This work provides evidence that centromere pairing occurs prior to telomere bouquet formation in early meiotic prophase in maize and is dependent upon centromere activity. Centromere association, in contrast with that in yeast and Drosophila, precedes ZYP1 installation. SMC6 is required for centromere pairing.
Abstract
Pairing of homologous chromosomes in meiosis is critical for their segregation to daughter cells. In most eukaryotes, clustering of telomeres precedes and facilitates chromosome pairing. In several species, centromeres also form pairwise associations, known as coupling, before the onset of pairing. We found that, in maize (Zea mays), centromere association begins at the leptotene stage and occurs earlier than the formation of the telomere bouquet. We established that centromere pairing requires centromere activity and the sole presence of centromeric repeats is not sufficient for pairing. In several species, homologs of the ZIP1 protein, which forms the central element of the synaptonemal complex in budding yeast (Saccharomyces cerevisiae), play essential roles in centromere coupling. However, we found that the maize ZIP1 homolog ZYP1 installs in the centromeric regions of chromosomes after centromeres form associations. Instead, we found that maize STRUCTURAL MAINTENANCE OF CHROMOSOMES6 homolog forms a central element of the synaptonemal complex, which is required for centromere associations. These data shed light on the poorly understood mechanism of centromere interactions and suggest that this mechanism may vary somewhat in different species.
INTRODUCTION
In meiosis, diploid cells undergo two rounds of chromosome segregation to yield daughter cells with haploid chromosome numbers. Accurate segregation of chromosomes during meiosis depends on the stable juxtaposition of homologs, known as the formation of bivalents. In most eukaryotic species, establishment of stable bivalents requires homologous chromosome pairing, synapsis, and recombination. To pair, homologous chromosomes must find and recognize each other and align along their lengths. Mechanisms governing chromosome pairing are poorly understood. However, studies in several species show that pairing along the entire length of chromosomes is preceded by interaction of specific chromosome regions, such as telomeres, centromeres, and specialized pairing centers (Scherthan, 2007; Stewart and Dawson, 2008; Ronceret and Pawlowski, 2010; Tsai and McKee, 2011).
In Arabidopsis thaliana, telomeres undergo pairing associated with the nucleolus before meiosis (Armstrong et al., 2001), and in most organisms, telomeres attach to the nuclear envelope in early meiotic prophase I and cluster to form a cytological structure called the telomere bouquet (Bass et al., 1997; Harper et al., 2004). It has been proposed that telomere bouquet formation facilitates the subsequent processes of homologous pairing by bringing chromosome ends together (Scherthan et al., 1994; Niwa et al., 2000). In maize (Zea mays), the plural abnormalities of meiosis1 mutant, which is deficient in telomere clustering, exhibits homologous pairing defects (Golubovskaya et al., 2002). However, chromosome pairing is only reduced in this mutant and not completely abolished, suggesting that the telomere bouquet facilitates, but is not absolutely required for, pairing. Furthermore, computational modeling suggests that telomere interactions alone may not be sufficient for pairing of large chromosomes, and simultaneous interactions at additional chromosome sites may be required to accomplish this (Penfold et al., 2012).
Pairing centers are specialized chromosome sites that facilitate pairing of homologous chromosomes during meiosis in Caenorhabditis elegans and in males of Drosophila melanogaster (McKim, 2005; Phillips and Dernburg, 2006). Pairing centers serve as binding sites for specialized proteins that bring chromosomes together to enable pairing interactions.
Interactions of centromeres that precede pairing along chromosome arms were first described in polyploid wheat (Triticum aestivum; Martínez-Pérez et al., 1999; Martinez-Perez et al., 2001). In wheat, centromeres associate premeiotically in a process dependent on the pairing homoeologous (Ph1) locus. Subsequently, coupling of centromeres has been described in a number of species, including budding yeast (Saccharomyces cerevisiae), Arabidopsis, barley (Hordeum vulgare), Brachypodium distachyon, and Drosophila (Kemp et al., 2004; Tsubouchi and Roeder, 2005; Tsubouchi et al., 2008; Ronceret et al., 2009; Takeo et al., 2011; Da Ines et al., 2012; Phillips et al., 2012; Wen et al., 2012). In these species, however, chromosomes enter meiosis with separate centromeres and centromere coupling takes place in early meiotic prophase I. In yeast, the initial interactions are mainly between centromeres of nonhomologous chromosomes and are dependent on installation of ZIP1, the central element of the synaptonemal complex (SC) (Tsubouchi and Roeder, 2005; Tsubouchi et al., 2008). SC dependence of centromere coupling was also observed in mouse and Drosophila (Takeo et al., 2011; Bisig et al., 2012). Centromere coupling is important for proper progression of chromosome interactions and, eventually, for correct segregation of chromosomes to daughter cells. However, the mechanisms directing centromere interactions remain poorly understood, and, in particular, it is not known how centromeric chromosome regions recognize each other to couple.
To further understand the dynamics of centromere interactions in early meiosis, we studied this process in maize. We found that maize centromeres begin associating at leptotene, prior to telomere bouquet formation. We discovered that centromere pairing requires active centromeres, suggesting that centromere interactions are not driven by the centromere-specific DNA sequences. Also, in maize, unlike in yeast, and Drosophila, a ZIP1 homolog does not exhibit obvious localization to centromeric regions during early prophase centromere interactions. By contrast, we found that centromere pairing requires STRUCTURAL MAINTENANCE OF CHROMOSOMES6 (SMC6), a novel component of the SC in maize.
