This study describes a screen for suppressors of peroxisomal defects that result from a dysfunctional peroxisomal protease in Arabidopsis and reveals that plant peroxisomes can be targeted by autophagy (pexophagy).
Abstract
Peroxisomes house critical metabolic reactions that are essential for seedling development. As seedlings mature, metabolic requirements change, and peroxisomal contents are remodeled. The resident peroxisomal protease LON2 is positioned to degrade obsolete or damaged peroxisomal proteins, but data supporting such a role in plants have remained elusive. Arabidopsis thaliana lon2 mutants display defects in peroxisomal metabolism and matrix protein import but appear to degrade matrix proteins normally. To elucidate LON2 functions, we executed a forward-genetic screen for lon2 suppressors, which revealed multiple mutations in key autophagy genes. Disabling core autophagy-related gene (ATG) products prevents autophagy, a process through which cytosolic constituents, including organelles, can be targeted for vacuolar degradation. We found that atg2, atg3, and atg7 mutations suppressed lon2 defects in auxin metabolism and matrix protein processing and rescued the abnormally large size and small number of lon2 peroxisomes. Moreover, analysis of lon2 atg mutants uncovered an apparent role for LON2 in matrix protein turnover. Our data suggest that LON2 facilitates matrix protein degradation during peroxisome content remodeling, provide evidence for the existence of pexophagy in plants, and indicate that peroxisome destruction via autophagy is enhanced when LON2 is absent.
INTRODUCTION
Peroxisomes are single membrane–bound organelles that compartmentalize certain oxidative reactions in eukaryotes, including fatty acid β-oxidation and hydrogen peroxide metabolism (reviewed in Hu et al., 2012). Peroxisomal proteins are imported into the organelle from the cytosol; defects in human peroxisomal biogenesis underlie the Zellweger syndrome spectrum disorders, which often are fatal (reviewed in Wanders and Waterham, 2005). Plant peroxisomes also are essential (reviewed in Hu et al., 2012); viable plant mutants lacking peroxisomes have not been identified.
Peroxisome formation and maintenance requires membrane recruitment, matrix protein import, organelle division, and distribution in daughter cells. Peroxisome biogenesis in yeast requires more than 30 peroxin (PEX) genes; about half of these are conserved in plants and mammals (reviewed in Hu et al., 2012; Nagotu et al., 2012). PEX5 and PEX7 are receptors that translocate matrix proteins into peroxisomes by binding cargo with peroxisome-targeting signals (PTSs) and docking with the PEX13 and PEX14 membrane peroxins (Nagotu et al., 2012). PEX5 recognizes proteins containing a PTS1 (McCollum et al., 1993; Van der Leij et al., 1993), a variant of a C-terminal Ser-Lys-Leu (SKL-COOH) motif. PEX7 recognizes proteins bearing a PTS2 (Marzioch et al., 1994), a nine–amino acid sequence embedded in an ∼30–amino acid N-terminal presequence. This presequence is removed upon peroxisome import in mammals and plants (Swinkels et al., 1991; Helm et al., 2007). After cargo translocation into the organelle, PEX5 and PEX7 are returned to the cytosol for reuse (Collins et al., 2000; Dammai and Subramani, 2001; Nair et al., 2004). PEX5 recycling requires the ubiquitin-conjugating enzyme PEX4, which is tethered to the peroxisome membrane by PEX22 (Collins et al., 2000; Zolman et al., 2005) and assisted by a complex of membrane peroxins with ubiquitin-protein ligase activity (Platta et al., 2009; Kaur et al., 2013).
In young seedlings, peroxisomes house fatty acid β-oxidation and the glyoxylate cycle (reviewed in Eastmond and Graham, 2001), which allow utilization of storage lipids for energy and fixed carbon before photosynthesis is established. Peroxisome-defective Arabidopsis thaliana mutants therefore often require external fixed carbon for normal development (Hayashi et al., 1998; Zolman et al., 2000). Peroxisomes also metabolize the protoauxin indole-3-butyric acid (IBA) (reviewed in Strader and Bartel, 2011) into the active auxin indole-3-acetic acid (IAA), which plays multiple critical roles in development (reviewed in Woodward and Bartel, 2005b). Mutants with dysfunctional peroxisomes often display impaired IBA-to-IAA conversion (Strader et al., 2010) and dampened IBA responsiveness (Zolman et al., 2000). IBA resistance provides an indirect measure of peroxisome function that facilitates the isolation and characterization of peroxisome-defective mutants (Zolman et al., 2000, 2005; Zolman and Bartel, 2004; Woodward and Bartel, 2005a; Ramón and Bartel, 2010; Ratzel et al., 2011).
As seedlings establish photosynthesis several days after germination, peroxisomes are remodeled, photorespiration enzymes like hydroxypyruvate reductase (HPR) are synthesized, and obsolete glyoxylate cycle enzymes, such as isocitrate lyase (ICL) and malate synthase (MLS), are degraded. Immunolabeling experiments in greening cucurbit cotyledons directly demonstrate the presence of both photorespiration enzymes and glyoxylate cycle enzymes in the same peroxisomes (Titus and Becker, 1985; Nishimura et al., 1986; Sautter, 1986). Moreover, MLS is stable following in vitro import into early pumpkin (Cucurbita sp Amakuri Nankin) seedling peroxisomes but degraded after import into transitional peroxisomes (Mori and Nishimura, 1989). These data provide support for the one-population hypothesis, in which individual peroxisomal proteins are degraded and new enzymes are imported into existing peroxisomes during seedling maturation, rather than a two-population hypothesis in which entire peroxisomes are degraded and resynthesized with new content during developmental transitions (reviewed in Nishimura et al., 1996).
Although glyoxylate cycle enzymes and photorespiration enzymes have not been directly colocalized in Arabidopsis peroxisomes, these enzymes are present together in cotyledons during the ∼2-d window during seedling development when ICL and MLS are degraded and HPR begins accumulating (Lingard et al., 2009). Peroxisome entry is a prerequisite for efficient ICL and MLS degradation; pex mutants with matrix protein import defects stabilize ICL and MLS (Lingard et al., 2009; Burkhart et al., 2013). Moreover, PEX4 and PEX22 promote ICL and MLS turnover, hinting at a role for ubiquitination in matrix protein degradation (Zolman et al., 2005; Lingard et al., 2009). However, because PEX4 and PEX22 also are implicated in PEX5 recycling and thus indirectly promote matrix protein import, this finding does not provide a definitive mechanism for matrix protein degradation.
Several peroxisomal proteases have been examined for possible matrix protein degradation roles (Lingard and Bartel, 2009). LON proteases were first characterized in bacteria (reviewed in Tsilibaris et al., 2006) and contain an N-terminal substrate binding domain, a central ATPase domain containing Walker A and B ATP-binding motifs, and a C-terminal proteolytic domain containing a Ser-Lys catalytic dyad (Tsilibaris et al., 2006). LON proteases are predicted to form ring-shaped oligomers and often degrade misfolded proteins (Tsilibaris et al., 2006). Eukaryotic LON isoforms are found in mitochondria, chloroplasts, and peroxisomes (Kikuchi et al., 2004; Ostersetzer et al., 2007; Lingard and Bartel, 2009). The peroxisomal Lon from the methylotrophic yeast Hansenula polymorpha promotes degradation of misfolded peroxisomally targeted dihydrofolate reductase (Aksam et al., 2007), and in Penicillium chrysogenum, peroxisomal Lon degrades oxidatively damaged catalase-peroxidase (Bartoszewska et al., 2012). By contrast, mammalian cells expressing a dominant-negative form of rat liver peroxisomal Lon mislocalize catalase to the cytosol (Omi et al., 2008), indicating that peroxisomal LON function somehow promotes matrix protein import. Arabidopsis encodes four LON isoforms; LON2 is the only isoform bearing a canonical PTS1 and localized in peroxisomes (Ostersetzer et al., 2007; Eubel et al., 2008; Reumann et al., 2009). All examined Arabidopsis lon2 alleles are resistant to IBA-induced lateral root formation but respond normally to auxins not requiring β-oxidation (Lingard and Bartel, 2009; Burkhart et al., 2013), suggesting reduced IBA-to-IAA conversion. Additionally, lon2 mutants display age-dependent defects in PTS2 processing that are accompanied by defects in peroxisomal matrix protein import (Lingard and Bartel, 2009; Burkhart et al., 2013), indicating that LON2 is necessary for sustained import of matrix proteins. This age-dependent defect in matrix protein import contrasts with import defects in pex14 mutants, which appear less severe as seedlings age (Hayashi et al., 2000; Monroe-Augustus et al., 2011). In spite of these defects in peroxisome physiology, disrupting Arabidopsis LON2 does not appear to result in matrix protein stabilization (Lingard and Bartel, 2009; Burkhart et al., 2013).
A second peroxisomal protease that has been tested for a role in Arabidopsis matrix protein degradation is DEG15. DEG15 processes PTS2 proteins into their mature forms by removing the PTS2-containing N-terminal region (Helm et al., 2007; Schuhmann et al., 2008), but DEG15 is not required for ICL or MLS degradation (Lingard and Bartel, 2009). Similarly, the peroxisomal M16 metalloprotease PXM16 is not required for degradation of glyoxylate cycle enzymes; pxm16 and lon2 pxm16 mutants efficiently degrade ICL and MLS (Lingard and Bartel, 2009).
Beyond possible protease involvement in degrading individual peroxisomal matrix proteins, yeast and mammals can degrade peroxisomes using a specialized form of autophagy termed pexophagy. Interestingly, Aspergillus nidulans ICL appears to be degraded via pexophagy when cells are moved from fatty acid to Glc carbon sources (Amor et al., 2000). Autophagy functions in bulk degradation and recycling of cytosolic constituents, often in response to nutrient limitation or other stresses (reviewed in Bassham, 2007; Xie and Klionsky, 2007; Reumann et al., 2010; Li and Vierstra, 2012). Preautophagosomal structures are formed by a subset of the AUTOPHAGY-RELATED (ATG) proteins and assist an isolation membrane in engulfing cytoplasmic constituents in autophagosomes. Autophagosomes can envelop diverse substrates, including ribosomes, organelles, and protein aggregates (reviewed in Xie and Klionsky, 2007; Li and Vierstra, 2012). Mature autophagosomes are delivered to the lytic vacuole by fusion of the outer autophagosome membrane with the vacuolar membrane. Once in the vacuole, the inner membrane is dissolved, and autophagosome contents are degraded by vacuolar hydrolases. Although the core autophagy machinery is conserved in plants (reviewed in Bassham, 2007; Reumann et al., 2010; Li and Vierstra, 2012), it is unclear whether peroxisome turnover by pexophagy occurs in plants.
We undertook a forward-genetic screen for lon2 suppressors to elucidate the molecular functions and targets of LON2 in Arabidopsis. We isolated multiple alleles of several pivotal ATG genes that suppressed the suite of lon2 physiological and molecular defects. At the same time, disrupting autophagy revealed an additional molecular defect in lon2; lon2 atg double mutants failed to degrade obsolete glyoxylate cycle enzymes in a timely fashion during seedling development. Our results imply that LON2 normally functions in the removal of obsolete matrix proteins during development. Moreover, the absence of LON2 accelerates pexophagy, which degrades peroxisomes that otherwise would carry out important metabolic functions in developing seedlings.
