Survey sequences of rye chromosomes were integrated with a new transcript map and a comparative genomics model to linearly order 22,426 gene loci. The rye genome exhibits five consecutive rearrangements in comparison to barley. Sequence and phylogenetic analysis reveal characteristics of introgressive hybridization and reticulated evolution of the rye genome.
Abstract
Rye (Secale cereale) is closely related to wheat (Triticum aestivum) and barley (Hordeum vulgare). Due to its large genome (∼8 Gb) and its regional importance, genome analysis of rye has lagged behind other cereals. Here, we established a virtual linear gene order model (genome zipper) comprising 22,426 or 72% of the detected set of 31,008 rye genes. This was achieved by high-throughput transcript mapping, chromosome survey sequencing, and integration of conserved synteny information of three sequenced model grass genomes (Brachypodium distachyon, rice [Oryza sativa], and sorghum [Sorghum bicolor]). This enabled a genome-wide high-density comparative analysis of rye/barley/model grass genome synteny. Seventeen conserved syntenic linkage blocks making up the rye and barley genomes were defined in comparison to model grass genomes. Six major translocations shaped the modern rye genome in comparison to a putative Triticeae ancestral genome. Strikingly dissimilar conserved syntenic gene content, gene sequence diversity signatures, and phylogenetic networks were found for individual rye syntenic blocks. This indicates that introgressive hybridizations (diploid or polyploidy hybrid speciation) and/or a series of whole-genome or chromosome duplications played a role in rye speciation and genome evolution.
INTRODUCTION
Rye (Secale cereale) is a member of the Triticeae tribe of the Pooideae subfamily of grasses. It is closely related to wheat (Triticum aestivum) and barley (Hordeum vulgare) and provides a main cereal for food and feed in Eastern and Northern Europe. Rye, in contrast with wheat and barley, is allogamous, and reproduction is controlled by a bifactorial self-incompatibility system promoting outcrossing (Lundqvist, 1956). A combination of male sterility inducing cytoplasms and nuclear-encoded fertility-restorer genes forms the basis of efficient hybrid breeding in rye for improved exploitation of heterosis (Geiger and Miedaner, 2009). Elevated abiotic stress tolerance to frost, drought, and marginal soil fertility make rye a perfect model for functional analyses and consequently improvement of cereal crops like wheat and barley, which are less tolerant to abiotic stress.
Rye has a large (1C = 8.1 Gb; Doležel et al., 1998) diploid genome (2n = 2x = 14), nearly 50% bigger than the barley genome. It is unknown whether this results from higher amounts of repetitive DNA only or if rye also contains more genes than other diploid Triticeae species. Similar to wheat and barley, the center of origin of genus Secale is in the Near East. Rye was domesticated during the Neolithic Era (7000 years ago) in Anatolia and later in Europe, where it first spread as a weed in wheat and barley fields (Sencer and Hawkes, 1980; Willcox, 2005). Rye and wheat diverged seven million years ago, and both lineages and the barley lineage diverged from a common Triticeae ancestor around 11 million years ago (Huang et al., 2002).
Despite extensive synteny to barley (H genome) and wheat (A, B, and D genomes), the rye genome (R) has undergone a series of rearrangements, as revealed by comparative restriction fragment length polymorphism (RFLP) mapping (Devos et al., 1993). Collinearity to wheat was disturbed by a series of translocations involving all chromosomes but 1R. It was postulated that a translocation involving the long arms of linkage groups 4 and 5 (4L/5L) occurred before the split of the wheat and rye lineages, since it is present in various Triticeae species and in the A genome of wheat (Moore et al., 1995; Mayer et al., 2011). Subsequent reorganization events involving several other chromosome arms were proposed (Devos et al., 1993). Comprehensive genome-wide analysis of the level of conserved synteny and extension of rearrangements between rye and other Triticeae genomes has so far been hampered by lack of genomic resources in rye.
High-density gene-based marker maps are important prerequisites for studying genome organization and evolution. Such maps in barley (Stein et al., 2007; Close et al., 2009; Sato et al., 2009) and wheat (Qi et al., 2004) allowed detailed comparisons to sequenced model grass genomes like rice (Oryza sativa), Brachypodium distachyon, and sorghum (Sorghum bicolor) (International Rice Genome Sequencing Project, 2005; Paterson et al., 2009; International Brachypodium Initiative, 2010). A dense gene-based genetic map of barley together with conserved synteny information of the above mentioned three model grass genomes provided the framework to integrate a linear gene order model comprising more than 21,000 barley genes. The gene content information of barley was obtained by survey sequencing of amplified DNA from individually sorted chromosomes (Mayer et al., 2009; Mayer et al., 2011). Thus genome size, which hampered systematic sequencing of Triticeae genomes for long time, could be turned into an advantage in Triticeae genome analysis since chromosomes can be sorted and enriched from different Triticeae species including rye (Kubaláková et al., 2003; Doležel et al., 2012).
For rye, existing genetic maps comprised limited numbers of gene-based markers (Gustafson et al., 2009; Hackauf et al., 2009) or were composed of anonymous genomic Diversity Arrays Technology markers (Milczarski et al., 2011). Recently, a large data set of gene-based single nucleotide polymorphisms (SNPs) could be data-mined from RNA sequencing data of rye, providing the basis for developing a high-throughput SNP genotyping assay comprising 5234 markers (Haseneyer et al., 2011). In this study, this SNP assay was employed to build a high-density transcript map of rye. Together with chromosomal survey sequences (CSSs) generated from flow-sorted and amplified rye chromosomes, a high-density linear gene-order map could be established. This provided the basis for in-depth comparative genetic analysis between rye and other grass genomes, leading us to propose a revised model of rye genome evolution. Global sequence conservation and synteny and phylogenetic network analysis revealed a heterogeneous composition of the rye genome, indicating its reticulate evolution (evolutionary relationships do not fit a simple bifurcate tree but instead fit a network structure), which can be linked to a series of translocations that shaped the rye genome. We postulate that this was the result of introgressive hybridization and/or allopolyploidization events. The outbreeding lifestyle of rye might have facilitated interspecies introgressive hybridization, thus providing an important prerequisite for the formation of the modern rye genome.
RESULTS
A High-Density Transcript Map of Rye
A high-density gene-based marker map of rye was developed by genotyping 495 recombinant inbred lines (RILs) from four mapping populations with a previously published Rye5K Infinium Bead Chip (Haseneyer et al., 2011) comprising 5234 SNP markers (Table 1). In addition, 271 Expressed Sequence Tag (EST)-SSR (for simple sequence repeat) markers were genotyped in two of the populations. Between 782 and 2158 SNP and SSR markers were mapped in the four individual mapping populations (Table 1). An integrated high-density genetic map comprising 3543 gene-based markers and 45 anchor markers (providing links to previous work in rye; Hackauf et al., 2012) was established, encompassing a cumulative map length of 1947 centimorgans (Figure 1; see Supplemental Figure 1 online).
Table 1. Molecular Marker Statistics for Transcript Mapping in Rye.
| Mapping Populationa | EST-SNP | EST-SSR | Anchor Markers | No. of Mapped Markers | No. of Mapped Genes | Map Length (cM)b |
|---|---|---|---|---|---|---|
| Lo7xLo225 | 1952 | 206 | – | 2158 | 1825 | 1428 |
| P87xP105 | 1813 | – | – | 1813 | 1504 | 1347 |
| Lo90xLo115 | 717 | 65 | – | 782 | 677 | 1084 |
| L2039-NxDH | 1200 | – | 45 | 1245 | 1038 | 1369 |
| Consensus | 3272 | 271 | 45 | 3588 | 2886 | 1947 |
Maps generated with JoinMap v4.0, except P87xP105, which has been calculated with MSTMap.
cM, centimorgans. –, not available.
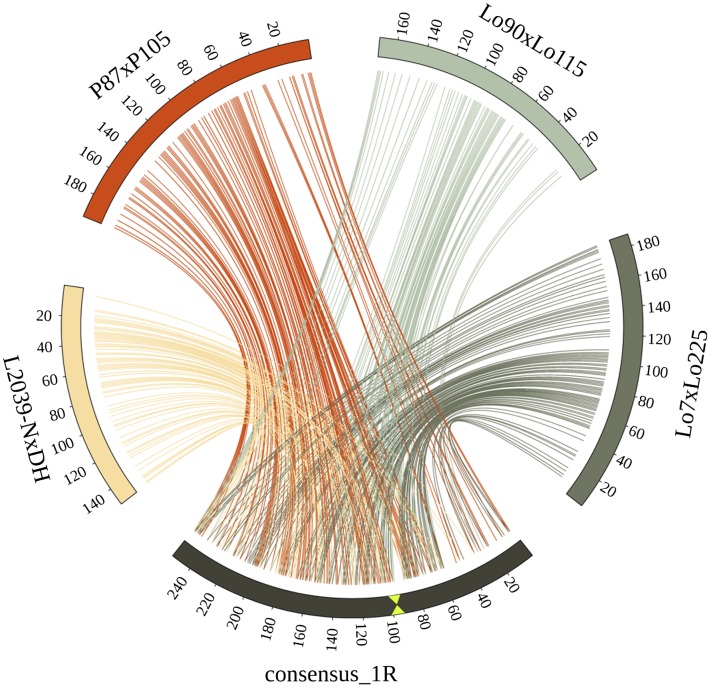
Figure 1.
