentiation following DYRK1A and DSCR1 overexpression is rescued by coexpression of CA-NFATc4. (A–C) Plasmids expressing DYRK1A, DSCR1, and CA-NFATc4 were electroporated, together with the GFP-expression plasmid, in E11 embryos, and the E13 brain sections were immunostained with antibodies against Pax6 (A) and Tbr1 (B). Images of the entire cerebral wall are shown. Bar, 50 μm. Quantification of a fraction of GFP-positive cells that was also positive for Pax6 and Tbr1 is plotted in C. Similar quantification was performed in control plasmid-introduced brains (see Fig. 1), and the data are also presented in the graph. Data are presented as mean ± SEM (n = 3–5 embryos for each group). (D–F) Plasmids expressing DYRK1A, DSCR1, and CA-NFATc4 were electroporated, together with the GFP-expression plasmid, in E11 embryos, and the E16 brain sections were immunostained with antibodies against Cux1 (D) and Tbr1 (E). Images of the entire cerebral wall are shown. Bar, 50 μm. Quantification of a fraction of GFP-positive cells that was also positive for Cux1 and Tbr1 is plotted in F. Similar quantification was performed in control plasmid-introduced brains (see Fig. 2), and the data are also presented in the graph. Data are presented as mean ± SEM (n = 3–5 embryos for each group). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, by a two-tailed Student's t-test.
Modulation of dysregulated DYRK1A/DSCR1/NFATc ameliorates the delayed neuronal differentiation observed in a mouse model of DS
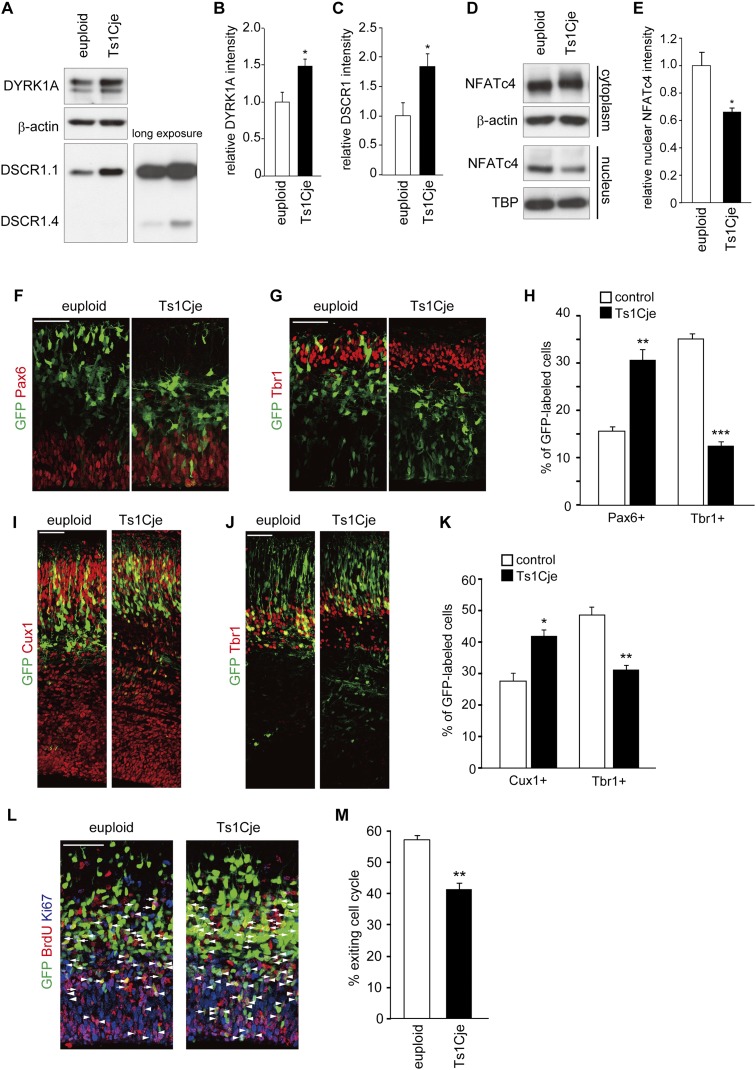
Ts1Cje mice, a widely used mouse model of DS, are trisomic for large segments of mouse chromosome 16, which is orthologous to human chromosome 21. The large segments contain DYRK1A and DSCR1 genes (Sago et al. 1998). As expected, the protein levels of DYRK1A and DSCR1 are increased to 1.49-fold ± 0.10-fold and 1.85-fold ± 0.21-fold, respectively, in the E13 neocortices of Ts1Cje embryos (Fig. 6A–C). We then sought to inspect the nuclear level of NFATc4 to examine potential dysregulation of the DYRK1A/DSCR1–NFATc pathway in Ts1Cje mice. The nuclear level of NFATc4 was decreased to 0.66-fold ± 0.03-fold, and the phosphorylation level of cytoplasmic NFATc4 was increased (Fig. 6D,E). These results underscore significant attenuation of the NFATc pathway in the developing neocortices of Ts1Cje mice.
Figure 6.
