The biosynthesis of secologanin in Madagascar periwinkle involves iridoid assembly in specialized internal phloem-associated parenchyma (IPAP) and final elaboration in leaf epidermal cells. We report the molecular cloning, functional characterization, and IPAP-associated localization of 7-deoxyloganetic acid glucosyltransferase.
Abstract
Iridoids form a broad and versatile class of biologically active molecules found in thousands of plant species. In addition to the many hundreds of iridoids occurring in plants, some iridoids, such as secologanin, serve as key building blocks in the biosynthesis of thousands of monoterpene indole alkaloids (MIAs) and many quinoline alkaloids. This study describes the molecular cloning and functional characterization of three iridoid glucosyltransfeases (UDP-SUGAR GLYCOSYLTRANSFERASE6 [UGT6], UGT7, and UGT8) from Madagascar periwinkle (Catharanthus roseus) with remarkably different catalytic efficiencies. Biochemical analyses reveal that UGT8 possessed a high catalytic efficiency toward its exclusive iridoid substrate, 7-deoxyloganetic acid, making it better suited for the biosynthesis of iridoids in periwinkle than the other two iridoid glucosyltransfeases. The role of UGT8 in the fourth to last step in secologanin biosynthesis was confirmed by virus-induced gene silencing in periwinkle plants, which reduced expression of this gene and resulted in a large decline in secologanin and MIA accumulation within silenced plants. Localization studies of UGT8 using a carborundum abrasion method for RNA extraction show that its expression occurs preferentially within periwinkle leaves rather than in epidermal cells, and in situ hybridization studies confirm that UGT8 is preferentially expressed in internal phloem associated parenchyma cells of periwinkle species.
INTRODUCTION
Madagascar periwinkle (Catharanthus roseus) is biochemically specialized for the production of numerous monoterpenoid indole alkaloids (MIAs), which are a feature of thousands of species from the Apocynaceae, Gentianaceae, Loganiaceae, and Rubiaceae plant families. The medical relevance of periwinkle is attributed to their trace presence in the important anticancer drugs vinblastine and vincristine, which are derived from the coupling of the more abundant MIAs, vindoline and catharanthine, found in aboveground parts of the plant. These dimeric MIAs, as well as a series of chemically modified derivatives, act by disrupting cell division through their binding to microtubules and continue to be used today for the effective treatment of various cancers. Studies to understand why only low levels of dimeric MIAs are produced in periwinkle plants have shown that the catharanthine molecule is exported to the surfaces of leaves and is spatially separated from vindoline, which accumulates within specialized cells in the leaf mesophyll (Roepke et al., 2010).
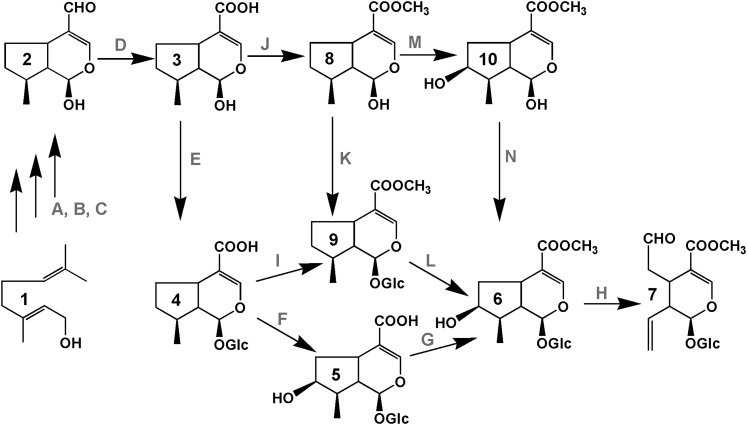
The biosynthetic pathway for assembly of MIAs is also organized so that different cell types are specialized for various parts of the biosynthetic pathway. Biochemically specialized internal phloem-associated parenchyma (IPAP) cells preferentially express the methyl erythritol phosphate pathway (Burlat et al., 2004), which provides geraniol through the action of an IPAP-associated geraniol synthase (Simkin et al., 2013). Within these cells, geraniol is then converted to 10-hydroxygeraniol via the action of geraniol-10-hydroxylase (Burlat et al., 2004) (Figure 1), and after further oxidation to the dialdehyde, a novel iridoid synthase recruited from a short-chain reductase gene family converts this intermediate into cis-trans-nepetalactol/iridoidial (Geu-Flores et al., 2012). The remaining steps involve uncharacterized enzymes (Figure 1D) that convert this intermediate into 7-deoxyloganetic acid (3, Figure 1), followed by glucosylation (Figure 1E) to form 7-deoxyloganic acid (4, Figure 1), hydroxylation (Figure 1F), and carboxy-O-methylation (Figure 1G) to form loganin (6, Figure 1). An alternative pathway has been proposed that involves carboxy-O-methylation (Figure 1J) of 7-deoxyloganetic acid (3, Figure 1) to form 7-deoxyloganetin (8, Figure 1) before glucosylation (Figure 1K) and hydroxylation (Figure 1L). A recent study that supports this latter pathway involved an iridoid glucosyltransferase (Nagatoshi et al., 2011) from gardenia (Gardenia jasminoides) that preferentially glucosylated the 1-O-hydroxyl group of 7-deoxyloganetin (8, Figure 1K) and had no activity toward 7-deoxyloganetic acid (3, Figure 1E). However, the preference of native and recombinant loganic acid O-methyltransferase (LAMT) from periwinkle (Murata et al., 2008) for loganic acid (5, Figure 1G) but not 7-deoxyloganic acid (4, Figure 1I) makes the alternative pathway less likely in periwinkle plants. The last two steps to form secologanin involve LAMT and secologanin synthase (SLS) (Figure 1H), which are preferentially expressed in the leaf epidermis of periwinkle plants, as determined by carborundum abrasion extraction (Murata et al., 2008) and in situ hybridization methods (Guirimand et al., 2011). The formation of secologanin in the leaf epidermis suggests that an undefined iridoid, possibly loganic acid or some earlier intermediate, must be transported to the leaf epidermis for its elaboration, and this cell type is also the site of expression of enzymes such as Trp decarboxylase and strictosidine synthase, which are required to elaborate the formation of strictosidine from which all of the MIAs of periwinkle are derived (Facchini and De Luca, 2008; Guirimand et al., 2011).
Figure 1.
Possible Secologanin Biosynthesis Pathways.
Intermediates in the secologanin biosynthesis pathway: 1, geraniol; 2, iridotrial; 3, 7-deoxyloganetic acid; 4, 7-deoxyloganic acid; 5, loganic acid; 6, loganin; 7, secologanin; 8, 7-deoxyloganetin; 9, 7-deoxyloganin; and 10, loganetin. Enzymes involved in Secologanin biosynthesis: A, Geraniol 10-hydroxylase; B, 10-hydroxygeraniol oxidoreductase; C, iridoid synthase/monoterpene cyclase; D, iridodial oxidoreductase; E, 7-deoxyloganetic acid glucosyltransferase; F, 7-deoxyloganic acid hydroxylase; G, loganic acid methyltransferase; H, SLS; I, 7-deoxyloganic acid methyltransferase; J, 7-deoxyloganetic acid methyltransferase; K, 7-deoxyloganetin glucosyltransferase; L, 7-deoxyloganin hydroxylase; M, 7-deoxyloganetin hydroxylase; N, Loganetin glucosyltransferase. The genes that have been cloned and functionally characterized are in italics in this list and include UGT8 described in this study.
The glucosylation of a range of natural plant products are well known to be catalyzed by family 1 plant secondary product glycosyltransferases (PSPGs) defined by the presence of a 44–amino acid C-terminal motif known as a PSPG box (Vogt and Jones, 2000), which functions as a sugar donor binding pocket. The genipin glucosyltransferase (UDP-SUGAR GLYCOSYLTRANSFERASE2 [UGT2]) gene from gardenia was cloned by isolating a number of genes containing this conserved PSPG box and by its functional characterization in Escherichia coli (Nagatoshi et al., 2011). Using the same approach, this article further elucidates the pathway responsible for formation of secologanin by describing the isolation, biochemical, and molecular characterization of three separate UGTs (UGT6, -7, and -8) that carry out iridoid glucosyltransferase reactions with remarkably different efficiencies. The study identifies the role of UGT8 in secologanin biosynthesis by showing that virus-induced gene silencing (VIGS) reduces expression of this gene and results in a large decline in secologanin and MIA accumulation within silenced plants. Localization studies of UGT8 by the carborundum abrasion method (Murata et al., 2008) that preferentially extracts RNA from the epidermis of periwinkle leaves shows that its expression occurs preferentially within leaves, and in situ hybridization confirms that UGT8 is preferentially expressed of in IPAP cells of periwinkle, where iridoid biosynthesis is initiated.
