SnRK1 protein kinases are negatively regulated by the same PP2C phosphatases that repress the action of the central phytohormone abscisic acid (ABA). During environmental stress, ABA releases PP2C repression, thereby activating two key and complementary pathways and providing better protection against stress through the joined modulation of metabolism and gene expression.
Abstract
Plant survival under environmental stress requires the integration of multiple signaling pathways into a coordinated response, but the molecular mechanisms underlying this integration are poorly understood. Stress-derived energy deprivation activates the Snf1-related protein kinases1 (SnRK1s), triggering a vast transcriptional and metabolic reprogramming that restores homeostasis and promotes tolerance to adverse conditions. Here, we show that two clade A type 2C protein phosphatases (PP2Cs), established repressors of the abscisic acid (ABA) hormonal pathway, interact with the SnRK1 catalytic subunit causing its dephosphorylation and inactivation. Accordingly, SnRK1 repression is abrogated in double and quadruple pp2c knockout mutants, provoking, similarly to SnRK1 overexpression, sugar hypersensitivity during early seedling development. Reporter gene assays and SnRK1 target gene expression analyses further demonstrate that PP2C inhibition by ABA results in SnRK1 activation, promoting SnRK1 signaling during stress and once the energy deficit subsides. Consistent with this, SnRK1 and ABA induce largely overlapping transcriptional responses. Hence, the PP2C hub allows the coordinated activation of ABA and energy signaling, strengthening the stress response through the cooperation of two key and complementary pathways.
INTRODUCTION
Changes in water and nutrient availability, soil salinity, and extreme temperatures, among others, generate signals in plants that need to be finely integrated with metabolic activity and development for optimal growth and survival (Smith and Stitt, 2007). One such signal is energy deficiency derived from impaired carbon assimilation and/or respiration in situations of stress, which triggers the activation of the SnRK1 protein kinases to restore homeostasis and elaborate adequate longer term responses through a vast metabolic and transcriptional reprogramming (Radchuk et al., 2006; Schwachtje et al., 2006; Baena-González et al., 2007; Baena-González and Sheen, 2008; Lee et al., 2009). The Arabidopsis thaliana genome encodes 38 SnRKs, of which three, SnRK1.1 (KIN10/AKIN10), SnRK1.2 (KIN11/AKIN11), and SnRK1.3 (KIN12/AKIN12), represent the orthologs of the budding yeast (Saccharomyces cerevisiae) sucrose-nonfermenting1 (Snf1) and mammalian AMP–activated protein kinase (AMPK) metabolic sensors (Halford et al., 2003; Polge and Thomas, 2007; Hardie, 2011). An increasing body of evidence suggests that SnRK1s act as convergence points for various metabolic, hormonal and stress signals during growth and development, linking it to key hormonal pathways and in particular to abscisic acid (ABA; Németh et al., 1998; Bhalerao et al., 1999; Bradford et al., 2003; Radchuk et al., 2006; Baena-González et al., 2007; Lu et al., 2007; Rosnoblet et al., 2007; Ananieva et al., 2008; Baena-González and Sheen, 2008; Lee et al., 2008; Jossier et al., 2009; Radchuk et al., 2010; Coello et al., 2012; Tsai and Gazzarrini, 2012). SnRK1 is a heterotrimeric complex composed of an α-catalytic subunit (SnRK1.1/1.2/1.3 in Arabidopsis) and two regulatory subunits, β and γ (Polge and Thomas, 2007). Similarly to its mammalian and yeast counterparts, SnRK1 activity requires phosphorylation of a highly conserved T-loop residue (T175 in SnRK1.1) (Estruch et al., 1992; Hawley et al., 1996; Stein et al., 2000; McCartney and Schmidt, 2001; Baena-González et al., 2007; Shen et al., 2009; Crozet et al., 2010). Under normal energy conditions in mammalian cells, MgATP is bound to the γ-subunit of the AMPK complex resulting, through the joint action of the constitutively active upstream liver kinase B1 and the still unknown upstream phosphatase, in a basal T-loop phosphorylation:dephosphorylation cycle with no net AMPK activation (Hardie, 2011). Under energy deficiency conditions, the replacement of MgATP by AMP/ADP triggers a conformational change that promotes AMPK phosphorylation and, most importantly, protects AMPK from dephosphorylation by rendering it a poor substrate for phosphatases (Oakhill et al., 2011; Xiao et al., 2011). Despite the rate of dephosphorylation being a primary determinant of AMPK activity, the identity of the AMPK phosphatase(s) remains unclear and may differ between tissues and conditions of cell stimulation (Steinberg and Kemp, 2009; Carling et al., 2012). In budding yeast, Reg1, a regulatory subunit of the type 1 protein phosphatase Glc7, interacts with Snf1 and is required to maintain Snf1 in an inactive state during growth on Glc (Sanz et al., 2000; Hong et al., 2005). The metabolic signal underlying Snf1 regulation remained enigmatic for a long time, but recent work demonstrated that Snf1 is also regulated by ADP at the substrate level, preventing its dephosphorylation by phosphatases (Mayer et al., 2011). In plants, SnAK1/2 (also called Geminivirus Rep interacting kinase 2/1) have been identified as upstream SnRK1 kinases (Shen et al., 2009; Crozet et al., 2010), but the phosphatases responsible for resetting SnRK1 signaling are unknown.
In Arabidopsis, at least seven of the nine type 2C protein phosphatases (PP2Cs) from clade A (Schweighofer et al., 2004) act as negative regulators of the ABA pathway (Gosti et al., 1999; Merlot et al., 2001; Leonhardt et al., 2004; Saez et al., 2004, 2006; Kuhn et al., 2006; Yoshida et al., 2006; Nishimura et al., 2007; Rubio et al., 2009; Antoni et al., 2012) through their interaction with SnRK2s, more divergent members of the SnRK family and specific to plants (Halford et al., 2003; Cutler et al., 2010). Arabidopsis contains 10 SnRK2s, of which three, SnRK2.2/2.3/2.6, are specifically activated by ABA and play a central role in the ABA pathway (Gómez-Cadenas et al., 1999; Li et al., 2000; Mustilli et al., 2002; Boudsocq et al., 2004, 2006; Yoshida et al., 2006; Fujii et al., 2007, 2009). Clade A PP2Cs regulate SnRK2.2/2.3/2.6 through physical obstruction and direct dephosphorylation of a conserved Ser residue in the T-loop (S175 in SnRK2.6) (Umezawa et al., 2009; Vlad et al., 2009; Soon et al., 2012). In the presence of ABA, the Pyrabactin Resistance1/Pyrabactin Resistance1-Like (PYL)/Regulatory Components of ABA Receptors family of ABA receptors (hereafter PYL) inhibit PP2Cs, resulting in SnRK2 activation and downstream gene expression (Ma et al., 2009; Park et al., 2009; Soon et al., 2012).
Considering that clade A PP2Cs, through interaction with a wide array of targets, act as a regulatory hub for different abiotic stress responses (Sheen, 1996; Chérel et al., 2002; Guo et al., 2002; Himmelbach et al., 2002; Ohta et al., 2003; Miao et al., 2006; Yang et al., 2006; Umezawa et al., 2009; Vlad et al., 2009; Geiger et al., 2010) and taking into account the role of SnRK1 as a convergence point for multiple types of stress (Baena-González et al., 2007), we postulated that clade A PP2Cs might function as SnRK1 phosphatases. An additional hint came from data mining on a high-throughput proteomics screen for yellow fluorescent protein (YFP)-ABI1-interacting proteins, which inadvertently identified SnRK1s as putative ABI1-interacting proteins (Nishimura et al., 2010) (see below).
Here, we provide molecular, genetic, and physiological evidence for the role of two clade A PP2Cs, ABI1 and PP2CA, as negative regulators of SnRK1 signaling in Arabidopsis through their direct interaction with the SnRK1 α-catalytic subunit, its dephosphorylation, and subsequent inactivation, hence contributing to resetting SnRK1 signaling upon the remittance of stress. In contrast, PP2C inhibition allows ABA to promote SnRK1 activity, potentiating the stress response through the interplay of two complementary pathways and providing an explanation for the extensive genetic interactions reported between ABA and sugar signaling (Rolland et al., 2006).
RESULTS
ABI1 and PP2CA Interact with the SnRK1 Catalytic Subunit
A high-throughput screen using green fluorescent protein (GFP)–affinity purification and mass-spectrometric analyses was performed by Nishimura and colleagues to identify proteins interacting with YFP-ABI1 (Nishimura et al., 2010). Data mining of their results revealed the presence of peptides corresponding to both SnRK1s in several of their replicate experiments with YFP-ABI1 (SnRK1.1 in experiments 1, 3, and 8 and SnRK1.2 in experiments 1 and 3), whereas neither of the two SnRK1s was identified in any of the YFP control experiments.
