Abstract
The evolutionarily conserved target of rapamycin (TOR) kinase controls fundamental metabolic processes to support cell and tissue growth. TOR functions within the context of two distinct complexes, TORC1 and TORC2. TORC2, with its specific component Rictor, has been recently implicated in aging and regulation of growth and metabolism. Here, we identify rict-1/Rictor as a regulator of embryonic development in C. elegans. The transcription factor skn-1 establishes development of the mesendoderm in embryos, and is required for cellular homeostasis and longevity in adults. Loss of maternal skn-1 function leads to misspecification of the mesendodermal precursor and failure to form intestine and pharynx. We found that genetic inactivation of rict-1 suppressed skn-1-associated lethality by restoring mesendodermal specification in skn-1 deficient embryos. Inactivation of other TORC2 but not TORC1 components also partially rescued skn-1 embryonic lethality. The SGK-1 kinase mediated these functions downstream of rict-1/TORC2, as a sgk-1 gain-of-function mutant suppressed the rict-1 mutant phenotype. These data indicate that TORC2 and SGK-1 antagonize SKN-1 during embryonic development.
Keywords: TOR signaling, RICTOR, SGK-1, SKN-1, Development, C. elegans
Introduction
The mammalian target of rapamycin (TOR) kinase is highly conserved in eukaryotes and integrates nutrient and anabolic signals to promote growth (Laplante and Sabatini, 2009; Wullschleger et al., 2006; Zoncu et al., 2011). TOR participates in two biochemically and functionally distinct kinase complexes, TORC1 and TORC2. TORC1 consists of TOR, Raptor (regulatory associated protein of mTOR), and LST8 (lethal with Sec13 protein8). Growth signals, nutrients, oxygen, and energy levels have been shown to activate TORC1. TORC1 regulates a set of well-characterized substrates to promote growth-related processes including protein synthesis, ribosome biosynthesis, and transcription, and to inhibit autophagy (reviewed in Loewith and Hall, 2011).
TORC2 includes the conserved proteins Rictor (rapamycin-insensitive companion of TOR), LST8, and Sin1 (stress activated protein kinase interacting protein 1), along with TOR. In contrast to TORC1, the upstream inputs and downstream effectors of TORC2 are poorly understood. Recent studies demonstrated that growth signals and association with ribosomes activate TORC2 (Oh et al., 2010; Zinzalla et al., 2011). TORC2 phosphorylates AGC kinases such as Akt and SGK (serum- and glucocorticoid-induced protein kinase) to activate downstream signaling (Zoncu et al., 2011). Phosphorylation of Akt at the hydrophobic site by TORC2 is important for its functions in cellular processes including cell proliferation and survival (Hresko and Mueckler, 2005; Sarbassov et al., 2005). Similarly to Akt, SGK1 is phosphorylated at its hydrophobic motif site by TORC2, which thereby regulates its activity (Garcia-Martinez and Alessi, 2008).
Rictor is required for TORC2 integrity and substrate specificity. Previous studies have established a conserved role for TORC2 in cell growth and metabolism (Cybulski et al., 2009; Hietakangas and Cohen, 2007; Shiota et al., 2006). Knockdown of Rictor/rict-1 in Caenorhabditis elegans resulted in an overall reduction of body size, developmental delay, and defects in fat metabolism (Jones et al., 2009; Soukas et al., 2009). Interestingly, these effects of rict-1 signaling were largely mediated through SGK-1, rather than AKT-1/2 in C. elegans. Moreover, genetic studies in C. elegans have shown that inhibition of rict-1 during adulthood confers longevity (Robida-Stubbs et al., 2012). Importantly, the transcription factor skn-1 was required for lifespan to be increased by rict-1 knockdown. SKN-1 and its mammalian ortholog Nrf have conserved functions in stress detoxification, and contribute to longevity (Sykiotis and Bohmann, 2010). SKN-1/Nrf orchestrates gene expression programs involved in stress resistance, longevity, protein homeostasis, and metabolism (Oliveira et al., 2009; Paek et al., 2012; Wang et al., 2010). The AKT and SGK-1 kinases phosphorylate SKN-1 and thereby prevent its accumulation in intestinal nuclei and inhibit its functions (Tullet et al., 2008).
SKN-1 was originally identified as an essential regulator of mesendodermal development during the earliest stages of C. elegans embryogenesis (Bowerman et al., 1992). Maternally expressed SKN-1 specifies the fate of the EMS blastomere. The E cell defines the endoderm comprising the intestine, while the MS cell gives rise to the mesoderm including the posterior portion of the pharynx, body muscle cells, and coelomocytes (for review see Maduro, 2010). Loss of skn-1 leads to a complete lack of MS-derived tissues and to most, but not all, embryos also lacking intestinal cells (Bowerman et al., 1992,1993). SKN-1 directly induces expression of the transcription factors MED-1 and MED-2, which are required for mesendodermal differentiation of the EMS lineage (Coroian et al., 2006; Maduro et al., 2001). In MS, the MEDs activate the T-box transcription factor tbx-35 and initiate a gene network for mesodermal specification (Broitman-Maduro et al., 2006). Specification of the E fate results from activation of two GATA factor-encoding genes, end-1 and end-3, by the MEDs and SKN-1 (Maduro et al., 2005; Owraghi et al., 2010; Zhu et al., 1997). These pathways finally converge on the activation of the GATA factor elt-2, which is essential for E lineage development and maintenance (Fukushige et al., 1999).
The regulatory cascade initiated by SKN-1 collaborates with the Wnt/β-catenin asymmetry pathway to distinguish MS and E identity. EMS receives an induction signal from its posterior sister P2 that results in differential localization and activity of POP-1, a TCF/Lef-related factor within MS and E. In the MS blastomere, POP-1 represses the endoderm-specific gene program allowing mesoderm development to occur. Inside the E cell, this repression is relieved and POP-1 is converted to an activator of the endoderm gene expression network (Lin et al., 1995; Maduro and Rothman, 2002; Rocheleau et al., 1997; Thorpe et al., 1997).
Given that SKN-1 seemed to be required for effects of TORC2 on longevity, we considered whether TORC2 might influence SKN-1 functions more broadly. Here, we define a new function for C. elegans TORC2 during early embryonic stages. We show that rict-1 mutation decreased the embryonic lethality associated with loss of skn-1 function by allowing mesendodermal development to be reestablished in a proportion of skn-1 deficient embryos. A gain-of-function mutation in sgk-1 suppressed these rict-1 developmental effects, indicating that sgk-1 is a primary effector of rict-1 activity in this context. In contrast to TORC2, TORC1 did not influence SKN-1 activity in the embryo. Our results define TORC2-SGK-1 as a pathway that may broadly influence SKN-1 functions.
Results
Loss of rict-1 rescues lethality of skn-1 deficient embryos
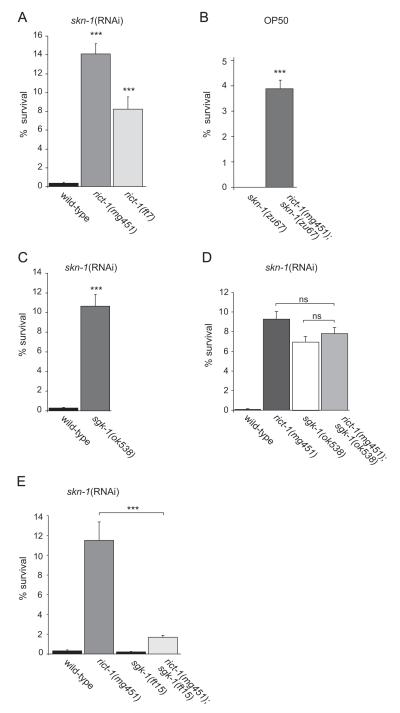
The transcription factor SKN-1 is well established as an initiator of mesendodermal development in embryos, and controls oxidative stress response. Since TORC2 has been recently implicated in the regulation of SKN-1 dependent longevity, we wondered if it interferes with SKN-1 functions more generally and might be involved in embryonic development. Rictor (RICT-1) is an essential component of TORC2. We initiated our study by analyzing the effects of rict-1 mutation on skn-1-dependent developmental processes. The rict-1 alleles mg451 and ft7 contain early stop mutations, and are likely to be strong loss-of-function and null alleles, respectively (Jones et al., 2009; Soukas et al., 2009). Embryos from skn-1 deficient mothers undergo developmental arrest, lack pharynx and endoderm, and die (Bowerman et al., 1992). First, we used RNAi feeding to inactivate maternal skn-1 function and to sensitize our ability to detect genetic interactions. We scored the portion of the skn-1(RNAi) progeny that hatched and progressed past the first larval stage (L1). Consistent with the crucial role of skn-1 during development, skn-1(RNAi) resulted in 0.4% viable embryos. Surprisingly, we found that combining skn-1(RNAi) with mutational rict-1 inactivation strongly increased the viability of the progeny: 8.2 and 14.1% of eggs produced by rict-1(ft7) and rict-1(mg451) mutants, respectively, fed with skn-1(RNAi) give rise to viable offspring (Fig. 1A and Table 1). Here, rict-1(mg451) and rict-1(ft7) survival was not statistically different. We also injected dsRNA corresponding to the skn-1 gene into the gonad of rict-1(mg451) mutants and wild-type worms and again observed that about 6% of rict-1(mg451);skn-1(RNAi) embryos hatched while 100% of skn-1(RNAi) embryos arrested development (data not shown). To test whether rict-1 specifically influenced skn-1 function and rule out that inactivation of rict-1 interferes with sensitivity to dsRNA per se, we tested two embryonic-lethal genes that to our knowledge are not defective in functions related to skn-1: let-423 and cdk-2. The complete embryonic lethality that was associated with RNAi-mediated knockdown of these genes was not altered in rict-1 mutants (data not shown).