RESULTS
Centromere Interactions Precede Telomere Bouquet Formation in Early Prophase I of Meiosis in Maize
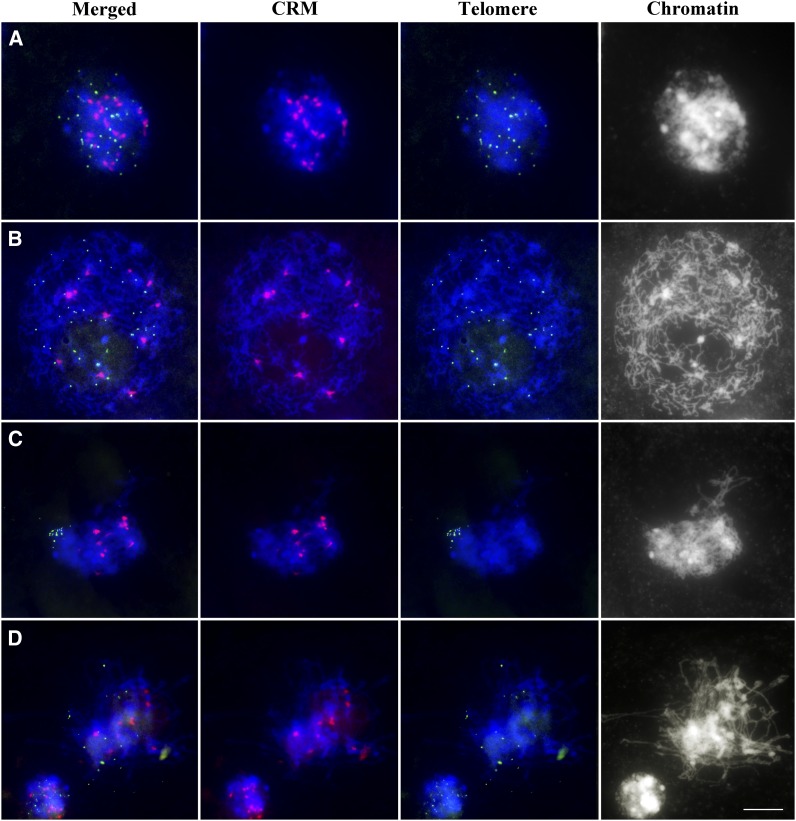
To study centromere dynamics in early meiotic prophase, we performed fluorescence in situ hybridization (FISH) experiments using centromeric retrotransposon of maize (CRM) and telomere sequences as probes. In premeiotic interphase cells, we found an average of 18.1 ± 1.5 (n = 45) centromere signals per cell (Figure 1A). At the leptotene stage, this dropped to 11.1 ± 1.3 (n = 107) (Figure 1B) and remained at a similar level during zygotene (9.5 ± 0.9; n = 124) (Figure 1C) and pachytene (9.5 ± 0.8; n = 101) (Figure 1D). As maize has 10 chromosome pairs, these results indicate that centromeres associate in maize, beginning at leptotene, and these associations persist until pachytene. The centromere associations precede telomere clustering to form the bouquet, which takes place during zygotene.
Figure 1.
Centromere and Telomere Behavior during Early Prophase I in Maize.
Premeiotic interphase (A), leptotene (B), zygotene (C), and early pachytene (D). Centromeres (CRM) are red, and telomeres are green. Blue indicates chromosomes counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Bar = 10 μm.
To confirm our observations, we conducted a three-dimensional immunostaining-FISH analysis using the telomere FISH probe and anti-CENH3 antibody, a marker for functional centromeres. Results of this experiment corroborated our CRM/telomere FISH results. We observed that centromeres began to associate in leptotene, but telomeres remained dispersed throughout the nucleus (see Supplemental Figure 1A online). During the leptotene–zygotene transition, centromeres were paired while telomeres attached to the nuclear envelope and began to cluster (see Supplemental Figure 1B online). Centromere association continued throughout zygotene, during which the telomere bouquet was present (see Supplemental Figure 1C online). Together, the FISH and immuno-FISH experiments provide strong evidence that centromeres in maize associate in early prophase I and prior to telomere bouquet formation.
Distal Regions of Chromosome Arms Are Not Paired When Centromeres Associate
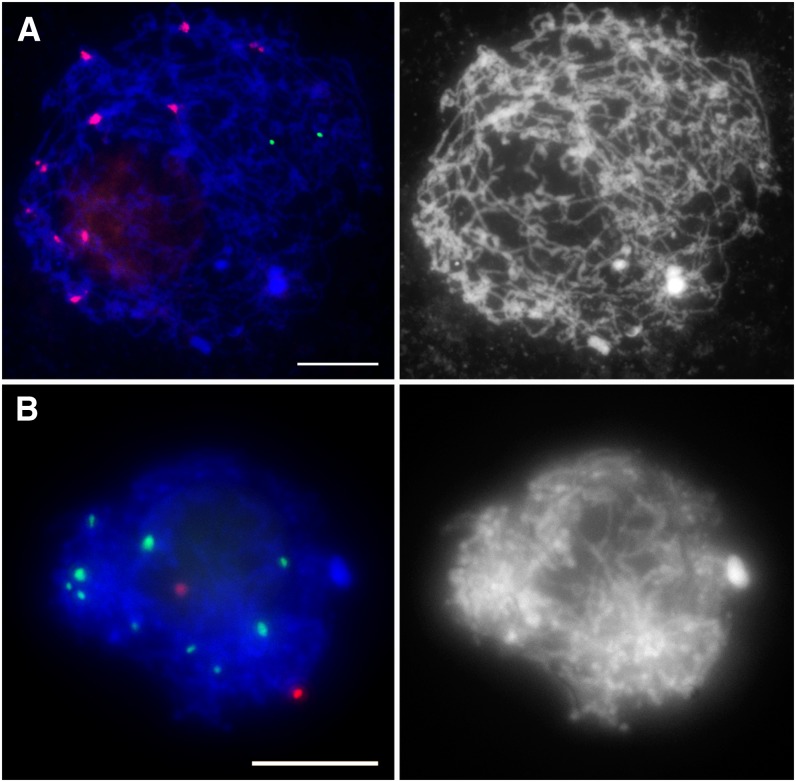
To elucidate the dynamics of centromere pairing, we examined pairing of chromosome arms at the time when centromeres begin to associate. To perform this analysis, we used the 5S rRNA gene sequence as probe, which recognizes a distal region on the long arm of chromosome 2. In leptotene, we always observed two separate 5S rRNA signals (n = 50) (Figures 2A and 2B), indicating that the 5S loci were unpaired. We detected only a single 5S signal per cell at later stages of prophase I when homologous chromosomes are expected to be paired (see Supplemental Figure 2 online). Similar results were obtained using the 45S rRNA gene probe, which recognizes the nucleolus organizer region on the short arm of chromosome 6. All leptotene meiocytes showed two unpaired 45S signals (n = 32) (see Supplemental Figure 3 online). These observations indicate that centromere association occurs prior to pairing of chromosome arms.
Figure 2.
Chromosome Arms Are Not Paired at the Leptotene Stage.
(A) FISH with CRM (red) and the 5S rRNA gene (green) probes.
(B) Immuno-FISH with the anti-CENH3 antibody (green) and the 5S rRNA gene FISH probe (red).
Blue indicates chromosomes counterstained with DAPI. Bars = 10 μm.