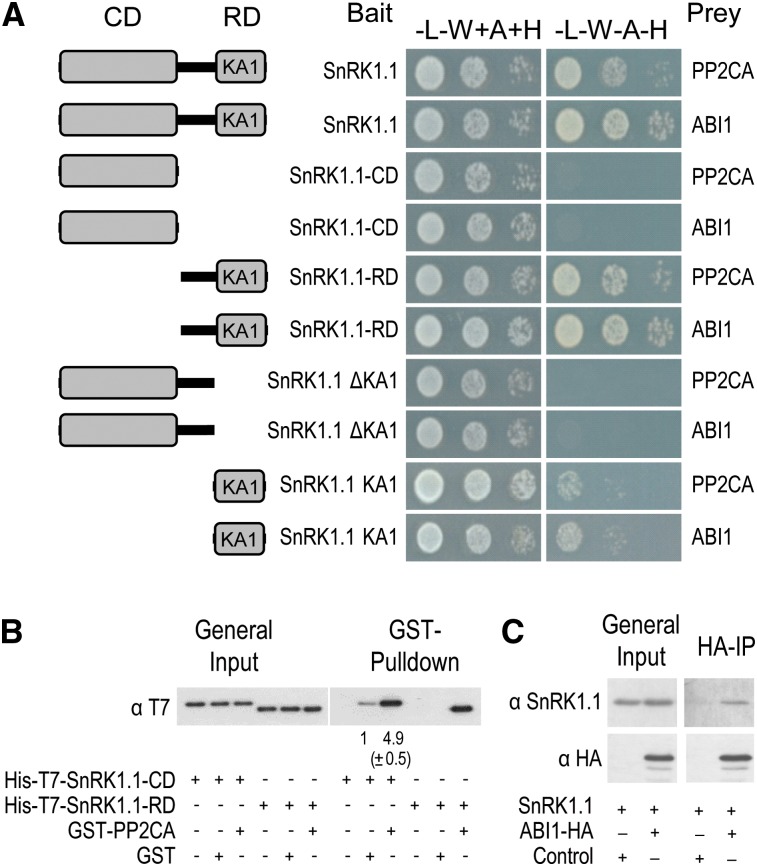
As a first step to validate these data and investigate the possible regulation of SnRK1 by clade A PP2Cs, we tested in yeast two-hybrid (Y2H) assays the interaction between the SnRK1 catalytic subunit and ABI1 or PP2CA, representative members of the two clade A branches in the PP2C family (Schweighofer et al., 2004). SnRK1.1 interacted with ABI1 and PP2CA in yeast cells, and deletion of its regulatory domain (RD) abolished this interaction (Figure 1A; see Supplemental Figure 1A online). The N terminus harbors the kinase catalytic domain (CD), whereas the C terminus harbors the RD that binds the β- and γ-subunits (Polge and Thomas, 2007). The SnRK1 RD contains a subdomain of unknown function, the kinase-associated1 (KA1) domain, that was reported in the SnRK3.11/Salt Overly Sensitive2 (SOS2) protein kinase to closely superimpose on the protein phosphatase interaction domain (Sánchez-Barrena et al., 2007), a docking site for the clade A PP2C ABI2 (Ohta et al., 2003). Modeling SnRK1.1 with the structures resolved for the KA1 domain in SnRK3.11 (Sánchez-Barrena et al., 2007), the AMPK-related microtubule-affinity–regulating kinase3 (Tochio et al., 2006), and for AMPKα (Xiao et al., 2011), revealed that in SnRK1.1, this subdomain spans residues 390 to 512 (see Supplemental Figure 2 online). As shown, the KA1 domain was both required and sufficient for the interaction with the phosphatase (Figure 1A). Nevertheless, colony growth when using the KA1 domain alone was weaker than with SnRK1.1-RD or the full-length protein, suggesting that other regions may play a role in the PP2C interaction.
Figure 1.
ABI1 and PP2CA Interact with SnRK1.1 in Vitro and in Vivo.
(A) SnRK1.1 interacts with ABI1 and PP2CA in Y2H assays. Protein interaction was determined by growth assay in medium lacking Leu, Trp, adenine and His (−L−W−A−H) compared with control medium lacking Leu and Trp but supplemented with adenine and His (−L−W+A+H).
(B) In vitro interaction between GST-PP2CA and His-T7-SnRK1.1 detected by GST pull down and T7 immunodetection of SnRK1.1 preys. Numbers below immunoblot denote band intensities compared with GST-alone control (=1); values represent means ±sd (n = 3).
(C) HA immunoprecipitation pulls down SnRK1.1 from protoplasts coexpressing SnRK1.1 (untagged) with ABI1-HA, but not with control DNA.
[See online article for color version of this figure.]
To further validate the Y2H data, we performed an in vitro pull-down assay (Figure 1B). Purified recombinant His-SnRK1.1-CD or His-SnRK1.1-RD was incubated with glutathione S-transferase (GST)—PP2CA, GST, or the beads and the interacting proteins were pulled down using a glutathione–agarose matrix. SnRK1.1-RD was recovered only when using GST-PP2CA as bait. In the case of SnRK1.1-CD, a fivefold enrichment was observed when using GST-PP2CA compared with GST alone, suggesting that even though not detected in the Y2H assay, PP2Cs interact also to some degree with the SnRK1.1-CD. No SnRK1-RD or SnRK1.1-CD was recovered from the beads alone. To determine whether a SnRK1.1-PP2C interaction occurs also in planta, SnRK1.1 was transiently coexpressed in Arabidopsis protoplasts with control DNA or with a plasmid expressing ABI1-hemagglutinin (HA). Immunoprecipitation with an anti-HA antibody revealed a specific interaction between SnRK1.1 and ABI1-HA (Figure 1C), demonstrating that ABI1 also interacts with SnRK1.1 in vivo.
ABI1 and PP2CA Dephosphorylate and Inactivate SnRK1.1
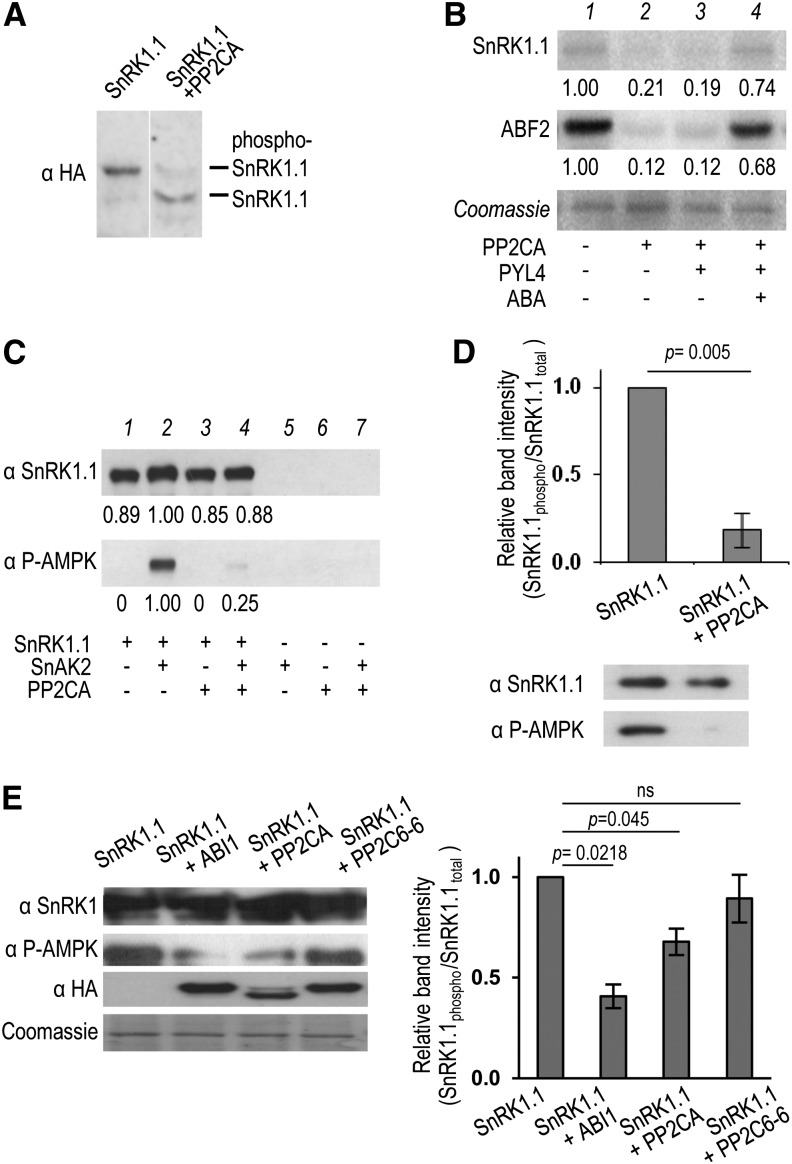
To evaluate whether the detected PP2C-SnRK1.1 interaction results in SnRK1.1 dephosphorylation and inactivation, we immunoprecipitated SnRK1.1 from plants overexpressing an HA-tagged version (35S:SnRK1.1-HA) (Baena-González et al., 2007) and treated with recombinant His-PP2CA. PP2CA treatment caused a clear dephosphorylation of SnRK1.1, as assessed by a faster mobility in a Phos-Tag SDS-PAGE that selectively retards phosphorylated proteins (Kinoshita et al., 2009) (Figure 2A). To investigate the effect of this dephosphorylation on SnRK1 activity, we performed in vitro kinase assays. In agreement with previous reports, active SnRK1.1 could efficiently autophosphorylate and phosphorylate the Abscisic acid responsive elements-Binding Factor2 (ABF2) transcription factor in vitro (Bhalerao et al., 1999; Zhang et al., 2008; Shen et al., 2009) (Figure 2B, lane 1). No ABF2 phosphorylation could be observed in control HA pull downs from wild-type (WT) plants, confirming that the measured activity corresponds to SnRK1-HA (see Supplemental Figure 3A online). Addition of PP2CA to the reaction caused a substantial decrease in the phosphorylation of both SnRK1.1 and ABF2 (Figure 2B, lane 2). The PYL receptors inhibit clade A PP2Cs in the presence of ABA, resulting in SnRK2 activation (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009). Adding the PYL4 receptor in the absence of ABA did not change the ability of PP2CA to inactivate SnRK1 (Figure 2B, lane 3), whereas in the presence of ABA, PYL4 fully blocked SnRK1.1 inactivation by PP2CA (Figure 2B, lane 4). To rule out the possibility that decreased ABF2 phosphorylation in the presence of PP2CA results from direct ABF2 dephosphorylation by PP2CA rather than from lower SnRK1 activity, SnRK1.1 was preincubated with PP2CA and PYL4 in the absence (PP2CA active) or presence (PP2CA inactive) of ABA (see Supplemental Figure 3B, lanes 2 and 3, online). Following this incubation, ABA was added to block further PP2CA action before the addition of ABF2. Preincubation of SnRK1 with PP2CA in the absence of ABA resulted in undetectable SnRK1 activity and ABF2 phosphorylation, suggesting that the effect of PP2CA on ABF2 phosphorylation was at least partly due to a reduction in SnRK1 activity rather than to a direct dephosphorylation of ABF2 by the phosphatase.
Figure 2.
ABI1 and PP2CA Inhibit SnRK1.1 by Dephosphorylation.
Immunoprecipitated SnRK1.1-HA is dephosphorylated (A) and inactivated (B) in vitro by PP2CA.
(A) HA immunoblot following Phos-Tag-SDS-PAGE (Kinoshita et al., 2009).
(B) Autoradiograms showing that SnRK1.1 activity on itself and ABF2 (lane 1) is lost following His-PP2CA-treatment (lane 2) but rescued by PYL4 and ABA (lane 4). GST-PP2CA dephosphorylates T175 in recombinant SnRK1.1, phosphorylated or not with SnAK2 (C), and in immunoprecipitated SnRK1.1 (n = 3) (D) in vitro. Numbers below autoradiograms and immunoblots denote band intensities relative to SnRK1.1 control (=1). At least three independent experiments were performed in (A) to (C) with similar results.