Alteration of DYRK1A/DSCR1 levels, NFATc activity, and neuronal differentiation in neocortical progenitors in Ts1Cje mice. (A–E) Whole-cell lysates (A), cytoplasmic fractions, and nuclear fractions (D) of the E13 neocortex of Ts1Cje mice and euploid littermates were subjected to immunoblotting with the antibodies indicated. (TBP) TATA-binding protein. The quantified band intensity was plotted in B, C, and E (mean ± SEM; n = 4 for euploid; n = 3 for Ts1Cje). The mean values of the band intensities of euploid mice were set to 1. (F–K) The plasmids expressing GFP were introduced into E11 embryos of Ts1Cje and euploid littermates, and the E13 (F–H) and E16 (I–K) brain sections were immunostained with the various antibodies indicated. (F,G) E13 brain sections immunostained with antibodies against Pax6 (F) and Tbr1 (G). Images of the entire cerebral wall are shown. Bars, 50 μm. (H) Quantification of a fraction of GFP-positive cells that was also positive for Pax6 and Tbr1. Data are presented as mean ± SEM (n = 3 embryos for each group). (**) P < 0.01; (***) P < 0.001 versus control by a two-tailed Student's t-test. (I,J) E16 brain sections immunostained with antibodies against Cux1 (I) and Tbr1 (J). Images of the entire cerebral wall are shown. Bars, 50 μm. (K) Quantification of a fraction of GFP-positive cells that was also positive for Cux1 and Tbr1. Data are presented as mean ± SEM (n = 3 embryos for each group). (*) P < 0.05; (**) P < 0.01 versus control by a two-tailed Student's t-test. (L) The plasmids expressing GFP were introduced into E13 embryos of Ts1Cje and euploid littermates. BrdU was administrated at E14, and the brains were fixed 24 h after the BrdU administration. Brain sections were then stained with antibodies against BrdU, Ki67, and GFP. GFP-positive cells labeled with both BrdU and Ki67 (arrowheads) are progenitor cells remaining in the cell cycle. GFP-positive cells labeled with BrdU but not Ki67 (arrows) are cells that exited the cell cycle. Bar, 50 μm. (M) Cell cycle exit index. Data are presented as mean ± SEM (n = 3 embryos for each group). (**) P < 0.01 versus euploid by a two-tailed Student's t-test.
It has been demonstrated that neuronal differentiation is delayed in the neocortices of Ts1Cje mice (Ishihara et al. 2010). We therefore inspected delayed neuronal differentiation in the mice by several methods. First, we electroporated plasmids encoding GFP into the VZ of Ts1Cje brains at E11 and harvested the embryos at E13 and E16. In E13 neocortices of Ts1Cje mice, a significantly larger fraction of GFP-labeled cells was positive for Pax6 when compared with euploid, whereas a smaller fraction of the cells was Tbr1-positive (Fig. 6F–H). On the other hand, in E16 cortices, a significantly larger fraction of GFP-labeled cells was positive for Cux1 when compared with euploid (Fig. 6I,K). In addition, the fraction of Tbr1-positive cells significantly decreased (Fig. 6J,K). These results suggest a delay in neuronal differentiation of progenitors in the Ts1Cje neocortices. We also inspected cell cycle exit of progenitors. To this end, embryos were electroporated with the GFP construct at E13, pulse-labeled with BrdU at E14, and harvested at E15 followed by immunostaining with antibodies to GFP, BrdU, and Ki67 (a marker for proliferating cells). We estimated the cell cycle exit index of GFP-labeled cells as the ratio of GFP+/BrdU+/Ki67− cells to GFP+/BrdU+ cells (Chenn and Walsh 2002). Consistent with the previous study (Ishihara et al. 2010), we observed a significant decrease in the cell cycle exit index (population of GFP-labeled cells exiting the cell cycle) in the Ts1Cje neocortices (Fig. 6L,M). Thus, neural progenitors in the Ts1Cje neocortices show delayed cell cycle exit and thereby neuronal differentiation, confirming the previous study (Ishihara et al. 2010).
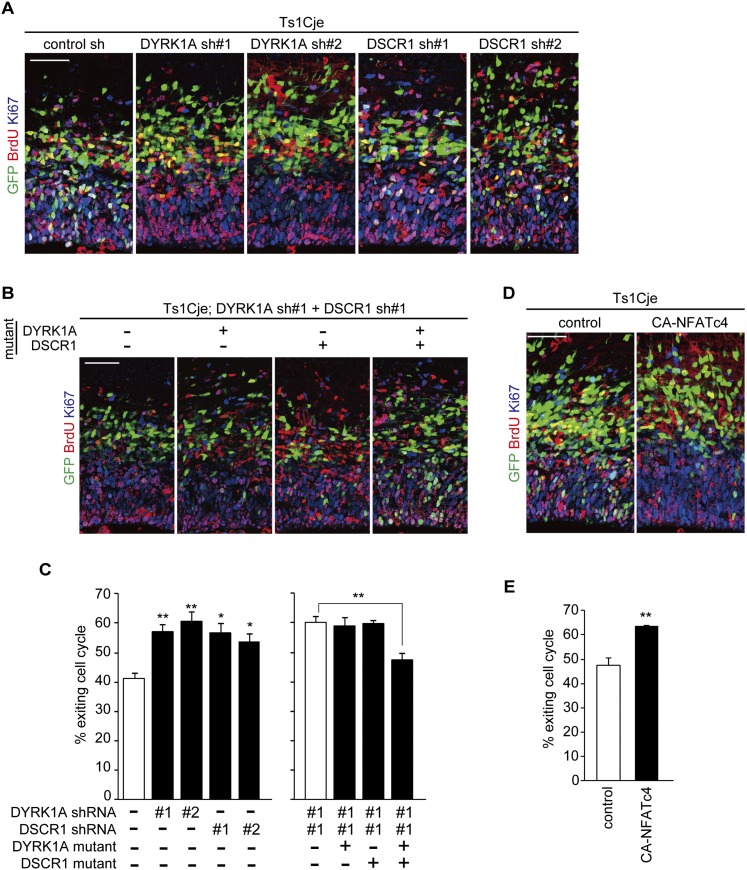