Rye Consensus Transcript Map.
Comparison of the integrated genetic map of chromosome 1R with the 1R maps of four individual mapping populations (Lo7xLo225, P87xP105, Lo90xLo115, and L2039-NxDH). Colored lines connect markers between the integrated map and each individual genetic linkage map. Complete collinearity could be observed between all individual maps and the integrated consensus. Centromere position in the consensus map is indicated by green triangles.
Composition of Rye Chromosomes Revealed by Survey Sequencing
Individual rye chromosomes were purified and used as template for CSS using Roche/454 technology. We obtained between 1.02 (chromosome 1R) and 1.43 (4R) Gb of sequence per chromosome. In total, 8.25 Gb provided sequence coverage between 0.93- and 1.17-fold (average 1.04-fold) for each individual chromosome fraction (Table 2). The expected base pair coverage was calculated to range between 60.5 and 68.9% (average 64.6%; Table 2). The estimated values were tested by comparing the CSS data sets against the available genetically anchored sequence markers. An average marker detection rate (sensitivity) of 78.7% was observed, and for all individual chromosomes, the theoretically expected Lander-Waterman values were significantly exceeded. The average specificity of 92.6% (Table 2) correlated well with cytological estimates of the average individual chromosome fraction purity of 93.5% obtained by fluorescence in situ hybridization on specimens prepared from sorted chromosome fractions.
Table 2. Sequence and Coverage Statistics from CSSs of Individual Rye Chromosomes.
| Chromosome | Size (Mb)a | Sequences (Mb) | Coverage (x-Fold) | Expectationb | Observed Marker Detection Rate (Sensitivity) | Anchored Reads (Specificity) |
|---|---|---|---|---|---|---|
| 1R | 1005 | 1023 | 1.02 | 63.9 | 75.4 | 84.7 |
| 2R | 1315 | 1253 | 0.95 | 61.3 | 80.2 | 95.7 |
| 3R | 1047 | 1226 | 1.17 | 68.9 | 77.4 | 93.0 |
| 4R | 1242 | 1435 | 1.16 | 68.6 | 80.7 | 93.4 |
| 5R | 1119 | 1229 | 1.10 | 66.7 | 80.9 | 93.9 |
| 6R | 1134 | 1060 | 0.93 | 60.5 | 76.4 | 94.3 |
| 7R | 1055 | 1027 | 0.97 | 62.1 | 79.9 | 93.1 |
| Total | (∑) 7917 | (∑) 8253 | (Ø) 1.04 | (Ø) 64.6 | (Ø) 78.7 | (Ø) 92.6 |
Calculated based on 2C DNA amount = 16.19 pg (Doležel et al., 1998), relative chromosome lengths according to Schlegel et al. (1987), and 1 pg = 0.978 Mb (Doležel et al., 2003).
Expectation was calculated using the Lander Waterman expectation (Lander and Waterman, 1988).
To identify the fraction of CSS reads containing gene and/or exon sequence, we masked all repetitive DNA sequences. About 74% of the CSS sequences consisted of repetitive DNA elements (see Supplemental Table 1 online). The remaining 2.2 Gb of sequence was distributed among the individual rye chromosomes resulting in a range between 275 Mb assigned to 7R and 437 Mb assigned to 4R. This repeat-masked CSS fraction was compared with a recently published set of barley genes (International Barley Genome Sequencing Consortium, 2012) and full gene sets of the sequenced genomes of rice, B. distachyon, and sorghum (International Rice Genome Sequencing Project, 2005; Paterson et al., 2009; International Brachypodium Initiative, 2010). Overall, sequence similarity was obtained for a nonredundant set of 31,008 genes. On the basis of the previously determined sensitivity of the sequence data sets, more than 39,400 genes thus can be estimated for the rye genome.
Virtual Linear Order of 22,426 Rye Genes (Genome Zipper)
Previously, we introduced the concept of developing virtual linear gene order maps (genome zippers) by integrating CSS data with dense gene-based marker maps and conserved synteny information from sequenced model grass genomes (i.e., B. distachyon, rice, and sorghum) (Mayer et al., 2009, 2011). We followed this approach for the rye CSS data. In the first step, a comparison of genes constituting the transcript map of rye established the putatively orthologous (conserved syntenic) regions of the model grass genomes. Subsequently, all coding sequences from CSS data were compared against genes from these reference genomes. Based on genes located in corresponding syntenic blocks of the respective model grass genomes and identified with rye CSS data, it was postulated that the putatively orthologous genes are present in a conserved order in rye as well. Hence, the high-density transcript map of rye provided the scaffold to position and orient blocks of conserved syntenic genes between rye and the model grass genomes. A total of 10,833 barley cDNAs, 20,370 nonredundant rye ESTs, and between 11,869 and 14,086 genes from reference genomes (see above) were unambiguously associated with rye CSS sequences (Table 3). Between 2693 (6R) and 3595 (2R) genes were assigned in linear order along individual rye chromosomes (Table 3; see Supplemental Data Sets 1 to 7 online). Overall, 22,426 rye genes were positioned along the genome. Thus, we were able to position 72% of all detected rye genes (22,426/31,008).
Table 3. Genome Zipper Statistics: Genes, ESTs, and Associated 454 Reads.
| Data Sets | 1R | 2R | 3R | 4R | 5R | 6R | 7R | ∑ |
|---|---|---|---|---|---|---|---|---|
| No. of SNP markers | 390 | 469 | 381 | 394 | 486 | 398 | 422 | 2,940 |
| No. of markers with orthologous gene in reference genome(s) | 224 | 270 | 223 | 215 | 276 | 199 | 236 | 1,643 |
| No. of barley fl-cDNAs | 1,386 | 1,663 | 1,567 | 1,437 | 1,697 | 1,370 | 1,713 | 10,833 |
| No. of nonredundant sequence reads | 23,720 | 29,907 | 24,948 | 36,818 | 33,671 | 21,436 | 24,304 | 194,804 |
| No. of matched rye ESTs | 2,489 | 3,121 | 2,849 | 2,892 | 3,382 | 2,877 | 2,760 | 20,370 |
| No. of B. distachyon genes | 1,761 | 2,291 | 2,146 | 1,960 | 2,391 | 1,750 | 1,787 | 14,086 |
| No. of rice genes | 1,469 | 2,060 | 1,825 | 1,510 | 1,767 | 1,444 | 1,794 | 11,869 |
| No. of sorghum genes | 1,538 | 1,818 | 2,015 | 1,644 | 2,050 | 1,439 | 1,740 | 12,244 |
| No. of nonredundant anchored gene loci in genome zipper | 2,806 | 3,595 | 3,201 | 3,299 | 3,751 | 2,693 | 3,081 | 22,426 |
Conserved Synteny between the Genomes of Rye and Barley
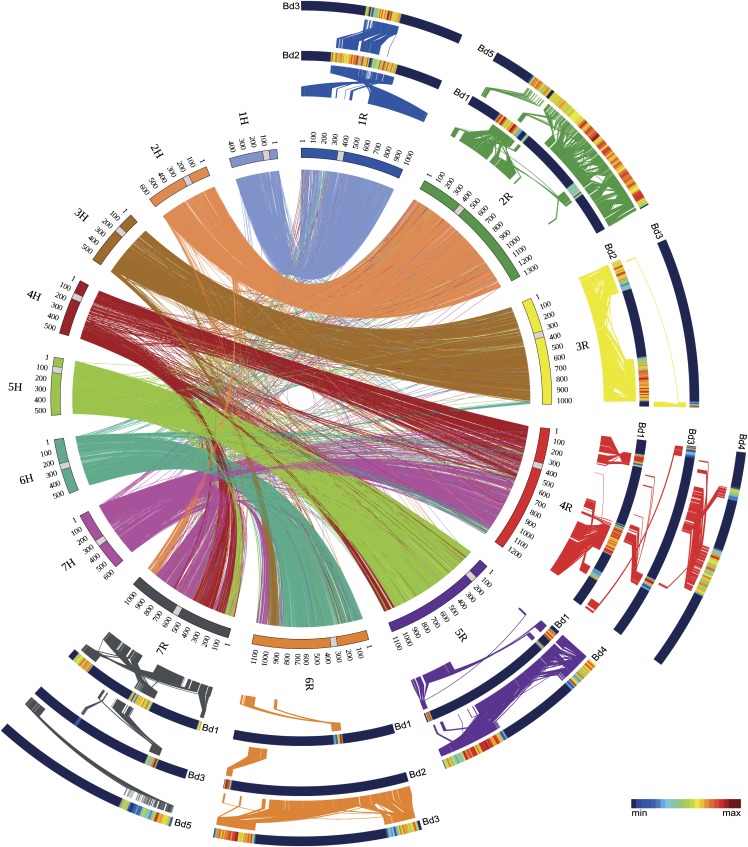
The close evolutionary relationship between rye and barley is reflected in extensively conserved synteny. On the basis of the above presented linear gene-order map of rye, structural differences, translocations, and the overall extent of conserved synteny could now be addressed at unprecedented resolution between rye and barley or the other reference grass genomes, respectively. Comparisons of the dense genetic rye map provided in this study and the physical/genetic barley genome assembly (International Barley Genome Sequencing Consortium, 2012) revealed numerous rearrangements in rye chromosomes (Figure 2; see Supplemental Figure 2 online). Only rye chromosome 1R exhibited collinearity over its entire length to a single barley chromosome (1H). All other rye chromosomes were composed of a mosaic pattern with two to four conserved syntenic segments of individual barley chromosomes (Figure 2; see Supplemental Figure 2 online). The 2R markers and 454 sequences of the genome zipper identified a small part corresponding to barley chromosome 7HL and almost the entire chromosome 2H. The 3R marker corresponded to almost the entire chromosome 3H and a region on 6HL, while 4R-tagged regions on 4H and segments from the short arms of 6H and 7H. Chromosome 5R tagged regions on 5H and 4HL. Chromosome 6R is homoeologous with most, but not all, of chromosome 6H and with the long arms of 3H and 7H. Chromosome 7R is composed of segments with homoeology to parts of 4HL, 5HL, and 7HL as well as to parts of 2HS and 7HS. All seven genetic centromeres in rye and barley (Figure 2) are conserved at syntenic positions and were not involved in translocations in rye. They thus remained conserved since the divergence of a common ancestor. Overall, we identified 17 conserved syntenic segments between rye and barley that make up both genomes and allow us to propose a revised model of rye genome evolution (Figure 3). This model describes a series of six translocation events that account for the major pattern of rearrangements between rye and barley.
