This work reports quantitative membrane proteomic analysis and provides insights into the mechanism of PHO2-mediated Pi acquisition via posttranslational regulation of PHT1s. The results highlight the fundamental role of PHO2 in orchestrating Pi acquisition at the root surface and Pi loading at the xylem of root stele to maintain Pi homeostasis.
Abstract
MicroRNA399-mediated regulation of the ubiquitin-conjugating enzyme UBC24/PHOSPHATE2 (PHO2) is crucial for Pi acquisition and translocation in plants. Because of a potential role for PHO2 in protein degradation and its association with membranes, an iTRAQ (for isobaric tags for relative and absolute quantitation)- based quantitative membrane proteomic method was employed to search for components downstream of PHO2. A total of 7491 proteins were identified from Arabidopsis thaliana roots by mass spectrometry, 35.2% of which were predicted to contain at least one transmembrane helix. Among the quantifiable proteins, five were significantly differentially expressed between the wild type and pho2 mutant under two growth conditions. Using immunoblot analysis, we validated the upregulation of several members in PHOSPHATE TRANSPORTER1 (PHT1) family and PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) in pho2 and demonstrated that PHO2 mediates the degradation of PHT1 proteins. Genetic evidence that loss of PHF1 or PHT1;1 alleviated Pi toxicity in pho2 further suggests that they play roles as downstream components of PHO2. Moreover, we showed that PHO2 interacts with PHT1s in the postendoplasmic reticulum compartments and mediates the ubiquitination of endomembrane-localized PHT1;1. This study not only uncovers a mechanism by which PHO2 modulates Pi acquisition by regulating the abundance of PHT1s in the secretory pathway destined for plasma membranes, but also provides a database of the membrane proteome that will be widely applicable in root biology research.
INTRODUCTION
P is one of the nutrients essential for all forms of life. It is a component of fundamental macromolecules, including nucleic acids and phospholipids, and participates in energy transfer, regulation of enzyme reactions, and metabolic pathways. Despite limited availability of Pi, a major acquired form of P in the soil, plants have to maintain cellular Pi homeostasis; this is achieved by coordination of Pi sensing and signaling, Pi uptake, and allocation and recycling coupled with the regulation of growth and development (Raghothama, 1999; Poirier and Bucher, 2002; Chiou and Lin, 2011).
Several mutants with abnormal cellular Pi concentrations have been identified, including those impaired in Pi uptake and translocation or the upstream regulatory factors (Lin et al., 2009). Among them, phosphate2 (pho2) mutants accumulate considerably higher levels of Pi in leaves (Delhaize and Randall, 1995). PHO2, which encodes a ubiquitin-conjugating E2 enzyme (UBC24) presumably implicated in protein degradation, functions as a repressor that prevents excessive accumulation of Pi by controlling Pi uptake and root-to-shoot Pi translocation (Aung et al., 2006; Bari et al., 2006). Expression of PHO2 is indispensable for maintenance of Pi homeostasis, especially when the Pi supply is ample. Upon Pi limitation, PHO2 is suppressed by upregulated levels of microRNA399 (miR399), which directs the cleavage of PHO2 transcripts and thereby activates Pi uptake and root-to-shoot translocation (Fujii et al., 2005; Chiou et al., 2006). Upregulation of miR399 by Pi deficiency is partially a result of the activation of PHOSPHATE STARVATION RESPONSE1 (PHR1), a key transcriptional regulator of Pi starvation responses (Rubio et al., 2001; Bari et al., 2006). Loss-of-function of PHO2, attributable to either a nonsense mutation or a T-DNA insertion, as well as suppression of PHO2 by overexpression of miR399, results in Pi toxicity (Aung et al., 2006; Chiou et al., 2006). Because of the identification of homologs of miR399 and PHO2 in diverse plant species and the inverse relationship of their expression patterns in response to Pi deficiency, the regulatory pathway mediated by miR399 and PHO2 is thought to be evolutionarily conserved and of biological importance (Kuo and Chiou, 2011). Results of reciprocal grafting further revealed that miR399 in the shoots serves as a long-distance signal of Pi starvation, moving through the phloem to suppress PHO2 that functions in the roots (Lin et al., 2008; Pant et al., 2008). This systemic regulation represents an ideal strategy for long-distance communications between shoots and roots to balance the demand and supply of Pi (Liu et al., 2009).
Despite the crucial role of miR399 and PHO2 in regulating Pi uptake and root-to-shoot translocation, the regulatory pathway downstream of PHO2 is not fully understood. To uncover downstream components of the PHO2-dependent regulatory pathway, we have undertaken two approaches. One is to identify pho2 suppressors; the other is to employ quantitative membrane proteomics. Our recent analysis of the pho2 suppressors revealed PHOSPHATE1 (PHO1) as a direct downstream target of PHO2 by showing that PHO2 mediates the degradation of PHO1 in the endomembrane (EM) (Liu et al., 2012). PHO1, which encodes an integral membrane protein, is involved in Pi loading into the xylem (Hamburger et al., 2002). We found that the PHO1 protein level is radically increased in pho2, which explains the increased activity of root-to-shoot translocation of Pi. Given that PHO1 is predominately expressed in the pericycle of roots and not responsible for direct Pi uptake from the rhizosphere (Hamburger et al., 2002; Liu et al., 2012) and that overexpression of PHO1 is not sufficient to reproduce the Pi toxicity phenotype of pho2 (Liu et al., 2012), there must be additional components regulated by PHO2.
In this article, we present the results from a quantitative proteomics analysis, in which the membrane proteins differentially expressed between Arabidopsis thaliana wild-type plants and pho2 mutants were identified. This approach was motivated by several observations. First, PHO2 potentially plays a role in regulating protein degradation. Second, PHO2 suppresses the activities of Pi uptake and translocation, the biological events in which membrane proteins are thought to be engaged. Third, PHO2 is a membrane-associated protein (Liu et al., 2012; Figure 1). We attempted not only to reveal downstream effectors of PHO2 but also to improve the inventory of the existing membrane proteome databases of Arabidopsis.
Figure 1.
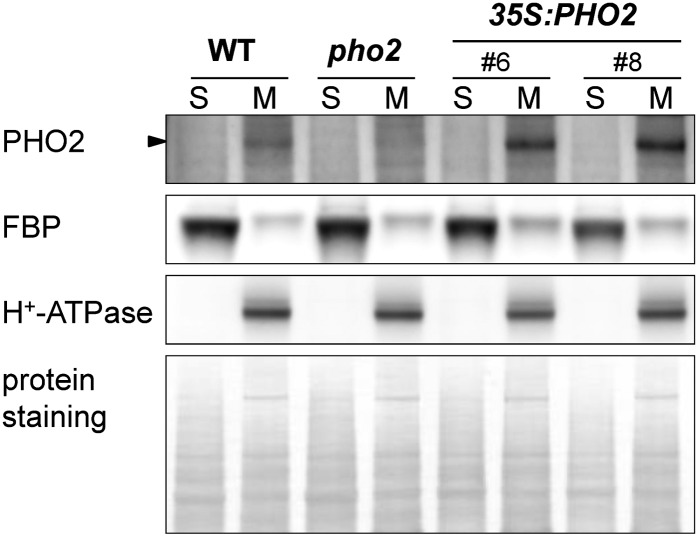
PHO2 Is a Membrane-Associated Protein.
Immunoblot analysis of PHO2 protein in the soluble (S) and membrane (M) fractions isolated from the roots of wild-type (WT), pho2, and two independent PHO2-overexpressing lines (35S:PHO2). Fructose-1,6-bisphosphatase (FBP) and H+-ATPase served as markers for soluble and membrane proteins, respectively. The protein staining of the blot is shown below the immunoblot.
We identified 7491 proteins from Arabidopsis roots, of which 3713 (49.6%) are annotated or predicted to be membrane proteins. Notably, among the 5539 quantified proteins, several Pi transporters in the PHOSPHATE TRANSPORTER1 (PHT1) family (PHT1;1, PHT1;2, PHT1;3, and PHT1;4) and PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) are upregulated in pho2 roots. Our subsequent validation by immunoblot along with analyses of protein stability, protein–protein interaction, and in vivo ubiquitination revealed that PHO2 operates at the EM to promote the degradation of PHT1 proteins and thereby regulates the trafficking of PHT1 protein toward the plasma membrane (PM), where Pi uptake takes place. The results from our two complementary approaches elucidate the molecular mechanism by which PHO2 regulates both activities of Pi uptake and root-to-shoot Pi translocation.
RESULTS
PHO2 Is a Membrane Protein Predominately Associated with the Endoplasmic Reticulum and Golgi
In order to minimize the complexity of proteome data and improve the detection of target proteins of low abundance, we first examined in which cellular fraction PHO2 protein is preferentially located. By immunoblot analysis using the anti-PHO2 antibody (Aung et al., 2006), we detected the native PHO2 protein in the microsomal fraction isolated from the roots of the wild type but not from pho2 (Figure 1). By contrast, no signal was observed in the soluble fraction. In addition, we observed an increased level of PHO2 protein in the membrane but not the soluble fraction isolated from the roots of PHO2-overexpressing transgenic plants (Figure 1). As PHO2 is not predicted to possess any transmembrane helix domain, our results showing PHO2 enrichment in the membrane fraction suggest that PHO2 is a peripheral membrane protein.
Our previous analysis of the subcellular localization of PHO2 by examining the C-terminal fluorescent protein–tagged PHO2 showed both reticular and spotted patterns (Liu et al., 2012). To confirm this, we further examined the colocalization of the N-terminal fluorescent protein–tagged PHO2 in Arabidopsis protoplasts and performed statistical analysis of the subcellular colocalization of PHO2 and several subcellular markers (see Supplemental Figure 1A online). Similarly to the C-terminal fluorescent protein–tagged PHO2, the N-terminal fluorescent fusion protein was not observed at the PM or tonoplast. While the reticular pattern of PHO2 signal largely overlapped with the signals from the endoplasmic reticulum (ER) markers At-WAK2 (Nelson et al., 2007) and PHF1 (González et al., 2005; Bayle et al., 2011) (Pearson’s correlation coefficient, r = 0.74 to 0.72), the punctate signal of PHO2 mainly colocalized with the markers for Golgi (r = 0.59 to 0.53) but to a lesser extent with the markers for the trans-Golgi network (r = 0.40), early endosomes (r = 0.44), and late endosomes (r = 0.31). Hence, we conclude that PHO2 is a peripheral membrane protein predominately associated with the ER and Golgi.
Interestingly, although both the N- and C-terminal fluorescent protein fusions of PHO2 are functional proteins capable of complementation of pho2 by reducing the shoot Pi concentration when expressed under its native promoter (see Supplemental Figure 1B online; Liu et al., 2012), we failed to observe a fluorescent signal for PHO2 in these complemented lines. The difficulty of detection of the PHO2 fluorescent fusions in planta has also been reported even when the inducible system was used (Eifler, 2010). We speculate that the PHO2 fluorescent fusion protein may be present at a very low level below the detection limit owing to its rapid protein degradation, and this possibility remains to be further examined.