We then sought to determine whether the deficit in neuronal differentiation is due to dysregulation of the DYRK1A/DSCR1–NFATc pathway. To test this, we examined the effect of reduction of the DYRK1A and DSCR1 expression on cell cycle exit of progenitors in Ts1Cje brains. For this purpose, DNA-based RNAi plasmids that express shRNAs against DYRK1A and DSCR1 were generated. We identified two different shRNA constructs capable of silencing the expression of DYRK1A and DSCR1 that was transiently expressed in HEK293T cells (Supplemental Fig. S6A,B). Also, these shRNA constructs silenced endogenous proteins in cultured neural progenitor cells (Supplemental Fig. S6C,D). When the shRNA construct of either DYRK1A or DSCR1 was introduced into the Ts1Cje brains, the fraction of GFP-positive cells that exited the cell cycle was increased to levels comparable with that of control GFP-labeled cells in euploid mice (Fig. 7A,C). In addition, coelectroporation of the two shRNA constructs showed no additive effect (Fig. 7B,C). On the other hand, introduction of the DYRK1A shRNA-resistant mutant or the DSCR1 shRNA-resistant mutant (Supplemental Fig. S6E,F) did not reverse the increased cell cycle exit index induced by depletion of both DYRK1A and DSCR1, whereas introduction of both the DYRK1A and DSCR1 mutants did rescue the phenotype (Fig. 7B,C). Together, these data suggest that increased dosage of both DYRK1A and DSCR1 contributes to a neuronal differentiation deficit in Ts1Cje mice. Of note, the effect of depletion of DYRK1A and DSCR1 was specific to the Ts1Cje brains, as neither DYRK1A shRNA nor DSCR1 shRNA had a significant effect on the cell cycle exit index in the brains of euploid littermates (Supplemental Fig. S7A,C).
Figure 7.
Modulation of DYRK1A/DSCR1 and NFATc ameliorates reduced cell cycle exit in progenitors in the Ts1Cje neocortex. (A,B) E13 Ts1Cje embryos were electroporated with plasmids encoding GFP, DYRK1A shRNA, and DSCR1 shRNA with or without plasmids expressing shRNA-resistant DYRK1A and DSCR1 mutants, as indicated. BrdU was administrated at E14, and brains were fixed 24 h after the BrdU administration. Brain sections were then stained with antibodies against BrdU, Ki67, and GFP. Bar, 50 μm. (C) Cell cycle exit index. Data are presented as mean ± SEM (n = 3–5 embryos for each group). (D) E13 Ts1Cje embryos were electroporated with plasmids encoding GFP and CA-NFATc4. BrdU was administrated at E14, and the brains were fixed 24 h after the BrdU administration. Brain sections were then stained with antibodies against BrdU, Ki67, and GFP. Bar, 50 μm. (E) Cell cycle exit index. Data are presented as mean ± SEM (n = 3–4 embryos for each group). (*) P < 0.05; (**) P < 0.01, by a two-tailed Student's t-test.
Finally, we attempted to rescue the neuronal differentiation deficit by expressing CA-NFATc. We found that a decrease in the fraction of cells exiting the cell cycle in the Ts1Cje neocortices was reversed when CA-NFATc4 was electroporated (Fig. 7D,E). Furthermore, when the expression plasmid encoding CA-NFATc4 was electroporated into euploid brains, no significant effect on fractions of GFP-labeled cells exiting the cell cycle was observed (Supplemental Fig. S7B,D). Altogether, dysregulation of the NFATc pathway by an increased dosage of both DYRK1A and DSCR1 is a key mechanism underlying delayed neuronal differentiation in Ts1Cje mice.
Discussion
DYRK1A and DSCR1 lie in the human chromosome 21 and are candidate genes for trisomy 21, DS. In this study, we examined the effect of DYRK1A and DSCR1 overexpression on neurogenesis. We found that modest (∼1.5-fold to 2.0-fold) overexpression of DYRK1A and DSCR1 in cortical progenitors caused a delay in neuronal differentiation of progenitors, resulting in alteration of their laminar fate (i.e., reduced proportion of early-born neurons and increased proportion of later-born neurons generated). Importantly, these abnormalities are exacerbated by increasing amounts of DYRK1A and DSCR1 in progenitors. Furthermore, we linked increased dosage of DYRK1A and DSCR1 to reduced nuclear levels of transcription factor NFATc. It is known that nuclear export of the NFATc family of transcription factors is accelerated upon its phosphorylation by DYRK1A (Arron et al. 2006; Gwack et al. 2006) and that their nuclear import is suppressed by DSCR1 through deactivation of calcineurin (Ryeom et al. 2008), an important regulator for nuclear entry of NFATc. Consistently, we found that overexpression of DYRK1A and DSCR1 diminished nuclear occupancy of NFATc in progenitors. Furthermore, decreased neuronal differentiation and thus reduced early-born neurons upon DYRK1A and DSCR1 overexpression were phenocopied by inhibition of NFATc activity. Importantly, the abnormalities induced by DYRK1A and DSCR1 overexpression are almost completely reversed by expression of the constitutive active form of NFATc. Taken together, the cooperative actions of DYRK1A and DSCR1 are dose-sensitive, and proper maintenance of the activity of the DYRK1A/DSCR1-regulated NFATc pathway in progenitors is critical for normal neuronal differentiation.