Figure 2.
Conserved Synteny between Rye, Barley, and B. distachyon.
Collinearity of the rye and barley genomes is depicted by the inner circle of the diagram. Rye (1R to 7R) and barley (1H to 7H) chromosomes were scaled according to the rye genetic and barley physical map, respectively. Lines (colored according to barley chromosomes) within the inner circle connect putatively orthologous rye and barley genes. The outer partial circles of heat map colored bars illustrate the density of B. distachyon genes hit by the 454 chromosome survey sequencing reads of the corresponding rye chromosomes. Conserved syntenic blocks are highlighted by yellow-red-colored regions of the heat maps. Putatively orthologous genes between rye and B. distachyon are connected with lines (colored according to rye chromosomes), and centromere positions are highlighted by gray rectangles.
Figure 3.
Rye Genome Reorganization and Translocation Events.
Rye genome reorganizations occurring in the common ancestor of rye and wheat (translocation between chromosomes 4 and 5) and divergence of the two lineages are postulated. Three of the five translocations that occurred after the split of wheat can be ordered, while for two the order cannot be deduced. They may have occurred in parallel or consecutively.
Conserved Synteny to Model Grass Genomes Is Nonuniform between Rye and Barley
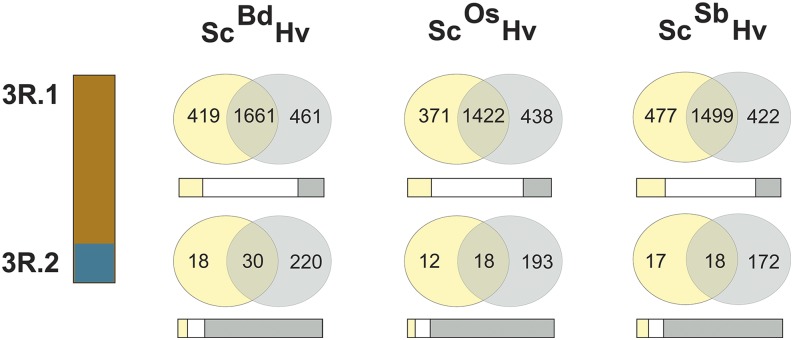
Based on the extent of conserved synteny between rye and barley, we compared the global pattern of conserved synteny to sequenced model grass genomes. Overall, rye and barley contain very similar numbers of conserved syntenic genes when compared with B. distachyon, rice, and sorghum (see Supplemental Table 2 and Supplemental Figures 3 and 4 online; Figure 2). Comparing the rye (this study) and barley (Mayer et al., 2011) genome zippers, which are established by integrating synteny information with regard to the same three model grass genomes, both species share 64 to 66% (14,408) of the 22,426 and 21,766 respective genome zipper loci. Given the large number of rearrangements between the rye and barley genomes, we addressed the question whether all conserved syntenic blocks between both genomes contain proportional numbers of conserved syntenic genes in comparison to the three model grass genomes. We surveyed all 17 conserved syntenic regions between rye and barley individually. In most cases, barley and rye segments carried similar or equal numbers of conserved syntenic genes when compared with the three model genomes (Figure 4; see Supplemental Figure 5 online). Additionally, most segments contained also a similar fraction of conserved genes that were uniquely shared between either rye or barley and any of the three model genomes. However, four out of the 17 segments revealed pronounced deviations from this equilibrium. As an example, the distal conserved syntenic segment of chromosome 3R (denoted as 3R.2 in Figure 4) contained 10 to 16 times fewer conserved syntenic genes (30 to 48 genes) to B. distachyon, rice, and sorghum than the putative orthologous segment of barley 6H (190 to 250 genes). Opposite examples were found for the most proximal segments of 7R (7R.4) or 4R (4R.1) (see Supplemental Figure 5 online) carrying up to 8 times more conserved syntenic genes to B. distachyon, rice, and sorghum than the respective segments of barley chromosomes 2H and 4H. The observed patterns could be due to differential retention of paralogs in rye and barley, differential evolutionary fate of conserved syntenic chromosome segments, or, in part, different evolutionary origins of the corresponding segments and/or their parts. We found significant differences between the syntenic segments of rye and barley regarding the number of conserved syntenic genes for each of the three reference genomes (Pearson’s χ2 test; 32 df; P < 1 × 10−6).
Figure 4.
Conserved Synteny Statistics of Rye Chromosome 3R and the Corresponding Barley Regions to Reference Genomes.
Venn diagrams show the absolute number of conserved syntenic rye (yellow) and barley (gray) genes in comparison to the reference grass genomes of B. distachyon, rice, and sorghum. The bars below depict the percentage of distribution of reference genes shared by barley and rye (white), or rye (yellow) and barley alone (gray), respectively. While the 3R.1 fragment shows a balanced conserved syntenic pattern, the second fragment 3R.2 showed 10-fold less conserved syntenic genes in comparison to the corresponding barley segment.
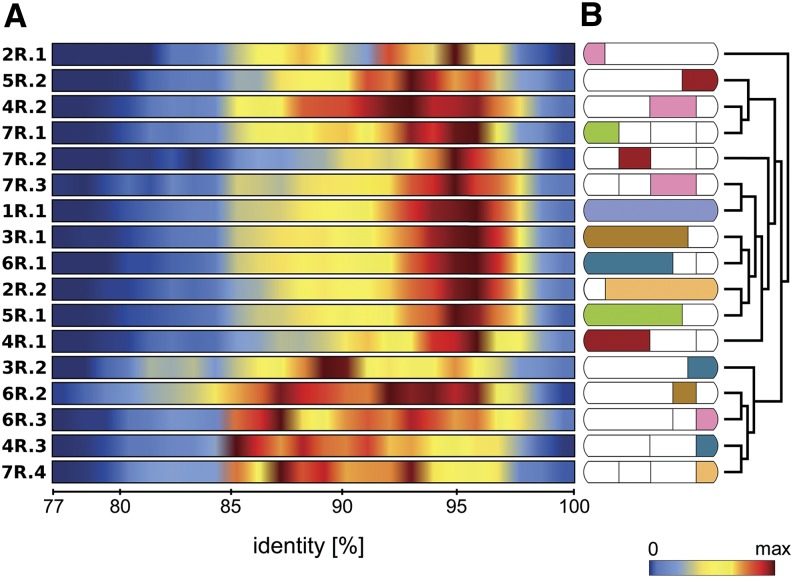
Varying Sequence Identity Thresholds in Conserved Syntenic Segments Indicate Reticulate Evolution of the Rye Genome
The observation of unbalanced conserved syntenic gene content of orthologous genome segments of rye and barley in comparison to model grasses prompted us to expand our analysis toward testing for sequence conservation of the involved genes. We assessed sequence conservation of all anchored genic sequence reads assigned to the 17 rye genome segments against a set of 28,622 full-length cDNAs (fl-cDNAs) of barley (Matsumoto et al., 2011). Corresponding orthologous genes and gene segments were selected using a first best hit criterion, and matching sequence regions had to exceed 100 nucleotides (≥30 amino acids). We plotted the sequence identity distribution for the 17 rye genomic fragments as heat map distributions (Figure 5A) and performed hierarchical clustering including 10,000-fold bootstrap resampling of sequence identity distributions for the respective segments. A broad distribution of sequence identity profiles was observed. Many segments (7R.3, 5R.1, 6R.1, 3R.1, 1R.1, 2R.2, and 4R.1) revealed overall sequence similarity in a relatively narrow range grouped around a maximum at 95% sequence identity. However, several individual segments (e.g., 2R.1, 3R.2, 6R.2, 6R.3, 4R.3, and 7R.4) exhibited a significant shift toward lower maximum sequence identity (Figure 5A). Statistical significance of sequence identity values was tested for segment-specific distributions also considering the amount of genes in the respective segment using a permutation test. For segment 2R.1, results were inconclusive, similar to previous results from the bootstrap clustering, most likely due to its small size. Strikingly, most segments involved in rye lineage specific translocations (Figures 3 and 5) showed deviating identity profiles and grouped more distantly by hierarchical clustering (Figure 5B).
