This study reports the identification and functional characterization of two stress-inducible R2R3-MYB–type transcription factors, termed MYB14 and MYB15, which regulate the stilbene biosynthetic pathway in grapevine.
Abstract
Plant stilbenes are phytoalexins that accumulate in a small number of plant species, including grapevine (Vitis vinifera), in response to biotic and abiotic stresses and have been implicated in many beneficial effects on human health. In particular, resveratrol, the basic unit of all other complex stilbenes, has received widespread attention because of its cardio-protective, anticarcinogenic, and antioxidant properties. Although stilbene synthases (STSs), the key enzymes responsible for resveratrol biosynthesis, have been isolated and characterized from several plant species, the transcriptional regulation underlying stilbene biosynthesis is unknown. Here, we report the identification and functional characterization of two R2R3-MYB–type transcription factors (TFs) from grapevine, which regulate the stilbene biosynthetic pathway. These TFs, designated MYB14 and MYB15, strongly coexpress with STS genes, both in leaf tissues under biotic and abiotic stress and in the skin and seed of healthy developing berries during maturation. In transient gene reporter assays, MYB14 and MYB15 were demonstrated to specifically activate the promoters of STS genes, and the ectopic expression of MYB15 in grapevine hairy roots resulted in increased STS expression and in the accumulation of glycosylated stilbenes in planta. These results demonstrate the involvement of MYB14 and MYB15 in the transcriptional regulation of stilbene biosynthesis in grapevine.
INTRODUCTION
Plant stilbenes are a small group of phenylpropanoid (PP) compounds that have been detected in a limited number of unrelated plant species and accumulate in response to biotic and abiotic stresses. Stilbenes have been detected in at least 72 plant species distributed among 31 genera and 12 families, including the Fagaceae, Liliaceae, Moraceae, Myrtaceae, Papilionaceae, Pinaceae, and Poaceae (Sotheeswaran and Pasupathy, 1993; Yu et al., 2005; Counet et al., 2006). Recently, stilbenes, and in particular trans-resveratrol (3,5,4’-trihydroxy-trans-stilbene), synthesized in grapevine (Vitis vinifera), cranberry (Vaccinium oxycoccos), blueberry (Vaccinium myrtillus), and peanut (Arachis hypogaea), have elicited particular interest within the scientific community, not only because of their role in plant defense, but also because of their pharmacological properties. The role of stilbenes in the prevention of and protection from cardiovascular and neurodegenerative diseases, cancer, and diabetes have been studied and validated in many laboratory models, and a growing body of evidence indicates that these compounds have health-protecting properties in mammals, although the bioavailability and long-term effects of their supplementation in the human diet have not yet been confirmed (Baur and Sinclair, 2006; Kalantari and Das, 2010). The interest in stilbenes has been growing since these compounds were found to accumulate in significant amounts not only in vegetative parts of plants, such as leaves and roots, but also in fruits, which in turn, are widely used in the production of mass industrial derivatives, such as juices and, in the case of grapevine, wines (Keen and Ingham, 1976; Langcake and Pryce, 1976; Oleszek et al., 2001; Yu et al., 2005; Counet et al., 2006; Chong et al., 2009).
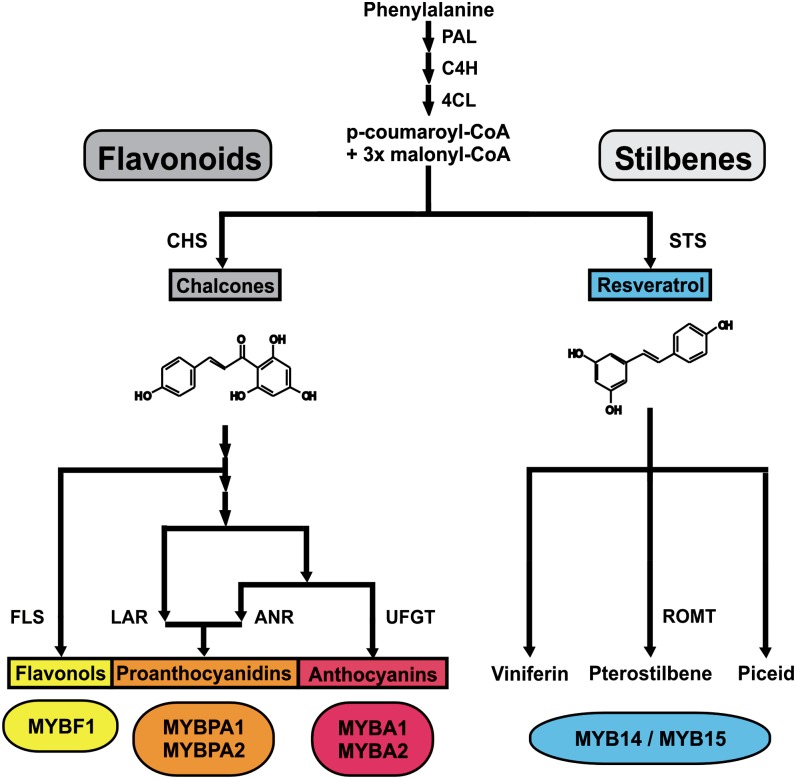
Most stilbene-producing plants, including grapevine, contain only low concentrations of these PPs under normal growth conditions, but these compounds rapidly accumulate in response to a wide range of abiotic and biotic stresses, as a result of an increased transcription of stilbene biosynthetic genes and the coordinate activation of upstream genes belonging to the PP pathway, such as phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H). A vast range of abiotic stress treatments lead to stilbene biosynthesis, including mechanical damage (Chiron et al., 2000; Pezet et al., 2003), exposure to UV-C light (Bais et al., 2000), treatment with chemicals, such as aluminum ions, cyclodextrins, and ozone (Rosemann et al., 1991; Adrian et al., 1996; Zamboni et al., 2009), and the application of plant hormones like ethylene and jasmonates (Belhadj et al., 2008a, 2008b; D'Onofrio et al., 2009). In terms of biotic stresses, the synthesis of stilbenes is particularly well documented in grapevine, where it has been shown to be induced upon infection with various pathogens, including Plasmopara viticola (Langcake and Pryce, 1976), Erisyphe necator (Fung et al., 2008; Schnee et al., 2008), and Botrytis cinerea (Adrian et al., 1997). The biosynthetic pathway leading to the production of stilbenes is a side branch of the general PP pathway and can be considered as an extension of the flavonoid pathway (Vannozzi et al., 2012). All higher plants are able to accumulate compounds like p-coumaroyl-CoA and cinnamoyl-CoA through the action of ubiquitous enzymes such as PAL, C4H, and 4-coumarate:CoA ligase, and in all plants, these compounds represent substrates for chalcone synthases, fundamental enzymes leading to the biosynthesis of the flavonoids. However, in stilbene-synthesizing plant species, these compounds are also substrates for stilbene synthases (STSs), which compete with chalcone synthases and represent the key enzymes responsible for the biosynthesis of resveratrol and its derivatives (Figure 1) (Schöppner and Kindl, 1984; Lanz et al., 1991).
Figure 1.
Simplified Representation of the Grapevine Phe/Polymalonate Pathway, Which Leads to the Biosynthesis of Flavonoids and Stilbenes.
Transcriptional regulation of the general enzymes is conducted by the R2R3 MYB TFs MYBPA1, MYBA, MYBF1, MYB14, and MYB15. 4CL, 4-coumarate-CoA ligase; CHS, chalcone synthase; ROMT, resveratrol-O-methyltransferase.
To date, STS genes have been cloned from peanut (Schröder et al., 1988), Scots pine (Pinus sylvestris) (Preisig-Müller et al., 1999), Eastern white pine (Pinus strobes) (Raiber et al., 1995), Japanese red pine (Pinus densiflora) (Kodan et al., 2002), grapevine (Sparvoli et al., 1994), and sorghum (Sorghum bicolor) (Yu et al., 2005); apart from the latter, for which only one STS has been identified, these genes appear to exist as families of closely related members. In grapevine, a recent analysis of the STS multigenic family based on both the PN40024 and PN ENTAV 115 genomes (Jaillon et al., 2007; Velasco et al., 2007) led to the identification of 48 putative Vv-STS gene sequences, with at least 33 of these encoding full-length proteins (Vannozzi et al., 2012). To date, transcription factors (TFs) regulating the expression of structural genes for stilbene synthesis have not yet been identified. However, recently, a growing number of TFs have been demonstrated to be responsible for the regulation of PP biosynthesis in a range of plant species, including grapevine, which represents one of the most studied crop plants in terms of the regulation of the flavonoid biosynthetic pathway (Figure 1; reviewed in Czemmel et al., 2012). For example, the grapevine MYB TFs MYB5a and MYB5b were found to modulate several branches of the flavonoid pathway, regulating structural genes, such as LEUCOANTHOCYANIDIN REDUCTASE (LAR), ANTHOCYANIDIN REDUCTASE (ANR), FLAVONOID 3′-HYDROXYLASE, and CHALCONE ISOMERASE (Deluc et al., 2006, 2008). MYBA1 and MYBA2 regulate UDP-Glc:FLAVONOID-3-0-GLUCOSYLTRANSFERASE (UFGT), which encodes an enzyme responsible for the conversion of anthocyanidins to anthocyanins (Kobayashi et al., 2002; Walker et al., 2007). MYBPA1 regulates the expression of the flavonoid branch pathway, leading to the production of proanthocyanidins (PAs; condensed tannins) (Bogs et al., 2007), and the grape At-MYB12–like gene, MYBF1, was shown to complement the flavonol-deficient mutant myb12, underlining its role in the regulation of this branch of the flavonoid pathway and in the transcriptional regulation of the flavanol aglycone biosynthetic gene FLAVONOL SYNTHASE1 (FLS1) (Czemmel et al., 2009). Besides MYBF1, all of the MYB TFs shown to date to regulate the flavonoid biosynthetic pathway in grapevine, belong to the R2R3-MYB subgroup, and require interaction with a basic helix-loop-helix (bHLH) protein (enhancer of glabra3 [Arabidopsis thaliana] and MYC1 [grapevine]) to activate gene expression. Based on a recent analysis of the PN40024 8.4X coverage assembly performed by Matus et al. (2008), grapevine appears to possess at least 108 R2R3-MYB TFs (Matus et al., 2008; Hichri et al., 2010). We now demonstrate that two closely related R2R3-MYB–type TFs, designated MYB14 and MYB15, play an important role in the transcriptional regulation of the stilbene biosynthetic pathway in grapevine. Transcript analyses revealed a close correlation between the expression of MYB14 and MYB15 and selected members of the STS gene family and the subsequent accumulation of stilbenes under normal and stressed conditions. Transient gene reporter assays and ectopic expression of MYB15 in grapevine hairy roots also strongly supported their key roles in the transcriptional control of STS expression and accumulation of stilbene derivatives in grapevine.
RESULTS
Identification of MYB Genes Coexpressed with STSs in Stressed Grapevine Tissues
To identify TFs potentially involved in the regulation of STS genes in grapevine, mRNA-Seq transcriptional data obtained from V. vinifera cv Pinot Noir leaf discs exposed to UV-C, which was shown to induce high levels of STS gene expression (Vannozzi et al., 2012), were used to analyze the expression patterns of all putative grape R2R3-MYB TFs (see Supplemental Figure 1A online). Of the 108 grape R2R3-MYB TFs examined, only two genes displayed similar expression patterns to that of the inducible STS genes. These two genes, corresponding to the identifiers VIT_07s0005g03340 and VIT_05s0049g01020 on the 12X V1 genome assembly of the PN40024 genotype (http://genomes.cribi.unipd.it/gb2/gbrowse/public/vitis_vinifera/) (Jaillon et al., 2007), were previously assigned the gene names MYB14 and MYB15, respectively, by Matus et al. (2008) based on their homology with the Arabidopsis genes At-MYB14 and At-MYB15, respectively (Stracke et al., 2001).