Mouse models of DS have been generated recently by duplications of parts of mouse chromosomes that are orthologous to human chromosome 21. These mice display variable abnormalities seen in DS. Studies on neocortical development in Ts1Cje and Ts65Dn mouse models, which are trisomic for large segments (including Dyrk1a and Dscr1 genes) of mouse chromosome 16, revealed multiple defects in neural progenitors, such as delayed neuronal differentiation, as evidenced by a decreased cell cycle exit index, and fewer early-born neurons generated (Chakrabarti et al. 2007; Ishihara et al. 2010; this study). The present study linked the neurogenic defects observed in the DS mouse model to dysregulation of DYRK1A/DSCR1 and NFATc. We found increased levels of DYRK1A/DSCR1 and attenuation of NFATc activity in the developing neocortices in Ts1Cje mice. Furthermore, the reduced cell cycle exit of progenitor cells in Ts1Cje mice was reversed to a level comparable with euploid littermates upon depletion of DYRK1A/DSCR1 and activation of NFATc. Thus, attenuation of NFATc by increased dosage of DYRK1A and DSCR1 is likely a key mechanism responsible for delayed neuronal differentiation in cortical progenitors in the DS mouse model. During development of the mammalian neocortex, the neurogenic phase is followed by the gliogenic phase, during which progenitors transform into astrocytes. In this context, delayed neuronal differentiation by overexpression of DYRK1A and DSCR1 may postpone the onset of neuronal production, and, if glial differentiation takes place in a timely manner, this may shorten the neurogenic phase and lead to reduced neuronal production. Together, the present study uncovers a potential mechanism underlying neurodevelopmental defects in the DS brain: delayed neuronal differentiation and reduced production of neurons by a pathway that is mediated by dysregulation of NFATc upon increased dosage of two genes on human chromosome 21.
Besides decreased neuronal differentiation in Ts65Dn and Ts1Cje mice, lengthening of the cell cycle, a lower BrdU-labeling index, and the smaller progenitor pool were observed (Chakrabarti et al. 2007; Ishihara et al. 2010). These observations together imply that retardation of cell cycle progression and decreased cell cycle exit take place simultaneously in these model mice. In the present study, we found that a BrdU-positive fraction of GFP-labeled, Sox2-positive progenitor cells was not significantly changed by DYRK1A and DSCR1 overexpression (50.0% ± 4.6% vs. 47.1% ± 3.1% in control and 0.5 μg/μL DYRK1A/DSCR1, respectively; n = 3–4 embryos; P = 0.58 by a Student's t-test) (see Fig. 1F), suggesting that cell cycle progression of progenitors was not significantly affected by DYRK1A and DSCR1 overexpression. In this regard, it is likely that a modest increase in DYRK1A and DSCR1 affects progenitor differentiation, but not cell cycle progression of progenitors, and that a defect in progenitor proliferation in DS model mice may result from triplication of genes other than DYRK1A and DSCR1. In conjunction with delayed neuronal differentiation exerted by increased dosage of DYRK1A and DSCR1, impaired proliferation (Chakrabarti et al. 2007; Ishihara et al. 2010) as well as enhanced glial differentiation (Moldrich et al. 2009; Lu et al. 2011) may also influence the total number of neurons and neuronal density by mechanisms that remain to be explored.
Of note, a previous study has demonstrated that, in contrast to our study, DYRK1A electroporation in the mouse neocortex suppressed progenitor proliferation via misregulation of cell cycle regulators and leads to precocious neuronal differentiation that is not seen in DS model mice (Yabut et al. 2010). Since expression levels of DYRK1A were not examined in progenitors in Yabut et al. (2010), we presumed that a much higher dosage of DYRK1A might be expressed in their study, thereby causing a different phenotype. To test this idea, we electroporated a higher concentration (5 μg/μL) of the DYRK1A-expressing plasmids in progenitors. As evidenced by a decreased BrdU-positive fraction and increased Tbr2-positive fraction, we found precocious differentiation of progenitors (Supplemental Fig. S8). Our findings are in agreement with the study by Yabut et al. (2010) and imply that levels of DYRK1A dosage influence downstream pathways differently.
Considering the present results, it is assumed that in certain DS phenotypes, increased dosage of single genes on human chromosome 21 is not sufficient for DS pathogenesis and that cooperative actions of multiple genes are required. Further analyses of cross-talk of molecular pathways elicited by increased dosage of genes in human chromosome 21 may provide novel insights into the molecular/cellular basis underlying DS.
Materials and methods
Animals
The Ts1Cje mice were a kind gift from Dr. Kazuhiro Yamakawa (RIKEN, Brain Science Institute). The Ts1Cje mice were maintained by crossing carrier males with C57BL/6J females. A genotyping of a Ts1Cje mouse was performed by PCR as previously described (Amano et al. 2004). All mice were housed under a 12-h light–12-h dark cycle and ad libitum access to food and water. All animal experiments were conducted in accordance with guidelines set by The University of Tokyo and approved (permit no. 21-01) by the Committee on Animal Care and Use of the Graduate School of Science at The University of Tokyo.