RESULTS
Molecular Cloning of UGTs from Periwinkle Cell Cultures and Leaves
Total RNA prepared from periwinkle cell cultures was used as the template for RT-PCR cloning of UGTs using primers based on of the conserved amino acid sequence within the PSPG box. Six partial cDNA fragments were obtained with deduced amino acid sequences similar to the C-terminal sequences of various PSPGs in the database but not identical to the UGTs previously isolated from periwinkle (Kaminaga et al., 2004; Masada et al., 2009). Using these partial cDNA fragments, we obtained two full-length cDNAs by 5′-rapid amplification of cDNA ends (RACE) and designated them as periwinkle UGT6 and UGT7. In addition, EST database mining of a periwinkle PlantGDB (Resources for Plant Comparative Genomics, http://www.plantgdb.org/) database with an iridoid-specific glucosyltransferase from gardenia (Nagatoshi et al., 2011; Gj-UGT2) identified 30 putative UGT cDNA contigs. Among these, eight contigs (see Supplemental Table 1 online) associated with group G PSPGs, where Gj-UGT2 belongs. Based on the sequences of these eight contigs, we tried to obtain additional UGT cDNAs by RACE from RNA isolated from periwinkle leaves. This approach led to the isolation of a full-length cDNA clone (UGT8) corresponding to the 596-bp contig Cr8440 (see Supplemental Table 1 online, bold). Sequence analysis of UGT8 revealed that contigs Cr9886 and Cr441 (see Supplemental Table 1 online, bold) encoded different parts of UGT8. This approach did not lead to the cloning of other full-length UGT cDNAs corresponding to the other five contigs described in Supplemental Table 1 online. An identical contig (CROWL1VD) to Cr8440 was also identified from the Phytometasyn periwinkle database (http://www.phytometasyn.ca), but sequences corresponding to UGT6 and UGT7 were not found. The sequences in the Phytometasyn database were generated from RNA isolated from the youngest first leaf pairs of periwinkle leaves.
Phylogenetic analyses based on the deduced amino acid sequences of Gr-UGT6, Cr-UGT7, and Cr-UGT8 suggested that while Cr-UGT6 and Cr-UGT8 both belonged to group G, Cr-UGT7 belonged to group H of Family 1 PSPGs (see Supplemental Figure 1 online). Functionally characterized members of group G are involved in the biosynthesis of the iridoid geniposide (Gj-UGT2) in gardenia, cyanogenic glucosides (UGT85B1) in sorghum (Sorghum bicolor), cytokinins (UGT85A1) in Arabidopsis thaliana, and in an undisclosed reaction in Lonicera japonica (Lj-UGT12). Members of group H are involved in the biosynthesis of steviosides (UGTG1) in Stevia rebaudiana, cytokinins (UGTC1) in Arabidopsis, and benzoaxizones (Zea Bx8) in maize (Zea mays). The amino acid sequence comparisons showed 78, 33, and 40% sequence identity of Cr-UGT6, Cr-UGT7, and Cr-UGT8, respectively with the gardenia iridoid-specific glucosyltransferase (Gj-UGT2 or UGT85A24). While these results might suggest that the best candidate for an iridoid-specific GT was UGT6, we decided to functionally characterize all three GTs in order to compare their biochemical properties and their substrate specificities.
Functional Characterization of Recombinant UGTs
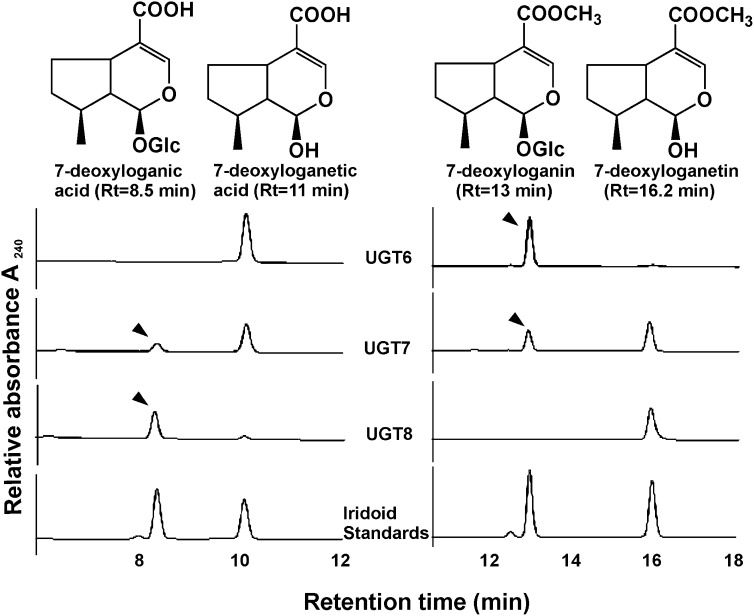
To examine the catalytic function of UGT6-8, their open reading frames were expressed in E. coli as N-terminal fusion proteins with a His6-tag. After purifying each recombinant UGT by nickel-nitrilotriacetic acid affinity chromatography, they were assayed for their O-glucosyltransferase activity using 7-deoxyloganetic acid and 7-deoxyloganetin as acceptor substrates in the presence of the UDP-Glc donor (Figure 2). UGT6 rapidly and efficiently converted 7-deoxylaganetin to a product with an identical retention time and UV spectrum as 7-deoxyloganin, whereas no reaction product was detected with 7-deoxyloganetic acid as substrate. UGT8 effectively produced 7-deoxyloganic acid from 7-deoxylaganetic acid, while no such conversion was detected when 7-deoxyloganetin was used as a sugar accepting substrate. By contrast, small amounts of products were detected when UGT7 was incubated with either 7-deoxyloganetic acid or 7-deoxyloganetin.
Figure 2.
Differential Conversions of 7-Deoxyloganetic Acid and 7-Deoxyloganetin by Recombinant UGT6, UGT7, and UGT8 to 7-Deoxyloganic Acid and 7-Deoxyloganin.
Iridoid substrates (1 mM) were incubated with each rUGT in the presence of 5 mM UDP-Glc for 2 h at 30°C, and the reaction mixture was subjected to HPLC analysis as described in Methods. The arrowheads indicate the respective reaction products (7-deoxyloganic acid or 7-deoxyloganin) being made when recombinant enzymes were incubated with their respective iridoid substrates.
The glucosyl acceptor specificities of UGT6-8 were tested against a broader range of iridoids, phenolics, flavonoids, and one hormone as glucosyl acceptor substrates (see Supplemental Figure 2 online). UGT6 and UGT8 exhibited strict substrate specificity toward 7-deoxyloganetin and 7-deoxyloganetic acid, respectively. By contrast, UGT7, which is a weak iridoid glucosyltransferase (Figure 2), did glucosylate a broader range of substrates, including curcumin, genistein, luteolin, and kaempferol. Finally, kinetic parameters of UGT6-8 for 7-deoxyloganetic acid and 7-deoxyloganetin were determined based on pseudo-single substrate kinetics using UDP-Glc as a sugar donor substrate (Table 1). The apparent Km, kcat values for 7-deoxloganetin of UGT6 and UGT7 were 0.142 mM, 0.0355 s−1, and 1.99 mM, 0.00493 s−1, respectively, giving a 76-fold higher kcat/Km ratio for UGT6 than for UGT7. The apparent Km, kcat values for 7-deoxloganetic acid of UGT8 were 0.088 mM, 0.130 s−1 giving an 18-fold higher kcat/Km ratio of UGT8 for 7-deoxyloganetic acid compared with that of UGT6 for 7-deoxyloganetin. By contrast, the kcat/Km ratios of UGT6, UGT7, and UGT8 in the presence of their respective iridoid substrates (Table 1) were 291.8, 42, and 64.4, respectively. Together, these analyses suggest that while UGT8 had an 8.5-fold higher catalytic efficiency toward its exclusive iridoid substrate, 7-deoxyloganetic acid, than those of the other two UGTs (Table 1), UGT6 had a 4.5-fold higher catalytic efficiency toward its Glc donor. Unfortunately, we were not able to obtain the 7-deoxyloganetic acid kinetic parameters for UGT7 because of the weak activity of the recombinant enzyme toward this substrate.
