Assembly factors are auxiliary proteins that promote the biogenesis of multiprotein complexes without being subunits of those. PHOTOSYNTHESIS-AFFECTED MUTANT68-LIKE (PAM68L) operates as an assembly factor of the NAD(P)H dehydrogenase complex in chloroplasts of flowering plants and derives evolutionarily from PAM68, an assembly factor of photosystem II in cyanobacteria, mosses, and plants.
Abstract
In vascular plants, the chloroplast NAD(P)H dehydrogenase complex (NDH-C) is assembled from five distinct subcomplexes, the membrane-spanning (subM) and the luminal (subL) subcomplexes, as well as subA, subB, and subE. The assembly process itself is poorly understood. Vascular plant genomes code for two related intrinsic thylakoid proteins, PHOTOSYNTHESIS-AFFECTED MUTANT68 (PAM68), a photosystem II assembly factor, and PHOTOSYNTHESIS-AFFECTED MUTANT68-LIKE (PAM68L). As we show here, inactivation of Arabidopsis thaliana PAM68L in the pam68l-1 mutant identifies PAM68L as an NDH-C assembly factor. The mutant lacks functional NDH holocomplexes and accumulates three distinct NDH-C assembly intermediates (subB, subM, and subA+L), which are also found in mutants defective in subB assembly (ndf5) or subM expression (CHLORORESPIRATORY REDUCTION4-3 mutant). NDH-C assembly in the cyanobacterium Synechocystis sp PCC 6803 and the moss Physcomitrella patens does not require PAM68 proteins, as demonstrated by the analysis of knockout lines for the single-copy PAM68 genes in these species. We conclude that PAM68L mediates the attachment of subB- and subM-containing intermediates to a complex that contains subA and subL. The evolutionary appearance of subL and PAM68L during the transition from mosses like P. patens to flowering plants suggests that the associated increase in the complexity of the NDH-C might have been facilitated by the recruitment of evolutionarily novel assembly factors like PAM68L.
INTRODUCTION
Chloroplasts and their evolutionary relatives, the cyanobacteria, contain molecular machines referred to as the NAD(P)H dehydrogenase complex (NDH-C), which shares many subunits with complex I in the mitochondrial respiratory chain (Friedrich and Scheide, 2000). In cyanobacteria, NADPH indeed appears to serve as the electron donor to the NDH-C (reviewed in Battchikova et al., 2011a), but the soluble stromal protein ferredoxin (Fd) has recently been shown to play this role in chloroplasts (Yamamoto et al., 2011). Therefore, the NDH-Cs in chloroplasts and cyanobacteria actually represent Fd:plastoquinone and NADPH:plastoquinone oxidoreductases, respectively, in contrast with their mitochondrial counterpart, which is an NADH:ubiquinone oxidoreductase. In the strain PCC6803 of the cyanobacterium Synechocystis sp, NAD(P)H dehydrogenase (NDH) subunits form at least three types of NDH-Cs (including NDH-1L, -1S, and -1M) with different subunit compositions (Zhang et al., 2004; Ogawa and Mi, 2007). NDH-1L is most closely related to the chloroplast NDH-C and is required for heterotrophic growth, probably via respiration and cyclic electron flow (CEF), while NDH-1M and NDH-1S form the NDH-1MS complex, which functions in CO2 uptake (Zhang et al., 2004; reviewed in Battchikova et al., 2011a). In angiosperms, chloroplast NDH-C is located in the stroma lamellae of thylakoids and also participates in chlororespiration and CEF around PSI, while transferring electrons to plastoquinone (reviewed in Rumeau et al., 2007; Shikanai, 2007). The physiological role of the NDH-C in chloroplasts is still enigmatic. Although its total abundance and overall contribution to CEF around PSI are comparatively low, it seems to alleviate stromal overreduction under stress conditions, becoming important for plant growth when alternative routes for CEF around PSI are unavailable (Burrows et al., 1998; Sazanov et al., 1998; Endo et al., 1999; Horvath et al., 2000; Li et al., 2004; Munekage et al., 2004; Munne-Bosch et al., 2005; Wang et al., 2006; Okegawa et al., 2008; Peng et al., 2008).
The cyanobacterial NDH-C has an L-shaped structure and comprises subunits also found in heterotrophic bacteria (NdhA to NdhK) and newly identified subunits that it shares with the chloroplast NDH (NdhL to NdhO and NdhS) (Battchikova et al., 2011b; reviewed in Ifuku et al., 2011). The membrane subcomplex that forms the horizontal element of the “L” contains NdhA to NdhG, whereas the peripheral arm is made of subunits NdhH to NdhO and NdhS (Battchikova et al., 2011b; reviewed in Ifuku et al., 2011). The three subunits associated with NADH binding in nonphototrophic bacterial NDH-1 complexes are missing in cyanobacterial NDH.
A structural model for the chloroplast NDH-C in Arabidopsis thaliana, which characteristically consists of five subcomplexes, has been proposed on the basis of extensive genetic, proteomic, and bioinformatic data and supporting studies in which the pattern of subunits accumulated in different mutant backgrounds was determined (Suorsa et al., 2009; Ifuku et al., 2010, 2011; Peng et al., 2011a; Yamamoto et al., 2011). In this model, the fully assembled complex consists of a membrane subcomplex (subM), two stroma-exposed subcomplexes (subA and subB), one lumen-exposed subcomplex (subL), and the electron donor binding subcomplex (subE). Of these, only subM and subA are conserved in cyanobacteria; subM is composed of seven plastid-encoded subunits (NdhA to NdhG), while subA contains four plastid-encoded (NdhH to NdhK) and four nucleus-encoded (NdhL to NdhO) subunits (Peng et al., 2009). The subunits present in subB, subL, and subE are all encoded by nuclear genes specific to angiosperms (Peng et al., 2009; Yamamoto et al., 2011). Moreover, chloroplast NDH interacts with PSI to form the NDH-PSI supercomplex in Arabidopsis, which is required for the stabilization of NDH, especially when light intensities are high (Peng et al., 2008, 2009; Peng and Shikanai, 2011). Two minor light-harvesting complex I proteins, Lhca5 and Lhca6, form a linker domain required for the interaction between NDH and PSI (Peng et al., 2009). In the liverwort Marchantia polymorpha, chloroplast NDH apparently does not interact directly with PSI (Ueda et al., 2012), implying that the NDH-PSI supercomplex represents a relatively recent evolutionary acquisition.
Detailed analysis of the assembly of the chloroplast NDH-C is hampered by its low abundance and fragility, but several nucleus-encoded auxiliary factors required for NDH assembly have been identified in genetic screens for mutants defective in NDH activity. These gene products are required for NDH activity but do not represent NDH subunits, and they fall into two classes. The first comprises RNA binding proteins that are specifically required for the expression of plastid genes encoding NDH subunits (reviewed in Suorsa et al., 2009) and so affect NDH assembly only indirectly. For instance, the CHLORORESPIRATORY REDUCTION2 protein (CRR2) is essential for the expression of the chloroplast ndhB gene (Hashimoto et al., 2003), whereas CRR4 is needed for editing of ndhD transcripts (Kotera et al., 2005). The second class consists of genuine assembly factors. These include the stromal proteins CRR1, which exhibits weak similarity to dihydrodipicolinate reductase (Shimizu and Shikanai, 2007), and CRR6 (Munshi et al., 2006), CRR7 (Kamruzzaman Munshi et al., 2005), CRR41, and CRR42 (Peng et al., 2012), all of which are specifically required for subA assembly. NDF5, identified on the basis of in silico coexpression analysis, is a thylakoid membrane protein and may be involved in the biogenesis, or required for the stability, of subcomplexes subA and subB (Ishida et al., 2009). This rich collection of assembly mutants, together with interactive proteomic studies, has allowed the construction of a model for subA assembly, which involves the sequential action of auxiliary factors with several subA assembly intermediates (Peng et al., 2012). However, the mode of assembly of the other NDH subcomplexes remains largely unclear.
Here, we report a detailed analysis of the integral thylakoid protein PHOTOSYNTHESIS-AFFECTED MUTANT68-LIKE (PAM68L), which acts as an auxiliary factor in the assembly of the chloroplast NDH-C. We employed comparative mutant analysis in Arabidopsis to define the function of the protein as promoting the formation of an assembly intermediate containing subM and subB, as well as subA and subL. Analysis of Physcomitrella lines lacking the single-copy PHOTOSYNTHESIS-AFFECTED MUTANT68 (PAM68) gene suggests that the involvement of PAM68 proteins in NDH assembly emerged only after duplication of the PAM68 gene in flowering plants.
RESULTS
The Genomes of Vascular Plants Code for Two PAM68 Proteins
It has previously been shown that the thylakoid membrane protein PAM68 is required for efficient PSII subunit PsbA (D1) biogenesis and PSII assembly in Arabidopsis (Armbruster et al., 2010). The corresponding nuclear gene, PAM68, is of cyanobacterial origin and is conserved in photosynthetic organisms (Armbruster et al., 2010; Figure 1A). Accordingly, in Synechocystis mutants defective for the PAM68 ortholog SLL0933, PSII assembly is perturbed (Armbruster et al., 2010). All sequenced nuclear genomes of land plants, except that of the moss Physcomitrella patens, code for a second PAM68-related protein, designated as PAM68L (Figure 1A). In Arabidopsis, PAM68L shares 38%/58% sequence identity/similarity with PAM68 (Armbruster et al., 2010), and, like PAM68, it is a member of the thylakoid proteome (Peltier et al., 2004). However, the two PAM68 proteins are apparently not functionally redundant in Arabidopsis, as the corresponding single and double mutants differ with respect to growth rate, leaf coloration, and accumulation of D1 (Armbruster et al., 2010).
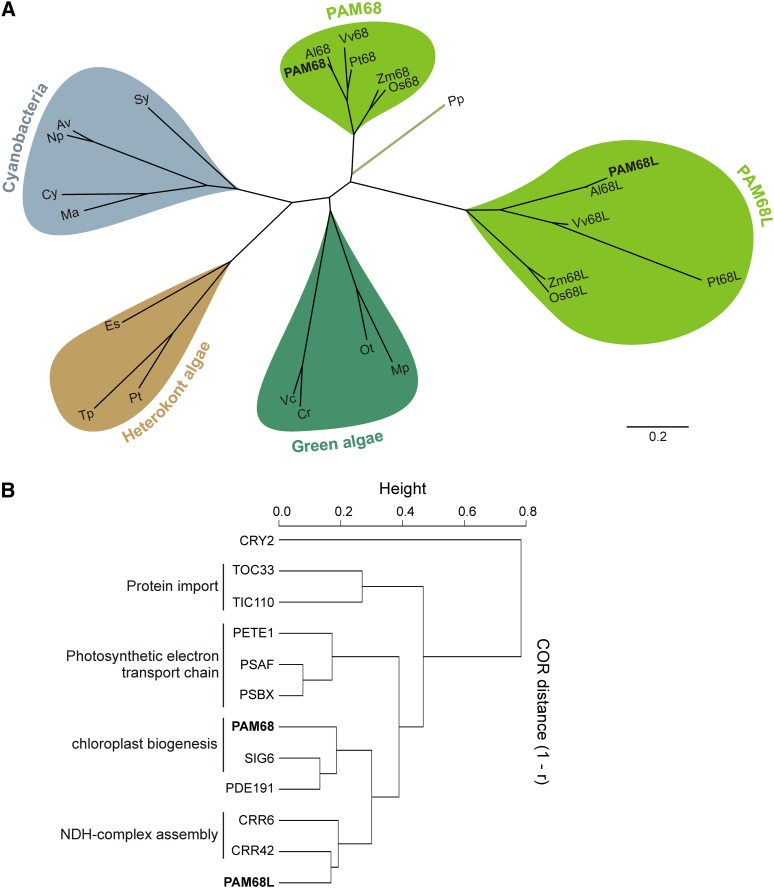
Figure 1.
Sequences and Expression Characteristics of PAM68 Proteins.
(A) Phylogenetic analysis of PAM68 and PAM68L sequences from cyanobacteria (shaded in blue), green algae (dark green), heterokont algae (brown), and land plants (light green). Full species names and accession numbers are provided in Methods; an alignment of the sequences used for tree construction is shown in Supplemental Figure 4 online. Branch lengths reflect the estimated number of substitutions per 100 sites.
(B) Expression characteristics of PAM68 and PAM68L. Expression profiling of the genes coding for PAM68 and PAM68L revealed tight coregulation with SIG6, PDE191, CRR42, and CRR6, respectively. Additional correlations with expression of genes for the envelope proteins TIC110 and TOC33, the photosynthetic electron transport components PSBX, PSAF, and plastocyanin (PETE1), and the nonchloroplast protein CRY2 were hierarchically clustered based on profile similarities using the single linkage method (gene coexpression database ATTEDII; http://atted.jp/). The degree of coexpression (COR) is measured by Pearson's correlation (r). Low distance (height) values (1-r) indicate high expression similarities.