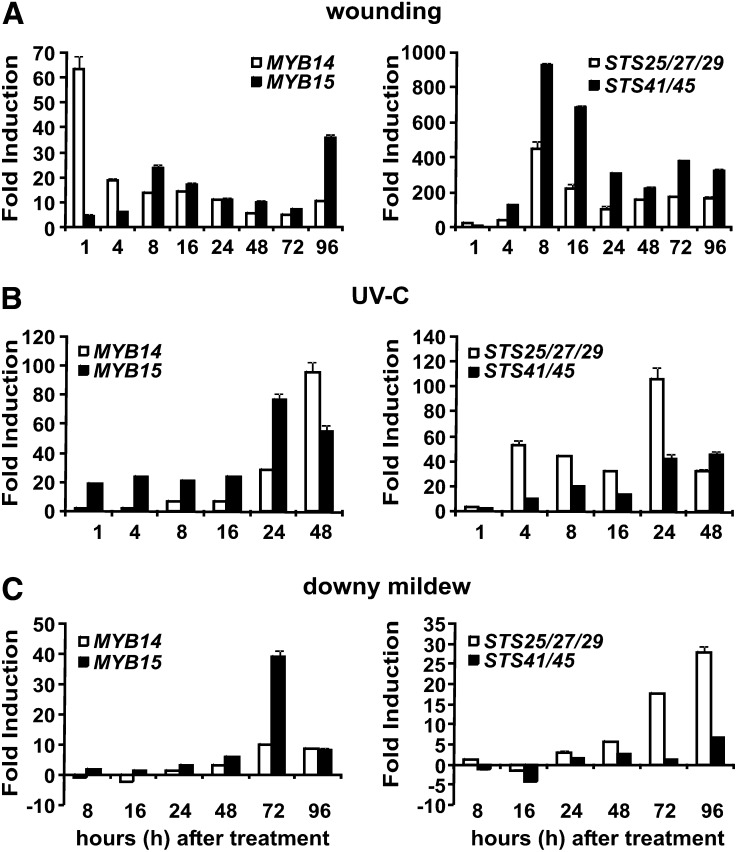
To confirm these mRNA-Seq observations, the transcript levels of two highly responsive group-B–type STS genes, STS29 and STS41 (Vannozzi et al., 2012), and of the MYB14 and MYB15 TFs, were monitored using quantitative RT-PCR in wounded, UV-C–irradiated, and P. viticola–infected V. vinifera cv Shiraz leaf discs (Figure 2). As the STS family contains 48 closely related genes (Vannozzi et al., 2012), it was not possible to design sequence-specific primers for the detection of only one STS isoform. Thus, the primers STS41-F/R detect the isoforms STS41 and STS45, while the primers STS29-F/R quantify the combined expression levels of STS29, STS25, and STS27.
Figure 2.
Expression of MYB14, MYB15, and Selected STS Group B-Type Genes (STS25/27/29 and STS41/45) in Response to Different Abiotic and Biotic Stresses.
(A) V. vinifera cv Shiraz leaf discs were subjected to wounding,
(B) UV-C light irradiation.
(C) Downy mildew infection.
Rapidly expanding leaves collected from nodes 5 to 8 of new shoots harvested at different time points following treatments (hours after treatment). Transcript levels were normalized to the expression of elongation factor EF1-α and plotted as fold change. Fold change for wounded discs was calculated relative to the untreated sample (0 h), whereas fold change for UV-C–treated and downy mildew infected discs was obtained by calculating the ratio between treated (UV-C or downy infected) and untreated (i.e., wounded discs) samples at the same time point. The experiment was repeated twice with the same results. Data show the results of one of these experiments. Bars indicate se of three technical replicates.
The results of the quantitative PCR analysis confirmed the findings of the mRNA-Seq analysis, showing a strong coinduction between the STS genes and MYB14/15 in response to different stress treatments (Figure 2). In all treatments analyzed, MYB14 and MYB15 expression was found to correlate with the expression of the highly responsive group B STS genes (Figures 2A to 2C). Thus, whenever STS genes were induced by biotic or abiotic stress, MYB14 and/or MYB15 were also found to be induced. In a second wounding experiment, the expression of other R2R3-MYB genes known to be involved in the regulation of flavonoid synthesis (i.e., MYBA, MYB5a, MYB5b, and MYBPA1) were also monitored along with MYB14 and MYB15 (see Supplemental Figure 1B online). Apart from MYB14 and MYB15, only MYBA showed a transient increase in transcription 1 h after wounding.
The level of MYB14 and MYB15 transcription was also correlated with the observed level of STS transcription. For example, exposure of grape leaf discs to UV-C light (Figure 2B) increased the transcription of MYB14 and MYB15 by 7- and 20-fold, respectively, within 8 h of treatment compared with untreated discs (wounding only). Similarly, STS41/45 and STS25/27/29 expression was increased by 20- and 40-fold, respectively, 8 h after UV-C treatment compared with transcript levels in untreated discs (Figure 2B). It is also important to note that the induction of MYB14 and MYB15 expression preceded the initial induction of STS transcription after wounding and UV-C treatment (Figures 2A and 2B). Taken together, these results confirm a strong coinduction of selected STS and MYB14/MYB15 genes in stressed grapevine tissues. Furthermore, the temporal expression profiles strongly suggest that these TFs might be involved in the induction of STS transcripts in grape leaves in response to biotic and abiotic stress.
Expression of MYB14 and MYB15 Correlates with Stilbene Accumulation in Skins and Seeds of Developing Grape Berries
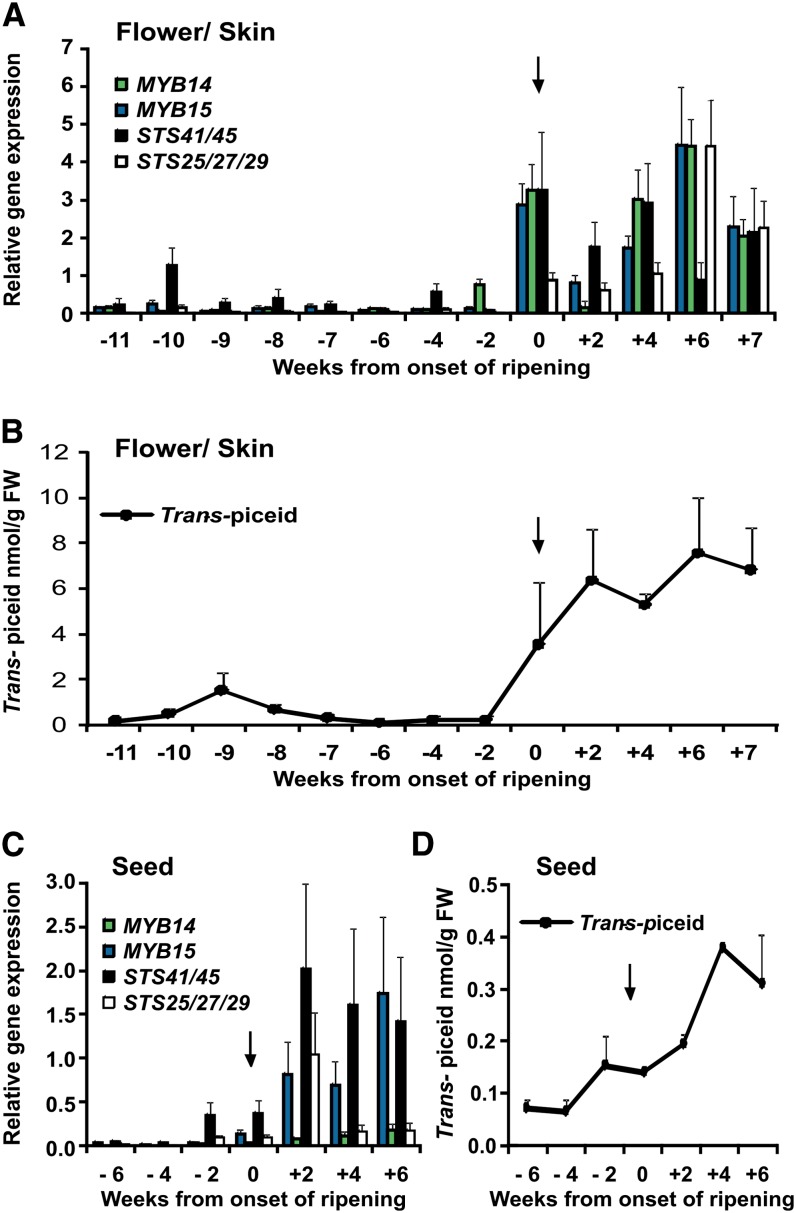
Although the majority of previous studies on stilbene biosynthesis in grapevine have focused on their accumulation in response to stress treatments, several studies have also reported that healthy grape berries synthesize stilbenes under natural growing conditions (Versari et al., 2001; Hall and De Luca, 2007; Gatto et al., 2008). To confirm if the observed correlation between MYB14/MYB15 and STS expression also holds for expression during the period of stilbene accumulation in developing grape berries, we compared transcript levels of selected STS genes, MYB14 and MYB15 TFs, and the accumulation of stilbene metabolites during berry development (Figure 3).
Figure 3.
Expression of MYB14, MYB15, STSs, and Stilbene Accumulation during Grape Berry Development of V. vinifera cv Pinot Noir.
(A) Columns represent MYB and STS transcript profiles in flower/skins.
(B) Stilbene accumulation in flower/skins.
(C) MYB and STS transcript profiles in seeds.
(D) Stilbene accumulation in seeds.
Data points are given as weeks from the onset of ripening (véraison; labeled with an arrow), with expression values as means of three replicate PCRs (n = 6) and error bars indicating se. Note that from 6 weeks before véraison (-6), berry skin has been separated from seeds. Flowering occurred 8 weeks before véraison. Accumulation of stilbenes was measured as methanol extracts of developing grape tissues using reverse-phase HPLC analysis. (B) and (D) show relative stilbene contents of trans-piceid in developing berries and in seeds as the mean of three different experiments, with error bars indicating se. Note that each time point during developmental series is a pool of >100 berries collected from ∼20 plants growing as described in Methods. FW, fresh weight.
Quantitative RT-PCR analyses revealed both MYB14 and MYB15 to be expressed in berry skins and seeds and their strong induction coincided with the onset of ripening (véraison) (Figures 3A and 3C), which correlated closely with the pattern of transcript accumulation of selected STS transcripts. In agreement with these observations, HPLC fluorometric determination indicated a significant accumulation of the glycosylated resveratrol derivative, trans-piceid, in skins and seeds with the onset of ripening (Figures 3B and 3D). Stilbene accumulation, therefore, closely correlates with the expression of MYB14/MYB15 and STS isoforms during grape berry development. Interestingly, trans-piceid accumulation was not only detected in berry skins after véraison, but also at flowering and fruit set (9 to 10 weeks before véraison), which correlates with an early increase in the expression of STS41, but not STS29, MYB14, or MYB15 (Figures 3A and 3B).
To confirm the presence of trans-piceid and search for other stilbenoid compounds in seeds, we performed a liquid chromatography–hybrid quadrupole time-of-flight–mass spectrometry (LC-QTOF-MS) analysis. Supplemental Table 1 online lists the range of stilbenoid compounds that were confirmed to be present in grape seeds.
