Abstract
The level of sequence heterogeneity among rrn operons within genomes determines the accuracy of diversity estimation by 16S rRNA-based methods. Furthermore, the occurrence of widespread horizontal gene transfer (HGT) between distantly related rrn operons casts doubt on reconstructions of phylogenetic relationships. For this study, patterns of distribution of rrn copy numbers, interoperonic divergence, and redundancy of 16S rRNA sequences were evaluated. Bacterial genomes display up to 15 operons and operon numbers up to 7 are commonly found, but ∼40% of the organisms analyzed have either one or two operons. Among the Archaea, a single operon appears to dominate and the highest number of operons is five. About 40% of sequences among 380 operons in 76 bacterial genomes with multiple operons were identical to at least one other 16S rRNA sequence in the same genome, and in 38% of the genomes all 16S rRNAs were invariant. For Archaea, the number of identical operons was only 25%, but only five genomes with 21 operons are currently available. These considerations suggest an upper bound of roughly threefold overestimation of bacterial diversity resulting from cloning and sequencing of 16S rRNA genes from the environment; however, the inclusion of genomes with a single rrn operon may lower this correction factor to ∼2.5. Divergence among operons appears to be small overall for both Bacteria and Archaea, with the vast majority of 16S rRNA sequences showing <1% nucleotide differences. Only five genomes with operons with a higher level of nucleotide divergence were detected, and Thermoanaerobacter tengcongensis exhibited the highest level of divergence (11.6%) noted to date. Overall, four of the five extreme cases of operon differences occurred among thermophilic bacteria, suggesting a much higher incidence of HGT in these bacteria than in other groups.
rRNA sequences play a central role in the study of microbial evolution and ecology. Particularly, the 16S rRNA genes have become the standard for the determination of phylogenetic relationships, the assessment of diversity in the environment, and the detection and quantification of specific populations (14, 16). Indeed, the rRNAs combine several properties which make them uniquely suited for such diverse applications. First, they are universally distributed, allowing the comparison of phylogenetic relationships among all extant organisms and thus the construction of a “tree of life.” Second, the rRNAs are generally thought to be part of a core of informational genes which are only weakly affected by horizontal gene transfer (HGT) (1, 8), so their relationships provide a solid framework for the assessment of evolutionary changes in lineages. Third, the rRNAs are functionally highly constrained mosaics of sequence stretches ranging from conserved to more variable. This enables the design of PCR primers and hybridization probes with various levels of taxonomic specificity and is exploited in microbial ecology when the number and distribution of different rRNA genes are taken as a measure of diversity (14). A testimony to the significance of these approaches is the vast and growing database of 16S rRNA genes, of which an increasing number are derived from the large majority of uncultured Bacteria and Archaea. Although many of these organisms appear to dominate in the environment, their distribution and relationships are only known from clone libraries derived from nucleic acids recovered from the environment (16).
The interpretation of microbial ecology and evolution via 16S rRNA sequences has been complicated in recent years by the realization that many bacteria harbor multiple, heterogeneous rRNA operons. The three rRNAs, namely 16S, 23S, and 5S rRNAs, are typically linked together into an operon, which frequently contains an internal transcribed spacer and at least one tRNA. It has been shown that bacterial genomes can contain between 1 and 15 such operons and that 16S rRNA sequences can differ up to several percent between operons (28, 40, 41, 43). Such sequence heterogeneity within single genomes creates a significant problem for culture-independent analysis of microbial communities since it can lead to a severe overestimation of microbial diversity based on 16S rRNA approaches (6). This has led many authors to omit small-scale sequence differences encountered in environmental 16S rRNA clone libraries from estimates of diversity (7, 17, 39). However, such “microdiversity” has been reported to make up a significant fraction of the sequence composition of clone libraries (4, 10, 13, 24). That it potentially signifies functional differences is suggested by comparisons between closely related 16S rRNA sequences and physiological properties of isolates (12, 32, 36) or overall genome architectures (2, 18, 31, 35). In addition, reports of relatively large divergence among rRNA genes between operons have suggested that HGT may affect rRNA genes to a larger extent than was previously assumed (28, 40, 41, 43). If this effect is indeed widespread, then phylogenetic relationships among bacteria may become significantly blurred.