Figure 5.
Sequence Conservation between Rye and Barley in 17 Conserved Syntenic Genome Segments.
(A) Rye gene-based chromosome survey sequences of the 17 conserved syntenic genome segments were compared with the putative barley orthologs (on the basis of fl-cDNAs) and the distribution of percentage of sequence identity is depicted by heat maps for each conserved block (max = highest no. of reads per segment with the given identity value; each block has its own maximum). The segments showed nonuniform sequence conservation patterns.
(B) The obtained sequence identity values were grouped by hierarchical clustering (average linkage, Euclidean distance) with the aim to find similarities between segments that could indicate their origin from the same progenitor genome and translocation or introgression event.
We expanded this analysis and measured synonymous (Ks) and nonsynonymous (Ka) substitution rates between rye/barley orthologs that were identified in the 17 conserved syntenic genome segments (see Supplemental Figure 6 online). Similar to the findings reported above, chromosomes 2R to 7R, all of which are composed of different syntenic segments with respect to barley, showed heterogeneous Ks mean and median values. The Ks distribution between the groups was significantly different (Kruskal-Wallis-test; P < 0.004351). However Ka/Ks values for the individual segments did not reveal pronounced differences; hence, no pattern of potential positive selection on individual genomic segments could be observed that might have caused the pronounced shifts in sequence similarities found for the individual rye segments.
Phylogenetic Analysis of Rye Chromosome Segments Indicates Variable Phylogenetic Networks
In a subsequent step, we analyzed the similarities and differences in phylogenetic networks for the 17 syntenic segments found in the rye genome. For each segment, we selected corresponding genes from five grass genomes for which either complete or draft genome sequences in different depth and resolution are available. Besides the rice genome that served as an outgroup, we also used the genome of B. distachyon, the barley genome, and the recently published genome sequences of the two diploid wheat subgenome progenitor species Aegilops tauschii and Triticum urartu (Jia et al., 2013; Ling et al., 2013). Corresponding genes were selected using a bidirectional best BLAST hit criterion, and a total of 705 gene clusters were generated and analyzed for phylogenetic networks (see Supplemental Figure 7 online). This analysis revealed that, consistent with the clustering results obtained using sequence conservation (Figure 5), rye genomic segments group differently in the phylogenetic networks. For eight rye segments (1R.1, 2R.2, 3R.1, 4R.2, 5R.1, 6R.1, 7R.2, and 7R.3), results indicate phylogenetic positioning of rye between barley and the wheat lineage (Ae. tauschii and T. urartu), but for other segments, the network structure was different, with varying relationship differences (e.g., 4R.1 found to group distant from the Triticeae). In addition, even within segments we found evidence for reticulate evolution for several segments (4R.3, 5R.2, 6R.2, and 7R.1). Thus, in summary, the phylogenetic networks for the 17 rye segments showed pronounced differences and even within some of the segments evidence for reticulate evolution was found.
DISCUSSION
Rye Genome Unlocked by Chromosomal Genomics
Wheat, barley, and rye are very closely related cereal crop species that were domesticated during a very narrow time span during the Neolithic Era. Their domestication was of critical importance for the establishment of early civilizations of the Fertile Crescent area in Near East and the spread of agriculture to Europe and Asia. For understanding evolution and domestication of the three species, as well as for any molecular genomic crop improvement strategy, it is a prerequisite to have access to (complete) genome sequence information. Significant progress has recently been reported for barley (Mayer et al., 2011; International Barley Genome Sequencing Consortium, 2012), wheat (Brenchley et al., 2012), and diploid wheat progenitor species (Jia et al., 2013; Ling et al., 2013). In this study, the rye genome could be unlocked by a combined approach of chromosomal genomics and conserved synteny analysis, providing comprehensive access to gene content as well as linear gene order information of about two thirds of the predicted rye genes.
We adopted an in silico method to establish so-called genome zippers to develop virtual linear gene order models that comprise considerable proportions of the genes of the ∼8-Gb rye genome. This advance delivered an enabling platform for future genome-based rye research and improvement but also for high-resolution comparative analysis of related Triticeae species and grass genomes in general. The procedure integrated gene content information with a dense genetic map and conserved synteny information provided by reference sequences of related model grass genomes. The method has been proven successful and powerful for barley (Mayer et al., 2011), Lolium (Pfeifer et al., 2013), and wheat chromosome 4A (Hernandez et al., 2012). We used DNA amplified from flow-sorted rye chromosomes to generate CSS data, and ∼31,000 genes were detected by sequence comparisons. Based on the measured sensitivity, ∼40,000 genes can be postulated for the entire rye genome. However, this number might be overestimated since gene fragments and pseudogenes are abundant in Triticeae genomes (Mayer et al., 2011; Wicker et al., 2011; International Barley Genome Sequencing Consortium, 2012), and due to the limited sequence coverage of the presented data sets, conclusions about the total gene set remain preliminary. Overall, this number is higher than, but comparable to, previous gene counts reported for other Triticeae genomes and rye chromosomes (Mayer et al., 2011; Martis et al., 2012; International Barley Genome Sequencing Consortium, 2012), suggesting that haploid gene content is similar in rye, barley, and wheat. A total of 22,426 genes (72% of the postulated genes) could be integrated into the rye genome zippers on the basis of the newly developed high-density gene-based genetic map of rye and conserved synteny information of the sequenced genomes of B. distachyon, rice, and sorghum. This number is similar to previous work, which identified 21,766 genes using the genome zipper approach for barley (Mayer et al., 2011).
Genome Collinearity between Rye and Barley
Synteny of grass genomes has been intensively studied, starting about two decades ago, on the basis of comparative RFLP mapping. Grass genomes share extensively conserved synteny and a circular model to visualize collinearity between smaller (i.e., rice) and larger grass genomes (i.e., Triticeae) was introduced (Moore et al., 1995). This model has been repeatedly revised as higher density maps became available for individual species (Devos, 2005) and recently has been enriched for information on ancient whole-genome duplication events leading to a refined model of grass karyotype evolution (Murat et al., 2010). We used the rye genome zippers developed in this work to reassess Triticeae genome collinearity and identified 17 segments representing the rye genome and exhibiting conserved synteny to the barley genome (International Barley Genome Sequencing Consortium, 2012). Rye chromosome 1R was the only linkage group that was collinear over its entire length to a single barley chromosome (1H). All other rye chromosomes were composed of between two and four segments corresponding to individual regions on the barley genome. However, our findings largely confirm earlier studies at unprecedented density and resolution since previous descriptions relied on mapping of 150 RFLP markers (Devos et al., 1993) in comparison to wheat. The major patterns of rearrangement between rye and barley can be described as a series of six subsequent translocation events, which we illustrate in a revised model of rye genome evolution. Starting from a set of seven ancestral Triticeae chromosomes that most closely resemble in organization the modern barley (HH) and Ae. tauschii (DD) genomes, four translocation events in rye can be sequentially ordered while the succession of two additional events remains uncertain. The initial translocation between ancestral chromosomes a4 and a5 is very similar and possibly homologous to a reciprocal translocation reported for the 4A and 5A chromosomes of wheat (Naranjo et al., 1987; Liu et al., 1992). In this scenario, three subsequent translocations between the ancestral chromosomes a3 and a6, a6 and a7, and a7 and a4 would have occurred. The two remaining translocations (a2/a7 and a6/a4) have likely taken place after the three preceding translocations. However, their sequential order remains unclear and both events may have occurred at the same time.
What Mechanisms Have Shaped the Modern Rye Genome?
The unprecedented access to rye genomic sequence information provided with this study as well as the detailed genome sequence information recently published for barley (International Barley Genome Sequencing Consortium, 2012) allowed a detailed comparative analysis of conserved orthologous genomic segments between both genomes. This revealed that individual conserved syntenic genomic segments of rye and barley carried strikingly different numbers of putatively conserved orthologous genes in comparison to the model grass genomes of rice, B. distachyon, and sorghum. Furthermore, the genes of defined conserved syntenic rye genome segments exhibited significantly different signatures of sequence conservation if compared with their putatively orthologous barley gene sequences.
Analysis of synonymous and nonsynonymous substitutions did not provide any evidence of different selective pressure among the different genomic regions of rye, but phylogenetic analysis of individual rye genomic segments revealed pronounced differences in their relationships to the five compared grass species. The observed network structures are largely consistent with the results obtained by comparison of global sequence similarities of genes found in specific genomic segments. For eight of the segments, the consensus tree/network structure positions rye between barley and the wheat lineage, but for the other segments, differing phylogenetic networks were found. It is noteworthy that patterns of reticulate evolution were found in four of the segments. Thus, overall, we conclude that the rye genome represents a concatenation of genomic segments with, in part, differing evolutionary origins. Hence, the rye genome, to some extent, was likely shaped by introgressive hybridization or reticulate evolution.