RESULTS
Isolating Suppressors of lon2 Defects
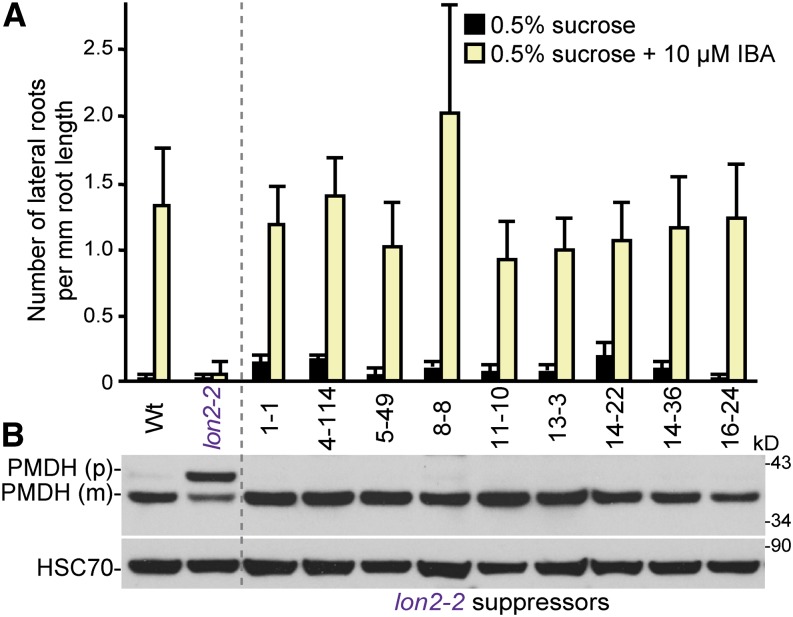
Because of the unusual assortment of defects in peroxisome physiology that result when Arabidopsis LON2 is mutated, we sought to identify LON2 regulators or substrates among LON2 genetic interactors. We screened for mutations that suppressed the IBA resistance and PTS2-processing defects of lon2-2, which contains a T-DNA insertion in the last exon of LON2 that would remove the PTS1 from the protein and prevent its import into the peroxisome (Lingard and Bartel, 2009). We mutagenized lon2-2 seeds carrying a cauliflower mosaic virus 35S promoter–driven version of green fluorescent protein (GFP) bearing an N-terminal PTS2 peptide (35S:PTS2-GFP) (Woodward and Bartel, 2005a) and screened ∼80,000 M2 seedlings for suppressors in a two-part screen. First, we identified M2 seedlings that produced approximately wild-type numbers of lateral roots in the presence of IBA, thereby rescuing the lon2-2 resistance to IBA-induced lateral root formation. Approximately 800 putative mutants were transferred to soil and allowed to self-fertilize, and surviving M3 progeny were rescreened for IBA sensitivity. Next, the 61 suppressors displaying the most robust IBA sensitivity were examined by immunoblotting for suppression of the lon2 PTS2-processing defect, revealing nine strong suppressors of both lon2 phenotypes from eight different M1 pools: 1-1, 4-114, 5-49, 8-8, 11-10, 13-3, 14-22, 14-36, and 16-24. Whereas 8-d-old lon2 seedlings rarely made a lateral root after growth on 10 μM IBA, IBA-induced lateral root numbers were comparable to those of the wild type in these suppressors (Figure 1A), suggesting rescued IBA-to-IAA conversion. Additionally, the suppressors fully restored the ability of lon2-2 to process the PTS2 protein peroxisomal malate dehydrogenase (PMDH) from its precursor into its mature form (Figure 1B), suggesting that these suppressors also rescued the lon2-2 peroxisomal matrix protein import defects.
Figure 1.
Suppressors of lon2 Are IBA Sensitive and Lack PTS2-Processing Defects.
(A) Number of lateral roots per millimeter root length of 8-d-old wild-type (Wt) 35S:PTS2-GFP, lon2-2 35S:PTS2-GFP, and lon2-2 suppressor seedlings (M5 progeny of original isolates) that were grown under yellow-filtered light on Suc-supplemented medium for 4 d and then transferred to Suc-supplemented medium with or without 10 μM IBA for an additional 4 d. Error bars show sd (n ≥ 12).
(B) Extracts from 8-d-old wild-type 35S:PTS2-GFP, lon2-2 35S:PTS2-GFP, and each suppressor line grown in yellow light on Suc- and IBA-supplemented medium were processed for immunoblotting and serially probed with antibodies raised against the indicated proteins. PMDH is expressed as a precursor (p); the PTS2 region is cleaved to the mature (m) form in the peroxisome. Protein loading was monitored by probing with antibodies against HSC70. The positions of molecular mass markers (in kilodaltons) are shown on the right.
[See online article for color version of this figure.]
Mutations in the Autophagy Gene ATG7 Suppress lon2 Defects
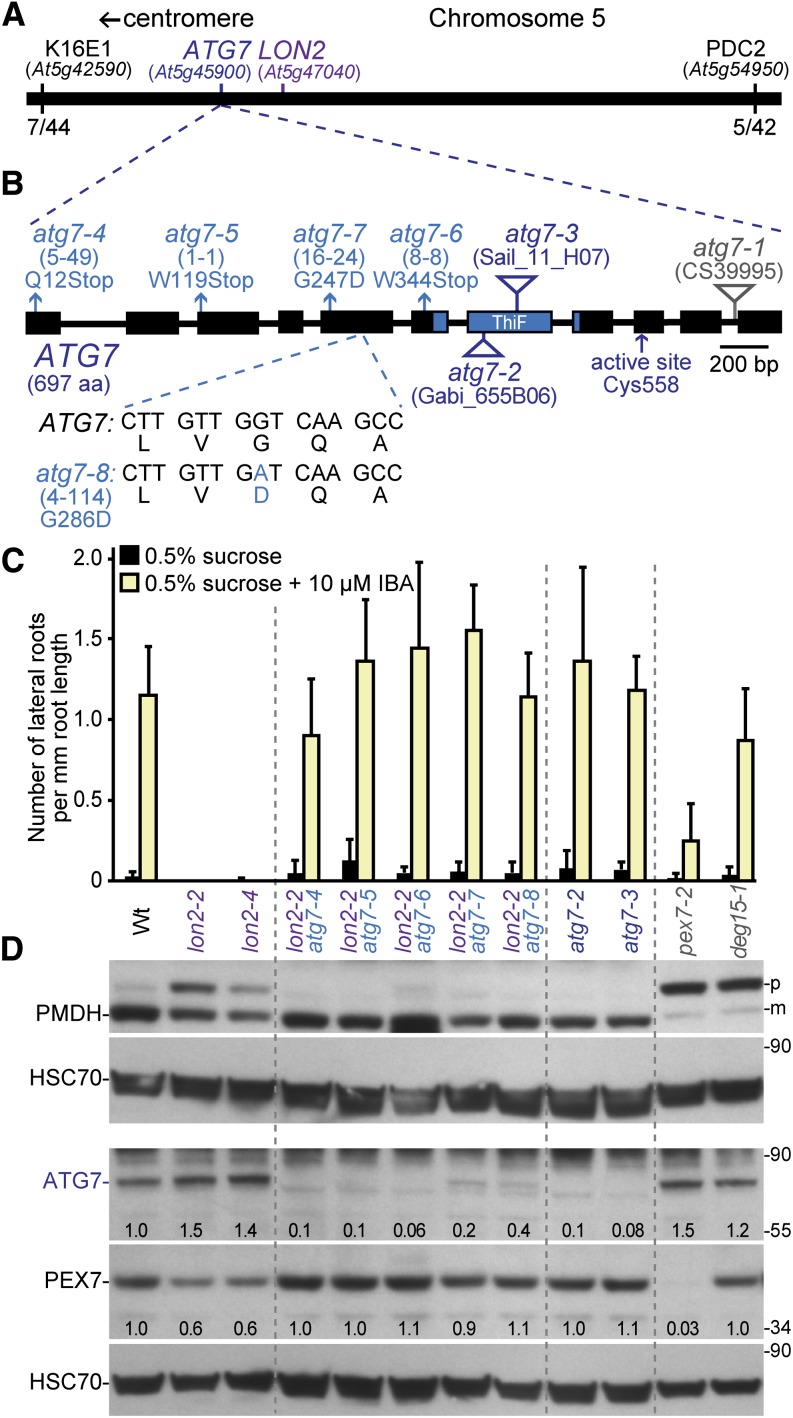
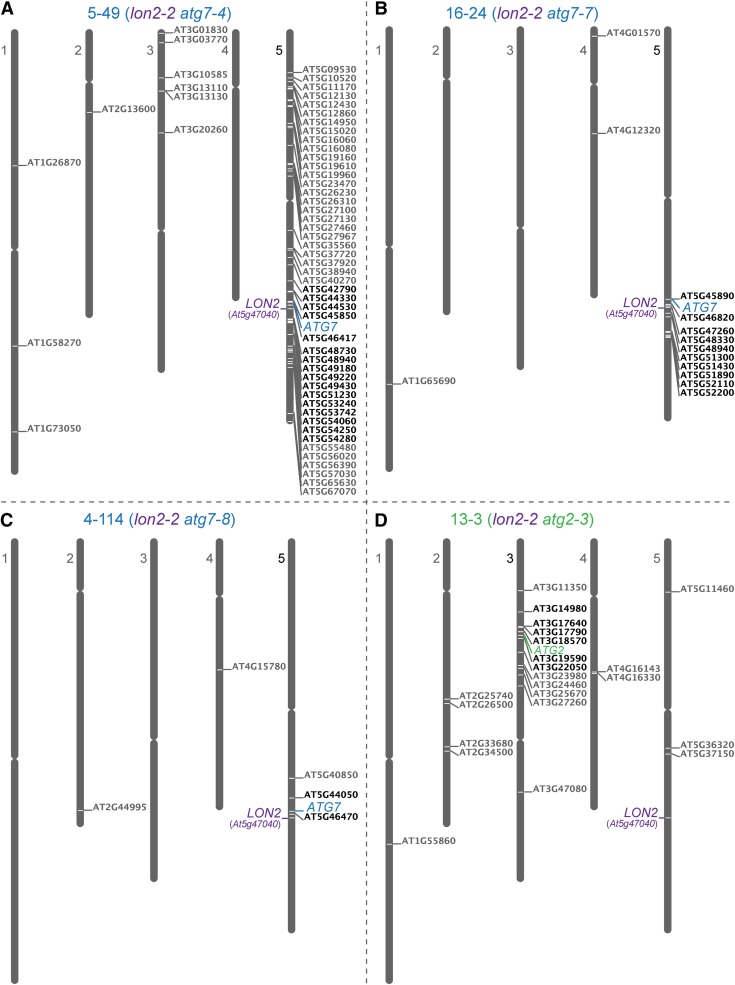
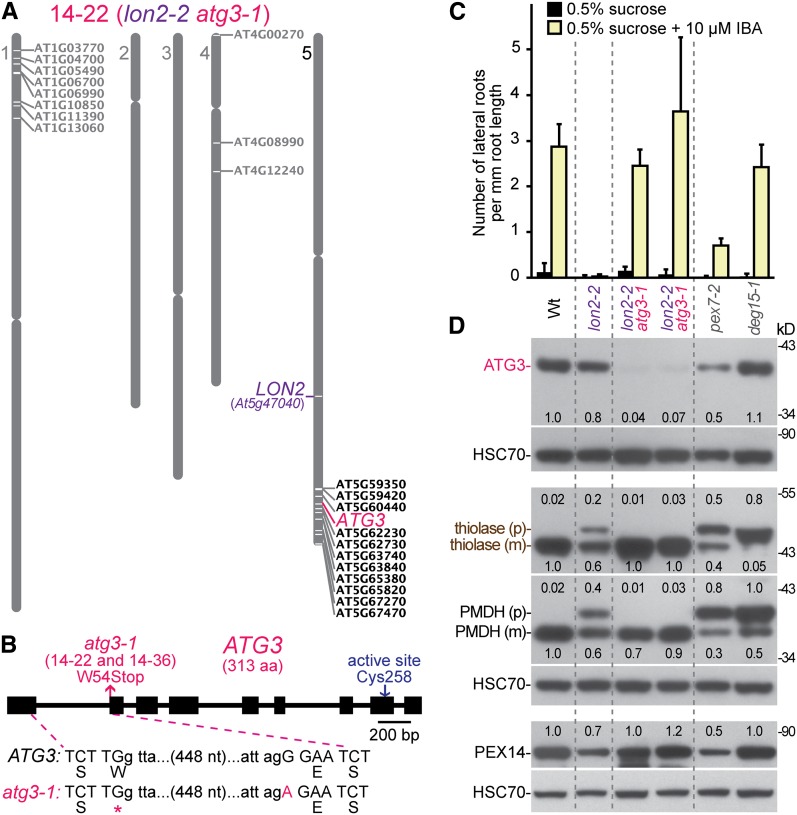
We used recombination mapping to localize the causal lesions in four suppressors (1-1, 4-114, 5-49, and 16-24) to the bottom of chromosome 5 between At5g42590 and At5g54950, near the LON2 locus (At5g47040) (Figure 2A). This linkage complicated our ability to identify the causal lesions by traditional mapping methods. Therefore, we used whole-genome sequencing of backcrossed suppressor lines to identify mutations in three suppressors (4-114, 5-49, and 16-24). Among the homozygous mutations present in these suppressors (Figures 3A to 3C; see Supplemental Data Set 1 online), each carried an independent mutation in ATG7 (At5g45900; Figure 2B). In addition, we identified atg7 lesions in the 1-1 and 8-8 suppressors (Figure 2B) following PCR amplification and sequencing of ATG7 genomic DNA from these mutants (see Supplemental Table 1 online). All five atg7 alleles identified in our lon2 suppressors are expected to impair ATG7 function; three were nonsense alleles, which we renamed atg7-4 (suppressor 5-49; Gln-12 to stop), atg7-5 (suppressor 1-1; Trp-119 to stop), and atg7-6 (suppressor 8-8; Trp-344 to stop), while two changed conserved Gly residues (see Supplemental Figure 1 online) to Asp residues, which we renamed atg7-7 (suppressor 16-24; Gly-247 to Asp) and atg7-8 (suppressor 4-114; Gly-286 to Asp).