Centromere Associations Are Mostly Homologous
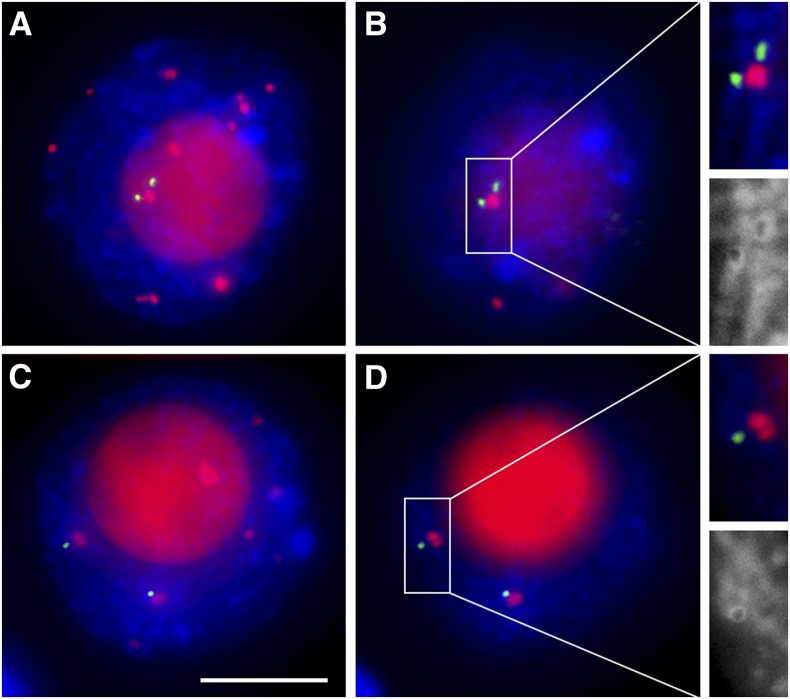
An important question regarding centromere interactions in early prophase I is whether they involve centromeres of homologous or nonhomologous chromosomes. To address this question, we used the centromere 4 (Cent4) FISH probe, which specifically detects a repeat present near the centromere of maize chromosome 4 (see Supplemental Figure 4 online) (Kato et al., 2004; Lamb et al., 2007). We scored meiocytes containing paired Cent4 signals (Figures 3A and 3B) as exhibiting homologous centromere associations and meiocytes with two separate Cent4 signals (Figures 3C and 3D) as exhibiting nonhomologous associations. To avoid confusion between chromosome 4 centromeres that have associated with nonhomologous centromeres and those that have not associated with any centromeres, we only included in this analysis cells showing 10 or fewer centromere signals, which indicated that all centromeres became associated. Approximately 65% of meiocytes (n = 31) showed homologous association of centromere 4 and 35% showed nonhomologous associations. Thus, centromere associations in meiocytes exhibiting complete centromere pairing at leptotene are mostly homologous. At pachytene, all meiocytes (n = 55) exhibited homologous centromere association (see Supplemental Figure 4 online).
Figure 3.
Homologous and Nonhomologous Centromere Associations at Leptotene.
(A) and (B) Homologous centromere associations.
(C) and (D) Nonhomologous centromere associations.
(A) and (C) Flat projections of three-dimensional images of whole nuclei.
(B) and (D) Single optical section from nuclei in (A) and (C), respectively.
Close-ups are shown in insets. CENH3 is red, and Cent4 is green. Blue indicates chromosomes counterstained with DAPI. Bar = 10 μm.
To further explore whether centromere pairing involves chromosome homology, we used haploid maize plants. In haploid meiocytes, we found that the average number of centromere signals per cell was 8.3 ± 0.9 (n = 33) at leptotene and 8.4 ± 1.0 (n = 51) at pachytene (see Supplemental Figure 5 online). This result confirms our observations from diploid maize that the majority of centromere associations are homologous.
Centromere Pairing Requires Functional Centromeres
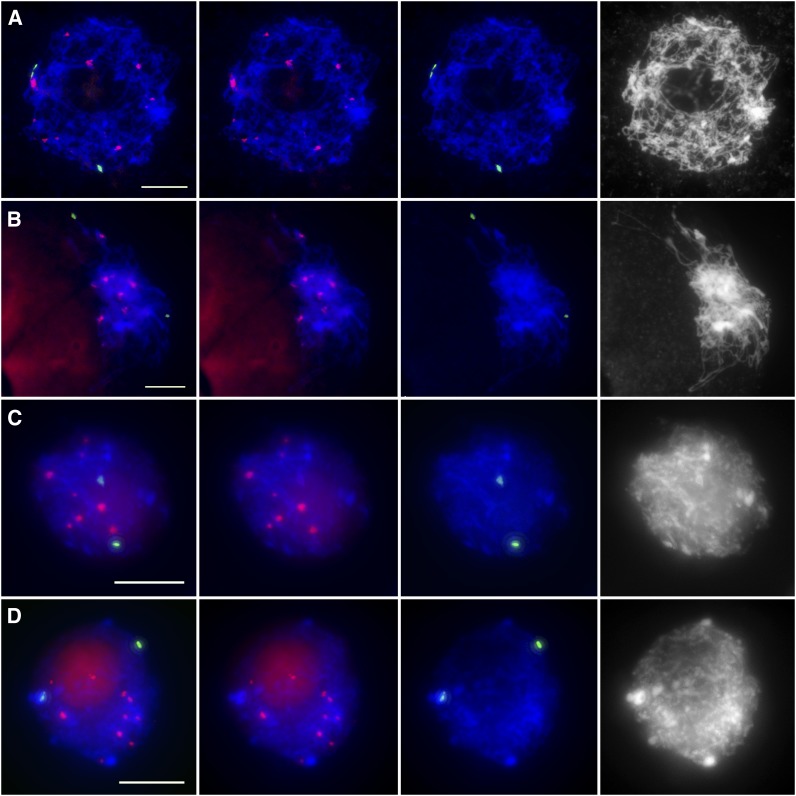
To elucidate the mechanism of centromere pairing, we investigated whether centromere sequences mediate centromere associations. To address this question, we examined whether inactive centromeres could associate in a manner similar to functional centromeres by studying two maize lines containing stable dicentric chromosomes, 7Bic-1 and T1-5. 7Bic-1 carries a segment of a B chromosome containing an inactive centromere fused to maize chromosome 7. Thus, 7Bic-1 contains two centromeres, an active A centromere and an inactive B centromere (Han et al., 2007). We used a B centromere–specific sequence to identify the inactive B centromere. Our FISH and immuno-FISH results indicated that inactive B centromeres never associated with each other at leptotene (n = 45) (Figures 4A and 4C). At zygotene, 68% of meiocytes showed two unpaired B-repeat signals (n = 77) (Figures 4B and 4D), and 32% of meiocytes showed paired signals. Thus, inactive B centromeres do not associate at leptotene.
Figure 4.
Inactive Centromeres Do Not Associate at the Leptotene Stage in the 7Bic-1 Dicentric Line.
(A) and (C) Leptotene.
(B) and (D) Zygotene.
(A) and (B) FISH with the CRM (red) and centromere B repeat (green) probes.
(C) and (D) Immuno-FISH with the anti-CENH3 antibody (red) and the centromere B repeat FISH probe (green).
Blue indicates chromosomes counterstained with DAPI. Bars = 10 μm.
To examine the possibility that inactive B centromeres fail to associate not because they are inactive but because B centromeres are different from A centromeres sequence-wise, we used a reciprocal translocation between chromosomes 1 and 5 (T1-5), which possesses an inactive A centromere at or near the breakpoint of the two chromosomes (Gao et al., 2011). Immuno-FISH using the anti-CENH3 antibody and the CRM FISH probe showed that inactive A centromeres also did not pair at the leptotene stage (n = 28) (Figure 5). Together, our data indicate that centromere associations do not rely on the centromere DNA sequence, but rather on centromere activity.