Here we present an in-depth comparison of 16S rRNA genes of bacterial and archaeal operons largely based on published complete genome sequences, focusing on the following questions. (i) What is the range and average of divergence among 16S rRNA genes coexisting in genomes? (ii) What is the redundancy of identical 16S rRNA sequences within multiple operons? (iii) How widespread does HGT appear to be, as evidenced by the number of genomes with highly divergent 16S rRNA genes? This work is intended as a complement to the recently compiled ribosomal copy number database (22) and pursues the overall goal of providing bounds of accuracy and reliability for 16S rRNA sequence-based estimations of diversity and phylogeny.
MATERIALS AND METHODS
Ribosomal operon sequences.
Information on 16S rRNA variation within 81 genomes with multiple operons was retrieved from several sources between 10 June and 5 August 2003 (Table 1). The majority of the genomes came from the National Center for Biotechnology Information (NCBI) Microbial Genome Database (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html), from which a total of 57 bacterial and 2 archaeal genomes with multiple rrn operons were recovered. In addition, rrn operons from two bacterial genomes were obtained from The Institute for Genomic Research (TIGR) Microbial Genomic Database (www.tigr.org/tdb/mdb/mdbcomplete.html). The rRNA Operon Copy Number Database (rrndb) (http://rrndb.cme.msu.edu) and multiple literature sources served as sources of information for 12 and 10 microorganisms, respectively (Table 1).
TABLE 1.
Number of operons, heterogeneity and redundancy of 16S rRNA genes within bacterial and archaeal genomes with multiple operonsa
| Domain or microorganism | Phylogenetic affiliation | No. of operons | No. of identical operonsb | No. of different ribotypesc | Nucleotide divergence (% 16S)d | No. of poly- morph- ismse | Source or referencef |
|---|---|---|---|---|---|---|---|
| Bacteria (76 genomes) | |||||||
| Aquifex aeolicus VF5 | Aquificales | 2 | 2 | 1 | 0 | 0 | rrndb |
| Chlorobium tepidum TLS | Chlorobiales | 2 | 2 | 1 | 0 | 0 | This paper |
| Synechocystis sp. PCC 6803 | Cyanobacteria | 2 | 2 | 1 | 0 | 0 | This paper |
| Treponema pallidum ATCC 25870 | Spirochaetales | 2 | 2 | 1 | 0 | 0 | rrndb |
| Leptospira interrogans serovar Lai 56601 | Spirochaetales | 2 | 2 | 1 | 0 | 0 | This paper |
| Caulobacter crecentus CB15 | Alpha proteobacteria | 2 | 2 | 1 | 0 | 0 | This paper |
| Xanthomonas axonopodis pv. citri 306 | Gamma proteobacteria | 2 | 2 | 1 | 0 | 0 | This paper |
| Xylella fastidiosa 9a5c | Gamma proteobacteria | 2 | 2 | 1 | 0 | 0 | rrndb |