MYB14 and MYB15 Contain a Putative C-Terminal Stress-Response Domain
Analysis of the deduced amino acid sequences of MYB14 and MYB15 revealed that the two genes encode proteins of 272 and 253 amino acids in length, respectively, and confirmed that both of them belong to the R2R3-MYB subfamily, based on the presence of the highly conserved R2R3-MYB DNA binding domains within the N-terminal region of the protein (see Supplemental Figure 2A, Supplemental Methods 1, and Supplemental References 1 online) (Matus et al., 2008). In contrast with the highly conserved N terminus, C-terminal regions of R2R3-MYBs are less conserved with only a few motives allowing subgrouping of proteins into separate clades (Stracke et al., 2001). Alignment of the predicted protein products of Vv-MYB14 and Vv-MYB15 with the R2R3-MYB family from Arabidopsis, together with several TFs functionally characterized from tobacco (Nicotiana tabacum) and carrot (Daucus carota), shows that both MYB14 and MYB15 cluster with a group of proteins including At-MYB13, At-MYB14, At-MB15, Nt-LBM1/Nt-LBM2, Nt-MYBJS1, and Dc-MYB1 (see Supplemental Figure 2B, Supplemental Data Set 1, Supplemental Methods 1, and Supplemental References 1 online), which share one of these conserved domains within the C terminus of the protein. This domain, designated as SG2 DxSFW–MxFWFD (see Supplemental Figure 2A, Supplemental Methods 1, and Supplemental References 1 online) appears to be associated with stress responses in Arabidopsis (Stracke et al., 2001). Indeed, all of the Arabidopsis, tobacco, and carrot TFs belonging to the Vv-MYB14/15 cluster have been shown to respond to abiotic stresses (Maeda et al., 2005; Chen et al., 2006; Gális et al., 2006). Interestingly, homologous MYB-factors from other stilbene-producing plants, such as Poaceae (Sb_06g022660, Sb_04g026480, and Sb_01g019270) and Pinaceae (Pg-MYB11 and Pg-MYB12), have been identified and also cluster with Vv-MYB14/MYB15 (see Supplemental Figure 2A, Supplemental Methods 1, and Supplemental References 1 online).
Although lacking any obvious nuclear localization signal motif, analysis of the predicted cellular localization of Vv-MYB14 and Vv-MYB15 using WOLF PSORT (Horton et al, 2006) predicted these TFs to be targeted to the nucleus, and this was confirmed for Vv-MYB14 by bombardment of a MYB14:GFP (for green fluorescent protein) fusion into onion (Allium cepa) epidermal cells (see Supplemental Figure 3 online).
MYB14 and MYB15 Activate STS Promoter Activity
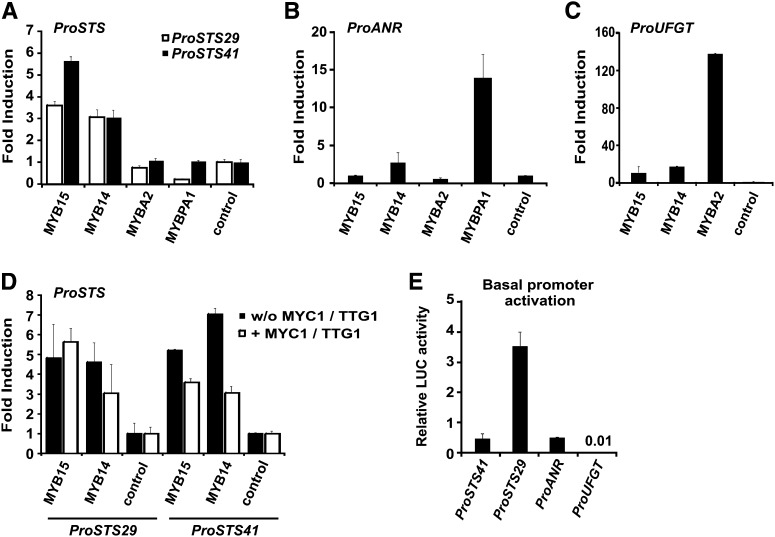
To determine the capacity of both MYB14 and MYB15 to regulate the transcription of structural genes of the stilbene biosynthetic pathway, transient expression assay were conducted using Chardonnay grapevine suspension cell cultures and a dual luciferase reporter system (Horstmann et al., 2004). Promoter fragments corresponding to ∼2 kb upstream of the start codon of STS29 and STS41 (Vannozzi et al., 2012) were first isolated and sequenced and found to contain several putative MYB binding and stress-related cis-elements using the plant DNA cis-element database (http://www.dna.affrc.go.jp/PLACE/signalscan.html) (Higo et al., 1998).
To confirm the in silico analysis and investigate the specificity of MYB14 and MYB15 for the activation of stilbene biosynthetic genes, the activation potentials of both TFs with the promoters of STS29, STS41, and selected flavonoid pathway branch genes UFGT (anthocyanins) and ANR (PAs) were analyzed and compared with previously characterized positive acting flavonoid-specific MYB TFs (Boss et al., 1996; Kobayashi et al., 2002; Bogs et al., 2007). Chardonnay cell suspensions transiently expressing STS luciferase reporter constructs showed significant increases in the promoter activity of 5.6-fold for STS41 and 3.5-fold for STS29 when cotransformed with MYB15 and 3.0-fold for STS41 and STS29 when cotransformed with MYB14 in comparison to the control bombardments in the absence of the MYB TFs (Figure 4A; see Supplemental Figure 4 and Supplemental References 1 online). By contrast, the flavonoid-specific MYB TFs MYBA2 and MYBPA1 had little or no effect on STS promoter activity. Conversely, coexpression of MYB14 or MYB15 with the promoters of ANR or UFGT resulted in only minor increases in promoter activity when compared with that obtained with the TFs MYBA2 and MYBPA1 (Figures 4B and 4C; see Supplemental Figure 4 and Supplemental References 1 online). For example, although the UFGT promoter was induced ∼17-fold by MYB14 and 11-fold by MYB15, this is much lower than the 137-fold induction obtained with the anthocyanin pathway-specific MYB factor MYBA2 (Figure 4C; see Supplemental Figure 4 and Supplemental References 1 online) and is comparable to activation rates observed with the UFGT promoter with other nonspecific MYB TFs such as MYBPA1 (2.5-fold; Czemmel et al., 2009).
Figure 4.
MYB14 and MYB15 Specifically Induce the Promoter Activity of STS41 and STS29, Which Are Involved in the Biosynthesis of Stilbenes.
(A) to (C) Fold induction of promoter activity in the presence of a MYB TF relative to promoter activity in the absence of the MYB TF. Control columns show a fold induction of 1.0, indicating no effect on promoter activity.
(D) Fold induction of STS promoter activity by MYB14 and MYB15 with and without (w/o) bHLH and WD40 cofactors Vv-MYC1 and At-TTG1.
(E) Basal activity of V. vinifera promoters in suspension cell culture in the absence of added MYB TFs.
Transient expression in V. vinifera cv Chardonnay suspension cell culture following particle bombardment. Specific promoters linked to a firefly luciferase gene were cobombarded into cells with pART7-MYB TF constructs or with a pART7 (empty vector) control. Each transfection contained the Renilla luciferase plasmid pRluc as an internal control (Horstmann et al., 2004) and the cofactors Vv-MYC1 (bHLH type) and At-TTG1 (WDR type) (except for the experiment shown in [D]). The columns represent relative LUC activity (Firefly/Renilla) of the corresponding promoter plus MYB factor relative to the respective control (without the MYB factor) of six independent experiments with error bars indicating se.
Hichri et al. (2010) previously showed that MYBA and MYBPA1 were not able to induce their target gene promoters without the presence of bHLH and WD40 cofactors (Figure 4D; see Supplemental Figure 4 and Supplemental References 1 online). By contrast, the induction of STS29 promoter activity by MYB14 and MYB15 was not dependent on the presence of the bHLH factor MYC1. Analysis of the deduced protein sequences of MYB14 and MYB15 indicates that these TFs lack the (D/E)L×2(R/K)×3L×6L×3R amino acid motif in their R3 repeat (see Supplemental Figure 2A, Supplemental Methods 1, and Supplemental References 1 online), which is necessary for the interaction with bHLHs (Grotewold et al., 2000; Zimmermann et al., 2004).
To exclude any grape cultivar–specific effects on the induction of STS promoters by MYB14/MYB15, transient expression assays were also undertaken in Pinot Noir grape suspension cell cultures (see Supplemental Figure 5A online). MYB15 was found to increase STS29 and STS41 promoter activity by 19- and 6.8-fold, respectively, confirming the results obtained with Chardonnay cell cultures.
One factor contributing to the lower apparent induction of the STS promoters by MYB14 and MYB15 (Figure 4A) compared with that observed for MYB TF activation of the ANR and UFGT promoters (Figures 4B and 4C) is the much higher background activity of the STS promoters in this transient assay–system (Figure 4E). To investigate this further, we measured the endogenous transcript levels of MYB14, MYB15, and MYBA2 in Chardonnay and Pinot Noir grape suspension cell cultures. Quantitative PCR analysis revealed that the basal transcript level of MYB14 and MYB15 was 20- to 35-fold higher than the level of expression of MYBA (see Supplemental Figure 5B online), which could explain the much higher levels of background activity of the STS promoters compared with the UFGT promoter. The high level of endogenous MYB14 and MYB15 expression in suspension cell cultures significantly limits the dynamic range of further STS promoter induction by MYB14/15 in this transient promoter assay.
The promoter induction assay was also conducted in non-grape tissue, lacking an endogenous stilbene biosynthetic pathway, in an attempt to reduce the background STS promoter activity. Transient coexpression of MYB14 or MYB15 with the STS29 promoter in Nicotiana benthamiana leaf discs was found to increase STS29 promoter activity by 3- and 13-fold, respectively, compared with control bombardments in the absence of the MYB TFs (see Supplemental Figure 5C online), which was not significantly different to the fold increase observed in grape suspension cells (Figure 4; see Supplemental Figure 4A and Supplemental References 1 online). However, in this assay system, the fold induction of the STS29 promoter by MYB15 approximated the fold induction of UFGT promoter by MYBA2 (see Supplemental Figure 5D online).
Taken together, the results of these transient promoter experiments, in grapevine suspension cells and tobacco leaves, strongly support the hypothesis that MYB14 and MYB15 have a role in the transcriptional regulation of stilbene biosynthesis in grapevine.
Overexpression of MYB15 in Grapevine Hairy Roots Leads to Increased STS Expression and Stilbene Accumulation
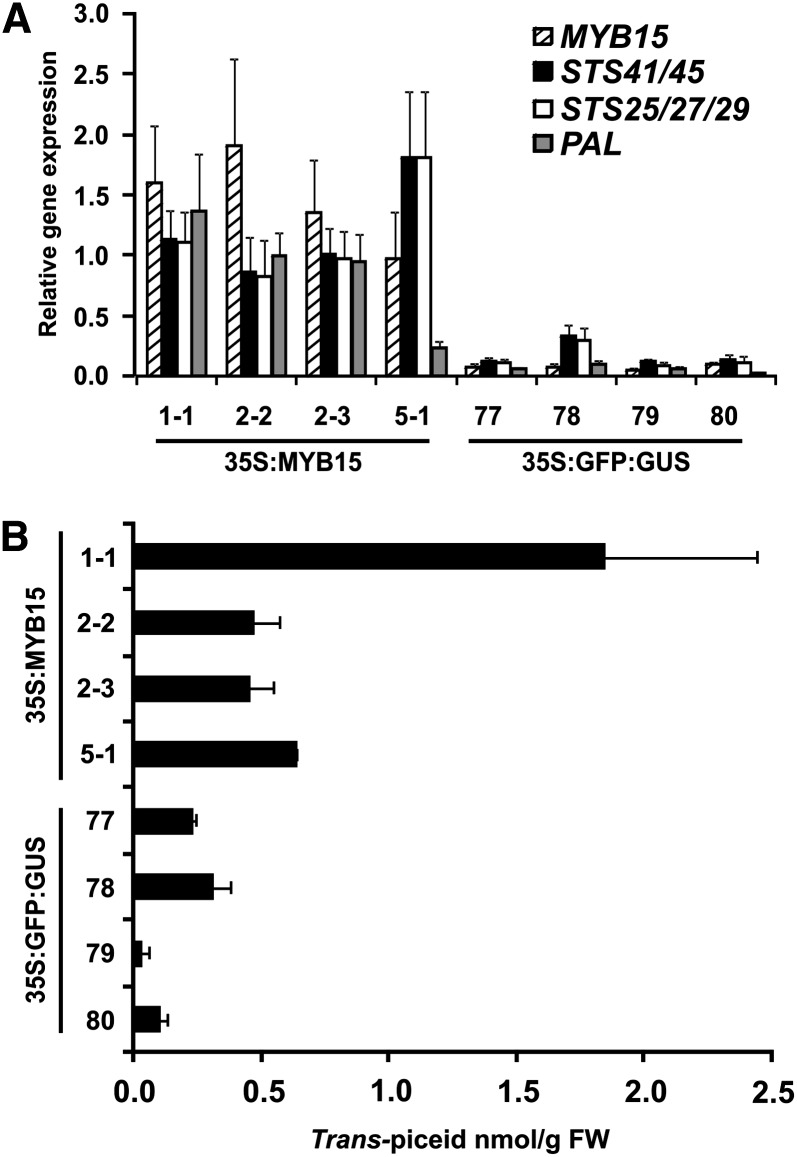
To confirm the role of MYB14/15 in the regulation of the stilbene biosynthetic pathway in planta, MYB15 was ectopically expressed in V. vinifera cv Chardonnay hairy roots under the control of the constitutively active cauliflower mosaic virus (CaMV) 35S promoter. Grapevine hairy roots expressing GFP coupled to β-glucuronidase (GUS) (Karimi et al., 2002) were used as a negative control. Four MYB15-overexpressing transgenic lines (MYB15 1-1, 2-2, 2-3, and 5-1) were identified by quantitative PCR, based on comparison with the level of endogenous MYB15 transcripts in various GFP:GUS-expressing control lines (GFP:GUS77, GFP:GUS78, GFP:GUS79, and GFP:GUS80) (Figure 5A). Selected MYB15 transgenic lines displayed increased levels of transcription of STS genes (Figure 5A) and accumulated higher levels of stilbenes compared with the GFP:GUS transformed lines (Figure 5B; see Supplemental Figure 6 online). On average, transgenic MYB15 transgenic lines accumulated a fivefold higher level of trans-piceid compared with GFP:GUS transformed lines (Figure 5B) and smaller increases in the level of trans-resveratrol, 2,3,4′,5-tetrahydroxystilbene-2-glucoside and other complex stilbenes (see Supplemental Figure 6 online) when measured by fluorimetric HPLC detection. The presence of trans-piceid and other stilbenoid compounds in MYB15 transgenic hairy roots was confirmed by LC-QTOF-MS (see Supplemental Table 1 online).