Large-Scale Profiling of the Membrane Proteome in Arabidopsis Roots
Given the membrane localization of PHO2 and potential engagement of membrane proteins in PHO2-mediated activities, we focused on the comparison analysis of the membrane proteomes of the wild type and pho2 by applying the method that we previously developed based on gel-assisted digestion: isobaric tags for relative and absolute quantitation (iTRAQ) technology combined with off-line two-dimensional liquid chromatography–tandem mass spectrometry (LC-MS/MS) (Han et al., 2008). Since the function of PHO2 in the root is responsible for the Pi toxicity phenotype (Bari et al., 2006; Lin et al., 2008), root samples were used for analysis. The 12-d-old and 17-d-old roots grown under solid agar and hydroponic media containing 250 μM KH2PO4 were collected for isolation of membrane proteins. The elevated Pi level in the shoots of pho2 was confirmed for both growth regimes (see Supplemental Figures 2A and 2B online). After gel-assisted digestion, peptides from different samples were extracted and labeled with iTRAQ reagents, combined, and separated by strong cation exchange chromatography into 38 fractions. Each strong cation exchange fraction was desalted and analyzed in duplicate by mass spectrometry. Three biological replicates were conducted for each growth condition. The raw data generated from the liquid chromatography–mass spectrometry analysis were processed to detect MS/MS peaks using UniQua (Chang et al., 2013) followed by the protein identification and quantification using MASCOT version 2.3 (Matrix Science). The processed spectra were searched against the TAIR10_pep_20110103_representative_gene_model database (December 2011; 27,416 sequences; ftp://ftp.arabidopsis.org/home/tair/Proteins/TAIR10_protein_lists/). Details of the liquid chromatography–mass spectrometry data processing are described in the Supplemental Methods 1 online.
After combining total protein of four samples (agar-grown and hydroponic-grown wild-type and pho2 roots) from three biological replicates, we identified 659,511 peptide spectra corresponding to 7491 proteins with confidence (criteria are described in Supplemental Methods 1 online). Next, we employed various tools to annotate the membrane proteins identified. First, Transmembrane Hidden Markov Model (TMHMM) Server version 2.0 algorithm (Krogh et al., 2001) was used to predict the proteins containing transmembrane helices. A total of 2639 (35.2%) proteins were predicted to contain at least one transmembrane helix and are likely to be integral membrane proteins. In addition, subcellular localization was classified using the Gene Ontology (GO) database (GO:TermFinder) (Boyle et al., 2004) and SUBA (Arabidopsis Subcellular Database) (Heazlewood et al., 2007; Tanz et al., 2013). The posttranslational modifications associated with membrane anchoring, such as myristoylation, palmitoylation, prenylation, and glycosylphosphatidylinositol anchors, were also predicted. Collectively, these analyses yielded 4890 (65.3%) membrane or putative membrane proteins (see Supplemental Data Set 1 online). To increase confidence for those proteins without a transmembrane helix, we considered the prediction reliable only if the protein was present in at least two analyses. Using this principle, a total of 3713 (49.6%) proteins were classified as membrane or membrane-associated proteins (Figure 2A). As the genome frequency of annotated membrane proteins predicted by TMHMM and GO is 24.5 and 16.7%, respectively, our results displayed a higher frequency of annotated membrane proteins (to 35.2% for TMHMM and 38.8% for GO), suggesting enrichment of membrane proteins in our analysis. Nevertheless, validation of membrane localization for those noncharacterized proteins will be required. Furthermore, we searched our data set against the most updated database of proteomic and green fluorescent protein (GFP) localization sets (April, 2013) in SUBA (Heazlewood et al., 2007; Tanz et al., 2013) and observed that 3489 (46.6%) proteins do not exist in the mass spectrometry database and, therefore, likely represent newly identified proteins. Identification of such large numbers of membrane proteins demonstrated the depth of our data set and the uniqueness of our methodology. The detailed descriptions including the protein scores and mass, annotation, predicted transmembrane helices, and quantification for the proteins identified are listed in Supplemental Data Set 1 online.
Figure 2.
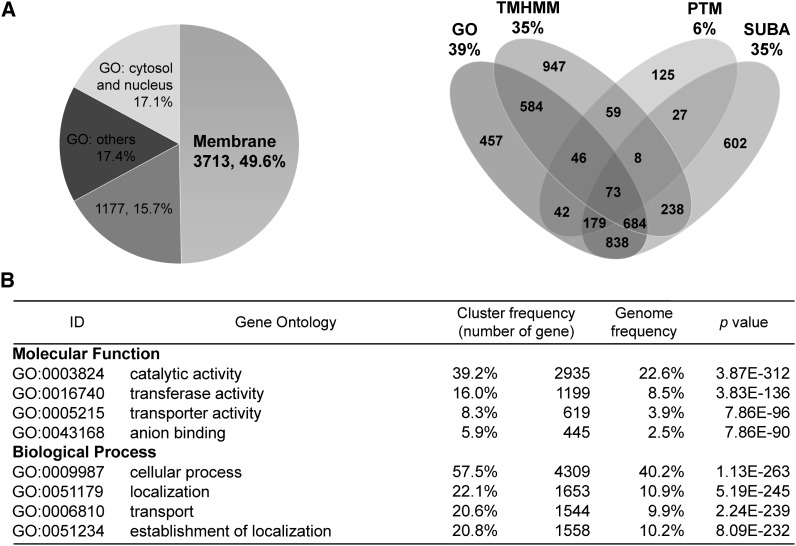
Analysis of 7491 Identified Proteins from Membrane Proteomics.
(A) The left panel illustrates the distribution of subcellular localization of total identified proteins. Based on our criterion, a total of 3713 (49.6%) proteins containing at least one transmembrane helix or present in at least two databases or analyses are classified as membrane or membrane-associated proteins. A total of 1177 (15.7%) proteins are present only in one database or analysis. The number and percentage of annotated or predicted membrane proteins revealed by the analysis of GO category, TMHMM, SUBA, and posttranslational modification (PTM) associated with membrane anchoring are shown on the right.
(B) The significantly enriched GO terms in the categories of molecular function and biological process. The cluster frequency and genome frequency indicate the proportion of identified proteins and total proteins annotated in the specific group versus the total annotated Arabidopsis proteins, respectively. The Bonferroni adjustment was applied to correct the P values.
We next subjected the 7491 identified proteins to GO analysis. Several GO terms were significantly enriched, including catalytic activity (GO:0003824), transferase activity (GO:0016740), transporter activity (GO:0005215), and anion binding (GO:0043168) in the molecular function category as well as cellular process (GO:0009987), localization (GO:0051179), transport (GO:0006810), and establishment of localization (GO:0051234) in the biological process category (Figure 2B). Many of these categories are tightly associated with membrane proteins.
Membrane Proteins Differentially Expressed in pho2 Roots
For quantitative proteomic comparison between the wild-type and pho2 roots, a total of 5539 proteins with at least two identified peptides were quantified. Among them, 2081 proteins were identified from all three biological replicates and subjected to statistical analysis by one-tailed Student’s t test. We considered the difference in protein amount to be significant when the P value was <0.05 and there was at least a 1.3-fold change (Table 1). Using these criteria, we identified 59 and 20 proteins differentially expressed between the wild-type and the pho2 roots grown in solid agar and in hydroponic culture, respectively (see Supplemental Data Set 2 online).
Table 1. Differentially Expressed Proteins in pho2 Roots Analyzed from Two Sets of Samples.
| Accession No. | MASCOT Scorea | Ratio 1 (pho2/Wild Type)b |
Ratio 2 (pho2/Wild Type)b |
Description | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | Meanc | P Valued | R1 | R2 | R3 | Meanc | P Valued | |||
| AT5G43350.1 | 9467 | 1.34 | 2.17 | 1.79 | 1.73 | 0.043 | 1.79 | 3.42 | 3.29 | 2.72 | 0.030 | PHT1;1 phosphate transporter |
| AT5G43370.1 | 8915 | 2.21 | 3.14 | 3.79 | 2.97 | 0.015 | 2.33 | 1.96 | 4.49 | 2.74 | 0.041 | PHT1;2 phosphate transporter |
| AT5G43360.1 | 3357 | 1.90 | 1.70 | 1.51 | 1.70 | 0.012 | 1.76 | 2.88 | 2.53 | 2.34 | 0.021 | PHT1;3 phosphate transporter |
| AT2G38940.1e | 745 | 1.79 | 2.62 | 2.20 | 2.18 | 0.014 | 0.66 | 1.78 | 1.63 | 1.24 | 0.317 | PHT1;4 phosphate transporter |
| AT3G52190.1e | 1019 | 1.45 | 1.38 | 1.22 | 1.35 | 0.021 | 1.17 | 1.30 | 1.27 | 1.25 | 0.016 | PHF1 |
| AT3G23430.1f | 87 | − | − | 1.57 | − | − | − | − | 1.47 | − | − | PHO1 |
| AT1G01580.1 | 48 | 0.57 | 0.52 | 0.50 | 0.53 | 0.003 | 0.65 | 0.45 | 0.63 | 0.57 | 0.031 | FRO2 |
| AT1G12110.1 | 1543 | 0.59 | 0.69 | 0.48 | 0.58 | 0.025 | 0.69 | 0.59 | 0.55 | 0.61 | 0.014 | NRT1.1/CHL1 nitrate transporter |
Average MASCOT score from replicates.
Ratio 1 and Ratio 2 represent the protein ratios of pho2 divided by the wild type derived from the solid agar and the hydroponic systems, respectively.
Geometric mean of ratios from three biological replicates. Significant changes with P value < 0.05 and ratio > 1.3 or < 0.77 are marked in bold.
P value is calculated based on a one-tailed Student’s t test.
PHT1;4 and PHF1 meet the criteria only in Ratio 1.
PHO1 was identified only in the third replicate (R3). — indicates undetected or undefined.
We found five proteins displaying a reproducible trend of change across the two different samples grown in the solid agar and hydroponic systems in all three biological replicates (Table 1). Among them, three proteins were upregulated and two were downregulated in the pho2 mutant. The protein levels of several Pi transporters, PHT1;1, PHT1;2, and PHT1;3, in the PHT1 family were increased in the pho2 roots. Members of the PHT1 family localize to the PM and are thought to account for Pi uptake directly from the rhizosphere (Muchhal et al., 1996; Chiou et al., 2001; Poirier and Bucher, 2002). When examining other annotated Pi transporters or Pi transport–related proteins in the list, we found that PHT1;4 and PHF1 proteins were upregulated in the agar-grown pho2 roots, yet did not show significant changes in the hydroponic samples (Table 1). PHF1 functions to facilitate the exit of PHT1 protein out of the ER and subsequent targeting to the PM (González et al., 2005; Bayle et al., 2011). Because the upregulation of PHT1s and PHF1 proteins may be closely associated with the increased Pi uptake activity in pho2 mutants, we analyzed PHT1;4 and PHF1 along with other Pi transporters in the following analyses. It is worth noting that although PHO1 was identified only in the third biological replicate; its amount was increased in pho2 roots grown under both growth conditions (Table 1). This result reconfirms our previous finding that PHO2 directs the degradation of PHO1 (Liu et al., 2012).
Next, we investigated whether the alteration in PHT1 protein abundance in pho2 (Table 1) resulted from changes in gene expression at the transcript level. By quantitative RT-PCR analysis, we found that the transcriptional levels of PHT1;2, PHT1;3, and PHT1;4 were increased in younger seedlings, likely attributable to the low Pi concentration in the roots of younger seedlings of pho2 (see Supplemental Figures 2A and 2C online). A detailed comparison between the transcript and protein levels of PHT1s in hydroponically grown older seedlings is described below.
