The evolution of bacteria started about 3.5 billion years ago, and since that time these microorganisms have conquered all available ecological niches. This was possible only because of the astonishing ability of bacteria to adapt to a multitude of environmental conditions and stress situations. This adaptation included development of highly specific cell envelope structures, synthesis of metabolites which could be released into the cells' surroundings, and other strategies to secure the survival of the given species in a hostile, competitive environment. Among other macromolecular features, the ability to glycosylate proteins developed during bacterial evolution. Indeed, the assumption described until recently in many textbooks that bacteria do not contain or synthesize glycoproteins is misleading and has hampered prokaryotic glycoprotein research for decades. While research on glycoproteins of higher organisms has flourished since Neuberger's glycopeptide preparations of ovalbumin in the late 1930s, the question of whether carbohydrates are integral components of prokaryotic proteins was not convincingly approached until the work of Strominger and coworkers on surface layer (S-layer) glycoproteins of the gram-negative halophile Halobacterium salinarium (11). With the affiliation of halobacteria with the domain Archaea, however, another misleading assumption took hold; it was assumed that glycoprotein formation is limited to archaea, although at about the same time glycosylated S-layer proteins of several clostridia were described (for details see references 12, 15, and 18a). The fact that in the prokaryotes that are most frequently studied, such as Escherichia coli, Salmonella sp., and Bacillus subtilis, no glycosylated proteins had been identified contributed to the lack of awareness of the presence of glycoproteins in organisms in the domain Bacteria. Workers have defined principles of prokaryotic glycoprotein structure that are valid not only for S-layer glycoproteins but presumably also for any other type of prokaryotic glycoprotein (Fig. 1) (for details see references 5, 11, 12, and 20).
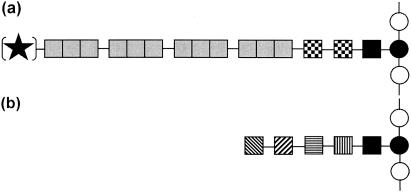
FIG. 1.
Schematic representation of prokaryotic glycoproteins. (a) Typical long-chain S-layer glycoproteins of organisms belonging to the domain Bacteria. (b) Typical non-S-layer glycoprotein or short-chain S-layer glycoprotein of organisms belonging to the domain Archaea. In either case the glycan chains can be linear or branched (modified from reference 15). Symbols: solid star, possible modification at the nonreducing end; shaded boxes, identical repeating unit; solid box, linkage glycose; checkered boxes, core glycose; boxes with horizontal, diagonal, and vertical lines, individual glycoses of the glycan; open circles, individual amino acids of the polypeptide; solid circle, linkage amino acid.
An additional problem with the acceptance of bacterial glycoproteins stems from the considerable structural differences between the glycans and linkage regions of common eukaryotic and viral glycoproteins (14) and the glycans and linkage regions of prokaryote glycoproteins, which have made any direct comparison difficult. Since the level of understanding bacterial glycoprotein structure is still limited, any detailed analysis of a glycoconjugate contributes importantly to an increase in the general awareness in the scientific community. In this issue of Journal of Bacteriology, Schirm and colleagues (16), in a thorough structural study, provide basic principles of the glycosylation of type a flagellins in different Pseudomonas aeruginosa strains.
What do we know today about prokaryotic glycoproteins? Much of the current knowledge was derived from initial analyses of halobacterial S-layer glycoproteins by Strominger's group (11). That work was then expanded in great detail by Sumper, Wieland, and coworkers in the 1980s. These authors, in a series of excellent publications, reported not only the structures but also biosynthetic aspects of halobacterial S-layer glycoproteins; furthermore, they provided the first sequence data for the gene encoding a glycosylated archaeal S-layer protein (for details see reference 20). Despite that ground work, an additional reason for the limited interest in early prokaryotic glycoprotein research may have been the fact that S-layer glycoproteins originate from nonpathogenic organisms that have no medical relevance. The situation changed, however, when protein glycosylation was correlated with prokaryotic pathogenicity (for reviews see references 13 and 18). Early reports of Kawamura and Shockman (9) and Erickson and Herzberg (6) described a glycoenzyme and a platelet aggregation-associated glycoprotein of Streptococcus sp. strains, respectively. A detailed structural analysis of the 45-kDa soluble antigen of Mycobacterium tuberculosis recently revealed that in this glycoprotein, short α-linked mannose chains are linked to threonine residues near the N and C termini (4).