Fig. 1.
Loss of rict-1 rescues lethality in skn-1 deficient embryos and acts in a pathway with sgk-1. (A) skn-1 embryonic lethality is suppressed by mutation of rict-1. RNAi feeding was used to inactivate skn-1 function in wild-type and rict-1 mutants starting from L1. Their total progeny and the portion that survived and progressed past the first larval stage (L1) was scored. The progeny of n>19 adult hermaphrodites for each condition was analyzed. Error bars indicate SEM. ***p<0.0001 versus wild-type on skn-1 (RNAi). See also Table 1. (B) Quantification of the fraction of survivors of rict-1;skn-1 double mutants. rict-1(mg451);skn-1(zu67) and skn-1(zu67) mutants were grown on OP50 and the fraction of embryos that survived beyond L1 was counted. n>39 for each strain. Error bars, SEM. ***p<0.0001 versus skn-1(zu67). See also Table 2. (C) sgk-1 mutants suppress lethality of skn-1 deficient embryos. RNAi feeding starting from L1 in wild-type and sgk-1(ok538) mutants. Quantification of larval survival per total progeny. n>45 for each strain. Error bars indicate SEM. ***p<0.0001 compared to skn-1(RNAi). See also Table 1. (D) Quantification of larval survival of wild-type, rict-1(mg451), sgk-1(ok538), and rict-1(mg451);sgk-1(ok538) fed with skn-1(RNAi). n>18 for each strain. Error bars indicate SEM. ns, not significant. See also Table 1. (E) sgk-1(ft15) gain-of-function mutants suppress the viability of rict-1;skn-1(RNAi) embryos. Quantification of the fraction of survivors of wild-type, rict-1(mg451), sgk-1(ft15), and rict-1(mg451) ;sgk-1(ft15) fed with skn-1(RNAi). n>20 for each strain. Error bars indicate SEM. ***p<0.0001. See also Table 1.
Table 1.
Loss of rict-1 rescues lethality in skn-1 deficient embryos and acts in a pathway with sgk-1. Corresponds to Fig. 1A, C–E. RNAi feeding was used to inactivate skn-1 function in the indicated wild-type and mutant strains starting from L1. Their total progeny and the portion that survived and progressed past the first larval stage (L1) was scored. N represents the number of adult hermaphrodites used for analysis of their progeny.
| Strain | RNAi |
n (Adult worms) |
Mean number of Eggs±SEM |
% Survival±SEM |
P value versus control |
Figure |
|---|---|---|---|---|---|---|
| N2 | skn-1 | 20 | 237±8 | 0.4±0.1 | 1A | |
| rict-1(mg451) | skn-1 | 19 | 120±3 | 14.1±1.1 | <0.0001a | 1A |
| rict-1(ft7) | skn-1 | 19 | 90±3 | 8.2±1.3 | <0.0001a | 1A |
| N2 | skn-1 | 45 | 256±4 | 0.3±0.1 | 1C | |
| sgk-1(ok538) | skn-1 | 48 | 122±3 | 10.7±1.2 | <0.0001a | 1C |
| N2 | skn-1 | 20 | 245±6 | 0.1 | 1D | |
| rict-1(mg451) | skn-1 | 20 | 141±2 | 9.3±0.8 | <0.0001a | 1D |
| sgk-1(ok538) | skn-1 | 18 | 139±4 | 6.8±0.5 | <0.0001a | 1D |
| rict-1(mg451);sgk-1(ok538) | skn-1 | 19 | 149±3 | 7.8±0.6 | nsb, nsc, <0.0001a | 1D |
| N2 | skn-1 | 20 | 238±12 | 0.3±0.1 | 1E | |
| rict-1(mg451) | skn-1 | 24 | 109±6 | 11.5±1.9 | <0.0001a | 1E |
| sgk-1(ft15) | skn-1 | 20 | 203±18 | 0.2±0.1 | nsa | 1E |
| rict-1(mg451);sgk-1(ft15) | skn-1 | 24 | 154±5 | 1.7±0.2 | <0.0001b | 1E |
| sgk-1(ft15) | control | 5 | 243±11 | 100 | ||
| rict-1(mg451);sgk-1(ft15) | control | 5 | 122±10 | 100 | ||
| N2 | skn-1 | 10 | 275±6 | 0.6±0.1 | ||
| akt-1(ok525) | skn-1 | 10 | 198±8 | 0.6±0.2 | nsa | |
| akt-2(ok393) | skn-1 | 10 | 210±14 | 0.5±0.1 | nsa | |
| rict-1(mg451) | skn-1 | 10 | 113±6 | 8.8±0.8 | <0.0001a |
ns, not significant. SEM, standard error of the mean.
p value was calculated versus N2 skn-1(RNAi).
p value was calculated versus rict-1(mg451), skn-1(RNAi).
p value was calculated versus sgk-1(ok538):skn-1(RNAi).
Next, we generated rict-1;skn-1 double mutants and analyzed the fraction of survivors. The skn-1 mutations zu67 and zu135 introduce premature stop codons and are predicted to be strong loss-of-function alleles based on the removal of the conserved DNA binding domain (Supplementary Fig. S1). Adult hermaphrodites homozygous for mutations in skn-1 produced no viable offspring, while double knockout of rict-1(mg451) and skn-1(zu67) significantly suppressed embryonic lethality. 3.9% of embryos produced by rict-1;skn-1 double mutants were viable and hatched (Fig. 1B). This finding was confirmed by several alleles for rict-1 and skn-1 (Table 2). Thus, rict-1 inactivation suppressed the embryonic lethality caused by skn-1 deficiency. Interestingly, rict-1;skn-1 mutant embryos that hatched developed into reproductive adults. They did not show obvious morphological defects and even produced progeny, although they grew slower and were smaller in body size than was characteristic of rict-1 mutants (Supplementary Fig. S2). The observation that the embryonic lethality associated with skn-1 can be partially suppressed by inhibition of rict-1 suggests that rict-1 and TORC2 antagonize skn-1 functions during development.
Table 2.
Quantification of the fraction of survivors of rict-1;skn-1 and skn-1;sgk-1 double mutants. Indicated strains were grown on OP50 and the fraction of embryos that survived beyond L1 was counted.
| Strain |
n (Adult worms) |
Mean number of Eggs±SEM |
% Survival ±SEM |
P value versus control |
Figure |
|---|---|---|---|---|---|
| N2 | 5 | 272±12 | 100 | ||
| rict-1(mg451) | 5 | 116±4 | 100 | ||
| skn-1(zu67) | 40 | 232±10 | 0 | 1B | |
| rict-1(mg451);skn-1(zu67) | 39 | 119±4 | 3.9±0.3 | <0.0001a | 1B |
| skn-1(zu135) | 24 | 272±13 | 0 | ||
| rict-1(mg451);skn-1(zu135) | 19 | 155±13 | 2.4±0.7 | <0.0001b | |
| rict-1(ft7) | 5 | 91±2 | 100 | ||
| skn-1(zu67) | 10 | 273±6 | 0 | ||
| rict-1(ft7);skn-1(zu67) | 13 | 154±7 | 3.0±0.5 | <0.0001a | |
| sgk-1(ok538) | 5 | 139±4 | 100 | ||
| skn-1(zu67) | 23 | 259±9 | 0 | ||
| skn-1(zu67);sgk-1(ok538) | 24 | 145±6 | 1.8±0.2 | <0.0001a |
p value was calculated versus skn-1(zu67).
p value was calculated versus skn-1(zu135).
rict-1 acts in a pathway with sgk-1 to effect skn-1 embryo survival
TORC2 has been reported to activate several AGC family kinases, including AKT and the serum- and glucocorticoid-induced kinase SGK (Garcia-Martinez and Alessi, 2008; Sarbassov et al., 2005). To investigate the mechanisms through which rict-1 regulates embryonic development, we first focused on Akt signaling. We analyzed the effect of akt-1 and akt-2 knockdown on skn-1(−) embryo survival. However, inactivation of akt-1 or akt-2 did not restore viability of skn-1(RNAi) embryos ((0.6% viable embryos in akt-1(ok525);skn-1(RNAi) and 0.5% in akt-2(ok393);skn-1(RNAi) compared to 8.8% in rict-1(mg451);skn-1(RNAi)) (Table 1). We conclude that the increased viability of rict-1;skn-1 deficient embryos is not a consequence of compromised AKT signaling.