| Xanthomonas campestris ATCC 33913 | Gamma proteobacteria | 2 | 2 | 1 | 0 | 0 | This paper |
| Helicobacter pylori 26695 | Epsilon proteobacteria | 2 | 2 | 1 | 0 | 0 | rrndb |
| Helicobacter pylori J99 | Epsilon proteobacteria | 2 | 0 | 2 | ND | ND | 27 |
| Ureaplasma urealyticum serovar 3 | Mycoplasmatales | 2 | 0 | 2 | 0.07 | 1 | rrndb |
| Mycobacterium celatum | Actinomycetales | 2 | 0 | 2 | ND | 4/5 | 34 |
| Mycobacterium strain “X" | Actinomycetales | 2 | 0 | 2 | 1.20 | 18 | 30 |
| Desulfotomaculum kuznestovii | Clostridales | 2 | 0 | 2 | 8.3 | ND | 40 |
| Chlamydia trachomatis | Chlamydiae | 2 | 2 | 1 | 0 | 0 | 27 |
| Geobacter sulfurreducens | Delta proteobacteria | 2 | 0/2 | 1/2 | 0/0.2 | 0/3 | This paper |
| Deincoccus radiodurans ATCC 13939 | Deinococcales | 3 | 2 | 2 | 0.13 | 2 | rrndb |
| Rhodobacter sphaeroides | Alpha proteobacteria | 3 | 3 | 1 | 0 | 0 | 10 |
| Brucella suis 1330, biovar 1 | Alpha proteobacteria | 3 | 3 | 1 | 0 | 0 | This paper |
| Brucella melitensis 16M | Alpha proteobacteria | 3 | 3 | 1 | 0 | 0 | This paper |
| Sinorhizobium meliloti | Alpha proteobacteria | 3 | 3 | 1 | 0 | 0 | This paper |
| Ralstonia solanacearum | Beta proteobacteria | 3 | 3 | 1 | 0 | 0 | This paper |
| Campylobacter jenuni ATCC 700819 | Epsilon proteobacteria | 3 | 3 | 1 | 0 | 0 | rrndb |
| Nostoc sp. PCC 7120 | Cyanobacteria | 4 | 3 | 2 | 0.07 | 1 | This paper |
| Agrobacterium tumefaciens C58 | Alpha proteobacteria | 4 | 4 | 1 | 0 | 0 | This paper |
| Neisseria meningitidis MC58 | Alpha proteobacteria | 4 | 4 | 1 | 0 | 0 | rrndb |
| Pseudomonas aeruginosa PAO1 | Gamma proteobacteria | 4 | 2 + 2 | 2 | 0.07 | 1 | This paper |
| Enterococcus faecalis V583 | Firmicutes | 4 | 3 + 1 | 2 | 0.07 | 1 | This paper |
| Streptococcus pneumoniae R6 | Firmicutes | 4 | 4 | 1 | 0 | 0 | This paper |
| Streptococcus pneumoniae TIGR4 | Firmicutes | 4 | 4 | 1 | 0 | 0 | This paper |
| Thermobispora bispora R51 | Actinomycetales | 4 | 0 | 4 | 6.4 | 98 | 41 |
| Thermoanaerobacter tengcongensis | Thermoanaerobacteriales | 4 | 0 | 4 | 6.5/11.6 | 99/188 | This paper |
| Pseudomonas syringae pv tomato DC3000 | Gamma proteobacteria | 5 | 5 | 1 | 0 | 0 | This paper |
| Fusobacterium nucleatum subsp. nucleatum | Fusobacteria | 5 | 2 + 1 + 1 + 1 | 4 | 0.20 | 3 | This paper |
| Corynebacterium efficiens YS-314 | Actinomycetales | 5 | 4 + 1 | 2 | 0.21/0.42 | 3/6 | This paper |
| Staphylococcus epidermidis ATCC 12228 | Firmicutes | 5 | 0 | 5 | 0.84 | 13 | This paper |
| Staphylococcus aureus N315 | Firmicutes | 5 | 2 + 1 + 1 + 1 | 4 | 0.25 | 4 | This paper |