Figure 5.
Expression of STSs and PAL and Accumulation of trans-Piceid in Grapevine Hairy Roots Ectopically Expressing MYB15.
(A) Transcript levels of MYB15, STS, and PAL were determined by quantitative PCR in V. vinifera cv Chardonnay hairy root lines transformed with pART27-MYB15 compared with control lines transformed with GFP:GUS (pKGWFS7; Karimi et al., 2002). Each column in shows transcript levels corrected to Ubiquitin1, EF1-α, and GAPDH (Pfaffl et al., 2002), expressed as mean values of three replicate PCRs (n = 6) and error bars indicating se. Note that expression levels can only be compared between MYB15 and GFP:GUS control lines and not between the individual genes using this method.
(B) Accumulation of trans-piceid MYB15-overexpressing hairy root lines compared with GFP transformed controls expressed as relative content in nmol/g fresh weight. Trans-piceid levels shown represent the mean of three different experiments with error bars indicating se. FW, fresh weight.
In addition to activation of STS expression, transgenic MYB15-overexpressing hairy root lines were also found to have elevated levels of PAL expression (Figure 5A). The average level of PAL expression in the four MYB15 transgenic lines was found to be 13-fold higher than the average level of PAL expression in the GFP:GUS transformed lines
The increase in STS expression and stilbene production in these transgenic hairy roots confirms the role of MYB15 as a regulator of stilbene biosynthesis in grapevine cells. Our data also demonstrate that MYB15 may have a role in the transcriptional regulation of the core PP pathway.
DISCUSSION
STS Transcription in Grapevine Is Correlated with MYB14 and MYB15 Expression
It is well known that stilbenes are synthesized in plant tissues as a result of the induction of STS genes in response to biotic and abiotic stresses (Versari et al., 2001; Borie et al., 2004; Vannozzi et al., 2012). But to date, no information is available regarding the identity of the TFs responsible for the transcriptional regulation of structural genes of the stilbene biosynthetic pathway. Considering the close relationship between the stilbene and flavonoid biosynthetic pathways and considering that the latter appears to be regulated exclusively by TFs belonging to the R2R3-MYB subgroup, we postulated that stilbene biosynthesis may also be under similar transcriptional control. The analysis of transcriptional data from grapevine leaf discs treated with various abiotic and biotic stresses led to the identification of two R2R3-MYB TFs, MYB14 and MYB15, that were coinduced with STS genes across a range of stress conditions. In all treatments analyzed, MYB14 and MYB15 expression was found to correlate with the expression of highly responsive STS genes in terms of pattern and timing. Thus, whenever STS genes were induced, either MYB14 and/or MYB15 was also found to be induced, and, most importantly, their induction preceded the initial induction of STS genes. By contrast, no evidence was found for any significant involvement of other flavonoid-specific R2R3-MYB TFs in the regulation of stilbene biosynthesis (see Supplemental Figure 1B online).
Our results indicate that MYB14 and MYB15 are not only involved in the induction of STS transcription in grape tissues in response to biotic and abiotic stress but are also responsible for stilbene biosynthesis observed in developing grape berries (Figure 3). Gatto et al. (2008) previously reported that stilbenes accumulate in red and white grapevine varieties after the onset of véraison. While the levels of stilbenes produced in ripening berries are only 1 to 5% that observed in both berries and leaves subjected to biotic and abiotic stress (Langcake, 1981; Douillet-Breuil et al., 1999; Bais et al., 2000), our data confirm that the induction of STS transcription in berry skins at véraison also coincides with an increase in MYB14/15 transcription (Figure 3). The induction of stilbenes with the onset of ripening may provide some protection to the berry from fungal pathogens, such as B. cinerea.
Stilbene biosynthesis shares common substrates with the synthesis of the three major flavonoid subgroups (i.e., PAs, flavonols, and anthocyanins; Figure 1), raising the question as to whether common MYB TFs might regulate both the stilbene and flavonoid pathway. However, previous studies have demonstrated that PA and flavonol accumulation during grape berry development does not correlate with stilbene accumulation. PAs accumulate in developing Shiraz grape berry skin and seeds until véraison (Downey et al., 2003a), correlating with the expression of the major PA pathway genes LDOX, ANR, LAR1, and LAR2 and their transcriptional regulators MYBPA1/2 (Bogs et al., 2005, 2007; Terrier et al., 2009), whereas stilbenes are induced after véraison. Downey et al. (2003b) reported that flavonols are present in high levels at flowering and early berry development of cv Chardonnay and Shiraz and in the late-maturation phase. Thus, the expression of FLS1 (Downey et al., 2003b) and its regulating TF MYBF1 correlates with flavonol synthesis (Czemmel et al., 2009) but not with the synthesis of stilbenes. Anthocyanin accumulation and the regulating TFs MYBA1/2 are induced in grape berry skin at véraison in red berry cultivars, and this does correlate with stilbenes biosynthesis (Figure 3). However, in white berry cultivars in which anthocyanins are not synthesized at véraison because of mutations in MYBA1/2 (Walker et al., 2007), the biosynthesis of stilbenes is still observed (Gatto et al., 2008), which argues against a direct control of STS genes by MYBA1/2. When considered together with the results of our transient promoter assays (Figure 4), these results suggest that STS transcription is regulated by MYB14 and MYB15 and that there is little or no direct involvement of the respective MYB regulators of flavonol, anthocyanin, and PA synthesis in the regulation of stilbene production during grape berry development.
MYB14 and MYB15 Contain a C-Terminal Amino Acid Motif Conserved in R2R3-MYB TFs Involved in Biotic and Abiotic Stress Responses
MYB14 and MYB15 not only share the highly conserved N-terminal R2R3 domain, which characterizes all R2R3-MYB TFs, but also share a small amino acid motif, designated as SG2, within the C-terminal region (see Supplemental Figure 2A, Supplemental Methods 1, and Supplemental References 1 online). This amino acid motif has been previously described as a stress-related motif (Stracke et al., 2001; Dubos et al., 2010) and is also a common feature of R2R3-MYB TFs from other plant species, such as At-MYB13, At-MYB14, and At-MYB15 from Arabidopsis and Nt-LBM1-4 from tobacco, which cluster with Vv-MYB14/15 and respond to environmental stresses (see Supplemental Figure 2B, Supplemental Methods 1, and Supplemental References 1 online). Little information is available regarding the function of At-MYB13, At-MYB14, or At-MYB15, apart from a few reports, which showed that ectopic expression of At-MYB15 improved drought and salt tolerance in Arabidopsis by enhancing the expression of genes involved in abscisic acid biosynthesis and signaling (Ding et al., 2009; Dubos et al., 2010). While conserved amino acid motifs suggest proteins have a common evolutionary origin, they do not always indicate a conserved functional similarity. For example, Ph-PH4, the homolog of the At-MYB5 and Vv-MYB5 TFs, is not involved in activation of the general flavonoid pathway in petunia (Petunia × hybrida) (Quattrocchio et al., 2006).
Nt-LBM2, Nt-MYBJS1, and Dc-MYB1 have also been shown to be involved in regulating upstream genes in the core PP pathway, such as PAL and C4H (Sugimoto et al., 2000; Gális et al., 2006), which are key genes in the production of p-coumaroyl-CoA, the main substrate for flavonoid and stilbene biosynthesis. Grapevine hairy roots ectopically expressing Vv-MYB15 showed a marked increase in the expression of Vv-PAL (Figure 5A). This suggests that, in addition to their role in the regulation of stilbene biosynthesis, Vv-MYB15 (and possibly Vv-MYB14) may also be involved in the regulation of upstream PP pathway genes to ensure the availability of substrates for stilbene biosynthesis.
The introduction of Vv-STS41 (formerly pVst1) into other plant species, such as tobacco, under the control of its own promoter, induced the production of stilbenes, suggesting there are common TFs present in these other plant species, which do not contain STS genes (Hain et al., 1990; Stark-Lorenzen et al., 1997; Thomzik et al., 1997; Zhu et al., 2005). A number of MYB factors orthologous to Vv-MYB14 and Vv-MYB15, which have been detected in other known stilbene-producing plants, such as sorghum and Picea glauca (see Supplemental Figure 2B, Supplemental Methods 1, and Supplemental References 1 online; i.e., SORBIDRAFT_06g022660; PgMYB12), would also be interesting candidates for studying the regulation of stilbene biosynthesis in these plants.
It has previously been reported that STS transcription in grape cells following stress or elicitor treatment is often biphasic in nature (Wiese et al., 1994; Borie et al., 2004; Wang et al., 2010). Vannozzi et al. (2012) demonstrated that this is most likely a result of the differential timing of the stress response of members of the different subgroups within the STS gene family. Similarly, our results demonstrate that MYB14 and MYB15 are differentially expressed in response to various biotic and abiotic stresses (Figure 2), which mirrors the STS subgroup–specific expression, indicating the specificity of the interaction of the two TFs (MYB14 or MYB15) with certain STS genes. The observed differences in expression patterns of MYB14 and MYB15 also raises the question as to whether their own transcription is regulated by different upstream stress response pathways, such as the jasmonic-acid and ethylene-signaling pathways (Belhadj et al., 2006, 2008a, 2008b; Vezzulli et al., 2007).
MYB14 and MYB15 Specifically Activate STS Transcription in the Absence of bHLH
The transcriptional control of the anthocyanin and PA pathway genes is accomplished, in most plant species, by a complex in which R2R3-MYB TFs, bHLH proteins, and WD40 proteins all interact to activate gene transcription (Mol et al., 1998; Winkel-Shirley, 2001; Hichri et al., 2010). By contrast, the induction of the Vv-STS promoter activity by Vv-MYB14 and Vv-MYB15 was independent of the Vv-MYC1 bHLH cofactor (Figure 4D) as was previously observed for Vv-MYBF1, a TF involved in the regulation of flavonol biosynthetic genes (Czemmel et al., 2009). In agreement with these observations, Vv-MYB14, Vv-MYB15, and Vv-MYBF1 lack the bHLH interaction amino acid motif [D/E]Lx2[R/K]x3Lx6Lx3R (see Supplemental Figure 2A, Supplemental Methods 1, and Supplemental References 1 online) (Grotewold et al., 2000; Zimmermann et al., 2004), indicating that these R2R3-MYB TFs do not require bHLH TFs for their activity.