It is important to note that reticulate genome evolution was postulated recently for rye by a multigenic phylogeny analysis (one chloroplast gene, 26 nuclear genes) of different Triticeae species (Escobar et al., 2011). Reticulate evolution or hybrid speciation was postulated to have occurred frequently during plant evolution (Kellogg and Bennetzen, 2004; Linder and Rieseberg, 2004; Mallet, 2005). In the Triticeae, it may have occurred in diploid species (Kellogg et al., 1996; Escobar et al., 2011), but it has been most frequently postulated for allopolyploid Triticeae genera (Kellogg et al., 1996; Mason-Gamer, 2004; Mason-Gamer et al., 2010; Mahelka et al., 2011). Reticulate or hybrid speciation can occur (reviewed in Linder and Rieseberg, 2004) as a consequence of allopolyploidization, which involves fusion of unreduced gametes, or instant genome duplication after fusion of haploid gametes, giving rise to a fertile hybrid species in which diploid parental genomes are maintained. This mechanism has been documented in a number of taxa, including Brassica and Triticum (Snowdon, 2007; Feldman and Levy, 2012). Reticulate speciation can also occur by diploid (homoploid) hybrid speciation, which involves fusion of reduced gametes of parental species (reviewed in Rieseberg, 1997; Linder and Rieseberg, 2004). Allopolyploid formation had a major impact on wheat evolution and provided advantages to new plant species to colonize new niches (Levy and Feldman, 2002; Matsuoka, 2011). Diploid hybrid species of sunflower (Helianthus annuus) exhibited a selective advantage over their parental species in more extreme habitats, as demonstrated by resynthesized hybrid species (Rieseberg et al., 2003). In the sedge species Carex curvula, it has been postulated that interspecies hybrid formation could have provided an advantage under changing environmental conditions (Choler et al., 2004). Furthermore, chromosomal aberrations and spontaneous aneuploidy were observed to occur at higher frequency in Aegilops speltoides populations in marginal environments (Belyayev and Raskina, 2013).
Whether allopolyploid or diploid hybrid speciation provided more likely mechanisms shaping the modern rye genome remains speculative. Given the diploid nature of today’s rye, it seems more intuitive to propose that rye underwent one or more diploid hybrid speciation events. The obligate outbreeding nature of rye may support that diploid hybrid speciation played a role in rye evolution since there is a strong correlation between outcrossing and diploid hybrid speciation in plant species with a confirmed reticulate evolutionary history (reviewed in Rieseberg, 1997). In this study, we found no obvious evidence of the allopolyploid nature of the rye genome. We identified no traces of additional whole-genome duplication (data not shown), besides the one shared by rice and other Triticeae species (Salse et al., 2008; Thiel et al., 2009). However, in comparison to the closely related barley and wheat genomes, rye has a 50% bigger monoploid genome, and it carries the highest number of translocations in comparison to a postulated ancestral Triticeae progenitor genome. It is tempting to speculate that rye genome evolution involved one (or more) episode(s) of polyploidization and/or interspecific hybridization between as yet unknown species leading to allopolyploidization. Thus, modern rye genome structure with seven chromosomes would be the outcome of extensive karyotype repatterning and diploidization. Cytological studies of interspecific hybrids in the genus Secale indicated that cultivated rye differs by three reciprocal translocations from its putative wild ancestors (Stutz, 1972; Singh and Röbbelen, 1977). It was hypothesized that cultivated rye S. cereale evolved from Secale vavilovii possibly after multiple introgressions from Secale montanum/Secale strictum. This is consistent with the idea of reticulate evolution of the genome of S. cereale with multiple introgression events and could also explain the different levels of sequence homology to barley for the individual corresponding genomic segments. Reciprocal translocations in combination with dysploid chromosome number reduction could explain how rye returned to a diploid status with extensive collinearity to the present day diploid Triticeae genomes (mechanism reviewed in Schubert and Lysak, 2011). In this scenario, the increased monoploid genome size of rye and the slightly increased gene content in comparison to diploid barley and wheat genomes may represent remnants of the allopolyploid origin of rye. The presence of B chromosomes in rye provides more support for the hypothesis that interspecies hybridization played a role in rye genome evolution (B chromosomes are absent in barley and wheat). B chromosomes are supernumerary chromosomes that do not follow Mendelian inheritance and may origin from standard A chromosomes after interspecific hybridization (reviewed in Camacho et al., 2000); however, they may also form without the need of hybridization. Survey sequenced flow-sorted rye B chromosomes carried thousands of gene signatures with homology to rye chromosomes 3R and 7R (Martis et al., 2012). Thus, rye B chromosomes can also be interpreted as side products of reorganization of the genome after hybridization or whole-genome duplication and subsequent rediploidization. In this scenario, the B chromosomes and their apparent correspondence to regions of the A genome can be seen as indicative for genomic segments that got eliminated from the A genome during the reshaping/diploidization process.
Outlook
Next-generation sequencing and chromosome flow sorting allowed us to greatly improve the genomic resources for rye genome analysis. This will facilitate future work toward molecular crop improvement as well as the more targeted characterization and utilization of genetic resources and crop wild relatives in rye breeding. The global analysis of conserved synteny and sequence conservation to related grass species provided a comprehensive novel insight into current state rye genome organization and indicates a history of the rye genome possibly involving reticulate evolution. With the recent relatively easy access to genome-wide sequence information, even from large genomes like those of the Triticeae, a much more fine-grained picture of grass species evolution can be expected for the near future that will provide us with novel insights into the dynamics of grass genome evolution over time.
METHODS
Plant Material
Four mapping populations, Lo7xLo225, P87xP105, Lo90xLo115, and L2039-NxDH, were employed for high-throughput genotyping. Lo7xLo225 was derived from an interpool cross between two inbred lines Lo7 and Lo225 by KWS LOCHOW, and 131 RILs (F4) from this cross were developed at the Julius Kühn-Institut. For P87xP105, 69 RIL F6 lines were derived from a pair of reciprocal crosses of the two inbred parents P87 and P105. The population was developed at the Institute of Genetics and Cytology, Minsk, Belarus, by T.S. Schilko (Korzun et al., 1998). For Lo90xLo115, 220 RIL F4 lines were obtained from a cross between two inbred lines Lo90 and Lo115 by KWS LOCHOW. For L2039-NxDH, 100 RIL F9 lines that originate from an interpool cross between an elite inbred nonrestorer inbred line (L2039-N source: HYBRO) as female parent and a doubled haploid (DH) recombinant line (L285xL290, developed at the University of Hohenheim, Germany) were established at the Julius Kühn-Institut.
Molecular Marker Resources
A custom rye (Secale cereale) 5k Illumina iSelect array comprising 5234 EST-derived SNP markers (Haseneyer et al., 2011) was used for high-throughput genotyping. Furthermore, 1385 gene-based SSRs were data-mined and evaluated for their use as SSR markers from previously published rye EST resources (Haseneyer et al., 2011) by applying the software tool Misa (Thiel et al., 2003). In addition, 45 more markers (SSR and STS) previously mapped in different rye populations (Hackauf et al., 2009, 2012) provided anchoring information to other published genetic maps of rye and to assign the obtained L2039-NxDH-linkage groups to the seven rye chromosomes and for orienting chromosome maps. The marker TC427 (ALDH2b) was derived from a rye mitochondrial aldehyde dehydrogenase mRNA sequence (GenBank accession number AB084896.1) and assayed using the primer pair 5′-TGTCCCTGGTTGAAAAACAG-3′ and 5′-TGATGTATGGCTGGAAAGTTG-3′ as previously described (Hackauf and Wehling, 2005).
SNP Genotyping and Data Processing
A total of 300 ng of genomic DNA per plant was used for genotyping on the Illumina iScan platform with the Infinium HD assay following manufacturer’s protocols. The fluorescence images of an array matrix carrying Cy3- and Cy5- labeled beads were generated with the two-channel scanner. Raw hybridization intensity data processing, clustering, and genotype calling (AA, AB, and BB) were performed using the genotyping module in the GenomeStudio software V2009.1 (Illumina). Genotyping data were cleaned by excluding SNP markers with (1) a GenTrain score < 0.6, (2) >10% missing data, or (3) monomorphic pattern.
Genotyping EST-Derived SSR Markers
A total of 688 EST-derived rye SSR markers were screened for polymorphism in four parents (Lo7, Lo90, Lo115, and Lo225) of two mapping populations (Lo7xLo225 and Lo90xLo115). The respective progenies were genotyped with 271 polymorphic markers. PCR was conducted in a total volume of 20 μL (20 ng of genomic DNA, 1× HotStar Taq PCR buffer, 250 nM each primer, 200 μM deoxynucleotide triphosphates, and 0.5 units of HotStar Taq DNA polymerase [Qiagen]). A touch-down PCR profile was applied (initial denaturation: 15 min at 95°C, 45 cycles: denaturation at 94°C for 1 min, annealing for 1 min [1°C incremental reduction from 65 to 55°C in the first 10 cycles and then 55°C] and extension at 72°C for 1 min [10 min at final extension]). PCR products were resolved on 1.5% agarose gels. Only markers with <10% missing values were used for mapping. Primer sequences of 688 tested and 271 mapped EST-SSRs are given in Supplemental Data Set 8 online.