To pinpoint processes in which PAM68L is involved, transcripts that are coexpressed with PAM68L RNA were identified (Arabidopsis thaliana trans-factor and cis-element prediction database II, the ATTED-II database; Obayashi et al., 2009). Both PAM68 and PAM68L were found to be coregulated with a group of genes involved in photosynthesis (Figure 1B). PAM68 showed the highest degree of coexpression with PDE191 and SIG6 (the latter is known to be required for chloroplast development; Ishizaki et al., 2005), while PAM68L expression coincided with CRR6 (Munshi et al., 2006; Peng et al., 2010) and CRR42, which are essential for assembly of the NDH-C (Peng et al., 2012). Thus, like other elements of the NDH-C previously identified by transcript profiling (Ishida et al., 2009; Takabayashi et al., 2009; Peng et al., 2012), PAM68L might be involved in its function or assembly.
Arabidopsis PAM68L Is Required for NDH-C Formation and Activity
A previously obtained T-DNA insertion Arabidopsis mutant (pam68l-1) lacking PAM68L (see Supplemental Figures 1A to 1F online; Armbruster et al., 2010) was therefore tested for NDH activity by monitoring chlorophyll a fluorescence during a light-to-dark transition (Figure 2A). This shift induces a transient increase in chlorophyll a fluorescence, which has been ascribed to reduction of plastoquinone by the NDH-C in the dark and is characteristically lacking in mutants defective in NDH activity (Shikanai et al., 1998). The pam68l-1 mutant showed no such increase in fluorescence, neither in young nor in mature leaves (Figure 2A). Leaf coloration, growth, electron transport rates (ETRs), and nonphotochemical quenching (NPQ) were as in wild-type plants (see Supplemental Figures 1F to 1H online). Two independent pam68l-1 lines (35S:PAM68L pam68l-1 or oePAM68L) in which the PAM68L cDNA was expressed under control of the cauliflower mosaic virus (CaMV) 35S promoter were generated (see Supplemental Figures 1D to 1H online) and tested for NDH activity as above. Because the transient increase in chlorophyll a fluorescence was restored in both oePAM68L lines (Figure 2A), we concluded that PAM68L is required for NDH activity. Chlorophyll a fluorescence kinetics were also analyzed in the PSII assembly mutants pam68-2 and pam68l-1 pam68-2. The rise in postillumination fluorescence observed in pam68-2 and pam68-1 pam68-2 seems to be characteristic of mutants with abnormally low levels of PSII (see also the fluorescence kinetics of pam68 mutants and lpa1 in Armbruster et al. [2010]). However, postillumination fluorescence increases much faster in pam68-2 than in pam68l-1 pam68-2, which might indicate that a functional NDH complex is required for the steep increase. To investigate how the lack of PAM68L interferes with NDH activity, thylakoid complexes from the wild type (Columbia-0 [Col-0]), pam68l-1, and the two oePAM68L lines were separated by blue native (BN)-PAGE according to Peng et al. (2008) and subjected to immunoblot analysis with an antibody raised against the L subunit of the NDH-C (NdhL) (Figure 2B; see Supplemental Figure 1I online). This showed that the pam68l-1 mutant lacks the PSI-NDH supercomplex (left panel of Figure 2B; see Supplemental Figure 1I online) in contrast with pam68-2, in which it is present in wild-type amounts (Armbruster et al., 2010). Accordingly, levels of NdhL were also severely reduced in pam68l-1 (see Supplemental Figure 1J online). Moreover, the mobility of the residual amounts detected in the BN-PAGE corresponded to those of assembly intermediates of NDH (Figure 2B, right panel). As expected, the oePAM68L lines accumulated wild-type amounts of PSI-NDH (Figure 2B; see Supplemental Figures 1I and 1J online).
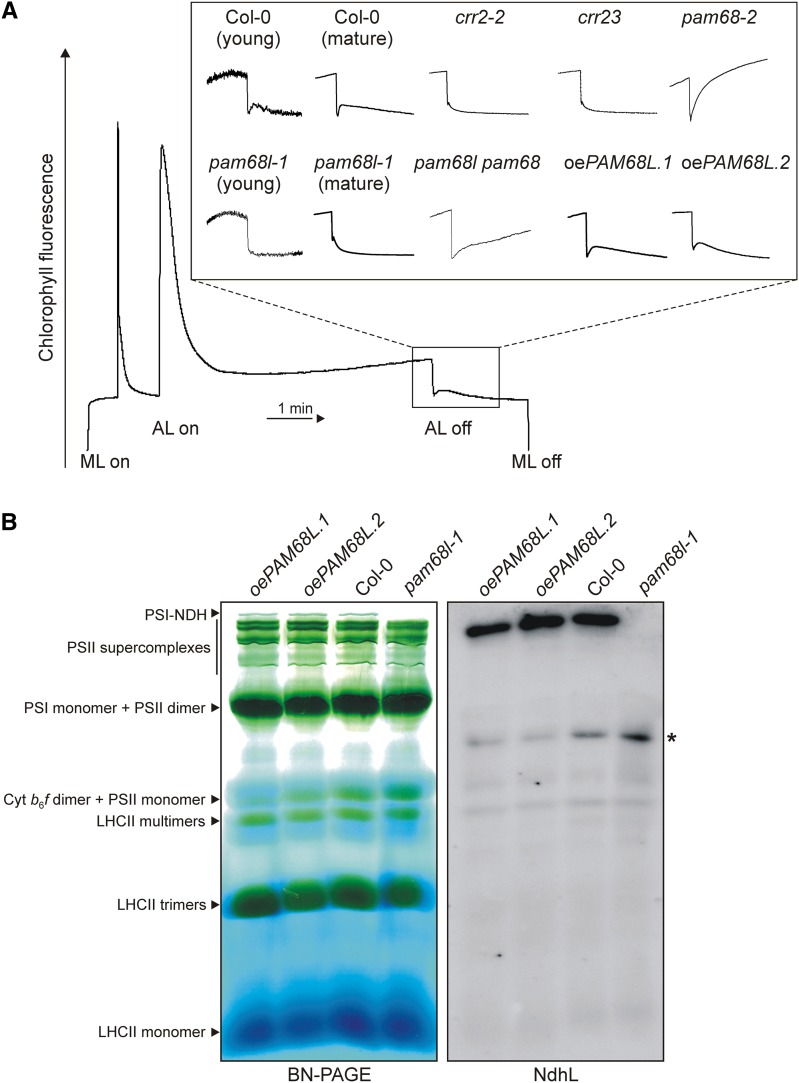
Figure 2.
PAM68L Is Required for NDH Activity and PSI-NDH Supercomplex Accumulation.
(A) NDH activity of 4-week-old wild-type (Col-0), pam68l-1, and two independent 35S:PAM68L pam68l-1 (oePAM68L) lines, as well as crr2-2, crr23 (ndhl), pam68-2, and pam68l-1 pam68-2 as controls, was determined by chlorophyll fluorescence analysis during a light-to-dark transition. For the wild type (Col-0) and pam68l-1, NDH activity was measured with 2-week-old leaves (young) and 4-week-old leaves (mature). Leaves were exposed to a saturating light flash to obtain maximum fluorescence (Fm) and then to low actinic light (AL) for 5 min. AL was then turned off and the subsequent transient rise in fluorescence ascribed to NDH activity was monitored using a pulse amplitude modulation chlorophyll fluorometer. The main panel shows the typical course of wild-type chlorophyll fluorescence under these conditions. Insets are magnified traces from the boxed area normalized to Fm. ML, measuring light.
(B) Thylakoid membranes from wild-type (Col-0) and pam68l-1 plants and two independent oePAM68L lines were solubilized in 1% (w/v) β-DM at a chlorophyll concentration of 1 µg μL−1, and the samples were fractionated by 5 to 12% BN-PAGE according to Peng et al. (2008). The major protein complexes were assigned to individual bands as described (Peng et al., 2008; Armbruster et al., 2010). Denatured protein complexes were then transferred to PVDF membrane and probed with antibodies raised against the NdhL subunit. The asterisk indicates the position of an NdhL-containing assembly intermediate that is particularly prominent in the pam68l-1 sample.
PAM68L Is an Intrinsic Thylakoid Protein
The thylakoid localization of PAM68L was confirmed by immunoblot analysis of chloroplast subfractions (Figure 3A) with an antibody raised against PAM68L (see Supplemental Figure 1C online; Armbruster et al., 2010). Furthermore, subfractionation of thylakoid membranes by ultracentrifugation as described (Hertle et al., 2013) revealed that it accumulates in the fraction enriched for stroma lamellae, where the NDH-C also collects (Lennon et al., 2003), and to a much lesser extent also in the grana margin-enriched fraction (Figure 3B). As demonstrated by its response to alkaline and chaotropic salts (which denature proteins by disturbing their intramolecular interactions) (Figure 3C), PAM68L, like PAM68 (Armbruster et al., 2010), is an intrinsic thylakoid membrane protein, which is compatible with the predicted presence of two transmembrane domains in the mature protein (Figure 3D). Moreover, tryptic digestion of isolated thylakoids strongly decreased the detectability of PAM68L with our antibody, which is directed against an N-terminal portion of the mature protein. This, together with the absence of a digestion product of 5.5 kD (Figure 3D), indicates that PAM68L adopts the same conformation in the thylakoid membrane as PAM68 (Armbruster et al., 2010), with its N and C termini facing the stroma (Figures 3D and 3E). Thus, in its thylakoid localization and topology, PAM68L resembles PAM68.
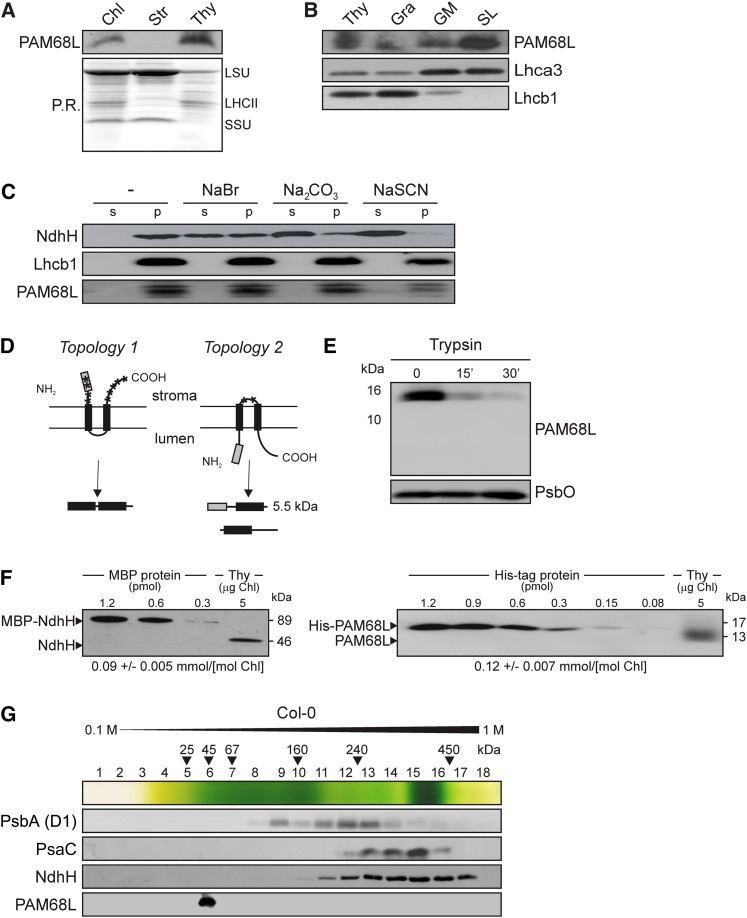
Figure 3.
Characteristics of the PAM68L Protein.
(A) Suborganellar localization of PAM68L. Chloroplasts, stroma, and thylakoids were isolated from wild-type (Col-0) plants, fractionated by SDS-PAGE, transferred to PVDF membrane, and subjected to protein gel blot analysis using an antibody raised against an epitope of PAM68L (see also [D]). Ponceau Red (P.R.) staining of the membrane prior to immunodetection is shown as loading control. Protein bands corresponding to the large and small subunits of ribulose-1,5-bis-phosphate carboxylase/oxygenase (LSU and SSU, respectively) and LHCII are indicated. Chl, chlorophyll.
(B) Localization of PAM68L in thylakoid subfractions. Thylakoid vesicles were formed by brief exposure to digitonin and separated by size using differential centrifugation at 10,000g, 40,000g, and 150,000g to obtain subfractions enriched for grana (Gra), grana margins (GM), and stroma lamellae (SL), respectively. Equal amounts of proteins from each subfraction were size-separated by SDS-PAGE and analyzed with PAM68L-specific antibodies, as well as with antibodies raised against Lhca3 (mainly localized in stroma lamellae) or Lhcb1 (preferentially localized in grana).