In C. elegans, rict-1 regulates growth, larval development, and metabolism largely through sgk-1 (Jones et al., 2009; Soukas et al., 2009). To evaluate the possible role of sgk-1 in embryonic development we analyzed the effect of sgk-1 knockdown on skn-1(−) embryo survival. The sgk-1(ok538) deletion removes most of the region encoding the kinase domain, and is likely a null allele (Hertweck et al., 2004). We found that sgk-1 mutants rescued the lethality of skn-1 deficient embryos: 10.7% of sgk-1(ok538);skn-1(RNAi) embryos were viable compared to 0.3% skn-1(RNAi) (Fig. 1C and Table 1). Moreover, skn-1(zu67);sgk-1(ok538) double mutants produced 1.8% viable embryos, while all embryos from skn-1(zu67) mutant underwent developmental arrest and died (Table 2). Hence, loss of sgk-1 function mimics rict-1, in that it partially rescued skn-1(−) embryonic lethality, suggesting a genetic relationship.
To assess whether rict-1 and sgk-1 define a genetic pathway in the embryo, we created rict-1(mg451);sgk-1(ok538) double mutants. 7.8% of rict-1;sgk-1 double mutants fed with skn-1(RNAi) gave viable progeny (Fig. 1D and Table 1). There was no significant difference in fraction of survivors compared to either single mutant (9.3% for rict-1(mg451);skn-1(RNAi) and 6.8% for sgk-1(ok538);skn-1(RNAi)). These results suggest that rict-1 and sgk-1 act in the same pathway.
This hypothesis was further strengthened by the analysis of a sgk-1 gain-of-function mutant. If sgk-1 acts downstream of rict-1 to regulate skn-1 embryonic development then a gain-of-function mutation in sgk-1 should suppress the viability of rict-1(−);skn-1(−). The sgk-1(ft15) gain-of-function mutant has been previously shown to suppress many phenotypes associated with rict-1 (Jones et al., 2009). We indeed found that sgk-1(ft15) greatly suppressed the fraction of viable embryos of rict-1;skn-1(RNAi), 1.7% of rict-1(mg451);sgk-1(ft15);skn-1(RNAi) progeny were viable, compared to 11.5% in rict-1(mg451);skn-1(RNAi) (Fig. 1E and Table 1). sgk-1(ft15) mutants displayed normal development and broodsize and did not influence skn-1(−) embryo survival. Taken together, these obser vations indicate that rict-1 functions through sgk-1 to regulate embryonic development mediated by skn-1.
TORC1 inhibition cannot suppress skn-1 embryonic lethality
TOR exists in two distinct complexes: association with RICT-1/CeRictor defines TORC2, while DAF-15/CeRaptor is the essential component of TORC1. We wondered whether the effect of rict-1 inhibition on embryonic development might be phenocopied by components of TORC1.
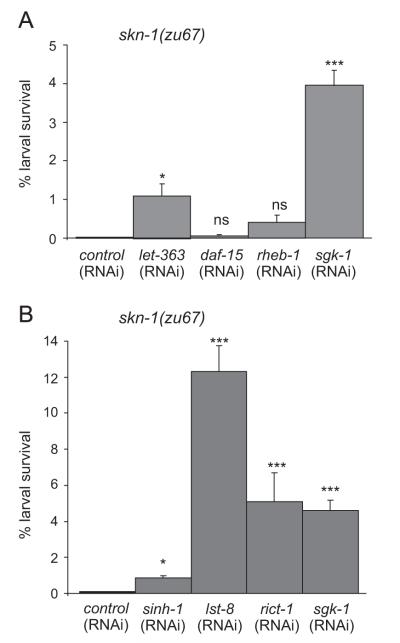
The rheb-1 GTPase is the key activator of TORC1 (Honjoh et al., 2009; Long et al., 2005; Yang and Guan, 2007). We found that knockdown of rheb-1 by RNAi did not suppress lethality of skn-1(zu67) mutant embryos. Although rheb-1(RNAi) efficiently reduced the RHEB-1 levels as evidenced by greatly abolished expression of a rheb-1::GFP reporter construct (data not shown), fewer than 0.4% of skn-1(zu67);rheb-1(RNAi) embryos were viable, while sgk-1(RNAi) significantly rescued the lethality of skn-1 deficient embryos (Fig. 2A and Table 3). Likewise, simultaneous RNAi-mediated knockdown of rheb-1 and skn-1 did not increase the fraction of viable embryos as compared to skn-1(RNAi) alone (Table 3). daf-15 encodes the C. elegans homolog of Raptor, and is essential for TORC1 activity. We found that knockdown of daf-15 failed to rescue embryonic lethality of skn-1 mutants (Fig. 2A), while daf-15(RNAi) strongly reduced daf-15 activity and caused a dauer-like larval arrest (data not shown) (Jia et al., 2004). Consistently, combined inhibition of daf-15 and skn-1 by RNAi did not promote embryonic viability (Table 3). The TOR kinase let-363 is at the center of both TOR complexes and therefore affects function of TORC1 and TORC2. As would be predicted, RNAi against let-363 suppressed the lethality of skn-1 mutants. 1.1% of eggs produced by skn-1(zu67);let-363(RNAi) gave rise to viable offspring compared to 0% of skn-1(zu67) (Fig. 2A and Table 3).
Fig. 2.
TORC2 but not TORC1 can suppress lethality of skn-1 deficient embryos. (A) Inhibition of TORC1 components cannot suppress skn-1(−) embryonic lethality. skn-1(zu67) mutants were fed with daf-15/CeRaptor(RNAi), rheb-1(RNAi), let-363/CeTOR(RNAi), and sgk-1(RNAi). Quantification of the fraction of survivors. n>20 per each condition. Error bars indicate SEM. *p<0.05, ***p<0.001. ns, not significant. See also Table 3. (B) Inhibition of TORC2 promotes skn-1(−) embryonic viability. Quantification of survival of skn-1(zu67) mutant embryo fed with rict-1(RNAi), sinh-1(RNAi), lst-8(RNAi), and sgk-1(RNAi). n>20 per each condition. Error bars indicate SEM. *p<0.05, ***p<0.001. See also Table 3.
Table 3.
TORC2 but not TORC1 can suppress lethality of skn-1 deficient embryos. Indicated strains were fed with RNAi and the fraction of embryos that survived beyond L1 was counted.
| Strain | RNAi | n (Adult worms) | Mean number of Eggs±SEM | % Survival±SEM | P value versus control | Figure |
|---|---|---|---|---|---|---|
| skn-1(zu67) | control | 34 | 211±10 | 0 | 2A | |
| skn-1(zu67) | let-363 | 42 | 65±5 | 1.1±0.3 | <0.05a | 2A |
| skn-1(zu67) | daf-15 | 33 | 132±9 | 0.05±0.04 | nsa | 2A |
| skn-1(zu67) | rheb-1 | 23 | 118±11 | 0.4±0.2 | nsa | 2A |
| skn-1(zu67) | sgk-1 | 35 | 100±6 | 3.9±0.5 | <0.001a | 2A |
| skn-1(zu67) | control | 38 | 203±9 | 0 | 2B | |
| skn-1(zu67) | sinh-1 | 39 | 152±6 | 0.8±0.1 | <0.05a | 2B |
| skn-1(zu67) | lst-8 | 35 | 58±4 | 12.3±1.5 | <0.001a | 2B |
| skn-1(zu67) | rict-1 | 20 | 89±6 | 5.2±1.6 | <0.001a | 2B |
| skn-1(zu67) | sgk-1 | 40 | 98±5 | 4.6±0.6 | <0.001a | 2B |
| N2 | skn-1 | 29 | 227±8 | 0.2±0.1 | Suppl.3 | |
| let-363(h111)/+ | skn-1 | 30 | 166±12 | 1.9±0.3 | <0.05c | Suppl.3 |
| daf-15(m81)/+ | skn-1 | 30 | 207±8 | 0.7±0.2 | nsc | Suppl.3 |
| rict-1(mg451) | skn-1 | 29 | 87±3 | 10.5±1.0 | <0.001c | Suppl.3 |
| Strain | Combined RNAi | n (Adult worms) | Mean number of Eggs±SEM | % Survival±SEM | P value versus control | |
|
| ||||||
| rrf-3(pk1426) | control;skn-1 | 30 | 68±3 | 0.2±0.1 | ||
| rrf-3(pk1426) | rheb-1;skn-1 | 21 | 53±4 | 0.1±0.1 | nsb | |
| rrf-3(pk1426) | rict-1;skn-1 | 29 | 65±5 | 3.2±0.9 | <0.001b | |
| rrf-3(pk1426) | sgk-1;skn-1 | 29 | 53±4 | 9.0±1.1 | <0.001b | |
| rrf-3(pk1426) | control;skn-1 | 39 | 72±3 | 0.4±0.2 | ||
| rrf-3(pk1426) | daf-15;skn-1 | 36 | 48±3 | 0.5±0.2 | nsb | |
| rrf-3(pk1426) | rict-1;skn-1 | 39 | 71±5 | 2.7±0.7 | <0.001b | |
| rrf-3(pk1426) | sgk-1;skn-1 | 39 | 53±4 | 14.0±2.3 | <0.001b | |
| rrf-3(pk1426) | control;skn-1 | 28 | 70±3 | 0.3±0.1 | ||
| rrf-3(pk1426) | sinh-1;skn-1 | 29 | 68±3 | 4.1±0.8 | <0.001b | |
| rrf-3(pk1426) | rict-1;skn-1 | 28 | 65±4 | 3.7±0.8 | <0.001b | |
| rrf-3(pk1426) | sgk-1;skn-1 | 19 | 68±3 | 8.5±1.4 | <0.001b | |
ns, not significant.
p value was calculated versus skn-1(zu67) on control(RNAi).
p values wase calculated versus rrf-3(pk1426) on control;skn-1(RNAi).
p values was calculated versus N2 on skn-1(RNAi).