Ectopic Expression of MYB15 in Grapevine Hairy Roots Results in the Induction of STSs and Their Metabolic Products
In grapevine, ectopic gene expression in hairy roots has previously been used for functional characterization and target gene identification of the flavonoid TFs MYBA1, MYBPA1, and MYBPA2 (Cutanda-Perez et al., 2009; Terrier et al., 2009). This approach was also used in this study to provide in planta evidence for the role of MYB15 in regulating stilbene biosynthesis in grapevine. Transgenic grapevine hairy root lines ectopically expressing MYB15 exhibited increased levels of transcription of STS41 and STS29 and an increased accumulation of stilbenes (Figure 5). Surprisingly, the level of stilbene production in hairy roots overexpressing MYB15, measured as trans-piceid accumulation, was found to be only fivefold higher, on average, than the control lines (Figure 5B). One explanation for this observation might be the stress-related induction of endogenous MYB14/15 and STS genes in control hairy root lines. Indeed, similar results were reported for grapevine hairy roots ectopically expressing MYBPA2, which regulates the PA pathway. Accumulation of PAs in the MYBPA2 overexpressing lines was found to be only fivefold higher than the control lines because of a high PA background level in the control roots (Terrier et al., 2009). Another explanation may be the possible phytotoxic effect of stilbene accumulation on hairy-root growth and development as reflected in the relatively low number of regenerating MYB15 hairy-root transformants and the loss of several lines showing high MYB15 expression and putative high stilbenoid levels during the culturing process (see Methods).
The ectopic expression of STS genes in a range of plant species, including Arabidopsis, rice (Oryza sativa), tomato (Solanum lycopersicum), alfalfa (Medicago sativa), apple (Malus domestica), barley (Hordeum vulgare), wheat (Triticum spp), kiwifruit (Actinidia spp), and strawberry (Fragaria spp), has revealed that, together with an increased resistance to pathogen attack, there is commonly an accumulation of trans- and cis-piceid or trans-resveratrol (reviewed in Delaunois et al., 2009). Similar results were observed in MYB15 overexpressing hairy roots (Figure 5B; see Supplemental Figure 6 online). It has been demonstrated that stilbenes, and in particular δ-viniferin, an oxidative resveratrol dimer, and pterostilbene, the 3,5-dimethoxy analog of resveratrol, have fungitoxic properties (Pezet et al., 2003, 2004; Schmidlin et al., 2008) and that the glycosylation of stilbenes may occur in plant cells both to protect cells from their potential toxic effects and to protect resveratrol from degradation (Jeandet et al., 1997; Hipskind and Paiva, 2000). This might explain the high amount of trans-piceid in the MYB15 transgenic hairy roots in comparison with the relatively low amounts of other modified stilbenes (see Supplemental Figure 6 online).
The vast majority of the genes responsible for stilbene diversity, which has been predicted to influence bioavailability, antioxidant capacity underlying phytoalexin function, and stability of the resulting polyphenols, are still unknown. The accumulation of different modified stilbenes in MYB15 transgenic lines suggests that the MYB factors MYB15, and presumably its paralog MYB14, regulate enzymes catalyzing downstream modification and transport processes of the stilbene biosynthetic pathway. Therefore, a comparative transcriptome analysis of transgenic MYB15 and GFP:GUS grapevine hairy roots could lead to the identification of genes involved in stilbene modification and transport. The analysis of these unknown enzymes would expand our knowledge of the stilbene biosynthetic pathway in plants and would be valuable tools for the pharmacological industry exploring modified resveratrol derivatives and their beneficial effect on human health (Mikulski and Molski, 2010; Agarwal and Baur, 2011; Kapetanovic et al., 2011). Quantitative variation of polyphenol levels, including anthocyanins and stilbenes, has been detected for different grapevine cultivars and susceptibility to fungal diseases has been correlated to the content and composition of the stilbene derivatives (Pezet et al., 2004). The variation in anthocyanin content in grape cultivars was recently explained by polymorphisms of a single gene cluster of three MYBA genes (Fournier-Level et al., 2010). Similarly, investigating the genetic differences in MYB14 and MYB15 expression and/or transcriptional activity in different grapevine cultivars might lead to the development of molecular markers for breeding of grapevines with optimized stilbene content and composition.
METHODS
Plant Material
For mRNA-Seq analysis, leaves of grape (Vitis vinifera cv Pinot Noir) were obtained from field-grown vines at the Lucio Toniolo experimental farm of the University of Padova (Legnaro, Italy) as described by Vannozzi et al. (2012). The quantitative RT-PCR analyses after stress treatment were conducted using V. vinifera cv Shiraz rapidly expanding leaves collected from nodes 5 to 8 of new shoots and samples from potted glasshouse vines at the Waite Campus (Adelaide, South Australia, latitude 34° 56’ south, longitude 138° 36’ east). Grapevine tissues of the developmental series (V. vinifera cv Pinot Noir) were collected from a commercial vineyard during the 2007 to 2008 season in Schrießheim, Germany (49° 28’ 40’’ south, 8° 40’ 22’’ east) with a moderate climate and mean daily temperature of 22°C during grape berry development. Sampling was performed from the start of floral initiation until harvest, with ∼100 berries from 20 different plants and bunches collected at weekly intervals, as previously described (Downey et al., 2003a, 2003b). Nicotiana benthamiana plants used for particle bombardment were grown under green house conditions for 6 to 8 weeks.
The grape cell suspension cultures used in the luciferase transient transfection assay were established from Chardonnay and Pinot Noir petiole callus culture and maintained in Grape Cormier medium as previously described (Czemmel et al., 2009). Transgenic hairy roots were generated as described below.
Analysis of mRNA-Seq Expression Data Sets for Identifying R2R3-MYB TFs Coexpressed with Vv-STS Genes
Generation and analysis of mRNA-Seq data from UV-C–treated V. vinifera cv Pinot noir leaf discs was performed as described previously (Vannozzi et al., 2012). STS and coexpressed MYB TFs expression patterns were analyzed and graphically expressed using mean of Multi Experiment Viewer software (Saeed et al., 2006). The mRNA-Seq data were deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) as series GSE37743.
Bioinformatic Analysis
Multiple sequence alignments were conducted using the ClustalW program (Chenna et al., 2003), and the editing of aligned sequences was performed using GeneDoc 1.6 software. An unrooted phylogenetic tree showing selected plant R2R3-MYB TFs was generated with a neighbor joining method using MEGA 3.1 software (Kumar et al., 2004). Protein sequences were identified after a BLASTp search with the protein sequence from Vv-MYB14/15. Selected protein sequences from TFs were downloaded from the GenBank or EMBL databases (http://www.ensembl.org/index.html; The EMBL Nucleotide Sequence Database), and their accession numbers are listed below.
The Vv-STS29 and Vv-STS41 promoter regions were screened for plant cis-acting regulatory DNA elements both manually and using the Plant cis-Element database (http://www.dna.affrc.go.jp/PLACE/) (Higo et al., 1999).
Quantitative RT-PCR Expression Analyses of Vv-MYB14, Vv-MYB15, and Selected Vv-STSs
Total RNA extraction and cDNA synthesis from V. vinifera cv Shiraz leaf discs subjected to stress treatments was performed as described previously (Vannozzi et al., 2012). Total grapevine RNA from the various tissues of the developmental stages was isolated according to Downey et al. (2003b). RNA of grapevine hairy roots and suspension cell cultures was isolated with the EURx GeneMATRIX Universal RNA purification kit (Roboklon) following the manufacturer’s protocol. For cDNA synthesis, 1 µg of grapevine total RNA was reverse transcribed in 20 μL for 1 h at 42°C using oligo(dT)18 primer, RNase inhibitor, and thermo-stable avian myeloblastosis virus reverse transcriptase (dART cDNA synthesis kit; Roboklon).
Expression analysis of MYB and STS genes after treatment of V. vinifera cv Shiraz leaves with abiotic (UV-C irradiation, wounding) and biotic (Plasmopara viticola infection) stress was performed by quantitative PCR as described previously (Vannozzi et al., 2012). Transcript levels of MYB14, MYB15, and selected STS genes during grape berry development and in transgenic hairy roots were determined by quantitative PCR with the SYBR Green method on a Rotor-Gene Q (Qiagen). The PCR reaction mix (12.5 µL) contained 1 μL of cDNA (diluted 1:10), 6.25 μL of Maxima SYBR Green qPCR Master Mix (2×) (Fermentas), 5 µM each primer, and sterile water. The thermal cycling conditions used were 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 58°C (Vv-MYBs) or 60°C (Vv-STS) for 30 s, and 72°C for 20 s, followed by a melt cycle with 1°C increments (5 s) from 56 to 96°C. The primer efficiency was tested in preliminary experiments with dilutions of pART7-MYB14/pART7-MYB15 plasmid or cDNAs of samples maintaining a value of r2 ≥ 0.90. The cycle threshold values of the developmental series and the transgenic hairy roots are corrected to Vv-Ubiquitin1, Vv-EF1-α, and Vv-GAPDH, which were previously shown to be appropriate reference genes during berry development (Reid et al., 2006). Normalization against three reference genes was conducted as described previously (Pfaffl et al., 2002). The analysis of melting curves, measurement of primer pair efficiencies, and determination of cycle threshold values, including the calculation of the mean normalized expression of the genes, were conducted using the Rotor-Gene Q Series Software Q 2.0.2 (Qiagen) and the Q-Gene software (Muller et al., 2002). The expression of Vv-MYB TFs was determined using the following gene-specific primers: MYB14-F (5′-TCTGAGGCCGGATATCAAAC-3′), MYB14-R (5′-GGGACGCATCAAGAGAGTGT-3′), MYB15-F2 (5′-CAAGAATGAACAGATGGAGGAG-3′), MYB15-R2 (5′-TCTGCGACTGCTGGGAAA-3′), Vv-MYB5a-F (5′-CTAGAACTGTCTGGGAACCT-3′), Vv-MYB5a-R (5′-TGCAAGGATCCATTTCACATAC-3′), Vv-MYB5b-F (5′-TGACAGCCGGTGTTCTTTAAT-3′), Vv-MYB5b-R (5′-AGCATACTAACACAACAACACAACC-3′), Vv-MYBPA1-F (5′-AGATCAACTGGTTATGCTTGCT-3′), and Vv-MYBPA1-R (5′-AACACAAATGTACATCGCACAC-3′). To measure the expression of the STS genes Vv-STS41 and Vv-STS29, it was not possible to design gene-specific quantitative PCR primers because of the high sequence homology between different family members (Vannozzi et al., 2012). Thus, STS primers Vv-STS41-F (5′-GAGTACTATTTGGTTTTGGACCT-3′) and Vv-STS41R (5′-AACTCCTATTTGATACAAAACAACGT-3′) also detect the isoforms Vv-STS41 and Vv-STS45, and the primers Vv-STS29-F (5′-GGTTTTGGACCAGGCTTGACT-3′) and Vv-STS29-R (5′-GAGATAAATACCTTACTCCTATTCAAC-3′) quantify the expression levels of Vv-STS29, Vv-STS25, and Vv-STS27. Expression of PAL in grapevine hairy roots was measured with the primers Vv-PAL_qF (5′-GTTGTCGTGAAAAACCAGCTT-3′) and Vv-PAL_qR (5′-GGATCACTCACGACGAAACTC-3′). The primers for Vv-MYBA, Vv-MYBA-F (5′-CTTTTCGGCTTCTGGAGAGA-3′), and Vv-MYBA-R (5′-CTGTGTTGCAGTTTCTTCTGTC-3′) detect the isoforms Vv-MYBA1, Vv-MYBA2, and Vv-MYBA3. All quantitative PCR products were verified by agarose gel electrophoresis and by determination of melting curves at the end of each run. Amplified gene products were sent for sequence analysis to Eurofins MWG Operon.