Table 1. Kinetic Parameters of rUGT6∼8 toward Iridoid Aglycones and UDPG.
| Kinetic Parameters |
|||
|---|---|---|---|
| Protein | Km (mM) | kcat (s−1) | kcat/Km (M−1 s−1) |
| 7-Deoxyloganetic Acid | |||
| UGT6 | – | – | – |
| UGT7 | * | * | * |
| UGT8 | 0.088 ± 0.022 | 0.130 ± 0.015 | 1523.3 ± 222.7 |
| 7-Deoxyloganetin | |||
| UGT6 | 0.202 ± 0.030 | 0.0355 ± 0.0068 | 179.9 ± 54.5 |
| UGT7 | 1.99 ± 0.74 | 0.00493 ± 0.00303 | 2.35 ± 0.58 |
| UGT8 | – | – | – |
| UDP-Glc | |||
| UGT6 | 0.117 ± 0.025 | 0.0320 ± 0.0172 | 291.8 ± 103.3 |
| UGT7 | 0.120 ± 0.013 | 0.00512 ± 0.00169 | 42.0 ± 10.4 |
| UGT8 | 5.38 ± 1.99 | 0.325 ± 0.051 | 64.4 ± 16.2 |
Each data set represents the mean ± sd from triplicate measurements. The purity of the UGTs used in the kinetic analyses was confirmed by Coomassie blue staining of gels after SDS-PAGE, which made it possible to estimate kcat. The alternative substrate concentrations used for UDP-Glc or iridoids were 5 and 0.5 mM, respectively, for saturation curves. –, No activity; *, activity too weak for kinetic assay.
Preferential Expression of UGTs in Plant Organs and Leaf Cells
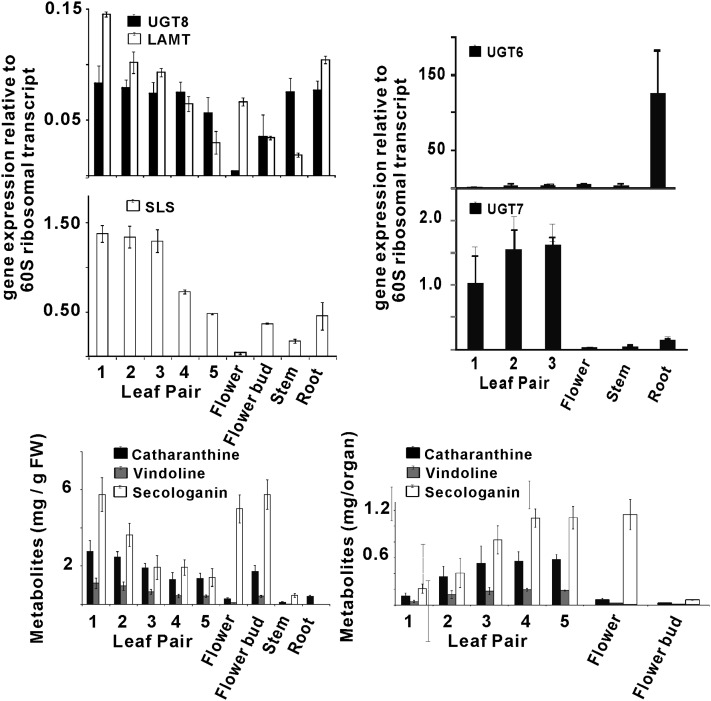
The transcript levels of UGT6-8 were determined in periwinkle plant organs (Figure 3) by real-time RT-PCR, and their relative abundance was compared with the levels of secologanin, catharanthine, and vindoline (Figure 3) present in each organ. Expression of UGT6 occurs preferentially in roots, while UGT7 transcripts were substantially more abundant in leaf pairs 1, 2, and 3 compared with the low levels observed in roots, stems, and flowers. The expression of UGT8 transcripts was detected mostly in leaves, roots, and stems with lower levels occurring in flowers. The transcript level of UGT8 was highest in the youngest leaves (Figure 3, leaf pairs 1 to 3) and gradually decreased in the older leaves (Figure 3, leaf pairs 4 and 5). The patterns of expression of UGT8 were very similar to those of LAMT and SLS (Figure 3), which is consistent with the significant levels of secologanin found in most periwinkle organs (Figure 3). The results also confirm that only part of the secologanin produced is incorporated into MIAs, which are preferentially biosynthesized in younger tissues (leaf pairs 1 and 2 and in root tips; Murata et al., 2008; Roepke et al., 2010) The accumulation of MIAs (catharanthine and vindoline) reaches a maximum in leaf pair 3 (Figure 3, compare mg/g fresh weight and mg/organ; Roepke et al., 2010), while secologanin continues to accumulate, reaching a maximum in leaf pair 4 (Figure 3, see mg/organ). Similarly, the levels of secologanin increase at least 10-fold in fully open flowers compared with flower buds (Figure 3, see mg/organ).
Figure 3.
Differential Expression of UGT8 Correlates with That of the Last Two Steps in Secologanin Biosynthesis, along with Iridoid and MIA Metabolite Profiles in Periwinkle Plant Organs.
Relative gene expression of UGT6, UGT7, UGT8, LAMT, and SLS was determined by quantitative RT-PCR analyses performed on total RNA extracted from periwinkle leaf pairs 1 to 5, from open flowers and flower buds, and from root tissues. Each point represents the mean of relative transcript abundance to RPPOC (gene encoding 60S acidic ribosomal protein P0-C) ± sd from at least triplicate measurements of biological and technical replicates. Metabolite (catharanthine, vindoline, and secologanin) levels are plotted as mg/gram fresh weight (FW) and as mg/organ. Each box and bar represents an average value and a se, respectively, from four different plant samples.
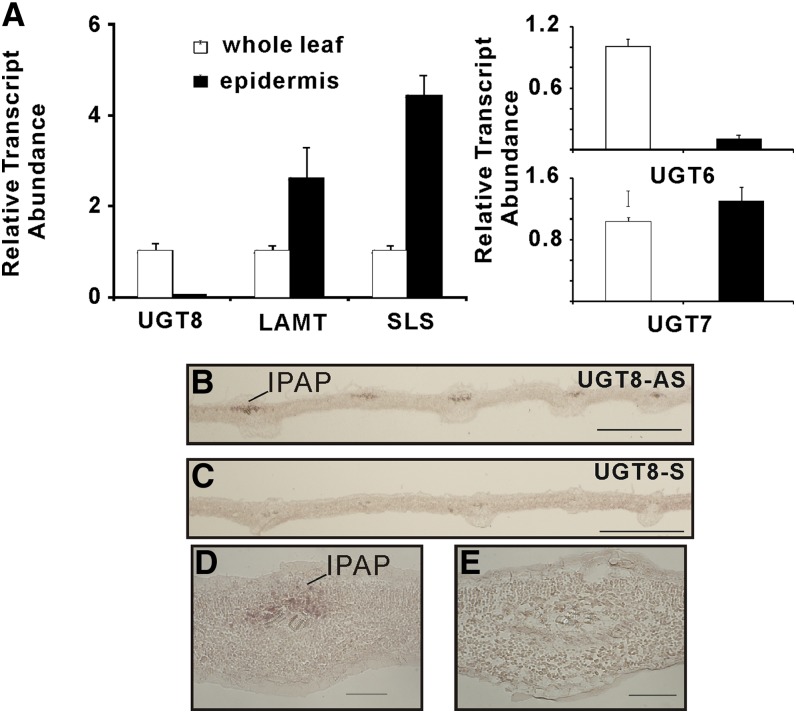
The carborundum abrasion technique has been used successfully to extract leaf epidermis-enriched RNA from periwinkle and to corroborate MIA pathway gene expression in these cells (Levac et al., 2008; Murata et al., 2008). RT-PCR analysis of leaf epidermis enriched transcripts showed that the levels of UGT6 and UGT8 were at least 10-fold lower than those found in whole leaves, while UGT7 transcripts were more equally distributed (Figure 4A). By contrast, transcripts for LAMT and SLS, well known to be preferentially expressed in the leaf epidermis (Murata et al., 2008; Guirimand et al., 2011), were two- to fourfold more enriched in these cells. Together, these results suggested that both UGT-6 and -8 are preferentially expressed within periwinkle leaves rather than in leaf epidermal cells, where the last two steps in secologanin assembly are expressed.
Figure 4.
Localization of UGT8 Transcripts in IPAP Cells of Young Developing Leaves of Periwinkle.
(A) Relative expression of UGT6, UGT7, UGT8, LaMT, and SLS in relation to RPP0C reference gene in leaf epidermis enriched transcript extracted by carborundum abrasion compared with those found in whole-leaf extracts. Each point represents the mean of relative transcript abundance to RPPOC ± sd from at least triplicate measurements of biological and technical replicates.
(B) to (E) Localization by in situ hybridization of UGT8 mRNA in young developing leaves of C. longifolius. Serial longitudinal 10-µm sections made from young leaves (10 to 15 mm long) were hybridized with UGT8-antisense ([B] and [D]) and UGT8-sense ([C] and [E]) probes. Bars = 500 µm in (B) and (C) and 50 µm in (D) and (E).
[See online article for color version of this figure.]