(E) Coexpression in protoplasts of SnRK1.1 with clade A PP2Cs ABI1 and PP2CA, but not with clade E PP2C6-6, results in SnRK1.1(T175) dephosphorylation. PP2Cs and SnRK1.1 bear HA and GFP tags, respectively. SnRK1.1(T175) phosphorylation was detected by immunodetection with anti-phospho-AMPKα(T172) antibodies (n = 6). Error bars = se; P values, two-tailed paired Student’s t test (D) and one-way ANOVA with Tukey test (E) on the nonnormalized ratio of SnRK1.1(T175) phosphorylation relative to total SnRK1.1.
SnRK1 requires phosphorylation of the T-loop T175 residue for activity (Baena-González et al., 2007; Shen et al., 2009; Crozet et al., 2010). To test whether T175 could be a substrate for ABI1 and PP2CA, we first performed in vitro dephosphorylation experiments. Recombinant SnRK1.1 is not phosphorylated and hence is barely active but it can be strongly activated by the upstream kinases SnAK1/2 through the specific phosphorylation of T175 (Shen et al., 2009; Crozet et al., 2010). GST-PP2CA treatment of recombinant GST-SnRK1.1, prephosphorylated with GST-SnAK2, resulted in significant T175 dephosphorylation, as detected with an anti-phospho-AMPKα(T172) (T172) antibody (Sugden et al., 1999; Baena-González et al., 2007) (Figure 2C) that specifically recognizes SnRK1.1 and SnRK1.2 phosphorylated in the T-loop (T175 for SnRK1.1; see Supplemental Figure 4 online). A similar effect was observed when SnRK1.1 was immunoprecipitated from 35S:SnRK1.1-HA plants and treated with GST-PP2CA (Figure 2D), altogether showing that T175 is efficiently dephosphorylated by PP2Cs in vitro.
To determine whether T175 is a PP2C substrate in vivo, we used Arabidopsis mesophyll protoplasts to transiently express SnRK1.1-GFP alone or in combination with various PP2Cs. As shown in Figure 2E, coexpression of SnRK1.1-GFP with either ABI1 or PP2CA (from clade A) resulted in a significant reduction in T175 phosphorylation levels, while coexpression with the unrelated PP2C6-6 from clade E (Schweighofer et al., 2004) did not have an impact on T175 phosphorylation. These results suggest that T175 is a substrate for ABI1 and PP2CA also in vivo.
ABI1 and PP2CA Repress SnRK1 Signaling
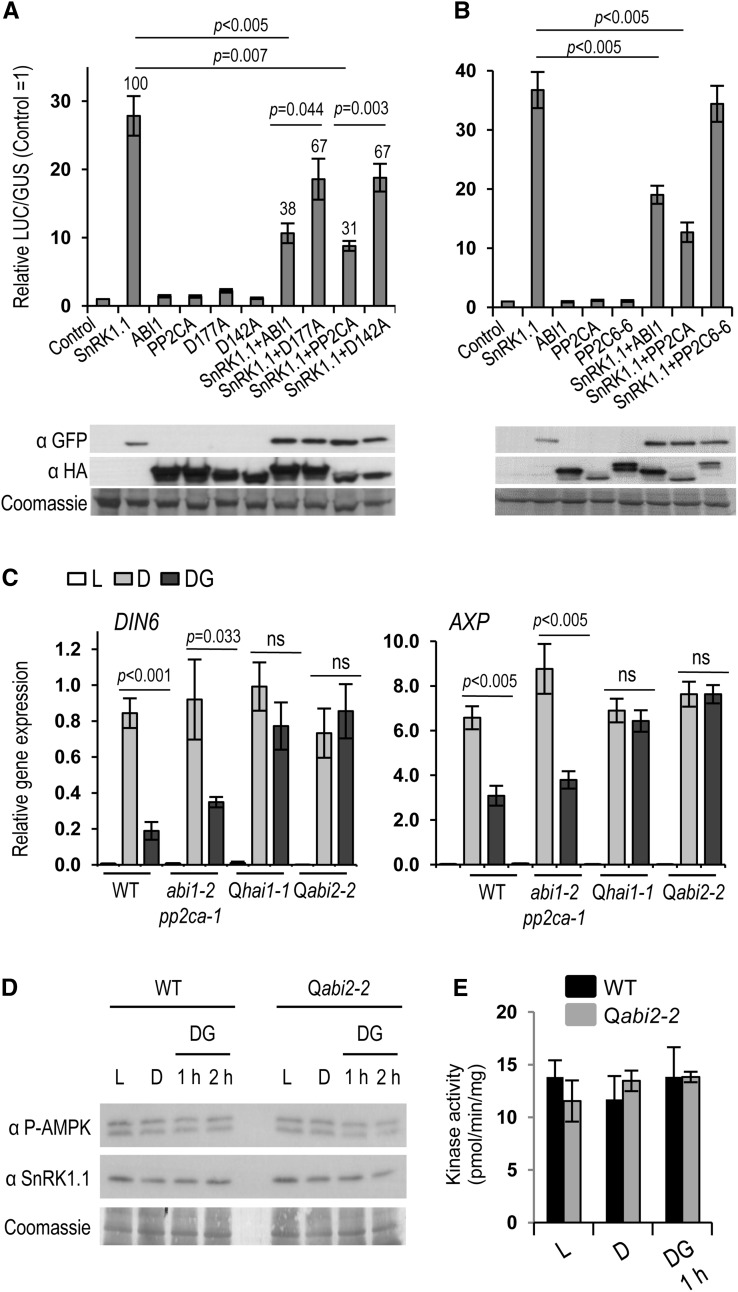
To further explore the functional implications of SnRK1 regulation by PP2Cs, we employed a transient cell-based assay that uses luciferase (LUC) induction from the DIN6:LUC reporter as a readout of SnRK1 activity (Baena-González et al., 2007). In transfected mesophyll protoplasts, SnRK1.1 overexpression is sufficient to induce strong LUC activity under control conditions (Figure 3A) (Baena-González et al., 2007). Coexpression with the ABI1 or PP2CA phosphatases reduced SnRK1.1-mediated DIN6:LUC induction by 60% without affecting SnRK1.1 levels (Figure 3A). Importantly, the ability of these phosphatases to repress reporter gene induction by SnRK1.1 was strongly diminished in the corresponding catalytically inactive variants (ABI1_D177A and PP2CA_D142A; Figure 3A), suggesting that repression of SnRK1 signaling by ABI1 and PP2CA occurs to a large extent through dephosphorylation. As a negative control, coexpression with the unrelated PP2C6-6 from clade E (Schweighofer et al., 2004) had no significant effect on the ability of SnRK1.1 to induce the reporter (Figure 3B), altogether supporting the specific repressive role of ABI1 and PP2CA on the SnRK1 pathway.
Figure 3.
ABI1 and PP2CA Repress SnRK1 Signaling.
(A) SnRK1.1 activity, measured as the induction of the DIN6:LUC reporter in protoplasts is severely reduced by clade A PP2Cs ABI1 and PP2CA, but to a much lesser extent by the corresponding catalytically inactive mutants ABI1_D177A and PP2CA_D142A (n = 9). Numbers above columns designate the percentage of SnRK1.1 inhibition as compared with 100% activity in the absence of PP2Cs.
(B) An unrelated clade E PP2C6-6 does not impinge on SnRK1.1 activity (n = 8).
(C) Reduced SnRK1 inactivation in double and quadruple pp2c knockout mutants Qhai1-1 and Qabi2-2. Relative gene expression of SnRK1.1 marker genes (DIN6, AXP) in control ( L), activating ( D), and inactivating (DG) conditions (n = 4). P values, one-way ANOVA with Tukey (A) and (B) and two-way ANOVA with Sidak test (C). Error bars = se. Analyses of SnRK1(T175) phosphorylation (D) and SnRK1 activity (E) from total cellular extracts reveal no differences in various conditions and between wild-type and Qabi2-2 mutant plants.
(D) SnRK1.1(T175) phosphorylation was detected by immunodetection with anti-phospho-AMPKα(T172) antibodies at the indicated time points. (E) SnRK1 activity was measured using SnRK1 immunoprecipitated from wild-type or Qabi2-2 leaves using the AMARA peptide assay. Values represent means ± sd (n = 2).
To investigate the influence of ABI1, PP2CA, and other clade A PP2Cs on endogenous SnRK1 signaling, we treated detached Arabidopsis leaves of the wild type, the double abi1-2 pp2ca-1 (Rubio et al., 2009), and two different quadruple pp2c knockout mutants (hai1-1 pp2ca-1 hab1-1 abi1-2, hereafter Qhai1-1; abi2-2 pp2ca-1 hab1-1 abi1-2, hereafter Qabi2-2; see Supplemental Figure 5 online; Antoni et al., 2013) under control (3 h of light [L]), activating (3 h of darkness [D]) and inactivating conditions (3 h of darkness followed by 1 h of darkness in 50 mM Glc [DG]), and analyzed SnRK1 target gene expression (Baena-González et al., 2007) by quantitative RT-PCR (qRT-PCR). Exposure to darkness triggered a strong induction of SnRK1 target genes in all genotypes (Figure 3C), in agreement with the current view that the conformation adopted by AMPK and Snf1 under conditions of low energy renders the kinases resistant to phosphatase action (Mayer et al., 2011; Oakhill et al., 2011; Xiao et al., 2011). In marked contrast, SnRK1 inactivation in response to subsequent Glc addition was deficient in abi1-2 pp2ca-1 plants (for DIN6) and completely blocked in the quadruple pp2c mutants (Figure 3C), demonstrating that clade A PP2Cs are essential components for the poststress inactivation of SnRK1 signaling.
In agreement with previous work (Baena-González et al., 2007), and despite the clear effect of PP2Cs on SnRK1 signaling under L, D, and DG conditions, analyses of total protein extracts of wild-type and Qabi2 leaves revealed no clear differences with regard to T175 phosphorylation or total SnRK1 activity (Figures 3D and 3E). This suggests that subtle changes in SnRK1 phosphorylation and activity are sufficient to trigger significant downstream effects in gene expression, and that neither immunodetection with phospho-AMPKα(T172) antibodies nor SnRK1 kinase assays on total cellular SnRK1 are sensitive enough to monitor these changes.