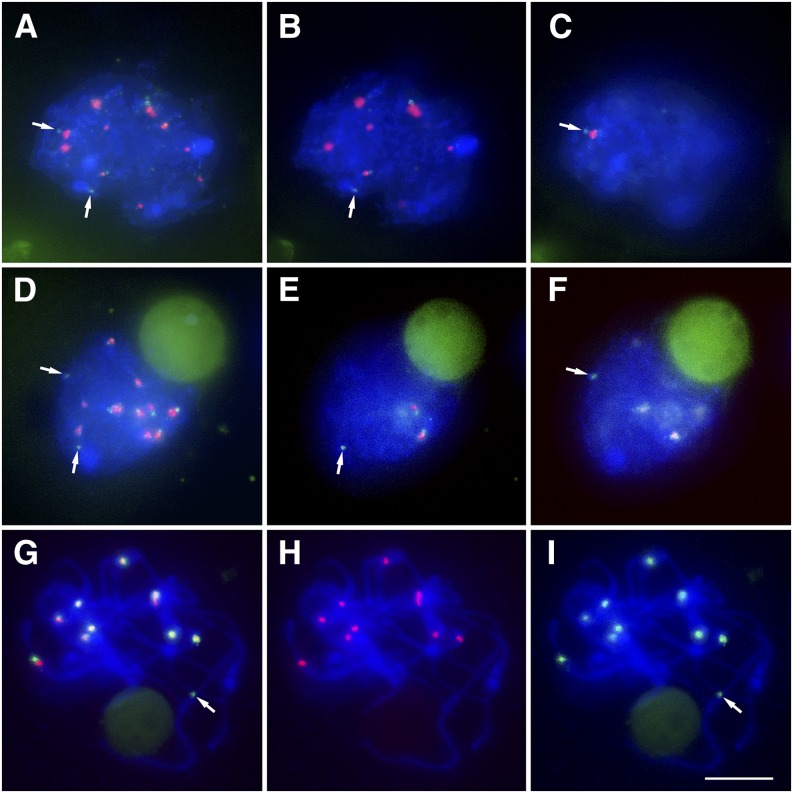
Figure 5.
Inactive Centromeres Do Not Associate at the Leptotene Stage in the T1-5 Dicentric Line.
(A) to (C) Leptotene.
(B) and (C) Images are single optical sections of the whole nucleus shown in (A).
(D) to (F) Zygotene.
(E) and (F) Images are single optical sections of the whole nucleus shown in (D).
(G) to (I) Pachytene.
CENH3 is red, and CRM is green. Blue indicates chromosomes counterstained with DAPI. Arrows indicate inactive A centromeres. Bar = 10 μm.
Sister-Chromatid Cohesion Is Required for Centromere Associations
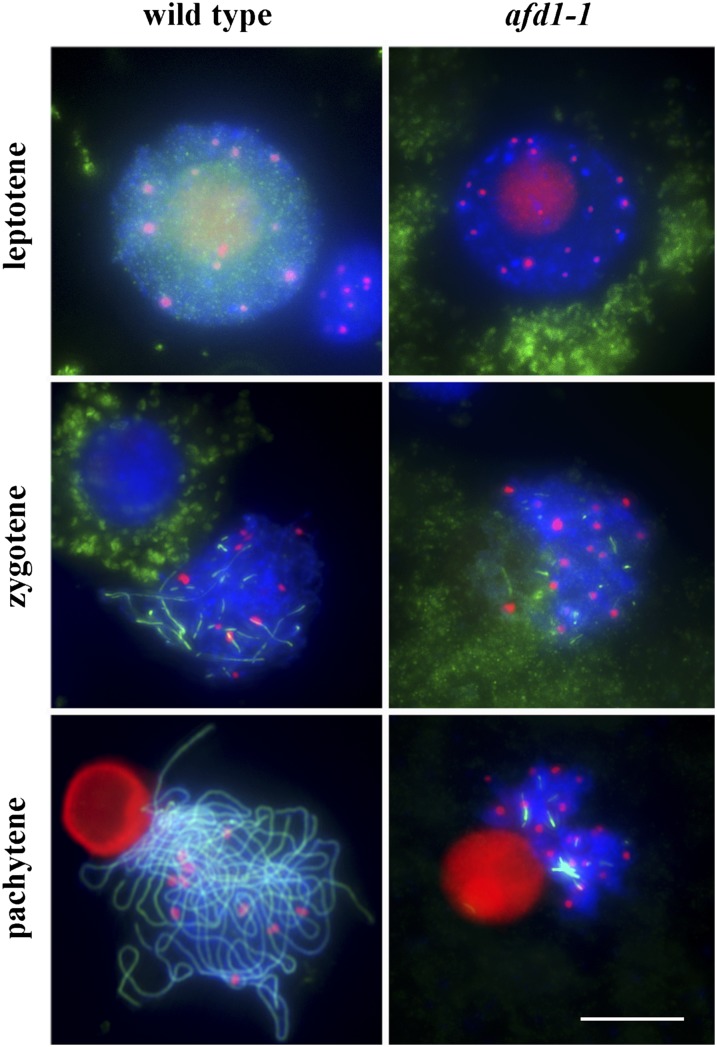
To investigate whether centromere associations require installation of the sister-chromatid cohesion complex and proper meiotic chromosome structure, we examined centromere pairing in the maize absence of first division1 (afd1) mutant, which carries a deletion of the afd1 gene encoding a homolog of the meiosis-specific α-kleisin Rec8. This mutant exhibits defective chromosome structure and lacks sister-chromatid cohesion (Golubovskaya et al., 2006). In the afd1 mutant, we observed 17.5 ± 1.3 centromere signals at leptotene (n = 39), 18.6 ± 1.3 signals at zygotene (n = 36), and 19.0 ± 1.0 signals at pachytene (n = 39) (Figure 6). These data indicate that centromeres do not associate at early prophase I in the absence of AFD1 and consequently suggest that sister-chromatid cohesion and proper meiotic chromosome structure conveyed by AFD1 are required for centromere pairing.
Figure 6.
Centromeres Do Not Pair in afd1-1 Mutant Meiocytes.
CENH3 is red, and ZYP1 is green. Blue indicates chromosomes counterstained with DAPI. Bar = 10 μm.