TORC1 plays an essential role during development. Consistent with published findings, inactivation of TORC1-specific genes by RNAi starting from L1 caused a dauer-like larval arrest of the F1 progeny (Jia et al., 2004; Long et al., 2002). Similarly, homozygous TORC1 mutants arrest larval development. To exclude that complete TORC1 inactivation might mask a rescue effect on skn-1 by interfering with larval development, we analyzed heterozygous TORC1 component mutants. Previous studies have shown that heterozygous daf-15 mutants display no obvious developmental defects but show significantly extended lifespan, indicating that loss of gene dose might decrease daf-15 activity enough to be effective (Jia et al., 2004). Nevertheless, we found that daf-15(m81) heterozygousity did not suppress skn-1(RNAi) embryonic lethality (Supplementary Fig. S3 and Table 3). Consistent with the results on let-363(RNAi), heterozygous let-363 mutants slightly rescued the lethality of skn-1 deficient embryos, 1.9% of let-363(h111)/+;skn-1(RNAi) embryos were viable compared to 0.2% skn-1(RNAi) (Supplementary Fig. S3 and Table 3).
Taken together these results indicate that inhibition of TORC1-specific components rheb-1 and daf-15/Raptor cannot suppress lethality of skn-1 deficient embryos, suggesting that TORC1 does not influence skn-1-mediated regulation of embryonic development. Since inhibition of the TOR kinase let-363 would eliminate TORC1 as well as TORC2 knockdown of let-363/TOR might at least partially promotes viability of skn-1 embryos.
Inhibition of TORC2 prevents skn-1 embryonic lethality
The TORC2 complex includes the LST8 and Sin1 proteins, in addition to Rictor and TOR. To investigate further whether TORC2 regulates skn-1 embryonic functions, we examined the role of these two proteins. sinh-1 encodes the C. elegans ortholog of mammalian Sin1, which plays a key role in Akt phosphorylation and signaling (Yang et al., 2006). Inactivation of sinh-1 by RNAi only slightly rescued embryonic lethality of skn-1 mutants (Fig. 2B and Table 3). However, combined knockdown of sinh-1 and skn-1 by RNAi significantly suppressed skn-1 embryonic lethality, 4.1% of sinh-1(RNAi);skn-1(RNAi) embryos were viable compared to 0.3% of skn-1(RNAi) (Table 3).
Next, we analyzed the effect of lst-8. LST8 is present in both TOR complexes, but recent data indicate that mLST8 is critical for TORC2 function (Guertin et al., 2006; Wang et al., 2012). We found that loss of lst-8 activity by RNAi resulted in increased fraction of 12.3% viable skn-1(zu67) embryos (Fig. 2B and Table 3). Surprisingly the effect of lst-8(RNAi) on skn-1(−) viability was significantly stronger than rict-1 or sgk-1.
Taken together, inhibition of TORC2-specific components, rict-1, lst-8, and sinh-1 can partially restore embryonic viability of skn-1 deficient embryos. These findings indicate new physiological roles of TORC2 in the regulation of embryonic development mediated by skn-1.
Inactivation of rict-1 promotes mesodermal specification
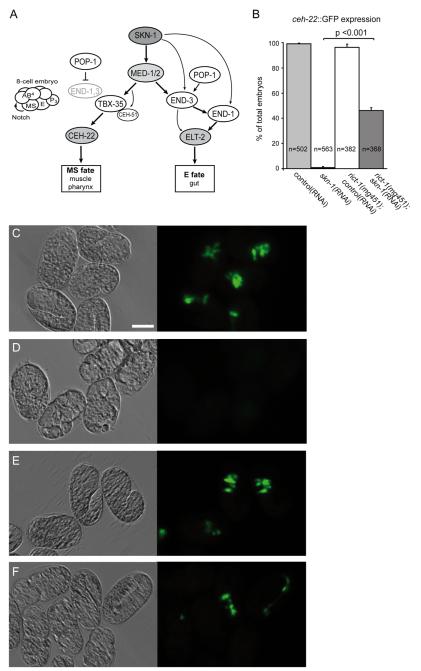
SKN-1 activates a complex gene network that specifies the development of endoderm (intestine) and part of the mesoderm (including pharynx and body muscle). Embryos from skn-1 deficient mothers arrest development and fail completely to specify the pharynx, while endoderm is absent in approximately 70% of the embryos (Bowerman et al., 1992, 1993). In the EMS blastomere SKN-1 directly activates expression of the two transcription factors, med-1 and med-2 (Coroian et al., 2006; Maduro et al., 2001). To promote specification of the endoderm, SKN-1 and the MEDs initiate a short signaling cascade by activating the GATA factor genes end-1 and end-3. Either END transcription factor can then activate the GATA factor elt-2, which is a key regulator of intestinal development (Fukushige et al., 1998; Goszczynski and McGhee, 2005; Maduro et al., 2007). The fate of the mesodermal precursor cell MS is specified primarily by activation of the T-box transcription factor tbx-35 by the MEDs (Broitman-Maduro et al., 2006, 2009). TBX-35 then regulates tissue-specific factors for pharynx and muscle development (Fig. 3A).
Fig. 3.
Rescue of defective mesodermal development in skn-1 embryos by inactivation of rict-1. (A) The C. elegans gene network for specification of mesoderm and endoderm through SKN-1 and the Wnt-effector POP-1, modified from Maduro (2009). (B) Embryos lacking rict-1 and skn-1 function make MS-type pharynx. Wild-type and rict-1(mg451) mutants bearing the integrated cuIs1[ceh-22::GFP] marker were fed with control(RNAi) or skn-1(RNAi) as indicated. Bars show percentage of terminally arrested embryos positive for ceh-22::GFP signal. Error bars, SEM. n, number of embryos analyzed. Mann–Whitney rank sum test. ***p<0.001. (C–F) Paired DIC (left) and ceh-22::GFP-fluorescence images (right) of representative embryos. Scale bar represents 20 μm. (C) In wild-type embryos the ceh-22::GFP marker shows broad induction of mesodermal tissue. (D) skn-1(RNAi) embryos arrest development and completely lack ceh-22::GFP expression. (E) In rict-1(mg451) mutants ceh-22::GFP expression is similar to wild-type. (F) rict-1(mg451);skn-1(RNAi) embryos show pharynx specification evident by ceh-22::GFP expression.
Given the impact of TORC2 on regulation of skn-1 embryonic development, we wondered whether rict-1 interferes with the gene network for mesendodermal specification. First, we assessed the production of MS-derived cell types. The MS blastomere generates diverse cell types, including pharyngeal cells. ceh-22 is expressed exclusively in the pharyngeal muscles, and together with pha-4 it is the earliest known marker of pharyngeal muscle differentiation (Okkema and Fire, 1994). To score the production of pharyngeal muscle cells we used a chromosomally integrated ceh-22::GFP reporter. As expected, skn-1 deficient embryos did not show any significant expression of ceh-22::GFP. In contrast, we found that 46% of rict-1(mg451);skn-1(RNAi) embryos produced pharyngeal muscle cells (Fig. 3B, D and F). rict-1 mutants alone showed normal pharynx development and ceh-22::GFP expression similar to wild-type (Fig. 3B, C and E). Interestingly, three times more rict-1;skn-1 embryos displayed induction of pharyngeal development (46%) while only one third completed development, hatched and were viable. The majority of rict-1;skn-1 embryos arrested at varying stages. Of note, we did not detect ectopic ceh-22::GFP expression in rict-1;skn-1 embryos. Hence, inactivation of rict-1 seems to restore pharyngeal specification to a greater extent than is apparent based upon scoring for viability.
E-derived tissues are made in rict-1;skn-1 Embryos
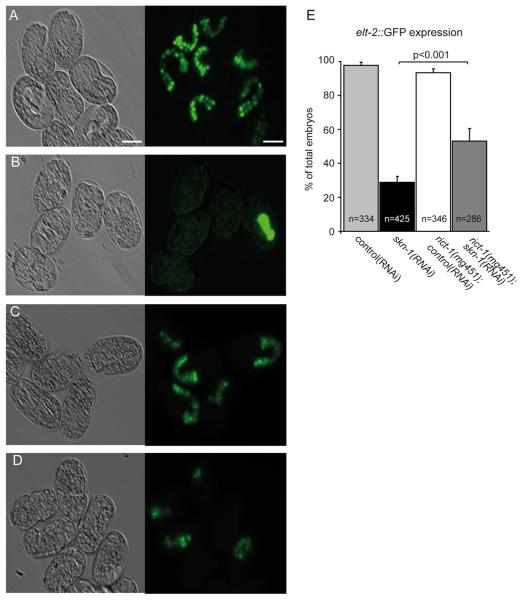
The principal target of the E-specification gene network initiated by SKN-1 is the transcription factor elt-2 (Fukushige et al., 1998). ELT-2 is required for embryonic gut development, and maintenance of the gut throughout larval development and adulthood (Fukushige et al., 1998; McGhee et al., 2007, 2009). To assess endoderm development, we introduced a chromosomally integrated elt-2::GFP reporter into rict-1 mutants. Expression of elt-2::GFP was detected in 55% of rict-1(mg451);skn-1(RNAi) embryos, while inactivation of skn-1 alone resulted in 29% of embryos with intestinal cells (Fig. 4B, D, and E). rict-1 mutants expressed elt-2::GFP in almost 100% of embryos similarly to wild-type (Fig. 4A, C, and E). This result suggests that inhibition of rict-1 partially rescues impaired gut formation in skn-1 deficient embryos. The observation that a fraction of skn-1 deficient embryos still generates intestinal cells is in line with the literature (Bowerman et al., 1992; Maduro et al., 2007) and might be the consequence of endoderm specification from a number of parallel activities.