(C) Extraction of thylakoid-associated proteins with chaotropic salts or at alkaline pH. Thylakoid membranes from wild-type (Col-0) plants were resuspended (at 0.5 mg) chlorophyll/mL in 10 mM HEPES/KOH, pH 7.5, containing either 2 M NaCl, 0.1 M Na2CO3, 2 M NaSCN, or no additive. After incubation for 30 min on ice, supernatants containing the extracted proteins (s) and membrane fractions (p) were fractionated by SDS-PAGE and probed with antibodies raised against PAM68L, NdhH (a peripheral membrane protein), or Lhcb1 (an integral membrane protein).
(D) Schematic representation of the two possible topologies of PAM68, with diagnostic proteolytic fragments indicated below. The predicted transmembrane domains are indicated by black boxes, the antigenic epitope as a gray box, and trypsin cleavage sites by asterisks.
(E) Immunoblot analysis of thylakoid membrane preparations with antisera specific for PAM68L or PsbO (present in the thylakoid lumen) before and after trypsination. Samples were analyzed before (time 0) and after 15 and 30 min of treatment with trypsin. Note that, in intact thylakoids, only stroma-exposed polypeptides are accessible to the enzyme.
(F) NdhH and PAM68L proteins were expressed in E. coli as C-terminal fusions to MBP in the case of NdhH or the His tag (PAM68L), purified by affinity chromatography, and quantified. Known amounts of the fusions were titrated against thylakoid membrane protein preparations and quantified by immunoblot analysis. The data shown are representative of the results obtained in four independent experiments. The calculated concentration of each of the two proteins in thylakoid preparations is noted below the blots.
(G) Distribution of PAM68L after Suc gradient centrifugation. Thylakoids (1 mg chlorophyll/mL) from wild-type (Col-0) plants were solubilized with 1% (w/v) β-DM and fractionated by centrifugation in a linear 0.1 to 1 M Suc gradient. Eighteen fractions were collected (numbered from top to bottom) from the gradient. Proteins were precipitated from each fraction, separated by SDS-PAGE, blotted onto PVDF membranes, and subjected to protein gel blot analysis with antibodies raised against PAM68L, NdhH, the D1 protein of PSII, and PsaC. The positions of molecular mass markers in the gradient are indicated.
In order to quantify the amount of PAM68L in the thylakoid membrane and analyze the stoichiometric relationship between PAM68L and the NDH-C, the mature PAM68L (without the predicted chloroplast transit peptide) and the chloroplast-encoded NdhH protein were expressed in Escherichia coli as His6- and maltose binding protein (MBP)–tagged fusions, respectively. These were affinity purified and titrated against thylakoid protein preparations. The amounts of NdhH detected were equivalent to ∼0.09 mmol/mol chlorophyll (Figure 3F), corresponding to ∼5% of the PSI or PSII level on a molar basis (Kirchhoff et al., 2002). This is higher than reported before (∼1.5% of the level of PSII; Burrows et al., 1998) but still in the range found by Peng et al. (2008) for the abundance of PSI in PSI-NDH complexes (0.5 to 5%). The concentration of PAM68L was ∼0.12 mmol/mol chlorophyll. This is close to equimolar relative to the NDH-C, suggesting that PAM68L might be an NDH subunit.
To test this possibility, thylakoids were isolated from wild-type (Col-0) and pam68-2 plants and solubilized with n-dodecyl-β-d-maltoside (β-DM). The samples were then subjected to ultracentrifugation on a linear 0.1 to 1 M Suc gradient to separate the different thylakoid protein complexes; 18 fractions (numbered from top to bottom) were collected, and proteins were precipitated and subjected to immunoblot analysis (Figure 3G). PAM68L clearly did not comigrate with NDH complexes, which were detected in Fractions 10 to 17 with an NdhH-specific antibody. Instead, PAM68L was found in Fraction 6, indicating that it migrates either as a monomer or in a small complex and thus could represent either a weakly bound NDH subunit (i.e., readily removable by mild detergents) or an auxiliary protein transiently involved in the assembly of the NDH-C or the PSI-NDH supercomplex.
PAM68L Is Required for NDH Assembly
The absence of individual subunits in a multiprotein complex often perturbs or precludes the attachment of others, as shown by the analysis of mutants defective for specific subunits of PSI (Pesaresi et al., 2003) or the different subcomplexes of the NDH-C (Yabuta et al., 2010). Therefore, to test whether the accumulation of PAM68L depends on the presence of the intact NDH-C or of any of its subcomplexes, immunoblot analyses were performed on thylakoid membranes from wild-type plants and mutants specifically defective for certain NDH subcomplexes (Figures 4A and 4B). The mutants examined were crr2-2 and crr4-3, which are defective in expression of NdhB and NdhD, respectively, subunits normally found in the membrane subcomplex subM (Hashimoto et al., 2003; Kotera et al., 2005); ndhl, in which levels of the stroma-exposed subcomplex A (subA) are strongly reduced (Shimizu et al., 2008; Peng et al., 2009); pnsb2 (ndf2 and ndh45), pnsb3 (ndf4), ndf5, and pnsb4 (ndf6), all of which accumulate less of the stroma-exposed subcomplex B (subB) (Ishikawa et al., 2008; Ishida et al., 2009; Sirpio et al., 2009; Takabayashi et al., 2009); ndhs (crr31), ndht (crrj), and ndhu (crrl) (Yamamoto et al., 2011), in which accumulation of the electron donor binding subcomplex (subE) is perturbed; and pnsl1 (ppl2), in which the lumen subcomplex (subL) is underrepresented (Ishihara et al., 2007; Peng et al., 2009) (Figure 4C). In all of these mutants, PAM68L was present in higher amounts than in the wild type (Figures 4A and 4B; see Supplemental Table 1 online), implying that it does not represent a structural subunit of the NDH-C.
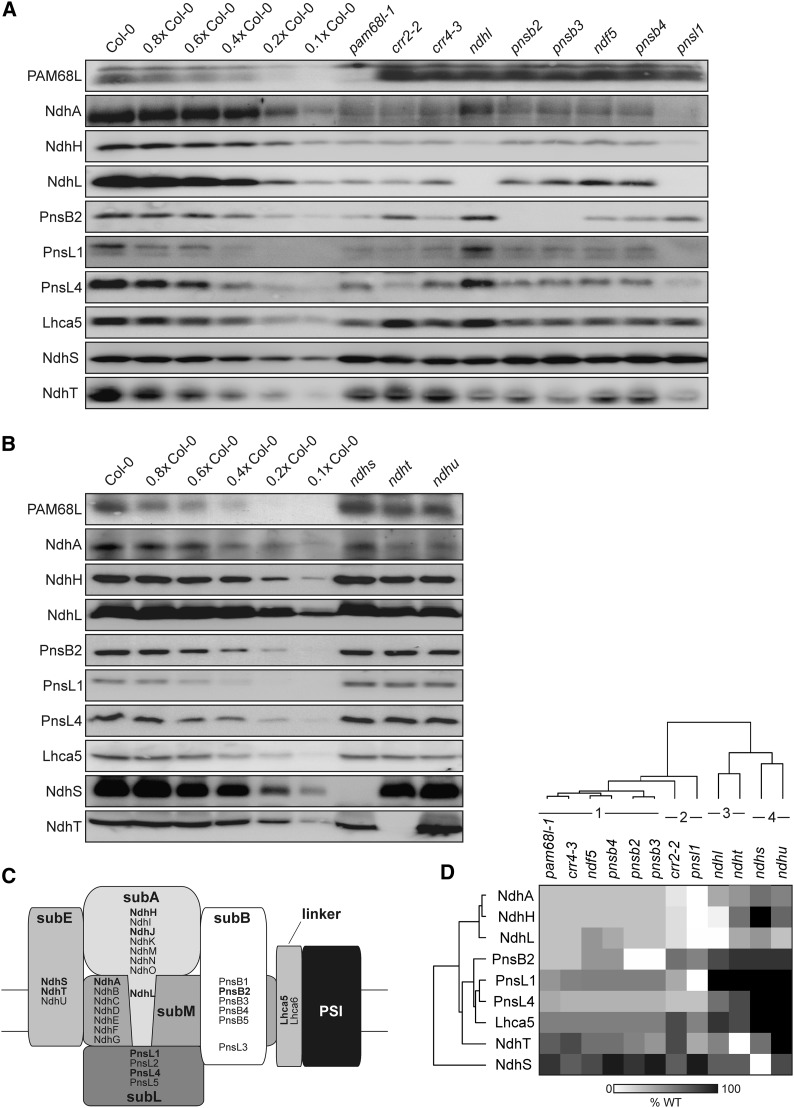
Figure 4.
Absence of PAM68L or CRR4 Has Similar Effects on the Composition and Assembly of NDH Subcomplexes.
(A) and (B) Immunoblot analysis of thylakoid proteins from the wild type, the pam68l-1 mutant, and other mutants that are known to affect the composition and/or assembly of the NDH-C. Thylakoid proteins (5 µg chlorophyll) from the wild type (Col-0) and the different mutants were loaded, together with a dilution series of the wild-type sample as indicated.
(C) Schematic overview of the subcomplex composition of NDH in the chloroplasts of land plants (Ifuku et al., 2011). For designations of subcomplexes, see main text. The marker subunits for individual subcomplexes that were analyzed here (and in Figure 7) are indicated in bold.
(D) Hierarchical clustering of the NDH subunit patterns found in the different genotypes analyzed in (A) and (B). The original average values determined from immunoblot analyses (representatives of which are shown in [A] and [B]) are provided in Supplemental Table 1 online. The four classes discussed in the main text are indicated. WT, the wild type.
To assess the potential function of PAM68L in NDH assembly, the pattern of accumulation of NDH subunits in pam68l-1 thylakoids was compared with those observed in the above-mentioned subcomplex mutants. To this end, protein blots were probed with antibodies against subunits representative of each NDH subcomplex: NdhA (subM), NdhH and NdhL (for subA), PnsB2/NDF2 (subB), PnsL1/PPL2 and PnsL4/FKBP16 (subL), NdhS/CRR31 and NdhT/CRRJ (subE), and Lhca5 for the PSI NDH linker (Figures 4A to 4C). The profiles of NDH subunit accumulation in each of the mutants were then ranked in terms of their similarity to the pattern seen in the wild type by hierarchical clustering (Figure 4D; see Supplemental Table 1 online). Four distinct classes of profile could be distinguished. (1) The first class included the subM mutant crr4-3, previously reported to be required for subB stability (Peng et al., 2009), and the subB mutants pnsb2/ndf2, pnsb3/ndf4, ndf5, and pnsb4/ndf6. In all of these mutants, levels of subA and subM subunits are strongly depleted, while subL and subE subunits, as well as Lhca5, are less affected. (2) The second class contained the subL mutant pnsl1/ppl2 and the subM mutant crr2-2. Indeed, the pnsl1 mutant exhibited the most severe effects on NDH assembly, with amounts of most of the tested NDH proteins being very low. Moreover, pnsl1/ppl2 is the only mutant that completely lacks the subM subunit NdhA, indicating that PnsL1/PPL2 is crucial for the incorporation of NdhA into subM. In the crr2-2 mutant, which lacks NdhB, levels of the other subunits are less depressed than in pnsl1/ppl2. Relative to crr4-3 from class 1, subB accumulation is less perturbed in crr2-2, but the amounts of NdhA (subM) are lower, which explains why the two profiles are assigned to different classes. (3) The third class included the subA mutant ndhl, which showed very low accumulation of the subA protein NdhH as described previously (Shimizu et al., 2008), but little change in markers for the other subcomplexes, indicating that the absence of subA only marginally affects the accumulation of the NDH-C. The ndht/crrj profile also falls into this class. (4) Mutants lacking the subE subunits NdhS/CRR31 or NdhU/CRRL comprised the fourth class of profiles and were characterized by an almost wild-type-like accumulation of all subcomplexes, except for subA proteins, as previously reported (Yamamoto et al., 2011; Figures 4C and 4D; see Supplemental Table 1 online). The profile of NDH subunits detected in pam68l-1 was similar to class 1 profiles, particularly that of crr4-3. This suggests that the defect in the pam68l-1 mutant lies either in the expression or assembly of the NdhD protein (which would correspond to the primary defect in crr4-3) or of subB (which would correspond to the secondary effect of crr4-3 and the primary consequence of the subB mutations pnsb2/ndf2, pnsb3/ndf4, ndf5, and pnsb4/ndf6) or in the assembly of an intermediate containing both subcomplexes.