Figure 2.
Multiple atg7 Alleles Suppress lon2-2 Peroxisomal Defects.
(A) Several lon2 suppressors mapped near ATG7 and LON2 on the southern arm of chromosome 5. The fractions of recombinant chromosomes identified at two mapping markers (K16E1 and PDC2) in the 4-114 (lon2-2 atg7-8) suppressor mapping population are shown.
(B) Diagram of the ATG7 gene. Boxes and lines denote protein coding regions and introns, respectively. The ThiF-like adenylation domain is indicated. Triangles mark the locations of previously described T-DNA insertion alleles. Arrows indicate the ATG7 active-site Cys and the positions of the nonsense (atg7-4, atg7-5, and atg7-6) and missense (atg7-7 and atg7-8) mutations that were identified as lon2-2 suppressors. The sequence of the atg7-8 missense allele relative to the wild-type ATG7 sequence is shown. The original isolation number of each mutation is listed in parentheses. aa, amino acids.
(C) Number of lateral roots per millimeter root length of 8-d-old wild type (Wt), lon2-2, lon2-4, lon2-2 atg7, atg7-2, atg7-3, pex7-2, and deg15-1 seedlings grown under yellow-filtered light on Suc-supplemented medium for 4 d and then transferred to Suc-supplemented medium with or without 10 μM IBA for an additional 4 d. Error bars show sd (n ≥ 8).
(D) Extracts prepared from 10-d-old seedlings grown in white light on Suc-supplemented medium were processed for immunoblotting with antibodies to the indicated proteins. PMDH is expressed as a precursor (p); the PTS2 region is cleaved to a mature (m) form in the peroxisome. pex7-2 is a control displaying reduced PMDH processing because of reduced matrix protein import (Ramón and Bartel, 2010); deg15-1 displays reduced processing because it is a null allele of the PTS2 processing protease (Schuhmann et al., 2008; Lingard and Bartel, 2009). Membranes from duplicate gels were serially probed with the indicated antibodies to obtain the top two and bottom three panels. Protein loading was monitored by probing with antibodies against HSC70. Numbers below bands indicate the ATG7/HSC70 or PEX7/HSC70 ratios in the labeled lanes, with the wild-type ratio normalized to 1.0. The positions of molecular mass markers (in kilodaltons) are shown on the right.
[See online article for color version of this figure.]
Figure 3.
Whole-Genome Sequencing Reveals atg7 and atg2 Lesions in lon2-2 Suppressors.
Genomic DNA prepared from pooled F3 seedlings from three backcrossed lines was sequenced and examined for homozygous single nucleotide polymorphisms typical of EMS mutagenesis (G/C to A/T transitions) in splice sites and coding sequences (nonsynonymous changes). Locus identifiers of mutated genes were placed to the right of their positions on the five Arabidopsis chromosomes using the Chromosome Map Tool at The Arabidopsis Information Resource (www.Arabidopsis.org). Mutations within the mapping intervals are depicted in black, other lesions are depicted in gray, and the LON2 locus is indicated. Mutations in ATG7 were found in mutants 5-49 (A), 16-24 (B), and 4-114 (C); a mutation in ATG2 was found in suppressor 13-3 (D).
[See online article for color version of this figure.]
ATG7 is a 697–amino acid ATP-dependent ubiquitin-activating (E1)-like enzyme that activates two ubiquitin-like modifiers acting in the autophagy conjugation pathway, resulting in ATG8 lipidation with phosphatidylethanolamine (PE). ATG8-PE in the inner autophagosomal membrane docks via adaptor proteins with cytosolic components to be engulfed, and ATG8-PE in the outer membrane is tethered to the microtubule-based transport machinery to allow autophagosome delivery to the vacuole (reviewed in Li and Vierstra, 2012). To determine whether blocking autophagy affected peroxisome physiology, we compared the IBA responsiveness and PTS2 processing in atg7 null mutants to two lon2 alleles and our lon2-2 atg7 suppressors. The atg7-2 and atg7-3 null alleles harbor T-DNA insertions in the 7th exon and abolish autophagy (Hofius et al., 2009; Chung et al., 2010; Lai et al., 2011; Wang et al., 2011). lon2-4 carries a splice site mutation at the 3′ end of intron 3 and generates a premature stop codon, likely truncating the protein. Both atg7-2 and atg7-3 single mutants responded to IBA similarly to the wild type and fully processed PMDH, whereas lon2-2 and lon2-4 mutants were completely resistant to the promotive effects of IBA on lateral root production and incompletely processed PMDH (Figures 2C and 2D).
All five of our atg7 alleles suppressed lon2-2 IBA resistance (Figure 2C) and rescued lon2 defects in PMDH processing (Figure 2D). We examined ATG7 protein levels in the suppressor mutants and did not detect ATG7 protein in the atg7-4, atg7-5, or atg7-6 nonsense alleles. Although the atg7-7 and atg7-8 missense alleles retained low levels of atg7 protein (Figure 2D), this protein is likely dysfunctional as atg7-7 and atg7-8 suppressed lon2-2 phenotypic defects as fully as the nonsense alleles (Figures 2C and 2D). As ATG7 activity is essential for autophagosome formation (reviewed in Li and Vierstra, 2012), we concluded that an intact autophagy system is required for lon2 deficiencies in IBA responsiveness and PTS2 processing to be apparent.
Mutations in the Autophagy Gene ATG2 Suppress lon2 Defects
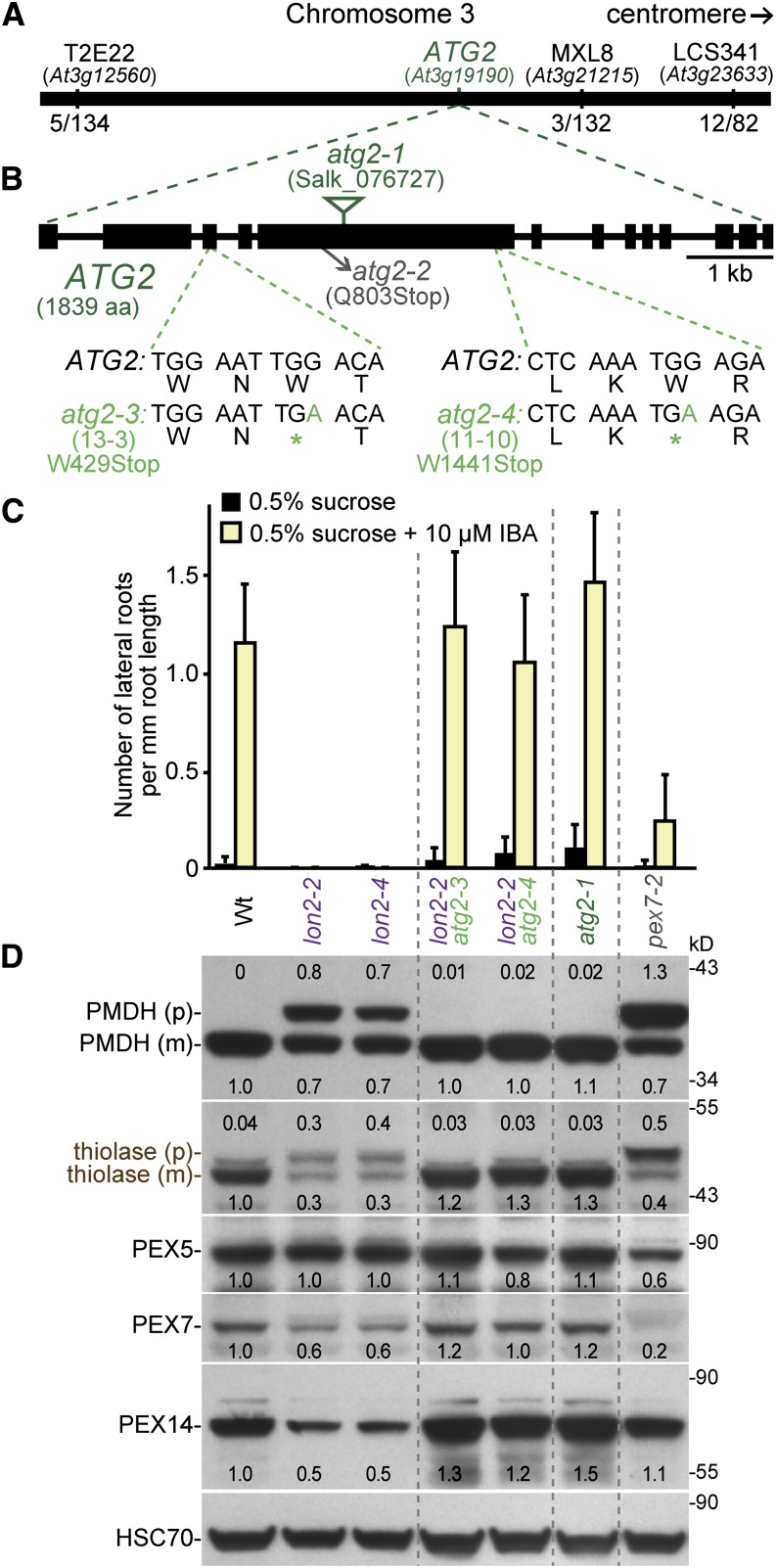
We used recombination mapping to localize suppressors 11-10 and 13-3 to the northern arm of chromosome 3 between At3g12560 and At3g23633 (Figure 4A). We sequenced the genome of a backcrossed line of suppressor 13-3, and among the seven homozygous mutations in coding sequences in the mapping interval (Figure 3D; see Supplemental Data Set 1 online), we identified a nonsense mutation (Trp-429 to stop) in the ATG2 locus (At3g19190), which we named atg2-3 (Figure 4B). Because the 11-10 suppressor mapped to a similar region, we amplified and sequenced ATG2 in this mutant and discovered a second nonsense allele (Trp-1441 to stop), which we named atg2-4 (Figure 4B; see Supplemental Table 1 online). In yeast, ATG2 associates peripherally with preautophagosomal structures during expansion and closure of the vesicle through association with ATG18 and the multi-membrane-spanning ATG9 (Suzuki et al., 2007; Xie and Klionsky, 2007).
Figure 4.
Multiple atg2 Alleles Suppress lon2-2 Peroxisomal Defects.
(A) Two lon2-2 suppressors (11-10 and 13-3) mapped near ATG2 on the northern arm of chromosome 3. The fraction of recombinant chromosomes identified at three mapping markers (T2E22, MXL8, and LCS341) in the 11-10 (lon2-2 atg2-4) suppressor mapping population are shown.
(B) Diagram of the ATG2 gene. Boxes and lines denote protein coding regions and introns, respectively. The locations of the previously characterized T-DNA insertion (atg2-1) and nonsense (atg2-2) alleles are shown. The sequences generated by the atg2-3 and atg2-4 nonsense mutations identified as lon2-2 suppressors are shown. The original number of each mutation is listed. aa, amino acids.
(C) Number of lateral roots per millimeter root length of 8-d-old wild-type (Wt), lon2-2, lon2-4, lon2-2 atg2, atg2-1, and pex7-2 seedlings that were grown under yellow-filtered light on Suc-supplemented medium for 4 d and then transferred to Suc-supplemented medium with or without 10 μM IBA for an additional 4 d. Error bars show sd (n ≥ 8). This experiment was conducted at the same time as the Figure 2C experiment; control data are duplicated on the two graphs for clarity.
(D) Extracts from 10-d-old seedlings grown in white light on Suc-supplemented medium were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. Thiolase and PMDH are synthesized as precursors (p) that are processed in the peroxisome to mature forms (m) lacking the PTS2 region. Protein loading was monitored by probing with antibodies against HSC70. Numbers below bands (or above for the PMDH and thiolase precursor bands) indicate the ratio of the band to HSC70, normalized such that the ratio from the wild type was set to 1.0. The positions of molecular mass markers (in kilodaltons) are shown on the right.
