Abstract
The polar regions of the Escherichia coli murein sacculus are metabolically inert and stable in time. Because the sacculus and the outer membrane are tightly associated, we investigated whether polar inert murein could restrict the mobility of other cell envelope elements. Cells were covalently labeled with a fluorescent reagent, chased in dye-free medium, and observed by microscopy. Fluorescent material was more efficiently retained at the cell poles than at any other location. The boundary between high and low fluorescence intensity areas was rather sharp. Labeled material consisted mostly of cell envelope proteins, among them the free and murein-bound forms of Braun's lipoprotein. Our results indicate that the mobility of at least some cell envelope proteins is restrained at regions in correspondence with underlying areas of inert murein.
The peptidoglycan (murein) sacculus of the bacterial cell wall plays a key role in bacterial morphogenesis (21, 34, 45, 46, 54). In gram-negative bacteria such as Escherichia coli, the sacculus occupies the periplasmic space between the cytoplasmic membrane (IM) and the outer membrane (OM) and interacts with both of them (24, 34, 35, 44, 45). Interaction with the OM is mediated by a number of proteins able to link murein to components of the OM. Among them, Braun's lipoprotein (Lpp) is remarkable as the only known protein that binds covalently to murein in E. coli (5, 6). Lpp is the most abundant protein in the cell envelope and coexists in free (60 to 70%) and murein-bound (30 to 40%) forms (4, 22). The attachment of Lpp to the OM presumably occurs through the insertion of the acyl chains into the inner phospholipid leaflet of the OM (4, 34, 57). Free Lpp molecules form trimers which may contribute to the strength of the murein-OM complex by additional noncovalent interactions (8, 9, 57). The sacculus and the OM also interact by means of noncovalently bound molecules such as the peptidoglycan-associated lipoprotein (PAL) and OmpA proteins. The binding between PAL and the sacculus is very strong and involves a murein-binding-specific sequence (26). PAL anchors to the OM primarily by the acylated N terminus, but direct protein-protein interactions with other OM proteins have been demonstrated (1, 26, 33). One of the major OM proteins is OmpA. It is an integral membrane protein with domains protruding from both the external and internal leaflets. On the outer side, it functions as a phage receptor, and on the internal side, it interacts directly with murein (18, 36). The interaction of OmpA with murein is strong enough to stabilize the cell envelope in the absence of Lpp (48, 56). In addition to Lpp, PAL, and OmpA, a number of other proteins able to interact with the OM and the sacculus, such as OmpF and OmpC, may contribute to the total interaction as well (38).
The turgor pressure of the cell forces a physical contact between the IM and the sacculus (23). However, no specific attaching elements have been demonstrated up to now. Interestingly the PAL-TOL complex might play such a role since three of its constituent proteins, TolA, TolQ, and TolR, are IM proteins (3).
An investigation of murein segregation in E. coli demonstrated domains of inert murein at the polar regions of sacculi. Lateral wall elongation is accomplished by the intercalation of new and preexisting murein over the cylindrical surface of the sacculus. At the time of septation, an area of localized murein synthesis develops at the division site and remains active until septum completion when zonal synthesis stops, and the new poles become inert and stable in time for not less than five generations (10, 11, 20, 55).
The complex and strong connections between the OM and the sacculus suggest that the mobility of OM elements may be restrained in the polar areas by interaction with the underlying inert polar caps of the sacculus. The results below substantiated our hypothesis showing that (some) proteins in the polar regions of the outer membrane are essentially immobile.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains used in this work were MC6RP1 (K-12 F− dra drm leuA lysA proA thi thr) (42), JE5509 (K-12 F− argH1 dapA his1 lacY1 lysA malA1 man1 mtl2 pps str9 thi1 xyl7), and JE5510 (K-12 F− argH1 dapA his1 lacY1 lpo5508 lysA malA1 man1 mtl2 pps str9 thi1 xyl7) (52). Cultures were routinely grown in Luria-Bertani (LB) medium (27) at 37°C in gyratory water baths. Growth was monitored by measuring the turbidity of the cultures at 550 nm (A550).
Labeling of bacterial surface with fluorescent probes.
Culture samples of an appropriate size were collected by centrifugation. Cells were resuspended at a cell density of ca. 1010 cells/ml in 0.1 M NaHCO3 and mixed 1:50 with a 2.5-mg/ml solution of Texas Red-X succinimidyl ester (TRSE; Molecular Probes Europe BV, Leiden, The Netherlands) in dimethyl sulfoxide. Reaction mixtures were kept in the dark at room temperature for 5 min. The reaction was stopped by adding 50 μl of lysine (50 mg/ml) in water per ml of labeling mixture. Samples were centrifuged as described above, and cell pellets were resuspended in equal volumes of LB medium and centrifuged once again. The pellets were resuspended in LB medium prewarmed at 37°C and diluted as required for individual experiments. Manipulation of Texas Red (TR)-labeled cells was done under dim light, and flasks with labeled cultures were wrapped in thick aluminum foil for further incubation.
Purification and fractionation of cell envelopes from TR-labeled cells.
Bacterial cells (ca. 1010) were labeled with TRSE and resuspended in 1 ml of ice-cold phosphate (10 mM)-buffered saline (PBS) (140 mM NaCl, 10 mM KCl), pH 7.4. Labeled cells were washed twice by centrifugation (6,000 rpm for 2 min with a Mikroliter centrifuge; Hettich Zentrifugen, Tuttlingen, Germany) and resuspension in the same buffer. The cell suspension was subjected to ultrasonic disruption on ice for 2 min in 30-s bursts with a Labsonic 2000U probe sonicator on the low setting (B. Braun Biotech Inc., Allentown, Pa.), and intact cells were removed by low-speed centrifugation (2 min at 4,000 rpm with a Hettich Mikroliter centrifuge). The supernatant was centrifuged at high speed (80,000 rpm at 4°C for 10 min with a Beckman [Palo Alto, Calif.] TL ultracentrifuge with a TL 100.3 rotor), and the particulate fraction was washed by resuspension (3 ml of PBS) and centrifugation as before. The pellet was resuspended in 100 μl of PBS. Further fractionation was performed by selective detergent extraction. Purified envelopes were mixed 1:1 with 1% (wt/vol) sodium dodecyl sarcosinate, kept at 30°C for 60 min, and subjected to high-speed centrifugation as described above. The supernatant (sarcosyl fraction) enriched in IM components was removed and kept at 4°C. The pellets were resuspended in 1% (wt/vol) sodium dodecyl sulfate (SDS) and incubated 5 min at 95°C to solubilize remaining proteins. Samples were centrifuged as before but at 30°C to remove insoluble peptidoglycan. The supernatant (SDS fraction) enriched in OM proteins was removed and kept at 4°C. The pellet was washed with water twice by resuspension and centrifugation as before. The last pellet was resuspended in 20 mM phosphate buffer, pH 4.9, and digested overnight with Cellosyl muramidase (20 μg/ml; Aventis, Frankfurt, Germany) at 37°C. The undigested material was removed by centrifugation, and the supernatant (murein fraction), together with the previous fractions, was further processed for gel electrophoresis.