Centromere Pairing in Maize Largely Precedes ZYP1 Installation in Centromeric Regions
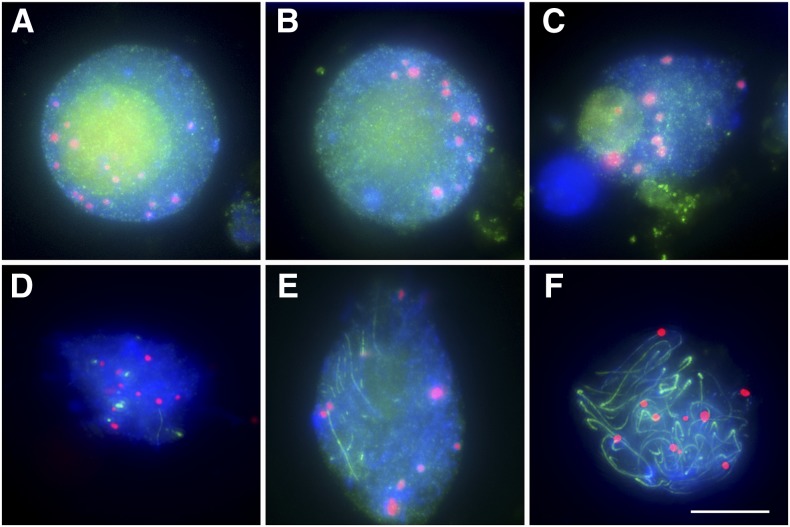
Studies in budding yeast and Drosophila indicate that early prophase centromere coupling requires ZIP1, a protein forming the central element of the SC (Tsubouchi and Roeder, 2005; Tsubouchi et al., 2008; Takeo et al., 2011). To elucidate whether this is also the case in maize, we used an antibody against ZYP1 (Golubovskaya et al., 2011), the maize homolog of budding yeast ZIP1. ZYP1 is first detected as diffuse staining in the nucleus in early leptotene (Figures 7A and 7B). In early zygotene, it forms foci and then stretches along chromosomes (Figures 7C to 7E). However, very few stretches of ZYP1 could be seen in the centromeric regions (Figures 7D and 7E). Only at pachytene, when ZYP1 became installed along entire chromosomes, was the protein present in centromeric regions of all chromosomes (Figure 7F). Consequently, in maize, in contrast with yeast, and Drosophila, ZYP1/ZIP1 does not exhibit a strong presence at centromeres at the time when they begin associating. These data suggest that ZYP1 may not participate in establishing early prophase centromere associations in maize.
Figure 7.
ZYP1 Is Not Detected as Strong Signals at Centromere Regions of Maize Chromosomes When Centromeres First Associate.
(A) Early leptotene.
(B) Mid-leptotene.
(C) Early zygotene.
(D) and (E) Mid-zygotene.
(F) Pachytene.
CENH3 is red, and ZYP1 is green. Blue indicates chromosomes counterstained with DAPI. Bar = 10 μm.
SMC6 Is a Component of the Maize SC Required for Centromere Pairing
As we concluded that the causal link between the installation of the central element of the SC and early prophase centromere interactions is not strong in maize, we embarked on a search for novel SC components affecting centromere pairing. We used BLAST searches with the SMC domain of ZIP1 and found that it is highly similar to the SMC domain of SMC6, a subunit of the SMC5/6 complex (see Supplemental Figure 6 and Supplemental Data Set 1 online). Using the sequence of the annotated rice (Oryza sativa) SMC6 gene, we conducted a BLAST search to find a maize SMC6 homolog. We identified a single open reading frame, encoding a hypothetical protein LOC100383441, which we called maize SMC6. Maize SMC6 is 1040 amino acids long and contains ATP binding domains at both the N and C termini as well as two extended coiled-coil domains separated by a hinge located in the middle of the protein. RT-PCR results showed that SMC6 is expressed mainly in leaves, shoot, and tassel (see Supplemental Figure 7A online), and its expression is not meiosis specific. We expressed in Escherichia coli the N-terminal region (amino acids 89 to 532) of maize SMC6, which is the domain thought to be responsible for homodimer formation and used it to develop a polyclonal antibody in mouse (see Supplemental Figure 7B online). Immunolocalization experiments on maize meiocytes showed that SMC6 appeared first as small dots in early zygotene (Figure 8A) and then formed linear signals later in zygotene (Figure 8B). In pachytene, SMC6 formed continuous dotted lines between homologous chromosomes (Figure 8C) and colocalized with ZYP1 (Figures 8D to 8F). These results suggest that SMC6 may be a component of the central element of the maize SC.
Figure 8.
Distribution of SMC6 in Early Prophase I.
(A) to (C) Dual immunolocalization of CENH3 and SMC6. CENH3 is red, and SMC6 is green.
(A) and (B) Zygotene.
(C) Pachytene.
(D) to (F) Colocalization of SMC6 (red) and ZYP1 (green) at the pachytene stage.
(E) A single optical section from the nucleus in (D) showing only SMC6 staining.
(F) The same optical section showing only ZYP1 staining.
Blue indicates chromosomes counterstained with DAPI. Bar = 10 μm.