PAM68L Is Dispensable for Accumulation of Chloroplast ndh mRNAs and the Association of ndhD Transcripts with Polysomes
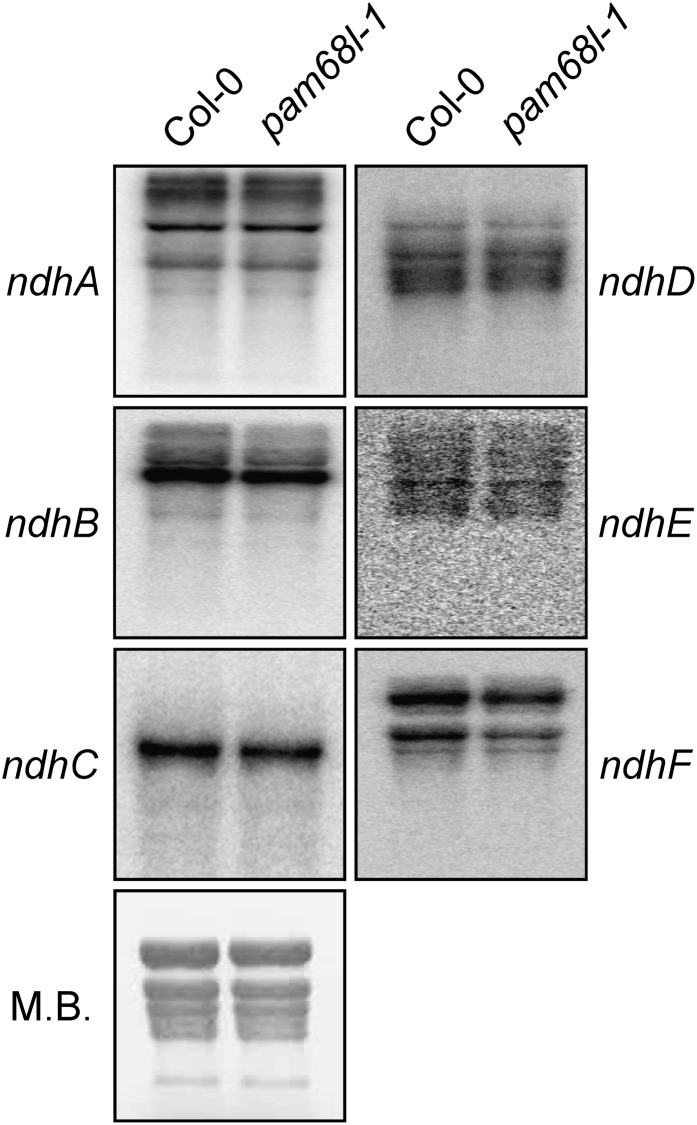
In order to analyze whether altered accumulation of chloroplast-encoded transcripts for NDH subunits might account for the defects seen in pam68l-1, RNA gel blot analysis was performed with probes specific for ndhA, B, C, D, E, and F, which together detect all chloroplast transcriptional units that code for NDH subunits. In the pam68l-1 mutant, transcript levels and processing patterns did not significantly differ from the wild type (Col-0) (Figure 5).
Figure 5.
Analysis of Plastid ndh Transcript Accumulation.
Analysis of transcripts encoding plastid NDH subunits in wild-type (Col-0) and mutant (pam68l-1) plants. Aliquots (20 µg) of total leaf RNA from 4-week-old plants were fractionated by denaturing agarose gel electrophoresis, blotted onto membrane, and hybridized with probes specific for the transcripts indicated on the left. Methylene blue (M.B.) staining of the membrane is shown as control.
Because pam68l-1 displays a pattern of accumulation of NDH subunits similar to the one in the crr4-3 mutant (Figure 4D), which cannot edit the ndhD start codon (Kotera et al., 2005), the association of ndhD transcripts with polysomes was analyzed to test for possible defects in the translation of this NDH subunit in pam68l-1 (see Supplemental Figure 2A online). As a control, polysomally bound ndhF mRNAs were also quantified (see Supplemental Figure 2A online). No dramatic variation of pam68l-1 or crr4-3 from wild-type plants with respect to polysome association of the tested mRNAs was observed (see Supplemental Figures 2A and 2B online).
In summary, these data indicate that PAM68L is dispensable for normal accumulation of chloroplast-encoded transcripts for NDH subunits and for the expression of NdhD at the level of transcription and translational initiation.
A Characteristic Set of NDH Assembly Intermediates Accumulates in pam68l-1, crr4-3, and ndf5 Mutants
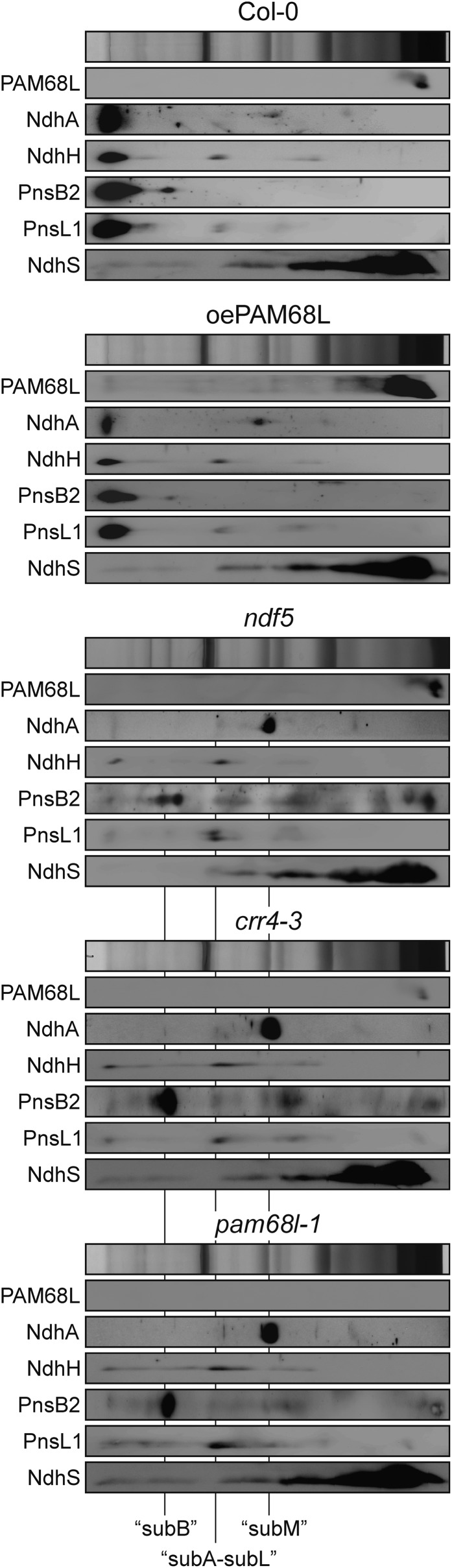
The data presented above argue against an effect of PAM68L on ndh transcript accumulation and on the expression of NdhD, a component of subM. Therefore, the most plausible explanation for the pattern of accumulation of the other NDH subunits seen in both pam68l-1 and crr4-3 is that PAM68L is necessary for an assembly step that requires factors that are also missing in crr4-3. To facilitate analysis of the small quantities of NDH assembly intermediates that accumulate in the wild type (Col-0), mutants (pam68l-1, crr4-3, and the subB assembly mutant ndf5), and the PAM68L overexpressor line (oePAM68L), larger amounts of thylakoid complexes (corresponding to 150 μg chlorophyll) from all these genotypes were fractionated by BN-PAGE in the first dimension, separated into individual subunits by SDS-PAGE in the second dimension, and subjected to protein gel blot analyses with antibodies raised against NdhA (a marker for subM), NdhH (subA), PnsB2 (subB), PnsL1 (subL), and NdhS (subE) (Figure 6). Immunodetection of PAM68L with a PAM68L-specific antibody confirmed that PAM68L is present in monomeric form, or in a small complex, in wild-type, crr4-3, and ndf5 thylakoids (Figure 6), supporting the results obtained by Suc gradient fractionation of thylakoid complexes (Figure 3G).
Figure 6.
NDH Complex Assembly Is Similarly Perturbed in pam68l-1 and crr4-3 Mutants.
Thylakoid membranes from the wild type (Col-0), mutants (pam68l-1, ndf5, and crr4-3), and the PAM68L overexpressor line (oePAM68L) were solubilized with 1% (w/v) β-DM, and the protein samples were fractionated on 4 to 12% BN-PAGE gels. Individual lanes from BN-PAGE gels were then subjected to electrophoresis in the presence of SDS on Tris/Tricine gels (10%, 4 M urea) and immunoblot analyses with antibodies raised against NdhA, NdhH, PnsB2, PnsL1, and NdhS. The positions of the intermediates containing subB, subM, and subA + subL are indicated.
In all three mutants, pam68l-1, ndf5, and crr4-3, the NDH holocomplex was barely detectable, whereas abnormally strong signals were observed for three assembly intermediates. The first was detected by the anti-NdhA antibody (NdhA representing a subunit of subM) but not by any of those recognizing subunits representative of the other subcomplexes and migrates between PSII monomers and light-harvesting complex II (LHCII) multimers in BN gels. The second, larger intermediate that accumulates in crr4-3 and pam68l-1 (and to a lesser extent in ndf5) was detected by the antibody raised against the subB component PnsB2 only. In ndf5, an additional signal at a slightly lower molecular weight was detected by the PnsB2 antibody, which might result from the destabilization of subB, which is characteristic of ndf2, 4, 5, and 6 mutants (Figure 4D). In addition, a further intermediate, which runs between subM and subB, is detected by NdhH- and PnsL1-specific antibodies in all three mutant genotypes. This result implies that the buildup of subB and subM might be linked to the accumulation of an intermediate that contains both subA and subL. As expected, overexpression of PAM68L in the pam68l-1 mutant background rescues assembly of the NDH-C and prevents accumulation of the three intermediates relative to the NDH holocomplex.
Thus, a characteristic set of three NDH assembly intermediates accumulates in the mutants pam68l-1, crr4-3, and ndf5, which also share a very similar profile of NDH subunits (Figure 4D). One of these intermediates appears to represent subM (without NdhD in crr4-3, whereby the intermediate appears to have approximately the same size in all three mutants), and another is likely to be subB. The third intermediate contains subA and subL and has previously been described in lines deficient in subB that have strongly reduced levels of subA and low levels of subL and accumulate an ∼500-kD intermediate that lacks PSI but contains subA and subL (Peng et al., 2009). Moreover, the three intermediates do not represent artificial complexes formed only in genotypes with a block in NDH subunit synthesis/assembly. They can also be detected, in smaller amounts, in wild-type samples (see top panel of Figure 6) and are thus, most probably, intermediates in the normal assembly pathway. Taken together, these findings suggest that absence of NdhD (in crr4-3) or PAM68L (in pam68l-1) or impaired assembly of subB (in ndf5 and other subB mutants) impede the same assembly step (see Discussion).
The PAM68 Proteins in Synechocystis and P. patens Are Not Required for NDH Assembly
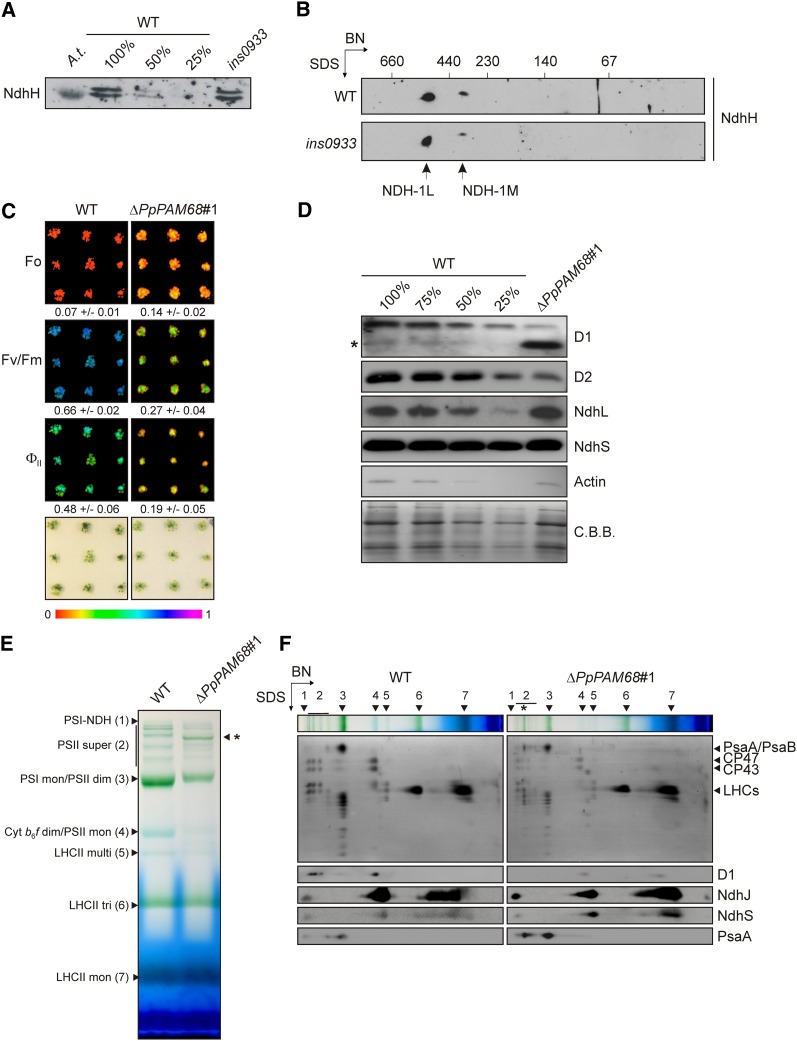
The roles of Arabidopsis PAM68 and PAM68L in the assembly of PSII (Armbruster et al., 2010) and NDH (this study), respectively, raise the question of whether both functions were already mediated by their common ancestor in the cyanobacterium that was the progenitor of chloroplasts. An ancient function in PSII assembly has already been demonstrated for the Synechocystis PAM68 (Armbruster et al., 2010), but the NDH assembly function might have evolved only after the duplication of PAM68 in vascular plants (Figure 1A) and therefore be restricted to PAM68L. To clarify this issue, the assembly of NDH was investigated in the previously described Synechocystis strain ins0933, in which the gene for PAM68 (SLL0933) is inactivated (Armbruster et al., 2010). Rates of respiration in the ins0933 mutant are normal (Armbruster et al., 2010), and the H subunit of NDH is present at wild-type levels (Figure 7A). Together with the observation that the two assembly products NDH-1L and NDH-1M (Herranen et al., 2004) accumulate in a wild-type-like manner in ins0933 (Figure 7B), this strongly suggests that Synechocystis PAM68 is not required for NDH assembly.