During the past decade, glycosylated cell surface appendages, such as pili and flagella, have been increasingly investigated (for reviews see references 12, 13, 18, and 23). The pili of the pathogenic bacteria Neisseria gonorrhoeae, Neisseria menigitidis, and Pseudomonas aeruginosa are composed mainly of the glycoprotein pilin (1, 2). Recently, several pilin glycosylation genes were discovered in these bacteria (13). Some of these genes encode enzymes that transfer rare sugars, such as a modified version of pseudaminic acid, a nine-carbon sugar that resembles sialic acid (for details see references 2 and 22). The pilins of gonococci and meningococci were found to be glycosylated at Ser63, but instead of GlcNAc, 2,4-diacetamido-2,4,6-trideoxyhexose was identified as the O-linked sugar residue (for details see references 1 and 19). Recently, in the Campylobacter jejuni NCTC 11168 protein PEB3, a similar trideoxyhexose has been identified as bacillosamine (21). Comparison of the C. jejuni pgl locus with the neisserial pgl locus suggested that most of the homologous genes are involved in the biosynthesis of this rare sugar. Due to these findings, glycans of gonococcal and meningococcal pilins now appear to be more complex than initially thought. In pili of pathogenic Neisseria sp. strains, alternative glycoforms can be produced as a result of phase variation of the pilin glycosylation genes (for details see references 1, 13, and 18).
The other important glycosylated cell surface appendages of bacteria are glycosylated flagella. These structures have been found not only in gram-negative pathogens (13, 18) but also in methanogenic archaea, such as Methanococcus voltae (8) and Halobacterium halobium (for details see reference 20). For the latter organism, the authors demonstrated that the N-linked glycans of the individual flagellins are similar but not identical to the Asn-Glc-linked glycan chains present in the halobacterial S-layer glycoprotein. Motility is a key factor in the adaptation of many bacterial pathogens. Complex flagella assembled from two or more structural flagellin proteins have been found in a diverse range of gram-negative bacterial genera, including Caulobacter, Campylobacter, Aeromonas, Vibrio, Pseudomonas, and Helicobacter (for details see references 2 and 17). As in the pilin of P. aeruginosa, various derivatives of pseudaminic acid have been identified (2, 7, 22). Evidence is growing that glycosylation is involved in the flagellar assembly process. Expression of flagellar motility is an energetically expensive process, involving the expression of a large number of different genes. The encoded proteins include regulatory proteins, structural components of the flagellar secretion and assembly apparatus, and proteins involved in generating the rotational motor force and the chemosensory machinery that controls flagellum-based chemotaxis (for details see references 3, 16, and 17).
P. aeruginosa is classified into two groups on the basis of different posttranslational modifications of flagellins (types a and b). Recently, the type a flagellins but not the type b flagellins were shown to be glycosylated. In P. aeruginosa PAK and very likely in all strains expressing type a flagellin, there is a unique genomic island that encodes proteins involved in synthesis, activation, or polymerization of sugars that are necessary for flagellin glycosylation (for details see reference 3). In a continuation of this work, Schirm and coworkers (16) investigated the structures of type a flagellins from two strains of P. aeruginosa, strains PAK and JJ692. In both strains, two O-glycosylation sites, with rhamnose as the linkage sugar, were identified in each flagellin monomer. Although both strains express type a flagellin, the glycan chain structures of the organisms are different. Therefore, the role of the glycosylation island gene cluster was investigated. Interestingly, the absence of flagellar glycosylation leads to neither a loss of flagellar filament assembly nor a loss of motility. Since the biological significance of flagellar glycans is unknown, determination of their structures is very important in order to assess their role, if any, in the clinical outcomes of Pseudomonas infections.
The examples described in this commentary do not represent all the known prokaryotic glycoproteins. There are, however, two more reports which, besides their medical relevance, may have a tremendous impact on biotechnology in the future. According to the commonly held view, E. coli, the standard organism for bacterial protein expression, is unable to glycosylate proteins. This view is in contrast to observations of Lindenthal and Elsinghorst (10), who identified clinical isolates of E. coli which were able to synthesize prokaryotic glycoproteins. The glycan structures of these proteins, however, differ from those of common, eukaryotic glycoproteins. Recently, Szymanski et al. (21) described a general glycosylation pathway in C. jejuni which resulted in the elaboration of at least 30 glycoprotein species. In these glycoproteins, heptasaccharide glycans are linked in an unusual N-glycosidic linkage to bacillosamine (2,4-diacetamido-2,4,6-trideoxyglucose). Wacker et al. (24) confirmed the glycan structure and demonstrated that the pgl gene cluster can be transferred and functions in E. coli to glycosylate proteins. The conclusion from this work is that given the proper set of glycosylation enzymes, bacterial protein glycosylation may be performed in a construct other than the initial host organism.
In summary, the observations described above demonstrate that important strides are being made toward new protein glycosylation systems. Indeed, carbohydrate engineering may help solve the problem of producing tailor-made glycoproteins in bacteria on a practical scale. Undoubtedly, further progress in the detailed understanding of prokaryotic glycobiology will support and accelerate this endeavor.