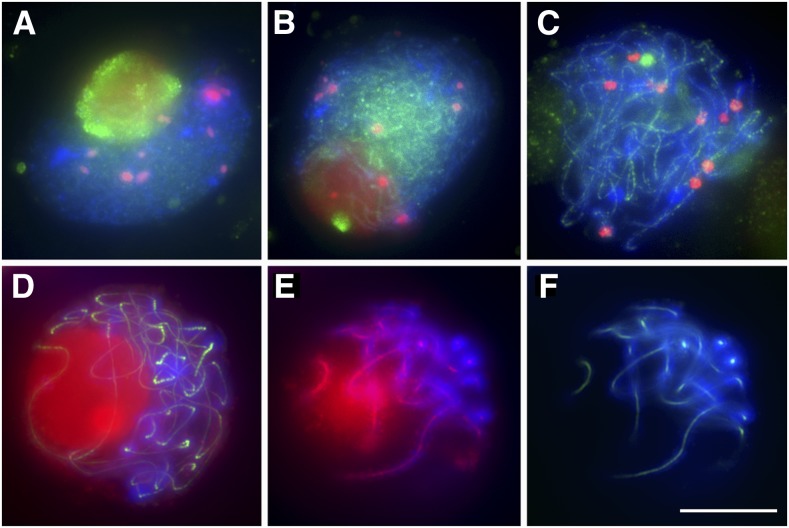
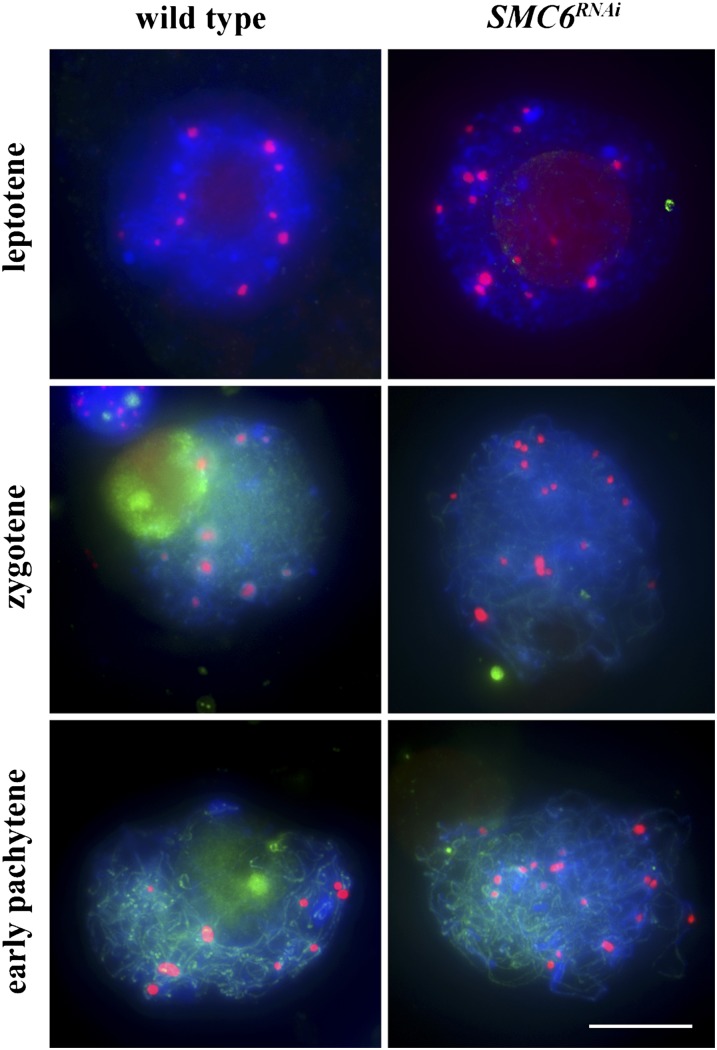
To study the role of SMC6, we generated RNA interference (RNAi) knockdown lines. To do this, we used a 411-bp cDNA fragment of maize SMC6 cloned into the pUCCRNAi vector. Among 10 SMC6RNAi transgenic lines, five exhibited severe sterility and a dwarf phenotype. Using RT-PCR, we determined that the RNAi construct caused a substantial decrease of SMC6 expression (see Supplemental Figure 8 online). Cytological examination of SMC6RNAi meiocytes revealed that, in contrast with the wild type, centromeres were not associated from leptotene to pachytene stage (Figure 9). These data suggest that SMC6 is required for early prophase centromere pairing in maize.
Figure 9.
Behavior of Centromeres in Wild-Type and SMC6RNAi Meiocytes from Leptotene to Pachytene Stage.
CENH3 is red, and SMC6 is green. Blue indicates chromosomes counterstained with DAPI. Bar = 10 μm.
DISCUSSION
Our investigation revealed that in maize, similar to several other eukaryotes, centromeres form persistent interactions during early meiotic prophase I (Martínez-Pérez et al., 1999; Tsubouchi and Roeder, 2005; Ronceret et al., 2009; Takeo et al., 2011; Da Ines et al., 2012; Phillips et al., 2012; Wen et al., 2012). Like in other species, maize centromere pairing precedes clustering of telomeres to form the bouquet as well as homologous pairing of chromosome arms. However, in contrast with previous reports from yeast and Drosophila, we found that centromere interactions in maize are present before ZYP1 installation. Using dicentric chromosomes, in which one of the centromeres is inactive, we discovered that only functional centromeres have the ability to pair.
Molecular Mechanism of Centromere Interactions
Even though centromere coupling has been reported in a number of diverse species of eukaryotes, the molecular mechanism of this phenomenon remains poorly understood. It has been shown in wheat, and postulated in Arabidopsis, that meiotic centromere interactions are controlled by heterochromatin (Prieto et al., 2004, 2005; Colas et al., 2008; Da Ines et al., 2012). Heterochromatin is ubiquitous in centromeres of most eukaryotes, although interestingly, not those of budding yeast (Sullivan et al., 2001). Results from our study demonstrate that centromere pairing requires functional centromeres, and the presence of centromeric DNA alone is not sufficient for the chromosome region to associate prior to the general chromosome arm pairing. However, our data do not exclude the possibility that centromeric heterochromatin is the basis of the meiotic centromere interactions. There are several features distinguishing heterochromatin of active centromeres from other heterochromatic chromosome regions, which could limit the early prophase interactions to active centromeres. These features include transcription of DNA repeats (Topp et al., 2004), the presence of the centromere-specific histone variant CENH3 (Allshire and Karpen, 2008), as well as phosphorylation of histones H2A and CENH3 (Peterson and Laniel, 2004). It is likely that one or several of these features directly facilitate centromere interactions. It might be tempting to speculate that, for example, transcripts of centromeric repeats, which become bound with the centromeric regions (Topp et al., 2004), provide guidance for centromere-centromere recognition. However, the features of centromeric heterochromatin are interdependent, connections between them are not well understood, and determining the exact nature of the centromere recognition and association process will require further studies.
Centromere Pairing and the SC
Centromere coupling in budding yeast and Drosophila is mediated by ZIP1, a central element of the SC (Tsubouchi and Roeder, 2005; Tsubouchi et al., 2008; Takeo et al., 2011). However, we found that in maize, ZYP1, is not present at all centromeric regions at the time when centromeres associate. Although we cannot state categorically that the amount of ZYP1 needed to mediate centromere interactions is below the detection limit of our immunolocalization experiments, this observation may suggest that the mechanism involved of centromere pairing in maize is somewhat different from these in budding yeast and Drosophila. In these two species, the strong presence of ZIP1 homologs is observed at centromeric regions in early prophase I. We discovered that maize SMC6 is a component of the SC. Interestingly, SMC6 is also required for centromere pairing in maize. Proteins of the SMC family are essential for genomic integrity through their functions in sister-chromatid cohesion, chromosome condensation, and DNA repair (Losada and Hirano, 2005; Nasmyth and Haering, 2005). There are three main SMC protein types: SMC1/3, which are involved in sister-chromatid cohesion (Nasmyth and Haering, 2009); SMC2/4, which play a role in chromosome condensation (Hirano, 2005); and SMC5/6. The exact function of the SMC5/6 complex is unknown, but it has been suggested that in yeast and Arabidopsis they are involved in DNA repair and replication, cell cycle checkpoint control, and telomere length maintenance (Lindroos et al., 2006; Watanabe et al., 2009; Farmer et al., 2011; Roy et al., 2011). In this study, we demonstrate that SMC6 takes part in early prophase of meiosis I and colocalizes with ZYP1. These data suggest that SMC6 is a component of the central element of the SC. Interestingly, when SMC6 expression was reduced using RNAi, centromeres did not pair from leptotene to pachytene stage. Multiple central element proteins have been identified recently in C. elegans and mouse and have been shown to have somewhat different functions (Bolcun-Filas et al., 2007; Schild-Prüfert et al., 2011). This may also be the case in maize.