Gel electrophoresis of TR-labeled membrane proteins.
Protein analysis was performed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli (25). Proteins were resolved in 16% (wt/vol) acrylamide slab (17 cm long, 2 mm thick) gels in the dark. Gels were briefly (20 min) washed in 5% (vol/vol) methanol in water. Fluorescent components were visualized by transillumination in a Fotodyne 3-3200 DNA UV transilluminator (Fotodyne Incorporated, New Berlin, Wis.) and photographed with a Kodak DC40 digital camera (Eastman Kodak Co., Rochester, N.Y.). Fluorescence was rather stable for as long as the gels were kept in water or 5% (vol/vol) methanol but was completely lost in the presence of acetic acid. Once pictures were taken, gels were fixed in 5% (vol/vol) acetic acid and 15% (vol/vol) methanol in water and stained overnight in a 0.01% (wt/vol) solution of Coomassie blue R in the same fixative mixture, and pictures of the hydrated gels were taken. Prestained proteins of known molecular weight (prestained SDS-PAGE standards, broad range; Bio-Rad, Hercules, Calif.) were used as standards.
Purification and gel electrophoresis of murein from TR-labeled cells.
Bacterial cells (ca. 1010) were labeled with TRSE and resuspended in 3 ml of PBS. Cells were washed by centrifugation and resuspension in the same buffer, slowly mixed with 6 ml of 6% (wt/vol) SDS in a boiling water bath, and kept for 3 h under these conditions. Samples were left overnight at 65°C. Insoluble material was washed free of SDS by repeated cycles of centrifugation (80,000 rpm, 15 min, 30°C; Beckman TL ultracentrifuge with a TL100.3 rotor) and resuspension in water. The last pellets were resuspended in 20 mM phosphate buffer, pH 4.9, and digested for 6 h with Cellosyl muramidase (50 μg/ml) at 37°C. Insoluble material was removed by centrifugation (80,000 rpm, 3 min, 30°C; Beckman TL ultracentrifuge with a TL100.1 rotor). Solubilized material was mixed with Laemmli sample buffer (25) and incubated 1 min in a boiling water bath. For comparative purposes, Lpp-free and Lpp-containing purified sacculi were also labeled with TRSE. E. coli MC6RP1 sacculi were purified as described previously (17), except that in the case of Lpp-containing sacculi, pronase E digestion was omitted. Samples (500 μg) of Lpp-free and Lpp-containing murein in 0.1 M NaHCO3 were mixed 1:100 with a 2.5-mg/ml solution of TRSE in dimethyl sulfoxide and incubated in the dark for 10 min. Murein was recovered by centrifugation (80,000 rpm, 10 min, 30°C; Beckman TL ultracentrifuge with a TL100.3 rotor) and washed dye-free by repeated cycles of resuspension in water and centrifugation. Final pellets were resuspended in 20 mM phosphate buffer, pH 4.9, and digested for 6 h with Cellosyl. Insoluble material was removed as described above. The supernatants were mixed with Laemmli sample buffer and briefly (2 min) incubated in a boiling water bath. The Cellosyl digestion products were subjected to SDS-PAGE in 22% (wt/vol) polyacrylamide slab gels (17 cm long, 2 mm thick). Fluorescent muropeptides were visualized as described above for protein gels.
Fractionation of membranes from TR-labeled cells by isopycnic sucrose density gradient centrifugation.
Purified membranes from TR-labeled and control unlabeled cells were prepared as described previously by Osborn et al. (37). Cells (ca. 2 × 1010) were resuspended in 5 ml of 10 mM Tris-HCl (pH 7.8) and 0.75 M sucrose containing 100 μg of lysozyme/ml cooled to 0°C in an ice-water bath. After 3 min, the cell suspension was mixed 1:2 with 1.5 mM EDTA, pH 7.4, and the formation of spheroplasts was followed by periodic observations under the microscope. After 30 min, most cells showed round or irregular shapes indicative of spheroplast formation. The cell suspension was subjected to ultrasonic disruption on ice (three 30-s bursts with a Labsonic 2000U probe sonicator on the low setting), and intact cells were removed by low-speed centrifugation (8,000 × g, 10 min, 4°C). The supernatant was centrifuged at high speed (80,000 rpm, 4°C, 20 min; Beckman TL ultracentrifuge with a TL 100.3 rotor), and the particulate fraction was resuspended in 1 ml of 25% (wt/wt) sucrose in 5 mM EDTA, pH 7.4. Samples were subjected to isopycnic centrifugation on discontinuous sucrose gradients on an SW40 rotor (Beckman) consisting of the following layers (bottom to top) of sucrose (percent is wt/wt) solutions in 5 mM EDTA (pH 7.4): 1 ml, 55%; 2.2 ml, 50%; 2.8 ml, 45%; 2.5 ml, 40%; 1.7 ml, 35%; and 1.7 ml, 30%. After centrifugation (22,000 rpm, 36 h, 4°C; Beckman L8-701 ultracentrifuge with an SW40 rotor), the gradients were fractionated, and each fraction was assayed for protein content by the bicinchoninic acid assay (BCA protein assay kit, product no. 23225; Pierce, Rockford, Ill.) for 2-keto-3-deoxyoctonate (KDO) according to the method of Waravdekar and Saslaw (53) and for TR by measuring A583.
Extraction and analysis of membrane lipids from TR-labeled cells.
TR-labeled cells (ca. 1010) were resuspended in 2 ml of PBS and washed twice by centrifugation (4,000 rpm, 2 min; Hettich centrifuge) and resuspension in the same buffer. The final pellet was resuspended in 0.5 ml of PBS, thoroughly mixed with 5 ml of CHCl3:CH3OH (2:1) and left standing for 20 min. The organic phase was removed and flushed with N2 until dry. The sediment was dissolved in 100 μl of CHCl3 and divided into two parts which were dried as before. One of the aliquots was kept dry at 4°C in the dark for further analysis. The second aliquot was resuspended in 200 μl of 0.1 M NaHCO3, mixed with 5 μl of 2.5 mg of TRSE/ml in dimethyl sulfoxide, and left to react for 20 min. A control sample without any lipid added was prepared in parallel to identify traces of the fluorescent dye and degradation products. Samples were mixed with 2 ml of CHCl3:CH3OH (2:1) left standing for 20 min, and the organic phases were removed, dried, and stored as described above. Thin-layer chromatography was performed on CHCl3-washed and activated (60 min, 120°C) 10-cm-long silica gel G-60 plates (E. Merk, Darmstat, Germany). Lipid fractions were dissolved in 15 μl of CHCl3, and 2 μl of each fraction was deposited with glass capillaries on the origin. Chromatograms were developed with CHCl3:CH3OH:H2O (65:25:4) and dried at 42°C. Fluorescent spots were detected in a Fotodyne 3-3200 DNA UV transilluminator and photographed with a Kodak DC40 digital camera. Total lipids were stained with I2 vapor for 30 min, and stained plates were photographed as before.