Loss of Function of PHF1 and PHT1;1 Alleviates the Pi Toxicity of pho2
To examine whether the increased levels of PHT1 and PHF1 proteins are responsible for Pi toxicity of pho2, we employed genetic approaches by analyzing the pht1;1 pho2 and phf1 pho2 double mutants. In contrast with the elevated Pi concentration of the pho2 shoots, the pht1;1 and phf1 single mutants displayed reduced shoot Pi concentrations compared with the wild type (Figure 3A) (Shin et al., 2004; González et al., 2005). Introduction of the mutation of PHT1;1 or PHF1 into pho2, giving rise to the pht1;1 pho2 or phf1 pho2 mutant, significantly reduced the shoot Pi concentration of pho2 by 14 or 76%, respectively (Figure 3A). Compared with PHT1;1, loss of function of PHF1 had a greater impact on reduction of shoot Pi in the pho2 mutant. The shoot Pi level in the phf1 mutant was also lower than that of the pht1;1 mutant. This could be attributed to the regulatory role of PHF1 in controlling the expression of multiple PHT1 proteins besides PHT1;1 (Bayle et al., 2011). Interestingly, pht1;1 pho2 and phf1 pho2 mutants had higher shoot Pi concentrations than the respective pht1;1 and phf1 single mutants, suggesting that additional factors, such as other PHT1 members revealed by our quantitative proteomic data (Table 1) or PHF1 homologs, may contribute to shoot Pi accumulation of pho2. Taken together, the genetic interaction of PHO2 with PHF1 and PHT1;1 supports the role of PHF1 and PHT1;1 in contributing to the high shoot Pi level of pho2.
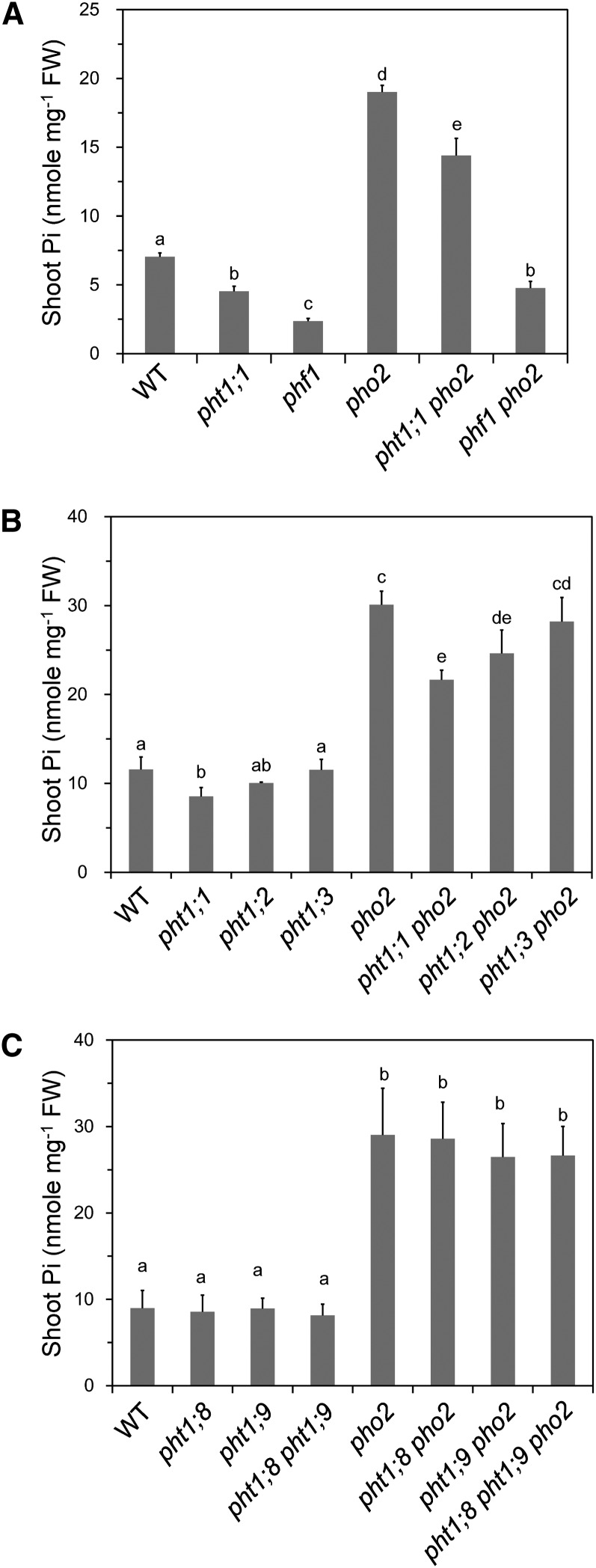
Figure 3.
Loss of Function of PHF1 and PHT1;1 Alleviates the Pi Toxicity of pho2.
(A) The shoot Pi concentrations of 12-d-old wild type (WT), pht1;1, phf1, pho2, pht1;1 pho2, and phf1 pho2 (n = 4).
(B) PHT1;1 has a greater contribution than PHT1;2 or PHT1;3 to the Pi accumulation in pho2. The shoot Pi concentrations of 13-d-old wild-type, pht1;1, pht1;2, pht1;3, pho2, pht1;1 pho2, pht1;2 pho2, and pht1;3 pho2 seedlings grown under Pi-sufficient conditions (n = 3).
(C) PHT1;8 and PHT1;9 do not contribute to the Pi accumulation in pho2. The shoot Pi concentration of 13-d-old wild-type, pht1;8, pht1;9, pho2, pht1;8 pho2, pht1;9 pho2, and pht1;8 pht1;9 pho2 seedlings grown in Pi-sufficient media (n = 4).
Ten seedlings were pooled as one replicate. The different letters indicate the statistical significance of variations analyzed by Duncan’s multiple range test (P < 0.05), and error bars represent sd. FW, fresh weight.
To investigate to what extent the other major PHT1 members, PHT1;2, PHT1;3, and PHT1;4, may contribute to the phenotype of pho2, we generated the pht1;2 pho2 and pht1;3 pho2 double mutants; generation of pht1;4 pho2 by crossing is not feasible because of the close proximity of the PHT1;4 and PHO2 genes on chromosome 2. Under Pi-sufficient conditions, we found that the shoot Pi concentrations were pht1;1 pho2 < pht1;2 pho2 < pht1;3 pho2 (Figure 3B), indicating that PHT1;1 makes a greater contribution than PHT1;2 or PHT1;3 to the Pi accumulation in the pho2 mutant. This may be explained by a much higher transcript level of PHT1;1 relative to PHT1;2 and PHT1;3 when Pi is adequate (see below).
Based on the previously observed increased mRNA level of PHT1;8 and PHT1;9 in pho2 roots, it was postulated that PHT1;8 and PHT1;9 are downstream components of PHO2 that contribute to Pi toxicity of the pho2 mutant (Aung et al., 2006; Bari et al., 2006). We did not identify these two proteins as candidates in our proteomic analysis and decided to examine their possible involvement using a genetic approach. Under Pi-sufficient conditions, the pht1;8 and pht1;9 single mutants did not display any apparent phenotype distinguishable from the wild type and had similar shoot Pi levels as the wild type. Notably, the shoot Pi concentrations of the pht1;8 pho2, pht1;9 pho2, and pht1;8 pht1;9 pho2 mutants remained as high as that of pho2 (Figure 3C). This result strongly argues against the involvement of PHT1;8 and PHT1;9 in the PHO2 regulatory pathway. We speculate that the upregulation of PHT1;8 and PHT1;9 transcripts is likely a result of secondary effects.
PHO2 Negatively Regulates the Protein Abundance of PHF1 and PHT1s under Pi-Sufficient Conditions
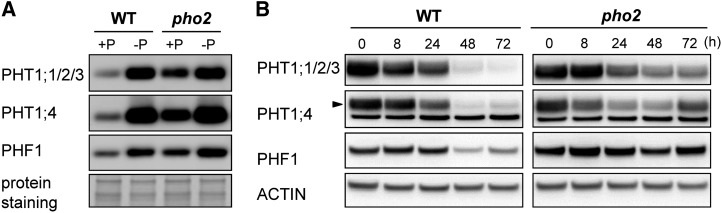
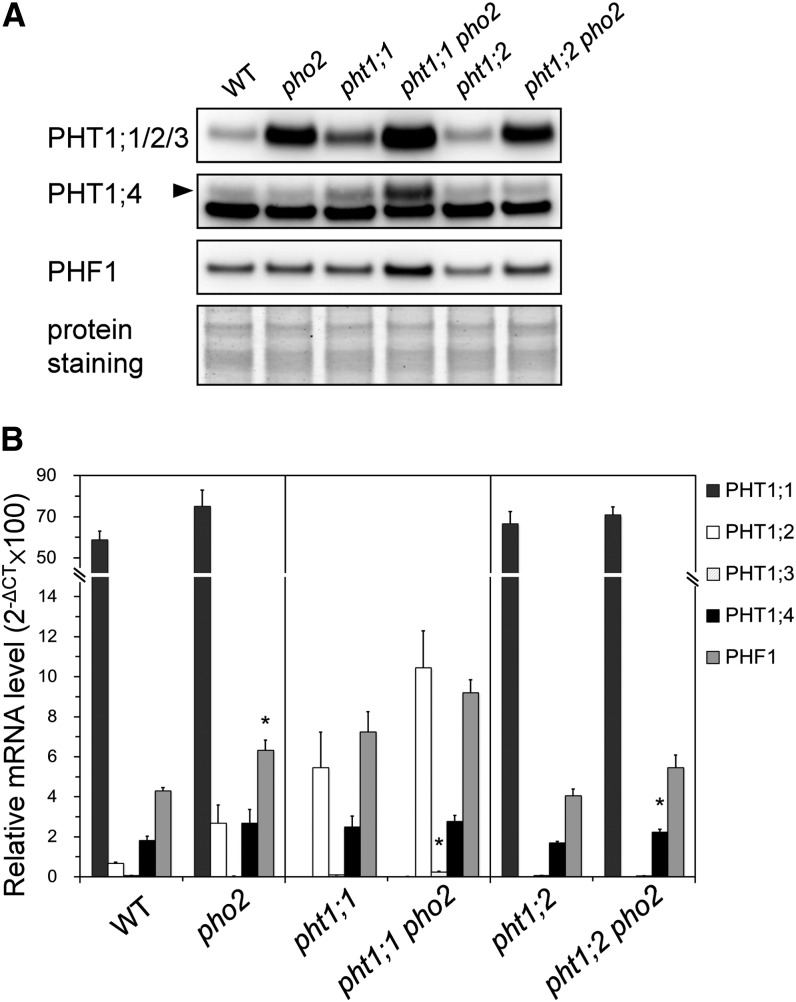
In order to validate the results obtained from membrane proteomics, we raised antibodies against PHT1;1, PHT1;2, PHT1;3, PHT1;4, and PHF1 using synthetic peptides. Because of the high amino acid sequence similarity among PHT1;1, PHT1;2, and PHT1;3, a common peptide was used to generate an antibody recognizing all three of them (Liu et al., 2011). The protein signals detected by immunoblot using these antibodies are specific because signals corresponding to the same size of protein in the T-DNA insertion mutants were not detected (phf1 and pht1;4) or reduced (pht1;1) compared with wild-type plants (see Supplemental Figures 3A and 3B online). Consistent with the data from quantitative membrane proteomics of 12-d-old seedlings, we found that the protein levels of PHT1;1/2/3, PHT1;4, and PHF1 in pho2 roots were higher than those in the wild-type controls under Pi-sufficient conditions (250 μM KH2PO4) (Figure 4A). The increased amount of PHT1 proteins in pho2 is in agreement with our previous observation of the increased Vmax of Pi uptake activity in pho2 (Aung et al., 2006). Under Pi-deficient conditions (10 μM KH2PO4), the amounts of these proteins were increased compared with the Pi-sufficient conditions, and the level was similar in pho2 and wild-type plants (Figure 4A). In other words, the increase of these proteins in pho2 was observed only under Pi-sufficient conditions; the differences between pho2 and the wild type became negligible when Pi was limited. This is in line with our previous finding that the difference of Pi uptake activity between pho2 and the wild type was drastic only under Pi-sufficient conditions (Aung et al., 2006).