UGT8 Is Preferentially Expressed in Internal Phloem Parenchyma of Periwinkle Leaves
The high catalytic efficiency of UGT8 toward its exclusive iridoid substrate, 7-deoxyloganetic acid (Table 1), and its expression within periwinkle leaves rather than in leaf epidermis prompted in situ hybridization studies to localize where this gene is preferentially expressed. Very young developing leaves of periwinkle and Catharanthus longifolius were harvested, fixed, embedded, sectioned, and prepared for in situ RNA hybridization analysis to localize transcripts of UGT8 (Figure 4). Labeling with antisense UGT8 probes was restricted to the adaxial phloem region in C. longifolius tissues surrounding the vasculature on longitudinal leaf sections (Figures 4B and 4D), while no significant labeling was observed when sense UGT8 probes were used in negative control experiments (Figures 4C and 4E). Unfortunately, the same experiments performed with periwinkle did not produce any labeling of the same sections, perhaps because of the severalfold lower abundance of UGT8 transcript found compared with the levels occurring young leaves of C. longifolius. It should be noted that the DNA sequence of C. longifolius UGT8 is 100% identical to that of Cr-UGT8 (see Supplemental Figure 1 online). These results strongly suggested that that UGT8 is expressed in internal phloem parenchyma cells, which also preferentially express the 2-C-methyl-d-erythritol 4-phosphate pathway (Burlat et al., 2004), geraniol synthase (Simkin et al., 2013), geraniol-10 hydroxylase (Burlat et al., 2004), and iridoid synthase (Geu-Flores et al., 2012).
VIGS of UGT8, LAMT, and SLS Triggers Large Declines in Accumulation of Secologanin and of MIAs in Silenced Periwinkle Leaves
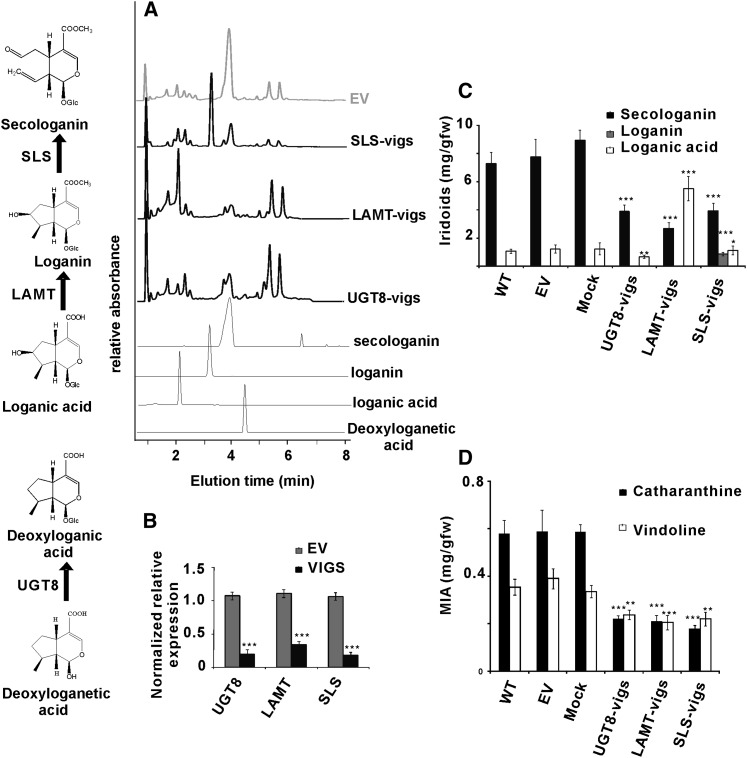
The strict specificity of UGT8 for 7-deoxyloganetic acid (Figure 2; see Supplemental Figure 2 online), its higher catalytic efficiency than the other UGTs (Table 1), its coordinate expression with LAMT and with SLS together with the accumulation of metabolites in relevant periwinkle tissues (Figure 3), and its preferred expression in IPAP cells of periwinkle leaves (Figure 4) prompted us to use VIGS for validating the role of UGT8 in iridoid biosynthesis in periwinkle (Figure 5). Fragments of UGT8, LAMT, and SLS genes (see Supplemental Table 2 online) were cloned independently into Tobacco Rattle Virus 2 (TRV2) vectors. A mixed culture of Agrobacterium tumefaciens harboring pTRV1 and either pTRV2 empty vector (pTRV2-EV) or pTRV2 constructs (pTRV2-UGT8, pTRV2-LAMT, and pTRV2-SLS) was infiltrated into the apical meristem of young periwinkle plants (De Luca et al., 2012a). Separate experiments suppressing the phytoene desaturase gene (see Supplemental Table 2 online) produced a visible photobleached phenotype, which was monitored as a visible marker for determining the timing in which to perform transcript and metabolite analyses in plants with suppressed UGT8, LAMT, and SLS expression. In order to confirm the success of Agrobacterium infiltration, plants were selected based on the detection of a 134-bp fragment derived from the presence of the pTRV2-derived TRV coat protein transcript (see Supplemental Figure 3 online). Periwinkle plants suppressed for each transcript showed large declines in secologanin accumulation as determined by ultra performance liquid chromatography-mass spectrometry (UPLC-MS) (Figure 5A). Plants suppressed for UGT8 did not show an increase in detectable deoxyloganetic acid (Figure 5A), while those suppressed for LAMT or for SLS both showed large increases in detectable loganic acid and loganin accumulation, respectively. These results were verified by monitoring the relative transcript levels of UGT8, LAMT, and SLS by quantitative RT-PCR. The transcript levels declined by 70 to 80% (P value of <0.001) in UGT8-, LAMT-, and SLS-vigs leaf tissues compared with EV controls (Figure 5B). More detailed metabolite analyses of UGT8-, LAMT-, and SLS-silenced tissues showed that they responded with a >50% decline of secologanin accumulation (Figure 5C), with LAMT- and SLS-silenced tissues responding with increased accumulation of their respective substrates loganic acid and loganin (Figure 5C), compared with periwinkle control, EV, and mock-inoculated tissues. More detailed MIA analyses of UGT8-, LAMT-, and SLS-silenced tissues revealed that they all responded with a >50% decline in catharanthine accumulation (Figure 5D), along with smaller (30 to 40%) declines in vindoline levels, compared with periwinkle control, EV, and mock-inoculated tissues.
Figure 5.
Downregulation of UGT8, LAMT, and SLS Affects the Accumulation of Iridoids and MIAs in Periwinkle.
(A) Silencing of UGT8, LAMT, and SLS was conducted by monitoring the iridoid metabolite profiles by UPLC-MS in silenced plants (UGT8-vigs, LAMT-vigs, and SLS-vigs) compared with the profiles obtained with plants treated with EV controls. UPLC-MS analysis of iridoid profiles were detected at A240 and by RTs related to iridoid standards: deoxyloganetic acid (RT = 4.68 min; m/z = 197), loganic acid (RT = 1.90 min; m/z = 377), loganin (RT = 3.1 min; m/z =391), and secologanin (RT = 3.88 min; m/z = 389).
(B) Silencing of UGT8, LAMT, and SLS was measured by monitoring relative transcript abundance of each iridoid pathway gene by quantitative RT-PCR. Differences in transcript levels for each silenced gene were measured relative to those obtained in EV and mock treatments and are represented as mean ± se. Gene-specific primers for each UGT8, LAMT, and SLS were used for comparison of transcript abundance between EV and for each VIGS treatment. The data represent measurements performed with six biological replicates (with three technical replicates per biological replicate) of mock, EV, UGT8-vigs, LAMT-vigs, and SLS-vigs treatments.
(C) and (D) Measurements of iridoids (loganic acid, loganin, and secologanin) (C) and MIAs (catharanthine and vindoline) (D) in untreated (wild type [WT]), EV, mock, UGT8-vigs, LAMT-vigs, and SLS-vigs treated periwinkle plants were performed with the same six biological replicates used for transcript analysis in (B). Significant differences were considered with *P < 0.05, **P < 0.01, and ***P < 0.001 by Student’s t test for the transcript analysis and metabolite contents of EV-infected plants and in each of the silenced lines. fw, fresh weight.