Microscopy and image densitometry.
Samples from bacterial cultures were mixed 1:1 with 0.4% (vol/vol) formaldehyde in PBS and conserved at 4°C in the dark. Small drops (2 to 5 μl) of bacterial suspension were spread on poly-lysine-covered glass slides and allowed to air dry. Slides were extensively washed with water, air dried, and mounted with Sigma (St. Louis, Mo.) mounting medium. Phase-contrast and epifluorescence microscopy were performed with an Olympus BX50 microscope fitted with a 60× NA 1.25 Olympus UPlan Fl objective and AD photographic system. Pictures were taken on Fuji Provia 1600 film and digitized on a Coolscan III slide scanner (Nikon Co., Tokyo, Japan). Confocal microscopy was performed in a Radiance 2000 (Bio-Rad) confocal microscope fitted with a 100× NA 1.3 Zeiss Plan-Neofluar objective and Laser Sharp image acquisition software (Bio-Rad). Further image editing was done with Adobe Photoshop software. Pictures for densitometry were obtained under identical exposure conditions, avoiding film saturation, and were digitized on a Nikon Coolscan III slide scanner. Images were converted from RGB (red-green- blue; 8 bits per channel) to 8-bit gray without any further modification. Scanned images were then analyzed by using the TINA version 2.09e software (Isotopenmeßgeräte GmbH, Munich, Germany). Straight cells were selected, and density profiles along the cell axis were obtained.
Plasmolysis of TR-labeled MC6RP1 filament cells.
Cells were TR labeled and chased in LB medium plus 1 μg of aztreonam/ml to inhibit cell division. At the end of the chase period, cells were subjected to plasmolysis by osmotic shock in 25% (wt/vol) sucrose. After 10 min, plasmolysis was stopped by adding the same volume of 4% (vol/vol) formaldehyde, 0.1% (vol/vol) glutaraldehyde, and 25% (wt/vol) sucrose in PBS to the cell suspension (15). Phase-contrast microscopy showed clearly detectable plasmolysis bays in most of the cells. Longer plasmolysis times did not improve the size or number of plasmolysis bays.
Immunoelectron microscopy of cryosections of plasmolyzed cells.
Fixed cells were washed with PBS, embedded in 2% (wt/vol) agarose, cut in small blocks, and infiltrated with 20% (vol/vol) polyvinylpyrrolidone-1.8 M sucrose in 100 mM phosphate buffer, pH 7.2. Specimen blocks were mounted on copper stubs, frozen in liquid nitrogen, and trimmed and sectioned (at 60 to 70 nm) with a Leica Ultracut S/FCS cryoultramicrotome. Cryosections were thawed and labeled with rabbit anti-TR antibodies (A-6399; Molecular Probes) diluted 1:100 and 6-nm protein A-gold complexes. Labeled sections were embedded in 1.4% (wt/vol) methyl cellulose and 0.3% (wt/vol) uranyl acetate and analyzed with a Philips CM10 transmission electron microscope at 60 kV.
RESULTS
Segregation of fluorescent elements in TR-labeled E. coli MC6RP1 cells.
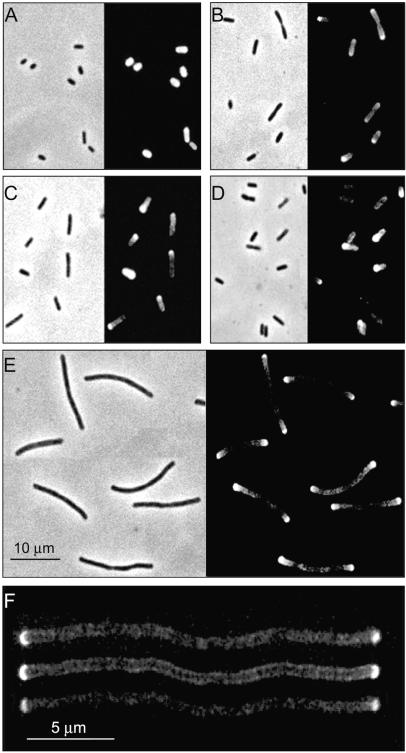
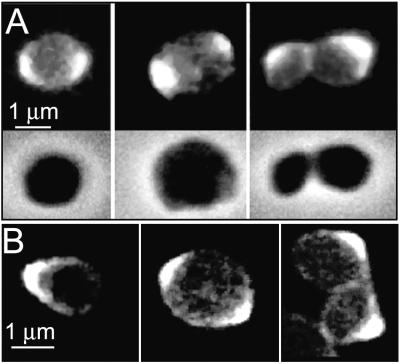
E. coli MC6RP1 cells growing in LB medium were labeled with TRSE and chased in dye-free LB medium, and samples were periodically removed and prepared for microscopy. TR-stained cells restarted growth readily, indicating that only minor damage was inflicted by the labeling procedure. Nonchased cells were homogeneously labeled (Fig. 1A). However, cells chased for a twofold increase of A550 exhibited a heterogeneous distribution of label. Constricting cells showed maxima at both poles with a darker central region. However most of the shorter cells had only one strongly labeled pole (Fig. 1B). In the samples chased for longer times, cells had either one strongly labeled end or none, with the frequency of the latter increasing with chase time (Fig. 1C and D). These results suggested that TR-labeled molecules had a longer residence time at the poles than elsewhere in the cell. Otherwise, labeled material should have diffused homogeneously over the cell as a consequence of molecular diffusion and increased surface area because of cell growth.
FIG. 1.
Segregation of fluorescent elements in TR-labeled cells of E. coli MC6RP1. Cells were labeled with TRSE and allowed to grow in LB medium (A to D) or in LB medium plus 1 μg of aztreonam/ml (E and F). Samples were removed and observed either by a combination of phase-contrast and epifluorescence microscopy (A to E) or by confocal scanning microscopy (F). Nonchased control cells (A) and cells chased for a twofold increase (B), a fourfold increase (C), a sixfold increase (D), a sixfold increase in the presence of aztreonam (E), and a sixfold increase in the presence of aztreonam observed by confocal scanning microscopy (F) at an optical density at 550 nm are shown. Optical sections had 0.4-μm nominal width. The upper and lower sections in the image were 0.5 μm above and below the central section, respectively.