Acknowledgments
I thank my colleagues in the nanoglycobiology group for their valuable contributions, Uwe B. Sleytr for his interest, and Frank M. Unger for his input during preparation of this commentary.
Work in my laboratory was supported by Austrian Science Fund projects P15612 and P15840 and by the Federal Ministry of Education, Science and Culture.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Banerjee, A., and S. K. Gosh. 2003. The role of pilin glycan in neisserial pathogenesis. Mol. Cell. Biochem. 253:179-190. [DOI] [PubMed] [Google Scholar]
- 2.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479-26485. (Author's correction, 276:36058.) [DOI] [PubMed] [Google Scholar]
- 3.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 4.Dobos, K. M., K.-H. Khoo, K. Swiderek, P. J. Brennan, and J. T. Belisle. 1996. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J. Bacteriol. 178:2498-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichler, J. 2003. Facing extremes: archaeal surface-layer (glyco)proteins. Microbiology 149:3347-3351. [DOI] [PubMed] [Google Scholar]
- 6.Erickson, P. R., and M. C. Herzberg. 1993. Evidence for the covalent linkage of carbohydrate polymers to a glycoprotein from Streptococcus sanguis. J. Biol. Chem. 268:23780-23783. [PubMed] [Google Scholar]
- 7.Goon, S., J. F. Kelly, S. M. Logan, C. P. Ewing, and P. Guerry. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50:659-671. [DOI] [PubMed] [Google Scholar]
- 8.Jarrell, K. F., D. P. Bayley, and A. S. Kostyukova. 1996. The archaeal flagellum: a unique motility structure. J. Bacteriol. 178:5057-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamura, T., and G. D. Shockman. 1983. Purification and some properties of the endogenous, autolytic N-acetylmuramoylhydrolase of Streptococcus faecium, a bacterial glycoenzyme. J. Biol. Chem. 258:9514-9521. [PubMed] [Google Scholar]
- 10.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mescher, M. F., and J. L. Strominger. 1976. Purification and characterization of a prokaryotic glycoprotein from the cell envelope of Halobacterium salinarium. J. Biol. Chem. 251:2005-2014. [PubMed] [Google Scholar]
- 12.Messner, P., and C. Schäffer. 2003. Prokaryotic glycoproteins, p. 51-124. In W. Herz, H. Falk, and G. W. Kirby (ed.), Progress in the chemistry of organic natural products, vol. 85. Springer-Verlag, Vienna, Austria. [DOI] [PubMed] [Google Scholar]
- 13.Power, P. M., L. F. Roddam, K. Rutter, S. Z. Fitzpatrick, Y. N. Srikhanta, and M. P. Jennings. 2003. Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol. Microbiol. 49:833-847. [DOI] [PubMed] [Google Scholar]
- 14.Reuter, G., and H.-J. Gabius. 1999. Eukaryotic glycosylation: whim of nature or multipurpose tool? Cell. Mol. Life Sci. 55:368-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schäffer, C., and P. Messner. 2001. Glycobiology of surface layer proteins. Biochimie 83:591-599. [DOI] [PubMed] [Google Scholar]
- 16.Schirm, M., S. K. Arora, A. Verma, E. Vinogradov, P. Thibault, R. Ramphal, and S. M. Logan. 2004. Structural and genetic characterization of glycosylation of type-a flagellin in Pseudomonas aeruginosa. J. Bacteriol. 186:2523-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schirm, M., E. C. Soo, A. J. Aubry, J. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579-1592. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11:554-561. [DOI] [PubMed] [Google Scholar]
- 18a.Sleytr, V. B., P. Messner, D. Pum, and M. Sará. 1999. Crystalline bacterial cell surface layers (S-layers): from supramolecular cell structure to biomimetrics and nanotechnology. Angew. Chem. Int. Ed. 38:1034-1054. [DOI] [PubMed] [Google Scholar]
- 19.Stimson, E., M. Virji, K. Makepeace, A. Dell, H. R. Morris, G. Payne, J. R. Saunders, M. P. Jennings, S. Barker, M. Panico, I. Blench, and E. R. Moxon. 1995. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17:1201-1214. [DOI] [PubMed] [Google Scholar]
- 20.Sumper, M., and F. T. Wieland. 1995. Bacterial glycoproteins, p. 455-473. In J. Montreuil, J. F. G. Vliegenthart, and H. Schachter (ed.), Glycoproteins. Elsevier, Amsterdam, The Netherlands.
- 21.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter--a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 22.Thibault, P., S. M. Logan, J. F. Kelly, J. R. Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276:34862-34870. [DOI] [PubMed] [Google Scholar]
- 23.Upreti, R. K., M. Kumar, and V. Shankar. 2003. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3:363-379. [DOI] [PubMed] [Google Scholar]
- 24.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]