Figure 4.
Increased Accumulation of PHF1 and PHT1 Proteins in the pho2 Mutant.
(A) Immunoblot analysis of PHT1;1/2/3, PHT1;4, and PHF1 proteins in the roots of 12-d-old wild-type and pho2 plants grown under Pi-sufficient (+P) and -deficient (−P) conditions. The bottom panels show the protein staining on the blot.
(B) Immunoblot analysis of PHT1;1/2/3, PHT1;4, and PHF1 proteins in the roots of Pi-starved wild-type and pho2 seedlings replenished with Pi over 72 h. The bands corresponding to PHT1;4 are indicated by an arrowhead. Actin is used as a loading control.
It has been shown that suppression of PHO2 by miR399 under Pi deficiency is reversed when Pi is resupplied to the medium (Bari et al., 2006). To examine the role of PHO2 in regulating the accumulation of PHT1 and PHF1 proteins upon Pi resupply, we examined the expression of these proteins following 3 d of Pi replenishment (250 μM KH2PO4) after 8 d of Pi starvation (10 μM KH2PO4). While the increased amounts of PHT1 proteins and PHF1 were comparable between the wild type and pho2 after the eighth day of starvation, a more rapid reduction of these proteins was observed in wild-type plants than in pho2 mutants after Pi replacement (Figure 4B). This suggests that PHO2 plays a role in downregulating the accumulation of these proteins once the Pi-starved plants sense the adequate resupply of Pi.
PHO2 Mediates Protein Degradation of PHT1 Proteins
In order to elucidate the role of PHO2 in downregulating the protein level of PHT1s and PHF1, we compared the mRNA and protein levels of PHT1s and PHF1 concurrently in the same sample of roots. In addition, because of the ambiguity resulting from transcriptional activation by reduced Pi concentrations in the roots of young pho2 seedlings (see Supplemental Figures 2A and 2C online), we focused on the analysis of hydroponically grown older seedlings [18 d old]) whose root Pi concentrations were comparable between the wild type and pho2. While the mRNA levels of PHT1;1, PHT1;2, and PHT1;3 showed no difference between wild-type and pho2 roots, these proteins increased significantly in the pho2 root (Figures 5A and 5B). Because the transcript level of PHT1;1 was much higher than those of PHT1;2 and PHT1;3 (more than 20-fold assuming that the amplification efficiency of different primer pairs are comparable), the protein signal detected by anti-PHT1;1/2/3 antibody in the wild type and pho2 may be contributed mainly by PHT1;1 (Figure 5A).
Figure 5.
PHO2 Negatively Regulates the Protein Level of PHF1 and PHT1s.
(A) Immunoblot analysis of PHT1;1/2/3, PHT1;4, and PHF1 proteins in the roots of 18-d-old wild-type (WT) and pho2, pht1;1, pht1;1 pho2, pht1;2, and pht1;2 pho2 plants grown under Pi-sufficient conditions. The bottom panel shows the protein staining on the blot.
(B) Quantitative RT-PCR analysis of PHT1;1, PHT1;2, PHT1;3, PHT1;4, and PHF1 mRNA in the roots of the wild type and mutants. Error bars represent se (n = 3). Data significantly different from the corresponding controls are marked by an asterisk (pho2 versus the wild type, pht1;1 pho2 versus pht1;1, or pht1;2 pho2 versus pht1;2, 0.01 < *P < 0.05, Student’s t test). CT, cycle threshold.
Interestingly, the protein signals detected by anti-PHT1;1/2/3 antibody in pht1;1 pho2 and pht1;2 pho2 double mutants remained relatively high when compared with the pht1;1 and pht1;2 single mutants, respectively (Figure 5A), supporting that PHO2 mediates the protein abundance of more than one PHT1 member as revealed by proteomic analysis. It is also worthwhile to mention that the signal detected by the anti-PHT1;1/2/3 antibody in the pht1;1 was stronger than in the wild type (Figure 5A). We suspected that this might result from upregulation of PHT1;2 and PHT1;3 in compensation for the loss of PHT1;1 (Figure 5B). However, such effect seems to be developmental stage dependent because it was not observed in younger seedlings (12 d old) (see Supplemental Figure 3A online).
Consistent with the proteomics result, immunoblot analysis of PHT1;4 showed no difference between the wild type and pho2 in the hydroponically grown samples. However, more PHT1;4 protein was accumulated in the pht1;1 pho2 than pht1;1 without any significant changes of the transcript level (Figures 5A and 5B). Additionally, without the elevated transcript level, we also detected an increase in PHF1 proteins in pht1;1 pho2 compared with pht1;1. On the basis of this comparison, we conclude that PHO2 regulates the expression of PHT1s and likely PHF1 as well at the protein level.
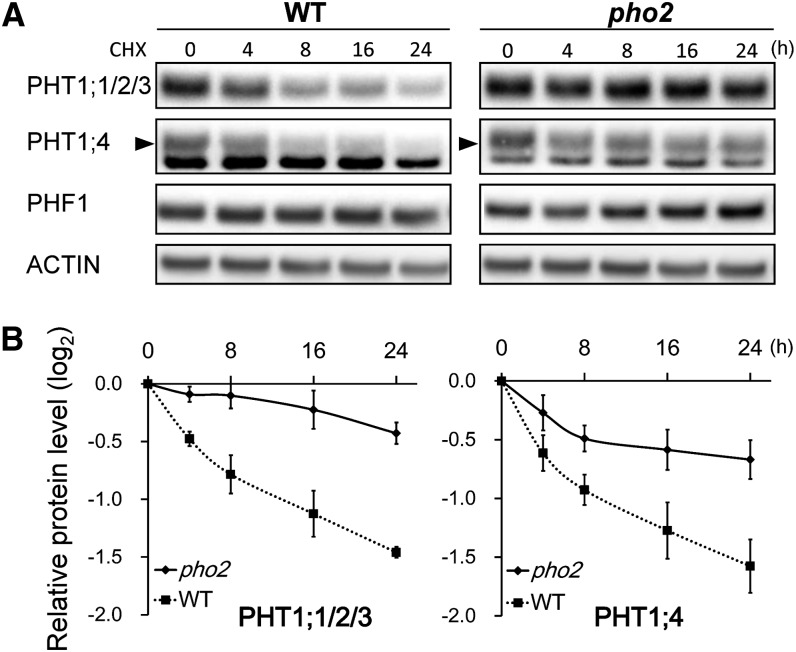
We next examined the protein stability of PHT1s and PHF1 by applying cycloheximide (CHX), an inhibitor of protein translation, to wild-type and pho2 seedlings grown under Pi-sufficient conditions. The expression of PHT1;1/2/3, PHT1;4, and PHF1 proteins in the root was examined at 4, 8, 16, and 24 h after CHX treatment. Notably, the CHX treatment resulted in a more rapid decline of PHT1;1/2/3 and PHT1;4 proteins in the wild type than in pho2 (Figures 6A and 6B), suggesting that PHO2 posttranslationally regulates these PHT1 proteins, probably via degradation. The half-life of PHT1;1/2/3 and PHT1;4 is estimated to be ∼12 h in the wild-type plant (Figure 6B). By contrast, the degradation rate of PHT1;1/2/3 and PHT1;4 in pho2 was much slower. To our surprise, the PHF1 protein was not affected by 24 h of CHX treatment in either wild-type or pho2 plants (Figure 6A). We speculate that either PHF1 proteins are relatively stable or degradation of a negative regulator of PHF1 during CHX treatment is involved.
Figure 6.
PHO2 Facilitates the Degradation of PHT1 Proteins.
(A) Time-course analysis of PHT1;1/2/3, PHT1;4, and PHF1 proteins in the roots of 12-d-old wild-type (WT) and pho2 seedlings subjected to the treatment with 200 µM CHX under Pi-sufficient conditions. Actin is used as a loading control. The bands corresponding to PHT1;4 are indicated by an arrowhead. One representative image out of 4 replicates is shown.
(B) The relative remaining amount of PHT1;1/2/3 and PHT1;4 proteins upon CHX treatment was calculated from (A) and plotted on a semilog graph. The protein level was normalized with the corresponding actin controls. Error bars represent se (n = 4).
PHO2 Interacts with PHT1 Proteins at the EM
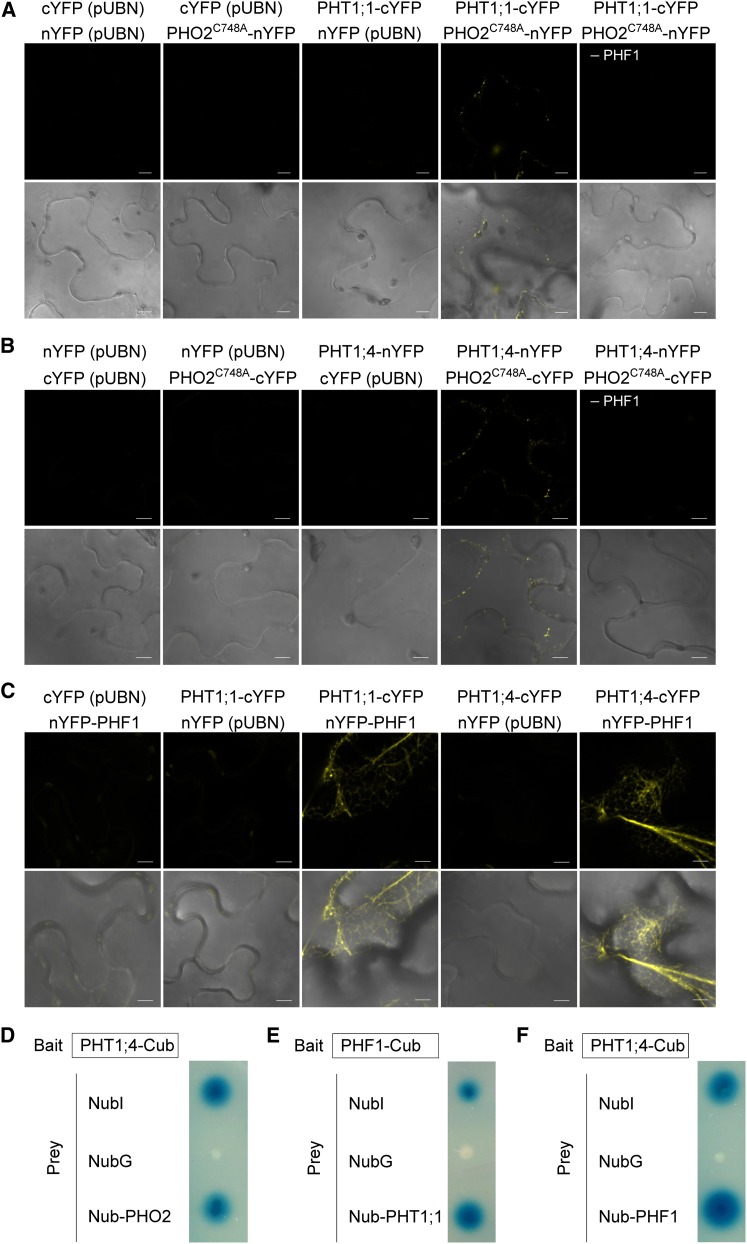
Because of the subcellular localization of PHO2 in the ER and Golgi compartments, through which PHT1s pass prior to their final destination of the PM, we next examined the physical interaction between these proteins. Bimolecular fluorescence complementation (BiFC) in tobacco (Nicotiana benthamiana) leaves using Agrobacterium tumefaciens infiltration and the split-ubiquitin yeast two-hybrid (Y2H) system were employed. To prevent protein degradation presumably mediated by the ubiquitin conjugase activity of PHO2, we generated a PHO2 variant (PHO2C748A) with a mutation in the conserved catalytic Cys residue in the UBC domain (Liu et al., 2012). When the interaction between PHO2 and PHT1;1 or PHT1;4 was examined, the reconstitution of yellow fluorescent protein signal was observed as intracellular dots only when coexpressed with PHF1, while no signal was observed in the corresponding negative controls (Figures 7A and 7B). This indicated that PHO2 is in close proximity to these PHT1s, hinting at a possible physical interaction. A previous study showed that coexpression of PHF1 with PHT1;2 in Agrobacterium-infiltrated tobacco leaves is critical to enhancing or stabilizing the expression of PHT1;2 as well as facilitating its targeting to the PM (Bayle et al., 2011). It is possible that PHF1-mediated ER exit of the PHT1s is a prerequisite to the potential interaction of PHO2 with the PHT1s at the post-ER compartments. The interaction of PHO2 and PHT1;4 was also validated by the split-ubiquitin Y2H system (Figure 7D). Unfortunately, we were unable to confirm the interaction of PHO2 and PHT1;1 in yeast cells for unknown reasons. We also attempted to examine the interaction between PHO2 and PHF1, but we could not draw any conclusion because of inconsistent results. Instead, we detected the interaction between PHF1 and the PHT1s in both BiFC and Y2H analyses (Figures 7C, 7E, and 7F), which corroborates the role of PHF1 in facilitating targeting of PHT1s to the PM (González et al., 2005; Bayle et al., 2011). The observation of interaction of PHF1 and the PHT1s in the ER (Figure 7C) was consistent with the subcellular localization of PHF1 in the ER and its function in assisting the exit of PHT1s out of the ER (González et al., 2005; Bayle et al., 2011).