Similar experiments made with cells blocked for division by the inhibition of PBP3, an enzyme necessary for septation (2), confirmed these observations. Bacteria were labeled with TRSE as described above and chased in medium containing 1 μg of aztreonam, an inhibitor of PBP3 (16), per ml. The results (Fig. 1E) confirmed that labeling was more persistent in the polar regions than in the cylindrical part of the cell. Analysis of the same sample by confocal scanning microscopy (Fig. 1F) showed a rather sharp boundary separating the areas of high and low fluorescence. In addition to the poles, low-fluorescence labeled material was detected all along the cell but was confined to the cell envelope, indicating a preferential reaction of the fluorescent dye with the cell surface.
Similar results were obtained with the NH2-specific fluorescent reagents Oregon Green 514 carboxylic acid succinimidyl ester 5-isomer and Oregon Green 488 carboxylic acid succinimidyl ester 5-isomer (Molecular Probes Europe BV), suggesting that the patterns observed are independent of the nature of the fluorescent probe (data not shown).
Slow decay of fluorescence intensity in the polar regions of TR-labeled E. coli.
Photographic images from TR-labeled cells chased for a sixfold increase of mass in medium with and without aztreonam (1 μg/ml) were subjected to densitometric analysis for a better estimation of signal strength variation with chase time. Signal strength is expressed in terms of optical density arbitrary units as calculated by the TINA software, without any further correction. The results indicated that fluorescence intensity decayed very slowly in the polar regions but was just as expected from growth dilution in the rest of the cell. Fluorescence intensity in poles of dividing cells and aztreonam-induced filaments was around 65% (1,650 ± 226 U, n = 22 cells) and 85% (2,200 ± 260 U, n = 18 cells) of the intensity in control, nonchased cells (2,553 ± 84 U, n = 25 cells), respectively. The mean fluorescence intensity along aztreonam-induced filaments represented about 17% (433 ± 82 U, n = 18 cells) of the control, precisely the value expected from growth dilution (16%) on the assumption that no material was lost in the course of growth.
TRSE reacts with surface-exposed elements of the cell envelope.
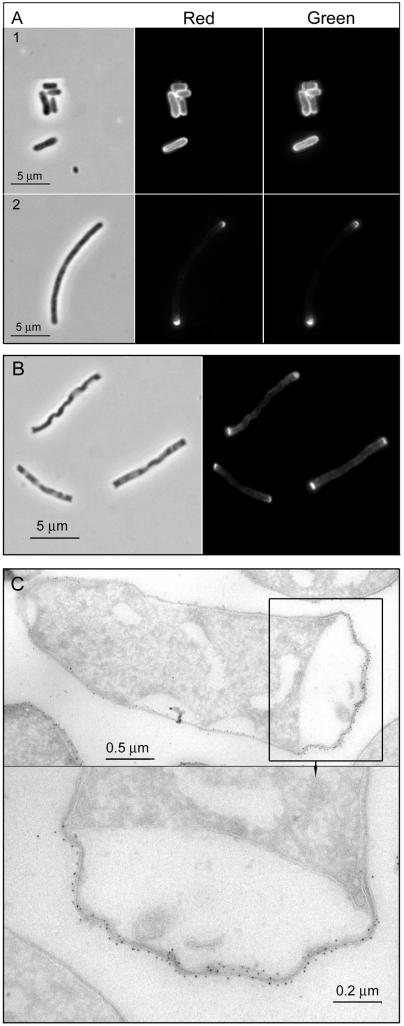
Under our labeling conditions, TRSE was expected to preferentially react with free amino groups in the cell surface. To confirm this point, we made use of anti-TR antibodies. In nonpermeabilized cells, antibodies reacted only with surface-exposed residues. Overlapping signals for TR itself and anti-TR antibody would indicate that TRSE reacts mostly with environment-exposed groups. On the contrary, poor overlapping would support a reaction with internal groups, too. Therefore, MC6RP1 cells were labeled with TRSE and chased in dye-free medium for appropriate times in the presence of 1 μg of aztreonam/ml to block division. Samples were subjected to immunolabeling with anti-TR rabbit antibody and Alexa488 goat anti-rabbit antibody and observed by epifluorescence microscopy. As shown in Fig. 2A, the distributions of green (antibody) and red (TR) fluorescence were almost identical. Hence, a substantial number of TR-labeled molecules must be exposed to the environment. The ability of the dye to reach deeper layers of the envelope was also checked by immunoelectron microscopy of cryosections from plasmolyzed TR-labeled cells. Cells were labeled and chased as described above, but at the time of sample collection, cells were plasmolyzed to force a separation of the OM-murein and IM layers of the envelope before processing for cryosectioning and immunolabeling. Sections were labeled with the same anti-TR antibody as described above, which was detected by using 6-nm protein A-gold. The results showed a strict association of gold particles with the OM-murein layer (Fig. 2C). Indeed, hardly any gold grain could be observed in the IM layer underlying heavily labeled OM-murein areas, both in polar regions and in the lateral wall. Therefore, under our conditions, TRSE exhibited a high preference for OM components exposed to the external environment.
FIG. 2.
Immunolocalization of TR dye in TR-labeled and chased MC6RP1 filament cells. Cells were labeled with TRSE and chased for a fivefold increase of the A550 in LB medium plus 1 μg of aztreonam/ml to block cell division. At this time, aliquots were either fixed and subjected to immunolabeling with anti-TR rabbit antibody and Alexa488 goat anti-rabbit antibody (A) or subjected to plasmolysis (B and C). After 10 min of osmotic shock, plasmolysed cells were fixed and further processed for fluorescence microscopy (B) or for cryosectioning and immunoelectron microscopy with the same primary antibody as described above and 6-nm protein A-gold conjugates (C). (A) Upper panels, nonchased cells; lower panels, chased cells. Leftmost panels show phase-contrast images, TR panels show the red fluorescence of TR itself, and anti-TR panels show the green fluorescence after TR immunodetection. (B) Chased and plasmolysed filaments observed by phase-contrast microscopy (left panel) and fluorescence microscopy in the red channel (right panel). (C) Immunoelectron microscopy of chased and plasmolysed filaments. TR-labeled molecules were detected with anti-TR rabbit antibody and 6-nm protein A-gold conjugates. The lower panel shows a framed region of the upper panel at a higher magnification.
Cell envelope elements accessible to TRSE labeling.
To define the chemical nature of the species reacting with TRSE, experiments were conducted to check for binding to the major components of the envelope. The lipopolysaccharide of E. coli has a free NH2 group in an N-acetylglucosamine that forms part of the core region. Because K-12 strains like MC6RP1 lack the O antigen, core sugars could be accessible enough for TRSE to react. However, gel electrophoresis (19) of lipopolysaccharide from TR-labeled MC6RP1 failed to show any labeling of TRSE to lipopolysaccharide.