Altered Sugar Responses in pp2c Mutants
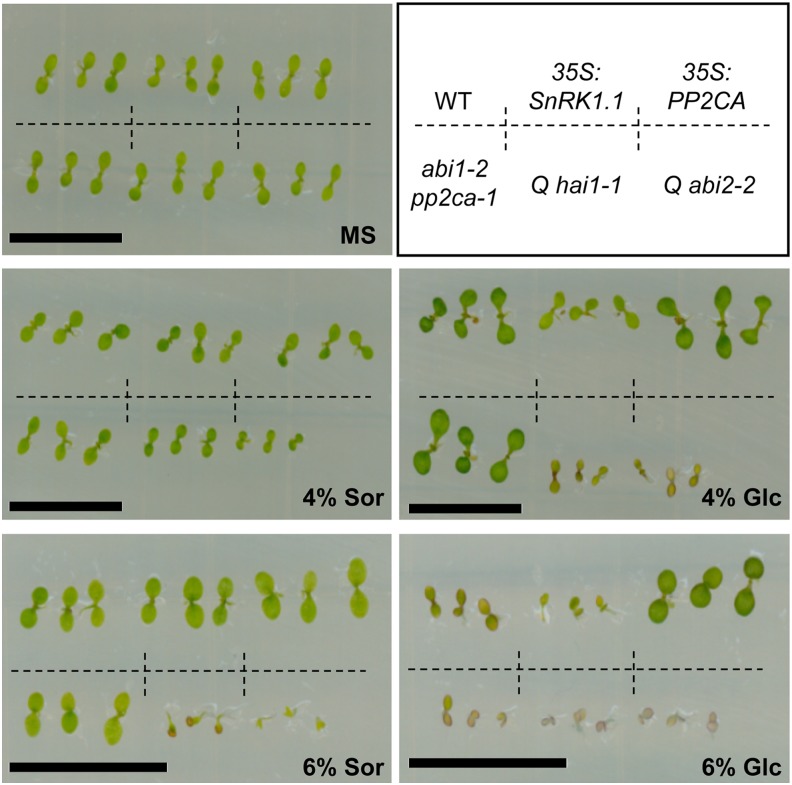
High concentrations of sugars (6% Glc, ∼330 mM) induce a developmental arrest characterized, for instance, by repression of cotyledon greening and expansion (Rolland et al., 2006). Wild-type seedlings grow well on plates containing 4% Glc but cotyledon greening and expansion are clearly impaired on higher sugar concentrations (Figure 4). Such adverse conditions trigger SnRK1 activation, leading to sugar hypersensitivity in 35S:SnRK1.1 seedlings (Jossier et al., 2009) (Figure 4). The abi1-2 pp2ca-1 double mutant displays Glc hypersensitivity visible only in 6% Glc, but this is markedly enhanced in the quadruple pp2c mutants, which exhibit a clear phenotype in 4% Glc (Figure 4). Even though the ABA hypersensitivity of these mutants (see Supplemental Figure 5 online) renders them more sensitive to increased osmolarity in the 4% sorbitol control plates (Antoni et al., 2012), a clear impact on development can be observed on 4% Glc plates. In 6% sorbitol and Glc plates, the growth of these mutants is so compromised that a distinction between osmotic and sugar effects is not possible. Consistent with the loss-of-function phenotype, plants overexpressing PP2CA are sugar insensitive (Figure 4), altogether genetically supporting the role of PP2Cs as negative regulators of SnRK1 signaling.
Figure 4.
Altered Glc Response in pp2c Knockout Mutants and PP2C Overexpressors.
Glc hypersensitivity of SnRK1.1 overexpressors (35S:SnRK1.1; 4-6% glc), double (abi1-2 pp2ca-1; 6% glc) and quadruple pp2c knockout mutants (Qhai1-1 and Qabi2-2; 4% glc), and Glc insensitivity of PP2CA overexpressors (35S:PP2CA; 6% glc) in early seedling development. Sor, sorbitol osmotic control; MS, control media without Glc or sorbitol. Bar = 1 cm.
ABA Promotes SnRK1 Signaling via PP2Cs
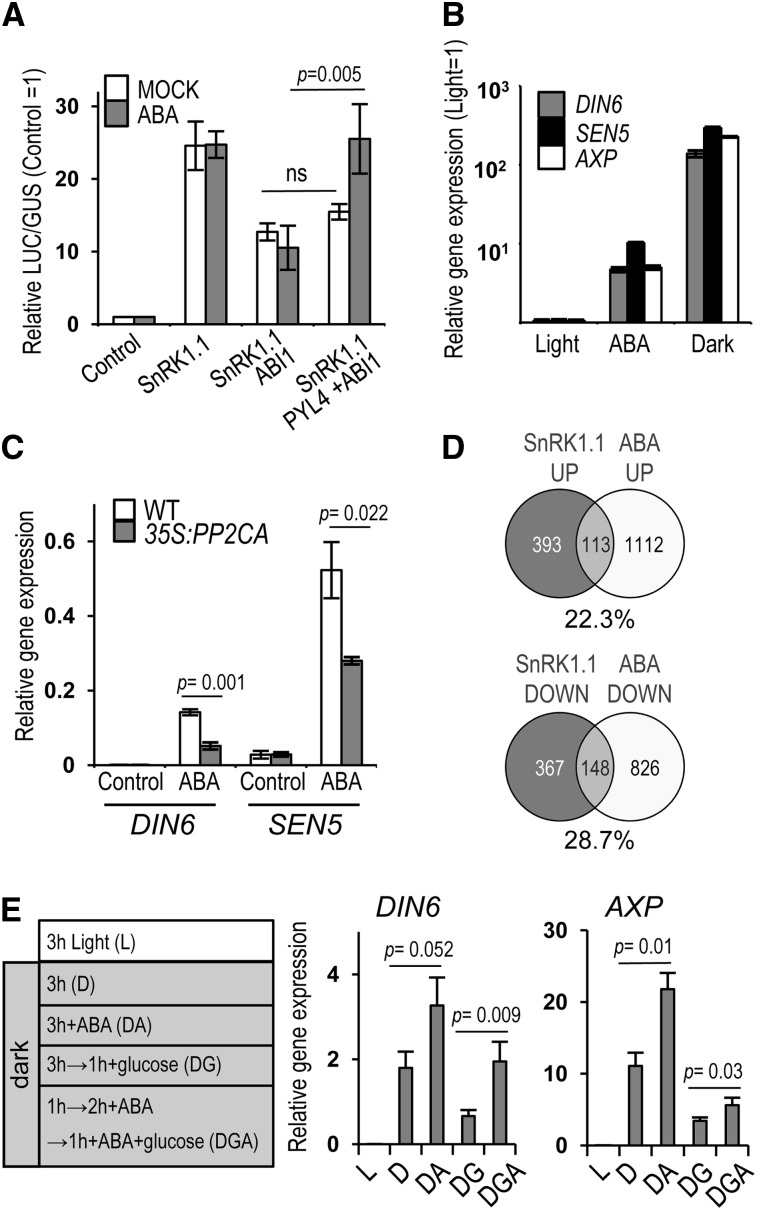
We next wanted to assess whether PP2C regulation of the SnRK1 pathway could allow ABA to modulate SnRK1 activity. The transient coexpression of PYL receptors with ABI1 in ABA-treated mesophyll protoplasts is enough to efficiently repress ABI1 action and to trigger the activation of an ABA signaling reporter (Fujii et al., 2009). Similarly, coexpression of ABI1 with PYL4 in the presence of ABA fully restored SnRK1.1 ability to induce the DIN6:LUC reporter in protoplasts (Figure 5A), presumably through ABI1 sequestration in the ABA-PYL-PP2C ternary complex. We observed an overall twofold increase in LUC activity when comparing mock- and ABA-treated samples (see Supplemental Figure 6 online), further suggesting that ABA can induce SnRK1 signaling. To further explore this possibility and to examine the effect of ABA on other SnRK1 target genes (Baena-González et al., 2007), we treated Arabidopsis leaf discs with or without ABA (100 µM) for 5 h and quantified downstream gene expression changes by qualitative qRT-PCR. ABA treatment did activate SnRK1, albeit to an extent 1 order of magnitude lower than that triggered by darkness (Figure 5B). Most importantly, the impact of ABA on SnRK1 target genes was reduced in plants overexpressing PP2CA (35S:PP2CA; Figure 5C) (Antoni et al., 2012), indicating that the effect of ABA on SnRK1 activity is via PP2C inhibition. To investigate this connection at the whole genome level, we compared the transcriptional profile associated with SnRK1.1 activation in protoplasts (Baena-González et al., 2007) with that of seedlings treated with ABA http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp, AtGenExpress Consortium; Nemhauser et al., 2006). Despite differences in tissue type and developmental stage in the two data sets, there was a significant overlap between the transcriptional changes triggered by SnRK1.1 and by ABA (Figure 5D; see Supplemental Data Set 1 online). More than 22 and 28% of the total number of genes upregulated and downregulated by SnRK1.1, respectively, were similarly regulated by ABA, in marked contrast with the negligible overlap with other hormone treatments or when comparing genes oppositely regulated in the SnRK1.1 and ABA data sets (see Supplemental Figure 7 online). Despite the wide impact of both SnRK1 and ABA on the transcriptome, the probability of obtaining such an overlap of similarly regulated genes by chance is very low (hypergeometric test, P < 9.2−42).
Figure 5.
ABA Promotes SnRK1 signaling.
(A) PP2C repression of SnRK1 signaling in protoplasts is blocked by coexpression of the PYL4 receptor in the presence of ABA (n = 3).
(B) Induction of SnRK1 target genes by ABA (n = 10) and energy stress (D; n = 12).
(C) Reduced induction of SnRK1 target genes by ABA in 35S:PP2CA plants (n = 3).
(D) SnRK1 activation and ABA treatment induce largely overlapping transcriptional responses. Percentage of upregulated or downregulated SnRK1.1 targets similarly regulated by ABA.
(E) ABA enhances SnRK1 activation by darkness and diminishes its Glc-triggered inactivation. SnRK1 target gene expression in L, DA, or D. Following dark activation, SnRK1 repression triggered by Glc was examined with (DGA) or without (DG) ABA pretreatment (n = 4). Error bars = se. P values, two-way ANOVA with Fisher's least significant difference test. DIN6, SEN5, AXP, SnRK1, target genes.