Figure 7.
Protein–Protein Interactions between PHO2 and PHT1s and between PHF1 and PHT1s.
(A) to (C) BiFC analysis of PHF1-dependent interaction between PHO2 and PHT1;1 (A) or between PHO2 and PHT1;4 (B) in tobacco leaves. The PHF1 construct was coinfiltrated in all experiments except where indicated. BiFC analysis of the interaction between PHF1 and PHT1;1 or PHT1;4 in tobacco leaves (C). Coexpression of N-terminal yellow fluorescent protein (nYFP) or C-terminal yellow fluorescent protein (cYFP) with the corresponding PHT1s, PHO2C748A, or PHF1 constructs was used as negative controls. Bars = 10 µm.
(D) to (F) Split-ubiquitin Y2H analysis of the interaction between PHO2 and PHT1;4 (D), PHF1 and PHT1;1 (E), or PHF1 and PHT1;4 (F). Coexpression of NubI or NubG with PHT1;4 or PHF1 was used as positive and negative controls, respectively.
PHO2-Mediated Ubiquitination of PHT1;1 Proteins at the EM
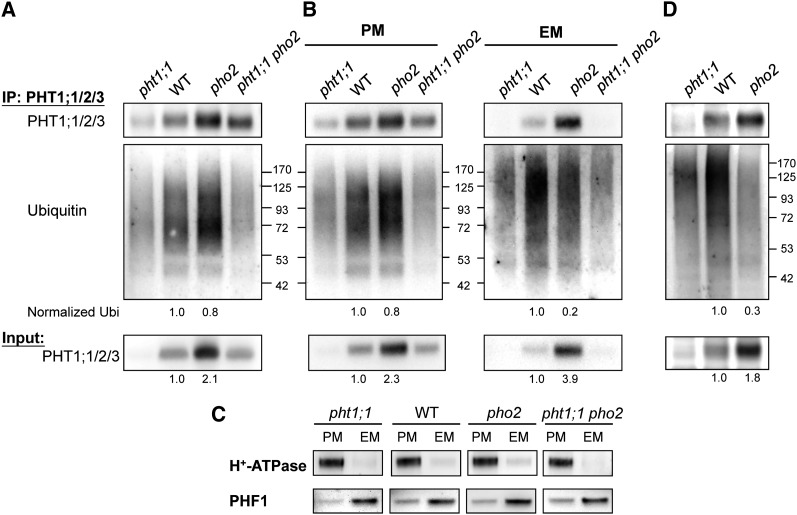
To examine whether PHO2 mediates ubiquitination of the PHT1s, we attempted to detect in vivo ubiquitinated proteins following immunoprecipitation. Total membrane proteins isolated from the Pi-replete roots of 12-d-old wild-type, pht1;1, and pho2 seedlings were subjected to immunoprecipitation analysis. Whereas immunoprecipitation using anti-PHT1;1/2/3 antibody was successful, anti-PHT1;4 and anti-PHF1 antibodies produced low signal or high background after immunoprecipitation. When probed with the antiubiquitin antibody, the immunoprecipitated proteins showed a smear pattern at the higher molecular weights in the wild-type sample, indicating that the presence of polyubiquitinated or multimonoubiquitinated PHT1;1/2/3 (Figure 8A). As the smeared signal was diminished in the samples of pht1;1 and pht1;1 pho2, loss of PHT1;1 greatly contributed to the reduced intensity (Figures 8A, 8B, and 8D). The remaining signal in pht1;1 and pht1;1 pho2 may result from the ubiquitination of PHT1;2 and PHT1;3. Interestingly, we did not observe a reduction in the level of ubiquitinated PHT1;1/2/3 in pho2 compared with the wild type. Considering that the EM rather than the PM is the location where PHO2 interacts with the PHT1s (see Supplemental Figure 1A online; Figure 7), we separated the total microsomal membranes into PM and EM fractions for immunoprecipitation analysis (Figure 8B). The PM and EM fractions were separated and enriched as shown by immunoblot using their respective protein markers (Figure 8C). Whereas the protein level of PHT1;1/2/3 was increased in both the PM and EM fractions of pho2 compared with the wild type, the level of ubiquitinated PHT1;1/2/3 protein was moderately decreased in the EM but not in the PM of pho2 (Figure 8B). When we normalized the intensity of ubiquitination signals in the PM and EM fractions of the wild type or pho2 with the corresponding protein level of PHT1;1/2/3, a more prominent reduction of ubiquitination of PHT1;1/2/3 was detected in the EM of pho2 (20% of wild-type signal) than in the PM (Figure 8B). These results support the notion that PHO2 mediates the ubiquitination of PHT1;1/2/3 at the EM rather than the PM.
Figure 8.
PHO2-Mediated Ubiquitination of PHT1;1 at the EM.
(A) and (B) Immunoblot analysis by anti-PHT1;1/2/3 or antiubiquitin antibody following immunoprecipitation (IP) of total membrane proteins (A) or enriched PM and EM proteins (B) with anti-PHT1;1/2/3 antibody. Membrane proteins were isolated from the roots of 12-d-old wild-type (WT), pho2, and pht1;1 seedlings grown under Pi-sufficient conditions.
(C) Assessment of the enrichment of isolated PM and EM fractions used in (B) by immunoblot analysis. H+-ATPase and PHF1 served as markers for the PM and EM protein, respectively.
(D) Analysis was performed similar to in (A), except 6-d-old seedlings were used.
The quantification of PHT1;1/2/3 in the input samples and ubiquitinated proteins normalized with the corresponding PHT1;1/2/3 is indicated.
We reasoned that the ubiquitination of PHT1;1/2/3 in the PM of pho2 is independent of PHO2 because of the absence of PHO2 in the PM. It is possible that other unidentified factors in the PM are involved in the augmented ubiquitination of PHT1;1/2/3 to compensate for the loss of PHO2 activity in the EM as the excessive Pi accumulates throughout the growth of pho2. To minimize this secondary effect, we analyzed the root samples from much younger seedlings (6 d old). As expected, the ubiquitination of PHT1;1/2/3 was clearly reduced in total membranes isolated from pho2 (Figure 8D). Altogether, these results reveal a role for PHO2 in mediating the ubiquitination of PHT1;1/2/3 in the EM compartment where PHO2 and the PHT1s interact.
DISCUSSION
As a component of the protein ubiquitination machinery, PHO2 negatively regulates the activities of Pi uptake and translocation. To elucidate how PHO2 mediates these activities in parallel, we undertook two approaches: pho2 suppressor screening and quantitative membrane proteomics. The pho2 suppressor screening revealed PHO1 to be a direct target of PHO2, which supports the role of PHO2 in modulating root-to-shoot translocation of Pi (Liu et al., 2012). Here, we report our quantitative membrane proteomic findings, which provide insights into the mechanism of PHO2-mediated Pi acquisition via posttranslational regulation of PHT1s.
Contributions to the Inventory of the Membrane Proteome
Recent advancements in proteomic technologies have provided tools to analyze the protein expression profiles of different organelles and specific subcellular structures. Many studies have focused on the analysis of the membrane proteome because of the pivotal role of membrane proteins in signal perception and transduction as well as the transport of essential substances (Groen et al., 2008; Sadowski et al., 2008; Nilsson et al., 2010). However, this is challenging because of the low abundance and hydrophobic nature of membrane proteins. Several modifications of protocols for protein extraction, solubilization, labeling, and enrichment have been made to improve the identification, coverage, and quantitation accuracy of membrane proteomics. Most of the large-scale plant membrane proteomic analyses conducted in the past used samples prepared from suspension-cultured Arabidopsis cells mainly because of advantages of the relatively uniform conditions and abundance of materials. Although several hundreds of membrane proteins were identified from each attempt, the resulting number was still far below expectation. Membrane proteins generally constitute 30% of the typical proteome (Bertone and Snyder, 2005). Based on the prediction of transmembrane helices, 25% (∼6500 proteins) of the Arabidopsis proteome are classified as putative membrane proteins in the ARAMEMNON, an Arabidopsis membrane protein database (http://aramemnon.uni-koeln.de/). (Schwacke et al., 2003). In this study, our aim was to contribute to the inventory of the current plant membrane proteome as well as to address an important biological question. We adapted the technique that we previously developed (Han et al., 2008). It was initially optimized for the mammalian system with high coverage, high accuracy, and precision, and we were able to retain the same levels of identification and quantitation performance for the Arabidopsis membrane proteins. We identified 7491 proteins, of which 3713 (49.6%) were categorized as membrane proteins (Figure 2A). After comparing against the most updated SUBA database (Heazlewood et al., 2007; Tanz et al., 2013), there are still 3498 (46.6%) proteins not previously identified in mass spectrometry studies. This large data set provides a basis for biochemical and functional characterizations of these proteins in the future.
It is important to note that our proteomics analysis is relatively comprehensive, as we were able to discriminate between proteins with high amino acid sequence identity, which is usually difficult to achieve by other conventional methods. For example, the amino acid sequences of the PHT1;1, PHT1;2, PHT1;3, and PHT1;4 proteins show strong similarity, especially among PHT1;1, PHT1;2, and PHT1;3 (up to 98.9% of identity). While no specific peptides assigned to PHT1;1, PHT1;2, and PHT1;3 were identified in the recent proteome analysis of Pi-deficient roots (Lan et al., 2012), our method was able to distinguish between these proteins and quantify them using specific peptides to each individual protein (see Supplemental Figure 4 online).