Fig. 4.
Inactivation of rict-1 promotes endoderm development in skn-1 deficient embryos. (A–E) Wild-type and rict-1(mg451) mutants carrying the integrated marker wIs84[elt-2::NLS::GFP] for intestinal differentiation were fed with control(RNAi) or skn-1(RNAi) as indicated. Paired DIC (left panel) and fluorescence images (right panel) of embryos expressing elt-2::GFP. Scale bar represents 20 μm. (A) Expression of the elt-2::GFP in the developing gut of wild-type embryos. (B) A large fraction of skn-1(RNAi) embryos fails to express elt-2::GFP and lacks endoderm. (C) In rict-1(mg451) mutants elt-2::GFP expression does not differ from wild-type. (D) Inactivation of rict-1(mg451) in skn-1(RNAi) embryos induces elt-2::GFP expression. (E) Quantification of elt-2::GFP positive embryos from A-D. Significantly more rict-1 mutants fed with skn-1(RNAi) develop elt-2::GFP positive gut cells compared to skn-1(RNAi). Mann–Whitney rank sum test, p<0.001. Error bars, SEM. n, number of embryos analyzed.
Next, we wondered if the induction of endoderm development by loss of rict-1 might reflect changes in the number of gut cells. We therefore quantified the number of gut cells using the integrated elt-2::GFP reporter, as it has been performed in previous studies (Kostic and Roy, 2002; Maduro et al., 2007). The E cell normally divides 4–5 times to produce 20 intestinal cells by the end of embryogenesis (Sulston et al., 1983). As expected, wild-type embryos produced an average of 20 elt-2 expressing cells with little variation (Supplementary Fig. S4). We found that rict-1 mutant embryos generate a nearly identical mean number of endodermal cells as detected in wild-type (18.9±0.2), although with a slightly higher variance. The increased variance is likely to have resulted from the delayed development in rict-1 mutants (Soukas et al., 2009) rather than differences in gut cell number. In rict-1 mutant embryos we did not detect elt-2::GFP expression in more than 20 cells. Among skn-1(RNAi) embryos that still made endoderm, the average number of gut cells was dramatically reduced to 8.9±0.5, with a range of 2–18 gut cells. rict-1(mg451);skn-1(RNAi) embryos showed a higher mean number of gut cells (15.1±0.5, range of 4–20) and a higher portion of embryos (10%) with a normal number of gut cells compared to skn-1(RNAi) (Supplementary Fig. S4). We conclude therefore that loss of rict-1 function can promote normal development of the endoderm in skn-1 embryos.
Loss of rict-1 induces med-1 in embryos lacking skn-1
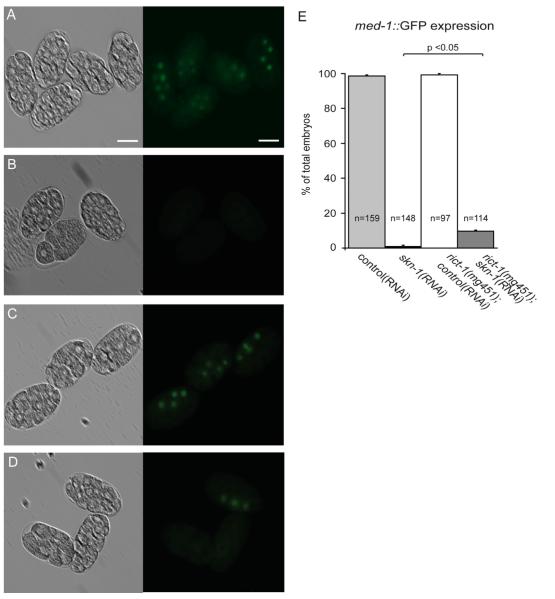
Our finding that E- and MS-specific genes are expressed in rict-1;skn-1 embryos supports the hypothesis that the canonical pathway for E and MS specifications is active. Two transcription factors, med-1 and med-2, are immediately activated by SKN-1 in EMS and essential for endomesodermal development (Maduro et al., 2001, 2007). To further analyze whether intestinal and pharyngeal cells made in rict-1;skn-1 embryos result from normal EMS specification, we introduced the translational fusion reporter med-1::GFP. In wild-type embryos, med-1::GFP expression is first detectable in EMS at the six-cell stage, and this expression is transient and becomes weaker in E and MS descendents (Maduro et al., 2001, 2007). Inactivation of skn-1(RNAi) alone resulted in nearly complete abrogation of med-1 expression (0.8%) while expression of med-1::GFP was observed in 9.7% of rict-1(mg451);skn-1(RNAi) embryos (Fig. 5). Together, these results show that inactivation of rict-1 induces mesendodermal specification in skn-1 deficient embryos involving activation of the canonical genetic network including med-1.
Fig. 5.
Loss of rict-1 activates med-1 in embryos lacking skn-1. (A–E) Wild-type and rict-1(mg451) mutants carrying the integrated marker wIs93[med-1::GFP] were fed with control(RNAi) or skn-1(RNAi) as indicated. Paired DIC (left) and fluorescence images (right). Scale bar represents 20 μm. (A) med-1::GFP expression in wild-type embryos. (B) skn-1(RNAi) embryos arrest development and completely lack med-1::GFP expression. (C) In rict-1(mg451) mutants med-1::GFP expression is similar to wild-type. (D) Embryos grown on rict-1(mg451);skn-1(RNAi) show an increased med-1::GFP expression. (E) Quantification of med-1::GFP positive embryos from A–D. Substantially more rict-1 mutants fed with skn-1(RNAi) display med-1::GFP positive cells compared to skn-1(RNAi) embryos. Mann–Whitney rank sum test. p<0.05. Error bars, SEM. n, number of embryos scored.
sknr-1 cannot compensate for loss of skn-1 function in the embryo
We have established above that TORC2 inhibits SKN-1 functions in the embryo. The simplest explanation is that TORC2 acts on SKN-1 itself, but this would require that residual SKN-1 function be present in the skn-1 mutants we analyzed. Alternatively, rict-1 might affect processes in parallel to or downstream of SKN-1. First, we evaluated the possibility of residual SKN-1 functions in skn-1(zu67) mutants. Low levels of residual SKN-1 protein might derive from perdurance of the maternal gene product in the skn-1 (−/−) animal, or from read-through. rict-1(mg451);skn-1(zu67) homozygous animals generated about 4% viable embryos (Fig. 1B and Table 2). These “rescued” offspring themselves produced viable rescued offspring at a similar rate (not shown), arguing against perdurance of maternal SKN-1. Supposing that SKN-1 still bears residual function in skn-1 mutants, we hypothesized that knockdown of skn-1 by RNAi might suppress rescue of lethality in rict-1;skn-1 mutants. Using skn-1(RNAi), the fraction of survivors in rict-1(mg451);skn-1(zu67) was reduced to 2.8%, compared to 5.2% survivors in rict-1;skn-1;control(RNAi) (Table 4). Hence, residual SKN-1 function might at least partially contribute to the viability of rict-1;skn-1 embryos.
Table 4.
sknr-1 cannot compensate for loss of skn-1 function. Strains were fed with RNAi or OP50 as indicated and the fraction of embryos that survived beyond L1 was counted.
| Strain |
n (Adult worms) |
Mean number of Eggs±SEM |
% Survival±SEM | P value versus control |
|
|---|---|---|---|---|---|
| rict-1(mg451);skn-1(zu135);sknr-1(tm2386) | OP50 | 14 | 142±4 | 2.0±0.3 | nsa, <0.001b |
| rict-1(mg451);skn-1(zu135) | OP50 | 10 | 141±9 | 2.2±0.1 | <0.001b |
| skn-1(zu135);sknr-1(tm2386) | OP50 | 10 | 269±5 | 0 | |
| skn-1(zu135) | OP50 | 10 | 304±14 | 0 | |
| sknr-1(tm2386) | OP50 | 8 | 233±13 | 100 | |
| rict-1(mg451);skn-1(zu67);sknr-1(ok1216) | OP50 | 26 | 103±12 | 4.2±0.5 | nsc, <0.001d |
| rict-1(mg451);skn-1(zu67) | OP50 | 26 | 95±19 | 4.5±0.5 | <0.001d |
| skn-1(zu67);sknr-1(ok1216) | OP50 | 26 | 206±14 | 0 | |
| skn-1(zu67) | OP50 | 27 | 211±14 | 0 | |
| sknr-1(ok1216) | OP50 | 5 | 254±16 | 100 | |
| skn-1(tm3411) | HT115 | 5 | 200±14 | 0 | |
| rict-1(mg451);skn-1(tm3411) | HT115 | 10 | 139±6 | 2.4±0.1 | <0.001g |
|
| |||||
| Strain | RNAi |
n (Adult worms) |
Mean number of Eggs±SEM |
% Survival±SEM |
P value versus control |
|
| |||||
| N2 | skn-1 | 20 | 240±9 | 0.5±0.1 | |
| rict-1(mg451) | skn-1 | 19 | 113±4 | 14.5±1.3 | <0.001e |
| rict-1(mg451);skn-1(zu67) | control | 27 | 133±6 | 5.2±0.4 | <0.001e |
| rict-1(mg451);skn-1(zu67) | skn-1 | 27 | 112±6 | 2.8±0.2 | <0.01f, <0.01e |
| skn-1(zu67) | control | 15 | 218±6 | 0 | |
| skn-1(zu67) | wdr-23 | 20 | 1±3±8 | 0.1 | nsh |
| rict-1(mg451);skn-1(zu67) | control | 20 | 109±5 | 4.6±0.5 | <0.001h |
| rict-1(mg451);skn-1(zu67) | wdr-23 | 20 | 106±4 | 4.0±0.3 | nsi, <0.001h |
ns, not significant.