[See online article for color version of this figure.]
We examined an atg2 null allele in assays of peroxisome function and found that seedlings of the atg2-1 T-DNA allele (Inoue et al., 2006; Yoshimoto et al., 2009) responded to IBA similarly to the wild type (Figure 4C) and processed PTS2 proteins normally (Figure 4D). Like our atg7 alleles, both atg2 nonsense alleles recovered in our screen fully suppressed lon2-2, producing similar numbers of lateral roots as the wild type when treated with IBA (Figure 4C) and fully processing the PTS2 proteins PMDH and thiolase (Figure 4D). Our isolation of atg2 as a suppressor of lon2 physiological and molecular defects supports the conclusion that autophagy is required to observe lon2 IBA resistance and PTS2-processing defects.
A Mutation in the Autophagy Gene ATG3 Suppresses lon2 Defects
We used recombination mapping to localize suppressors 14-22 and 14-36 to the southern arm of chromosome 5 near the LON2 locus. We sequenced the genome of pooled backcrossed lines of the 14-22 suppressor. Among the 12 homozygous mutations on chromosome 5 (Figure 5A; see Supplemental Data Set 1 online), we identified a mutation in the ATG3 locus (At5g61500), which we named atg3-1 (Figure 5B). This mutation changes the G that is the first nucleotide of exon 2 to an A, which would change Trp-54 to a stop codon if the mRNA were still spliced correctly. We also sequenced a single backcrossed line of the 14-36 mutant and found the same atg3-1 lesion (see Supplemental Data Set 1 online). Because 14-22 and 14-36 arose from the same M2 pool and carried the same atg3 lesion, we concluded that these isolates were likely siblings. ATG3 functions as the E2-like conjugating enzyme that acts with ATG7 to lipidate ATG8 (Phillips et al., 2008; Yamaguchi et al., 2012); atg3 mutants are thus expected to display defects similar to those of atg7 mutants. We found that ATG3 immunoreactivity was reduced in the lon2-2 atg3-1 mutant, indicating that the atg3-1 splicing or nonsense lesion impaired ATG3 accumulation (Figure 5D). The atg3-1 nonsense allele fully suppressed lon2-2 defects; the lon2-2 atg3-1 double mutant induced wild-type numbers of lateral roots in the presence of IBA (Figure 5C) and fully processed the thiolase and PMDH PTS2 proteins (Figure 5D).
Figure 5.
An atg3 Mutant Suppresses lon2-2 Peroxisomal Defects.
(A) Whole-genome sequencing data for the backcrossed 14-22 suppressor was analyzed as described in the legend to Figure 3. A G-to-A mutation in the first nucleotide of ATG3 exon 2 was found. The same lesion was present in whole-genome sequencing data from the 14-36 suppressor (see Supplemental Data Set 1 online).
(B) The position of the atg3 mutation present in the 14-22 and 14-36 suppressors is shown on a diagram of the ATG3 gene along with the location of the ATG7 active-site Cys. Boxes and lines denote protein coding regions and introns, respectively. aa, amino acids.
(C) Number of lateral roots per millimeter root length of 8-d-old wild-type (Wt), lon2-2, lon2-2 atg3-1 (two isolates), pex7-2, and deg15-1 seedlings that were grown under white light on Suc-supplemented medium for 5 d and then transferred to Suc-supplemented medium with or without 10 μM IBA and grown under yellow-filtered light for an additional 3 d. Error bars show sd (n ≥ 5).
(D) Extracts from 10-d-old seedlings grown in white light on Suc-supplemented medium were processed for immunoblotting. Triplicate membranes were serially probed with antibodies to the indicated proteins. Thiolase and PMDH are synthesized as precursors (p) that are processed to a mature form (m) lacking the PTS2 region in the peroxisome. Protein loading was monitored by probing with antibodies against HSC70. Numbers above or below bands indicate the ratio of the band to HSC70, normalized such that the ratio from the wild type was set to 1.0. The positions of molecular mass markers (in kilodaltons) are shown on the right.
[See online article for color version of this figure.]
Disrupting Autophagy Reduces lon2 Defects in Peroxisome Size and Abundance
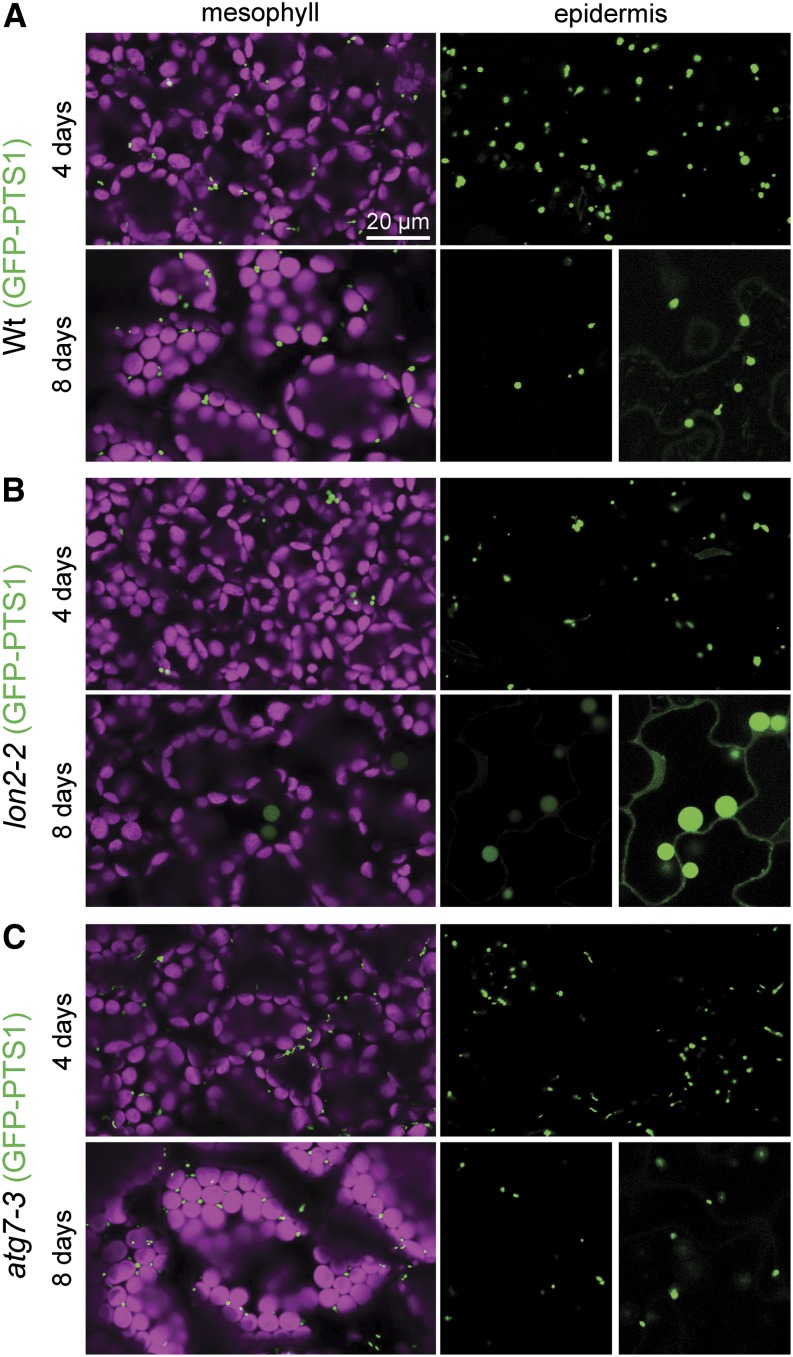
Enlarged peroxisomes can reflect defects in peroxisome division or metabolic defects in the organelle. For example, mutants disrupted in various peroxisome division factors (Mano et al., 2004; Lingard et al., 2008; Zhang and Hu, 2009), the peroxisomal NAD+ and CoA transporter (PXN) (Mano et al., 2011; Agrimi et al., 2012; Bernhardt et al., 2012), or the fatty acid β-oxidation enzyme 3-ketoacyl-CoA thiolase (KAT2/PED1) (Hayashi et al., 1998) have enlarged peroxisomes. Similarly, lon2 mutants carrying a peroxisomally targeted GFP derivative display larger fluorescent puncta than the wild type (Burkhart et al., 2013). We observed that the size of lon2-2 peroxisomes marked by GFP-PTS1 (Zolman and Bartel, 2004) resembled wild-type peroxisomes in epidermal and mesophyll cells of 4-d-old seedling cotyledons (Figures 6A and 6B). As the seedlings aged, however, lon2-2 puncta appeared larger and less abundant in both mesophyll and epidermal cells (Figure 6B). In addition, some cytosolic GFP-PTS1 became apparent in lon2-2 cotyledon cells (Figure 6B), concomitant with the appearance of PTS2-processing defects as lon2 seedlings age (Lingard and Bartel, 2009). In contrast with the enlarged GFP-PTS1 puncta, chloroplasts appeared similar to the wild type in the lon2-2 mutant (Figure 6B). Like lon2-2 expressing 35S:GFP-PTS1 (Figure 6B), cotyledons of 8-d-old lon2-2 seedlings expressing 35S:PTS2-GFP displayed large puncta that appeared to be less abundant than wild-type seedling peroxisomes (Figures 7A and 7B). Because older lon2 seedlings appeared to possess fewer peroxisomes than the wild type, we used immunoblotting to examine levels of several peroxins in 10-d-old lon2 seedlings. We observed that lon2 mutants displayed apparently normal levels of the PTS1 receptor PEX5 but slightly reduced levels of the PTS2 receptor PEX7 (Figure 4D) and the membrane peroxin PEX14 (Figures 4D and 5D), consistent with our observation that peroxisomes were less abundant in older lon2 seedlings.
Figure 6.
GFP-PTS1 Localization Reveals Import and Morphology Defects in lon2-2 and Normal Appearance of atg7-3 Peroxisomes.
Cotyledon mesophyll and epidermal cells in 4- and 8-d-old wild-type (Wt) (A), lon2-2 (B), and atg7-3 (C) seedlings expressing 35S:GFP-PTS1 were imaged for GFP fluorescence (green) using confocal microscopy. Mesophyll cells also were imaged for chlorophyll autofluorescence (magenta) to visualize chloroplasts. Epidermal cells from 8-d-old seedlings were imaged at both normal (left panels) and heightened (right panels) gain settings to allow clear visualization of the GFP-PTS1 import defect of the lon2-2 mutant. Bar = 20 μm.
Figure 7.
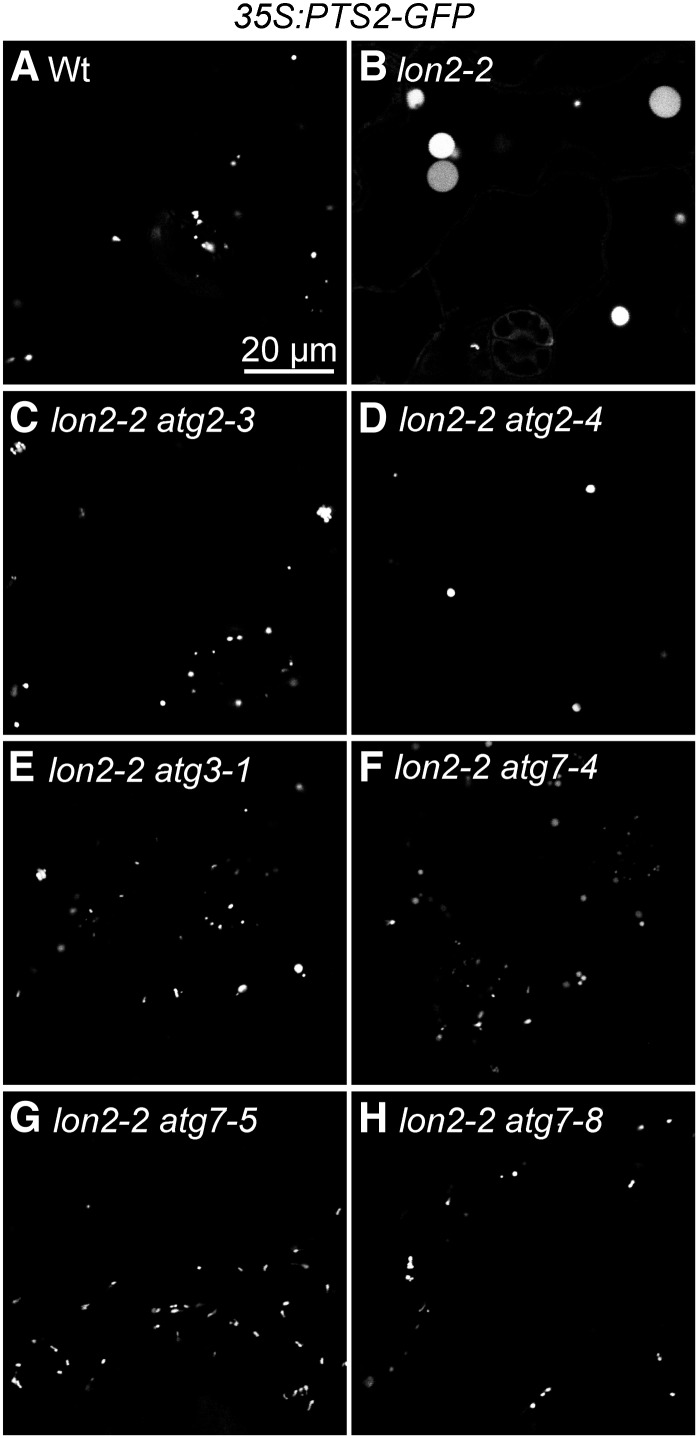
atg2, atg3, and atg7 Mutations Suppress lon2-2 Defects in Peroxisome Size.