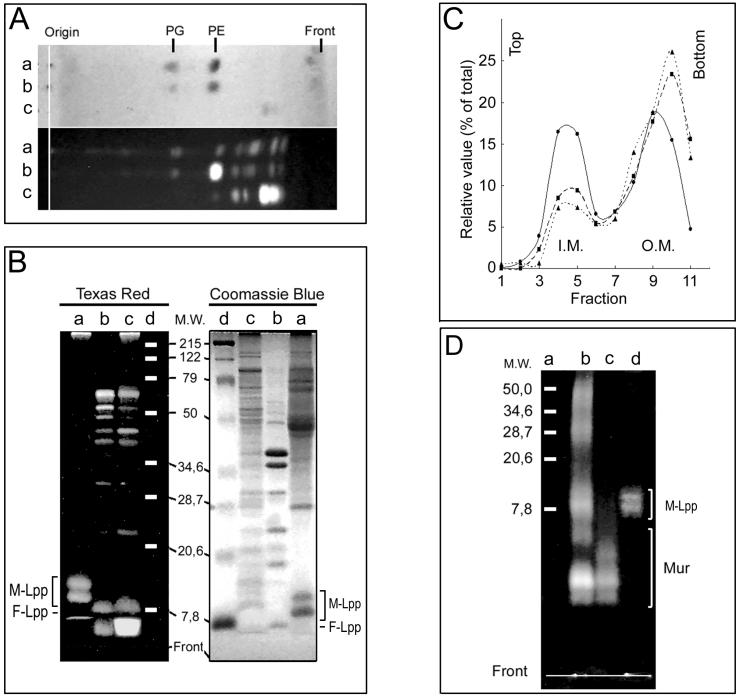
The most abundant phospholipid in E. coli cell membranes, phosphatidylethanolamine, also had the potential to react with TRSE through the free NH2 group of the polar moiety. To check this possibility, lipids were extracted from TR-labeled cells and analyzed by thin-layer chromatography. An aliquot of the extracted lipids was labeled again with TRSE and run in parallel as a positive control. All the phosphatidylethanolamine present in the sample should have been labeled after the second incubation. The result of the experiment indicated that TR labeling of lipids was at the most marginal under our conditions (Fig. 3A).
FIG. 3.
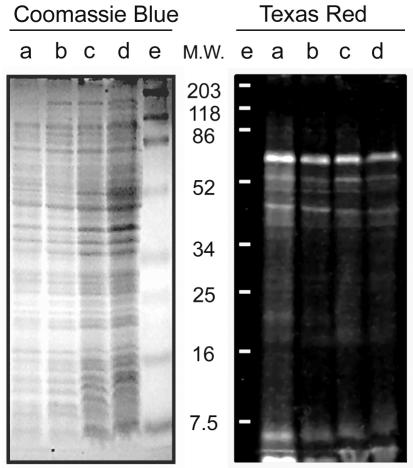
Identification of fluorescent molecules in TR-labeled MC6RP1 cells. (A) Lipids from TR-labeled cells were extracted and subjected to thin-layer chromatography either directly or after a second labeling with TRSE. Fluorescent lipids were visualized by illumination with UV light and photographed with a digital camera. Total lipids were stained with I2 vapor and photographed as before. Lane a, lipids from TR-labeled cells; lane b, lipids from TR-labeled cells subjected to a second labeling with TRSE; lane c, as panel b but without added lipids. PG, phosphatidylglycerol; PE, phosphatidylethanolamine. (B) Cell envelopes from TR-labeled cells were prepared and fractionated by selective detergent extraction to obtain fractions enriched in IM proteins, OM proteins, and murein-associated proteins. After SDS-PAGE, fluorescent proteins were detected by transillumination with UV light, and total proteins were detected by Coomassie blue staining. A set of prestained proteins of known molecular weight (prestained SDS-PAGE standards, broad range; Bio-Rad) was used as a reference to estimate protein mobility in the gels. Lane a, murein-associated proteins; lane b, OM proteins (sarcosyl-insoluble, SDS-soluble proteins); lane c, IM proteins (sarcosyl-solubilized proteins). M-Lpp, muropeptide-bound form of Braun's lipoprotein; F-Lpp, free form of Braun's lipoprotein. (C) Membranes from TR-labeled cells were subjected to sucrose density gradient centrifugation, and the distribution of protein (•), KDO (▪), and TR (▴) along the gradient was measured as indicated in Materials and Methods. (D) Murein extracted from TR-labeled cells was digested with muramidase, and solubilized materials were subjected to SDS-PAGE. As controls, samples of Lpp-free and Lpp-containing murein were labeled with TRSE, digested with muramidase, and run in parallel. Fluorescent components were detected by transillumination with UV light. Lane a, prestained proteins of known molecular weight (M.W.) (prestained SDS-PAGE standards, broad range; Bio-Rad); lane b, Lpp-containing control murein; lane c, Lpp-free control murein; lane d, murein from TR-labeled cells. M-Lpp, muropeptide-bound form of Braun's lipoprotein; Mur, murein-derived muropeptides.
Envelope proteins were the prime candidates to react with TRSE once lipopolysaccharide and phospholipids were discarded. To investigate this point, cell envelopes obtained from TR-labeled cells of E. coli MC6RP1 were fractionated by sequential detergent extraction and analyzed by gel electrophoresis. The results showed a preferential reaction of TRSE with a relatively small group of proteins which were mostly, but not exclusively, in the sarcosyl-insoluble (OM-enriched) fraction (Fig. 3B). To better define the subcellular location of TR-labeled components, envelopes purified from TR-labeled cells were subjected to isopycnic centrifugation in sucrose density gradients to separate the OM and IM (37). The result (Fig. 3C) confirmed that most (73%) of the TR label migrated to the OM fraction, while 14% was found in the IM fraction and the remaining 13% was in the “intermediate” fractions. These values were almost exactly matched by the distribution of the OM-specific marker KDO (70, 17, and 13% of the total in the OM, IM, and intermediate fractions, respectively), suggesting that most of the signal in the IM fraction might be accounted for by contaminating OM fragments.
Rather surprisingly, Lpp was able to react with TRSE in both the murein-bound and the free forms (Fig. 3B). Therefore, we checked whether some TRSE could reach the periplasmic space and react with the free NH2 groups in the sacculus. Murein purified from TR-labeled cells was muramidase (Cellosyl) digested, and the resulting mixture of muropeptides was analyzed by gel electrophoresis together with samples of Lpp-free and Lpp-containing purified murein that was TR labeled as described in Materials and Methods. The only fluorescent signals detected in the murein of TR-labeled cells corresponded to Lpp-bound muropeptides, with no visible fluorescence in the free muropeptide region (Fig. 3D).