We next analyzed SnRK1 target gene expression in wild-type leaf discs subjected to ABA at the beginning of the dark treatment to test the combined effect of ABA and energy stress or 2 h prior to Glc addition to test the impact of ABA on the sugar-induced inactivation of SnRK1. Addition of ABA enhanced SnRK1 activation by darkness (Figure 5E, samples D, and DA). Moreover, adding ABA prior to Glc diminished SnRK1 inactivation in response to sugar (Figure 5E, samples DG and DGA). Collectively, these results show that ABA positively regulates SnRK1 signaling by inhibiting clade A PP2Cs, thereby promoting SnRK1 signaling during stress and once energy deficiency remits.
DISCUSSION
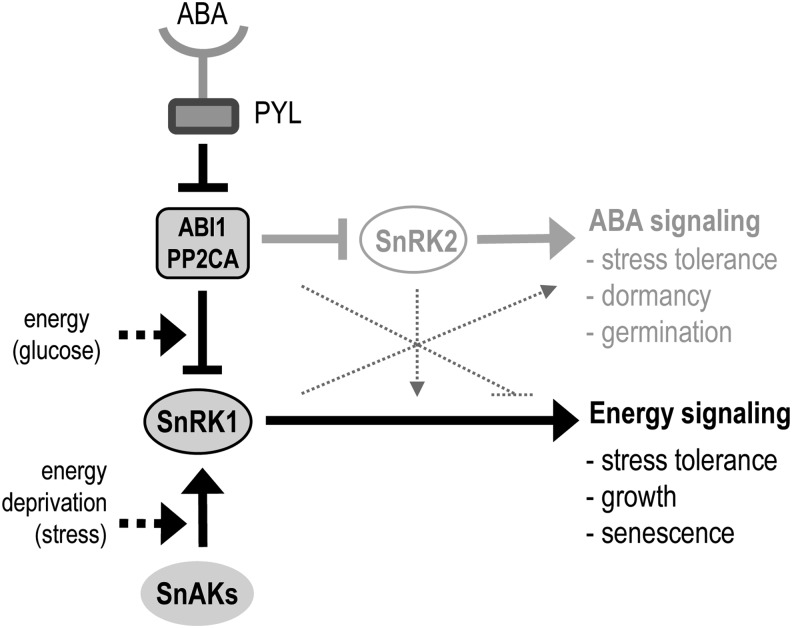
Despite the central role of SnRK1 kinases in the plant stress response, the regulatory mechanisms underlying SnRK1 function are poorly understood. We have demonstrated here that ABI1 and PP2CA are bona fide SnRK1 phosphatases that contribute to resetting SnRK1 activity upon restoration of energy levels and that allow ABA to induce and potentiate SnRK1 signaling during stress (Figure 6). Although our results indicate that several clade A PP2Cs, including ABI1 and PP2CA, are important for SnRK1 regulation, this may not be true for all members of this clade. Furthermore, even though clade E PP2C6-6 had no significant impact on SnRK1 phosphorylation and signaling (Figures 2E and 3B), we cannot exclude the possibility that other PP2Cs regulate SnRK1 in other tissues or under different conditions. A clear interaction of SnRK1.1 with ABI1 and PP2CA was observed both in vitro and in vivo (Figure 1), demonstrating that PP2Cs act through direct binding to the SnRK1 α-catalytic subunit, probably using the C-terminal RD of SnRK1 as a docking site, albeit interacting also with the catalytic region that harbors the T175 target residue. Based on Y2H experiments, the KA1 domain of SnRK1 may play a key role in the PP2C–SnRK1 interaction (Figure 1A). As previously noted (Sánchez-Barrena et al., 2007), the KA1 domain can be closely superimposed on the phosphatase interaction domain of SOS2/SnRK3.11 and, given its presence also in the related AMPK and microtubule-affinity regulating kinase 3 kinases, has been suggested to represent an ancient highly conserved scaffold for interaction with PP2Cs (Sánchez-Barrena et al., 2007) (see Supplemental Figure 2 online). SnRK2.2/2.3/2.6 also require their C-terminal region, namely the ABA box, for PP2C binding (Vlad et al., 2009; Soon et al., 2012), and additional regions of interaction exist within the N-terminal CD (Soon et al., 2012), some of which, such as the T-loop and the αG helix, correspond to conserved features of the protein kinase canonical fold (Hanks and Hunter, 1995) (see Supplemental Figure 2 online). Our in vitro pull-down assays suggested that the SnRK1.1-PP2CA interaction may not solely rely on the SnRK1 RD and that similarly to SnRK2s, some parts of the CD may also play a role in this interaction (Figure 1B). Interestingly, a high-throughput screen for YFP-ABI1 interactors employing affinity purification and liquid chromatography coupled with tandem mass spectrometry identified SnRK1s as candidate ABI1-interacting proteins, whereas peptides corresponding to SnRK2.6 were not retrieved and the ABI1–SnRK2.6 interaction could only be confirmed by coimmunoprecipitation of the transiently overexpressed proteins in tobacco (Nicotiana benthamiana) (Nishimura et al., 2010).
Figure 6.
SnRK1 Regulation by Energy Signals and ABA through ABI1 and PP2CA.
SnRK1 is activated by the energy deficiency triggered by stress and is inactivated by ABI1 and PP2CA once normal energy levels are restored. These PP2Cs repress also SnRK2s and ABA signaling but are inhibited by PYL receptors upon ABA binding. Via its effect on PP2Cs, the ABA-PYL complex induces SnRK1 signaling, potentiating the effect of energy stress, diminishing the effect of sugar on SnRK1 repression, and complementing the ABA response. The SnRK1 and ABA pathways are likely to undergo crosstalk also at other levels (dotted lines). SnAK, SnRK1-activating kinases.
As an outcome of the interaction with ABI1 and PP2CA, SnRK1 is dephosphorylated and inactivated (Figures 2 and 3). Nevertheless, disruption of the catalytic site in the ABI1_D177A and PP2CA_D142A mutants did not fully restore SnRK1 activity (Figure 3A), suggesting that, although dephosphorylation plays a major role in SnRK1 inactivation, physical blockage may, similarly to SnRK2s (Soon et al., 2012), also be important for SnRK1 repression. The mechanism of action also may differ between the various PP2Cs, as suggested by the fact that despite having a lower impact on SnRK1(T175) phosphorylation (Figure 2E), PP2CA had a consistently stronger effect than ABI1 on SnRK1 signaling (Figure 3A). Given that the SnRK1 RD is the major region of interaction with PP2Cs (Figure 1) and that this region is responsible for binding the regulatory subunits (Bhalerao et al., 1999; Kleinow et al., 2000), it is plausible that PP2C binding affects SnRK1 activity also by interfering with trimer formation.
PP2CA was able to efficiently dephosphorylate T175 in vitro and in vivo (Figure 2), consistent with the in vitro dephosphorylation of this residue by mammalian PP2C (Sugden et al., 1999). Nevertheless, despite the clear differences in gene expression observed between control, inducing, and inactivating conditions and between the wild-type and Qabi2-2 leaves (Figure 3C), we were unable to detect differences in T175 phosphorylation or SnRK1 activity in these conditions in the endogenous SnRK1 (Figures 3D and 3E), suggesting that the relatively short treatment times employed result in subtle changes in kinase phosphorylation and activity that are not possible to detect with the phospho-AMPKα(T172) antibodies or the kinase activity assays from total cellular SnRK1.1. Indeed, a much longer (24-h) starvation treatment of rice suspension cells resulted in mild (1.9-fold) differences in SnRK1 activity, as measured with the SAMS peptide (Lu et al., 2007). These results are in agreement with the view on cellular enzyme cascades in which slight changes in enzyme activity may trigger significant downstream effects by amplifying the signal (Chock et al., 1980). More sensitive and quantitative techniques like Mass Western (Lehmann et al., 2008) and/or the enrichment of specific SnRK1 subcellular pools may be required for accurately assessing changes in SnRK1 T-loop phosphorylation and activity in response to stress and nutrient signals.
Our results employing reporter gene assays and gene expression analyses in the wild type, pp2c knockout mutants, and PP2CA overexpressors show that PP2Cs are negative regulators of SnRK1 signaling (Figures 3 and 4). Transient coexpression of ABI1 and PP2CA with SnRK1 in protoplasts reduced by 60% the ability of SnRK1 to activate gene expression (Figure 3). Using a similar approach, Fujii and colleagues showed that the extent of repression by ABI1 was nearly 100% when coexpressing SnRK2.6 and its downstream ABF2 transcription factor to activate an ABA reporter (Fujii et al., 2009). However, the ability of PP2Cs to repress kinase activity varied depending on the SnRK2 and PP2C combination employed, and in the case of SnRK2.6 and HAB1, the repression was only 30%. Because some clade A PP2Cs have been shown to dephosphorylate ABF2 (Antoni et al., 2012), it is also possible that the difference in the extent of repression is due to a simultaneous effect of ABI1 on the kinase and on the transcription factor.
Most importantly, constitutive PP2C depletion in the quadruple pp2c mutants abrogates SnRK1 inactivation and downstream target gene repression after stress-derived energy deprivation subsides (Figure 3C, DG samples). However, the impact of PP2C depletion is less obvious under activating stress conditions (Figure 3C, D samples) presumably because, as for AMPK and Snf1 (Mayer et al., 2011; Oakhill et al., 2011; Xiao et al., 2011), the kinase is protected from dephosphorylation when energy levels are low (Sugden et al., 1999). Similarly to plants overexpressing SnRK1.1, double and quadruple pp2c knockout mutants showed varying degrees of a sugar hypersensitive phenotype, while PP2CA overexpressors displayed an opposite phenotype (Figure 4), all consistent with the conclusions from the molecular data that PP2Cs negatively regulate SnRK1.