Proteins Differentially Expressed in the pho2 Mutant
Using quantitative membrane proteomics, we identified 59 and 20 proteins that were differentially expressed in the wild type and the pho2 mutant grown in solid agar and in hydroponic culture, respectively (see Supplemental Data Set 2 online). However, only five proteins showed consistent upregulation or downregulation in both samples (Table 1). The small number of overlapping targets may be attributable to differences in the developmental stages of plants, the culture conditions, or the level of cellular Pi accumulation. We observed that Pi accumulation in the shoot of the pho2 mutant was exacerbated over time and with plant growth. By contrast, the root Pi concentration of the pho2 mutant was lower than that of the wild type at the early growth stage, but the difference became negligible when the plants were older (see Supplemental Figures 2A and 2B online). The alterations in the protein profile in the pho2 mutant shown here are the collective consequences of enhanced Pi accumulation and the indirect effects or secondary responses to toxic levels of cellular Pi.
In addition to the proteins related to Pi transport, nitrate transport (NRT1.1/CHL1) and FERRIC REDUCTION OXIDASE2 (FRO2) involved in Fe acquisition were downregulated in the pho2 mutant (Table 1). This implies the adjustment of homeostasis of other nutrients when Pi homeostasis is disturbed. Interactions between P and different nutrient elements have been reported. Changes of Pi availability affected the acquisition or accumulation of Fe, S, Zn, and Mn, while high nitrate supply reduced Pi accumulation (Huang et al., 2000; Misson et al., 2005; Ward et al., 2008; Kant et al., 2011; Pedas et al., 2011; Rouached et al., 2011). Although multilevel coordination among different nutrients is expected to maintain the physiological homeostasis, the underlying mechanism remains largely unknown. The elevated cellular Pi concentrations seen in pho2 may provide an additional aspect to investigate the interaction of P with other nutrients in vivo.
PHO2 Facilitates the Degradation of PHT1 Proteins at the EM
The upregulation of PHT1 transcription in response to Pi deficiency through PHR1 has been well documented (Rubio et al., 2001). Recent reports also disclosed the importance of several posttranslational steps in modulating the activity of PHT1s, including membrane trafficking, phosphorylation, and endocytosis from the PM (González et al., 2005; Bayle et al., 2011). Here, we revealed that PHO2 functions in promoting the protein degradation of the PHT1s and probably PHF1 as well. We showed that several PHT1 proteins and PHF1 were significantly upregulated in the roots of pho2 (Table 1, Figure 4A). The upregulation of these proteins contributed to the excessive shoot Pi in the pho2 mutant because disruptions to PHF1 or PHT1;1 alleviated the Pi toxicity of pho2 (Figure 3A). Systematic comparisons between the transcript and protein levels in different genetic backgrounds with or without functional PHO2 indicated that PHO2 regulates the quantity of the PHT1s and PHF1 proteins (Figures 5A and 5B). CHX treatment demonstrated that loss of PHO2 delays the degradation of PHT1s (Figures 6A and 6B), reinforcing that PHO2 negatively regulates the PHT1s at the posttranslational level. Our findings emphasize the importance of protein degradation of the PHT1s when Pi supply is adequate. Notably, PHR1 acts as an upstream transcriptional regulator to PHT1s, PHF1, and the miR399 in response to Pi limitation (Rubio et al., 2001; González et al., 2005; Bari et al., 2006). PHR1 seems to activate multiple Pi regulatory pathways to promote the activity of PHT1 Pi transporters through a combination of strategies, including increased transcription, proper targeting to the PM, and the circumvention of protein degradation.
A specific subset of Arabidopsis UBC enzymes, subgroup VI, which includes UBC8 to 11 and UBC28 to 30, was found to be recruited to the PM through interaction with membrane-anchored ubiquitin-fold proteins (Dowil et al., 2011). Despite our finding of membrane localization of PHO2, there is no transmembrane-helix domain or posttranslational modification sites predicted for membrane anchoring within PHO2. It is possible that the interaction of PHO2 with other proteins accounts for its membrane localization. In this regard, we showed that PHO2 can interact with the PHT1s (Figures 7A and 7B) and PHO1 (Liu et al., 2012), all of which are integral membrane proteins. When transiently expressed in protoplasts, the signals of fluorescent protein–tagged PHO2 were detected at the ER and Golgi, where PHO1 localizes and the PHT1s pass before being targeted to the PM (see Supplemental Figure 1A online; Liu et al., 2012). The evidence of the physical interaction of PHO2 with PHT1;1 and PHT1;4 at the post-ER compartments (Figures 7A and 7B) and the reduced ubiquitination of PHT1;1/2/3 in the EM fraction of pho2 (Figure 8B) reveals that PHO2 functions to modify PHT1s by ubiquitination in the early secretory pathway and thereby to redirect PHT1 proteins for degradation before their arrival at the PM. This conclusion is supported by a recent study using ubiquitin-tagged reporters in which ubiquitin was shown to act as a sorting signal to reroute Golgi-localized membrane proteins to the vacuole using the secretory pathway (Scheuring et al., 2012).
There are multiple surveillance systems controlling protein quality and quantity along the secretory pathway (Arvan et al., 2002; Anelli and Sitia, 2008; Hegde and Ploegh, 2010). The ER-associated degradation (ERAD) pathway is responsible for disposal of aberrant proteins as well as excessive normal proteins at the ER via the ubiquitin-proteasome system (Vembar and Brodsky, 2008; Hegde and Ploegh, 2010). Additionally, post-ER protein control systems operating at the Golgi, PM, and endosomal compartments have also been reported, in which the vacuolar and 26S proteasome proteolytic pathways are involved (Arvan et al., 2002; Okiyoneda et al., 2011). As PHO2 is associated with the ER and Golgi, it is reasonable to speculate that PHO2 may be a component of the ERAD pathway, or it may function at the Golgi to facilitate the degradation of PHT1 proteins. We favor the latter assumption because the interactions between PHO2 and the PHT1s occur at the vesicle-like post-ER compartments (Figures 7A and 7B), a location beyond the ERAD pathway. In addition, 26S proteasome machinery, an essential component of the ERAD pathway, is unlikely to be involved in the degradation of PHT1s because the application of a 26S proteasome inhibitor, MG132, affects neither the vacuolar degradation of PHT1;1-GFP protein (Bayle et al., 2011) nor the abundance of PHT1 proteins in our analysis (see Supplemental Figure 5 online).
The results from in vivo ubiquitination assay showed that PHO2 meditates the ubiquitination of PHT1;1/2/3 in the EM rather than the PM compartment (Figure 8B). The remaining ubiquitination signal in the EM of pho2 could be attributable to the residual contamination of PM or the involvement of other factors. The ubiquitination of PHT1;1/2/3 detected in the PM (Figure 8B) is likely independent of PHO2. A mechanism for continuous internalization of PHT1s from the PM to the endosome followed by recycling or vacuolar degradation in response to Pi availability was recently described (Bayle et al., 2011). This regulation appeared to be independent of PHO2 because it was not compromised by the loss of PHO2 activity (Bayle et al., 2011). Our results here indeed support this conclusion. In fact, our recent finding showing that NITROGEN LIMITATION ADAPTATION (NLA) mediates the ubiquitination of PM-localized PHT1s followed by endocytosis of PHT1s (Lin et al., 2013) further support this notion.
Potential Non-Cell-Autonomous Role of PHO2 in Regulating Pi Uptake
We have previously shown by promoter-reporter assays that the transcriptional activity of MIR399s and PHO2 are predominately expressed in the vascular tissue of roots (Aung et al., 2006). In particular, the expression of PHO2 is tightly restricted inside the central vascular tissue. However, PHT1;1/2/3/4 is strongly expressed in the epidermis, root hairs, and cortex where the primary uptake of Pi from the rhizosphere takes place (Mudge et al., 2002; Misson et al., 2004). This implies a spatial separation of the transcriptional expression of PHO2 from that of PHT1 genes. This raises several intriguing questions, including how the vascular tissue–expressed PHO2 interacts with and regulates the PHT1 proteins that are mainly expressed in the outer layer of cells and whether miR399 also moves from the vascular tissue to the outer layers of roots if PHO2 mRNA is mobile. We hypothesize that PHO2 mRNA or PHO2 protein may act in a non-cell-autonomous manner to regulate the PHT1s via cell-to-cell trafficking from the vascular tissue to the cortex or epidermis. Examples for cell-to-cell movement of proteins and small RNAs have been reported. In Arabidopsis roots, the reciprocal signaling pathway across a vascular boundary was demonstrated to control tissue patterning (Carlsbecker et al., 2010). In this case, SHORT ROOT transcription factor is synthesized in the vascular tissue but moves to the endodermis where it activates the expression of miR165/166. The miR165/166 is then moved to the vascular tissue where it downregulates the targets to specify differentiation to xylem cell types (Carlsbecker et al., 2010). Intriguingly, recent RNA-seq analysis identified the PHO2 transcript in the GFP-tagged root hair cells (Lan et al., 2013), implying the possibility of cell-to-cell movement of PHO2 mRNA.
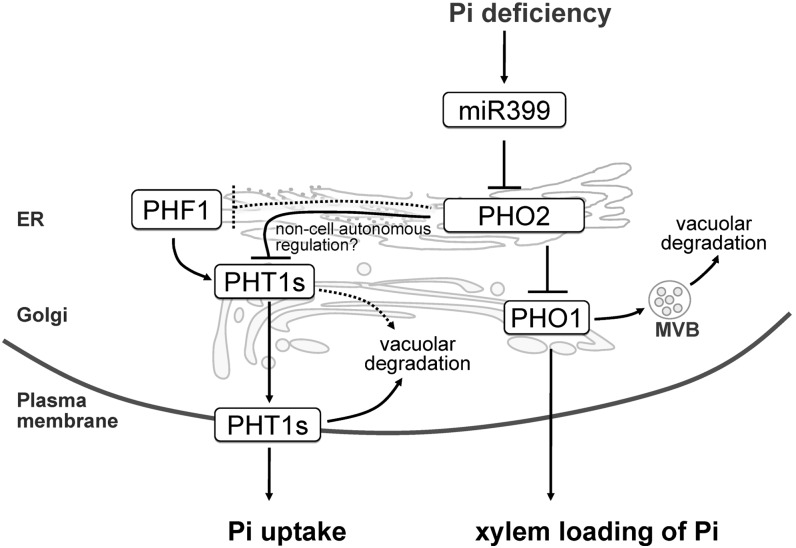
Role of PHO2 in Coordinating the Activities of Pi Acquisition and Xylem Loading
Here, we propose a working model illustrating the molecular mechanism of how miR399 and PHO2 regulate Pi homeostasis (Figure 9). PHO2 localizes to the ER and Golgi to mediate protein degradation and in turn suppress both Pi loading into the xylem and Pi acquisition, which take place in the stele and outer cell layer of roots, respectively. Previously, we found that PHO2 modulates the degradation of EM-localized PHO1 (Liu et al., 2012), a key player responsible for root-to-shoot Pi translocation via loading of Pi into the xylem (Hamburger et al., 2002). In this study, we showed that PHO2, acting as a checkpoint in the secretory pathway, facilitates the degradation of the PHT1s and possibly PHF1, thereby controlling the quantity of PHT1s accessing the PM. Once the PHT1s reach the PM, a PHO2-independent mechanism likely mediated by NLA (Lin et al., 2013) is involved in internalization of PHT1s from the PM for degradation.
Figure 9.
The Regulatory Mechanism of Pi Uptake and Root-to-Shoot Pi Translocation through PHT1s and PHO1 by PHO2.
PHO2 localizes to the ER and Golgi and suppresses Pi uptake and xylem loading of Pi by mediating the protein degradation of PHT1s and PHO1, which takes place in the outer cell layer and stele of roots, respectively. To simplify the illustration, these two activities are drawn in the same cell. The pathway indicated by the dashed line requires further evidence. See text for more details. MVB, multivesicular body.