p value was calculated versus rict-1(mg451);skn-1(zu135).
p value was calculated versus skn-1(zu135).
p value was calculated versus rict-1(mg451);skn-1(zu67).
p value was calculated versus skn-1(zu67).
p value was calculated versus N2 on skn-1(RNAi).
p value was calculated versus rict-1(mg451);skn-1(zu67);control(RNAi).
p value was calculated versus skn-1(tm3411).
p value was calculated versus skn-1(zu67);control(RNAi).
p value was calculated versus rict-1(mg451);skn-1(zu67);control(RNAi).
We considered whether TORC2 and SGK-1 might regulate skn-1 transcription, providing a possible mechanism for restoration of viability. When we performed quantitative PCR to measure endogenous amounts of skn-1 mRNA, we found that skn-1 expression was not changed in rict-1 and sgk-1 mutants compared to wild-type (Supplementary Fig. S5). The levels of skn-1 mRNA decreased in skn-1(zu67) and skn-1(tm3411) mutants consistent with a previous study (Tullet et al., 2008), and we observed similar skn-1 expression in rict-1;skn-1 double mutants. Hence, rict-1 and sgk-1 do not affect SKN-1 functions by modulating the abundance of skn-1 transcription.
We next investigated whether increased SKN-1 protein stability might influence rict-1;skn-1 embryonic viability. It was reported previously that WDR-23 represses SKN-1 activity by targeting SKN-1 for degradation (Choe et al., 2009; Hasegawa and Miwa, 2010). Accordingly, loss of wdr-23 increased SKN-1 protein levels and activity. Here, we tested whether knockdown of wdr-23 affects rict-1;skn-1 embryonic viability. However, wdr-23(RNAi) did not alter the fraction of viable rict-1;skn-1 embryos (Table 4). This suggests that wdr-23 might not influence SKN-1 activity in the embryo, or that increased levels of embryonic SKN-1 per se are not sufficient to enhance rescue of the embryonic skn-1 phenotype.
To address further whether residual SKN-1 might be required for TORC2 embryonic functions, we tested the effect of a skn-1 allele that is predicted to be null: tm3411 bears a deletion that should essentially destroy the DNA binding capacity of SKN-1 (Blackwell et al., 1994) (Supplementary Fig. S1). Interestingly, we still observed 2.4% viable embryos in rict-1(mg451);skn-1(tm3411), while all embryos of skn-1(tm3411) underwent developmental arrest and died (Table 4). This result suggests that the increased viability of rict-1;skn-1 mutants might not be solely the consequence of residual SKN-1 activity, and argues towards involvement of an alternative downstream process that works together with SKN-1.
Following the hypothesis that rict-1 could exert its functions independently of skn-1 and modulate another factor we considered the possibility that this might involve SKNR-1, a protein that is closely related to SKN-1 but so far has no known function (Blackwell et al., 1994; Bowerman et al., 1992) (Supplementary Fig. S1). To assess if sknr-1 can compensate for loss of skn-1 function and account for viability in rict-1;skn-1 embryos we created rict-1;skn-1; sknr-1 triple mutants. We found that mutation of sknr-1 did not affect rict-1;skn-1 viability: there was no difference in the fraction of survivors between rict-1(mg451);skn-1(zu135) mutants and rict-1;skn-1;sknr-1(tm2386) (Table 4). sknr-1 mutants displayed no obvious defects in development. We also tested a different sknr-1 allele, ok1216. Again, sknr-1(ok1216) did not change the viability of rict-1;skn-1 mutant embryos (Table 4). These observations indicate that sknr-1 does not contribute to the rescue of embryonic lethality by rict-1. Together, the data suggest that the embryonic functions of RICT-1/TORC2 involve SGK-1 and SKN-1, but also that RICT-1/TORC2 might regulate an alternative, as yet undefined process that acts in parallel to SKN-1 in the embryo.
Discussion
We have identified rict-1/CeRictor as a new regulator of embryonic development in C. elegans. Our findings revealed that mutation of rict-1 re-established mesendodermal specification in skn-1 deficient worms, and thereby suppressed skn-1-associated lethality. Moreover, inactivation of let-363/CeTOR, lst-8, and sinh-1, but not rheb-1 and daf-15/Raptor partially rescued skn-1 embryonic lethality. Thus, TORC2 but not TORC1 antagonizes skn-1 during embryonic development. Importantly, sgk-1 mediated these functions downstream of rict-1/TORC2 as the rict-1 mutant phenotype was phenocopied by loss of sgk-1 function, and suppressed by a sgk-1 gain-of-function mutant.
Inactivation of rict-1/TORC2 promotes skn-1 developmental processes
rict-1 deficient worms display pleiotropic phenotypes, including reduced growth and body size, metabolic changes of the fat content, and increased lifespan compared to wild-type (Jones et al., 2009; Robida-Stubbs et al., 2012; Soukas et al., 2009). Our previous analyses suggested that TORC2 opposed skn-1 to regulate aging (Robida-Stubbs et al., 2012). We now describe a function of rict-1/TORC2 in skn-1 dependent developmental processes. skn-1 mutant embryos lack mesodermal tissue, and most but not all embryos also fail to specify endoderm (Bowerman et al., 1992; Maduro et al., 2007; our own data). While essentially all skn-1 deficient embryos are dead, loss of rict-1 restored viability to a small but reproducibly detectable fraction of the embryos. We observed that a fraction of rict-1;skn-1-deficient embryos generated a complete pharynx and intestine (based upon morphology and staining of ceh-22 and elt-2). Furthermore, expression of med-1, which is a direct target of skn-1 and correlates with skn-1 function (Maduro et al., 2001, 2007), was induced by rict-1 deficiency indicating activation of the canonical SKN-1 network for mesendodermal specification by EMS. Interestingly, inactivation of rict-1 restored not only formation of the posterior, MS-derived portion of the pharynx but also of the anterior pharynx in skn-1 deficient embryos. The anterior portion of the pharynx is specified by cell-cell interaction between MS and ABa descendants (Fig. 3A). A skn-1 dependent signal is expressed in the MS cell that triggers anterior pharynx development in ABa. The identity of this factor remains unknown, though it is likely to be a DSL (Delta/Serrate/Lag) family ligand that induces anterior pharynx development through the GLP-1/Notch receptor (Bowerman et al., 1992; Mango et al., 1994; Priess et al., 1987). Thus, at least two distinct SKN-1 targets are restored to their correct spatiotemporal expression in rict-1 mutant embryos: med-1/2 assigning mesendodermal specification as well as Delta-like factor signaling from MS to ABa.
Our data suggest that genetic interference with rict-1/TORC2 might release skn-1 function from inhibition, and promote developmental processes. Consistent with the hypothesis that rict-1 inactivation enhances SKN-1 activity, knockdown of residual skn-1 functions by RNAi in rict-1(mg451);skn-1(zu67) mutants significantly abolished embryonic viability, albeit the effect was not complete. Of note, the skn-1 alleles zu67 and zu135 show incomplete penetrance of the mesendodermal differentiation phenotype and may represent strong loss-of-function rather than true null alleles (Bowerman et al., 1992; Raj et al., 2010). One might speculate that TORC2-SGK-1 directly influences SKN-1 activity at several levels, from transcription to protein degradation and posttranslational modifications. However, we observed that skn-1 mRNA levels were not affected by rict-1 and sgk-1, suggesting that TORC2-SGK-1 does not simply influence skn-1 expression. Notably, it was shown in previous studies that SKN-1 activity is regulated through phosphorylation in adult worms. SGK-1 and AKT-1/2 phosphorylated SKN-1, and repressed its nuclear accumulation and activation (Tullet et al., 2008). Alternatively the interaction could be more indirect, if both SKN-1 and TORC2 impinge on common processes. In agreement with this hypothesis, we observed that double knockout of rict-1(mg451) and skn-1(tm3411) predicted null mutants still resulted in a fraction of viable embryos. This result argues towards an alternative factor involved in mesendodermal development. TORC2 might provide a permissive environment for the specification of mesendoderm fate by SKN-1 and possibly other factors. Further work is needed to identify these factors and determine their functions in TORC2-signaling.