Construction of Individual and Consensus Linkage Maps
Map construction of populations Lo7xLo225, L2039-NxDH, and Lo90xLo115 was performed with JoinMap 4.0 (Kyazma). Grouping was performed at an independence logarithm (base 10) of odds score between 4.0 and 10.0. For locus ordering, the maximum likelihood algorithm was used. The genetic linkage map of the P87xP105 population was constructed using MSTMap (Wu et al., 2008) at the probability level 1E−7. The centimorgan distances were calculated by applying the Kosambi mapping function (Kosambi, 1944). In populations Lo7xLo225 and Lo90xLo115, SSR markers were distributed manually to the SNP-based linkage maps using the software MapManager QTX (Manly et al., 2001).
A draft consensus map based on the four individual linkage maps was constructed using MergeMap (Wu et al., 2008). The consensus linkage groups were then compared with the original four homologous linkage groups in order to identify conflicts in marker order. MapChart v2.2 (Voorrips, 2002) and Circos (Krzywinski et al., 2009) were used for graphical representation of the linkage maps. Genotyping and detailed map information of the individual and the consensus map are provided as Supplemental Data Sets 9 and 10 online.
Purification and Amplification of Chromosomal DNA for Sequencing
Aqueous suspensions of intact mitotic chromosomes were prepared from root tips of seedlings (‘Imperial’ rye for 1R and ‘Chinese Spring’–‘Imperial’ wheat [Triticum aestivum]–rye disomic chromosome addition lines for 2R to 7R; Driscoll and Sears, 1971), and rye chromosomes 1R to 7R were purified using FACSAria SORP flow sorter (BD Biosciences) as described earlier (Kubaláková et al., 2003). Approximately 20,000 copies of each rye chromosome were flow-sorted, and their DNA was purified and multiple-displacement amplified (MDA) by the Illustra GenomiPhi V2 DNA amplification kit (GE Healthcare) in three independent reactions as described before (Simková et al., 2008). MDA DNA samples from each chromosome were pooled prior to sequencing. The identity and purity of sorted chromosome fractions was determined using fluorescence in situ hybridization with pSc119.2 and 5S rDNA probes (Kubaláková et al., 2003) (see Supplemental Figures 8 and 9 online). The purity of flow-sorted chromosome fractions and resulting quantities of amplified chromosomal DNA are summarized in Supplemental Table 3 online.
Roche/454 Sequencing
DNA amplified from sorted chromosomes was used for Roche/454 shotgun sequencing. Five micrograms of individual chromosome MDA DNAs was used to prepare the 454 sequencing libraries with the GS Titanium General Library Preparation Kit following the manufacturer’s instructions (Roche Diagnostics). The 454 sequencing libraries were processed utilizing the GS FLX Titanium LV emPCR (Lib-L) and GS FLX Titanium Sequencing (XLR70) kits (Roche Diagnostics) according to the manufacturer's instructions. Statistics and details about the CSS data are summarized in Table 2 and Supplemental Table 1 online. Base pair coverage per chromosome was calculated according to Lander and Waterman (1988). The estimated values were tested by comparing the CSS data sets against the available genetically anchored sequence markers. The specificity (Sp) of individual rye chromosome data sets was determined as the proportion of false positive (FP) and true negative (TN) sequence matches with genetically anchored markers providing the reference ( ).
).
Bioinformatic Analyses: Identification of Repetitive Regions
The repetitive DNA content of CSS data was detected using Vmatch (http://www.vmatch.de) against the Munich Information Center for Protein Sequences-REdat Poaceae 8.6.2 repeat library (Nussbaumer et al., 2013). The following parameters were applied: 70% identity cutoff, 100-bp minimal length, seed length 14, exdrop 5, and e-value 0.001.
Analysis of Conserved Synteny
To assess the number of genes present in rye and to determine conserved syntenic regions between rye, barley (Hordeum vulgare; International Barley Genome Sequencing Consortium, 2012), and the three model grass genomes rice (Oryza sativa; International Rice Genome Sequencing Project, 2005), sorghum (Sorghum bicolor; Paterson et al., 2009), and Brachypodium distachyon (International Brachypodium Initiative, 2010), the repeat-filtered 454 sequence reads (with stretches of at least 100-bp nonmasked nucleotides) were compared against the protein sequences of the other grass species using BLASTX. Only homologs with at least 85% (barley), 75% (B. distachyon), or 70% (rice and sorghum) similarity and a minimum length of 30 amino acids were considered. Genes with multiple evidence were counted only once. The number of conserved genes was calculated using a sliding window approach (window size of 0.5 Mb; window shift of 0.1 Mb) and visualized by Circos heat maps (Krzywinski et al., 2009).
Generation of Rye Genome Zippers
Genetic map data, chromosomal gene content of rye, and conserved synteny information to model grass genomes were used for developing virtual gene order maps (genome zippers) of all seven rye chromosomes according to the earlier described approach (Mayer et al., 2011). This framework was substantiated by information based on rye EST assemblies (Haseneyer et al., 2011) and barley full-length cDNAs (Matsumoto et al., 2011). The genome zipper integration data sets are available as Supplemental Data Sets 1 to 7 online.
Analysis of Rye/Barley Synteny
The 2940 genetic markers of rye were compared via bidirectional BLASTN against 2785 genetic markers of barley (Close et al., 2009), and the homologous pairs were displayed in a scatterplot using matplotlib (Hunter, 2007). This comparison revealed syntenic segments and various chromosomal rearrangements. The same overall but higher density picture was obtained comparing the nonmasked 454 reads of the rye genome zippers against the physical/genetic barley genome scaffold (International Barley Genome Sequencing Consortium, 2012). The comparison was achieved using BLASTN (Altschul et al., 1990) with (1) the best match with minimum 85% identity and (2) a minimal alignment length of 100 bp. Subsequently, the conserved syntenic regions were detected using a sliding window approach (window size of 5 Mb; window shift of 1 Mb) and visualized by heat maps for each rye chromosome separately. The rye/barley orthologous pairs were defined using bidirectional BLASTN hits with the cutoff values mentioned above and plotted with the help of Circos (Krzywinski et al., 2009).
Assessment of Sequence Diversity and Conservation in Rye/Barley Conserved Syntenic Regions of the Rye Genome in Comparison to Other Grass Species
After manual inspection of the syntenic patterns between rye and barley, several distinct syntenic regions with a variable amount of reads (326 to 21,175) and genes (55 to 2,140) were defined. In the next step, these individual fragments were assigned to the virtual gene maps of barley and rye by investigating the rye reads and corresponding barley genes and their position in the genome zipper. To calculate the synonymous (Ks) and nonsynonymous (Ka) substitution rates between barley and rye, the 454 reads of the individual syntenic blocks were compared against the derived protein sequence from barley fl-cDNAs. The protein sequences of the barley fl-cDNAs were predicted using OrfPredictor (Min et al., 2005). The comparison and identification of protein alignments were done using BLASTX. All first best hits with at least 85% identity and a minimum of 50 amino acids without internal stop codon were filtered for further analysis. The Ka/Ks substitution rate was calculated using the YN00 module of the PAML 4 suite (Yang, 2007). In a last step, the average Ka and Ks values were calculated for those proteins that were tagged by multiple 454 reads. All Ks values up to 10 were used for statistical analysis. The Ks and Ka values were visualized by boxplots using the matplotlib library (MATLAB; MathWorks).
To test the sequence diversity in the syntenic fragments, the 454 reads assigned to the corresponding regions were compared using BLASTN against barley fl-cDNAs (28,622 sequences) (Matsumoto et al., 2011). The obtained sequence identities of all matches with at least 100-bp alignment length were summarized in bins and plotted. The individual blocks on particular chromosomes showed nonuniform distribution patterns. To group fragments with similar distribution, a hierarchical clustering of the identity bins was performed. We applied a hierarchical clustering, employing the Euclidean distance and average linkage.
Statistical Analysis
The syntenic conservation of both rye and barley against the three reference genomes (B. distachyon, rice, and sorghum) was tested for homogeneity with respect to the degree of syntenic conservation for each segment. For each reference organism, Pearson’s χ2 test was applied separately by comparing the numbers of barley and rye genes mapped against the reference across all syntenic fragments.
The significance of the identity values clustering was assessed using bootstrap resampling (B = 10,000) as implemented in the pvclust package in R (Suzuki and Shimodaira, 2006). The reported approximately unbiased P values indicate the significance of the observed cluster, with values close to 100 showing clusters that have the strongest support. As the segment size varied strongly (326 to 21,175), we tested whether the observed patterns were random by employing a permutation test. For each syntenic segment (sample size N), we randomly drew N identity values from the complete set of identity values and tested whether these were significantly different from the observed values using a Kolmogorov-Smirnov test (Massey, 1951). This was repeated 10,000 times. These analyses were performed using R (http://www.R-project.org).