Role of Centromere Associations in Progression of Meiosis
A universal role of early prophase centromere coupling has not emerged from studies of this process conducted in different organisms. However, the fact that this phenomenon has been observed in species that are distantly related and have genomes with very different structures and sizes suggests that centromere association may play a fundamental role in meiotic chromosome dynamics. It has been suggested that the main role of the telomere bouquet is in facilitating rapid chromosome movements that take place during early prophase I and are critical for homologous chromosome recognitions (Koszul and Kleckner, 2009; Penkner et al., 2009; Sato et al., 2009; Sheehan and Pawlowski, 2009). It is possible that centromere association plays a role in structuring chromosome arm interactions during zygotene that is somewhat similar to that of the bouquet. Associations formed by centromeres and telomeres prior to the onset of the vigorous chromosome motility in zygotene may act to provide physical structure to chromosome movements that facilitate pairing interactions along chromosome arms.
METHODS
Plant Materials
Maize (Zea mays) inbred lines KYS and B73 were used as the wild type in cytological analysis. Genotyping of afd1-1 mutants was conducted with the following primers: forward 5′- GGTGGCTCTGGATCGGCGTTTATAC-3′ and reverse 5′- GCCCCCTTTTGGACCTGTGTTCA -3′. After MboII (NEW ENGLAND BioLabs) digestion of the resulting PCR product, a 360-bp band was produced in the wild type, whereas a 280-bp band was present in homozygous mutants.
DNA Probe Preparation and Labeling
The following DNA probes were used: the centromere-specific retrotransposon of maize (CRM), the telomere repeat sequence, the B centromere–specific repeat sequence (Alfenito and Birchler, 1993), the 5S rRNA gene repeat, the 45S rRNA gene repeat, and the Cent4 satellite repeat. All probes were labeled with ChromaTide Alexa Fluor-5-dUTP using the nick translation method (Kato et al., 2004).
Immunolocalization and FISH
Immunolocalization and FISH were performed as described (Han et al., 2009). Images were acquired using an Olympus BX 61 microscope. Flat projection of multiple optical sections were created using MetaMorph and processed with Adobe Photoshop CS 3.0.
Cloning the Maize SMC6 cDNA
RNA from young tassels of the B73 inbred line was isolated using TRIzol (Invitrogen). Primers 5′-AAATCGCCACTCAACCTTTC-3′ and 5′-GTCTCAACACTGTCAACCGC-3′ were used to amplify the predicted coding region of the SMC6 transcript. The amplified fragment was cloned and sequenced.
SMC6 RNAi Lines
To produce the RNAi construct, we used a 411-bp cDNA fragment of the Zm-SMC6 gene, which was amplified with the following primers: 5′-CCGCTCGAGGGCCAGCGCTTAGAGAACTTGAT-3′ (adding an XhoI site); and 5′-GGAAGATCTCCACACGGGCATTCATTTTATTCA-3′ (adding a BglII site). This fragment was cloned into the pUCCRNAi vector. The RNAi construct was transformed followed the method described by Vain et al. (1993).
Antibody Production and Protein Gel Blot Analysis
To generate the anti-ZmSMC6 antibody, a partial cDNA corresponding to amino acids 89 to 532 of the SMC6 protein was cloned into the pMAL plasmid. The recombinant protein was expressed in Escherichia coli strain DE 3, and the resulting protein was used to produce a polyclonal antibody in mouse. For protein gel blot analysis, 30-mg protein samples were separated in 10% SDS-PAGE gels and transferred onto a polyvinylidene fluoride membrane (Millipore). The anti-ZMSMC6 protein was used at dilution 1:2000. An anti-mouse-IgG antibody produced in donkey and conjugated to horseradish peroxidase 1:20,000 was used to detect the recombinant protein. Protein bands were visualized using the enhanced chemiluminescence substrate.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number KC814181 (maize SMC6 coding sequence).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Dynamics of Centromeres and Telomeres during Prophase I in Maize Monitored Using an Antibody against Centromeric Histone CENH3 and a Telomere-Specific FISH Probe.
Supplemental Figure 2. Pairing of Centromeres and 5S rRNA Loci at the Pachytene Stage.
Supplemental Figure 3. Behavior of Centromeres and the 45S rRNA Loci at the Leptotene Stage.
Supplemental Figure 4. Pairing of Centromeres at the Pachytene Stage.
Supplemental Figure 5. Centromere Behavior at the Pachytene Stage in Haploid Maize.
Supplemental Figure 6. Bayesian Phylogeny Reconstruction of the SMC Domain from SMC Family Proteins and ZIP1 Homologs.
Supplemental Figure 7. The Maize SMC6 Gene.
Supplemental Figure 8. Transcription Analysis of SMC6 in Wild-Type and SMC6RNAi Lines.
Supplemental Data Set 1. Text File of Alignment Corresponding to Supplemental Figure 6.
Acknowledgments
We thank James Birchler (University of Missouri, Columbia) for comments and critical reading of the article as well as Ryan Douglas (University of Missouri, Columbia) and Nathan D. Han (Washington University, St. Louis) for critical reading of the article and valuable suggestions. We thank Zac Cande and Rachel Wang (University of California, Berkeley) for the ZYP1 antibody and the Maize Genetics Cooperation Stock Center for the afd1-1 mutant. This work was supported by the National Basic Research Program of China (973 Program 2011CB944601).
AUTHOR CONTRIBUTIONS
F.H. and J.Z. designed the research, J.Z. and F.H. performed the research, J.Z., W.P.P., and F.H. analyzed the data. J.Z., W.P.P., and F.H. wrote the article.