Segregation of fluorescent surface elements in filament cells of a TR-labeled Lpp mutant of E. coli.
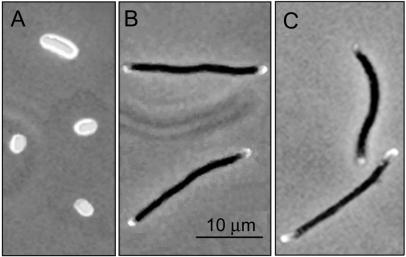
Because the strongest OM-sacculus interaction is mediated by Lpp, a dominant implication of this protein in the differential behavior of polar and cylindrical surface material was suspected. Therefore, the segregation of TR-labeled material was analyzed in the Lpp mutant JE5510 and its parental strain, JE5509. Cells of both strains were labeled, allowed to grow in LB medium plus 1 μg of aztreonam/ml to block division, and prepared for microscopy after two doublings in mass. The results (Fig. 4) indicated that the segregation of labeled material in both strains and in filaments of MC6RP1 (Fig. 1E) was identical.
FIG. 4.
Segregation of fluorescent elements in TR-labeled filament cells of the Lpp mutant strain JE5510. Cells of the E. coli Lpp mutant strain JE5510 and its parental strain, JE5509, were labeled with TRSE and allowed to grow in dye-free LB medium supplemented with 1 μg of aztreonam/ml until the A550 increased by sixfold. Samples were prepared and subjected to microscopical examination. Phase-contrast and epifluorescence photographic images of the specimens were digitized and merged with Photoshop software. (A) JE5510 nonchased control cells. (B) JE5510 (Lpp) chased cells. (C) JE5509 chased cells.
Effect of amdinocillin on the segregation of TR-labeled surface elements in E. coli MC6RP1.
The inhibition of PBP2 by the β-lactam antibiotic amdinocillin (mecillinam, or FL1060) induces round cell morphology and drastic modifications in the pattern of murein segregation in E. coli. The incorporation of murein precursors under those conditions occurs without the mixing of new and old murein. As a consequence, old murein is preserved for long periods of time as one or two large discrete areas (7, 10, 41, 47, 49-51). If the segregation of cell surface materials was indeed conditioned by the dynamics of underlying murein, it should have been affected by amdinocillin in a parallel way. To check this point, E. coli MC6RP1 cells were labeled with TRSE and chased in LB medium with 1.0 μg of amdinocillin/ml for a sixfold increase in mass. Under these conditions one or, more often, two large areas of the surface remained strongly labeled by TR after the chase, producing a fluorescence pattern resembling that described for murein segregation (Fig. 5A). Confocal microscopy of the same samples further confirmed the observations and showed that boundaries between the areas of conserved TR label and the rest of the surface were rather sharp (Fig. 5B).
FIG. 5.
Segregation of fluorescent elements in TR-labeled cells of MC6RP1 chased in the presence of amdinocillin. Cells of E. coli MC6RP1 were labeled with TRSE, chased for a fourfold increase of A550 in LB medium plus 1 μg of amdinocillin/ml, and subjected to microscopical examination. (A) Selected cells as observed by phase-contrast and epifluorescence microscopy. (B) Serial optical sections of selected cells as observed by confocal scanning microscopy. Optical sections had 0.4-μm nominal width, and spacing between successive sections was 0.5 μm.
Stability of TR-labeled proteins in growing cells of E. coli MC6RP1.
Knowledge of the stability of TR-labeled proteins was certainly important for any interpretation of the results observed in chased cells. Therefore, TR-labeled MC6RP1 cells were chased in LB medium, and samples of equal volume were withdrawn at appropriate times. Samples were further processed to prepare cell envelopes, and proteins were analyzed by SDS-PAGE. As all samples were of equal volume, the amount of any given TR-labeled protein should have remained constant, provided it was stable during the chase period. Otherwise, it should have decreased at a rate proportional to its half-life. The result of the experiment (Fig. 6) indicated that proteins preferentially labeled by TRSE were essentially stable. Indeed, after a fivefold increase in the A550, the amount of TR-labeled proteins in the gels was still about 70% of that of the control as estimated by densitometry. The calculated rate of dilution, estimated as a decrease in fluorescence intensity, was about 12% per generation, which for a generation time of 32 to 35 min would correspond to half-lives of around 150 min.
FIG. 6.
Stability of fluorescent proteins in the cell envelope of TR-labeled E. coli MC6RP1 cells. Cells were labeled with TRSE and allowed to grow in LB medium. After appropriate chase times, samples of equal volume were withdrawn and processed to obtain cell envelopes, which were in turn subjected to SDS-PAGE. Fluorescent proteins were visualized by transillumination with UV light, and total proteins were visualized by Coomassie blue staining. A set of prestained proteins of known molecular weight (prestained SDS-PAGE standards, broad range; Bio-Rad) was used to estimate protein mobility. Lane a, nonchased cells; lane b, cells chased for a twofold increase of A550; lane c, cells chased for a threefold increase of A550; lane d, cells chased for a fivefold increase of A550; lane e, molecular weight (M.W.) markers.
DISCUSSION
The identification of long-lived domains of inert murein at the poles of the sacculus (11) and the strong interaction between the OM and the sacculus (1, 6, 18, 34) suggested that the mobility of (some) OM molecules in the polar regions could be constrained. Interaction among OM elements and the sacculus might be homogeneous all over the cell. However, as murein is continuously remodeled due to biosynthesis and recycling in the lateral (cylindrical) area (20, 21, 32, 39, 40, 55), OM molecules that interact with the sacculus in this area may exhibit an apparent freedom of movement. To test this hypothesis, we investigated the fate of surface molecules amenable to in vivo labeling with the fluorescent probe TRSE upon subsequent growth.
The reaction of cells with TRSE resulted in the preferential labeling of a set of cell envelope proteins. The reagent showed a high reactivity with surface-exposed proteins, but neither membrane phospholipids nor murein was labeled to any significant degree. Although minor labeling of IM proteins could not be totally excluded, membrane fractionation and immunoelectron microscopy indicated that the bulk of TR-labeled materials was associated with the OM. Interestingly, Braun's Lpp was identified as one of the TR-labeled proteins. This was unexpected because Lpp is presumed to be anchored to the inner leaflet of the OM by the acylating fatty acids, with most of the polypeptide chain exposed to the periplasmic space (57). Hence, for Lpp to be labeled by TRSE, the dye would have to reach the periplasm, which is in contradiction with the lack of reaction with periplasm-exposed phospholipids and murein. Therefore, the labeling of Lpp was indicative of either an atypical accessibility to (some) externally added compounds or a high reactivity to TRSE (Lpp has nine NH2 groups in the free form and is the most abundant envelope protein) that could efficiently titer traces of the dye reaching the periplasm, thus preventing reactions with other periplasmic molecules.