Cotyledon epidermal cells in 8-d-old wild-type (Wt), lon2-2, and lon2-2 suppressor seedlings expressing 35S:PTS2-GFP were imaged for GFP fluorescence (white) using confocal microscopy. Bar = 20 μm.
In contrast with the large puncta observed in 8-d-old lon2-2 seedlings, the lon2-2 atg2, lon2-2 atg3, and lon2-2 atg7 mutants had peroxisomes that more closely resembled wild-type peroxisomes in both size and abundance; we no longer detected enlarged puncta in the suppressors (Figures 7C to 7H). Moreover, PEX14 levels were no longer reduced in lon2 atg2 (Figure 4D) or lon2 atg3 (Figure 5D) mutants compared with the wild type. Because ATG2, ATG3, and ATG7 are essential for autophagosome formation and closure, we concluded that mutations in these loci prevented deposition of lon2 peroxisomes into the lytic vacuole.
Because disrupting autophagy suppressed the size and abundance defects of lon2 peroxisomes (Figure 7), we also observed the effects of autophagy defects on peroxisome morphology in seedlings with wild-type LON2. We found that GFP-PTS1 fluorescence appeared similar to the wild type in cotyledon cells of 4- and 8-d-old atg7-3 seedlings, which both displayed abundant small peroxisomes (Figure 6C). Our observations that disrupting autophagy did not notably impair peroxisome morphology or abundance in these cells (Figure 6C) and that atg2 and atg7 mutant seedlings responded like the wild type to IBA (Figures 2C and 4C) are consistent with the possibility that autophagy does not dramatically limit peroxisome function in wild-type seedlings, at least in the tissues that we assessed.
Disrupting Autophagy Stabilizes Several Peroxisomal Matrix Proteins in lon2 Mutants
During early seedling development, the metabolism and utilization of stored fatty acids for energy is a critical function of peroxisomes. The contents of seedling peroxisomes shift 4 to 5 d after germination when the glyoxylate cycle becomes obsolete as oil stores are depleted and photosynthesis begins. During this remodeling, the glyoxylate cycle enzymes ICL and MLS are degraded, and photorespiration enzymes, including HPR, are synthesized. Ubiquitin-dependent degradation is implicated in peroxisome remodeling as mutations in both PEX4 and PEX22 result in partial ICL and MLS stabilization (Zolman et al., 2005; Lingard et al., 2009). By contrast, resident peroxisomal proteases have not been implicated in matrix protein turnover; ICL and MLS are not notably stabilized in deg15 or lon2 mutants (Lingard and Bartel, 2009; Burkhart et al., 2013).
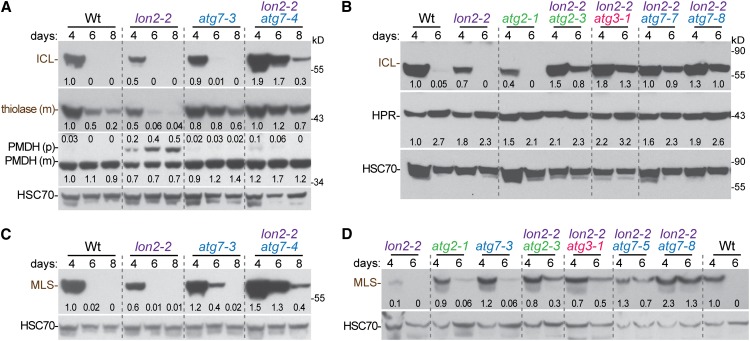
To determine whether autophagy plays a role in peroxisome remodeling via protein turnover during seedling maturation, we examined ICL levels in maturing seedlings carrying the atg7-3 and atg2-1 null alleles. We found that ICL appeared to be degraded similarly to the wild type in both mutants (Figures 8A and 8B), suggesting that autophagy is not a major pathway through which obsolete ICL is degraded. Moreover, we confirmed that ICL was not stabilized in the lon2-2 mutant (Figures 8A and 8B), consistent with previous reports (Lingard and Bartel, 2009; Burkhart et al., 2013). Surprisingly, however, we observed that ICL was markedly stabilized in our lon2-2 suppressors, including lon2-2 atg2-3, lon2-2 atg3-1, and several lon2-2 atg7 mutants (Figures 8A and 8B). These results suggest that autophagy must be blocked to observe matrix protein stabilization in a lon2 mutant background.
Figure 8.
Several Peroxisomal Enzymes Are Stabilized in lon2-2 atg Mutants.
Extracts from 4-, 6-, and 8-d-old wild-type (Wt), lon2, atg, and lon2 atg seedlings grown in white light were processed for immunoblotting and serially probed with antibodies against the indicated proteins. Protein loading was monitored by probing with antibodies against HSC70. Numbers below bands (or above for the PMDH precursor band) indicate the ratio of the band to HSC70, normalized such that the ratio from 4-d-old wild type was set to 1.0. The positions of molecular mass markers (in kilodaltons) are shown on the right.
[See online article for color version of this figure.]
We also examined degradation of MLS during seedling maturation. As previously reported (Lingard et al., 2009), MLS was similarly unstable in wild-type and lon2 seedlings (Figures 8C and 8D). Like ICL, MLS was dramatically stabilized in lon2-2 atg double mutants (Figures 8C and 8D). In addition, we found that MLS was slightly stabilized in the atg7-3 and atg2-1 single mutants (Figures 8C and 8D). These results suggest that both LON2 and autophagy contribute to MLS turnover in wild-type seedlings.
To examine whether LON2 substrates might extend beyond the ICL and MLS glyoxylate cycle enzymes, we monitored thiolase levels during seedling development. Although not as dramatic as the complete disappearance of ICL and MLS during seedling maturation, thiolase levels also decline as seedlings mature (Lingard and Bartel, 2009; Lingard et al., 2009). This decline is accelerated in lon2 mutants (Lingard and Bartel, 2009) (Figure 8A). As with MLS, we found that the rate of thiolase degradation was somewhat reduced in an atg7 mutant and further delayed in a lon2 atg7 mutant (Figure 8A).
Because the HPR photorespiration enzyme begins accumulating around 4 d after germination (Lingard et al., 2009), we also assessed HPR levels in these experiments to examine whether delayed development might contribute to the apparent stabilization of ICL, MLS, and thiolase in lon2 atg or atg seedlings. We found that HPR accumulated similarly to the wild type in these mutants (Figures 8B and 8D). Like HPR, the PMDH matrix protein was present at wild-type levels in lon2, atg7, and lon2 atg7 mutants (Figure 8A). We concluded that the prolonged presence of glyoxylate cycle and β-oxidation enzymes in lon2 atg mutants was due to a degradation defect rather than delayed development.
DISCUSSION
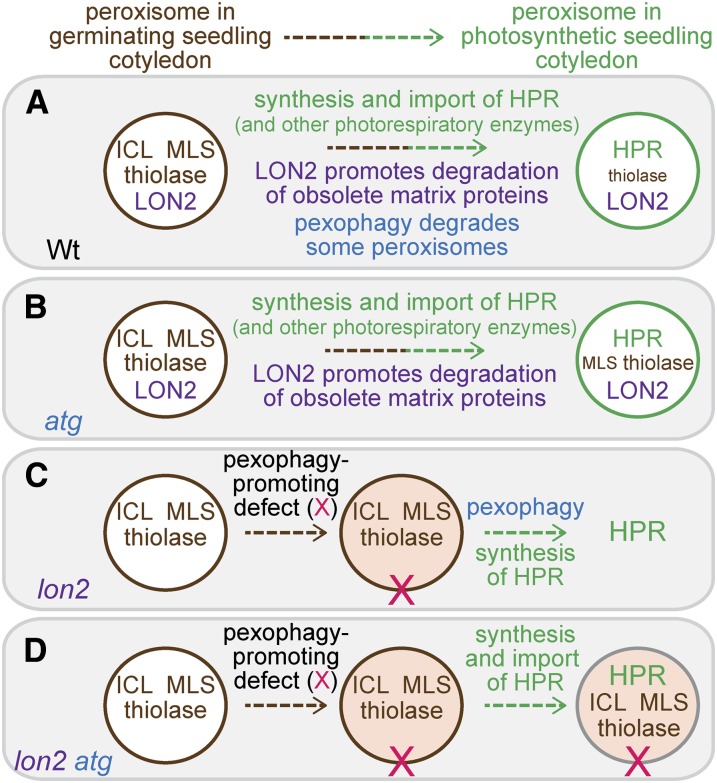
Arabidopsis LON2 is a peroxisomal protease that is necessary for sustained import of matrix proteins into peroxisomes (Lingard and Bartel, 2009). We screened for lon2 suppressors and found that the lon2 peroxisome-related defects, IBA resistance and inefficient PTS2 processing (Figures 1, 2, 4, and 5) as well as large peroxisomes (Figure 7), were suppressed by genetically preventing autophagy. Moreover, impairing autophagy in the lon2 background revealed an additional phenotype: clear stabilization of the β-oxidation enzyme thiolase and the glyoxylate cycle enzymes ICL and MLS (Figure 8). Although not as dramatic, we also observed slight stabilization of thiolase and MLS in atg single mutants (Figure 8), suggesting that some peroxisomes are degraded via autophagy in wild-type Arabidopsis seedlings. Our data are consistent with a model in which Arabidopsis LON2 assists in the degradation of obsolete matrix proteins (Figure 9A). When autophagy is prevented in atg mutants, some matrix proteins (e.g., MLS and thiolase) appear to be degraded slightly more slowly (Figure 9B). When matrix protein degradation is impaired in lon2 mutants, peroxisomes may be recognized as abnormal and targeted for destruction via autophagy at increased rates (Figure 9C). As lon2 cells age and peroxisome numbers decline, newly synthesized matrix proteins appear to be inefficiently imported, presumably leading to the observed lon2 defects in IBA-responsive lateral rooting and processing of PTS2 proteins. When key autophagy genes (e.g., ATG2, ATG3, or ATG7) also are nonfunctional, autophagy is prevented, allowing lon2 peroxisomes to persist and continue importing and processing matrix proteins and metabolizing IBA, despite the continued presence of obsolete matrix proteins in these peroxisomes (Figure 9D).
Figure 9.
A Working Model for LON2 Action during Maturation of Seedling Peroxisomes.
(A) As wild-type seedlings mature, LON2 acts to degrade obsolete matrix proteins, including glyoxylate cycle enzymes ICL and MLS and the β-oxidation enzyme thiolase. In addition, autophagy acts to remove some peroxisomes, contributing to matrix protein degradation. As photosynthesis is established, peroxisomal photorespiratory enzymes, exemplified by HPR, are synthesized and imported into peroxisomes.
(B) When autophagy is prevented, seedling peroxisomes function normally, but reduced peroxisome turnover results in slight stabilization of certain matrix proteins (MLS and thiolase).