Our results indicate that the ABA and energy signaling pathways interact through PP2Cs and that ABA can induce SnRK1 signaling through PP2C inhibition (Figure 5). This is in agreement with a recent study reporting enhanced SnRK1 activity in wheat (Triticum aestivum) roots in response to ABA (Coello et al., 2012), and provides a molecular explanation for the extensive interactions observed between ABA and sugar signaling in genetic screens (Rolland et al., 2006). SnRK1s were never identified among ABA-activated kinases, most probably because the extent of SnRK1 activation by ABA is 1 order of magnitude lower than that by energy stress (Darkness; Figure 5B), and would probably remain masked by the much stronger activities of SnRK2s. In contrast, these studies relied on in-gel kinase assays for detecting of kinase activities (Yoshida et al., 2002; Furihata et al., 2006; Fujii et al., 2007). Despite our current lack of knowledge regarding the exact subunit composition of functional SnRK1, and despite the fact that the catalytic subunit alone is active (Bhalerao et al., 1999; Shen et al., 2009; Crozet et al., 2010), in vivo SnRK1 most likely operates, similarly to Snf1 and AMPK, as a heterotrimeric complex (Polge and Thomas, 2007; Hedbacker and Carlson, 2008; Hardie, 2011; Ramon et al., 2013), whose dissociation under the denaturing conditions employed in the in-gel kinase assays may result in loss of kinase activity.
In addition to the interaction through PP2Cs, other points of crosstalk are likely to exist between ABA and energy signaling, and SnRK1 may regulate ABA transcription factors, such as ABF2 (Figure 2B) or FUS3 (Zhang et al., 2008; Tsai and Gazzarrini, 2012) that can also be directly dephosphorylated by PP2Cs (Antoni et al., 2012). It is conceivable that aberrant PP2C:SnRK1 ratios as well as the possible PP2C/SnRK1 coregulation of downstream factors could account for the altered ABA sensitivity and ABA-related phenotypes of plants with altered SnRK1 signaling (Radchuk et al., 2006; Lu et al., 2007; Rosnoblet et al., 2007; Jossier et al., 2009; Radchuk et al., 2010; Tsai and Gazzarrini, 2012).
We propose a dual role for the regulation of SnRK1 by ABI1 and PP2CA (Figure 6). On one hand, activation of the SnRK1 pathway through alternative signals like ABA, could support the ABA response with a more general one directed toward a metabolic and transcriptional reprogramming to cope with energy deficiency. Activation of SnRK1 by ABA could also serve to prime the SnRK1 system, potentiating a subsequent response to energy imbalance derived from stress. On the other hand, PP2C regulation appears to be an integral part of the SnRK1 signaling pathway, resetting the system once stress subsides or an energy balance is attained through the appropriate metabolic readjustments. Persistence of ABA under these conditions would in turn promote the maintenance of SnRK1 in an active state, similarly to how elevated interleukin-6 sustains high AMPK activity in skeletal muscle when energy levels are presumably no longer altered after exercise (Ruderman et al., 2006). With this scenario in mind, one could envision that in tissues directly exposed to stress, SnRK1 activation would be mainly dictated by the energy-dependent branch, whereas in distant tissues, this activation could be mediated by ABA. In addition to interleukin-6, AMPK responds to other inflammatory mediators and hormones, but the precise mechanisms underlying this regulation are in most cases unknown (Steinberg and Kemp, 2009; Lim et al., 2010). Interestingly, chronic Tumor Necrosis Factor α treatment in muscle cells suppresses the AMPK pathway by inducing the repressor PP2C (Steinberg et al., 2006), suggesting that a connection between hormone signals and energy signaling through the inhibitory PP2Cs might be conserved in multicellular eukaryotes.
In summary, we have identified ABI1 and PP2CA as upstream phosphatases of SnRK1, uncovering a mechanism through which ABA can stimulate SnRK1 action. Future work to further understand SnRK1 regulation and to unravel the interplay of these two central pathways may offer insight not only into the mechanisms of stress tolerance but also into fundamental developmental processes, such as seed maturation and germination.
METHODS
Primers and Constructs
A list of all primers, cloning steps, and vectors is provided in Supplemental Table 1 online.
Plant Material and Growth Conditions
All used Arabidopsis thaliana plants are in the Columbia (Col-0) background, except 35S:SnRK1.1-HA (Landsberg erecta) (Baena-González et al., 2007). The 35S:SnRK1.1 (35S:SnRK1.1-2) (Jossier et al., 2009), 35S:PP2CA (Antoni et al., 2012), and abi1-2 pp2ca-1 (Rubio et al., 2009) lines have been described. Quadruple pp2c knockout mutants were generated from pp2ca-1 hai1-1 (Antoni et al., 2012) and the corresponding triple pp2c mutants (Rubio et al., 2009).
Plants were grown in soil under a 12-h-light (100 µE)/12-h-dark regime. For in vitro culture, sterilized seeds were stratified in the dark at 4°C for 2 days and sowed on plates containing Murashige and Skoog medium with 0.1% MES, 0.8% phytoagar, and Glc (4 or 6%) or sorbitol (4 or 6%). Plates were sealed and incubated at 23°C under continuous light.
Antibodies and Protein Expression Analyses
The SnRK1.1 antibody was purchased from Agrisera (anti-AKIN10, AS10919). Phospho-SnRK1.1(T175) was detected with an anti–phospho-AMPKα(T172) antibody (referred to as αP-AMPK; Cell Signaling), which also detects phospho-SnRK1.2(T176) as a lower band (Baena-González et al., 2007). An anti-GST polyclonal antibody (Sigma), anti-HA (Roche), and anti-T7 (Novagen) monoclonal antibodies were used to detect the corresponding tagged proteins.
For analyses of protein expression from protoplast pellets and leaf tissue, the material was directly ground in 2× Laemmli solubilization buffer to maintain the phosphorylation status during protein extraction.
Protoplast Transient Expression Assays
Vectors for protoplast transient expression and assays were as described (Yoo et al., 2007), using the UBQ10-β-glucuronidase reporter as transfection efficiency control. For constructs for overexpression of SnRK1.1-GFP, ABI1-HA, PP2CA-HA, PP2C6-6-HA, and FLAG-PYL4, the corresponding coding sequences were cloned into a pHBT95 vector harboring the indicated C- or N-terminal tag. SnRK1 signaling was monitored using a DIN6:LUC reporter (Baena-González et al., 2007). ABA and Glc were added to a final concentration of 5 µM and 30 mM, respectively.
For coimmunoprecipitation assays, untagged SnRK1.1 was expressed with ABI1-HA or mER7 control DNA (Yoo et al., 2007) in 3 mL of protoplasts (6 × 105 cells) under standard conditions.
Frozen cell pellets were lysed in 500 μL of lysis buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 10 mM EDTA, 10% glycerol, 0.5% Triton X-100, and complete protease inhibitor cocktail [Roche]), 20 mM sodium fluoride, 1 mM orthovanadate, 1/500 (v/v) phosphatase inhibitor 2 (Sigma P044), and 1/500 (v/v) phosphatase inhibitor 3 (Sigma P5726)], incubated at 4°C for 10 min, and diluted to a final volume of 1.5 mL with lysis buffer without Triton X-100. The cleared lysate was incubated with 40 μL of anti-HA affinity matrix (Roche 11815016001) for 3 h at 4°C. Agarose beads containing immunoprecipitated proteins were washed five times with lysis buffer containing 0.05% Triton, eluted with 4× Laemmli solubilization buffer, and analyzed by immunoblotting with an anti-SnRK1.1 antibody.
Recombinant Protein Production
The coding sequence of PP2CA was cloned into pGEX-4T1. Recombinant GST-PP2CA was produced in Escherichia coli (BL21:DE3) and purified through S-linked glutathione agarose affinity chromatography as recommended by the manufacturer (Sigma G4510).
N- (residues 1 to 293, CD) and C-terminal (residues 294–512, RD) SnRK1.1 were cloned into pET28a (Novagen). Recombinant proteins were produced in E. coli (BL21:DE3) and purified using immobilized metal ion affinity chromatography (TALON, BD Clontech) following the manufacturer’s instructions. Successful protein production and purification were verified by immunoblotting with anti-GST and anti-T7 antibodies. Recombinant His-PYL4, His-PP2CA, and His-ΔC ABF2 (residues 1–173) were produced as described by Antoni et al. (2012), and recombinant GST-SnRK1.1 and GST-SnAK2 were produced as described by Crozet et al. (2010).
In Vitro Pull-Down Assays
Proteins (3 µg of each) were incubated for 1 h at room temperature in 100 μL of buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.05% Triton X-100, and 1/500 [v/v] plant-specific protease inhibitor cocktail [Sigma P9599]), mixed with 30 μL of glutathione–agarose beads and incubated one more hour. Beads were washed four times with buffer A, and bound proteins were analyzed by immunoblotting using anti-T7 antibodies.
SnRK1.1 Immunoprecipitation, Phosphatase Treatment, and in Vitro Kinase Assays
SnRK1.1 was immunoprecipitated from leaves of 35S:SnRK1.1-HA plants treated for 1 h in darkness. Plant material (∼1 g) was extracted in 3 volumes of 1× PBS supplemented with 1 mM EDTA, 0.05% Triton X-100, and 1/500 (v/v) plant-specific protease inhibitor cocktail (Sigma). After centrifugation (16,000g, 4°C, 15 min), the supernatant was recovered, and 1 mg of total protein was incubated overnight at 4°C with 30 μL of anti-HA affinity matrix. The matrix was washed three times with extraction buffer and resuspended in a total volume of 66 μL of buffer (50 mM Tris-HCl, pH 7.6, 250 mM KCl, 10% glycerol, and 0.1% Tween 20), of which 3 μL was used for each reaction.
To assess dephosphorylation of immunoprecipitated SnRK1.1 by PP2CA, SnRK1.1 was incubated with His-PP2CA (2 μg) in a 50-μL reaction containing 25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 1 mM DTT. The reaction was stopped with Laemmli solubilization buffer and analyzed by Phos-Tag SDS-PAGE (50 μM Phos-Tag ligand [Wako] and 100 μM MnCl2) (Kinoshita et al., 2009) and immunoblot with an anti-HA antibody. The Phos-Tag ligand selectively retards phosphorylated proteins. For assessing the effect of PP2CA on T-loop phosphorylation, immunoprecipitated SnRK1.1-HA (5 μL of beads) was treated or not with GST-PP2CA (1 µg) in 50 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 1 mM EDTA, and 1/1000 protease inhibitor cocktail (Sigma P9599) at 30°C for 30 min. The beads were then washed twice with the same buffer complemented with 150 mM NaCl and 0.05% Triton X-100. Finally, they were boiled in Laemmli solubilization buffer and analyzed by immunoblotting with anti–phospho-AMPKα(T172) and anti-SnRK1.1 antibodies.