PHO2-mediated suppression is alleviated by miR399 upon Pi starvation. The reduction of ubiquitination of the PHT1s can direct these proteins to the PM without being degraded in the secretory pathway. Such regulation allows for a rapid redistribution of the protein in response to changing nutrient or other conditions without involvement of de novo protein synthesis. For example, a RING membrane-anchor E3 ubiquitin ligase homolog (Rma1H1) was shown to mediate the ubiquitination of PIP2;1 aquaporin in the ER and thereby inhibit its trafficking to the PM in response to dehydration (Lee et al., 2009). Moreover, while the yeast amino acid permease, Gap1p, is sorted to the PM when N is limited, ubiquitination of Gap1p at the Golgi diverts the protein to vacuoles for degradation when the internal levels of amino acids are high (Helliwell et al., 2001; Soetens et al., 2001). Taken together, our results highlight the fundamental role of PHO2 in orchestrating two activities of Pi transport, namely, Pi acquisition at the root surface and Pi loading at the xylem of root stele, to maintain Pi homeostasis at a whole plant level.
METHODS
Plant Materials
Seeds of the Arabidopsis thaliana wild-type (Columbia-0), pho2 (Delhaize and Randall, 1995), and T-DNA insertion lines of pht1;1 (SALK_088586), pht1;2 (SALK_110194), pht1;3 (GABI_557C09), pht1;8 (SALK_056529), pht1;9 (SAIL_898_C11), and phf1 (SALK_037068) were obtained from the ABRC. Loss-of-function of these mutants was confirmed by RT-PCR analysis. Double mutants were generated by crossing with pho2. The double and corresponding single mutants were isolated from the F2 population for comparative analyses. Seedlings were grown under Pi-sufficient conditions using one-half modified Hoagland nutrient solution containing 250 µM KH2PO4 either on agar plates supplemented with 1% Suc or in a hydroponic culture system without Suc, as previously described (Aung et al., 2006). Pi starvation treatment was performed using 10 µM KH2PO4. Application of CHX (200 µM) was performed for indicated time periods on 12-d-old seedlings grown under Pi-sufficient conditions (Liu et al., 2012).
Membrane Proteomics Analysis
The 12-d-old seedlings grown on solid agar plates supplemented with 250 µM KH2PO4 were sampled or transferred to the hydroponic culture system to grow for an additional 5 d before harvest. Three biological replicates were prepared and employed in subsequent proteomics analysis. For membrane protein extraction, roots were collected and homogenized with ice-cold 0.22-μm filtered membrane extraction buffer containing 0.33 M Suc, 50 mM Tris-HCl, pH 7.5, 10 mM KCl, 5 mM EDTA, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× protease inhibitor cocktail (Roche). The crude extract was centrifuged at 2000g at 4°C for 10 min twice. The clear supernatant was then centrifuged at 400,000g at 4°C for 30 min. To exclude the nonspecific binding of soluble and loosely associated peripheral proteins, the pellet was resuspended with ice-cold 0.22-μm filtered 1 M KCl and placed on ice for at least 5 min, followed by centrifugation at 400,000g at 4°C for 30 min. The pellet was then resuspended with ice-cold 0.22-μm filtered 0.1 M Na2CO3, pH 11, and placed on ice for at least 5 min, followed by centrifugation at 400,000g at 4°C. After being washed with 0.1 M Na2CO3, the pellet was dried and stored at −20°C.
The gel-assisted digestion was performed following a previously described protocol (Han et al., 2008). The extracted peptides were combined, concentrated in a centrifugal evaporator, and subjected to iTRAQ labeling followed by liquid chromatography–MS/MS analysis. For details, please see the Supplemental Methods 1 online.
For analyzing the subcellular localization of those identified proteins, the TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) was used to predict the number of transmembrane helices, and their function and subcellular localization were categorized by GO (http://go.princeton.edu/cgi-bin/GOTermFinder) (Boyle et al., 2004). Subcellular localization of the proteins was also analyzed using SUBA (Arabidopsis Subcellular Database; http://suba.plantenergy.uwa.edu.au/) (Heazlewood et al., 2007; Tanz et al., 2013). For prediction of posttranslation modification associated with membranes, the following Web resources were applied: N-terminal myristoylation (Plant-Specific Myristoylation Predictor, http://plantsp.genomics.purdue.edu/myrist.html) (Podell and Gribskov, 2004), S-palmitoylation (TermiNator, http://www.isv.cnrs-gif.fr/Terminator/), C-terminal prenylation (PrePS, http://mendel.imp.univie.ac.at/sat/PrePS) (Maurer-Stroh et al., 2007), and glycosylphosphatidylinositol anchors (GPI-SOM, http://gpi.unibe.ch/).
Constructs for Gene Expression
All the inserted fragments of interest were amplified and cloned into pCR8/GW/TOPO (Invitrogen) for sequencing and then recombined into the desired Gateway destination vectors via Gateway®LR Clonase™II (Invitrogen). To examine subcellular localization of PHO2, the expression constructs (35S:CFP-PHO2 and 35S:Red Fluorescent Protein [RFP]-PHO2) were generated using p2CWG7 (Karimi et al., 2005) and pUBN-RFP (Grefen et al., 2010), respectively, as the Gateway destination vectors. The DNA fragment containing the coding sequence of C-terminal mCherry fusion of PHF1 was recombined into pMDC32 (Curtis and Grossniklaus, 2003) to generate the construct of 35S:PHF1-mCherry. For BiFC analysis, the coding sequences of full-length PHO2C748A, PHT1;1, PHT1.4, and PHF1 were cloned into appropriate vectors driven by the UBQ10 promoter (Grefen et al., 2010) as indicated in Figure 7. The full-length PHF1 coding sequence was cloned into pMDC32 (35S:PHF1), and the resulting construct was used for the coexpression with PHT1;1 and PHT1;4 in the BiFC assay. All primers used are listed in Supplemental Table 1 online.
Transient Expression in Mesophyll Protoplasts of Arabidopsis
Transformations of Arabidopsis mesophyll protoplasts for transient expression of fluorescence fusion proteins were performed as described (Wu et al., 2009). The subcellular fluorescent markers used were the same as previously described (Liu et al., 2012). Briefly, AtWAK2-mCherry and GmMAN1-mCherry constructs were derived from the multicolored organelle marker set (Nelson et al., 2007) and obtained from the ABRC. ARA7-RFP and VTI12-TFP constructs were derived from WAVE lines (Geldner et al., 2009) and obtained from the Nottingham Arabidopsis Stock Centre. N-ST-RFP and RFP-ARA6 were previously reported (Grebe et al., 2003). Confocal microscopy images were taken using a Zeiss LSM 510 META NLO DuoScan with LCI Plan-Neofluar ×63/1.3 Imm and Plan-Apochromat ×100/1.4 oil objectives. Excitation/emission wavelengths were 458 nm/465 to 510 nm for cyan fluorescent protein and TFP, 514 nm/520 to 550 nm for yellow fluorescent protein, and 561 nm/575 to 630 for red fluorescent protein and mCherry.
Antibody Production
The anti-PHT1;1 polyclonal rabbit antibody was raised and affinity purified against a peptide of PHT1;1 corresponding to the amino acid residues 266 to 285 (ELEERVEDDVKDPRQNYGLF) (Liu et al., 2011). The anti-PHT1;4 polyclonal rabbit antibody was raised and affinity purified against an internal fragment of PHT1;4 corresponding to the amino acid residues 265 to 283 (IEPEQQKLEEISKEKSKAF). The anti-PHF1 polyclonal rabbit antibody was raised and affinity purified against the C-terminal fragment of PHF1 corresponding to the amino acid residues 453 to 471 (GSSSTPSEDHSRWNLDL). The final concentration of affinity-purified antibodies used for immunoblot analysis is 20 to 100 ng/mL. Antibodies directed against fructose-1,6-bisphosphatase, ADP-RIBOSYLATION FACTOR1, and H+-ATPase were purchased from Agrisera, and the antibody against actin was from Millipore.
Total and Membrane Protein Isolation
For total protein extraction, roots were ground in liquid nitrogen and dissolved in protein lysis buffer containing 2% SDS, 60 mM Tris-HCl, pH 8.5, 2.5% glycerol, 0.13 mM EDTA, and 1× protease inhibitor cocktail (Roche). Membrane proteins were isolated as described above except without the wash of KCl and Na2CO3. The pelleted membrane proteins were resuspended in ice-cold extraction buffer for immunoblot analysis or in ice-cold storage buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM N-ethylmaleimide, 1 mM PMSF, and 1× protease inhibitor (Roche) for immunoprecipitation. Total microsomal membranes were separated into the PM and EM fractions by two-phase partitioning according to the previous description (Larsson et al., 1987). Briefly, the microsomal pellet was resuspended in 3 mL of resuspension solution (345 mM Suc and 4.8 mM K-PO4 buffer, pH 7.8). Then, 3.34 g of membrane sample was added to a phase separation solution containing 6.2% (w/w) Dextran T-500, 6.2% (w/w) polyethylene glycol 3350, 0.23 M Suc, 5 mM KCl, and 3.6 mM K-PO4 buffer, pH 7.8, mixed vigorously for 10 s, and equilibrated in an ice bath for 4 min. After centrifugation at 1500g for 8 min at 4°C, the upper polyethylene glycol and bottom Dextran phases were separated, resuspended (330 mM Suc, 10 mM KCl, 5 mM EDTA, 50 mM Tris, pH 7.5, and 1 mM PMSF), and centrifuged (100,000g for 1 h at 4°C) to yield the PM and EM fractions, respectively.
Immunoprecipitation and Immunoblot Analysis
A 100-µg sample of total microsomal protein or 20 µg of PM or EM protein was resuspended in 500 μL of ice-cold lysis buffer containing 20 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Tween 20, 0.5% deoxycholate, 0.1% SDS, 10 mM N-ethylmaleimide, 1 mM PMSF, and 1× protease inhibitor cocktail (Roche) and incubated with 20 μL of magnetic beads conjugated with the anti-PHT1;1/2/3 antibody at 4°C for 1.5 h with gentle shaking (Kasai et al., 2011). The immunoblot analysis was performed as described (Liu et al., 2011). To detect the immunoprecipitated PHT1;1/2/3 proteins, the EasyBlot anti-rabbit IgG (GeneTex) was used as the secondary antibody to reduce interference caused by heavy and light chains of the antibody used for immunoprecipitation. Monoclonal antiubiquitin antibody (P4D1; Santa Cruz Biotechnology) was used for detecting ubiquitinated proteins. The signals on immunoblots were detected by chemiluminescence, which was captured by a charge coupled device camera in a FluorChem HD2 image system and quantified by AlphaView SA Ver.3.3.1 (Cell Biosciences).
RNA Isolation and Quantitative RT-PCR
RNA isolation and quantitative RT-PCR were conducted as described (Liu et al., 2012).
Protein–Protein Interaction Analysis
For BiFC analysis, the Agrobacterium tumefaciens–mediated transient expression in Nicotiana benthamiana tobacco leaves was conducted as described (Voinnet et al., 2003) with minor modifications (Liu et al., 2012). The amount of p19, a gene silencing suppressor, coinfiltrated was carefully adjusted to eliminate the false positive signal in the negative controls containing only one interaction partner (Figures 7A to 7C).
Split-ubiquitin Y2H assay was performed according to the manufacturer’s instructions provided with the DUALmembrane kit (Dualsystems Biotech). The coding region of PHF1 and PHT1;4 were cloned in frame into the bait vector pTMBV4. The full-length PHO2, PHT1;1, and PHF1 coding sequences were cloned into the prey vector pDL-Nx via BamHI and SmaI sites. Yeast strain DSY-1 cells were cotransformed with the resulting constructs and plated onto synthetic medium lacking Leu, Trp, and His. The specificity of protein–protein interactions was confirmed by chloroform overlay β-galactosidase plate assay as described (Duttweiler, 1996).