DISCUSSION
Periwinkle Contains UGTs, Which Glucosylate Iridoids with Varying Efficiencies
Three functional UGT clones were obtained from cell suspension culture (UGT6 and UGT7) and leaf (UGT8) RNA preparations of periwinkle based on the conserved PSPG box of this family of glucosyltransferases and sequence information from the only characterized iridoid glucosyltransferase from gardenia (Nagatoshi et al., 2011; UGT2). While UGT6 and UGT8 belong to Group G of Family 1 PSPGs (see Supplemental Figure 1 online), UGT7 aligns with group H of this family. The biochemical function of UGT6, whose amino acid sequence is 78% identical to that of Gj-UGT2, is most similar to that of the gardenia enzyme, which also shows strict specificity for 7-deoxyloganetin rather than 7-deoxyloganetic acid. By contrast, UGT8, whose amino acid sequence is 40% identical to that of Gj-UGT2, only used deoxyloganetic acid as a substrate, and this enzyme possessed a high catalytic efficiency for its substrate compared with the other identified iridoid GTs (Table 1). Inspection of the Phytometasyn periwinkle database (http://www.phytometasyn.ca) also identified genes that are identical to Cr-UGT8 (see Supplemental Figure 1 online), for example, genes from Catharanthus ovalis (Co-UGT8) and C. longifolius (Cl-UGT8), as well as genes closely corresponding to Cr-UGT8, including genes from Tabernamonana elegans (Te-UGT8), Amsonia hubrichtii (Ah-UGT8), Rauvolfia serpentina (Rs-UGT8), and Vinca minor (Vm-UGT8). In addition, similar candidate genes were readily identified in databases of L. japonica (Lj-UGT8, which produces secologanin) and Cinchona ledgeriana (Cl-UGT8, which produces quinoline alkaloids). The presence of such highly similar UGTs in each plant species that produce iridoids and MIAs provides suggestive evidence for the key role played by UGT8 in secologanin biosynthesis in periwinkle and, more generally, in members of the Apocynaceae family. However, direct evidence for this should be obtained by functionally characterizing a few more UGT8 genes from some of these other species. Furthermore, this result does not preclude the involvement of a UGT6-based pathway that may play a role in iridoid biosynthesis under the appropriate environmental circumstances within these plant species, as was already shown in the biosynthesis of geniposide in gardenia, where the alternative pathway appears to predominate (Nagatoshi et al., 2011). The existence of independent iridoid UGTs that glucosylate 7-deoxyloganetic acid (UGT8) and 7-deoxyloganetin (UGT6) in periwinkle suggests that these functionalities have evolved by convergent evolution and raises the question of whether a 7-deoxyloganetic acid GT similar to UGT8 may also exist in gardenia. In this context, the biochemical characterization of UGT6, -7, and -8 showed that UGT6 may not be the functional homolog for the preferred pathway in periwinkle.
Transcriptional Downregulation of UGT8 by VIGS Suppresses Secologanin and MIA Accumulation in Periwinkle Plants
VIGS technology has been successfully exploited, together with modern bioinformatic approaches, to select and identify candidate genes that may be involved in the biosynthesis of particular metabolites in plants (De Luca et al., 2012b). The use of VIGS has been validated in periwinkle, where VIGS-mediated suppression of 16-methoxy-2,3-dihydro-3-hydroxytabersonine N-methyltransferase resulted in plants that stopped accumulating vindoline in favor of 16-methoxy-2,3-dihydro-3- hydroxytabersonine (Liscombe and O’Connor, 2011). VIGS suppression of iridoid cyclase also led to plants containing reduced levels of catharanthine and vindoline (Geu-Flores et al., 2012). In this study, the silencing of UGT8, LAMT, and SLS decreased the levels of each respective transcript by 70 to 80% (Figure 5B) and to general declines (Figure 5D) of >50% in cantharanthine levels and 30 to 40% declines in vindoline levels in silenced leaves. Silencing of UGT8, LAMT, and SLS also greatly decreased the levels of secologanin by more than 50% in silenced lines (Figure 5C). Silencing of LAMT and SLS resulted in the accumulation of considerable amounts of their respective substrates loganic acid (5.21 mg/g fresh weight) and loganin (0.77 mg/g fresh weight) (Figure 5C). This clearly illustrates how such silencing events could be used to supply substrates that may not be available commercially or that may be difficult to produce through organic chemistry. Silencing of UGT8 did not lead to the accumulation of its substrate, deoxyloganetic acid (Figure 5C), perhaps due to the increased reactivity of the aglycone or due to its conversion to other end products that were not detected under our assay conditions.
UGT8 Catalyzes the Fourth to Last Step in Secologanin Biosynthesis within IPAP Cells of Periwinkle
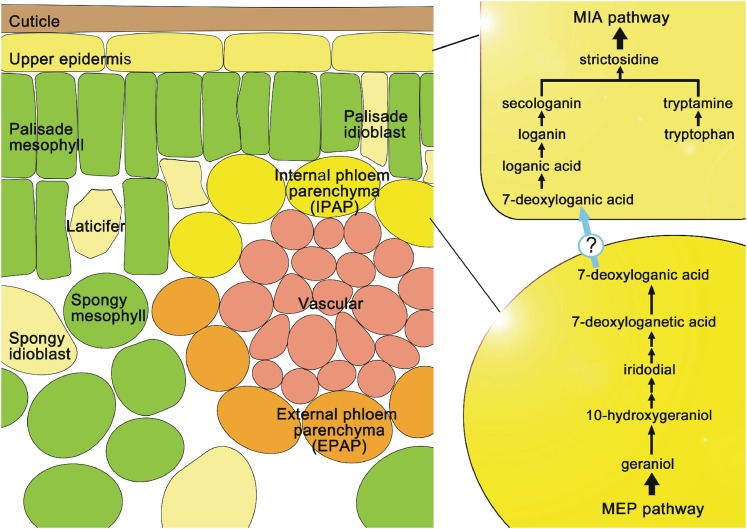
Many of the steps involved in secologanin biosynthesis from the 2-C-methyl-d-erythritol 4-phosphate/1-deoxy-d-xylulose 5-phosphate (MEP) pathway have been identified at the biochemical and molecular level, including GS, G10H (Figure 1A), a putative 10-hydroxygeraniol oxidoreductase (Figure 1B), iridoid synthase (Figure 1C), LAMT (Figure 1G), and SLS (Figure 1H). The most recent molecular and biochemical characterization of iridoid synthase showed it to be a progesterone-5β-reductase–like gene that catalyzed a unique reductive cyclization in iridoid biosynthesis and was preferentially expressed in IPAP cells (Geu-Flores et al., 2012). This study provides strong evidence that UGT8 catalyzes the fourth to last step in secologanin biosynthesis (Figure 1, reactions A to H) and is also preferentially expressed within IPAP cells (Figure 4). Although nothing is known about the hydroxylation step that converts 7-deoxyloganic acid to loganic acid, the model shown in Figure 6 suggests that this metabolite may be transported to the leaf epidermis to be converted into loganic acid followed by methylation (LAMT) and oxidative ring opening (SLS) in the leaf epidermis. The molecular cloning of the hydroxylase responsible for the third to last step in vindoline biosynthesis should provide tools to identify its presence in IPAP or epidermal cells of periwinkle leaves.
Figure 6.
Spatial Model of Iridoid Biosynthesis and Translocation in Periwinkle Leaves.
The MEP pathway and iridoid biosynthesis to 7-deoxyloganic acid occurs in IPAP cells, while the terminal LAMT and SLS reactions occur in the leaf epidermis. The model shows 7-deoxyloganic acid hydroxylase in leaf epidermis, but this reaction and its location still remain to be elucidated. The model shows that MIA assembly from secologanin and tryptamine also takes place in leaf epidermal cells. Solid lines represent a single enzymatic step, whereas double arrows indicate the involvement of multiple enzyme steps. The question mark indicates the putative transport system of 7-deoxyloganic acid from IPAP cells to leaf epidermal cells.
[See online article for color version of this figure.]
In conclusion, three separate UGTs with distinct substrate specificities and catalytic efficiencies have been described in this study. The optimal catalytic and biochemical properties of UGT8 and its preferred expression in IPAP leaf cells of periwinkle, together with the loss of iridoids and MIAs in VIGS silenced plants, strongly suggest its key role as a biosynthetic enzyme in the assembly of secologanin. These combined approaches of using bioinformatics, silencing technologies, and functional characterization of candidate genes is revolutionizing the gene discovery process for mining the chemical diversity of plants (De Luca et al., 2012b; Facchini et al., 2012; Góngora-Castillo et al., 2012; Xiao et al., 2013).
METHODS
Plant Materials
Madagascar periwinkle (Catharanthus roseus) and Catharanthus longifolius plants were grown in either a greenhouse or an incubator at 25°C under a 16-h photoperiod. Cell suspension cultures of periwinkle were originally established from seedling-derived callus and maintained in LS medium (Linsmaier and Skoog, 1965) supplemented with 3% Suc, 1 μM 2,4-D, and 1 μM kinetin. The cells were cultured on a rotary shaker (100 rpm) at 25°C in the dark and subcultured at 2-week-intervals.