(C) In lon2 seedlings, autophagy of peroxisomes (pexophagy) is triggered, perhaps due to a modification of the peroxisome (X). Although obsolete matrix proteins are no longer degraded by LON2, peroxisomes and their contents are degraded via pexophagy, resulting in ICL, MLS, and thiolase disappearance. As pexophagy continues, the number of import-competent peroxisomes declines, and cytosolic accumulation of newly synthesized matrix proteins, such as HPR, becomes apparent.
(D) In lon2 atg double mutants, autophagy is absent and lon2 peroxisomes are no longer subject to pexophagy. The continued import of matrix proteins into lon2 peroxisomes allows peroxisome functions to continue, restoring PTS2 processing and IBA-to-IAA conversion.
[See online article for color version of this figure.]
The model predicts that proteins delivered to the peroxisome matrix in lon2 mutants will be degraded via pexophagy whereas proteins synthesized after lon2 cells have few normal peroxisomes will be predominantly cytosolic. Whether the latter matrix proteins are stable or unstable in the cytosol can vary depending on the protein. For example, HPR and PMDH are abundant in the cytosol of 8-d-old lon2 cotyledon cells (Lingard and Bartel, 2009) and are present at wild-type levels in lon2 seedling extracts (Figure 8). By contrast, thiolase levels are reduced in lon2 seedlings (Figure 8) (Lingard and Bartel, 2009), implying that thiolase is degraded when not appropriately compartmentalized in the peroxisome.
Peroxisomal LON isoforms are found in plants (Ostersetzer et al., 2007; Lingard and Bartel, 2009), mammals (Kikuchi et al., 2004; Omi et al., 2008; Okumoto et al., 2011), and a subset of lower eukaryotes, including H. polymorpha (Aksam et al., 2007) and P. chrysogenum (Bartoszewska et al., 2012), but not yeast (Saccharomyces cerevisiae) or fruit fly (Drosophila melanogaster) (Lingard and Bartel, 2009). Unlike Arabidopsis lon2 mutants, mutants lacking the H. polymorpha peroxisomal LON isoform (Pln) display slightly more numerous peroxisomes than the wild type (Aksam et al., 2007). P. chrysogenum pln mutants accumulate inactive catalase-peroxidase, implying that Pln normally functions in the degradation of oxidatively damaged proteins (Bartoszewska et al., 2012). The apparently normal peroxisome numbers in P. chrysogenum pln mutants, combined with the normal peroxisomal GFP-PTS1 localization in P. chrysogenum and H. polymorpha pln mutants (Aksam et al., 2007; Bartoszewska et al., 2012), suggests that fungal peroxisomes lacking their resident LON protease are not targeted for pexophagy.
Peroxisomal LON (pLon) isoforms also are present in mammals (Kikuchi et al., 2004; Omi et al., 2008; Okumoto et al., 2011). RNA interference–mediated reduction of pLon levels in mammalian cells via transient transfection does not noticeably alter peroxisomal morphology or matrix protein import (Okumoto et al., 2011). By contrast, expressing a dominant-negative pLon derivative in tissue culture cells impairs peroxisomal matrix protein import, and overexpressing a tagged pLon derivative results in reduced numbers of enlarged peroxisomes (Omi et al., 2008), similar to our observations in the Arabidopsis lon2 mutant. It would be interesting to examine a role for pexophagy in these changes.
Our lon2 suppressor screen identified atg2, atg3, and atg7 alleles. Three atg7 null alleles (Figure 2B) have previously been characterized using reverse-genetic approaches: atg7-1 (apg7-1) is a T-DNA insertion in the 10th intron and displays premature senescence (Doelling et al., 2002; Thompson et al., 2005), atg7-2 is a T-DNA insertion in the 7th exon that lacks ATG8-PE adducts and confers hypersensitivity to darkness-induced carbon starvation (Hofius et al., 2009; Chung et al., 2010), and atg7-3 is a T-DNA insertion in the 7th exon that confers powdery mildew (Golovinomyces cichoracearum) resistance (Wang et al., 2011) and hypersusceptibility to necrotrophic fungal pathogens (Lai et al., 2011). Previously reported Arabidopsis ATG2 alleles include atg2-1, a T-DNA insertion in the 5th exon that disrupts autophagy and induces early senescence and excessive programmed cell death under nutrient-rich conditions (Inoue et al., 2006; Yoshimoto et al., 2009), and atg2-2, a nonsense mutation (Gln-803 to stop) that blocks autophagy, displays early senescence, and exhibits enhanced powdery mildew resistance via increased cell death and constitutive defense-related gene expression (Wang et al., 2011). Our lon2-suppressing atg7 alleles include three nonsense alleles, and all five alleles reduce ATG7 protein accumulation (Figures 2B and 2D). Similarly, our atg2 and atg3 alleles are all nonsense alleles (Figures 4B and 5B). Because ATG2 functions in a different step of the autophagy pathway than ATG3 and ATG7 (reviewed in Li and Vierstra, 2012), it is likely that any mutation abolishing autophagy would suppress lon2 defects. Our recovery of multiple mutations in ATG2 and ATG7 may reflect the fact that both are single-copy genes in Arabidopsis that are above average in size, thereby presenting unusually large targets for mutagenesis. ATG3 also is a single-copy gene in Arabidopsis but had not previously emerged from forward genetic screens. As atg3 alleles are not present in publically available T-DNA collections, the atg3-1 nonsense allele described here will provide a useful tool for analyzing ATG3 function in Arabidopsis. Moreover, we anticipate that further screening for lon2 suppressors will uncover additional factors required for efficient autophagy in seedlings, as well as genes that might be specifically required for pexophagy in plants.
The reduced numbers of peroxisomes in lon2 mutants and the suppression of lon2 defects by blocking autophagy are consistent with the possibility that pexophagy is induced in lon2 mutants (Figure 9). What might trigger pexophagy in lon2? Autophagy in plants is generally used under stress conditions including starvation and pathogen attack, as well as during programmed cell death (reviewed in Bassham, 2007; Reumann et al., 2010; Li and Vierstra, 2012). Perhaps metabolic stress in lon2 mutants triggers pexophagy as reactive oxygen species (ROS) are produced by peroxisomal metabolism. P. chrysogenum pln mutants display increased oxidative stress (Bartoszewska et al., 2012), and elevated ROS is accompanied by increased pexophagy in H. polymorpha (van Zutphen et al., 2011). However, H. polymorpha peroxisomal LON mutants display both increased ROS levels and more peroxisomes than the wild type, so a positive relationship between ROS and pexophagy is not universal (Aksam et al., 2007). H. polymorpha pexophagy also can be triggered by degradation of Pex3p, a membrane peroxin acting in early peroxisome biogenesis (van Zutphen et al., 2011), suggesting that alterations to peroxisome membrane proteins can trigger pexophagy. We detected reduced levels of the membrane peroxin PEX14 in lon2 seedlings, and it will be interesting to learn whether this alteration is a cause or consequence of the increase in pexophagy observed in lon2 mutants. Finally, accumulation of damaged or misfolded organellar proteins can trigger autophagy of the affected organelle. For example, treating Arabidopsis seedlings with tunicamycin, which blocks protein glycosylation and triggers the unfolded protein response, promotes autophagy of the endoplasmic reticulum (Liu et al., 2012). In addition, accumulation of protein aggregates in the peroxisome matrix is accompanied by heightened pexophagy in H. polymorpha (Manivannan et al., 2013). It is tempting to speculate that the oxidized or obsolete matrix proteins that accumulate as lon2 seedlings age trigger pexophagy.
Regardless of the specific pexophagy trigger in lon2, it does not seem that undegraded matrix proteins cause the apparent peroxisome expansion observed in lon2 as lon2 atg double mutants overaccumulate ICL, MLS, and thiolase (Figure 8) but have normally sized peroxisomes (Figure 7). This observation raises the question of whether the large spherical structures accumulating PTS2-GFP or GFP-PTS1 in older lon2 cells are in fact peroxisomes or whether these structures might be pexophagy intermediates. Future studies of seedlings in which autophagosome markers have been introduced into lon2 and lon2 atg mutants are needed to address this question.
Specialized forms of autophagy, such as pexophagy, require receptors targeting cargo destined for destruction to the inner isolation membrane. For example, Pichia pastoris Atg30 localizes to the peroxisomal membrane through interactions with the membrane peroxins Pex3 and Pex14, where phosphorylated Atg30 can recruit Atg11 and Atg17 and induce pexophagy (Farré et al., 2008). In S. cerevisiae, Atg36 binds to Pex3 and functions as a pexophagy receptor (Motley et al., 2012). However, Atg30 and Atg36 homologs are not found outside of fungi (Farré et al., 2008; Motley et al., 2012). A more widespread class of selective autophagy receptors is represented by p62 and NBR1, adaptors that tether ubiquitinated cargo to orthologs of the ATG8 ubiquitin-like protein on the isolation membrane (Bjørkøy et al., 2005; Kirkin et al., 2009). NBR1 functions as a pexophagy receptor in mammals (Deosaran et al., 2013). It will be interesting to learn whether Arabidopsis NBR1 (Svenning et al., 2011; Zientara-Rytter et al., 2011), which functions in autophagy of insoluble ubiquitinated aggregates (Zhou et al., 2013), similarly targets peroxisomes for pexophagy in Arabidopsis and whether any ubiquitinated proteins in the peroxisome membrane are required for pexophagy. It is noteworthy that several peroxisomal proteins are found in a recent proteomic analysis of ubiquitinated Arabidopsis proteins (Kim et al., 2013).
Three pathways by which obsolete or damaged peroxisomal matrix proteins might be turned over in Arabidopsis have been suggested (Burkhart et al., 2013): (1) degradation within the organelle by a resident protease, (2) degradation of the entire organelle by autophagy, and (3) retrotranslocation and ubiquitination for proteasomal degradation in the cytosol. The first and third pathways are consistent with the one-population hypothesis and detection of transitional peroxisomes containing both glyoxylate cycle and photorespiration enzymes (Titus and Becker, 1985; Nishimura et al., 1986; Sautter, 1986). Previous studies in Arabidopsis provide evidence for the third pathway (Zolman et al., 2005; Lingard et al., 2009; Burkhart et al., 2013), and our current data provide evidence for the first two pathways. Is it possible that all three mechanisms are in play? Peroxisomal protein import is unusual in that matrix proteins can be imported as fully folded, oligomeric complexes (McNew and Goodman, 1994; Lee et al., 1997). Rather than fully degrading matrix proteins, perhaps LON2 processes or disaggregates matrix proteins, receptor complexes, or receptor-matrix protein complexes to allow their ubiquitin-dependent retrotranslocation out of the organelle. In the absence of LON2 function, ubiquitinated matrix proteins might not efficiently retrotranslocate, resulting in ubiquitination at the surface of the peroxisome, which could trigger pexophagy. Future studies will be needed to resolve these possibilities.
Although pexophagy functions in mammals and yeast to degrade excess peroxisomes (Iwata et al., 2006; Till et al., 2012), plant peroxisomes are not known to undergo the dramatic shifts in abundance that facilitate the study of pexophagy in other systems. In addition, peroxisomal β-oxidation normally is required to mobilize carbon from fatty acids released during autophagy of other organelles. For example, autophagy allows plants to survive periods of carbon starvation, such as that induced by extended darkness (Thompson et al., 2005; Phillips et al., 2008). Peroxisomes also promote plant survival during carbon starvation (Dong et al., 2009; Contento and Bassham, 2010), suggesting that membrane lipids freed from extraneous organelles during autophagy are β-oxidized in peroxisomes. Gene expression studies suggest that peroxisomes also would mobilize carbon from fatty acids during senescence (Lopez-Huertas et al., 2000), another time when autophagy is important (Doelling et al., 2002; Hanaoka et al., 2002; Thompson et al., 2005; Xiong et al., 2005). In contrast with these instances in which the autophagy system collaborates with peroxisomes to recycle nutrients, our results show that autophagy also can dispose of damaged or abnormal peroxisomes and demonstrate a requirement for ATG2, ATG3, and ATG7 in this process. Our data indicate that pexophagy can dispose of seedling peroxisomes and provide tools for the future elucidation of pexophagy requirements and regulation in plant development.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana accession Columbia-0 (Col-0) or Col-0 transformed with 35S:PTS2-GFP (Woodward and Bartel, 2005a) were used as the wild type. All mutants were in the Col-0 accession. lon2-2/SALK_043857 (Lingard and Bartel, 2009), deg15-1/SALK_007184 (Schuhmann et al., 2008; Lingard and Bartel, 2009), atg2-1/SALK_076727 (Yoshimoto et al., 2009), atg7-2/GABI_655B06 (Chung et al., 2010), and atg7-3/SAIL_11_H07 (Lai et al., 2011; Wang et al., 2011) were previously described and were from the ABRC. lon2-2 35S:PTS2-GFP (Lingard and Bartel, 2009) and pex7-2 (Ramón and Bartel, 2010) were previously described. lon2-4 is a probable null allele that was isolated from a screen for Suc dependence and IBA resistance (by A.W. Woodward and M. Bjornson); it contains a G-to-A transition in the 3′ nucleotide of intron 3 that generates a frameshift followed by a premature stop codon. Col-0 transformed with 35S:GFP-PTS1 was described previously (Zolman and Bartel, 2004), and lon2-2 35S:GFP-PTS1 and atg7-3 35S:GFP-PTS1 lines were generated by crossing and genotyping the F2 progeny by PCR. Mutant genotypes were assayed using PCR-based markers (see Supplemental Table 2 online).