Plasmids
The pCAGIG plasmid (expressing GFP under the control of the CAG promoter) and pBS/U6 plasmid are kind gifts from Dr. Takahiko Matsuda (Kyoto University) and Dr. Yang Shi (Harvard Medical School), respectively. For generating a plasmid expressing mCherry under the control of the CAG promoter, the full-length ORF of mCherry was subcloned into a pCAGEN plasmid (a kind gift from Dr. Takahiko Matsuda). The full-length ORF of mouse Dyrk1a, Dscr1 isoform 1 (Dscr1.1), and Nfatc4 was amplified by PCR with Pfu Turbo polymerase (Stratagene) from cDNA derived from E13 mouse cortices and subcloned into the pCAGEN plasmid. The primers used for these were as follows: for Dyrk1a, 5′-CGAATTCGCCACCATGCATACAGGAGGAGAGAC-3′ (forward) and 5′-AAGCTTGCGGCCGCTCAATGTAGTCACGAGCTAG-3′ (reverse); for Dscr1, 5′-AGAATTCGCCACCATGGAGGACGGCGTGGCCGGGC-3′ (forward) and 5′-AAGCTTGCGGCCGCTCAGCTGAGGTGGATGGGTGTG-3′ (reverse); and for NFATc4, 5′-GAATTCGCCACCATGGGGGCCGCAAGCTGCG-3′ (forward) and 5′-AAGCTTGCGGCCGCTCAGGCAGGAGGCTCTTCTC-3′ (reverse). Plasmids encoding silent mutants of DYRK1A and DSCR1 were generated by the QuickChange mutagenesis technique. Plasmids encoding a constitutively active form and a dominant-negative form of NFATc4 were PCR-generated using the NFATc4/pCAGEN vector as a template and then subcloned into the pCAGEN plasmid. The constitutively active form and a dominant-negative form of NFATc4 encoded NFATc4 amino acids 317–901 and 1–191, respectively. For a GFP-NFATc4-expressing plasmid, an oligonucleotide encoding the GFP sequence was fused to the 5′ end of the full-length NFATc4 coding sequence and subcloned into the pCAGEN plasmid. Plasmids encoding DYRK1A and DSCR1 shRNA were generated by inserting the annealed oligonucleotides into a pBS/U6 plasmid (Sui et al. 2002) as described (Asada et al. 2007). The targets sequenced for RNAi were as follows: for DYRK1A shRNA #1, 5′-GAACUUAGUAUCAUUCACUGU-3′; for DYRK1A shRNA #2, 5′-GAGCUAUGGACGUUAAUUUGA-3′; for DSCR1 shRNA #1, 5′-GAAGAGAUGGAGAGAAUGAAG-3′; and for DSCR1 shRNA #2, 5′-GGGCCAGGAGAGAAGUAUGAA-3′.
Antibodies
The following antibodies were used for immunostaining: rabbit anti-DYRK1A (1:100; Santa Cruz Biotechnology), rabbit anti-DSCR1 (1:100; Sigma-Aldrich), rat anti-GFP (1:2000; Nakalai Tesque), rabbit anti-GFP (1:1000; MBL), rat anti-BrdU (1:500; Funakoshi), mouse anti-BrdU (1:500; Dako), goat anti-Sox2 (1:200; Santa Cruz Biotechnology), mouse anti-Neurogenin2 (1:10; a kind gift from Dr. David J. Anderson at the California Institute of Technology), rabbit anti-PH3 (1:1000; Upstate Biotechnology), rabbit anti-Pax6 (1:1000; Covance), rabbit anti-Tbr2 (1:1000; Chemicon), rabbit anti-Tbr1 (1:1000; Abcam), rabbit anti-Cux1 (1:200; Santa Cruz Biotechnology), and rabbit anti-Ki67 (1:250; Novocastra Laboratories).
The following antibodies were used for immunoblotting: rabbit anti-GFP (1:1000; MBL), rabbit anti-DYRK1A (1:2000; Santa Cruz Biotechnology), rabbit anti-DSCR1(1:5000; Sigma-Aldrich), rabbit anti-NFATc4 (1:1000, Santa Cruz Biotechnology), rabbit anti-TBP (1:2000; Santa Cruz Biotechnology), mouse anti-β-actin (1:20,000; Sigma-Aldrich), and mouse anti-dynein intermediate chain (1:50,000; Santa Cruz Biotechnology).
In situ hybridization
The full-length ORFs of Dyrk1a and Dscr1 were subcloned into the pBluescriptII KS− vector. Also, cDNA fragments of Nfatc1 (139–1140; PubMed NM_016791), Nfatc2 (1186–1948; PubMed NM_010899), Nfatc3 (289–1288; PubMed NM_010901), and Nfatc4 (243–1242; PubMed NM_023699) were subcloned into the pBluescriptII KS− vector. The cDNA-containing vectors were used as templates for sense or antisense cRNA probes, and in situ hybridization was carried out as described (Asada et al. 2007).