Figure 7.
PAM68 Is Not Required for NDH Assembly in Synechocystis or Physcomitrella.
(A) Immunodetection of NdhH in the Synechocystis mutant ins0933, which lacks PAM68. Protein samples from ins0933 and varying amounts of a similar wild-type (WT) extract (100, 50, and 25%) were fractionated on SDS gels and blotted onto a nitrocellulose membrane. As a positive control, an Arabidopsis (Col-0) thylakoid membrane sample (A.t.) was loaded. NdhH was visualized by immunodetection using an NdhH-specific antibody.
(B) Synechocystis wild-type and ins0933 membrane samples were solubilized with β-DM, and complexes were fractionated on 2D-BN/SDS gels. NdhH was immunodetected after protein transfer to a nitrocellulose membrane. The positions of NDH-1L and NDH-1M, the two NDH complexes in Synechocystis (Herranen et al., 2004), are indicated.
(C) Four-week-old P. patens plants (nine wild-type and nine ΔPpPAM68#1 individuals) were grown under continuous low light in a climate chamber (21 to 23°C) on solid growth medium. The photosynthetic parameters Fo, Fv/Fm, and ΦII (at actinic light of 35 μmol photons m−2 s−1) were determined using an Imaging PAM system as described in Methods. Fo, Fv/Fm, and ΦII values are indicated by the color scale from 0 to 1 at the bottom, and numbers ± sd (n = 9) are given below each picture. Note that ΔPpPAM68#1 is one of five independently generated ΔPpPAM68 lines, all of which display the same mutant photosynthetic phenotype.
(D) Immunoblot analysis of total protein extracts from wild-type (including several dilutions) and ΔPpPAM68#1 with antibodies raised against D1, D2, NdhL, and NdhS. As a control, actin was also detected with an appropriate antibody. Membranes were stained with Coomassie blue G 250 (C.B.B.) to control for equal loading. The asterisk marks the position of a putative degradation product of D1 in ΔPpPAM68#1.
(E) Accumulation of thylakoid membrane complexes in mutant (ΔPpPAM68#1) and wild-type P. patens strains. Membranes were isolated from lines grown under continuous light as described in Methods. Samples were solubilized by β-DM (1% [w/v] final concentration) for 10 min, and complexes were fractionated on a 5 to 12% BN-PAGE gel. Bands are annotated according to Armbruster et al. (2010) and on the basis of the fractionation patterns of the numbered complexes in the second dimension shown in (F): PSI-NDH supercomplex (PSI-NDH; band 1), PSII supercomplexes (PSII super, bands subsumed under 2), PSI-LHCII complexes (indicated by the asterisk), PSI monomers and PSII dimers (PSI mono/PSII dim, band 3), cytochrome b6/f dimers and PSII monomers (Cyt b6/f dim/PSII mono, band 4), multimeric LHCII (LHCII multi, band 5), trimeric LHCII (LHCII tri, band 6), and monomeric LHCII (LHCII mon, band 7).
(F) Complexes separated by BN-PAGE (see lanes at the top) were resolved into their constituent subunits on denaturing Tris/Tricine gels (10%, 4 M urea). Proteins were blotted onto PVDF membranes and visualized by Coomassie blue G 250 staining. Subunits characteristic of PSII (D1), PSI (PsaA), and NDH-C (NdhJ and NdhS) were then immunodetected. The position of the PSI-LHCII complex is indicated by an asterisk as in (E).
Like Synechocystis, the moss P. patens contains only one PAM68 gene, Pp-PAM68, which codes for a protein that is more similar to Arabidopsis PAM68 (56/71% identity/similarity over the stretch of the 115 most conserved amino acids) than to PAM68L (40/53% identity/similarity) and therefore branches from the PAM68 clade in the phylogenetic tree (Figure 1A). The corresponding knockout (ΔPpPAM68) was generated (see Methods and Supplemental Figure 3 online), and five independent knockout strains were generated (ΔPpPAM68#1 and #4 to #7). The five strains were tested for the accumulation and assembly of its PSII and NDH and were found to behave identically; therefore, if not mentioned otherwise, the results obtained from ΔPpPAM68#1 are provided in the following (Figures 7C to 7F). A clear increase in the minimum fluorescence (Fo) was noted in the mutant (ΔPpPAM68#1, 0.14 ± 0.02; the wild type, 0.07 ± 0.01), together with reductions in the maximum quantum yield of PSII (Fv/Fm: ΔPpPAM68#1, 0.27 ± 0.04; the wild type, 0.66 ± 0.02) and the effective quantum yield of PSII (ΦII: ΔPpPAM68#1, 0.19 ± 0.05; the wild type, 0.48 ± 0.06) (Figure 7C). This combination of a dramatic decrease in Fv/Fm and an increase in Fo is characteristic for mutants with abnormally low levels of PSII (see, e.g., pam68 [Armbruster et al., 2010] and hcf136 [Meurer et al., 1998]). Protein gel blot analysis of thylakoid proteins showed that amounts of the PSII core proteins D1 and D2 in ΔPpPAM68#1 are <25% of those in the wild type, and an additional signal was detected between 17 and 26 kD by the D1 antibody (Figure 7D), which probably represents a degradation product of D1 (Sun et al., 2010). In complete contrast, lack of P. patens PAM68 had no effect on the accumulation of the NDH subunit NdhL (Figure 7D). Moreover, the level of NdhS, which can accumulate independently of NDH-C in Arabidopsis (Figures 4 and 6), was similar in the two genotypes (Figure 7D).
Because these results indicate that P. patens PAM68 is required for accumulation of PSII but not of NDH complexes, the patterns of assembly intermediates of the two thylakoid membrane complexes were studied. Separation of thylakoid membranes of the wild type and ΔPpPAM68#1 by one-dimensional and two-dimensional (2D) PAGE analyses showed that, as in Arabidopsis lines lacking PAM68 (Armbruster et al., 2010), fewer PSII dimers and monomers accumulate in ΔPpPAM68#1 (Figures 7E and 7F). However, residual amounts of PSII supercomplexes (Figure 7E) were still detectable in ΔPpPAM68#1, in contrast with the Arabidopsis pam68-2 mutant (Armbruster et al., 2010). This indicates that subtle differences in the dependence of PSII assembly on PAM68 exist between Arabidopsis and P. patens. With respect to the NDH-C, an assembled NDH-PSI complex could be detected in both wild-type and knockout strains with antibodies raised against NdhJ, NdhS, and PsaA. In both genotypes, NdhS accumulated independently of NDH-C and appeared to colocalize with NdhJ. Moreover, in ΔPpPAM68 strains, essentially the same assembly intermediates as in wild-type strains were detected with these two antibodies, although smaller NdhJ-containing intermediates appeared to accumulate to higher concentrations than in the wild type (Figure 7F).
Taken together, these data indicate that only the PAM68L protein in vascular plants is required for NDH assembly. In species with only one PAM68 gene, its product is apparently involved in the assembly of PSII only.
DISCUSSION
The later steps in NDH assembly are relatively well understood. Thus, NdhS and NdhU are not required for the stability of the PSI-NDH supercomplex, indicating that subE is probably added last, whereas NdhS and NdhT can accumulate independently of NDH-C (Yamamoto et al., 2011). Moreover, NdhS accumulation depends on NdhT, whereas accumulation of NdhU requires NdhT and NDH-C (Yamamoto et al., 2011). Mutants without subA (i.e., lacking NdhL or NdhM) accumulate an intermediate that contains all other components of the PSI-NDH supercomplex (Peng et al., 2008, 2009, 2012). Therefore, it is very likely that integration of subA also occurs late in the assembly process. In plants lacking subM (i.e., without NdhB, NdhD, or NdhF), subA apparently fails to form (Peng et al., 2008).
The earlier steps in PSI-NDH assembly are much less clear, as they cannot be unequivocally deduced from the analysis of mutants lacking individual NDH subcomplexes. Thus, plants without subL (i.e., lacking PnsL1/PPL2 or PnsL4/FKB16-2) have less subB and no subA and accumulate two assembly intermediates, one without subL, the other also missing subA; the larger of these contains PSI, subB, and other components, and the smaller one probably represents PSI-subB (Peng et al., 2009). Lines lacking subB have very little subA but make low levels of subL and accumulate an ∼500-kD intermediate that lacks PSI but contains subA and subL (Peng et al., 2009). Similarly, in the crr4-3 mutant, destabilization of subM is associated with barely detectable levels of subA and subB proteins, but small amounts of subL do accumulate (Peng et al., 2009). At the level of assembly intermediates, in the crr4-3 mutant, NdhS/CRR31 accumulates in a 300-kD intermediate that lacks subA and PSI, while NdhT/CRRJ comigrates with NdhL and possibly PSI in BN-PAGE (Yamamoto et al., 2011). When the linker consisting of Lhca5 and Lhca6 is missing, an assembly intermediate accumulates that lacks PSI but contains subA, subL, and subB proteins (Peng and Shikanai, 2011). Taken together, this rather confusing picture suggests that assembly intermediates that do not normally accumulate in wild-type plants are formed in the absence of individual subcomplexes, greatly complicating the reconstruction of early assembly steps in wild-type plants on the basis of BN-PAGE analysis of lines lacking known NDH subunits.
PAM68L Promotes Formation of a Complex That Contains subM and subB
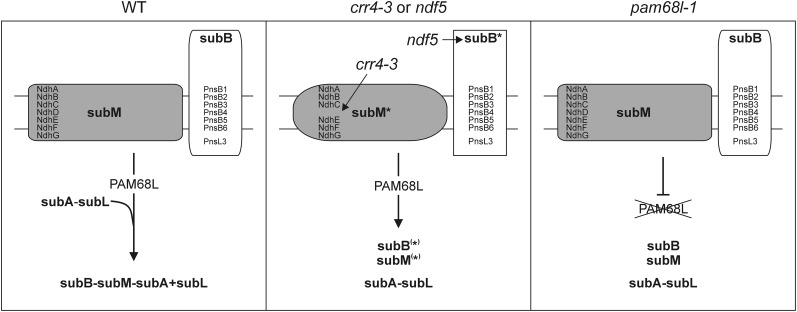
PAM68L can accumulate independently of the NDH-C (Figure 4) and is required for its correct assembly (Figures 4 and 6) but not for the accumulation of transcripts of plastid ndh genes in Arabidopsis (Figure 5). These findings, together with the involvement of its homolog PAM68 in PSII assembly (Armbruster et al., 2010), allow us to conclude that PAM68L is a genuine NDH assembly factor. In several respects, the molecular phenotype of the pam68l-1 mutant (which lacks PAM68L) resembles those of the subB assembly mutant ndf5 and, in particular, the crr4-3 mutant, in which the subM component NdhD is missing. All three mutants show the same pattern of accumulation of NDH subunits (Figure 4D) and the same subset of assembly intermediates (Figure 6). The three stable intermediates common to pam68l-1, ndf5, and crr4-3 plants appear to represent subcomplexes subB, subM, and subA+subL, respectively. Therefore, it can be concluded that PAM68L is essential for a step that leads to formation of a complex containing subA, subB, subL, and subM. In principle, therefore, PAM68L could be involved in (1) generation of an assembly-competent subM complex, analogous to the function of CRR4; (2) generation of an assembly-competent subB complex, like NDF5; or (3) assembly of a subB-subM-subA+subL–containing complex (Figure 8). The expression of chloroplast ndh mRNAs do not seem to require PAM68L (Figure 5). Hence, even though the profile of NDH subunit accumulation in pam68l-1 resembles that of crr4-3 more than those of the subB mutant ndf5 (Figure 4D), we favor the hypothesis that PAM68L functions not at the level of subunit expression, but in the assembly of a subB-subM-subA+subL–containing intermediate (Figure 8). This hypothesis is strongly supported by the observation that subB and subM are also found in P. patens, and this species does not require PAM68L for the assembly of NDH holocomplexes.