Chemicals
7-Deoxyloganin tetraacetate was kindly provided by K. Inoue (Yokohama College of Pharmacy). 7-Deoxyloganin and 7-deoxyloganic acid were prepared from 7-deoxyloganin tetraacetate according to the method described previously (Nagatoshi et al., 2011). Loganetin, 7-deoxyloganetin, and 7-deoxyloganetic acid were enzymatically prepared from loganin, 7-deoxyloganin, and 7-deoxyloganic acid, respectively, as follows. A 20-mg aliquot of each glucoside was dissolved in 10 mL of 50 mM phosphate-citrate buffer, pH 4.8, containing 50 units/mL almond β-glucosidase (Sigma-Aldrich) and incubated for 3 h at 37°C. After centrifugation, the reaction mixture was extracted with ethyl acetate three times. The combined ethyl acetate extract was concentrated under reduced pressure to yield the aglycone. The purity of each aglycone thus obtained was estimated by quantitative 1H NMR analysis (Hasada et al., 2011). Geniposide, genipin, and loganin were obtained from Wako Pure Chemicals and secologanin from Sigma-Aldrich. Iridotrial was synthesized by N. Kato (Nagoya City University). All other chemicals were of commercial reagent-grade quality unless otherwise stated.
Homology-Based Cloning of UGTs
Total RNA was isolated from periwinkle cultured cells or leaves using an RNeasy plant mini kit (Qiagen). RT-PCR was performed using a CapFishing full-length cDNA premix kit (Seegene). Two degenerate primers UGT2mFw (5′-TTYBTIWSICAYTGYGGITGGAA-3′) and PSPG2Fw (5′-TGYGGITGGAAYTCIRYIYTIGA-3′) were designed based on the highly conserved amino acid sequences Phe(Leu/Val)(Thr/Ser)HisCysGlyTrpAsn and CysGlyTrpAsnSer(Thr/Val)LeuGlu, respectively, in the PSPG box of plant glucosyltransferases (Vogt and Jones, 2000). A 5-μL aliquot of the cDNA was used as a template for PCR amplification in a 50-μL reaction mixture containing 1 μM primer UGT2mFw, 0.2 μM 3′-RACE primer from the CapFishing kit, and 25-μL SeeAmp TaqPlus Master Mix (Seegene). A portion of the first PCR product was used as the template for nested PCR using the PSPG2Fw and the 3′-RACE primers. PCR was performed under the following conditions: denaturation at 94°C for 3 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 45°C for 1 min, extension at 72°C for 1 min, and final extension at 72°C for 5 min. Amplified products of ∼500 bp were recovered from agarose gel and subcloned into the pMD20 T-vector (Takara). Randomly selected cloned inserts were sequenced using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) with a PRISM 3130 genetic analyzer (Applied Biosystems). The 5′-ends of these cDNAs were obtained using the gene-specific primers and the 5′-RACE primer from the CapFishing kit. The full-length cDNA clones (UGT6 and UGT7) were amplified and sequenced using the 5′- and 3′-sequences as specific primers.
Cloning of UGTs by EST Database Screening
Periwinkle EST assemblies on the PlantGDB server (http://plantgdb.org/cgi-bin/blast/PlantGDBblast) were mined using TBLASTN with the Gj-UGT2 amino acid sequence as a query. From an initial list of UGT candidates obtained by BLASTX search, the contigs belonging to the group G of PSPGs (Nagatoshi et al., 2011) were manually selected to provide the master candidate list, as shown in Supplemental Table 3 online. We designed gene-specific primers and isolated a full-length cDNA corresponding to the contig Cr8440 as UGT8.
The nucleotide sequences of PCR primers used for RACE PCR and amplification of full-length cDNAs of UGTs are shown in Supplemental Tables 4 and 5 online, respectively.
Heterologous Expression of UGTs
The UGT6, -7, and -8 cDNA fragments containing their respective open reading frames were amplified by PCR with Thermococcus kodakaraensis FX DNA polymerase (Toyobo) using the PCR primers containing appropriate restriction sites (see Supplemental Table 6 online). The PCR products were cloned into the pMD20 T-vector to confirm their sequences and then subcloned into pQE-30 vector (Qiagen) to create N-terminal fusion proteins with a His6 tag. Escherichia coli strain JM109 was used as the host for expression. Transformed cells were cultured at 37°C until the absorbance at 600 nm reached 0.6 and then further incubated overnight at 30°C before harvest. The recombinant proteins were affinity purified on a nickel-nitrilotriacetic acid agarose matrix (Qiagen) according to the manufacturer’s instructions. Protein content in the enzyme preparations was estimated using the method of Bradford (1976).
Enzyme Assays
The standard reaction mixture for the enzyme assays (total volume of 50 μL) contained 50 mM Tris-HCl, pH 8.0, 5 mM UDP-Glc, 1 mM acceptor substrates, and the enzyme preparation. The reaction was performed at 30°C and then terminated by the addition of 100 μL of methanol. After centrifugation at 12,000g for 5 min, the reaction products were analyzed by reversed-phase HPLC, and elution was monitored using a photodiode array detector. To separate the substrates and their glucosides, the following gradient elution programs were used: for iridoid glucosides, 0 to 16 min, 30 to 60% methanol and 16 to 19 min, 60% methanol; for flavonoids, coumarins, and various phenolic glucosides, 0 to 26 min, 15 to 52% acetonitrile, 26 to 29 min, 52 to 100% acetonitrile, and 29 to 33 min, 100% acetonitrile. To determine the kinetic parameters, enzyme assays were performed in triplicate at each substrate concentration with 10 to 25 μg of the purified enzyme at 30°C for 10 min. The substrate concentrations used were 0.01 to 5 mM sugar acceptors with UDP-Glc at 5 mM for acceptor kinetics and 0.25 to 10 mM UDP-Glc with 7-deoxyloganetic acid or 7-deoxyloganetin at 0.5 mM for donor kinetics. The initial velocity data were visualized by Lineweaver-Burk plots, and kinetic parameters were calculated based on linear regression analysis using Excel 2007 (Microsoft Japan).
Analysis of Gene Expression by Quantitative RT-PCR
Total RNA was prepared from leaves, stems, flowers, and roots of periwinkle using an RNeasy plant mini kit. Leaf epidermis–enriched RNA was prepared by the carborundum abrasion technique as described previously (De Luca et al., 2012b). Depending on the experiment, three to six biological replicates were extracted and three technical replicates were performed for measurement of gene expression. First-strand cDNAs for real-time PCR were synthesized from 0.5 µg of total RNA using SuperScript III RNase H− reverse transcriptase (Invitrogen). Real-time PCR was performed with the 7300 real-time PCR system (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions. Briefly, the reaction mixture consisted of cDNA template, 10 pmol primers, and 10 μL of Power SYBR Green PCR Master Mix in a total volume of 20 µL. The standard PCR condition was as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. Gene-specific primers for UGT6, -7, and -8 are listed in Supplemental Table 7 online.
Phylogenetic Analyses
Sequence alignments were generated based on a comparison of the amino acid sequences using the ClustalW program (Thompson et al., 1994) with the following values: 10 for gap opening penalty, 0.1 for gap extension penalty in pairwise alignment, 10 for gap opening penalty, and 0.2 for gap extension penalty in multiple alignment, Gonnet for protein weight matrix, 4 for gap separation distance, residue-specific penalties, hydrophilic penalties, and 30% delay divergent cutoff. These alignments were used to construct the neighbor-joining unrooted phylogenetic trees using MEGA 5.1 (Tamura et al., 2011) with scope of all selected taxa, amino acid substitution type following Poisson model, uniform rates, homogenous pattern among lineages, and complete deletion for gaps or missing data treatment. The scale bar of 0.1 indicates a 10% change, and each number shown next to the branches is the percentage of replicate trees in which the related taxa clustered in the bootstrap test with 10,000 replicates. The accession numbers of amino acid sequences used for this phylogenetic analysis are listed in Supplemental Table 3 online, and the alignments are shown in Supplemental Table 8 online.
VIGS
VIGS was performed as described previously (Liscombe and O’Connor, 2011) and as modified by De Luca et al. (2012b). Fragments of PHYTOENE DESATURASE (460 bp), Cr-UGT8 (349 bp), LAMT (373 bp), and SLS (359 bp) were amplified by PCR using gene-specific primers listed in Supplemental Table 2 online. Each amplicon was cloned into the pGEM-T easy vector (Promega) and mobilized to pTRV2 vector after digestion with appropriate restriction enzymes. Agrobacterium tumefaciens strain GV3101 harboring pTRV1, empty pTRV2 vector (EV), or the pTRV2 construct was cultured overnight at 28°C in 300 mL of Luria-Bertani medium containing 10 mM MES, 20 μM acetosyringone, and 50 μg mL−1 kanamycin. These cultures were centrifuged at 5000g for 10 min, and the bacterial pellets were resuspended in 5 mL of infiltration buffer (10 mM MES, 200 μM acetosyringone, and 10 mM MgCl2) and further incubated at 28°C for 3 h with shaking.