In utero electroporation
DNA solution in PBS containing 0.01% Fast Green was injected into the lateral ventricles of mouse embryos. Thereafter, electroporation (five 50-msec square pulse with 950-msec intervals; Nepa Gene, CUY21-EDIT) was carried out with forceps-type electrodes (Nepa Gene, CUY650P3). The electroporation voltage used was 42 V. Final concentrations of the plasmids used were as follows: 5 μg/μL plasmids expressing GFP (pCAGIG), DYRK1A (indicated in Figs. 1 and 2 and Supplemental Figs. S2, S3, and S8), DSCR1 (indicated in Figs. 1 and 2 and Supplemental Figs. S2 and S3), 1 μg/μL DYRK1A with silent mutation, 1 μg/μL DSCR1 with silent mutation, 3 μg/μL DN-NFATc4, 0.2 μg/μL CA-NFATc4, 2 μg/μL DYRK1A shRNA, and 2 μg/μL DSCR1 shRNA. For BrdU-labeling experiments, pregnant dams were injected with BrdU (50 μg per gram of body weight at E12 and 100 μg per gram of body weight at E14).
Immunohistochemistry
Brains were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and cryoprotected in 30% sucrose in PBS overnight at 4°C. Thereafter, the brains were embedded in a solution of a 2:1 mixture of 30% sucrose/PBS and OCT compound (Sakura), frozen by liquid nitrogen, and stored at −80°C until use. Thick cryosections (20 μm) were made. Brain sections were washed with PBS, incubated with blocking solution (3% [w/v] BSA, 5% [v/v] FBS, 0.2% [w/v] Triton X-100 in PBS), and then incubated with primary antibodies overnight at 4°C. The sections were then incubated with Alexa 488/Cy3/Cy5-conjugated secondary antibodies overnight at 4°C and mounted in a Prolong Gold mounting solution (Invitrogen). For staining with Cux1, brain sections were pretreated with HistoVT One solution (Nakalai Tesque) for 15 min at 70°C. For staining with BrdU, brain sections were pretreated with 4 N HCl for 25 min at room temperature. Images were obtained with a 63× objective (Plan-Apochromat, Zeiss) on a Zeiss LSM5 confocal microscope.
Cell culture and transfection
For evaluation of DYRK1A and DSCR1 knockdown by shRNA constructs, HEK293T cells maintained in 10% FBS/DMEM were transiently transfected using Lipofectamine 2000 (Invitrogen). Transfections were allowed to proceed for 4–5 h, and then cells were cultured in 10% FBS/DMEM for 48 h. The cells were then subjected to immunoblotting. For neocortical cell culture, E14 embryos were electroporated with plasmids encoding GFP, DYRK1A, DSCR1, GFP-NFATc4, DN-NFATc4, CA-NFATc4, and shRNAs for DYRK1A and DSCR1. Twenty-four hours after electroporation, neocortical cells were prepared and cultured as described earlier (Sanada and Tsai 2005). Neocortical cells were fixed at DIV1 or DIV2 with 4% paraformaldehyde in PBS for 30 min at 37°C, permeabilized with 0.5% Triton X-100 in PBS for 5 min, blocked with 3% BSA/0.2% Triton X-100 in PBS, and incubated with primary antibodies in the blocking solution overnight at 4°C. The coverslips were then incubated with Alexa 488/Cy3/Cy5-conjugated secondary antibodies for 2 h at room temperature and mounted in a Prolong Gold mounting solution (Invitrogen). Fluorescent images were obtained by using Zeiss LSM5 confocal microscope.
Preparation of nuclear and cytoplasmic fraction
E13 mouse neocortices were homogenized with 9 vol (volume/wet weight of tissue) of buffer A (10 mM HEPES-NaOH, 1 mM DTT, 1 mM EDTA, 50 mM NaF, 1 mM Na3VO4, protease inhibitor cocktail [Complete, EDTA-free; Roche Molecular Biochemicals] at pH 7.8 at 4°C) using a Dounce homogenizer. The homogenate was centrifuged at 700g for 5 min at 4°C, and the unsedimented material was collected as a cytoplasmic fraction. The resulting pellet (the source of the nuclear fraction) was washed with 9 vol (volume/wet weight of tissue) of buffer A and suspended with 2 vol (volume/wet weight of tissue) of buffer B (20 mM HEPES-NaOH, 2% [v/v] glycerol, 400 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 mM EDTA, 50 mM NaF, 1 mM Na3VO4, protease inhibitor cocktail [Complete, EDTA-free; Roche Molecular Biochemicals] at pH 7.8 at 4°C). The suspended pellet was gently rotated for 30 min at 4°C and centrifuged at 21,600g for 30 min at 4°C, and the resultant supernatant was collected as a nuclear fraction. These cytoplasmic and nuclear fractions were then subjected to immunoblotting.
Statistical analysis
All bar graphs were plotted as mean ± SEM. Direct comparisons were made using a two-tailed Student's or Welch's t-test. The significance level was set at P < 0.05 for all tests.
Acknowledgments
We thank Dr. Minh Dang Nguyen for critical reading of the manuscript. We also thank Dr. Kazuhiro Yamakawa for Ts1Cje mice, Dr. Takahiko Matsuda and Dr. Yang Shi for plasmids, and Dr. David J. Anderson for antibody. This work was supported in part by a Grant-in-Aid for Young Scientists (B) (to N.K.), a Grant-in-Aid for Scientific Research (B) (to K.S.), and a Grant-in-Aid for Scientific Research on Innovative Areas “Neural Diversity and Neocortical Organization” (to K.S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226381.113.