Figure 8.
Schematic Model for the Proposed Function of PAM68L.
In wild-type (WT) plants, subM and subB are assembled into an intermediate, which may also include subA and subL. Whether this takes place in two steps (formation of a subB-subM complex, followed by incorporation of subA-subL) or in one step (direct linkage of subB, subM, and subA-subL to form a single intermediate) is unclear. In crr4-3, the impairment in NdhD expression leads to the accumulation of the incomplete subM (as indicated by the accumulation of an assembly intermediate containing NdhA), which cannot be integrated into a larger intermediate. As a consequence, subB and subA-subL intermediates accumulate. Similarly, ndf5 plants with a primary defect in subB assembly also accumulate all three intermediates. Lack of PAM68L also leads to the accumulation of the three intermediates found in crr4-3 and ndf5. Because (1) ndhD expression does not require PAM68L (Figure 5), although the profile of residual NDH subunit accumulation in pam68l-1 is more similar to that of crr4-3 than that seen in the subB mutant ndf5 (Figure 4D), and (2) P. patens does not require a PAM68L protein for the assembly of subB and subM, we conclude that PAM68L is involved in the assembly of subB and subM into a larger intermediate that possibly also includes subA and subL.
Recruitment of PAM68L for NDH Assembly Is an Evolutionarily Recent Event
Two major lines of evidence indicate that the recruitment of PAM68L as an NDH assembly factor occurred during the evolution of flowering plants (i.e., relatively recently in evolutionary terms). (1) The parallel occurrence of PAM68 and PAM68L genes is restricted to flowering plants (Figure 1A). (2) In species with only one PAM68 gene, like Synechocystis and P. patens, its product is not required for NDH assembly (Figure 7). The obvious conclusion from these data is that, after duplication of the PAM68 gene in the progenitor of flowering plants, one copy retained its original function in PSII assembly, which can be traced back to the cyanobacterial endosymbiont (Armbruster et al., 2010), whereas the other gene copy evolved to mediate a function in NDH assembly. Moreover, the fact that PAM68L and true NDH subunits occur in equimolar amounts (Figure 3F) suggests that PAM68L might not yet be fully optimized for its role as a (transiently active) NDH assembly factor.
NDH Assembly: A Case of Evolutionary Tinkering?
The Arabidopsis NDH-C is made up of subM, subA, subB, subL, and subE, together with the linker proteins Lhca5 and 6 that mediate formation of the NDH-PSI supercomplex. The NDH-C of the liverwort M. polymorpha, which is considered to belong to the oldest surviving lineage of land plants, lacks subL and the linker proteins Lhca5 and Lhca6, and does not form NDH-PSI supercomplexes, while cyanobacterial genomes also lack genes for subB subunits (Ueda et al., 2012). This has prompted the conclusion that subL and NDH-PSI complexes, as observed in angiosperms, are evolutionary novelties invented by flowering plants (Ueda et al., 2012). The moss P. patens also lacks genes for subL (see Supplemental Table 2 online) but encodes and expresses the linker protein Lhca5 (Alboresi et al., 2008; Busch et al., 2013). Accordingly, PSI-NDH supercomplexes can be detected in P. patens, but only a small amount of P. patens NDH-C was detected in the supercomplex (Figure 7), whereas all of the Arabidopsis NDH-C is in the supercomplex (Figure 6). This clearly indicates that PSI-NDH formation predates the evolution of flowering plants. The smaller fraction of NDH-C present in the P. patens PSI-NDH supercomplex may represent the actual in vivo situation or the supercomplex may be more fragile in P. patens.
But given that P. patens possesses the same subcomplexes as Arabidopsis, apart from subL, and contains only one PAM68 gene (which is dispensable for NDH assembly; Figure 7), why is PAM68L required for NDH assembly in flowering plants? One obvious explanation is that subL makes the difference (i.e., that subB and subM are assembled into a larger complex that contains subL). Indeed, we found that accumulation of the intermediates subB and subM is associated with the buildup of an intermediate containing subA and subL, as described previously for lines lacking subB (Peng et al., 2009). Therefore, following duplication of the gene for the PSII assembly factor PAM68 in flowering plants, the copy that became PAM68L may have evolved to facilitate the incorporation of the novel subL subcomplex present in these species into the NDH-C (Figure 8). This represents an unusual situation in which a preexisting assembly factor has diversified sufficiently to take on a role in the assembly of a different multiprotein complex. Nonetheless, such functional recruitment is in line with the evolutionary history of several subunits of the vascular plant-specific subcomplex L, which can be traced back to PSII. Thus, the NDH complexes of vascular plants include subunits related to polypeptides that function in the PSII complex, including the PsbP family member PnsL1 (Ishihara et al., 2007) and the PsbQ family members PnsL2 and L3 (Majeran et al., 2008; Peng et al., 2009; Suorsa et al., 2010; Yabuta et al., 2010). Thus, PSII might have supplied both structural (PnsL1, L2, and L3) and regulatory proteins (PAM68L) for the establishment of subL in the NDH-C of vascular plants. Moreover, CRR42 (a homolog of prokaryotic proteins that interact with His kinases; Peng et al., 2012) and Cpn60β4/CRR27 (a vascular plant-specific subunit of the type I chaperonin Cpn60 in chloroplasts that is related to GroEL in bacteria and is essential for NdhH folding; Peng et al., 2011b) also represent instances of proteins that have been hijacked by the NDH-C in flowering plants.
METHODS
Computational Analyses
Protein sequences were retrieved from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), and homologs of the Arabidopsis thaliana PAM68 proteins were identified by NCBI-BLAST (http://blast.ncbi.nlm.nih.gov/). Amino acid sequences were aligned using ClustalW (v2.0.12, http://mobyle.pasteur.fr/; Thompson et al., 1994; default settings, gap extension: 50). Sequence identities and similarities were calculated using NCBI BLAST 2 sequences (http://blast.ncbi.nlm.nih.gov; Tatusova and Madden, 1999). Transmembrane domains were predicted by the program TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/; Krogh et al., 2001).
Identification of coexpression networks and hierarchical clustering of expression profiles were performed using the gene coexpression database ATTEDII (v6.0, http://atted.jp/; Obayashi et al., 2007). Hierarchical clustering of protein accumulation patterns was performed using Cluster 3.0 (http://www.falw.vu/~huik/cluster.htm) with Euclidean distance as similarity metric and average linkage.
Phylogenetic Analysis
For the phylogenetic analysis of PAM68 sequences, the C-terminal 115 amino acids of the proteins were aligned (see Supplemental Figure 4 online). The program Phylip (v3.67, http://mobyle.pasteur.fr/) was used, using as distance model the Jones-Taylor-Thornton matrix and bootstrapping (1000 repetitions) to compute a consensus tree. FigTree (v1.3.1, http://tree.bio.ed.ac.uk/software/figtree) was used for the unrooted phylogenetic tree visualization. The neighbor-joining method was employed for tree building.
Plant Material and Its Propagation
The Arabidopsis pam68l-1 mutant (SALK_143426), from the SALK T-DNA collection (http://signal.salk.edu/; Alonso et al., 2003), is in the Col-0 background (Armbruster et al., 2010). The crr2-2, crr4-3, ndhl, ppl2/pnsl1, crr31/ndhs, crrj/ndht, crrl/ndhu, ndf2/pnsb2, ndf4/pnsb3, ndf5, and ndf6/pnsb4 mutants have been described previously (Ishihara et al., 2007; Ishikawa et al., 2008; Peng et al., 2008; Shimizu et al., 2008; Ishida et al., 2009; Sirpio et al., 2009; Takabayashi et al., 2009; Yamamoto et al., 2011). Arabidopsis wild-type and mutant plants were propagated under controlled greenhouse conditions as described (Armbruster et al., 2010)
Plants overexpressing PAM68L (oePAM68L) were generated by introducing the PAM68L coding region into the plant expression vector pH2GW7 (Karimi et al., 2002) under the control of the 35S promoter of CaMV and then transforming flowers of pam68l-1 mutant plants with the PAM68L overexpression construct using the floral dip method as described (Clough and Bent, 1998). The plants were then transferred to the greenhouse, and seeds were collected after 3 weeks. Individual transgenic plants were selected on the basis of their resistance to hygromycin. The presence and expression of the transgene was confirmed by PCR and protein gel blot analyses.
Physcomitrella patens plants were cultivated in liquid medium containing 250 mg L−1 KH2PO4, 250 mg L−1 KCl, 250 mg L−1 MgSO4×7H2O, 1 g L−1 Ca(NO3)2, and 12.5 mg L−1 FeSO4, pH 5.8, as described (Reski and Abel, 1985). Suspension cultures in 250-mL Erlenmeyer flasks were kept on a rotary shaker at 150 rpm at 21 to 23°C under continuous light (70 µmol photons m−2 s−1). Every 2 weeks, suspension cultures were subcultured after disruption with a Disperser (Miccra D-9, 20 mm disperser, emulsion rotor variant, DS-20/PF EMR) at 16,000 rpm. For chlorophyll a fluorescence analyses, P. patens plants were cultivated on solid medium (12 g purified agar/L and nutrients as for liquid medium) in Petri dishes under the same growth conditions as for suspension cultures. The P. patens mutant ΔPpPAM68 was generated by targeted gene knockout.
Chlorophyll Fluorescence Analysis
In vivo chlorophyll a fluorescence of Arabidopsis leaves was measured using the pulse amplitude modulation 101/103 instrument (PAM 101/103; Walz) as described (Varotto et al., 2000; Tillich et al., 2009). Plants were dark-adapted for 30 min, and minimal fluorescence (Fo) was measured. Then, pulses (0.8 s) of white light (5000 μmol photons m−2 s−1) were used to determine the maximum fluorescence (Fm). The transient increase in fluorescence was recorded after a 5-min exposure to actinic light (80 μmol photons m−2 s−1; as previously described in Tillich et al., 2009). ETR and NPQ of Arabidopsis leaves and in vivo chlorophyll a fluorescence of whole P. patens plants were recorded using the Imaging PAM (Walz). Dark-adapted plants were exposed to a pulsed blue measuring beam (1 Hz, 2.5 µmol photons m−2 s−1; Fo) and a saturating light flash (1000 µmol photons m−2 s−1) to obtain Fv/Fm. ETR and NPQ values of Arabidopsis leaves were determined at 10 different actinic light phases, each lasting 10 min (1, 21, 56, 110, 186, 281, 336, 396, 461, and 531 µmol photons m−2 s−1) by applying saturating light flashes (1000 µmol photons m−2 s−1). The effective quantum yield (ΦII) of P. patens plants was determined at an actinic light intensity of 35 µmol photons m−2 s−1.
Nucleic Acid Analysis
Arabidopsis DNA was isolated according to a phenol- and chloroform-free method (Edwards et al., 1991) and T-DNA insertion-junction sites were recovered by amplification (36 cycles, annealing temperature 55°C) with gene- and T-DNA–specific primers using a DNA Engine Dyad Thermal Cycler (Bio-Rad). For RNA analysis, total leaf RNA was extracted from fresh tissue using the TRIzol reagent (Invitrogen). RT-PCR was performed by synthesizing first-strand cDNA using SuperScript reverse transcriptase (Invitrogen) and dT oligomers, followed by PCR with specific primers. RNA gel blot analyses were performed under stringent conditions, according to standard protocols (Sambrook and Russell, 2001). Probes complementary to ndhA, ndhB, ndhC, ndhD, ndhE, and ndhF transcripts (see Supplemental Table 3 online for primer information), labeled with 32P, were used for the hybridizations. Signals were quantified using a phosphor imager (Typhoon, GE Healthcare) and the program IMAGE QUANT (version 1.2; Molecular Dynamics).
Polysomes were isolated as described (Barkan, 1988). Leaf tissue (200 mg) was frozen with liquid nitrogen in a mortar and ground with a pestle. Subsequently, the microsomal membranes were solubilized with 1% (v/v) Triton X-100 and 0.5% (w/v) sodium deoxycholate. The solubilized material was layered onto 15/55% Suc step gradients (corresponding to 0.44/1.6 M) and centrifuged at 250,000g for 65 min at 4°C. The step gradient was fractionated, and the mRNA associated with polysomes was then extracted with phenol/chloroform/isoamyl alcohol (25:24:1), followed by precipitation at room temperature with 95% ethanol. All samples were then subjected to RNA gel blot analysis. The relative probe signal in each fraction was normalized with respect to the relative rRNA signal. To this end, the total signal (all 10 fractions) was determined for the specific probe, as well as for the rRNA (methylene blue stain), with IMAGE QUANT (version 1.2). The relative amounts of probe and rRNA signal were calculated for each polysome fraction, respectively. Experiments were repeated with three biological replicates.