Seeds were surface-sterilized, resuspended in sterile water or 0.1% agar, and stratified at 4°C for 1 to 3 d. Unless otherwise noted, plants were grown at 22°C on plant nutrient medium (Haughn and Somerville, 1986) supplemented with 0.5% (w/v) Suc (PNS) and solidified with 0.6 or 1.0% agar. After stratification, seeds were placed under continuous light. To quantify IBA sensitivity, seedlings were grown on PNS plates under light filtered with yellow long-pass filters (to slow photochemical breakdown of indolic compounds; Stasinopoulos and Hangarter, 1990) for 4 to 5 d, transferred to new PNS plates with or without 10 μM IBA, and grown vertically under yellow light for an additional 3 to 4 d. Lateral roots emerged from the primary root were counted using a dissecting microscope; primary root lengths were measured with a ruler or using Image J 3.0 (http://rsbweb.nih.gov/ij/) after imaging seedlings using a Gel Doc System (Bio-Rad).
Mutant Isolation and Recombination Mapping
Seeds of lon2-2 (Lingard and Bartel, 2009) carrying the 35S:PTS2-GFP transgene (Woodward and Bartel, 2005a) were mutagenized with ethyl methanesulfonate (EMS) (Normanly et al., 1997) and grown in 16 pools. Approximately 5000 M2 seeds from each pool were surface sterilized, stratified for 2 d at 4°C, and plated on PNS supplemented with 3 μM IBA. Seedlings were grown vertically under yellow-filtered light for 10 d, and seedlings with more than three to four lateral roots were moved to soil for seed production. M3 progeny were retested for lateral root production on 10 µM IBA, and seedlings from the 61 strongest suppressor lines were tested by immunoblot analysis with anti-PMDH antibodies (as described below) for rescue of the lon2 PTS2-processing defect. Lines exhibiting IBA sensitivity and lacking PTS2-processing defects were retained as lon2 suppressors. Suppressor lines were backcrossed to lon2-2 once prior to phenotypic analyses shown in Figures 2 and 4. Homozygous backcrossed suppressor lines were selected from the F2 generation for IBA-induced lateral root formation and were subsequently genotyped for lon2-2 homozygosity as described (Lingard and Bartel, 2009).
For recombination mapping, suppressor mutants isolated in the Col-0 background carrying the lon2-2 T-DNA insertion were outcrossed to a lon2-2 line that had been introgressed into Landsberg erecta by three successive outcrosses to Landsberg erecta. F2 seedlings from each outcross were selected on 10 µM IBA for suppression of lon2-2 IBA resistance. DNA was isolated from F2 plants exhibiting wild-type lateral root numbers by homogenizing leaf tissue in extraction buffer (200 mM Tris, pH 7.5, 250 mM NaCl, 25 mM EDTA, and 0.5% SDS) followed by chloroform extraction and isopropanol precipitation of the aqueous phase. DNA samples were assayed using PCR-based polymorphic markers (see Supplemental Table 3 online) as previously described (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). ATG2, ATG3, and ATG7 were PCR amplified from genomic DNA prepared from mutants with the primer pairs listed in Supplemental Table 1 online. Amplicons were sequenced directly (Lone Star Labs) with the primers used for amplification.
Genomic DNA Isolation and Whole-Genome Sequencing
For each suppressor mutant, F3 seedlings from three backcrossed lines (homozygous for both lon2-2 and the suppressing lesion) were pooled for DNA extraction to reduce the number of noncausal homozygous mutations identified. Approximately 2000 surface-sterilized seeds were grown under continuous white light on PNS overlaid with sterile filter paper for 7 or 9 d. Seedlings were ground in liquid nitrogen and homogenized in 7 mL of prewarmed (65°C) Buffer S (110 mM Tris, pH 8.0, 55 mM EDTA, 1.54 M NaCl, and 1.1% hexadecyltrimethylammonium bromide). Homogenates were vortexed, supplemented with 0.7 mL of 20% SDS, mixed end-over-end, and incubated at 65°C for 2 h with occasional inversion. Samples were allowed to cool at room temperature for 5 min, after which 4 mL of 24:1 chloroform:isoamyl alcohol was added. Samples were mixed by inversion for 15 min and then centrifuged at 3000 rpm for 20 min. The aqueous phase was extracted with 4 mL of 24:1 chloroform:isoamyl alcohol and DNA was precipitated by adding 0.6 volumes of isopropanol. DNA was collected by centrifugation and dissolved in 4 mL of 10 mM Tris-HCl, 1 mM EDTA, pH 8.0. Samples were treated with 10 µg/mL of RNase A (Sigma-Aldrich; R-4875) at 37°C for 1 h. After chloroform extraction, DNA in the aqueous phase was precipitated by adding 0.1 volumes of 3 M sodium acetate, pH 5.2, and 2 volumes of ice-cold 95% ethanol. After centrifugation for 20 min at 2000 rpm, the DNA pellet was washed with 3 mL 70% ethanol. Sample tubes were inverted and allowed to drain for 20 min, dried at 37°C for 20 min, and dissolved in 200 to 500 μL 10 mM Tris-HCl, 1 mM EDTA, pH 8.0.
Genomic DNA (∼20 µg/mutant) was submitted to the Genome Technology Access Center at Washington University in St. Louis for sequencing with an Illumina HiSequation 2000 sequencer and comparison to The Arabidopsis Information Resource 10 build of the Arabidopsis Col-0 genome. Because EMS causes G/C-to-A/T transitions in Arabidopsis (Greene et al., 2003), we disregarded insertions and deletions and retained single nucleotide polymorphisms consistent with EMS mutagenesis and located in splice sites or coding sequences. We also discarded heterozygous lesions, those that resulted in synonymous codon changes, those that were present in our lab stock of Col-0, and two lesions found in all sequenced suppressors that likely represented noncausal, fixed lesions present in the starting strain.
Immunoblot Analyses
Extracts were prepared from seedlings at indicated ages by homogenizing frozen seedlings in 2 volumes of 2× sample buffer (Invitrogen) containing 0.05 M DTT and heated at 100°C for 5 min. Samples were electrophoresed on NuPAGE 10% or 12% Bis-Tris gels (Invitrogen) using 1× MES running buffer (50 mM MES, 50 mM Tris base, 0.1% SDS, and 1 mM EDTA) or 1× MOPS running buffer (50 mM MOPS, 50 mM Tris base, 0.1% SDS, and 1 mM EDTA) and transferred to Hybond Nitrocellulose membrane (Amersham Pharmacia Biotech) using NuPAGE transfer buffer (Invitrogen). Membranes were blocked in 8% milk solution (or 5% BSA for the anti-ATG3 antibody) in 20 mM Tris, pH 7.5, 150 mM NaCl, and 0.1% Tween 20 and subsequently incubated overnight at 4°C with primary antibodies in blocking solution. Rabbit antibodies against ATG3 (1:10,000; Phillips et al., 2008), ATG7 (1:1000; Doelling et al., 2002), ICL (1:1000; Maeshima et al., 1988), HPR (1:10,000; Agrisera AS11 1797), MLS (1:25,000; Olsen et al., 1993), the PED1 isoform of thiolase (1:2500; Lingard et al., 2009), PEX5 (1:100; Zolman and Bartel, 2004), PEX7 (1:800; Ramón and Bartel, 2010), PEX14 (1:10,000; Agrisera AS08 372), and PMDH2 (1:2000; Pracharoenwattana et al., 2007) were diluted as indicated. Mouse antibodies against HSC70 (1:20,000 or 1:50,000; StressGen Bioreagents SPA-817) were used. Primary antibodies were visualized with horseradish peroxidase–conjugated goat anti-rabbit or anti-mouse IgG secondary antibodies (1:5000 dilution in blocking buffer; Santa Cruz Biotechnology, SC2030 or SC2031). Horseradish peroxidase was visualized by incubation with WesternBright ECL reagent (Advansta), and signal was detected using autoradiography film. Membranes were reblocked and sequentially probed with the indicated antibodies without stripping the membrane between incubations.
For quantification of immunoblot images, films were photographed on a light box, and the resulting TIFF images were analyzed using ImageJ Gel Analysis software. The areas under the density curves from selected bands were normalized by dividing by the area of the appropriate HSC70 bands (after subtraction of background density). These ratios were further normalized by dividing by the wild-type band for the protein of interest.
Confocal Fluorescence Microscopy
Cotyledons of light-grown seedlings were mounted in water, and fluorescence was visualized using a Carl Zeiss LSM 710 laser scanning confocal microscope equipped with a meta detector. lon2-2 suppressor seedlings were from self-fertilized progeny of the original isolates. Samples were imaged using a ×63 oil immersion objective. GFP and chlorophyll were excited with a 488-nm argon laser, GFP emission was collected between 493 and 572 nm, and chlorophyll autofluorescence was detected between 620 and 719 nm. Each image is an average of four different exposures using a 47.1-μm pinhole, corresponding to a 0.8-μm optical slice.
Protein Alignment
Protein sequences were aligned using Lasergene MegAlign (DNASTAR) using the ClustalW default settings with the Gonnet series protein weight matrix.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ATG2 (At3g19190), ATG3 (At5g61500), ATG7 (At5g45900), DEG15 (At1g28320), LON2 (At5g47040), and PEX7 (At1g29260).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ATG7 Protein Alignment Depicting atg7 Mutations.
Supplemental Table 1. Primers Used for Candidate Gene Sequencing.
Supplemental Table 2. PCR-Based Markers Used for Mutant Genotyping.
Supplemental Table 3. PCR-Based Markers Used for Recombination Mapping.
Supplemental Data Set 1. Mutations Identified in lon2-2 Suppressors by Whole-Genome Sequencing.
Supplementary Material
Acknowledgments
We thank Richard Vierstra (University of Wisconsin–Madison) for the anti-ATG3 and anti-ATG7 antibodies, John Harada (University of California, Davis) for the anti-MLS antibody, Steven Smith (University of Western Australia) for the anti-PMDH2 antibody, Masayoshi Maeshima (Nagoya University) for the anti-ICL antibody, and the ABRC for seeds from T-DNA insertion lines. We thank Andrew Woodward (University of Mary Hardin-Baylor) and Marta Bjornson (Univeristy of California, Davis) for sharing lon2-4 seeds prior to publication, Matthew Lingard (St. Louis) for crossing lon2-2 and atg7-3 to 35S:GFP-PTS1, Wendell Fleming (Rice University) for assistance with bioinformatics, and Lucia Strader (Washington University) for advice on genome sequencing. We thank Wendell Fleming, Kim Gonzalez (Rice University), Yun-Ting Kao (Rice University), and Andrew Woodward for critical comments on the article. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. This center is supported by National Cancer Institute Cancer Center Support (P30 CA91842), the National Institutes of Health (NIH) National Center for Research Resources (UL1RR024992), and NIH Roadmap for Medical Research. Confocal microscopy was performed on equipment obtained through a Shared Instrumentation Grant from the NIH (S10RR026399-01). This research was supported by the NIH (R01GM079177), the National Science Foundation (MCB-0745122), and the Robert A. Welch Foundation (C-1309).
AUTHOR CONTRIBUTIONS
L.M.F. and B.B. conceived and designed the experiments. L.M.F. executed most of the experiments. M.A.R. conducted the confocal microscopy and assisted with protein stability experiments. P.G.Y. assisted with genotyping and atg3 characterization. C.H.D. assisted with suppressor mapping and genotyping. S.E.B. assisted with protein stability experiments and genotyping. L.M.F. and B.B. wrote the article with input from the other authors. All authors approved the final version of the article.