Periwinkle (cv Little Delicata) seeds were germinated and grown in a greenhouse under a 16/8-h light/dark photoperiod at 28°C for 3 to 6 weeks to produce at least two true leaf pairs. Young plants were wounded using toothpicks through the stem just below the apical meristem and infiltrated with a 1:1 (v/v) mixture of Agrobacterium cultures harboring pTRV1 and either empty pTRV2 vector or pTRV2 constructs. Typically, the phytoene desaturase phenotype was observed 3 week after inoculation of seedlings to signal the stage of growth during which leaves from control uninoculated wild-type, mock, EV, UGT8, LAMT, and SLS inoculations were harvested. After recording fresh weights of harvested materials, one member of a leaf pair was used for RNA extraction, while the other was used for metabolite analysis. Leaf tissues were frozen in liquid nitrogen and submitted to extraction with a tissue lyser (TissueLyser II; Qiagen) for quick pulverization. Frozen 2-mL microfuge tubes containing leaf materials and 100 μL of 1- and 2-mm glass bead mixtures (4:1 ratio) were transferred to a frozen (−80°C for at least 2 h) TissueLyser Adapter Set that can accommodate 24 samples/plate for performing extractions. Tissue lysis was conducted at 30 Hz for 1 min, after which sample were cooled in liquid nitrogen for 1 to 2 min, and tissue lysis was repeated for another minute. Samples were then subjected to RNA extraction or metabolite analysis.
VIGS-Treated Plant RNA Isolation
Tissue-lysed leaf material was mixed with 1 mL Trizol reagent (Invitrogen) and incubated with shaking (100 rpm; Innova 2000 shaker; New Brunswick Scientific) at room temperature for 5 min. Trizol extracts were mixed with 200 μL of chloroform and incubated for 3 min at room temperature, and the phases were separated by centrifugation (15 min at 14,000 rpm at 4°C). The aqueous phase was harvested, mixed with 0.7 volume of isopropanol, and incubated for 1 h at room temperature, and the RNA pellet was harvested by centrifugation (14,000 rpm at 4°C for 30 min). The pellet was washed with 75% ethanol in diethylpyrocarbonate (DEPC)–treated water (1 mL), decanted, dried in an SPD Speed Vac Thermo Savant for 2 min, air dried for 5 min, and then resuspended in 200 μL DEPC water. The RNA was washed (200 μL of phenol:chloroform:isoamyl alcohol [25:24:1], saturated with 10 mM Tris, pH 8.0, and 1 mM EDTA, pH 8.0) by mixing for 3 min, and the phases were separated by centrifugation (14,000 rpm at 4°C for 15 min). The aqueous phase was washed with chloroform (200 µL) by mixing for 3 min, and the phases were separated by centrifugation (14,000 rpm at 4°C for 15 min). RNA in the aqueous layer was precipitated by overnight incubation at 4°C in the presence of 2.67 M LiCl followed by centrifugation (14,000 rpm at 4°C for 30 min). The pellet was washed with 75% ethanol in DEPC water (1 mL), decanted, dried in an SPD Speed Vac Thermo Savant for 2 min, air dried for 5 min, and then resuspended in 20 μL of MilliQ water. After a 10-min incubation at room temperature, 3 μL of each sample was incubated with DNase (1 μL of each 10× DNase buffer [New England BioLabs] + 1 μL DNase [New England Biolabs] + 5 μL of milliQ water) at 37°C for 30 min. The DNA digestion was stopped by the addition of 2 μL of 25 mM EDTA and heating in a dry bath at 75°C for 10 min. The samples were cooled for 15 min before performing RT-PCR. Single-stranded cDNA was synthesized from 2.0 µg of RNA using Avian Myeloblastosis Virus reverse transcriptase (Promega) and oligo(dT) (Alpha DNA) following the manufacturer’s protocol. Detection of TRV coat protein in plants infiltrated with pTRV vectors was performed as described previously (See Supplemental References 1.).
Metabolite Analysis by UPLC-MS
Tissue-lysed leaf material was extracted in 1 mL of methanol at room temperature with occasional mixing over a 2-h period. A 200-μL aliquot of the methanol extract was filtered through Acrodic syringe filters (0.22 µm; VWR International) before analysis by UPLC/single-quadrupole mass spectrometry. Iridoid analysis was performed using an Acquity UPLC system (Waters) equipped with an Acquity UPLC BEH C18 column (1.0 × 50 mm i.d., 1.7 μm; Waters) and a mobile phase consisting of solvent A (0.1% [v/v] formic acid) and solvent B (100% acetonitrile). Iridoids were eluted at a flow rate 0.3 mL/min with the following linear gradient: 0 to 0.5 min, 99% A, 1% B; 0.5-5.0 min 92% A, 8% B; 5.0-6.5 min 70% A, 30% B; 6.5-7.2 min 50% A, 50% B; 7.2-7.5 min 70% A, 30% B, 7.5 to 8.0 min, 92% A and 8% B; 8.0 min 99% A and 1% B. Deoxyloganetic acid, deoxyloganic acid, loganic acid, loganin, and secologanin reference standards were also analyzed by this method, and standard curves were generated to measure the levels of each iridoid in the extracts. The mass spectrometer was operated with an electrospray ionization source of positive ionization mode with a capillary voltage of 3.0 kV, cone voltage of 30 V, cone gas flow of 1 L/h, desolvation gas flow of 600 L/h, desolvation temperature of 350°C, and a source temperature of 150°C. All iridoids were detected at 240 nm with secologanin (mass-to-charge ratio [m/z] = 389, retention time [RT] = 3.88 min), deoxyloganic acid (m/z = 361, RT = 4.06 min), deoxyloganetic acid (m/z = 197, RT = 4.68 min), loganin (m/z = 391, RT = 3.10 min), and loganic acid (m/z = 377, RT = 1.90 min).
MIAs were separated using the same column in part as described previously (Roepke et al., 2010). Samples were maintained at 4°C and 5-μL injections were made into the column. The analytes were detected by photodiode array and mass spectrometry. The solvent systems for alkaloid analysis were as follows: solvent A, methanol:acetonitrile:5 mM ammonium acetate (6:14:80); solvent B, methanol:acetonitrile:5 mM ammonium acetate (25:65:10). The following linear elution gradient was used: 0 to 0.5 min 99% A and 1% B at 0.3 mL/min; 0.5 to 0.6 min 99% A and 1% B at 0.4 mL/min; 0.6 to 7.0 min 1% A and 99% B at 0.4 mL/min;7.0 to 8.0 min 1% A and 99% B at 0.4 mL/min; 8.0 to 8.3 min 99% A and 1% B at 0.4 mL/min; 8.3 to 8.5 min 99% A and 1% B at 0.3 mL/min; and 8.5 to 10.0 min 99% A and 1% B at 0.3 mL/min. The mass spectrometer was operated with a capillary voltage of 2.5 kV, cone voltage of 34 V, cone gas flow of 2 L/h, desolvation gas flow of 460 L/h, desolvation temperature of 400°C, and a source temperature of 150°C. Catharanthine was detected at 280 nm, and its identity was verified by its diode array profile, its mass (337 m/z), and RT (4.45 min). Vindoline was detected at 305 nm, and its identity was verified by its diode array profile, its mass (457 m/z), and RT (4.55 min). Chromatographic peaks were integrated and compared with standard curves for catharanthine and vindoline to give the total amount of alkaloids in each sample.
In Situ Hybridization
The in situ RNA hybridization was performed basically as described previously (St-Pierre et al., 1999) with some modifications. The first pair of leaves from C. longifolius were fixed in FAA (50% ethanol, 5% acetic acid, and 5% formaldehyde), dehydrated through an ethanol and tert-butanol series, and then embedded in Paraplast Xtra (Fisher Scientific). The embedded samples were sectioned into 10-µm thickness using a rotary microtome (Reichert Jung). The sections were carefully spread onto slides treated with 2% (v/v) 3-aminopropyltriethoxysilane (Sigma-Aldrich) in acetone, incubated for 24 h at 40°C, and stored at 4°C until use. Serial sections were deparaffinized by two incubations of 15 min each in xylene before rehydration at room temperature with separate 5-min incubations in 100% ethanol, 100% ethanol, 95% ethanol, 70% ethanol, 50% ethanol, 100%, DEPC water, and a final transfer to 100% DEPC water.
Full-length UGT8 clones in the pGEM-T easy vector (Promega) were used for the synthesis of sense and antisense digoxigenin-labeled RNA probes using a DIG RNA labeling kit (SP6/T7; Roche) according to the manufacturer’s instructions. The RNA probes were submitted to partial alkaline hydrolysis for 20 min at 60°C. After prehybridization, hybridization of the digoxigenin-labeled RNA probes, and washing, the slides were stained with alkaline phosphatase–conjugated antidigoxigenin antibodies (1:200 dilution) (Roche).