Differences between rye and barley distributions of the synonymous substitution rate (Ks) were tested with the Kruskal-Wallis test using the R software package (http://www.R-project.org).
Phylogenetic Analysis
To test for reticulate evolution/introgressive hybridization, the protein sequences of six distinct species (rye, barley, Aegilops tauschii, Triticum urartu, B. distachyon, and rice) that map to the 17 syntenic conserved regions were analyzed. For each segment, corresponding orthologous genes from the respective species were extracted using a bidirectional best BLAST hit criteria against the respective rye genes. To generate sufficient data points for all segments, either clusters of six corresponding genes (from rye, barley, rice, B. distachyon, Ae. tauschii, and T. urartu) or clusters of five corresponding genes (as before but without a corresponding gene from T. urartu) were extracted. A total of 705 gene clusters were generated. For each segment, the amount of gene clusters used varied between 1 and 160. The sequences of each cluster were aligned using MUSCLE (Edgar, 2004). The maximum likelihood phylogeny inference was constructed using FastTree2 (Price et al., 2010) with the JTT+CAT substitution model and the Shimodaira-Hasegawa test to compute the confidence values of tree branches. The trees were rooted by defining rice as outgroup. The level-k network consensus algorithm implemented in Dendroscope3 (Huson and Scornavacca, 2012) was used to combine and visualize the phylogenetic trees for each individual fragment into a single phylogenetic consensus network. Each network represents all clusters from all input trees, if the clusters appear in more than 30%.
Accession Numbers
Sequence data from this article were submitted to the European Bioinformatics Institute sequence read archive under study accession ID ERP001745, sample IDs ERS167396 to ERS167402, experiment IDs ERX140512 to ERX140518, run IDs ERR164635 to ERR164641.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Rye Consensus Transcript Map.
Supplemental Figure 2. Conserved Homologous Regions between Rye and Barley.
Supplemental Figure 3. Conserved Synteny between Rye, Barley, and Rice.
Supplemental Figure 4. Conserved Synteny between Rye, Barley, and Sorghum.
Supplemental Figure 5. Conserved Synteny between Rye and Barley with B. distachyon, Rice, and Sorghum Genomes.
Supplemental Figure 6. Sequence Conservation of Rye and Barley Genes in Corresponding Genome Segments.
Supplemental Figure 7. Phylogenetic Networks for Individual Segments of the Rye Genome.
Supplemental Figure 8. Flow Cytometric Sorting of Rye Chromosome 1R from cv Imperial.
Supplemental Figure 9. Example of the Use of Wheat-Rye Chromosome Addition Lines to Purify Chromosomes 2R to 7R Using Flow Sorting.
Supplemental Table 1. Sequence and Repeat Analysis Statistics for Individual Rye Chromosomes.
Supplemental Table 2. Genome Zipper Statistics for Rye/Barley Orthologous Genome Segments.
Supplemental Table 3. Purity of Flow-Sorted Rye Chromosome Fractions and DNA Amounts Obtained after Amplification of Chromosomal DNA.
Supplemental Data Set 1. Genome Zipper of Rye Chromosome 1R.
Supplemental Data Set 2. Genome Zipper of Rye Chromosome 2R.
Supplemental Data Set 3. Genome Zipper of Rye Chromosome 3R.
Supplemental Data Set 4. Genome Zipper of Rye Chromosome 4R.
Supplemental Data Set 5. Genome Zipper of Rye Chromosome 5R.
Supplemental Data Set 6. Genome Zipper of Rye Chromosome 6R.
Supplemental Data Set 7. Genome Zipper of Rye Chromosome 7R.
Supplemental Data Set 8. EST-Derived Rye SSR Markers.
Supplemental Data Set 9. Mapping Data of Four Populations.
Supplemental Data Set 10. Rye Consensus Transcript Map.
Supplementary Material
Acknowledgments
We thank Adam Lukaszewski for providing seeds of rye cv ‘Imperial’ and wheat-rye chromosome addition lines, Jarmila Číhalíková, Zdenka Dubská, and Romana Šperková for assistance with chromosome sorting and DNA amplification, and Heidrun Gundlach for help in repeat masking. We also thank Bjoern Usadel and Doreen Pahlke from Plant2030-PD for support with submission of sequence data sets to the European Bioinformatics Institute. This work was financially supported by the following grants: GABI Barlex 0314000 to N.S. and K.F.X.M.; GABI Rye-Express 0315063 from the German Ministry of Education and Research (BMBF) to N.S., K.F.X.M., and E.B.; FP7-212019 TriticeaeGenome from the European Union commission to N.S., K.F.X.M., and J.D.; SFB 924 grant of the Deutsche Forschungsgemeinschaft to K.F.X.M.; and Czech Science Foundation Award P501/12/G090 and the Ministry of Education, Youth, and Sports of the Czech Republic and the European Regional Development Fund (Operational Programme Research and Development for Innovations No. ED0007/01/01) to J.D., M.K., and J.V.
AUTHOR CONTRIBUTIONS
K.F.X.M., E.B., and N.S. designed the research. R.Z., G.H., S.K., B.H., V.K., M.K., and J.V. performed experiments. E.B., G.H., B.H., V.K., T.S., and U.S. contributed data sets and analytical/computational tools. M.M.M., G.H., R.Z., K.G.K., and T.S. performed data analysis. K.F.X.M., M.M.M., R.Z., G.H., C.-C.S., E.B., J.D., and N.S. wrote/edited the article. K.F.X.M. and N.S. contributed equally to this work as joint senior authors. All authors read and approved the article.
Glossary
- RFLP
restriction fragment length polymorphism
- SNP
single nucleotide polymorphism
- CSS
chromosomal survey sequence
- RIL
recombinant inbred line
- SSR
simple sequence repeat
- MDA
multiple-displacement amplified
- fl-cDNA
full-length cDNA
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Belyayev A., Raskina O. (2013). Chromosome evolution in marginal populations of Aegilops speltoides: Causes and consequences. Ann. Bot. (Lond.) 111: 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley R., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491: 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho J.P.M., Sharbel T.F., Beukeboom L.W. (2000). B-chromosome evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 163–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choler P., Erschbamer B., Tribsch A., Gielly L., Taberlet P. (2004). Genetic introgression as a potential to widen a species’ niche: Insights from alpine Carex curvula. Proc. Natl. Acad. Sci. USA 101: 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T.J., et al. (2009). Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10: 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M. (2005). Updating the ‘crop circle’. Curr. Opin. Plant Biol. 8: 155–162 [DOI] [PubMed] [Google Scholar]
- Devos K.M., Atkinson M.D., Chinoy C.N., Francis H.A., Harcourt R.L., Koebner R.M.D., Liu C.J., Masojc P., Xie D.X., Gale M.D. (1993). Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85: 673–680 [DOI] [PubMed] [Google Scholar]
- Doležel J., Bartoš J., Voglmayr H., Greilhuber J. (2003). Nuclear DNA content and genome size of trout and human. Cytometry A 51: 127–128, author reply 129 [DOI] [PubMed] [Google Scholar]
- Doležel J., Greilhuber J., Lucretti S., Meister A., Lysák M.A., Nardi L., Obermayer R. (1998). Plant genome size estimation by flow cytometry: Inter-laboratory comparison. Ann. Bot. (Lond.) 82: 17–26 [Google Scholar]
- Doležel J., Vrána J., Safář J., Bartoš J., Kubaláková M., Simková H. (2012). Chromosomes in the flow to simplify genome analysis. Funct. Integr. Genomics 12: 397–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll C., Sears E. (1971). Individual addition of the chromosomes of ‘Imperial’ rye to wheat. Agronomy Abstracts 6. [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar J.S., Scornavacca C., Cenci A., Guilhaumon C., Santoni S., Douzery E.J., Ranwez V., Glémin S., David J. (2011). Multigenic phylogeny and analysis of tree incongruences in Triticeae (Poaceae). BMC Evol. Biol. 11: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Levy A.A. (2012). Genome evolution due to allopolyploidization in wheat. Genetics 192: 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, H., and Miedaner, T. (2009). Rye breeding. In Handbook of Plant Breeding: Cereals, M. Carena, ed (New York: Springer Science + Business Media), pp. 157–181. [Google Scholar]
- Gustafson J.P., Ma X.-F., Korzun V., Snape J.W. (2009). A consensus map of rye integrating mapping data from five mapping populations. Theor. Appl. Genet. 118: 793–800 [DOI] [PubMed] [Google Scholar]
- Hackauf B., Korzun V., Wortmann H., Wilde P., Wehling P. (2012). Development of conserved ortholog set markers linked to the restorer gene Rfp1 in rye. Mol. Breed. 30: 1507–1518 [Google Scholar]
- Hackauf B., Rudd S., van der Voort J.R., Miedaner T., Wehling P. (2009). Comparative mapping of DNA sequences in rye (Secale cereale L.) in relation to the rice genome. Theor. Appl. Genet. 118: 371–384 [DOI] [PubMed] [Google Scholar]
- Hackauf B., Wehling P. (2005). Approaching the self-incompatibility locus Z in rye (Secale cereale L.) via comparative genetics. Theor. Appl. Genet. 110: 832–845 [DOI] [PubMed] [Google Scholar]
- Haseneyer G., Schmutzer T., Seidel M., Zhou R., Mascher M., Schön C.C., Taudien S., Scholz U., Stein N., Mayer K.F., Bauer E. (2011). From RNA-seq to large-scale genotyping - Genomics resources for rye (Secale cereale L.). BMC Plant Biol. 11: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez P., Martis M., Dorado G., Pfeifer M., Gálvez S., Schaaf S., Jouve N., Šimková H., Valárik M., Doležel J., Mayer K.F.X. (2012). Next-generation sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. Plant J. 69: 377–386 [DOI] [PubMed] [Google Scholar]