When the fate of TR-labeled molecules in growing cells was monitored, it became clear that fluorescence intensity remained higher at the polar regions than at any other location of the cell, irrespective of the chase time. Labeled cells divided normally, and after one division, each daughter cell had one strongly labeled and one unlabeled pole. Cells with one labeled pole were clearly discernible by fluorescence microscopy even after a prolonged (2.5 generations) chase period. At this time, fluorescence intensity of the labeled poles was still around 65% of the initial value, much higher than expected from the growth dilution (16%). These results were further confirmed in division-blocked filament cells. Polar regions retained fluorescence more efficiently than the body of the cell. After a sixfold increase in mass, fluorescence intensities were about 85 and 17% of the initial value for polar and cylindrical regions, respectively. The latter value indicated that labeled material was essentially stable, as it closely matched expectations from the growth dilution (15%). This observation found further support when the stability of labeled proteins was directly estimated by gel electrophoresis. Indeed, our results revealed rather slow turnover rates (≤12% per generation) for the more conspicuous TR-labeled proteins. Therefore, it seems that as cells grow, TR-labeled materials can mix well with new molecules over the cylindrical surface of the cell but not at the poles, where they are apparently immobilized.
The connection between the OM and the sacculus is mediated by multiple elements, but Lpp is the only one that interacts covalently with the sacculus (6). Therefore, we expected Lpp to play a key role in the immobilization of OM polar material. However, the fact that a null mutation for Lpp behaved as the wild type indicated that this was not the case (Fig. 4). Therefore, even though Lpp might play an important role, it seems that other interactions (PAL, OmpA, etc.) are strong enough to effectively tether OM proteins to the sacculus by themselves. The recent proposal by Dmitriev et al. (13) of a model for murein architecture in which glycan strands are perpendicular instead of parallel to the IM plane provides a particularly intriguing alternative. In fact, glycan strands with such an orientation could directly interact with OM molecules through their distal ends and exert a strong influence on their behavior without the intervention of linking molecules.
An investigation of the fate of TR-labeled surface elements in cells treated with the PBP2 inhibitor amdinocillin further supported our hypothesis. The inhibition of PBP2 leads to a change in the pattern of murein segregation (10). The most relevant feature is that old murein is conserved in the form of large areas which cover a substantial part of the spherical surface of the sacculus, not only the tips of the polar regions as happens in rod cells. A very similar change was observed for the segregation pattern of TR-labeled surface elements. In fact, chased round cells exhibited large areas with clearly distinct fluorescence intensity. Although direct proof for a strict overlapping of the areas of conserved old murein in the sacculus and of high fluorescence in the OM could not be obtained, a correlation between both phenomena looks rather plausible. Therefore, alterations in the distribution of inert murein in the sacculus are apparently able to trigger concomitant changes in the distribution of OM domains of reduced protein mobility. The association between inert murein and OM areas of low mobility is also supported by results from a study of branching E. coli mutants (12). Morphological abnormalities in branching cells were found to be associated with areas of inert murein and often with areas of reduced OM mobility as determined by TR label and chase experiments.
The results presented here constitute, in our opinion, convincing evidence in favor of a strong interaction between the sacculus and OM which conditions the mobility of OM elements to the local dynamics of murein metabolism. The inert nature of polar murein would therefore restrict diffusion of at least those macromolecules able to interact with the sacculus directly or through bridging molecules. On the cylindrical body of the cell, the random insertion of murein precursors and recycling of muropeptides could effectively favor the redistribution and mixing of murein-interacting OM molecules. The final result would be the apparent free diffusion of those elements at any location outside the poles. The reduced mobility of OM proteins may also result from the existence of a barrier between the polar and cylindrical parts of the cell instead of the direct murein-OM interaction as proposed above. Indeed, experimental data support the presence of a diffusion barrier for periplasmic elements at the edges of polar regions and developing septa (14, 43). Nevertheless, the parallelism observed between areas of inert murein and restricted OM protein mobility seems too strict to be purely accidental. A most interesting possibility is that all three aspects mentioned above, periplasmic diffusion barriers, OM restricted mobility, and inert murein areas, are manifestations of a single differentiation process which might also imply the generation of a stable physical barrier delimiting the poles.
The relative stability of the polar regions with respect to the lateral wall in the cell envelope may be instrumental in the determination of polarity in rod-shaped bacteria (28). Processes such as location of actin-polymerizing proteins, polar flagella, chemotactic receptors, cell division proteins, etc. (29-31) are clear manifestations of the importance of cell polarity in bacterial physiology.
Acknowledgments
This work was supported by grant BCM2001-2264 from the PGC and a MPG-CSIC bilateral cooperation grant to M.A.P. as well as by an institutional grant from Fundación Ramón Areces to the CBM “Severo Ochoa.”
REFERENCES
- 1.Abergel, C., A. Walburger, S. Chenivesse, and C. Lazdunski. 2001. Crystallization and preliminary crystallographic study of the peptidoglycan-associated lipoprotein from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 57:317-319. [DOI] [PubMed] [Google Scholar]
- 2.Ayala, J. A., T. Garrido, M. A. de Pedro, and M. Vicente. 1994. Molecular biology of bacterial septation, p. 73-101. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 3.Bouveret, E., H. Bénédetti, A. Rigal, E. Loret, and C. Lazdunski. 1999. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J. Bacteriol. 181:6306-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V. 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta 415:335-377. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., and K. Rehn. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur. J. Biochem. 10:426-438. [DOI] [PubMed] [Google Scholar]
- 6.Braun, V., and H. C. Wu. 1994. Lipoproteins, structure, function, biosynthesis and model for protein export, p. 319-341. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 7.Canepari, P., G. Botta, and G. Satta. 1984. Inhibition of lateral wall elongation by mecillinam stimulates cell division in certain cell division conditional mutants of Escherichia coli. J. Bacteriol. 157:130-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, D. S., H. Yamada, T. Mizuno, and S. Mizushima. 1986. Trimeric structure and localization of the major lipoprotein in the cell surface of Escherichia coli. J. Biol. Chem. 261:8953-8957. [PubMed] [Google Scholar]
- 9.Choi, D. S., H. Yamada, T. Mizuno, and S. Mizushima. 1987. Molecular assembly of the lipoprotein trimer on the peptidoglycan layer of Escherichia coli. J. Biochem. (Tokyo) 102:975-983. [DOI] [PubMed] [Google Scholar]
- 10.de Pedro, M. A., W. D. Donachie, J. V. Höltje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 183:4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Pedro, M. A., J. C. Quintela, J. V. Höltje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Pedro, M. A., K. D. Young, J. V. Höltje, and H. Schwarz. 2003. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dmitriev, B. A., F. V. Toukach, K. J. Schaper, O. Holst, E. T. Rietschel, and S. Ehlers. 2003. Tertiary structure of bacterial murein: the scaffold model. J. Bacteriol. 185:3458-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foley, M., J. M. Brass, J. Birmingham, W. R. Cook, P. B. Garland, C. F. Higgins, and L. I. Rothfield. 1989. Compartmentalization of the periplasm at cell division sites in Escherichia coli as shown by fluorescence photobleaching experiments. Mol. Microbiol. 3:1329-1336. [DOI] [PubMed] [Google Scholar]
- 15.Freudl, R., H. Schwarz, M. Klose, N. R. Movva, and U. Henning. 1985. The nature of information, required for export and sorting, present within the outer membrane protein OmpA of Escherichia coli K-12. EMBO J. 4:3593-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgopapadakou, N. H., S. A. Smith, and R. B. Sykes. 1982. Mode of action of azthreonam. Antimicrob. Agents Chemother. 21:950-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glauner, B., J. V. Höltje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088-10095. [PubMed] [Google Scholar]
- 18.Hancock, R. E. W., D. N. Karunaratne, and C. Bernegger-Egli. 1994. Molecular organization and structural role of outer membrane macromolecules, p. 263-279. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 19.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Höltje, J. V., and U. Schwarz. 1985. Biosynthesis and growth of the murein sacculus, p. 77-119. In N. Nanninga (ed.), Molecular cytology of Escherichia coli. Academic Press Inc., New York, N.Y.