Glossary
- IBA
indole-3-butyric acid
- IAA
indole-3-acetic acid
- HPR
hydroxypyruvate reductase
- ICL
isocitrate lyase
- MLS
malate synthase
- GFP
green fluorescent protein
- PMDH
peroxisomal malate dehydrogenase
- ROS
reactive oxygen species
- Col-0
Columbia-0
- PNS
plant nutrient medium supplemented with 0.5% (w/v) Suc
- EMS
ethyl methanesulfonate
References
- Agrimi G., Russo A., Pierri C.L., Palmieri F. (2012). The peroxisomal NAD+ carrier of Arabidopsis thaliana transports coenzyme A and its derivatives. J. Bioenerg. Biomembr. 44: 333–340 [DOI] [PubMed] [Google Scholar]
- Aksam E.B., Koek A., Kiel J.A., Jourdan S., Veenhuis M., van der Klei I.J. (2007). A peroxisomal lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy 3: 96–105 [DOI] [PubMed] [Google Scholar]
- Amor C., Domínguez A.I., De Lucas J.R., Laborda F. (2000). The catabolite inactivation of Aspergillus nidulans isocitrate lyase occurs by specific autophagy of peroxisomes. Arch. Microbiol. 174: 59–66 [DOI] [PubMed] [Google Scholar]
- Bartoszewska M., Williams C., Kikhney A., Opaliński L., van Roermund C.W., de Boer R., Veenhuis M., van der Klei I.J. (2012). Peroxisomal proteostasis involves a Lon family protein that functions as protease and chaperone. J. Biol. Chem. 287: 27380–27395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C. (2007). Plant autophagy—More than a starvation response. Curr. Opin. Plant Biol. 10: 587–593 [DOI] [PubMed] [Google Scholar]
- Bell C.J., Ecker J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Bernhardt K., Wilkinson S., Weber A.P., Linka N. (2012). A peroxisomal carrier delivers NAD⁺ and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J. 69: 1–13 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Øvervatn A., Stenmark H., Johansen T. (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart S.E., Lingard M.J., Bartel B. (2013). Genetic dissection of peroxisome-associated matrix protein degradation in Arabidopsis thaliana. Genetics 193: 125–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Collins C.S., Kalish J.E., Morrell J.C., McCaffery J.M., Gould S.J. (2000). The peroxisome biogenesis factors pex4p, pex22p, pex1p, and pex6p act in the terminal steps of peroxisomal matrix protein import. Mol. Cell. Biol. 20: 7516–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento A.L., Bassham D.C. (2010). Increase in catalase-3 activity as a response to use of alternative catabolic substrates during sucrose starvation. Plant Physiol. Biochem. 48: 232–238 [DOI] [PubMed] [Google Scholar]
- Dammai V., Subramani S. (2001). The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105: 187–196 [DOI] [PubMed] [Google Scholar]
- Deosaran E., et al. (2013). NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126: 939–952 [DOI] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Dong C.H., Zolman B.K., Bartel B., Lee B.H., Stevenson B., Agarwal M., Zhu J.K. (2009). Disruption of Arabidopsis CHY1 reveals an important role of metabolic status in plant cold stress signaling. Mol. Plant 2: 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P.J., Graham I.A. (2001). Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6: 72–78 [DOI] [PubMed] [Google Scholar]
- Eubel H., Meyer E.H., Taylor N.L., Bussell J.D., O’Toole N., Heazlewood J.L., Castleden I., Small I.D., Smith S.M., Millar A.H. (2008). Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 148: 1809–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré J.C., Manjithaya R., Mathewson R.D., Subramani S. (2008). PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E.A., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Comai L., Henikoff S. (2003). Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G.W., Somerville C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434 [Google Scholar]
- Hayashi M., Nito K., Toriyama-Kato K., Kondo M., Yamaya T., Nishimura M. (2000). AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19: 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M., Lück C., Prestele J., Hierl G., Huesgen P.F., Fröhlich T., Arnold G.J., Adamska I., Görg A., Lottspeich F., Gietl C. (2007). Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc. Natl. Acad. Sci. USA 104: 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Hu J., Baker A., Bartel B., Linka N., Mullen R.T., Reumann S., Zolman B.K. (2012). Plant peroxisomes: Biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Iwata J., Ezaki J., Komatsu M., Yokota S., Ueno T., Tanida I., Chiba T., Tanaka K., Kominami E. (2006). Excess peroxisomes are degraded by autophagic machinery in mammals. J. Biol. Chem. 281: 4035–4041 [DOI] [PubMed] [Google Scholar]
- Kaur N., Zhao Q., Xie Q., Hu J. (2013). Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins(F). J. Integr. Plant Biol. 55: 108–120 [DOI] [PubMed] [Google Scholar]
- Kikuchi M., Hatano N., Yokota S., Shimozawa N., Imanaka T., Taniguchi H. (2004). Proteomic analysis of rat liver peroxisome: Presence of peroxisome-specific isozyme of Lon protease. J. Biol. Chem. 279: 421–428 [DOI] [PubMed] [Google Scholar]
- Kim D.Y., Scalf M., Smith L.M., Vierstra R.D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66: 953–968 [DOI] [PubMed] [Google Scholar]
- Lee M.S., Mullen R.T., Trelease R.N. (1997). Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 9: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Vierstra R.D. (2012). Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Lingard M.J., Bartel B. (2009). Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 151: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard M.J., Gidda S.K., Bingham S., Rothstein S.J., Mullen R.T., Trelease R.N. (2008). Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20: 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard M.J., Monroe-Augustus M., Bartel B. (2009). Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Burgos J.S., Deng Y., Srivastava R., Howell S.H., Bassham D.C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Huertas E., Charlton W.L., Johnson B., Graham I.A., Baker A. (2000). Stress induces peroxisome biogenesis genes. EMBO J. 19: 6770–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M., Yokoi H., Asahi T. (1988). Evidence for no proteolytic processing during transport of isocitrate lyase into glyoxysomes in castor bean endosperm. Plant Cell Physiol. 29: 381–384 [Google Scholar]
- Manivannan S., de Boer R., Veenhuis M., van der Klei I.J. (2013). Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events. Autophagy 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Fukao Y., Araki M., Matsuda A., Kondo M., Nishimura M. (2011). A defect of peroxisomal membrane protein 38 causes enlargement of peroxisomes. Plant Cell Physiol. 52: 2157–2172 [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Kondo M., Hayashi M., Nishimura M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38: 487–498 [DOI] [PubMed] [Google Scholar]
- Marzioch M., Erdmann R., Veenhuis M., Kunau W.H. (1994). PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13: 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum D., Monosov E., Subramani S. (1993). The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells—The PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J. Cell Biol. 121: 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew J.A., Goodman J.M. (1994). An oligomeric protein is imported into peroxisomes in vivo. J. Cell Biol. 127: 1245–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe-Augustus M., Ramón N.M., Ratzel S.E., Lingard M.J., Christensen S.E., Murali C., Bartel B. (2011). Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol. Biol. 77: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H., Nishimura M. (1989). Glyoxysomal malate synthetase is specifically degraded in microbodies during greening of pumpkin cotyledons. FEBS Lett. 244: 163–166 [Google Scholar]
- Motley A.M., Nuttall J.M., Hettema E.H. (2012). Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31: 2852–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagotu S., Kalel V.C., Erdmann R., Platta H.W. (2012). Molecular basis of peroxisomal biogenesis disorders caused by defects in peroxisomal matrix protein import. Biochim. Biophys. Acta 1822: 1326–1336 [DOI] [PubMed] [Google Scholar]
- Nair D.M., Purdue P.E., Lazarow P.B. (2004). Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J. Cell Biol. 167: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M., Hayashi M., Kato A., Yamaguchi K., Mano S. (1996). Functional transformation of microbodies in higher plant cells. Cell Struct. Funct. 21: 387–393 [DOI] [PubMed] [Google Scholar]
- Nishimura M., Yamaguchi J., Mori H., Akazawa T., Yokota S. (1986). Immunocytochemical analysis shows that glyoxysomes are directly transformed to leaf peroxisomes during greening of pumpkin cotyledons. Plant Physiol. 81: 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Grisafi P., Fink G.R., Bartel B. (1997). Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K., Kametani Y., Fujiki Y. (2011). Two proteases, trypsin domain-containing 1 (Tysnd1) and peroxisomal lon protease (PsLon), cooperatively regulate fatty acid β-oxidation in peroxisomal matrix. J. Biol. Chem. 286: 44367–44379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L.J., Ettinger W.F., Damsz B., Matsudaira K., Webb M.A., Harada J.J. (1993). Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell 5: 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi S., Nakata R., Okamura-Ikeda K., Konishi H., Taniguchi H. (2008). Contribution of peroxisome-specific isoform of Lon protease in sorting PTS1 proteins to peroxisomes. J. Biochem. 143: 649–660 [DOI] [PubMed] [Google Scholar]
- Ostersetzer O., Kato Y., Adam Z., Sakamoto W. (2007). Multiple intracellular locations of Lon protease in Arabidopsis: Evidence for the localization of AtLon4 to chloroplasts. Plant Cell Physiol. 48: 881–885 [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Bäumer B.E., Schlee D., Girzalsky W., Erdmann R. (2009). Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol. Cell. Biol. 29: 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J.E., Smith S.M. (2007). Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Ramón N.M., Bartel B. (2010). Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol. Biol. Cell 21: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzel S.E., Lingard M.J., Woodward A.W., Bartel B. (2011). Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic 12: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., Linka N., Switzenberg R., Wilkerson C.G., Weber A.P., Olsen L.J., Hu J. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150: 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Voitsekhovskaja O., Lillo C. (2010). From signal transduction to autophagy of plant cell organelles: Lessons from yeast and mammals and plant-specific features. Protoplasma 247: 233–256 [DOI] [PubMed] [Google Scholar]
- Sautter C. (1986). Microbody transition in greening watermelon cotyledons. Double immunocytochemical labeling of isocitrate lyase and hydroxypyruvate reductase. Planta 167: 491–503 [DOI] [PubMed] [Google Scholar]
- Schuhmann H., Huesgen P.F., Gietl C., Adamska I. (2008). The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 148: 1847–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos T.C., Hangarter R.P. (1990). Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Bartel B. (2011). Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Culler A.H., Cohen J.D., Bartel B. (2010). Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 153: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. (2007). Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- Svenning S., Lamark T., Krause K., Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7: 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels B.W., Gould S.J., Bodnar A.G., Rachubinski R.A., Subramani S. (1991). A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10: 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till A., Lakhani R., Burnett S.F., Subramani S. (2012). Pexophagy: The selective degradation of peroxisomes. Int. J. Cell Biol. 2012: 512721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus D.E., Becker W.M. (1985). Investigation of the glyoxysome-peroxisome transition in germinating cucumber cotyledons using double-label immunoelectron microscopy. J. Cell Biol. 101: 1288–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilibaris V., Maenhaut-Michel G., Van Melderen L. (2006). Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 157: 701–713 [DOI] [PubMed] [Google Scholar]
- Van der Leij I., Franse M.M., Elgersma Y., Distel B., Tabak H.F. (1993). PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90: 11782–11786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen T., Veenhuis M., van der Klei I.J. (2011). Damaged peroxisomes are subject to rapid autophagic degradation in the yeast Hansenula polymorpha. Autophagy 7: 863–872 [DOI] [PubMed] [Google Scholar]
- Wanders R.J., Waterham H.R. (2005). Peroxisomal disorders I: Biochemistry and genetics of peroxisome biogenesis disorders. Clin. Genet. 67: 107–133 [DOI] [PubMed] [Google Scholar]
- Wang Y., Nishimura M.T., Zhao T., Tang D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68: 74–87 [DOI] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005a). The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005b). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. (2007). Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Matoba K., Sawada R., Fujioka Y., Nakatogawa H., Yamamoto H., Kobashigawa Y., Hoshida H., Akada R., Ohsumi Y., Noda N.N., Inagaki F. (2012). Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat. Struct. Mol. Biol. 19: 1250–1256 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57: 146–159 [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang J., Cheng Y., Chi Y.J., Fan B., Yu J.Q., Chen Z. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9: e1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K., Lukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., Liszewska F., Wawrzyńska A., Sirko A. (2011). Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Bartel B. (2004). An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc. Natl. Acad. Sci. USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Monroe-Augustus M., Silva I.D., Bartel B. (2005). Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Yoder A., Bartel B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.