Phosphate Analysis
Pi concentration and uptake activity were determined as described (Chiou et al., 2006).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: PHO2, At2g33770; PHT1;1, At5g43350; PHT1;2, At5g43370; PHT1;3, At5g43360; PHT1;4, At2g38940; PHT1;8, At1g20860; PHT1;9, At1g76430; and PHF1, At3g52190. The first set of replicates of proteome data can be downloaded from ProteomeCommons.org Tranche using the following hash: kwzun0TPfAMFQMDdTatkc8/T/MBvPWvydqj5BHHjizlkNedZLm/ih0DocqRLiVDsEM9ZkekktNP1vlqgIdE+w+lj2GoAAAAAAAAC3w==. The other two sets of replicates have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno, et al., 2013) with the data set identifier PXD000398.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PHO2 Resides at the Endomembrane.
Supplemental Figure 2. Analyses of Wild-Type and pho2 Seedlings Subjected to Membrane Proteomic Analysis.
Supplemental Figure 3. Evaluation of the Specificity of Antibodies.
Supplemental Figure 4. Identified Peptides Corresponding to PHT1;1, PHT1;2, PHT1;3, or PHT1;4 Proteins.
Supplemental Figure 5. Effect of MG132 on the Accumulation of PHT1;1/2/3 Proteins.
Supplemental Table 1. Sequences of Primers Used in This Study.
Supplemental Methods 1. iTRAQ Labeling and LC-MS/MS Analysis.
Supplemental Data Set 1. List of Total 7491 Identified Proteins from the Membrane Proteome of Arabidopsis Roots.
Supplemental Data Set 2. Differentially Expressed Proteins in the pho2 Roots Grown under Solid Agar or Hydroponic Medium.
Supplementary Material
Acknowledgments
This work was supported by Academia Sinica, Taiwan (Grant 98-CDA-L11 and a postdoctoral fellowship to S.-I.L. and T.-Y.L.) and by the National Science Council of the Republic of China (Grant NSC100-2321-B-001-005). We thank the Plant Tech Core Facility in the Agricultural Biotechnology Research Center of Academia Sinica for assistance in protoplast transformation, Wei-Hung Chang and Chi-Ying Lee for assistance with proteomic analysis, and Shu-Chen Shen from the Scientific Instrument Center of Academia Sinica for assistance with the confocal microscopy analysis. We thank Pei-Chi Wu and Ya-Shiuan Lai for analyzing pht1;8 and pht1;9 mutants, Chiung-Yun Chang for isolating pht1;1, pht1;2, and pht1;3 mutants, and Ya-Ni Chen for isolating the phf1 mutant. We also thank Laurent Nussaume (Institute for Biotechnology and Environmental Biology, France) for sharing the pht1;4 seed, the Samuel Roberts Noble Foundation for the seed of pht1;1 pht1;4, and Wolfgang Lugmayr (Research Institute of Molecular Pathology, Vienna, Austria) for the assistance with prenylation analysis.
AUTHOR CONTRIBUTIONS
Y.-J.C. and T.-J.C. designed the research. T.-K.H., S.-I.L., Y.-C.T., J.-W.C., W.-Y.L., Y.-S.C., and C.-M.S. performed the experiments. C.-L.H., Y.-C.T., Y.-R.C., P.-M.C., S.-I.L., and T.-K.H. analyzed the proteomics data. T.-K.H., C.-L.H., S.-I.L., Y.-J.C., T.-Y.L., Y.-R.C., and T.-J.C. wrote the article.
Glossary
- EM
endomembrane
- PM
plasma membrane
- ER
endoplasmic reticulum
- iTRAQ
isobaric tags for relative and absolute quantitation
- MS/MS
tandem mass spectrometry
- GO
Gene Ontology
- GFP
green fluorescent protein
- CHX
cycloheximide
- BiFC
bimolecular fluorescence complementation
- Y2H
yeast two-hybrid
- ERAD
ER-associated degradation
- PMSF
phenylmethylsulfonyl fluoride
References
- Anelli T., Sitia R. (2008). Protein quality control in the early secretory pathway. EMBO J. 27: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvan P., Zhao X., Ramos-Castaneda J., Chang A. (2002). Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic 3: 771–780 [DOI] [PubMed] [Google Scholar]
- Aung K., Lin S.-I., Wu C.-C., Huang Y.-T., Su C.-l., Chiou T.-J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Pant D.B., Stitt M., Scheible W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V., Arrighi J.-F., Creff A., Nespoulous C., Vialaret J., Rossignol M., Gonzalez E., Paz-Ares J., Nussaume L. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P., Snyder M. (2005). Prospects and challenges in proteomics. Plant Physiol. 138: 560–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. (2004). GO:TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W.-H., Lee C.-Y., Lin C.-Y., Chen W.-Y., Chen M.-C., Tzou W.-S., Chen Y.-R. (2013). UniQua: A universal signal processor for MS-based qualitative and quantitative proteomics applications. Anal. Chem. 85: 890–897 [DOI] [PubMed] [Google Scholar]
- Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.l. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.-J., Lin S.-I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Chiou T.-J., Liu H., Harrison M.J. (2001). The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 25: 281–293 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E., Randall P.J. (1995). Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowil R.T., Lu X., Saracco S.A., Vierstra R.D., Downes B.P. (2011). Arabidopsis membrane-anchored ubiquitin-fold (MUB) proteins localize a specific subset of ubiquitin-conjugating (E2) enzymes to the plasma membrane. J. Biol. Chem. 286: 14913–14921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttweiler H.M. (1996). A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet. 12: 340–341 [DOI] [PubMed] [Google Scholar]
- Eifler, K. (2010). The PHO2 Family of Ubiquitin Conjugating Enzymes in Arabidopsis thaliana and Its Contribution to Plant Programmed Cell Death. PhD dissertation (Koeln, Germany: Universität zu Köln). [Google Scholar]
- Fujii H., Chiou T.-J., Lin S.-I., Aung K., Zhu J.-K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.-D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E., Solano R., Rubio V., Leyva A., Paz-Ares J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M., Xu J., M^bius W., Ueda T., Nakano A., Geuze H.J., Rook M.B., Scheres B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Groen A.J., de Vries S.C., Lilley K.S. (2008). A proteomics approach to membrane trafficking. Plant Physiol. 147: 1584–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., MacDonald-Comber Petetot J., Somerville C., Poirier Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C.-L., Chien C.-W., Chen W.-C., Chen Y.-R., Wu C.-P., Li H., Chen Y.-J. (2008). A multiplexed quantitative strategy for membrane proteomics: Opportunities for mining therapeutic targets for autosomal-dominant polycystic kidney disease. Mol. Cell. Proteomics 7: 1983–1997 [DOI] [PubMed] [Google Scholar]
- Heazlewood J.L., Verboom R.E., Tonti-Filippini J., Small I., Millar A.H. (2007). SUBA: The Arabidopsis subcellular database. Nucleic Acids Res. 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R.S., Ploegh H.L. (2010). Quality and quantity control at the endoplasmic reticulum. Curr. Opin. Cell Biol. 22: 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell S.B., Losko S., Kaiser C.A. (2001). Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153: 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Barker S.J., Langridge P., Smith F.W., Graham R.D. (2000). Zinc deficiency up-regulates expression of high-affinity phosphate transporter genes in both phosphate-sufficient and -deficient barley roots. Plant Physiol. 124: 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Peng M., Rothstein S.J. (2011). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kasai K., Takano J., Miwa K., Toyoda A., Fujiwara T. (2011). High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. (2001). Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kuo H.-F., Chiou T.-J. (2011). The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 156: 1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, C., Widell, S., and Kjellbom, P. (1987). Preparation of high-purity plasma membranes. Methods Enzymol. 148: 558–568 [Google Scholar]
- Lan P., Li W., Lin W.-D., Santi S., Schmidt W. (2013). Mapping gene activity of Arabidopsis root hairs. Genome Biol. 14: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Li W., Schmidt W. (2012). Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics 11: 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-I., Chiang S.-F., Lin W.-Y., Chen J.-W., Tseng C.-Y., Wu P.-C., Chiou T.-J. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.-Y., Huang T.-K., Chiou T.-J. (2013). NITROGEN LIMITATION ADAPTATION, a target of microRNA827, mediates degradation of plasma membrane–localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.-Y., Lin S.-I., Chiou T.-J. (2009). Molecular regulators of phosphate homeostasis in plants. J. Exp. Bot. 60: 1427–1438 [DOI] [PubMed] [Google Scholar]
- Liu T.-Y., Aung K., Tseng C.-Y., Chang T.-Y., Chen Y.-S., Chiou T.-J. (2011). Vacuolar Ca2+/H+ transport activity is required for systemic phosphate homeostasis involving shoot-to-root signaling in Arabidopsis. Plant Physiol. 156: 1176–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.-Y., Chang C.-Y., Chiou T.-J. (2009). The long-distance signaling of mineral macronutrients. Curr. Opin. Plant Biol. 12: 312–319 [DOI] [PubMed] [Google Scholar]
- Liu T.-Y., Huang T.-K., Tseng C.-Y., Lai Y.-S., Lin S.-I., Lin W.-Y., Chen J.-W., Chiou T.-J. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S., Koranda M., Benetka W., Schneider G., Sirota F., Eisenhaber F. (2007). Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput. Biol. 3: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J., Thibaud M.-C., Bechtold N., Raghothama K., Nussaume L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Misson J., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal U.S., Pardo J.M., Raghothama K.G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge S.R., Rae A.L., Diatloff E., Smith F.W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nilsson R., Bernfur K., Gustavsson N., Bygdell J., Wingsle G., Larsson C. (2010). Proteomics of plasma membranes from poplar trees reveals tissue distribution of transporters, receptors, and proteins in cell wall formation. Mol. Cell. Proteomics 9: 368–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okiyoneda T., Apaja P., Lukacs G. (2011). Protein quality control at the plasma membrane. Curr. Opin. Cell Biol. 23: 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B., Buhtz A., Kehr J., Scheible W. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedas P., Husted S.r., Skytte K., Schjoerring J.K. (2011). Elevated phosphorus impedes manganese acquisition by barley plants. Front. Plant Sci. 2: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S., Gribskov M. (2004). Predicting N-terminal myristoylation sites in plant proteins. BMC Genomics 5: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier, Y., and Bucher, M. (2002). Phosphate transport and homeostasis in Arabidopsis The Arabidopsis Book 1: e0024.10.1199/tab.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama K.G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Rouached H., Secco D., Arpat B., Poirier Y. (2011). The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martin A.C., Iglesias J., Leyva A., Paz-Ares J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski P.G., Groen A.J., Dupree P., Lilley K. (2008). Sub-cellular localization of membrane proteins. Proteomics 8: 3991–4011 [DOI] [PubMed] [Google Scholar]
- Scheuring D., Kunzl F., Viotti C., San Wan Yan M., Jiang L., Schellmann S., Robinson D., Pimpl P. (2012). Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway. BMC Plant Biol. 12: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.-I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Shin H.S., Dewbre G.R., Harrison M.J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Soetens O., De Craene J.-O., André B. (2001). Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276: 43949–43957 [DOI] [PubMed] [Google Scholar]
- Tanz S., Castleden I., Hooper C., Vacher M., Small I., Millar H. (2013). SUBA3: A database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 41: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S.S., Brodsky J.L. (2008). One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., et al. (2013). The Proteomics Identifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 41: D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Ward J.T., Lahner B., Yakubova E., Salt D.E., Raghothama K.G. (2008). The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. (2009). Tape-Arabidopsis Sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.