Cloning of Vv-MYB14, Vv-MYB15, and Vv-STS Promoter Constructs
The complete coding sequences of MYB14 and MYB15 were amplified using proofreading Taq polymerase from Shiraz cDNA obtained from UV-C–irradiated grape berry skins (MYB14) or grape berry cDNA (MYB15). Sequence-specific oligonucleotides binding into the putative 5′ and 3′ untranslated regions of the target genes were designed using Primer 3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) on grape sequences provided by Genoscope (http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis). Gene-specific primers Vv-MYB14-ATG-Xho (5′-GCGCTCGAGAAAATGGGGAGAGCTCCTTG-3′) and Vv-MYB14-STOP-BamHI (5′-CGCGGATCCTTCTTCCCTCATATTTCTGATAATT-3′) introduced XhoI and BamHI restriction sites to the complete coding sequence of MYB14. Additional restriction sites for XbaI and BamHI were attached to MYB15 using sequence-specific primers MYB15-F (5′-TGATCTAGAGCCAGAGG TTAGAGTAGTGGTG-3′) and MYB15-R (5′-TGAGGATCCTTCCGCTTTTTGCTTTCTTG-3′). The generated PCR fragments were purified, digested with the above-mentioned restriction enzymes, and cloned in the vector pART7 (Gleave, 1992) to give pART7-MYB14 and pART7-MYB15, where both MYB factors were under the control of the CaMV 35S promoter. The nucleotide sequence of the MYB14 open reading frame (ORF) with 836 bp and MYB15 ORF with 926 bp in pART7 was determined by DNA sequencing. The expression cassette present in pART7-MYB14 and pART7-MYB15 was isolated as a NotI fragment and cloned into the NotI site of the binary vector pART27 (Gleave, 1992). For hairy root transformation, pART27-MYB15 was transferred into Agrobacterium rhizogenes strain ATCC 15834 by electroporation.
To conduct the transient expression assay, promoter regions related to the 8.4× grape genome sequences of the STSs STS29 (1255 bp) and STS41 (1436 bp) (Vannozzi et al., 2012) were isolated from genomic DNA (cv Shiraz) using Phusion DNA polymerase (Finnzymes). The specific primers STS41pF (5′-TATGAGCTCCGTCCATAGGAAGTAGAGTAAAATG-3′) and STS41pR (5′-TATCTCGAGATGCCAGATACGTTCTGAAATTG-3′) were used to introduce the restriction sites SacI-XhoI to the chosen promoter sequence of STS41 and STS29pF (5′-TATGAGCTCAAAATGTCGAAACACTTTGTATTAAA-3′) and STS29pR (5′-TATCTCGAGTGATCCC AGCTACGTACTCAA-3′) added this motive to STS29. The promoter regions of STS29 and STS41 were ligated into the luciferase vector pLuc (Horstmann et al., 2004) and confirmed by DNA sequencing. The cloning of control promoter fragments of ANR and UFGT into pLuc as well as the coding regions of MYBPA1 and MYBA2 into pART7 was described previously (Bogs et al., 2007).
Localization of Vv-MYB14 in Onion Epidermal Strips
A modified version of shuttle vector pART7 (Gleave, 1992), termed pN’gfp (Selth et al., 2005), was used to produce the MYB14:GFP fusion construct. The MYB14 ORF was amplified and cloned into the XhoI-BamHI sites of pN’gfp to generate an in-frame C-terminal fusion to the GFP gene downstream of the CaMV 35S promoter. Onion (Allium cepa) epidermal strips were prepared and bombarded with MYB14:GFP as described previously (Selth et al., 2005). Onion strips were sampled after 48 h and mounted in 1× PBS, and GFP was visualized with a Zeiss Axio Imager M1 microscope, GFP filter (470/40-nm excitation; 525/50-nm emission) under ×20 magnification. Nuclei were visualized using 1 µg/mL of 4',6-diamidino-2-phenylindole dissolved in 1× PBS, using a 4',6-diamidino-2-phenylindole filter (365-nm excitation; 445/50-nm emission).
Transformation and Induction of Transgenic Grapevine Hairy Roots
The A. rhizogenes strain ATCC 15834 containing either pART27-MYB15 or pKGWFS7 (Karimi et al., 2002), which both express their respective gene of interest, MYB15 or GFP:GUS, under control of the CaMV 35S promoter, was grown with shaking at 200 rpm and 28°C for 24 h on MGL medium (Mathews et al., 2003).
Transgenic hairy roots were generated as previously described by Torregrosa and Bouquet (1997) with the following modifications: Leaves from in vitro–grown Chardonnay plants with attached petioles were cut and placed upside down with the petiole facing upwards on hairy root (HR) medium agar plates (1% agar). HR medium was prepared with macroelements (1.6 g/L NH4NO3, 4.4 g/L CaCl2, 3.7 g/L MgSO4, and 23.25 g/L KNO3) and Murashige and Skoog microelements (3.1 g/L H3BO3, 8.45 g/L MnSO4, 4.3 g/L ZnSO4, 0.414 g/L KI, 0.125 g/L Na2MoO4, 12.4 mg/L CuSO4, and 12.5 g/L CoCl2) modified as previously described by Murashige and Skoog (1962), B5 vitamins (Gamborg et al., 1968), FeEDTA (7.44 g/L Na2EDTA and 1.86 g/L FeSO4), and 1.5% Suc, pH 5.7 (1 n KOH). The freshly cut petioles were inoculated with one drop of the overnight Agrobacterium tumefaciens culture on the top of the stem and were cocultivated for 5 d at 22°C in the dark. Leaves were transferred onto HR medium containing 400 μg/mL of timentin with the petioles and the lower surface of the leaf in contact with the agar. Leaves were incubated at 28°C in the light. Newly emerging root tips were removed once they reached ∼2 cm in length and were placed onto HR medium containing 400 μg/mL of timentin. Stable transgenic roots were cultivated at 24°C in the dark and subcultured every 4 to 5 weeks on the same medium.
Due to low transformation/regeneration and slow growth of MYB15 transgenic hairy roots and to avoid possible gene silencing effects, 1 to 2 cm of roots were sampled from each line during the subculturing process at weeks 4 and 5 and frozen at −80°C. The remaining root material was divided and transferred to new media. To harvest enough root material, each line was harvested over a period of 1 year and the samples combined for HPLC, quantitative RT-PCR, and LC-QTOF-MS analysis. In addition, several MYB15 transformation experiments were undertaken, each generating several independent lines showing ectopic MYB15 expression. However, those lines showing the highest levels of MYB15 expression were found to become necrotic during the culturing process, and it was not possible to sample enough root material to carry out a complete analysis. Therefore, the independent transgenic hairy root lines MYB15 1-1, 2-2, 2-3, and 5-1 described in Figure 5 represent those lines with intermediate levels of STS induction. Control root lines showed no necrotic phenotype during growth.
Transient Promoter Assays
Transient promoter assays were developed using a cell suspension of V. vinifera cv Chardonnay and Pinot Noir petiole callus cultures and N. benthamiana as previously described (Bogs et al., 2007; Walker et al., 2007). For transient expression in N. benthamiana, leaf discs (d = 3 cm) were used. The Dual Luciferase Assay protocol was modified based on Czemmel et al. (2009). The Renilla luciferase plasmid pRluc was used as an internal control in each transfection experiment (Horstmann et al., 2004). Transient assays were performed with and without the bHLH and WD40 cofactors Vv-MYC1 and At-TTG1 (Figure 4D). All transfection experiments were repeated six times. Mean values of firefly and Renilla luciferase ratios are reported as relative luciferase activity with error bars indicating se.
Analysis of Stilbenoids in Grapevine via HPLC
HPLC analysis of stilbene compounds was performed on a reverse-phase HPLC device (Kontron Instruments; 322 pump system/360 autosampler/335 HPLC detector) with a Symmetry C18 column (3.5 µm, 4.6 mm × 150 mm [WAT200632]; Waters) protected by a guard column (Czemmel et al., 2009). Buds, flowers, berries, and seeds of the developmental series and the transgenic hairy roots overexpressing Vv-MYB15 or GFP (pKGWFS7) were frozen in liquid nitrogen and homogenized, and 50-mg samples were extracted by adding 200 μL of 50% (v/v) methanol (HPLC grade) in water and sonicated for 20 min in an ice water bath. Extracts were incubated at room temperature for 15 min and centrifuged for 10 min at 13,000 rpm to obtain a clear supernatant, and 150 μL was used for HPLC analysis. Separation was performed with a binary gradient of solvent A (10% [v/v] formic acid in water) to solvent B (100% [v/v] methanol; HPLC grade). The gradient conditions were 0 min, 17% solvent B; 30 min, 35% solvent B; 41 min, 37% solvent B; 43 min, 100% solvent B; 51 min, 100% solvent B; 52 min, 17% solvent B; and 59 min, 17% solvent B. The column was maintained at 40°C, and the flow rate was 1.0 mL/min. Fluorimetric detection with a maximum excitation wavelength at 330 nm and emission at 374 nm was used to detect stilbenes as described previously (Pezet et al., 1994). Data acquisition and processing were performed using Kroma System 2000 software (Kontron) (Czemmel et al., 2009). Concentrations were calculated from calibration curves prepared from commercial stilbene standard trans-piceid (PhytoLab). The stilbene concentrations were quantified relative to the trans-piceid calibration curve in each sample and expressed in nmol/g fresh weight. Columns in Figures 3B, 3D, and 5B show three HPLC measurements, and error bars indicate se. Trans-piceid was identified and retention times were verified in all HPLC extracts by spiking with the commercial standard (0.4 µg trans-piceid), and acidic hydrolysis was performed to cleave the glycosyl group for a shift to the aglycon by the addition of 50 μL 3 n HCL and 150 μL 50% (v/v) methanol in water and incubation for 3 h at 95°C.
Identification of Stilbenoids via LC-QTOF-MS Analysis
LC-QTOF-MS was performed to identify flavonoids and stilbenoids in skin and seed during grape berry development and within grapevine hairy roots overexpressing MYB15 and GFP:GUS control lines. Extraction of flavonoids and stilbenoids was performed as described by for HPLC analysis as methanolic extracts from 50 mg of tissue. LC-QTO-MS was performed as previously described (Broecker et al., 2011a, 2011b) with the following modifications: Liquid chromatography was performed using an Agilent 1260 Infinity (Agilent Technologies) with 1260 degasser, 1260 high performance autosampler, binary pump 1260 (maximum 600 bar), and 1260 TCC column oven. The Agilent Zorbax Eclipse Plus RRHT C18 column (100 mm × 2.1 mm, 1.8 µm; Agilent Technologie) was used at 40°C with the following separation conditions: eluent (A) 94.8% water, 5% methanol, and 0.2% CH2O2 and eluent (B) 5% water, 94.8% methanol, and 0.2% CH2O2 with the time program: 0 min, 100% A; 2 min, 100% A; 10 min, 0% A; 15 min, 0% A; 15.5 min, 100% A; 19.5 min, 100% A (post) with the flow rate of 0.4 mL/min. The injection volume was 2 µL. For mass spectrometry analysis, a Q-TOF 6530 time-of-flight mass spectrometer (Agilent Technologies) was used with ion source electrospray ionization+ Agilent Jet Stream in positive (protonated molecules [M+H]+) and negative (deprotonated molecules [M-H]−) ionization modes. Quadrupole was used as described by previously (Broecker et al., 2011b) for selection of precursor ions within a mass window ∆m/z (mass-to-charge ratio) of 1.3 until 4 in tandem mass spectrometry (MS-MS) mode. MS-MS spectra were generated in product-ion scan at collision-induced dissociation energies of 10, 20, and 40 eV. The QTOF conditions are as follows: drying gas, 325°C; 6 L/min; sheath gas, 380°C; 11 L/min; nebulizer pressure, 35 p.s.i.; Vcap voltage, 3000 V; fragmentor voltage, 150 V; mass rage (mass spectrometry and MS-MS), 100 to 1700 m/z, acquisition rate 1 spectra/s; and reference ions for mass calibration 112.985587 [M-H]− and 966.000725 [M+H]+. Software tools for measurement, identification, and analysis were previously described by Broecker et al. (2011b). The stilbenoid database was based on over 100 forms reviewed by Pawlus et al. (2012). For trans-piceid identification, retention time was additionally verified using a commercial standard, and fragmentation of the precursor ion m/z 389.123 led to the identification of trans-resveratrol m/z 227.077 in skin and seed.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative (http://www.Arabidopsis.org), GenBank, or EMBL database (The EMBL Nucleotide Sequence Database) under the following accession numbers: AM489345 (Vv-STS41), AM463938 (Vv-STS29), Vv-PAL (AEX32784), TC32075 (Vv-Ubiquitin1), EC959059 (Vv-EF1-α), CB973647 (Vv-GAPDH), CAD91911 (Vv-ANR), AY955269 (Vv-UFGT), EU447172 (Vv-MYC1), AT5G24520 (At-TTG1; NM_180738), BAE54312 (Dc-MYB1), BAE93149 (Nt-MYBJS1), AT2G31180 (At-MYB14; AAD20663), ABW34392 (Vv-MYB14), AT1G06180 (At-MYB13; AAF80215), KC514110 (Vv-MYB15), XP_002448188 (hypothetical protein SORBIDRAFT_06g022660 [Sorghum bicolor]), AT3G23250 (At-MYB15; NP_188966), XP_002454197 (hypothetical protein SORBIDRAFT_04g026480 [sorghum]), XP_002464484 (hypothetical protein SORBIDRAFT_01g019270 [sorghum]), ACN40772 (unknown [Picea sitchensis]), ABQ51228 (Pg-MYB12), ABQ51227 (Pg-MYB11), ACK56131 (Vv-MYBPA2), AM259485 (Vv-MYBPA1), P27898 (Zm-P), AAS68190 (Vv-MYB5a), Q58QD0 (Vv-MYB5b), BAD18977 (Vv-MYBA1), BAD18978 (Vv-MYBA2), and FJ948477(Vv-MYBF1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Coinduction of Selected MYB TFs and STS Genes in Grapevine Leaves in Response to Abiotic Stress.