References
- Amano K, Sago H, Uchikawa C, Suzuki T, Kotliarova SE, Nukina N, Epstein CJ, Yamakawa K 2004. Dosage-dependent over-expression of genes in the trisomic region of Ts1Cje mouse model for Down syndrome. Hum Mol Genet 13: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S 2004. Chromosome 21 and Down syndrome: From genomics to pathophysiology. Nat Rev Genet 5: 725–738 [DOI] [PubMed] [Google Scholar]
- Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, et al. 2006. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441: 595–600 [DOI] [PubMed] [Google Scholar]
- Asada N, Sanada K, Fukada Y 2007. LKB1 regulates neuronal migration and neuronal differentiation in the developing neocortex through centrosomal positioning. J Neurosci 27: 11769–11775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfone A, Smiga SM, Shimamura K, Peterson A, Puelles L, Rubenstein JL 1995. T-brain-1a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron 15: 63–78 [DOI] [PubMed] [Google Scholar]
- Canfield MA, Honein MA, Yuskivm N, Xing J, Mai CT, Collins JS, Devine O, Petrini J, Ramadhani TA, Hobbs CA, et al. 2006. National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999–2001. Birth Defects Res A Clin Mol Teratol 76: 747–756 [DOI] [PubMed] [Google Scholar]
- Chakrabarti L, Galdzicki Z, Haydar TF 2007. Defects in embryonic neurogenesis and initial synapse formation in the forebrain of the Ts65Dn mouse model of Down syndrome. J Neurosci 27: 11483–11495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR 2004. A field of myocardial–endocardial NFAT signaling underlies heart valve morphogenesis. Cell 118: 649–663 [DOI] [PubMed] [Google Scholar]
- Chapman RS, Hesketh LJ 2000. Behavioral phenotype of individuals with Down syndrome. Ment Retard Dev Disabil Res Rev 6: 84–95 [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA 2002. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297: 365–369 [DOI] [PubMed] [Google Scholar]
- Chow CW, Rincon M, Davis RJ 1999. Requirement for transcription factor NFAT in Interleukin-2 expression. Mol Cell Biol 19: 2300–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR 1999. Generic signals and specific outcomes: Signaling through Ca2+, calcineurin, and NF-AT. Cell 96: 611–614 [DOI] [PubMed] [Google Scholar]
- Delabar JM, Theophile D, Rahmani Z, Chettouh Z, Blouin JL, Prieur M, Noel B, Sinet PM 1993. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet 1: 114–124 [DOI] [PubMed] [Google Scholar]
- Dominguez MH, Ayoub AE, Rakic P 2013. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex 23: 2632–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotaki V, Dierssen M, Alcantara S, Martine S, Marti E, Casas C, Visa J, Soriano E, Estivill X, Arbonés ML 2002. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol 22: 6636–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes JJ, Genescà L, Kingsbury TJ, Cunningham KW, Pérez-Riba M, Estivill X, de la Luna S 2000. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin-mediated signaling pathways. Hum Mol Genet 9: 1681–1690 [DOI] [PubMed] [Google Scholar]
- Golden JA, Hyman B 1994. Development of the superior temporal neocortex is anomalous in trisomy 21. J Neuropathol Exp Neurol 53: 513–520 [DOI] [PubMed] [Google Scholar]
- Götz M, Stoykova A, Gruss P 1998. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21: 1031–1044 [DOI] [PubMed] [Google Scholar]
- Graef IA, Mermelstein PG, Stankunas K, Neilson JR, Deisseroth K, Tsien RW, Crabtree GR 1999. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature 401: 703–708 [DOI] [PubMed] [Google Scholar]
- Graef IA, Chen F, Chen L, Kuo A, Crabtree GR 2001. Signals transduced by Ca2+/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 105: 863–875 [DOI] [PubMed] [Google Scholar]
- Guedj F, Pereira PL, Najas S, Barallobre MJ, Chabert C, Souchet B, Sebrie C, Verney C, Herault Y, Arbones M, et al. 2012. DYRK1A: A master regulatory protein controlling brain growth. Neurobiol Dis 46: 190–203 [DOI] [PubMed] [Google Scholar]
- Guimera J, Casas C, Estivill X, Pritchard M 1999. Human minibrain homologue (MNBH/DYRK1): Characterization, alternative splicing, differential tissue expression, and overexpression in Down syndrome. Genomics 57: 407–418 [DOI] [PubMed] [Google Scholar]
- Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, Okamura H, Bolton D, Feske S, Hogan PG, et al. 2006. A genome-wide Drosophila RNAi screen identifies DYRK-family kinase as regulators of NFAT. Nature 441: 646–650 [DOI] [PubMed] [Google Scholar]
- Hämmerle B, Elizalde C, Tejedor FJ 2008. The spatio-temporal and subcellular expression of the candidate Down syndrome gene Mnb/Dyrk1A in the developing mouse brain suggests distinct sequential roles in neuronal development. Eur J Neurosci 27: 1061–1074 [DOI] [PubMed] [Google Scholar]
- Hasle H, Clemmensen IH, Mikkelsen M 2000. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet 355: 165–169 [DOI] [PubMed] [Google Scholar]
- Ishihara K, Amano K, Takaki E, Shimohata A, Sago H, Epstein CJ, Yamakawa K 2010. Enlarged brain ventricles and impaired neurogenesis in the Ts1Cje and Ts2Cje mouse models of Down syndrome. Cereb Cortex 20: 1131–1143 [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Kawashima H, Pulst SM, Ikeuchi T, Ogasawara N, Yamamoto K, Schonberg SA, West R, Allen L, Magenis E, et al. 1990. Molecular definition of a region of chromosome 21 that causes features of the Down syndrome phenotype. Am J Hum Genet 47: 236–246 [PMC free article] [PubMed] [Google Scholar]