- Huang S., Sirikhachornkit A., Faris J.D., Su X., Gill B.S., Haselkorn R., Gornicki P. (2002). Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 48: 805–820 [DOI] [PubMed] [Google Scholar]
- Hunter J. (2007). Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9: 90–95 [Google Scholar]
- Huson D.H., Scornavacca C. (2012). Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61: 1061–1067 [DOI] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium; Mayer K.F., Waugh R., Brown J.W., Schulman A., Langridge P., Platzer M., Fincher G.B., Muehlbauer G.J., Sato K., Close T.J., Wise R.P., Stein N. (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jia J., et al. International Wheat Genome Sequencing Consortium (2013). Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Kellogg E.A., Appels R., Mason-Gamer R.J. (1996). When gene trees tell different stories: The diploid genera of Triticeae. Syst. Bot. 21: 312–347 [Google Scholar]
- Kellogg E.A., Bennetzen J.L. (2004). The evolution of nuclear genome structure in seed plants. Am. J. Bot. 91: 1709–1725 [DOI] [PubMed] [Google Scholar]
- Korzun V., Malyshev S., Kartel N., Westermann T., Weber W.E., Börner A. (1998). A genetic linkage map of rye (Secale cereale L.). Theor. Appl. Genet. 96: 203–208 [Google Scholar]
- Kosambi D. (1944). The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175 [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubaláková M., Valárik M., Barto J., Vrána J., Cíhalíková J., Molnár-Láng M., Doležel J. (2003). Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 46: 893–905 [DOI] [PubMed] [Google Scholar]
- Lander E.S., Waterman M.S. (1988). Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics 2: 231–239 [DOI] [PubMed] [Google Scholar]
- Levy A.A., Feldman M. (2002). The impact of polyploidy on grass genome evolution. Plant Physiol. 130: 1587–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder C.R., Rieseberg L.H. (2004). Reconstructing patterns of reticulate evolution in plants. Am. J. Bot. 91: 1700–1708 [PMC free article] [PubMed] [Google Scholar]
- Ling H.-Q., et al. (2013). Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496: 87–90 [DOI] [PubMed] [Google Scholar]
- Liu C., Atkinson M., Chinoy C., Devos K., Gale M. (1992). Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theor. Appl. Genet. 83: 305–312 [DOI] [PubMed] [Google Scholar]
- Lundqvist A. (1956). Self-incompatibility in rye. I. Genetic control in the diploid. Hereditas 42: 293–348 [Google Scholar]
- Mahelka V., Kopecký D., Paštová L. (2011). On the genome constitution and evolution of intermediate wheatgrass (Thinopyrum intermedium: Poaceae, Triticeae). BMC Evol. Biol. 11: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. (2005). Hybridization as an invasion of the genome. Trends Ecol. Evol. (Amst.) 20: 229–237 [DOI] [PubMed] [Google Scholar]
- Manly K.F., Cudmore R.H., Jr, Meer J.M. (2001). Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930–932 [DOI] [PubMed] [Google Scholar]
- Martis M.M., et al. (2012). Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 109: 13343–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason-Gamer R.J. (2004). Reticulate evolution, introgression, and intertribal gene capture in an allohexaploid grass. Syst. Biol. 53: 25–37 [DOI] [PubMed] [Google Scholar]
- Mason-Gamer R.J., Burns M.M., Naum M. (2010). Reticulate evolutionary history of a complex group of grasses: Phylogeny of Elymus StStHH allotetraploids based on three nuclear genes. PLoS ONE 5: e10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey F.J. (1951). The Kolmogorov-Smirnov test for goodness of fit. J. Am. Stat. Assoc. 46: 68–78 [Google Scholar]
- Matsumoto T., et al. (2011). Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 156: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y. (2011). Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 52: 750–764 [DOI] [PubMed] [Google Scholar]
- Mayer K.F.X., et al. (2011). Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23: 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.F.X., et al. (2009). Gene content and virtual gene order of barley chromosome 1H. Plant Physiol. 151: 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milczarski P., Bolibok-Brągoszewska H., Myśków B., Stojałowski S., Heller-Uszyńska K., Góralska M., Brągoszewski P., Uszyński G., Kilian A., Rakoczy-Trojanowska M. (2011). A high density consensus map of rye (Secale cereale L.) based on DArT markers. PLoS ONE 6: e28495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min X.J., Butler G., Storms R., Tsang A. (2005). OrfPredictor: Predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33 (Web Server issue): W677–W680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G., Devos K.M., Wang Z., Gale M.D. (1995). Cereal genome evolution. Grasses, line up and form a circle. Curr. Biol. 5: 737–739 [DOI] [PubMed] [Google Scholar]
- Murat F., Xu J.-H., Tannier E., Abrouk M., Guilhot N., Pont C., Messing J., Salse J. (2010). Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 20: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo T., Roca A., Gooicoechea P., Giraldez R. (1987). Arm homoeology of wheat and rye chromosomes. Genome 29: 873–882 [Google Scholar]
- Nussbaumer T., Martis M.M., Roessner S.K., Pfeifer M., Bader K.C., Sharma S., Gundlach H., Spannagl M. (2013). MIPS PlantsDB: A database framework for comparative plant genome research. Nucleic Acids Res. 41 (Database issue): D1144–D1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pfeifer M., Martis M., Asp T., Mayer K.F.X., Lübberstedt T., Byrne S., Frei U., Studer B. (2013). The perennial ryegrass GenomeZipper: Targeted use of genome resources for comparative grass genomics. Plant Physiol. 161: 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. (2010). FastTree 2—Aapproximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.L., et al. (2004). A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L.H. (1997). Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 28: 359–389 [Google Scholar]
- Rieseberg L.H., Raymond O., Rosenthal D.M., Lai Z., Livingstone K., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C. (2003). Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216 [DOI] [PubMed] [Google Scholar]
- Salse J., Bolot S., Throude M., Jouffe V., Piegu B., Quraishi U.M., Calcagno T., Cooke R., Delseny M., Feuillet C. (2008). Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Nankaku N., Takeda K. (2009). A high-density transcript linkage map of barley derived from a single population. Heredity (Edinb) 103: 110–117 [DOI] [PubMed] [Google Scholar]
- Schlegel R., Melz G., Nestrowicz R. (1987). A universal reference karyotype in rye, Secale cereale L. Theor. Appl. Genet. 74: 820–826 [DOI] [PubMed] [Google Scholar]
- Schubert I., Lysak M.A. (2011). Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27: 207–216 [DOI] [PubMed] [Google Scholar]
- Sencer H., Hawkes J. (1980). On the origin of cultivated rye. Biol. J. Linn. Soc. Lond. 13: 299–313 [Google Scholar]
- Simková H., Svensson J.T., Condamine P., Hribová E., Suchánková P., Bhat P.R., Bartoš J., Safár J., Close T.J., Doležel J. (2008). Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genomics 9: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.J., Röbbelen G. (1977). Identification by Giemsa technique of the translocations separating cultivated rye from three wild species of Secale. Chromosoma 59: 217–225 [Google Scholar]
- Snowdon R.J. (2007). Cytogenetics and genome analysis in Brassica crops. Chromosome Res. 15: 85–95 [DOI] [PubMed] [Google Scholar]
- Stein N., Prasad M., Scholz U., Thiel T., Zhang H., Wolf M., Kota R., Varshney R.K., Perovic D., Grosse I., Graner A. (2007). A 1,000-loci transcript map of the barley genome: New anchoring points for integrative grass genomics. Theor. Appl. Genet. 114: 823–839 [DOI] [PubMed] [Google Scholar]
- Stutz H. (1972). On the origin of cultivated rye. Am. J. Bot. 59: 59–70 [Google Scholar]
- Suzuki R., Shimodaira H. (2006). Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Thiel T., Graner A., Waugh R., Grosse I., Close T.J., Stein N. (2009). Evidence and evolutionary analysis of ancient whole-genome duplication in barley predating the divergence from rice. BMC Evol. Biol. 9: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T., Michalek W., Varshney R.K., Graner A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106: 411–422 [DOI] [PubMed] [Google Scholar]
- Voorrips R.E. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78 [DOI] [PubMed] [Google Scholar]
- Wicker T., et al. (2011). Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 23: 1706–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox G. (2005). The distribution, natural habitats and availability of wild cereals in relation to their domestication in the Near East: Multiple events, multiple centres. Veget. Hist. Archaeobot. 14: 534–541 [Google Scholar]
- Wu Y., Bhat P.R., Close T.J., Lonardi S. (2008). Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4: e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.