- 22.Inouye, M., J. Shaw, and C. Shen. 1972. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J. Biol. Chem. 247:8154-8159. [PubMed] [Google Scholar]
- 23.Koch, A. L. 1983. The surface stress theory of microbial morphogenesis. Adv. Microb. Physiol. 24:301-366. [DOI] [PubMed] [Google Scholar]
- 24.Labischinski, H., and H. Maidhof. 1994. Bacterial peptidoglycan: overview and evolving concepts, p. 23-38. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lazzaroni, J. C., and R. Portalier. 1992. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol. Microbiol. 6:735-742. [DOI] [PubMed] [Google Scholar]
- 27.Lennox, E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 28.Lybarger, S. R., and J. R. Maddock. 2001. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 183:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddock, J. 1994. The control of spatial organization during cellular differentiation. Cell. Mol. Biol. Res. 40:199-205. [PubMed] [Google Scholar]
- 30.Maddock, J. R., M. R. Alley, and L. Shapiro. 1993. Polarized cells, polar actions. J. Bacteriol. 175:7125-7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 32.Mengin-Lecreulx, D., J. van Heijenoort, and J. T. Park. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate: l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno, T., R. Kagiyama, and M. Kageyama. 1982. The peptidoglycan-associated lipoprotein (PAL) of the Proteus mirabilis outer membrane: characterization of the peptidoglycan-associated region of PAL. J. Biochem. (Tokyo) 91:19-24. [DOI] [PubMed] [Google Scholar]
- 34.Mizushima, S. 1987. Assembly of membrane proteins, p. 163-185. In M. Inouye (ed.), Bacterial outer membranes as model systems. John Wiley & Sons, New York, N.Y.
- 35.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osborn, M. J., J. E. Gander, E. Parisi, and J. Carson. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 247:3962-3972. [PubMed] [Google Scholar]
- 38.Osborn, M. J., and H. C. Wu. 1980. Proteins of the outer membrane of gram-negative bacteria. Annu. Rev. Microbiol. 34:369-422. [DOI] [PubMed] [Google Scholar]
- 39.Park, J. T. 1996. The convergence of murein recycling research with beta-lactamase research. Microb. Drug Resist. 2:105-112. [DOI] [PubMed] [Google Scholar]
- 40.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, J. T., and L. Burman. 1973. FL-1060: a new penicillin with a unique mode of action. Biochem. Biophys. Res. Commun. 51:863-868. [DOI] [PubMed] [Google Scholar]
- 42.Prats, R., and M. A. de Pedro. 1989. Normal growth and division of Escherichia coli with a reduced amount of murein. J. Bacteriol. 171:3740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothfield, L. I., and W. R. Cook. 1988. Periseptal annuli: organelles involved in the bacterial cell division process. Microbiol. Sci. 5:182-185. [PubMed] [Google Scholar]
- 44.Rothfield, L. I., T. J. MacAlister, and W. R. Cook. 1987. Murein-membrane interactions in cell division, p. 247-275. In S. Inouye (ed.), Bacterial outer membranes as model systems. John Wiley & Sons, New York, N.Y.
- 45.Salton, M. R. J. 1994. The bacterial cell envelope—a historical perspective, p. 1-22. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 46.Schwarz, U., and W. Leutgeb. 1971. Morphogenetic aspects of murein structure and biosynthesis. J. Bacteriol. 106:588-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Signoretto, C., F. Di Stefano, and P. Canepari. 1996. Modified peptidoglycan chemical composition in shape-altered Escherichia coli. Microbiology 142:1919-1926. [DOI] [PubMed] [Google Scholar]
- 48.Sonntag, I., H. Schwarz, Y. Hirota, and U. Henning. 1978. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 136:280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spratt, B. G. 1977. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur. J. Biochem. 72:341-352. [DOI] [PubMed] [Google Scholar]
- 51.Spratt, B. G., and A. B. Pardee. 1975. Penicillin-binding proteins and cell shape in E. coli. Nature 254:516-517. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, H., Y. Nishimura, S. Yasuda, A. Nishimura, M. Yamada, and Y. Hirota. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 167:1-9. [DOI] [PubMed] [Google Scholar]
- 53.Waravdekar, V. S., and L. D. Saslaw. 1959. A sensitive colorimetric method for the estimation of 2-deoxy sugars with the use of the malonaldehyde-thiobarbituric acid reaction. J. Biol. Chem. 234:1945-1950. [PubMed] [Google Scholar]
- 54.Weidel, W., and H. Pelzer. 1964. Bagshaped macromolecules—a new outlook on bacterial cell walls. Adv. Enzymol. 26:193-232. [DOI] [PubMed] [Google Scholar]
- 55.Woldringh, C. L., P. Huls, E. Pas, G. H. Brakenhoff, and N. Nanninga. 1987. Topography of peptidoglycan synthesis during elongation and polar cap formation in a cell division mutant of Escherichia coli. J. Gen. Microbiol. 133:575-586. [Google Scholar]
- 56.Yem, D. W., and H. C. Wu. 1978. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J. Bacteriol. 133:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, F., H. Furukawa, K. Nakamura, and S. Mizushima. 1984. Mechanism of localization of major outer membrane lipoprotein in Escherichia coli. Studies with the OmpF-lipoprotein hybrid protein. J. Biol. Chem. 259:6013-6018. [PubMed] [Google Scholar]