Accession Numbers
Sequence data from this article can be found in the DNA Data Bank of Japan/GenBank/European Bioinformatics Institute libraries under the following accession numbers: full-length clones, Cr-UGT6 (UGT85A23, GenBank accession number AB591741), Cr-UGT7 (UGT76A2, GenBank accession number AB733666), Cr-UGT8 (UGT709C2, GenBank accession number AB733667); clones used for VIGS, phytoene desaturase (460 bp) (GenBank accession number JQ655739), Cr-UGT8 (349 bp) (GenBank accession number KF415118), LAMT (373 bp) (GenBank accession number KF415116), and SLS (359 bp) (GenBank accession number KF415117).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Nonrooted Molecular Phylogenetic Trees of Plant UGTs.
Supplemental Figure 2. Substrate Specificity of Recombinant UGT6, UGT7, and UGT8.
Supplemental Figure 3. Detection of pTRV2-Derived TRV Coat Protein Transcript (134 bp) in Plants Infiltrated with pTRV Vectors Confirmed the Success of Agrobacterium Infiltration.
Supplemental Table 1. PSPG-Derived Contigs Found by Searching EST Database of Catharanthus roseus.
Supplemental Table 2. Primers for Cloning Partial Sequences of UGT8, LAMT, and SLS Used in VIGS Experiments.
Supplemental Table 3. Accession Numbers of PSPGs Used for Constructing the Phylogenetic Tree Shown in Supplemental Figure 1.
Supplemental Table 4. Primer Sequences Used for RACE PCR Cloning of UGT6∼8.
Supplemental Table 5. Primer Sequences Used for Full-Length Amplification of UGT6∼8.
Supplemental Table 6. Primer Sequences Used for Heterologous Expression of Cr-UGT6∼8.
Supplemental Table 7. Primer Sequences Used for Real-Time RT-PCR.
Supplemental Table 8. Fasta Files of Alignments Used for Phylogenetic Analysis.
Supplemental Reference 1. Supplemental Reference for Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank Peter J. Facchini (University of Calgary) for pTRV1 and pTRV2, Kenichiro Inoue (Yokohama College of Pharmacy) for 7-deoxyloganin, and Nobuki Kato (Nagoya City University) for iridotrial. We recognize the skilled technical work of next-generation sequencing personnel at the McGill University-Genome Québec-Innovation Centre. We thank Christoph Sensen, Mei Xiao, and Ye Zhang of the University of Calgary for their dedicated bioinformatic support and large-scale gene annotation efforts that that helped in the identification of various UGT8 genes from the Phytometasyn website. This work was supported by Japanese Society for the Promotion of Science KAKENHI Grants 22710218 (to K.T.), 22590009, and 25460127 (to H.M.). This work was also supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant (V.D.L.), Genome Canada, Genome Alberta, Genome Prairie, Genome British Columbia, the Canada Foundation for Innovation, the Ontario Ministry of Research and Innovation, the National Research Council of Canada, and other government and private sector partners. S.M.-A. is the recipient of postdoctoral funding from Genome Canada and the Ontario Ministry of Research and Innovation.
AUTHOR CONTRIBUTIONS
K.A. performed research, analyzed data, and wrote parts of the article. V.S. and S.M.-A. performed research, analyzed data, and wrote parts of the article. E.E. performed research and analyzed data. M.N. and K.T. performed research. H.M. and V.D.L. designed the research, analyzed data, and wrote the article.
Glossary
- MIA
monoterpenoid indole alkaloid
- IPAP
internal phloem-associated parenchyma
- PSPG
plant secondary product glycosyltransferase
- VIGS
virus-induced gene silencing
- RACE
rapid amplification of cDNA ends
- UPLC-MS
to be defined
- EV
empty vector
- DEPC
diethylpyrocarbonate
- m/z
mass-to-charge ratio
- RT
retention time
References
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burlat V., Oudin A., Courtois M., Rideau M., St-Pierre B. (2004). Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 38: 131–141 [DOI] [PubMed] [Google Scholar]
- De Luca V., Salim V., Atsumi S.M., Yu F. (2012a). Mining the biodiversity of plants: A revolution in the making. Science 336: 1658–1661 [DOI] [PubMed] [Google Scholar]
- De Luca V., Salim V., Levac D., Atsumi S.M., Yu F. (2012b). Discovery and functional analysis of monoterpenoid indole alkaloid pathways in plants. Methods Enzymol. 515: 207–229 [DOI] [PubMed] [Google Scholar]
- Facchini P.J., Bohlmann J., Covello P.S., De Luca V., Mahadevan R., Page J.E., Ro D.K., Sensen C.W., Storms R., Martin V.J. (2012). Synthetic biosystems for the production of high-value plant metabolites. Trends Biotechnol. 30: 127–131 [DOI] [PubMed] [Google Scholar]
- Facchini P.J., De Luca V. (2008). Opium poppy and Madagascar periwinkle: Model non-model systems to investigate alkaloid biosynthesis in plants. Plant J. 54: 763–784 [DOI] [PubMed] [Google Scholar]
- Geu-Flores F., Sherden N.H., Courdavault V., Burlat V., Glenn W.S., Wu C., Nims E., Cui Y., O’Connor S.E. (2012). An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492: 138–142 [DOI] [PubMed] [Google Scholar]
- Góngora-Castillo E., et al. (2012). Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS ONE 7: e52506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirimand G., Guihur A., Ginis O., Poutrain P., Héricourt F., Oudin A., Lanoue A., St-Pierre B., Burlat V., Courdavault V. (2011). The subcellular organization of strictosidine biosynthesis in Catharanthus roseus epidermis highlights several trans-tonoplast translocations of intermediate metabolites. FEBS J. 278: 749–763 [DOI] [PubMed] [Google Scholar]
- Hasada K., Yoshida T., Yamazaki T., Sugimoto N., Nishimura T., Nagatsu A., Mizukami H. (2011). Application of 1H-NMR spectroscopy to validation of berberine alkaloid reagents and to chemical evaluation of Coptidis Rhizoma. J. Nat. Med. 65: 262–267 [DOI] [PubMed] [Google Scholar]
- Kaminaga Y., Sahin F.P., Mizukami H. (2004). Molecular cloning and characterization of a glucosyltransferase catalyzing glucosylation of curcumin in cultured Catharanthus roseus cells. FEBS Lett. 567: 197–202 [DOI] [PubMed] [Google Scholar]
- Levac D., Murata J., Kim W.S., De Luca V. (2008). Application of carborundum abrasion for investigating the leaf epidermis: molecular cloning of Catharanthus roseus 16-hydroxytabersonine-16-O-methyltransferase. Plant J. 53: 225–236 [DOI] [PubMed] [Google Scholar]
- Linsmaier E.M., Skoog F. (1965). Organic growth factor requirement of tobacco tissue cultures. Physiol. Plant. 18: 100–127 [Google Scholar]
- Liscombe D.K., O’Connor S.E. (2011). A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry 72: 1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masada S., Terasaka K., Oguchi Y., Okazaki S., Mizushima T., Mizukami H. (2009). Functional and structural characterization of a flavonoid glucoside 1,6-glucosyltransferase from Catharanthus roseus. Plant Cell Physiol. 50: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Murata J., Roepke J., Gordon H., De Luca V. (2008). The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20: 524–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatoshi M., Terasaka K., Nagatsu A., Mizukami H. (2011). Iridoid-specific glucosyltransferase from Gardenia jasminoides. J. Biol. Chem. 286: 32866–32874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roepke J., Salim V., Wu M., Thamm A.M.K., Murata J., Ploss K., Boland W., De Luca V. (2010). Vinca drug components accumulate exclusively in leaf exudates of Madagascar periwinkle. Proc. Natl. Acad. Sci. USA 107: 15287–15292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg D., Thompson T.S., German T.L., Willis D.K. (2006). Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods 138: 49–59 [DOI] [PubMed] [Google Scholar]
- Simkin A.J., et al. (2013). Characterization of the plastidial geraniol synthase from Madagascar periwinkle which initiates the monoterpenoid branch of the alkaloid pathway in internal phloem associated parenchyma. Phytochemistry 85: 36–43 [DOI] [PubMed] [Google Scholar]
- St-Pierre B., Vazquez-Flota F.A., De Luca V. (1999). Multicellular compartmentation of catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell 11: 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T., Jones P. (2000). Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 5: 380–386 [DOI] [PubMed] [Google Scholar]
- Xiao M., et al. (2013). Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J. Biotechnol. 166: 122–134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.