For in vitro kinase assays, immunoprecipitated SnRK1.1 was preincubated (for 10 min) or not with His-PP2CA (0.6 μg) and His-PYL4 (2.0 μg) in 30 μL of kinase buffer (20 mM Tris-HCl, pH 7.8, 20 mM MgCl2, and 2 mM MnCl2) ± ABA (30 μM) and further incubated with GST-ΔC ABF2 (0.5 μg) for 1 h at room temperature in the presence of 3.5 μCi of [γ32P]ATP. The reaction products were resolved in an 8% SDS-PAGE gel, transferred to an Immobilon-P membrane (Millipore), and detected using a phosphor image system (FLA5100; Fujifilm; Antoni et al., 2012).
For preactivation of SnRK1.1, GST-SnRK1.1 and GST-SnAK2 (1 µg of each) were incubated in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 100 µM ATP, 1 mM DTT, and 1/1000 protease inhibitor cocktail (Sigma) at 30°C for 30 min. After adding or not GST-PP2CA (1 µg), the mix was further incubated for 30 min and analyzed by immunoblot employing anti-phospho-AMPKα(T172) and anti-SnRK1.1 antibodies.
For measurements of endogenous SnRK1 activity, SnRK1.1 was immunoprecipitated from leaves of 4-week-old plants of the indicated genotypes. Plant material (∼1 g) was extracted in 2 volumes of Buffer C (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.05% Triton X-100), and complete protease inhibitor cocktail (one tablet/50 mL, Roche) and 1/500 (v/v) phosphatase inhibitor 2 (Sigma P044) and 1/500 (v/v) phosphatase inhibitor 3 (Sigma). After two successive centrifugations (20,000g, 4°C, 10 min), the supernatant was recovered, and 1 mg of total protein was incubated with gentle shaking for 3 h at 4°C with 15-μL beads of protein A–antibody complex prepared as follows. For each immunoprecipitation, 15 μL (bed volume) of protein A–agarose (Roche) was equilibrated in 1× PBS (Sigma-Aldrich) and incubated with 1.5 µg of anti-SnRK1.1 antibody in 500 μL of 1× PBS for 1 h at room temperature with gentle shaking. After three washes in buffer C, the beads were used for immunoprecipitation. After the incubation for 3 h at 4°C under shaking, the beads were washed three times with buffer C, and one-third (5 µL) was kept for immunoblot analysis with an anti-SnRK1.1 antibody.
The remaining 10 μL was used to determine the specific activity of SnRK1 on the AMARA peptide as described previously (Crozet et al., 2010). Briefly, the beads were incubated for 1 h at 30°C in a kinase assay buffer (100 mM Tris-HCl, pH7.5, 10 mM MgCl2, 200 µM ATP, 1 mM EDTA, and 1/500 anti-protease and anti-phosphatase cocktails), 90 µM AMARA peptide (AMARAASAAALARRR), and 2 µCi [γ-32P]ATP. Ten microliters of the reaction was spotted three times on P81 filter (GE-Whatman), and the filters were subsequently washed three times for 5 min in 1% phosphoric acid. After a quick wash in acetone, radioactivity was measured using a scintillation counter. A positive control with recombinant SnRK1.1 and SnAK2 was always performed to confirm that the reaction was occurring.
Y2H Assays
Y2H assays were performed as described (Saez et al., 2008). The full-length coding sequence of SnRK1.1 and the various deletions, cloned into pGBKT7, were faced with constructs harboring full-length PP2CA and ABI1 in fusion with the GAL4 activation domain. To generate the GAL4 activation domain-PP2CA fusion, the PP2CA coding sequence was cloned into pGADT7. The pGADT7-ABI1 construct was described previously (Vlad et al., 2010).
Gene Expression Analyses
Fully expanded leaves of 5-week-old plants were used as such or to cut leaf discs (9-mm diameter) and incubated on sterile MilliQ water in Petri dishes. For examining SnRK1 regulation in wild-type and pp2c mutants, leaves were incubated for 3 h in L (control; 100 µE) or D or DG. Unexpected darkness is perceived as stress and activates SnRK1 (Baena-González et al., 2007). For assessing the effect of ABA, leaf discs of wild-type or 35S:PP2CA plants were incubated ± ABA under light for 5 h. For the effect of ABA on SnRK1 activation by stress and inactivation by sugar, leaf discs of wild-type plants were incubated for 3 h in light (L), in darkness with (DA) or without ABA (D), or for 1 h in darkness followed by 2 h in darkness with ABA and 1 h in darkness with ABA and Glc (DGA). Glc and ABA were added to a final concentration of 50 mM and 100 μM, respectively.
Following the indicated treatments, total RNA was extracted using TRIzol reagent (Life Technologies), treated with RNase-Free DNase (Promega), and reverse transcribed (1.5 μg) using SuperScript III Reverse Transcriptase (Life Technologies). qRT-PCR analyses were performed using a 7900HT fast real-time PCR System (Applied Biosystems) employing the Eva-Green fluorescent stain (Biotium), and the 2−ΔCT or comparative CT method (Livak and Schmittgen, 2001). Expression levels were normalized using the CT values obtained for EIF4. Efficient ABA uptake and signaling was confirmed by monitoring the induction of the ABA marker genes RAB18 and RD29.
Microarray Dataset Comparisons
The data set for the SnRK1.1-induced transcriptional profile corresponds to supplemental table 3 in Baena-González et al. (2007). The hormone treatment data sets, as compared in (Nemhauser et al., 2006), are from the Arabidopsis AtGenExpress consortium (http://Arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp). A twofold change filter was applied to all the hormone data sets and, given the 6-h incubation of the SnRK1.1 overexpression data set, only the 3-h (and not the 1-h) time points were considered for the comparisons. Overlap between the compared data sets was revealed using the Venny Venn diagram online application (http://bioinfogp.cnb.csic.es/tools/venny/index.html). The data set for the SnRK1.1-induced transcriptional profile corresponds to supplemental table 3 in Baena-González et al. (2007). For determining the significance of overlap between the two experiments, hypergeometric testing was applied using the dhyper function in R (http://www.r-project.org/).
Statistical Analyses
All statistical analyses were performed with the GraphPad Prism software. For analyses of qPCR data, the statistical significance of the indicated changes was assessed employing log2-transformed relative expression values (Rieu and Powers, 2009).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SnRK1.1, At3g01090; SnRK1.2, At3g29160; ABI1, At4g26080; PP2CA, At3g11410; ABI2, At5g57050; HAB1, At1g72770; HAI1, At5g59220; PYL4, At2g38310; PP2C6-6, At1g03590; DIN6, At3g47340; SEN5, At3g15450; and AXP, At2g33830.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Y2H Controls for the SnRK1.1 and PP2C Interaction.
Supplemental Figure 2. Alignment and Structural Comparison of SnRK1 and SnRK2.
Supplemental Figure 3. SnRK1.1 Is Inactivated by Recombinant His-PP2CA In Vitro.
Supplemental Figure 4. Specific Detection of Phosphorylated SnRK1.
Supplemental Figure 5. Clade A pp2c Quadruple Mutants Are ABA Hypersensitive.
Supplemental Figure 6. ABA Promotes SnRK1 Signaling in Protoplasts.
Supplemental Figure 7. Overlap between Transcriptional Changes Induced by SnRK1.1 and Various Hormone Treatments.
Supplemental Table 1. Primers and Cloning Strategies Used in This Study.
Supplemental Data Set 1. Genes Similarly Regulated by SnRK1.1 and ABA.
Supplementary Material
Acknowledgments
We thank Vera Nunes for plant management, Filip Rolland and Jörg Kudla for comments, and Júlia Costa for help with the scintillation counter. The 35S:SnRK1.1-2 line was provided by Martine Thomas. E.B.-G. was supported by grants from Marie Curie IRG, the EMBO Installation program, Marie Curie Actions FP7-People-2010-ITN, the Fundação para a Ciência e a Tecnologia (FCT-PTDC/AGR-AAM/104939/2008), and the Portugal-Spain Bilateral Collaboration program Ações integradas (Ação E-26/10). A.C. was supported by SFRH/BPD/47280/2008, C.M. was supported by SFRH/BD/33563/2008, L.M. was supported by SFRH/BD/51627/2011, and P.C. was supported by SFRH/BPD/79255/2011. A. Rabissi was supported by a Generalitat de Catalunya PhD grant (FI-AR067443). P.L.R. was supported by the Ministerio de Ciencia e Innovación (grants BIO2011-23446 and PT2009-0155), R.A. was supported by the Junta para Ampliación de Estudios e Investigaciones Científicas-Consejo Superior de Investigaciones Cientificas fellowship, and M.G.-G. was supported by a Juan de la Cierva contract.
AUTHOR CONTRIBUTIONS
A. Rodrigues and E.B.-G. conceived the project. A. Rodrigues, M.A., P.C., P.L.R., and E.B.-G. designed the research. A. Rodrigues, M.A., P.C., L.M., A.C., C.M., A.E., M.G.-G., R.A., A. Rabissi, and E.B.-G. performed the research. A. Rodrigues, M.A., P.C., L.M., A.C., C.M., A.E., M.G.-G., R.A., P.L.R., A. Rabissi, V.L., and E.B.-G. analyzed the data. A. Rodrigues, P.C., P.L.R., and E.B.-G. wrote the article.