- Lu J, Esposito G, Scuderi C, Steardo L, Delli-Bovi LC, Hecht JL, Dickinson BC, Chang CJ, Mori T, Sheen V 2011. S100B and APP promote a gliocentric shift and impaired neurogenesis in Down syndrome neural progenitors. PLoS ONE 6: e22126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DMA, Yates PO, Marcyniuk B, Ravindra CR 1987. Loss of neurons from cortical and subcortical areas in Down's syndrome patients at middle age: Quantitative comparisons with younger Down's patients and patients with Alzheimer's disease. J Neurol Sci 80: 79–89 [DOI] [PubMed] [Google Scholar]
- McCormick MK, Schinzel A, Petersen MB, Stetten G, Driscoll DJ, Cantu ES, Tranebjaerg L, Mikkelsen M, Watkins PC, Antonarakis SE 1989. Molecular genetic approach to the characterization of the ‘Down syndrome region’ of chromosome 21. Genomics 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Moldrich RX, Dauphinot L, Laffaire J, Vitalis T, Hérault Y, Beart PM, Rossier J, Vivien D, Gehrig C, Antonarakis SE, et al. 2009. Proliferation deficits and gene expression dysregulation in Down's syndrome (Ts1Cje) neural progenitor cells cultured from neurospheres. J Neurosci Res 87: 3143–3152 [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antosm CL, Markham B, Richardsonm J, Robbins J, Grant SR, Olson EN 1998. Calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller MR, Rao A 2010. NFAT, immunity and cancer: A transcription factor comes of age. Nat Rev Immunol 10: 645–656 [DOI] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Götz M, Walsh CA 2004. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol 479: 168–180 [DOI] [PubMed] [Google Scholar]
- Olson LE, Richtsmeier JT, Leszl J, Reeves RH 2004. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science 306: 687–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LE, Roper RJ, Sengstaken CL, Peterson EA, Aquino V, Galdzicki Z, Siarey R, Pletnikov M, Moran TH, Reeves RH 2007. Trisomy for the Down syndrome ‘critical region’ is necessary but not sufficient for brain phenotypes of trisomic mice. Hum Mol Genet 16: 774–782 [DOI] [PubMed] [Google Scholar]
- Petit TL, LeBoutillier JC, Alfano DP, Becker LE 1984. Synaptic development in the human fetus: A morphometric analysis of normal and Down's syndrome neocortex. Exp Neurol 83: 13–23 [DOI] [PubMed] [Google Scholar]
- Roizen NJ, Patterson D 2003. Down's syndrome. Lancet 361: 1281–1289 [DOI] [PubMed] [Google Scholar]
- Rothermel B, Vega RB, Yang J, Wu H, Bassel-Duby R, Williams RS 2000. A protein encoded within the Down syndrome critical region is enriched in striated muscles and inhibits calcineurin signaling. J Biol Chem 275: 8719–8725 [DOI] [PubMed] [Google Scholar]
- Ryeom S, Baek KH, Rioth MJ, Lynch RC, Zaslavsky A, Birsner A, Yoon SS, McKeon F 2008. Targeted deletion of the calcineurin inhibitor DSCR1 suppresses tumor growth. Cancer Cell 13: 420–431 [DOI] [PubMed] [Google Scholar]
- Sago H, Carlson EJ, Smith DJ, Kilbridge J, Rubin EM, Mobley WC, Epstein CJ, Huang TT 1998. Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc Natl Acad Sci 95: 6256–6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K, Tsai LH 2005. G protein βγ subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell 122: 119–131 [DOI] [PubMed] [Google Scholar]
- Schmidt-Sidor B, Wisniewski KE, Shepard TH, Sersen EA 1990. Brain growth in Down syndrome subjects 15 to 22 weeks of gestational age and birth to 60 months. Clin Neuropathol 9: 181–190 [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar EB, Gay F, Shi Y, Forrester WC, Shi Y 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci 99: 5515–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S, Becker LE, Armstrong DL, Chan F 1981. Abnormal neuronal development in the visual cortex of the human fetus and infant with Down's syndrome. A quantitative and qualitative golgi study. Brain Res 225: 1–21 [DOI] [PubMed] [Google Scholar]
- Tanzi RE 1996. Neuropathology in the Down's syndrome brain. Nat Med 2: 31–32 [DOI] [PubMed] [Google Scholar]
- Tuan N, Simone DG 2008. NFAT signaling in neural development and axon growth. Int J Dev Neurosci 26: 141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Doevendans PA, de Theije CC, Babiker FA, Molkentin JD, de Windt LJ 2002. Requirement of nuclear factor of activated T-cells in calcineurin-mediated cardiomyocyte hypertrophy. J Biol Chem 277: 48617–48626 [DOI] [PubMed] [Google Scholar]
- Weitzdoerfer R, Dierssen M, Fountoulakis M, Lubec G 2001. Fetal life in Down syndrome starts with normal neuronal density but impaired dendritic spines and synaptosomal structure. J Neural Transm Suppl 61: 59–70 [DOI] [PubMed] [Google Scholar]
- Yabut O, Domogauer J, D'Arcangelo G 2010. Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J Neurosci 30: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Yamaguchi Y, Nonomura K, Kawakami K, Takahashi Y, Miura M 2010. Simultaneous expression of different transgenes in neurons and glia by combining in utero electroporation with the Tol2 transposon-mediated gene transfer system. Genes Cells 15: 501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]