Glossary
- SC
synaptonemal complex
- FISH
fluorescence in situ hybridization
- CRM
centromeric retrotransposon of maize
- RNAi
RNA interference
- DAPI
4′,6-diamidino-2-phenylindole
References
- Alfenito M.R., Birchler J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R.C., Karpen G.H. (2008). Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat. Rev. Genet. 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong S.J., Franklin F.C., Jones G.H. (2001). Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 114: 4207–4217 [DOI] [PubMed] [Google Scholar]
- Bass H.W., Marshall W.F., Sedat J.W., Agard D.A., Cande W.Z. (1997). Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol. 137: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisig C.G., Guiraldelli M.F., Kouznetsova A., Scherthan H., Höög C., Dawson D.S., Pezza R.J. (2012). Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS Genet. 8: e1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E., Costa Y., Speed R., Taggart M., Benavente R., De Rooij D.G., Cooke H.J. (2007). SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J. Cell Biol. 176: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas I., Shaw P., Prieto P., Wanous M., Spielmeyer W., Mago R., Moore G. (2008). Effective chromosome pairing requires chromatin remodeling at the onset of meiosis. Proc. Natl. Acad. Sci. USA 105: 6075–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Ines O., Abe K., Goubely C., Gallego M.E., White C.I. (2012). Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana. PLoS Genet. 8: e1002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer S., San-Segundo P.A., Aragón L. (2011). The Smc5-Smc6 complex is required to remove chromosome junctions in meiosis. PLoS ONE 6: e20948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Fu S., Dong Q., Han F., Birchler J.A. (2011). Inactivation of a centromere during the formation of a translocation in maize. Chromosome Res. 19: 755–761 [DOI] [PubMed] [Google Scholar]
- Golubovskaya I.N., Hamant O., Timofejeva L., Wang C.J., Braun D., Meeley R., Cande W.Z. (2006). Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 119: 3306–3315 [DOI] [PubMed] [Google Scholar]
- Golubovskaya I.N., Harper L.C., Pawlowski W.P., Schichnes D., Cande W.Z. (2002). The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162: 1979–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya I.N., Wang C.J., Timofejeva L., Cande W.Z. (2011). Maize meiotic mutants with improper or non-homologous synapsis due to problems in pairing or synaptonemal complex formation. J. Exp. Bot. 62: 1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Gao Z., Birchler J.A. (2009). Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 21: 1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Lamb J.C., Yu W., Gao Z., Birchler J.A. (2007). Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. Plant Cell 19: 524–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L.C., Golubovskaya I.N., Cande W.Z. (2004). A bouquet of chromosomes. J. Cell Sci. 117: 4025–4032 [DOI] [PubMed] [Google Scholar]
- Hirano T. (2005). Condensins: Organizing and segregating the genome. Curr. Biol. 15: R265–R275 [DOI] [PubMed] [Google Scholar]
- Kato A., Lamb J.C., Birchler J.A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B., Boumil R.M., Stewart M.N., Dawson D.S. (2004). A role for centromere pairing in meiotic chromosome segregation. Genes Dev. 18: 1946–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Kleckner N. (2009). Dynamic chromosome movements during meiosis: A way to eliminate unwanted connections? Trends Cell Biol. 19: 716–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J.C., Danilova T., Bauer M.J., Meyer J.M., Holland J.J., Jensen M.D., Birchler J.A. (2007). Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175: 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos H.B., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. (2006). Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22: 755–767 [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano T. (2005). Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes Dev. 19: 1269–1287 [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E., Shaw P., Moore G. (2001). The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature 411: 204–207 [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez E., Shaw P., Reader S., Aragón-Alcaide L., Miller T., Moore G. (1999). Homologous chromosome pairing in wheat. J. Cell Sci. 112: 1761–1769 [DOI] [PubMed] [Google Scholar]
- McKim K.S. (2005). When size does not matter: Pairing sites during meiosis. Cell 123: 989–992 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. (2005). The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74: 595–648 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. (2009). Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Niwa O., Shimanuki M., Miki F. (2000). Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19: 3831–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold C.A., Brown P.E., Lawrence N.D., Goldman A.S. (2012). Modeling meiotic chromosomes indicates a size dependent contribution of telomere clustering and chromosome rigidity to homologue juxtaposition. PLoS Comput. Biol. 8: e1002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A.M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., Woglar A., Csaszar E., Pasierbek P., Ammerer G., Gruenbaum Y., Jantsch V. (2009). Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139: 920–933 [DOI] [PubMed] [Google Scholar]
- Peterson C.L., Laniel M.A. (2004). Histones and histone modifications. Curr. Biol. 14: R546–R551 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Dernburg A.F. (2006). A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11: 817–829 [DOI] [PubMed] [Google Scholar]
- Phillips D., Nibau C., Wnetrzak J., Jenkins G. (2012). High resolution analysis of meiotic chromosome structure and behaviour in barley (Hordeum vulgare L.). PLoS ONE 7: e39539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto P., Moore G., Reader S. (2005). Control of conformation changes associated with homologue recognition during meiosis. Theor. Appl. Genet. 111: 505–510 [DOI] [PubMed] [Google Scholar]
- Prieto P., Shaw P., Moore G. (2004). Homologue recognition during meiosis is associated with a change in chromatin conformation. Nat. Cell Biol. 6: 906–908 [DOI] [PubMed] [Google Scholar]
- Ronceret A., Doutriaux M.P., Golubovskaya I.N., Pawlowski W.P. (2009). PHS1 regulates meiotic recombination and homologous chromosome pairing by controlling the transport of RAD50 to the nucleus. Proc. Natl. Acad. Sci. USA 106: 20121–20126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A., Pawlowski W.P. (2010). Chromosome dynamics in meiotic prophase I in plants. Cytogenet. Genome Res. 129: 173–183 [DOI] [PubMed] [Google Scholar]
- Roy M.A., Siddiqui N., D’Amours D. (2011). Dynamic and selective DNA-binding activity of Smc5, a core component of the Smc5-Smc6 complex. Cell Cycle 10: 690–700 [DOI] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., Dernburg A.F. (2009). Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139: 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H. (2007). Telomere attachment and clustering during meiosis. Cell. Mol. Life Sci. 64: 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Bähler J., Kohli J. (1994). Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J. Cell Biol. 127: 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Prüfert K., Saito T.T., Smolikov S., Gu Y., Hincapie M., Hill D.E., Vidal M., McDonald K., Colaiácovo M.P. (2011). Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics 189: 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan M.J., Pawlowski W.P. (2009). Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc. Natl. Acad. Sci. USA 106: 20989–20994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M.N., Dawson D.S. (2008). Changing partners: Moving from non-homologous to homologous centromere pairing in meiosis. Trends Genet. 24: 564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Blower M.D., Karpen G.H. (2001). Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2: 584–596 [DOI] [PubMed] [Google Scholar]
- Takeo S., Lake C.M., Morais-de-Sá E., Sunkel C.E., Hawley R.S. (2011). Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr. Biol. 21: 1845–1851 [DOI] [PubMed] [Google Scholar]
- Tsai J.H., McKee B.D. (2011). Homologous pairing and the role of pairing centers in meiosis. J. Cell Sci. 124: 1955–1963 [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Macqueen A.J., Roeder G.S. (2008). Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 22: 3217–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T., Roeder G.S. (2005). A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308: 870–873 [DOI] [PubMed] [Google Scholar]
- Topp C.N., Zhong C.X., Dawe R.K. (2004). Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 101: 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vain P., McMullen M.D., Finer J.J. (1993). Osmotic treatment enhances particle bombardment-mediated transient and stable transformation of maize. Plant Cell Rep. 12: 84–88 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Pacher M., Dukowic S., Schubert V., Puchta H., Schubert I. (2009). The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21: 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R., Moore G., Shaw P.J. (2012). Centromeres cluster de novo at the beginning of meiosis in Brachypodium distachyon. PLoS ONE 7: e44681. [DOI] [PMC free article] [PubMed] [Google Scholar]