Supplemental Figure 2. Phylogenetic Analysis of Putative Phenylpropanoid Transcriptional Regulators.
Supplemental Figure 3. Targeting of a MYB14:GFP Fusion Protein within Onion Cells.
Supplemental Figure 4. MYB14 and MYB15 Specifically Induce the Activity of STS41 and STS29 Promoters Involved in the Biosynthesis of Stilbenes.
Supplemental Figure 5. STS Promoter Induction and Expression Analysis of MYBs in V. vinifera Cell Culture and Promoter Induction Analysis in N. benthamiana Leaf Discs.
Supplemental Figure 6. Chromatogram of Putative Stilbenoids Extracted from MYB15-Overexpressing Hairy Root Line 1-1 Compared with GFP:GUS Control Line 77.
Supplemental Table 1. Identification of Stilbenoids in V. vinifera Tissues via LC-QTOF-MS.
Supplemental Methods 1. Accession Numbers Used for Phylogenetic Analysis.
Supplemental References 1. References for Supplemental Figures 2 and 4.
Supplemental Data Set 1. Alignment Used to Generate the Phylogeny Presented in Supplemental Figure 2B.
Supplementary Material
Acknowledgments
We thank Marzia Salmaso and Sabrina Canova (University of Padova) for their technical assistance, Angela Feechan and Angelica Jermakow for their technical support at the Commonwealth Scientific and Industrial Research Organization Plant Industry and Werner Dachtler for LC-QTOF-MS analysis (Dienstleistungszentrum Laendlicher Raum). This research was financially supported by the Bundesministerium für Bildung und Forschung and its initiative Genomanalyse im biologischen System Pflanze (GABI) and by the Italian project AGER-SERRES, 2010-2105.
AUTHOR CONTRIBUTIONS
J.H., A.V., S.C., M.L., I.B.D., and J.B. designed the research. J.H. and A.V. performed research. J.H., A.V., C.D., T.R., P.K.B., I.B.D., and J.B. analyzed data. J.H., A.V., S.C., A.R.W., and P.K.B. contributed new analytic/computational tools. J.H., A.V., I.B.D., and J.B. wrote the article.
Glossary
- PP
phenylpropanoid
- STS
stilbene synthase
- TF
transcription factor
- LC-QTOF-MS
liquid chromatography–hybrid quadrupole time-of-flight–mass spectrometry
- GFP
green fluorescent protein
- CaMV
Cauliflower mosaic virus
- GUS
β-glucuronidase
- PA
proanthocyanidins
- bHLH
basic helix-loop-helix
- ORF
open reading frame
- HR
hairy root
- m/z
mass-to-charge ratio
References
- Adrian M., Jeandet P., Bessis R., Joubert J.M. (1996). Induction of phytoalexin (resveratrol) synthesis in grapevine leaves treated with aluminum chloride (AlCl3). J. Agric. Food Chem. 44: 1979–1981 [Google Scholar]
- Adrian M., Jeandet P., Veneau J., Weston L.A., Bessis R. (1997). Biological activity of resveratrol, a stilbenic compound from grapevines, against Botrytis cinerea, the causal agent for gray mold. J. Chem. Ecol. 23: 1689–1702 [Google Scholar]
- Agarwal B., Baur J.A. (2011). Resveratrol and life extension. Ann. N. Y. Acad. Sci. 1215: 138–143 [DOI] [PubMed] [Google Scholar]
- Bais A.J., Murphy P.J., Dry I.B. (2000). The molecular regulation of stilbene phytoalexin biosynthesis in Vitis vinifera during grape berry development. Funct. Plant Biol. 27: 425–433 [Google Scholar]
- Baur J.A., Sinclair D.A. (2006). Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 5: 493–506 [DOI] [PubMed] [Google Scholar]
- Belhadj A., Saigne C., Telef N., Cluzet S., Bouscaut J., Corio-Costet M.-F., Mérillon J.-M. (2006). Methyl jasmonate induces defense responses in grapevine and triggers protection against Erysiphe necator. J. Agric. Food Chem. 54: 9119–9125 [DOI] [PubMed] [Google Scholar]
- Belhadj A., Telef N., Saigne C., Cluzet S., Barrieu F., Hamdi S., Mérillon J.-M. (2008b). Effect of methyl jasmonate in combination with carbohydrates on gene expression of PR proteins, stilbene and anthocyanin accumulation in grapevine cell cultures. Plant Physiol. Biochem. 46: 493–499 [DOI] [PubMed] [Google Scholar]
- Belhadj A., Telef N.G., Cluzet S.P., Bouscaut J.R.M., Corio-Costet M.-F., Mérillon J.-M. (2008a). Ethephon elicits protection against Erysiphe necator in grapevine. J. Agric. Food Chem. 56: 5781–5787 [DOI] [PubMed] [Google Scholar]
- Bogs J., Downey M.O., Harvey J.S., Ashton A.R., Tanner G.J., Robinson S.P. (2005). Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 139: 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J., Jaffé F.W., Takos A.M., Walker A.R., Robinson S.P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borie B., Jeandet P., Parize A., Bessis R., Adrian M. (2004). Resveratrol and stilbene synthase mRNA production in grapevine leaves treated with biotic and abiotic phytoalexin elicitors. Am. J. Enol. Vitic. 55: 60–64 [Google Scholar]
- Boss P.K., Davies C., Robinson S.P. (1996). Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broecker S., Herre S., Wüst B., Zweigenbaum J., Pragst F. (2011a). Development and practical application of a library of CID accurate mass spectra of more than 2,500 toxic compounds for systematic toxicological analysis by LC-QTOF-MS with data-dependent acquisition. Anal. Bioanal. Chem. 400: 101–117 [DOI] [PubMed] [Google Scholar]
- Broecker S., Pragst F., Bakdash A., Herre S., Tsokos M. (2011b). Combined use of liquid chromatography-hybrid quadrupole time-of-flight mass spectrometry (LC-QTOF-MS) and high performance liquid chromatography with photodiode array detector (HPLC-DAD) in systematic toxicological analysis. Forensic Sci. Int. 212: 215–226 [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang X., Wu W., Chen Z., Gu H., Qu L.-J. (2006). Overexpression of the wounding-responsive gene AtMYB15 activates the shikimate pathway in Arabidopsis. J. Integr. Plant Biol. 48: 1084–1095 [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiron H., Drouet A., Claudot A.-C., Eckerskorn C., Trost M., Heller W., Ernst D., Sandermann H., Jr (2000). Molecular cloning and functional expression of a stress-induced multifunctional O-methyltransferase with pinosylvin methyltransferase activity from Scots pine (Pinus sylvestris L.). Plant Mol. Biol. 44: 733–745 [DOI] [PubMed] [Google Scholar]
- Chong J., Poutaraud A., Hugueney P. (2009). Metabolism and roles of stilbenes in plants. Plant Sci. 177: 143–155 [Google Scholar]
- Counet C., Callemien D., Collin S. (2006). Chocolate and cocoa: New sources of trans-resveratrol and trans-piceid. Food Chem. 98: 649–657 [Google Scholar]
- Cutanda-Perez M.-C., Ageorges A., Gomez C., Vialet S., Terrier N., Romieu C., Torregrosa L. (2009). Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 69: 633–648 [DOI] [PubMed] [Google Scholar]
- Czemmel S., Heppel S.C., Bogs J. (2012). R2R3 MYB transcription factors: Key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 249 (suppl. 2): S109–S118 [DOI] [PubMed] [Google Scholar]
- Czemmel S., Stracke R., Weisshaar B., Cordon N., Harris N.N., Walker A.R., Robinson S.P., Bogs J. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151: 1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunois B., Cordelier S., Conreux A., Clément C., Jeandet P. (2009). Molecular engineering of resveratrol in plants. Plant Biotechnol. J. 7: 2–12 [DOI] [PubMed] [Google Scholar]
- Deluc L., Barrieu F., Marchive C., Lauvergeat V., Decendit A., Richard T., Carde J.-P., Mérillon J.-M., Hamdi S. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140: 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L., Bogs J., Walker A.R., Ferrier T., Decendit A., Merillon J.-M., Robinson S.P., Barrieu F. (2008). The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147: 2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Li S., An X., Liu X., Qin H., Wang D. (2009). Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 36: 17–29 [DOI] [PubMed] [Google Scholar]
- D'Onofrio C., Cox A., Davies C., Boss P.K. (2009). Induction of secondary metabolism in grape cell cultures by jasmonates. Funct. Plant Biol. 36: 323–338 [DOI] [PubMed] [Google Scholar]
- Douillet-Breuil A.C., Jeandet P., Adrian M., Bessis R. (1999). Changes in the phytoalexin content of various Vitis spp. in response to ultraviolet C elicitation. J. Agric. Food Chem. 47: 4456–4461 [DOI] [PubMed] [Google Scholar]
- Downey M.O., Harvey J.S., Robinson S.P. (2003a). Analysis of tannins in seeds and skins of Shiraz grapes throughout berry development. Aust. J. Grape Wine Res. 9: 15–27 [Google Scholar]
- Downey M.O., Harvey J.S., Robinson S.P. (2003b). Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.). Aust. J. Grape Wine Res. 9: 110–121 [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Fournier-Level A., Lacombe T., Le Cunff L., Boursiquot J.M., This P. (2010). Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.). Heredity (Edinb) 104: 351–362 [DOI] [PubMed] [Google Scholar]
- Fung R.W.M., Gonzalo M., Fekete C., Kovacs L.G., He Y., Marsh E., McIntyre L.M., Schachtman D.P., Qiu W. (2008). Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol. 146: 236–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gális I., Šimek P., Narisawa T., Sasaki M., Horiguchi T., Fukuda H., Matsuoka K. (2006). A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J. 46: 573–592 [DOI] [PubMed] [Google Scholar]
- Gamborg O.L., Miller R.A., Ojima K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Gatto P., Vrhovsek U., Muth J., Segala C., Romualdi C., Fontana P., Pruefer D., Stefanini M., Moser C., Mattivi F., Velasco R. (2008). Ripening and genotype control stilbene accumulation in healthy grapes. J. Agric. Food Chem. 56: 11773–11785 [DOI] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Grotewold E., Sainz M.B., Tagliani L., Hernandez J.M., Bowen B., Chandler V.L. (2000). Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 97: 13579–13584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain R., Bieseler B., Kindl H., Schröder G., Stöcker R. (1990). Expression of a stilbene synthase gene in Nicotiana tabacum results in synthesis of the phytoalexin resveratrol. Plant Mol. Biol. 15: 325–335 [DOI] [PubMed] [Google Scholar]
- Hall D., De Luca V. (2007). Mesocarp localization of a bi-functional resveratrol/hydroxycinnamic acid glucosyltransferase of Concord grape (Vitis labrusca). Plant J. 49: 579–591 [DOI] [PubMed] [Google Scholar]
- Hichri I., Heppel S.C., Pillet J., Léon C., Czemmel S., Delrot S., Lauvergeat V., Bogs J. (2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 3: 509–523 [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Higo H. (1998). PLACE: A database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 26: 358–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipskind J.D., Paiva N.L. (2000). Constitutive accumulation of a resveratrol-glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol. Plant Microbe Interact. 13: 551–562 [DOI] [PubMed] [Google Scholar]