Of note, previous studies indicated that the TOR kinase let-363 and ruvb-1, an AAA+ATPase homolog, can suppress pha-4 associated lethality. Mutation of let-363/TOR and ruvb-1 restored pharynx development in a hypomorphic pha-4 strain, suggesting that TOR antagonizes pha-4 (Sheaffer et al., 2008; Updike and Mango, 2007). Whether this is primarily a TORC1 effect or also involves TORC2 is so far unknown. The transcription factor PHA-4 has an essential role in the embryonic development of the pharynx, but also acts later in life to regulate growth and longevity (Gaudet and Mango, 2002; Panowski et al., 2007). Here, we have described TORC2-mediated suppression of a more upstream component in the early EMS lineage. A speculative possibility is that there might be a common mechanism in which TORC2-mediated de-repression of a SKN-1 like activity increases pha-4 expression, thereby allowing pharynx development.
TORC2 but not TORC1 suppresses skn-1 associated embryonic lethality
We observed that inactivation of the TORC2 core components rict-1, let-363/TOR, lst-8, and sinh-1 rescued skn-1 embryonic lethality. Of note, LST-8 is part of both complexes, TORC1 and TORC2. Yet, previous studies have demonstrated that Lst8 functions exclusively in TORC2 and that mutation of Lst8 recapitulated the rictor knockout phenotype in mice, flies, and C. elegans (Guertin et al., 2006; Jones et al., 2009; Wang et al., 2012). Interestingly, genetic inhibition of the TOR kinase let-363 only had a mild effect on skn-1 associated lethality. LET-363 is part of both complexes, making it difficult to distinguish individual functions. However, our genetic analyses indicate that TORC1 appears not to be involved in control of early embryonic development. As very strong inhibition causes defects in larval developmental which might confound embryonic functions (Honjoh et al., 2009; Jia et al., 2004), we tested different genetic manipulations (RNAi and mutations) to gradually reduce TORC1 functions. We show that knockdown of the TORC1 specific genes daf-15/Raptor and rheb-1 did not promote skn-1 viability. Taken together, our data show that TORC2 but not TORC1 regulates skn-1-dependent developmental processes.
SGK-1 functions downstream of TORC2 to modulate skn-1 activity
Our results indicate that TORC2 and SGK-1 participate in a pathway that regulates embryonic development. In rescuing skn-1 embryonic lethality, inactivation of sgk-1 phenocopied inactivation of TORC2 components, and rict-1;sgk-1 double mutants showed no additive effect compared to single mutants. Additionally, a constitutive active sgk-1 mutant (Jones et al., 2009) suppressed the rict-1 embryonic phenotype, consistent with positioning sgk-1 downstream of rict-1/TORC2 in a common pathway.
TORC2 is believed to control cell survival and growth by phosphorylating several AGC kinases, including AKT and SGK. Emerging data emphasize the function of SGK-1 as an important downstream effector of TORC2. TORC2 signals predominantly through SGK-1 to regulate growth, fat metabolism, and reproduction, while AKT seems only be partially involved downstream of TORC2 under certain conditions in C. elegans (Jones et al., 2009; Soukas et al., 2009). In yeast, activation of the SGK homolog YPK2 required phosphorylation by TORC2, and seemed to mediate most of TORC2 functions (Aronova et al., 2008; Kamada et al., 2005; Niles et al., 2012). Similarly, in mammals it has been shown that TORC2 phosphorylates and activates SGK1 (Garcia-Martinez and Alessi, 2008). The physiological significance of this mTORC2-SGK interaction however is just beginning to be revealed. Our data support the hypothesis that SGK rather than AKT is a critical output of TORC2 in regulating growth and development in C. elegans.
Together, our data show that TORC2-SGK-1 signaling antagonizes SKN-1 to control mesendodermal embryonic development. It is particularly surprising that TORC2 influences SKN-1 in the context of both embryonic development and aging. TORC2 is involved in growth and metabolism (Cybulski and Hall, 2009; Jones et al., 2009; Robida-Stubbs et al., 2012; Soukas et al., 2009), and SKN-1 functions include response to environmental stres sors, regulation of development and cellular homeostasis (Bowerman et al., 1992; Wang et al., 2010). Perhaps under certain conditions it might be advantageous to mobilize these mechanisms regulated by SKN-1.
Experimental procedures
C. elegans growth conditions
C. elegans were raised at 20 °C on standard nematode growth media plates seeded with Escherichia coli OP50 as described (Brenner, 1974).
Strains used in this study
The following strains were used in this study: N2 Bristol (wild-type), NL2099 rrf-3(pk1426), ENH190 rict-1(mg451) (gift from A. Soukas & G. Ruvkun, outcrossed 4 times), KQ1366 rict-1(ft7), BR4774 sgk-1(ok538), ENH261 rict-1(mg451);sgk-1(ok538), KQ1564 sgk-1(ft15), ENH337 rict-1(mg451);sgk-1(ft15), EU1 skn-1(zu67)/nT1[unc-?(n754);let-?], ENH177 rict-1(mg451);skn-1(zu67)/nT1[unc-?(n754);let-?], ENH226 skn-1(zu67)/nT1[unc-?(n754);let-?];sgk-1(ok538), ENH256 rict-1(ft7);sgk-1(ft15), EU31 skn-1(zu135)/nT1[unc-?(n754);let-?], ENH197 rict-1(mg451);skn-1(zu135)/nT1[unc-?(n754);let-?], DR2381 let-363(h111)/dpy-5(e61), DR412 daf-15(m81)/unc-24(e138), RB759 akt-1(ok525), VC204 akt-2(ok393), ENH200 skn-1(tm3411)/nT1[unc-?(n754);let-?] (outcrossed 4x), ENH224 rict-1(mg451);skn-1(tm3411)/nT1[unc-?(n754);let-?], ENH284 sknr-1(tm2386) (outcrossed 4x), ENH297 rict-1(mg451);sknr-1(tm2386), ENH298 skn-1(zu135)/nT1[unc-?(n754);let-?];sknr-1(tm2386), ENH299 rict-1(mg451);skn-1(zu135)/nT1[unc-?(n754);let-?];sknr-1(tm2386). LD1151 sknr-1(ok1216) (4x out-crossed), ENH269 rict-1(mg451);sknr-1(ok1216), ENH278 skn-1(zu67)/nT1[qIs51];sknr-1(ok1216), ENH279 rict-1(mg451);skn-1(zu67)/nT1[qIs51];sknr-1(ok1216). JR1130 wIs84[elt-2::NLS::GFP::lacZ,rol-6D], MS94 cuIs1(ceh-22::GFP;rol-6), MS730 wIs93[med-1::GFP::MED-1]. Integrated GFP reporters for elt-2, ceh-22, and med-1 were obtained from M. Maduro. ENH271 rict-1(mg451);wIs84[elt-2::NLS::GFP::lacZ rol-6D], ENH272 rict-1(mg451);cuIs1(ceh-22::GFP;rol-6), ENH273 rict-1(mg451);wIs93[med-1::GFP::MED-1] .
Double mutants rict-1;skn-1 and skn-1;sgk-1 were created by mating male rict-1 or sgk-1 homozygous with skn-1/nT1[unc-?(n754);let-?] heterozygous hermaphrodites that have an Unc phenotype. After transgenic males were successfully crossed twice with skn-1/nT1 hermaphrodites F3 Unc progeny was selected for maintenance. Similarly, triple mutants rict-1;skn-1;sknr-1 were created by mating rict-1;sknr-1 males with skn-1/nT1[unc-?(n754);let-?]. All mutations were confirmed by PCR and sequencing.
RNA interference
RNAi by bacterial feeding was performed as described (Kamath et al., 2001). HT115 bacteria expressing the appropriate construct or pPD129.36 as empty vector control were grown in overnight cultures containing 12.5 μg/ml tetracycline and 50 μg/ml ampicillin. On the following day, cultures were diluted, grown to an OD600 of 1, and induced with 1 mM IPTG. This culture was seeded onto NGM plates containing 1 mM IPTG, ampicillin and tetracycline. For simultaneous knockdown of skn-1 and TOR complex components RNAi cultures were mixed 1:1. The rrf-3(pk1426) RNAi supersensitive strain was used to enhance the effectiveness of targeting multiple genes.
RNAi clones for rict-1, sgk-1, rheb-1, sinh-1, and wdr-23 were derived from the ORF-RNAi library v1.1. skn-1 was obtained from K. Blackwell (Tullet et al., 2008), let-363 was previously described (Vellai et al., 2003). For lst-8 and daf-15 RNAi partial cDNA was cloned into pPD129.36. All clones were confirmed by sequencing.
Brood size and larval survival
Hermaphrodites were allowed to lay eggs for 4–6 h on OP50 or RNAi, and progeny were grown to the L4 stage. To assess the brood size and viable offspring, individual L4 larvae were then placed onto fresh NGM or RNAi plates. Worms were transferred every other day onto new NGM or RNAi plates until egg-laying ceased. The total number of progeny and viable offspring of each animal were counted one to three days after removal of the parental generation. All experiments were performed at 20 °C.