BN- and 2D-PAGE
Leaves were harvested from 4- to 5-week-old Arabidopsis plants, and thylakoids were prepared as described (Bassi et al., 1985). BN-PAGE analysis was performed as previously described (Peng et al., 2008), and protein blot analysis was performed as described below. For 2D-PAGE, samples were subsequently fractionated by electrophoresis on denaturing Tricine-SDS gels (10%, 4 M urea). Second-dimension gels were subjected to protein blot analysis with antibodies against PAM68L, NdhA, NdhH, PnsB2, PnsL1, and NdhS as described below.
For BN/SDS-PAGE of P. patens thylakoid membranes, plant material propagated in liquid culture was harvested by filtering and homogenized with a pestle in T1 buffer (0.4 M sorbitol, 0.1 M Tricine, pH 7.8, 1 mM benzamidine, 5 mM aminocaproic acid, and 0.2 mM phenylmethylsulfonyl fluoride). The suspension was filtered through a nylon membrane (120 µm) and centrifuged at 4000g for 4 min. Thylakoid membranes in the pellet were resuspended in T2 buffer (20 mM HEPES, pH 7.5, and 10 mM EDTA) and incubated for 10 min. Subsequent steps in BN sample preparation and BN/SDS-PAGE were performed as described by Peng et al. (2008).
Immunoblot Analyses
For 2D-PAGE analysis, proteins were prepared as described above, whereas for conventional SDS-PAGE analysis, total proteins were prepared from 4- to 5-week-old Arabidopsis leaves as reported (Martinez-Garcia et al., 1999), then fractionated on SDS-PAGE gels (10% acrylamide, 4 M urea). P. patens proteins were extracted from plants grown on solid medium as described above. Plant material was homogenized with a pestle in extraction buffer (120 mM Tris-HCl, pH 6.8, 3.5% SDS, 10% glycerol, 1 M urea, 0.05 M DTT, and 0.5 M Na2CO3) and incubated for 1 h at room temperature. After centrifugation (16,000g, 10 min), solubilized proteins were fractionated on 10% Tris-Tricine gels. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes, and replicate filters were incubated with appropriate antibodies. Antibodies for PAM68L were generated as described by Armbruster et al. (2010) and used in a 1:100 dilution. Signals were detected by enhanced chemiluminescence (GE Healthcare) and quantified using ImageJ (http://rsb.info.nih.gov/ij/).
Determination of PAM68L Topology
Intact Arabidopsis chloroplasts were isolated and purified from leaves of 4- to 5-week-old plants as described (Kunst, 1998), ruptured by mixing with 10 volumes of lysis buffer (20 mM HEPES/potassium hydroxide (KOH), pH 7.5, and 10 mM EDTA), and incubated on ice for 30 min. Thylakoid and stroma phases were subsequently separated by centrifugation (42,000g, 30 min; 4°C).
For salt washes of thylakoids according to Karnauchov et al. (1997), isolated thylakoids were resuspended in 50 mM HEPES/KOH, pH 7.5, at a chlorophyll concentration of 0.5 mg/mL. Extraction with 2 M NaCl, 0.1 M Na2CO3, or 2 M sodium thiocyanate was performed for 30 min on ice, soluble and membrane proteins were separated by centrifugation for 10 min at 10,000g at 4°C, and protein gel blot analysis was performed using specific antibodies as described above.
For tryptic proteolysis experiments, thylakoid membranes were isolated and then resuspended in 50 mM HEPES/KOH, pH 8.0, and 300 mM sorbitol at a chlorophyll concentration of 1 mg/mL. Trypsin was added to a concentration of 10 µg/mL. Samples were taken before, and 15 and 30 min after addition of trypsin. Proteins were precipitated with 10 volumes of acetone and resuspended in SDS loading dye containing 5 mM of the Ser endopeptidase inhibitor PMSF.
Heterologous Expression and Protein Titration
Sequences coding for the mature PAM68L and NdhH proteins (the former without cTP) were amplified by RT-PCR and directionally cloned into the pet151-TOPO vector (Invitrogen; PAM68L) or the multiple cloning site of pMal-c2 (New England Biolabs; NdhH) downstream of the MBP coding sequence, using the restriction enzymes EcoRI (5′) and XbaI (3′) (see Supplemental Table 3 online and Hertle et al. [2013] for primer information). Fusion proteins were expressed in the Escherichia coli strain BL21 and purified under native conditions with Nickel-nitrilotriacetic acid (Ni-NTA) (Qiagen; His6-tag) or amylose resin (New England Biolabs; MBP) according to manufacturer's instructions. Purified proteins were quantified (Protein Assay Kit; Bio-Rad), and concentrations of recombinant proteins were calculated. Total amounts of the respective proteins in thylakoid membrane preparations (5 µg chlorophyll) were estimated using IMAGE QUANT to compare thylakoid protein signals, obtained by immunoblot analysis with the appropriate antibodies, with signals given by a dilution series of the recombinant proteins.
Suc Gradient Fractionation of Thylakoid Complexes
Thylakoids were washed twice with 5 mM EDTA, pH 7.8, and diluted in 20 mM Tricine/KOH, pH 7.5, to a chlorophyll concentration of 2 mg/mL. Solubilization of membrane complexes was performed by addition of 2% β-DM and incubation on ice for 10 min. Centrifugation at 16,000g for 5 min at 4°C removed nonsolubilized membranes. The supernatant was loaded onto a linear 0.1 to 1.0 M Suc gradient in 20 mM Tricine/KOH, pH 7.5, and 0.06% (w/v) β-DM and centrifuged at 191,000g for 21 h at 4°C. The gradient was divided into 18 fractions (numbered from the top). Proteins were precipitated by extraction with methanol-chloroform (Wessel and Flügge, 1984) and separated on denaturing gradient Tricine-SDS gels (10 to 16% acrylamide). Protein gel blot analyses were performed as described above. The molecular mass standard covered the range from 25 to 450 kD (Serva).
Cultivation, Immunoblot Analyses, and 2D-BN/SDS-PAGE of Synechocystis
Synechocystis sp PCC 6803 strains were grown at 30°C under continuous light (30 µmol photons m−2 s−1) in BG 11 medium supplemented with 5 mM Glc. The sll0933ˉ mutant (ins0933) was constructed as described previously (Armbruster et al., 2010). Isolation of Synechocystis whole-cell protein extracts was performed, and protein gel blot analyses were performed, as described previously (Schottkowski et al., 2009a). Equal amounts of proteins (40 µg) were loaded in the 100% lane of the wild type and ins0933. For 2D-PAGE analysis, Synechocystis cells were grown to an OD750 of 2 followed by membrane isolation as described (Dühring et al., 2006). Subsequent BN-SDS gel electrophoresis was performed as described (Schottkowski et al., 2009a, 2009b). Samples corresponding to 20 µg of chlorophyll were loaded into each lane in the first dimension.
Construction of P. patens Lines
A fragment of the Pp-PAM68 coding sequence was amplified from genomic DNA with the primers Pam_PP_KO_s2 and Pam_PP_KO_as (see Supplemental Table 3 online) and cloned into pGEM T-Easy (Promega). The selection cassette containing the kanamycin resistance-mediating aph3′II/NptII gene under the control of the 35S CaMV promoter and terminator was obtained by EcoRV digestion of a plasmid employed for the generation of the pGreen vector series (Hellens et al., 2000). The pGEM vector carrying the Pp-PAM68 fragment was linearized using EcoRV and subsequently ligated with the selection cassette. The orientation of the cassette was controlled by sequencing. Prior to transfection, the knockout construct was removed from the pGEM backbone by EcoRI digestion for efficient homologous recombination. Pp-PAM68 knockout constructs were transfected into P. patens protoplasts following standard procedures (Frank et al., 2005). Transgenic lines were analyzed by PCR to identify lines that had integrated the Pp-PAM68 knockout construct into the endogenous PAM68 locus (see Supplemental Table 3 and Supplemental Figure 3 online for primer information). Genomic DNA was extracted from positive clones as described (Edwards et al., 1991) and finally dissolved in 30 μL water. Each 25-μL PCR mixture (GENAXXON, Taq DNA-Polymerase E) contained 3 μL of P. patens DNA and 2 μL of 3 mM spermidine. Five independently generated lines with a knockout of the PAM68 gene were identified by PCR and analyzed by chlorophyll fluorescence analysis. All of them showed changes in photosynthetic parameters that are characteristic for altered PSII function.
For RT-PCR analysis, total RNA was extracted from plant material grown in suspension culture. Residual genomic DNA was removed by DNase I digestion (Fermentas). First-strand cDNA synthesis was performed on 2 µg total RNA using Superscript III (Invitrogen) according to the manufacturer’s protocol. To detect Pp-PAM68 transcripts, the primer pair Pam68_ver_5‘_s and Pam68_ver_3‘_as were used. As control, a primer pair specific for the LHCB4.1-4 family was also used (PpLhcb4.1-4.4for and PpLhcb4.1-4.4rev). For all RT-PCRs 100-ng aliquots of total RNA were used as templates.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative (http://www.Arabidopsis.org/), GenBank (http://www.ncbi.nlm.nih.gov), or EMBL (http://www.ebi.ac.uk/embl/) databases under the following accession numbers: for Arabidopsis: PAM68, At4g19100; PAM68L, At5g52780; SIG6, At2g36990; PDE191, At4g38160; CRR42, At5g20935; CRR6, At2g47910; TIC110, At1g06950; TOC33, At1g02280; PSBX, At2g06520; PSAF, At1g31330; PETE1, At1g76100; CRY2, At1g04400; for Arabidopsis lyrata: Al1, 297804232; Al2, 297796087; for grape (Vitis vinifera): Vv1, 225466099; Vv2, 225464604; for Populus trichocarpa: Pt1, 224138364; Pt2, 224079762; for maize (Zea mays): Zm1, 226491094; Zm2, 226491404; for rice (Oryza sativa subsp japonica): Os1, 297724785; Os2, 115483120; for P. patens subsp patens: Pp, Pp1s342_35V6.1; for Phaeodactylum tricornutum: Pt, 219116028; for Ectocarpus siliculosus: Es, 299470124; for Chlamydomonas reinhardtii: Cr, 159464827; for Volvox carteri: Vc, 302843515; for Micromonas sp RCC299: Ms, 255087933; for Synechocystis sp PCC 6803: Sy, 16329473; for Nostoc punctiforme PCC 73102: Np, 186681223; and for Anabaena variabilis: Av, 75910846.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of Mutants and Overexpressors of Arabidopsis PAM68L.
Supplemental Figure 2. Analysis of Plastid ndh Transcript Association with Polysomes.
Supplemental Figure 3. Generation and Characterization of the P. patens Knockout Mutant ΔPpPAM68.
Supplemental Figure 4. Alignment of PAM68 and PAM68L Protein Sequences for Phylogenetic Analyses.
Supplemental Table 1. Quantification of NDH Subunit Accumulation in Different Genotypes Relative to the Wild Type (Col-0) According to Figures 4A and 4B.
Supplemental Table 2. NDH Subunits in P. patens.
Supplemental Table 3. Overview of Oligonucleotides (5′ to 3′) Used in PCR Assays.
Supplementary Material
Acknowledgments
We thank Toshiharu Shikanai for lines crr2-2, crr4-3, ndhl, ndhs, ndht, and ndhu and antisera against NdhH, NdhL, PnsL4, NdhS, and NdhT; Tsuyoshi Endo for lines pnsb3, ndf5, and pnsb4; Kentaro Ifuku for line pnsl1 and antiserum against PnsL1; Eva-Mari Aro for line pnsb2 and antiserum against PnsB2; Peter Westhoff for antiserum against NdhA and NdhJ; and Dominique Rumeau for antiserum against NdhH. We thank Lina Makarenko for technical support.
AUTHOR CONTRIBUTIONS
U.A., T.R., J.N., W.F., and D.L. designed research. U.A., T.R., R.K., C.S., J.Z., L.T., T.B., A.P.H., Y.Q., and B.R. performed research. U.A., T.R., and D.L. prepared the article. D.L. supervised the whole study.