- Horstmann V., Huether C.M., Jost W., Reski R., Decker E.L. (2004). Quantitative promoter analysis in Physcomitrella patens: A set of plant vectors activating gene expression within three orders of magnitude. BMC Biotechnol. 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35: W485–W487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., et al. French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jeandet P., Breuil A.C., Adrian M., Weston L.A., Debord S., Meunier P., Maume G., Bessis R. (1997). HPLC analysis of grapevine phytoalexins coupling photodiode array detection and fluorometry. Anal. Chem. 69: 5172–5177 [Google Scholar]
- Kalantari H., Das D.K. (2010). Physiological effects of resveratrol. Biofactors 36: 401–406 [DOI] [PubMed] [Google Scholar]
- Kapetanovic I.M., Muzzio M., Huang Z., Thompson T.N., McCormick D.L. (2011). Pharmacokinetics, oral bioavailability, and metabolic profile of resveratrol and its dimethylether analog, pterostilbene, in rats. Cancer Chemother. Pharmacol. 68: 593–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Keen N.T., Ingham J.L. (1976). New stilbene phytoalexins from American cultivars of Arachis hypogaea. Phytochemistry 15: 1794–1795 [Google Scholar]
- Kobayashi S.K., Ishimaru M.I., Hiraoka K.H., Honda C.H. (2002). Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215: 924–933 [DOI] [PubMed] [Google Scholar]
- Kodan A., Kuroda H., Sakai F. (2002). A stilbene synthase from Japanese red pine (Pinus densiflora): Implications for phytoalexin accumulation and down-regulation of flavonoid biosynthesis. Proc. Natl. Acad. Sci. USA 99: 3335–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Langcake P. (1981). Disease resistance of Vitis spp. and the production of the stress metabolites resveratrol, epsilon-viniferin, alpha- viniferin and pterostilbene. Physiol. Plant Pathol. 18: 213–226 [Google Scholar]
- Langcake P., Pryce R.J. (1976). The production of resveratrol by Vitis vinifera and other members of the Vitaceae as a response to infection or injury. Physiol. Plant Pathol. 9: 77–86 [Google Scholar]
- Lanz T., Tropf S., Marner F.J., Schröder J., Schröder G. (1991). The role of cysteines in polyketide synthases. Site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J. Biol. Chem. 266: 9971–9976 [PubMed] [Google Scholar]
- Maeda K., Kimura S., Demura T., Takeda J., Ozeki Y. (2005). DcMYB1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol. Biol. 59: 739–752 [DOI] [PubMed] [Google Scholar]
- Mathews H., Clendennen S.K., Caldwell C.G., Liu X.L., Connors K., Matheis N., Schuster D.K., Menasco D.J., Wagoner W., Lightner J., Wagner D.R. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15: 1689–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus J.T., Aquea F., Arce-Johnson P. (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulski D., Molski M. (2010). Quantitative structure-antioxidant activity relationship of trans-resveratrol oligomers, trans-4,4′-dihydroxystilbene dimer, trans-resveratrol-3-O-glucuronide, glucosides: trans-piceid, cis-piceid, trans-astringin and trans-resveratrol-4′-O-β-D-glucopyranoside. Eur. J. Med. Chem. 45: 2366–2380 [DOI] [PubMed] [Google Scholar]
- Muller P.Y., Janovjak H., Miserez A.R., Dobbie Z. (2002). Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32: 1372–1374, 1376, 1378–1379 [PubMed] [Google Scholar]
- Mol J., Grotewold E., Koes R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3: 212–217 [Google Scholar]
- Murashige T., F. Skoog. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Oleszek W., Sitek M., Stochmal A., Piacente S., Pizza C., Cheeke P. (2001). Resveratrol and other phenolics from the bark of Yucca schidigera roezl. J. Agric. Food Chem. 49: 747–752 [DOI] [PubMed] [Google Scholar]
- Pawlus A.D., Waffo-Teguo P., Merillon J.M. (2012). Stilbenoid chemistry from wine and the genus Vitis, a review. J. Int. Sci. Vigne Vin. 45: 57–111 [Google Scholar]
- Pezet R., Gindro K., Viret O., Spring J.L. (2004). Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol. Mol. Plant Pathol. 65: 297–303 [Google Scholar]
- Pezet R., Pont V., Cuenat P. (1994). Method to determine resveratrol and pterostilbene in grape berries and wines using high-performance liquid chromatography and highly sensitive fluorimetric detection. J. Chromatogr. A 663: 191–197 [Google Scholar]
- Pezet R., Perret C., Jean-Denis J.B., Tabacchi R., Gindro K., Viret O. (2003). δ-Viniferin, a resveratrol dehydrodimer: One of the major stilbenes synthesized by stressed grapevine leaves. J. Agric. Food Chem. 51: 5488–5492 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W., Horgan G.W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisig-Müller R., Schwekendiek A., Brehm I., Reif H.J., Kindl H. (1999). Characterization of a pine multigene family containing elicitor-responsive stilbene synthase genes. Plant Mol. Biol. 39: 221–229 [DOI] [PubMed] [Google Scholar]
- Quattrocchio F., Verweij W., Kroon A., Spelt C., Mol J., Koes R. (2006). PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18: 1274–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiber S., Schröder G., Schröder J. (1995). Molecular and enzymatic characterization of two stilbene synthases from Eastern white pine (Pinus strobus). A single Arg/His difference determines the activity and the pH dependence of the enzymes. FEBS Lett. 361: 299–302 [DOI] [PubMed] [Google Scholar]
- Reid K.E., Olsson N., Schlosser J., Peng F., Lund S.T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemann D., Heller W., Sandermann H. (1991). Biochemical plant responses to ozone: II. Induction of stilbene biosynthesis in scots pine (Pinus sylvestris L.) seedlings. Plant Physiol. 97: 1280–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed, A.I., Bhagabati, N.K., Braisted, J.C., Liang, W., Sharov, V., Howe, E.A., Li, J., Thiagarajan, M., White, J.A., and Quackenbush, J. (2006). TM4 microarray software suite. Method Enzymol. 411: 134–193 [DOI] [PubMed] [Google Scholar]
- Schmidlin L., Poutaraud A., Claudel P., Mestre P., Prado E., Santos-Rosa M., Wiedemann-Merdinoglu S., Karst F., Merdinoglu D., Hugueney P. (2008). A stress-inducible resveratrol O-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol. 148: 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee S., Viret O., Gindro K. (2008). Role of stilbenes in the resistance of grapevine to powdery mildew. Physiol. Mol. Plant Pathol. 72: 128–133 [Google Scholar]
- Schöppner A., Kindl H. (1984). Purification and properties of a stilbene synthase from induced cell suspension cultures of peanut. J. Biol. Chem. 259: 6806–6811 [PubMed] [Google Scholar]
- Schröder G., Brown J.W.S., Schröder J. (1988). Molecular analysis of resveratrol synthase. cDNA, genomic clones and relationship with chalcone synthase. Eur. J. Biochem. 172: 161–169 [DOI] [PubMed] [Google Scholar]
- Selth L.A., Dogra S.C., Rasheed M.S., Healy H., Randles J.W., Rezaian M.A. (2005). A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 17: 311–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotheeswaran S., Pasupathy V. (1993). Distribution of resveratrol oligomers in plants. Phytochemistry 32: 1083–1092 [Google Scholar]
- Sparvoli F., Martin C., Scienza A., Gavazzi G., Tonelli C. (1994). Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Mol. Biol. 24: 743–755 [DOI] [PubMed] [Google Scholar]
- Stark-Lorenzen P., Guitton M.C., Werner R., Mühlbach H.P. (1997). Detection and tissue distribution of potato spindle tuber viroid in infected tomato plants by tissue print hybridization. Arch. Virol. 142: 1289–1296 [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Takeda S., Hirochika H. (2000). MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12: 2511–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier N., Torregrosa L., Ageorges A., Vialet S., Verriès C., Cheynier V., Romieu C. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149: 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomzik J.E., Stenzel K., Stöcker R., Schreier P.H., Hain R., Stahl D.J. (1997). Synthesis of a grapevine phytoalexin in transgenic tomatoes (Lycopersicon esculentum Mill.) conditions resistance against Phytophthora infestans. Physiol. Mol. Plant Pathol. 51: 265–278 [Google Scholar]
- Torregrosa L., Bouquet A. (1997). Agrobacterium rhizogenes and A. tumefaciens co-transformation to obtain grapevine hairy roots producing the coat protein of grapevine chrome mosaic nepovirus. Plant Cell Tissue Organ Cult. 49: 53–62 [Google Scholar]
- Vannozzi A., Dry I.B., Fasoli M., Zenoni S., Lucchin M. (2012). Genome-wide analysis of the grapevine stilbene synthase multigenic family: Genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 12: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versari A., Parpinello G.P., Tornielli G.B., Ferrarini R., Giulivo C. (2001). Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. J. Agric. Food Chem. 49: 5531–5536 [DOI] [PubMed] [Google Scholar]
- Vezzulli S., Civardi S., Ferrari F., Bavaresco L. (2007). Methyl jasmonate treatment as a trigger of resveratrol synthesis in cultivated grapevine. Am. J. Enol. Vitic. 58: 530–533 [Google Scholar]
- Walker A.R., Lee E., Bogs J., McDavid D.A.J., Thomas M.R., Robinson S.P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49: 772–785 [DOI] [PubMed] [Google Scholar]
- Wang W., Tang K., Yang H.-R., Wen P.-F., Zhang P., Wang H.-L., Huang W.-D. (2010). Distribution of resveratrol and stilbene synthase in young grape plants (Vitis vinifera L. cv. Cabernet Sauvignon) and the effect of UV-C on its accumulation. Plant Physiol. Biochem. 48: 142–152 [DOI] [PubMed] [Google Scholar]
- Wiese W., Vornam B., Krause E., Kindl H. (1994). Structural organization and differential expression of three stilbene synthase genes located on a 13 kb grapevine DNA fragment. Plant Mol. Biol. 26: 667–677 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 127: 1399–1404 [PMC free article] [PubMed] [Google Scholar]
- Yu C.K.Y., Springob K., Schmidt J., Nicholson R.L., Chu I.K., Yip W.K., Lo C. (2005). A stilbene synthase gene (SbSTS1) is involved in host and nonhost defense responses in sorghum. Plant Physiol. 138: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A., Gatto P., Cestaro A., Pilati S., Viola R., Mattivi F., Moser C., Velasco R. (2009). Grapevine cell early activation of specific responses to DIMEB, a resveratrol elicitor. BMC Genomics 10: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.J., Tang C.S., Moore P. (2005). Increased disease resistance in papaya by transforming a pathogen-inducible stilbene synthase gene. Acta Hortic. 692: 107–114 [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.