Glossary
- ABA
abscisic acid
- SnRK1
Snf1-related protein kinase1
- PP2C
clade A type 2C protein phosphatase
- Snf1
sucrose-nonfermenting1
- AMPK
AMP–activated protein kinase
- PYL
Pyrabactin resistance1-Like
- Y2H
yeast-two-hybrid
- RD
regulatory domain
- CD
catalytic domain
- KA1
kinase-associated1
- GST
glutathione S-transferase
- qRT-PCR
quantitative RT-PCR
References
- Ananieva E.A., Gillaspy G.E., Ely A., Burnette R.N., Erickson F.L. (2008). Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol. 148: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Peirats-Llobet M., Pizzio G.A., Fernandez M.A., De Winne N., De Jaeger G., Dietrich D., Bennett M.J., Rodriguez P. L. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 148: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Rodrigues A., Pizzio G.A., Rodriguez P.L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 158: 970–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baena-González E., Sheen J. (2008). Convergent energy and stress signaling. Trends Plant Sci. 13: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R.P., Salchert K., Bakó L., Okrész L., Szabados L., Muranaka T., Machida Y., Schell J., Koncz C. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA 96: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Barbier-Brygoo H., Laurière C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279: 41758–41766 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Droillard M.J., Barbier-Brygoo H., Laurière C. (2007). Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 63: 491–503 [DOI] [PubMed] [Google Scholar]
- Bradford K.J., Downie A.B., Gee O.H., Alvarado V., Yang H., Dahal P. (2003). Abscisic acid and gibberellin differentially regulate expression of genes of the SNF1-related kinase complex in tomato seeds. Plant Physiol. 132: 1560–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling D., Thornton C., Woods A., Sanders M.J. (2012). AMP-activated protein kinase: New regulation, new roles? Biochem. J. 445: 11–27 [DOI] [PubMed] [Google Scholar]
- Chérel I., Michard E., Platet N., Mouline K., Alcon C., Sentenac H., Thibaud J.B. (2002). Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chock P.B., Rhee S.G., Stadtman E.R. (1980). Interconvertible enzyme cascades in cellular regulation. Annu. Rev. Biochem. 49: 813–843 [DOI] [PubMed] [Google Scholar]
- Coello P., Hirano E., Hey S.J., Muttucumaru N., Martinez-Barajas E., Parry M.A., Halford N.G. (2012). Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2. J. Exp. Bot. 63: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P., Jammes F., Valot B., Ambard-Bretteville F., Nessler S., Hodges M., Vidal J., Thomas M. (2010). Cross-phosphorylation between Arabidopsis thaliana sucrose nonfermenting 1-related protein kinase 1 (AtSnRK1) and its activating kinase (AtSnAK) determines their catalytic activities. J. Biol. Chem. 285: 12071–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Estruch F., Treitel M.A., Yang X., Carlson M. (1992). N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., Liese A., Wellmann C., Al-Rasheid K.A., Grill E., Romeis T., Hedrich R. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A., Verhey S.D., Holappa L.D., Shen Q., Ho T.H., Walker-Simmons M.K. (1999). An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. USA 96: 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F., Beaudoin N., Serizet C., Webb A.A., Vartanian N., Giraudat J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Xiong L., Song C.P., Gong D., Halfter U., Zhu J.K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Halford N.G., Hey S., Jhurreea D., Laurie S., McKibbin R.S., Paul M., Zhang Y. (2003). Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J. Exp. Bot. 54: 467–475 [DOI] [PubMed] [Google Scholar]
- Hanks S.K., Hunter T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 9: 576–596 [PubMed] [Google Scholar]
- Hardie D.G. (2011). AMP-activated protein kinase: An energy sensor that regulates all aspects of cell function. Genes Dev. 25: 1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley S.A., Davison M., Woods A., Davies S.P., Beri R.K., Carling D., Hardie D.G. (1996). Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271: 27879–27887 [DOI] [PubMed] [Google Scholar]
- Hedbacker K., Carlson M. (2008). SNF1/AMPK pathways in yeast. Front. Biosci. 13: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Hoffmann T., Leube M., Höhener B., Grill E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.P., Momcilovic M., Carlson M. (2005). Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase alpha as Snf1-activating kinases in yeast. J. Biol. Chem. 280: 21804–21809 [DOI] [PubMed] [Google Scholar]
- Jossier M., Bouly J.P., Meimoun P., Arjmand A., Lessard P., Hawley S., Grahame Hardie D., Thomas M. (2009). SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 59: 316–328 [DOI] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Koike T. (2009). Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4: 1513–1521 [DOI] [PubMed] [Google Scholar]
- Kleinow T., Bhalerao R., Breuer F., Umeda M., Salchert K., Koncz C. (2000). Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J. 23: 115–122 [DOI] [PubMed] [Google Scholar]
- Kuhn J.M., Boisson-Dernier A., Dizon M.B., Maktabi M.H., Schroeder J.I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Terzaghi W., Gusmaroli G., Charron J.B., Yoon H.J., Chen H., He Y.J., Xiong Y., Deng X.W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.W., Chen P.W., Lu C.A., Chen S., Ho T.H., Yu S.M. (2009). Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lehmann U., Wienkoop S., Tschoep H., Weckwerth W. (2008). If the antibody fails—A mass western approach. Plant J. 55: 1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang X.Q., Watson M.B., Assmann S.M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287: 300–303 [DOI] [PubMed] [Google Scholar]
- Lim C.T., Kola B., Korbonits M. (2010). AMPK as a mediator of hormonal signalling. J. Mol. Endocrinol. 44: 87–97 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu C.A., Lin C.C., Lee K.W., Chen J.L., Huang L.F., Ho S.L., Liu H.J., Hsing Y.I., Yu S.M. (2007). The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19: 2484–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mayer F.V., et al. (2011). ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 14: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney R.R., Schmidt M.C. (2001). Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276: 36460–36466 [DOI] [PubMed] [Google Scholar]
- Merlot S., Gosti F., Guerrier D., Vavasseur A., Giraudat J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Miao Y., Lv D., Wang P., Wang X.C., Chen J., Miao C., Song C.P. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12: 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshik A., Nito K., Park S.Y., Wang A., Carvalho P.C., Lee S., Caddell D.F., Cutler S.R., Chory J., Yates J.R., Schroeder J.I. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Yoshida T., Kitahata N., Asami T., Shinozaki K., Hirayama T. (2007). ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 50: 935–949 [DOI] [PubMed] [Google Scholar]
- Oakhill J.S., Steel R., Chen Z.P., Scott J.W., Ling N., Tam S., Kemp B.E. (2011). AMPK is a direct adenylate charge-regulated protein kinase. Science 332: 1433–1435 [DOI] [PubMed] [Google Scholar]
- Ohta M., Guo Y., Halfter U., Zhu J.K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100: 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C., Thomas M. (2007). SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Radchuk R., Radchuk V., Weschke W., Borisjuk L., Weber H. (2006). Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 140: 263–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk R., Emery R.J., Weier D., Vigeolas H., Geigenberger P., Lunn J.E., Feil R., Weschke W., Weber H. (2010). Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 61: 324–338 [DOI] [PubMed] [Google Scholar]
- Ramon M., Ruelens P., Li Y., Sheen J., Geuten K., Rolland F. (2013). The hybrid four-CBS-domain KINbetagamma-subunit functions as the canonical gamma subunit of the plant energy sensor SnRK1. Plant J. 75: 11‑25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I., Powers S.J. (2009). Real-time quantitative RT-PCR: Design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rosnoblet C., Aubry C., Leprince O., Vu B.L., Rogniaux H., Buitink J. (2007). The regulatory gamma subunit SNF4b of the sucrose non-fermenting-related kinase complex is involved in longevity and stachyose accumulation during maturation of Medicago truncatula seeds. Plant J. 51: 47–59 [DOI] [PubMed] [Google Scholar]
- Rubio S., Rodrigues A., Saez A., Dizon M.B., Galle A., Kim T.H., Santiago J., Flexas J., Schroeder J.I., Rodriguez P.L. (2009). Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman N.B., et al. (2006). Interleukin-6 regulation of AMP-activated protein kinase. Potential role in the systemic response to exercise and prevention of the metabolic syndrome. Diabetes 55 (Suppl 2): S48–S54 [DOI] [PubMed] [Google Scholar]
- Saez A., Robert N., Maktabi M.H., Schroeder J.I., Serrano R., Rodriguez P.L. (2006). Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Rodrigues A., Santiago J., Rubio S., Rodriguez P.L. (2008). HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20: 2972–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Apostolova N., Gonzalez-Guzman M., Gonzalez-Garcia M.P., Nicolas C., Lorenzo O., Rodriguez P.L. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Sánchez-Barrena M.J., Fujii H., Angulo I., Martínez-Ripoll M., Zhu J.K., Albert A. (2007). The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 26: 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P., Alms G.R., Haystead T.A., Carlson M. (2000). Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J., Minchin P.E.H., Jahnke S., van Dongen J.T., Schittko U., Baldwin I.T. (2006). SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc. Natl. Acad. Sci. USA 103: 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A., Hirt H., Meskiene I. (2004). Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274: 1900–1902 [DOI] [PubMed] [Google Scholar]
- Shen W., Reyes M.I., Hanley-Bowdoin L. (2009). Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol. 150: 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Stitt M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Soon F.F., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S.C., Woods A., Jones N.A., Davison M.D., Carling D. (2000). The regulation of AMP-activated protein kinase by phosphorylation. Biochem. J. 345: 437–443 [PMC free article] [PubMed] [Google Scholar]
- Steinberg G.R., Kemp B.E. (2009). AMPK in Health and Disease. Physiol. Rev. 89: 1025–1078 [DOI] [PubMed] [Google Scholar]
- Steinberg G.R., et al. (2006). Tumor necrosis factor alpha-induced skeletal muscle insulin resistance involves suppression of AMP-kinase signaling. Cell Metab. 4: 465–474 [DOI] [PubMed] [Google Scholar]
- Sugden C., Crawford R.M., Halford N.G., Hardie D.G. (1999). Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J. 19: 433–439 [DOI] [PubMed] [Google Scholar]
- Tochio N., et al. (2006). Solution structure of the kinase-associated domain 1 of mouse microtubule-associated protein/microtubule affinity-regulating kinase 3. Protein Sci. 15: 2534–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A.Y., Gazzarrini S. (2012). AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 69: 809–821 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., Leung J., Rodriguez P.L., Laurière C., Merlot S. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Droillard M.J., Valot B., Khafif M., Rodrigues A., Brault M., Zivy M., Rodriguez P.L., Merlot S., Laurière C. (2010). Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 63: 778–790 [DOI] [PubMed] [Google Scholar]
- Xiao B., et al. (2011). Structure of mammalian AMPK and its regulation by ADP. Nature 472: 230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sulpice R., Himmelbach A., Meinhard M., Christmann A., Grill E. (2006). Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc. Natl. Acad. Sci. USA 103: 6061–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida R., Umezawa T., Mizoguchi T., Takahashi S., Takahashi F., Shinozaki K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281: 5310–5318 [DOI] [PubMed] [Google Scholar]
- Yoshida R., Hobo T., Ichimura K., Mizoguchi T., Takahashi F., Aronso J., Ecker J.R., Shinozaki K. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Andralojc P.J., Hey S.J., Primavesi L.F., Specht M., Koehler J., Parry M.A.J., Halford N.G. (2008). Arabidopsis sucrose non-fermenting-1-related protein kinase-1 and calcium-dependent protein kinase phosphorylate conserved target sites in ABA response element binding proteins. Ann. Appl. Biol. 153: 401–409 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.