For experiments with skn-1 mutants, heterozygous skn-1 hermaphrodites bearing the nT1 balancer and showing Unc phenotype were allowed to lay eggs. Homozygous F1 non-Unc skn-1 L4 larvae were then used for analysis of the brood size and embryonic viability. The zu67 and zu135 alleles behave as strict maternal-effect lethal mutations, all progeny produced by self-fertilization of homozygous hermaphodites arrested during embryogenesis (Bowerman et al., 1992).
Microscopy and immunofluorescence
For analysis of mesodermal and endodermal development ceh-22::GFP (Okkema and Fire, 1994), med-1::GFP (Maduro et al., 2001), and elt-2::GFP (Fukushige et al., 1998) transgenic worms were fed with skn-1(RNAi) or control(RNAi) starting from L1. Embryos were collected from adult hermaphrodites, transferred to an agar pad with M9 buffer and allowed to develop for an interval of time equivalent to normal embryogenesis at 20 °C.
Light microscopy was performed using a Zeiss Axioplan 2-microscope equipped with Nomarski differential interference contrast (DIC), an AxioCam-camera and the AxioVision-Software Rel.4.8. GFP was detected using appropriate EGFP-filter sets (480/20-nm excitation, 510/20-nm emission). Images were processed with Adobe Photoshop Version 12.0. and figures compiled with Adobe Illustrator CS5.
RNA isolation and quantitative PCR
For RNA preparation 300–500 young adult worms were picked to a clean NGM plate to minimize bacterial contamination and washed several times with M9 buffer. Total RNA extraction was performed with Trizol (Sigma) followed by purification with RNA Clean & Concentrator (Zymo Research) and DNAse treatment (Quiagen). The total concentration and purity of RNA was determined by absorbance at 260/280 nm (NanoDrop spectrophotometer). cDNA synthesis was performed with 250 ng RNA using Superscript III Kit (Invitrogen).
Quantitative PCR reactions were performed with qPCR Master-Mix Plus w/o UNG for SYBR® Assay ROX (Eurogentec) and the following primers: skn-1_for gaagttgtaccaaacgatgtgttcc and skn-1_rev tgctgttgacgtcctgaagatcca. Y45F10D.4_for gtcgcttcaatcagttcagc and Y45F10D.4_rev gttcttgtcaagtgatccgaca. cdc-42_for ctgctggacaggaagattacg and cdc-42_rev ctcggacattctcgaatgaag. Experiments were performed in duplicates and carried out using Roche Light-Cycler 480. The threshold cycle (Ct) values of the target genes were analyzed and normalized to the reference genes Y45F10D.4 and cdc-42 (Hoogewijs et al., 2008). Relative skn-1 expression of mutant strains was compared to wild-type using the 2−ΔΔCt method.
Statistical methods
Statistical analyses were carried out with SigmaStat 3.5. Comparing more than two groups, statistical analysis was performed with non-parametric Kruskal–Wallis one-way analysis of variance. Non-parametric Mann–Whitney (also called Wilcoxon rank-sum) test was used for pair-wise comparisons.
Supplementary Material
Acknowledgments
The authors would like to thank Morris Maduro for providing JR1130, MS94, and MS730, and Alex Soukas for rict-1 mutants. We are also grateful to the Caenorhabditis Genetic Center (University of Minnesota, Minneapolis) and the Japanese knockout consortium National BioResource Project for providing C. elegans strains. We thank A. Thien for critically reading the manuscript, P. Sekula for helpful discussions, and A. Schwierzock for excellent technical support.
This study was supported by grants from the DFG (KFO 201), the European Social fund and the Ministry of Science, Research, and Arts Baden-Württemberg to ENH, and by grant GM062891 from the NIH and an Ellison Medical Foundation Senior Scholar Award to TKB. GW was funded by BMBF (01ES0708).
Footnotes
Appendix A. Supporting information Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.08.011.
References
- Aronova S, Wedaman K, Aronov PA, Fontes K, Ramos K, Hammock BD, Powers T. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 2008;7:148–158. doi: 10.1016/j.cmet.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Draper BW, Mello CC, Priess JR. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–452. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman-Maduro G, Lin KT, Hung WW, Maduro MF. Specification of the C. elegans MS blastomere by the T-box factor TBX-35. Development. 2006;133:3097–3106. doi: 10.1242/dev.02475. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Owraghi M, Hung WW, Kuntz S, Sternberg PW, Maduro MF. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development. 2009;136:2735–2746. doi: 10.1242/dev.038307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell. Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coroian C, Broitman-Maduro G, Maduro MF. Med-type GATA factors and the evolution of mesendoderm specification in nematodes. Dev. Biol. 2006;289:444–455. doi: 10.1016/j.ydbio.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem. Sci. 2009;34:620–627. doi: 10.1016/j.tibs.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Cybulski N, Polak P, Auwerx J, Ruegg MA, Hall MN. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc. Natl. Acad. Sci. USA. 2009;106:9902–9907. doi: 10.1073/pnas.0811321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 1998;198:286–302. [PubMed] [Google Scholar]
- Fukushige T, Hendzel MJ, Bazett-Jones DP, McGhee JD. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc. Natl. Acad. Sci. USA. 1999;96:11883–11888. doi: 10.1073/pnas.96.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem. J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Goszczynski B, McGhee JD. Reevaluation of the role of the med-1 and med-2 genes in specifying the Caenorhabditis elegans endoderm. Genetics. 2005;171:545–555. doi: 10.1534/genetics.105.044909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Miwa J. Genetic and cellular characterization of Caenorhabditis elegans mutants abnormal in the regulation of many phase II enzymes. PloS One. 2010;5:e11194. doi: 10.1371/journal.pone.0011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Cohen SM. Re-evaluating AKT regulation: role of TOR complex 2 in tissue growth. Genes Dev. 2007;21:632–637. doi: 10.1101/gad.416307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol. Cell. Biol. 2007;27:5630–5638. doi: 10.1128/MCB.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2(1) doi: 10.1186/gb-2000-2-1-research0002. research0002.1–0002.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic I, Roy R. Organ-specific cell division abnormalities caused by mutation in a general cell cycle regulator in C. elegans. Development. 2002;129:2155–2165. doi: 10.1242/dev.129.9.2155. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Long X, Spycher C, Han ZS, Rose AM, Muller F, Avruch J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 2002;12:1448–1461. doi: 10.1016/s0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- Maduro MF. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim. Biophys. Acta. 2009;1789:250–260. doi: 10.1016/j.bbagrm.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF. Cell fate specification in the C. elegans embryo. Dev. Dynamics: Off. Publ. Am. Assoc. Anat. 2010;239:1315–1329. doi: 10.1002/dvdy.22233. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH. Maternal deployment of the embryonic SKN-1→MED-1,2 cell specification pathway in C. elegans. Dev. Biol. 2007;301:590–601. doi: 10.1016/j.ydbio.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Hill RJ, Heid PJ, Newman-Smith ED, Zhu J, Priess JR, Rothman JH. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev. Biol. 2005;284:509–522. doi: 10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell. 2001;7:475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Rothman JH. Making worm guts: the gene regulatory network of the Caenorhabditis elegans endoderm. Dev. Biol. 2002;246:68–85. doi: 10.1006/dbio.2002.0655. [DOI] [PubMed] [Google Scholar]
- Mango SE, Thorpe CJ, Martin PR, Chamberlain SH, Bowerman B. Two maternal genes, apx-1 and pie-1, are required to distinguish the fates of equivalent blastomeres in the early Caenorhabditis elegans embryo. Development. 1994;120:2305–2315. doi: 10.1242/dev.120.8.2305. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, Gaudet J, Kohara Y, Bossinger O, Zhao Y, Khattra J, Hirst M, Jones SJ, Marra MA, Ruzanov P, Warner A, Zapf R, Moerman DG, Kalb JM. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev. Biol. 2009;327:551–565. doi: 10.1016/j.ydbio.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, Goszczynski B, Tian H, Krich ND, Khattra J, Holt RA, Baillie DL, Kohara Y, Marra MA, Jones SJ, Moerman DG, Robertson AG. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. DevL. Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proceedings of the National Academy of Sciences of the United States of America. 2012;vol. 109:1536–1541. doi: 10.1073/pnas.1117563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Fire A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development. 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro MF. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev. Biol. 2010;340:209–221. doi: 10.1016/j.ydbio.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek J, Lo JY, Narasimhan SD, Nguyen TN, Glover-Cutter K, Robida-Stubbs S, Suzuki T, Yamamoto M, Blackwell TK, Curran SP. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 2012;16:526–537. doi: 10.1016/j.cmet.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sheaffer KL, Updike DL, Mango SE. The target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr. Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell. 2006;11:583–589. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Stress-activated cap ‘n’ collar transcription factors in aging and human disease. Sci. Signaling. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Mango SE. Genetic suppressors of Caenorhabditis elegans pha-4/FoxA identify the predicted AAA helicase ruvb-1/RuvB. Genetics. 2007;177:819–833. doi: 10.1534/genetics.107.076653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, Blackwell TK. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6(8):e1001048. doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Blumhagen R, Lao U, Kuo Y, Edgar BA. LST8 regulates cell growth via target-of-rapamycin complex 2 (TORC2) Mol. Cell. Biol. 2012;32:2203–2213. doi: 10.1128/MCB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17:666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.