Glossary
- NDH-C
NAD(P)H dehydrogenase complex
- CEF
cyclic electron flow
- NDH
NAD(P)H dehydrogenase
- ETR
electron transport rate
- NPQ
nonphotochemical quenching
- Col-0
Columbia-0
- BN
blue native
- β-DM
n-dodecyl-β-d-maltoside
- LHCII
light-harvesting complex II
- NCBI
National Center for Biotechnology Information
- 2D
two-dimensional
- PVDF
polyvinylidene difluoride
References
- Alboresi A., Caffarri S., Nogue F., Bassi R., Morosinotto T. (2008). In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: Identification of subunits which evolved upon land adaptation. PLoS ONE 3: e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Armbruster U., Zühlke J., Rengstl B., Kreller R., Makarenko E., Rühle T., Schünemann D., Jahns P., Weisshaar B., Nickelsen J., Leister D. (2010). The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7: 2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R., dal Belin Peruffo A., Barbato R., Ghisi R. (1985). Differences in chlorophyll-protein complexes and composition of polypeptides between thylakoids from bundle sheaths and mesophyll cells in maize. Eur. J. Biochem. 146: 589–595 [DOI] [PubMed] [Google Scholar]
- Battchikova N., Eisenhut M., Aro E.M. (2011a). Cyanobacterial NDH-1 complexes: Novel insights and remaining puzzles. Biochim. Biophys. Acta 1807: 935–944 [DOI] [PubMed] [Google Scholar]
- Battchikova N., Wei L., Du L., Bersanini L., Aro E.M., Ma W. (2011b). Identification of novel Ssl0352 protein (NdhS), essential for efficient operation of cyclic electron transport around photosystem I, in NADPH:plastoquinone oxidoreductase (NDH-1) complexes of Synechocystis sp. PCC 6803. J. Biol. Chem. 286: 36992–37001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows P.A., Sazanov L.A., Svab Z., Maliga P., Nixon P.J. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17: 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Petersen J., Webber-Birungi M.T., Powikrowska M., Lassen L.M., Naumann-Busch B., Nielsen A.Z., Ye J., Boekema E.J., Jensen O.N., Lunde C., Jensen P.E. (2013). Composition and structure of photosystem I in the moss Physcomitrella patens. J. Exp. Bot. 64: 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dühring U., Irrgang K.D., Lünser K., Kehr J., Wilde A. (2006). Analysis of photosynthetic complexes from a cyanobacterial ycf37 mutant. Biochim. Biophys. Acta 1757: 3–11 [DOI] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Shikanai T., Takabayashi A., Asada K., Sato F. (1999). The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 457: 5–8 [DOI] [PubMed] [Google Scholar]
- Frank W., Decker E.L., Reski R. (2005). Molecular tools to study Physcomitrella patens. Plant Biol. (Stuttg.) 7: 220–227 [DOI] [PubMed] [Google Scholar]
- Friedrich T., Scheide D. (2000). The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479: 1–5 [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Endo T., Peltier G., Tasaka M., Shikanai T. (2003). A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36: 541–549 [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Herranen M., Battchikova N., Zhang P., Graf A., Sirpiö S., Paakkarinen V., Aro E.M. (2004). Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803. Plant Physiol. 134: 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertle A.P., Blunder T., Wunder T., Pesaresi P., Pribil M., Armbruster U., Leister D. (2013). PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol. Cell 49: 511–523 [DOI] [PubMed] [Google Scholar]
- Horváth E.M., Peter S.O., Joët T., Rumeau D., Cournac L., Horváth G.V., Kavanagh T.A., Schäfer C., Peltier G., Medgyesy P. (2000). Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 123: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K., Endo T., Shikanai T., Aro E.M. (2011). Structure of the chloroplast NADH dehydrogenase-like complex: Nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 52: 1560–1568 [DOI] [PubMed] [Google Scholar]
- Ifuku K., Ishihara S., Sato F. (2010). Molecular functions of oxygen-evolving complex family proteins in photosynthetic electron flow. J. Integr. Plant Biol. 52: 723–734 [DOI] [PubMed] [Google Scholar]
- Ishida S., Takabayashi A., Ishikawa N., Hano Y., Endo T., Sato F. (2009). A novel nuclear-encoded protein, NDH-dependent cyclic electron flow 5, is essential for the accumulation of chloroplast NAD(P)H dehydrogenase complexes. Plant Cell Physiol. 50: 383–393 [DOI] [PubMed] [Google Scholar]
- Ishihara S., Takabayashi A., Ido K., Endo T., Ifuku K., Sato F. (2007). Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol. 145: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa N., Takabayashi A., Ishida S., Hano Y., Endo T., Sato F. (2008). NDF6: A thylakoid protein specific to terrestrial plants is essential for activity of chloroplastic NAD(P)H dehydrogenase in Arabidopsis. Plant Cell Physiol. 49: 1066–1073 [DOI] [PubMed] [Google Scholar]
- Ishizaki Y., Tsunoyama Y., Hatano K., Ando K., Kato K., Shinmyo A., Kobori M., Takeba G., Nakahira Y., Shiina T. (2005). A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 42: 133–144 [DOI] [PubMed] [Google Scholar]
- Kamruzzaman Munshi M., Kobayashi Y., Shikanai T. (2005). Identification of a novel protein, CRR7, required for the stabilization of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant J. 44: 1036–1044 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karnauchov I., Herrmann R.G., Klösgen R.B. (1997). Transmembrane topology of the Rieske Fe/S protein of the cytochrome b6/f complex from spinach chloroplasts. FEBS Lett. 408: 206–210 [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Mukherjee U., Galla H.J. (2002). Molecular architecture of the thylakoid membrane: Lipid diffusion space for plastoquinone. Biochemistry 41: 4872–4882 [DOI] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kunst L. (1998). Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol. Biol. 82: 43–48 [DOI] [PubMed] [Google Scholar]
- Lennon A.M., Prommeenate P., Nixon P.J. (2003). Location, expression and orientation of the putative chlororespiratory enzymes, Ndh and IMMUTANS, in higher-plant plastids. Planta 218: 254–260 [DOI] [PubMed] [Google Scholar]
- Li X.G., Duan W., Meng Q.W., Zou Q., Zhao S.J. (2004). The function of chloroplastic NAD(P)H dehydrogenase in tobacco during chilling stress under low irradiance. Plant Cell Physiol. 45: 103–108 [DOI] [PubMed] [Google Scholar]
- Majeran W., Zybailov B., Ytterberg A.J., Dunsmore J., Sun Q., van Wijk K.J. (2008). Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol. Cell. Proteomics 7: 1609–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Monte E., Quail P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Meurer J., Plücken H., Kowallik K.V., Westhoff P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y., Hashimoto M., Miyake C., Tomizawa K., Endo T., Tasaka M., Shikanai T. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Munné-Bosch S., Shikanai T., Asada K. (2005). Enhanced ferredoxin-dependent cyclic electron flow around photosystem I and alpha-tocopherol quinone accumulation in water-stressed ndhB-inactivated tobacco mutants. Planta 222: 502–511 [DOI] [PubMed] [Google Scholar]
- Munshi M.K., Kobayashi Y., Shikanai T. (2006). Chlororespiratory reduction 6 is a novel factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Physiol. 141: 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37 (Database issue): D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35 (Database issue): D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Mi H. (2007). Cyanobacterial NADPH dehydrogenase complexes. Photosynth. Res. 93: 69–77 [DOI] [PubMed] [Google Scholar]
- Okegawa Y., Kagawa Y., Kobayashi Y., Shikanai T. (2008). Characterization of factors affecting the activity of photosystem I cyclic electron transport in chloroplasts. Plant Cell Physiol. 49: 825–834 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Ytterberg A.J., Sun Q., van Wijk K.J. (2004). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279: 49367–49383 [DOI] [PubMed] [Google Scholar]
- Peng L., Cai W., Shikanai T. (2010). Chloroplast stromal proteins, CRR6 and CRR7, are required for assembly of the NAD(P)H dehydrogenase subcomplex A in Arabidopsis. Plant J. 63: 203–211 [DOI] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Fujiwara M., Shikanai T. (2012). Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 24: 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Fujiwara M., Takami T., Shikanai T. (2009). Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21: 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2011b). A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biol. 9: e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng L., Shikanai T. (2011). Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Physiol. 155: 1629–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Shimizu H., Shikanai T. (2008). The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 283: 34873–34879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng L., Yamamoto H., Shikanai T. (2011a). Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta 1807: 945–953 [DOI] [PubMed] [Google Scholar]
- Pesaresi P., Varotto C., Richly E., Lessnick A., Salamini F., Leister D. (2003). Protein-protein and protein-function relationships in Arabidopsis photosystem I: Cluster analysis of PSI polypeptide levels and photosynthetic parameters in PSI mutants. J. Plant Physiol. 160: 17–22 [DOI] [PubMed] [Google Scholar]
- Reski R., Abel W.O. (1985). Induction of budding on chloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta 165: 354–358 [DOI] [PubMed] [Google Scholar]
- Rumeau D., Peltier G., Cournac L. (2007). Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 30: 1041–1051 [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W.(2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Sazanov L.A., Burrows P.A., Nixon P.J. (1998). The chloroplast Ndh complex mediates the dark reduction of the plastoquinone pool in response to heat stress in tobacco leaves. FEBS Lett. 429: 115–118 [DOI] [PubMed] [Google Scholar]
- Schottkowski M., Gkalympoudis S., Tzekova N., Stelljes C., Schünemann D., Ankele E., Nickelsen J. (2009b). Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 284: 1813–1819 [DOI] [PubMed] [Google Scholar]
- Schottkowski M., Ratke J., Oster U., Nowaczyk M., Nickelsen J. (2009a). Pitt, a novel tetratricopeptide repeat protein involved in light-dependent chlorophyll biosynthesis and thylakoid membrane biogenesis in Synechocystis sp. PCC 6803. Mol. Plant 2: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2007). Cyclic electron transport around photosystem I: Genetic approaches. Annu. Rev. Plant Biol. 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Shikanai T., Endo T., Hashimoto T., Yamada Y., Asada K., Yokota A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H., Peng L., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2008). CRR23/NdhL is a subunit of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 49: 835–842 [DOI] [PubMed] [Google Scholar]
- Shimizu H., Shikanai T. (2007). Dihydrodipicolinate reductase-like protein, CRR1, is essential for chloroplast NAD(P)H dehydrogenase in Arabidopsis. Plant J. 52: 539–547 [DOI] [PubMed] [Google Scholar]
- Sirpiö S., Allahverdiyeva Y., Holmström M., Khrouchtchova A., Haldrup A., Battchikova N., Aro E.M. (2009). Novel nuclear-encoded subunits of the chloroplast NAD(P)H dehydrogenase complex. J. Biol. Chem. 284: 905–912 [DOI] [PubMed] [Google Scholar]
- Sun X., Fu T., Chen N., Guo J., Ma J., Zou M., Lu C., Zhang L. (2010). The stromal chloroplast Deg7 protease participates in the repair of photosystem II after photoinhibition in Arabidopsis. Plant Physiol. 152: 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M., Sirpiö S., Aro E.M. (2009). Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol. Plant 2: 1127–1140 [DOI] [PubMed] [Google Scholar]
- Suorsa M., Sirpiö S., Paakkarinen V., Kumari N., Holmström M., Aro E.M. (2010). Two proteins homologous to PsbQ are novel subunits of the chloroplast NAD(P)H dehydrogenase. Plant Cell Physiol. 51: 877–883 [DOI] [PubMed] [Google Scholar]
- Takabayashi A., Ishikawa N., Obayashi T., Ishida S., Obokata J., Endo T., Sato F. (2009). Three novel subunits of Arabidopsis chloroplastic NAD(P)H dehydrogenase identified by bioinformatic and reverse genetic approaches. Plant J. 57: 207–219 [DOI] [PubMed] [Google Scholar]
- Tatusova T.A., Madden T.L. (1999). BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247–250 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Hardel S.L., Kupsch C., Armbruster U., Delannoy E., Gualberto J.M., Lehwark P., Leister D., Small I.D., Schmitz-Linneweber C. (2009). Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. USA 106: 6002–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M., Kuniyoshi T., Yamamoto H., Sugimoto K., Ishizaki K., Kohchi T., Nishimura Y., Shikanai T. (2012). Composition and physiological function of the chloroplast NADH dehydrogenase-like complex in Marchantia polymorpha. Plant J. 72: 683–693 [DOI] [PubMed] [Google Scholar]
- Varotto C., Pesaresi P., Meurer J., Oelmüller R., Steiner-Lange S., Salamini F., Leister D. (2000). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22: 115–124 [DOI] [PubMed] [Google Scholar]
- Wang P., Duan W., Takabayashi A., Endo T., Shikanai T., Ye J.Y., Mi H. (2006). Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 141: 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D., Flügge U.I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Yabuta S., Ifuku K., Takabayashi A., Ishihara S., Ido K., Ishikawa N., Endo T., Sato F. (2010). Three PsbQ-like proteins are required for the function of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 51: 866–876 [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Peng L., Fukao Y., Shikanai T. (2011). An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Battchikova N., Jansen T., Appel J., Ogawa T., Aro E.M. (2004). Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16: 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.