| Staphylococcus aureus Mu50 | Firmicutes | 5 | 0 | 5 | 0.38 | 6 | This paper |
| Lactobacillus plantarum WCFS1 | Firmicutes | 5 | 2 + 2 + 1 | 3 | 0.13 | 2 | This paper |
| Streptococcus mutans UA159 | Firmicutes | 5 | 3 + 2 | 2 | 0.19 | 3 | This paper |
| Streptococcus pyogenes SSI-1 | Firmicutes | 5 | 5 | 1 | 0 | 0 | This paper |
| Clostridium tetani E88 | Clostridiales | 5 | 2 + 1 + 1 + 1 | 4 | 0.39 | 6 | This paper |
| Desulfovibrio vulgaris | Delta proteobacteria | 5 | 2 + 1 + 1 + 1 | 4 | 0.26 | 4 | This paper |
| Bacteroides thetaiotaomicron VPI-5482 | Bacteroidales | 5 | 2 + 2 + 1 | 3 | 1.23/1.3 | 17/18 | This paper |
| Haemophilus influenzae Rd (ATCC 51907) | Gamma proteobacteria | 6 | 6 | 1 | 0 | 0 | rrndb |
| Yersina pestis CO92 | Gamma proteobacteria | 6 | 2 + 1 + 1 + 1 + 1 | 5 | 0.27 | 4 | This paper |
| Corynebacterium glutamicum ATCC 13032 | Actinomycetales | 6 | 2 + 1 + 1 + 1 + 1 | 5 | 0.33/0.39 | 6 | This paper |
| Thermomonospora chromogena | Actinomycetales | 6 | 0 | 6 | 6 | ND | 43 |
| Staphylococcus aureus MW2 | Firmicutes | 6 | 3 + 1 + 1 + 1 | 4 | 0.32 | 5 | This paper |
| Listeria monocytogenes EGD-e | Firmicutes | 6 | 2 + 2 + 1 + 1 | 4 | 0.32 | 5 | This paper |
| Listeria innocua Clip 11262 | Firmicutes | 6 | 5 + 1 | 2 | 0.25 | 4 | This paper |
| Lactococcus lactis subsp. lactis | Firmicutes | 6 | 5 + 1 | 2 | 0.06 | 1 | This paper |
| Streptococcus pyogenes MGAS315 | Firmicutes | 6 | 6 | 1 | 0 | 0 | This paper |
| Streptococcus pyogenes M1 GAS (SF370) | Firmicutes | 6 | 3 + 2 + 1 | 3 | 0.075/0.15 | 1/2 | This paper |
| Streptococcus pyogenes MGAS8232 | Firmicutes | 6 | 6 | 1 | 0 | 0 | This paper |
| Pseudomonas putida KT2440 | Gamma proteobacteria | 7 | 2 + 2 + 2 + 1 | 4 | 0.19 | 3 | This paper |
| Shigella flexneri 2a 301 | Gamma proteobacteria | 7 | 4 + 2 + 1 | 3 | 0.51/0.58 | 8/9 | This paper |
| Escherichia coli K-12 | Gamma proteobacteria | 7 | 3 + 1 + 1 + 1 + 1 | 5 | 1.23/1.36 | 18/21 | This paper |
| Escherichia coli EDL933 (O157:H7) | Gamma proteobacteria | 7 | 2 + 1 + 1 + 1 + 1 + 1 | 6 | 0.45/0.97 | 7/15 | This paper |
| Escherichia coli Sakai (O157:H7) | Gamma proteobacteria | 7 | 3 + 2 + 1 + 1 | 4 | 0.45/0.58 | 7/9 | This paper |
| Escherichia coli ATCC 10798 | Gamma proteobacteria | 7 | 3 + 1 + 1 + 1 + 1 | 5 | 1.23 | 19 | rrndb |
| Salmonella entercia serovar Typhimurium LT2 | Gamma proteobacteria | 7 | 3 + 1 + 1 + 1 + 1 | 5 | 0.64/0.58 | 9/10 | This paper |
| Salmonella enterica subs P. enterica serovar Typhi | Gamma proteobacteria | 7 | 3 + 2 + 1 + 1 | 4 | 0.13/0.19 | 2/3 | This paper |
| Salmonella enterica subs P. enterica serovar Typhi Ty2 | Gamma proteobacteria | 7 | 3 + 3 + 1 | 3 | 0.13/0.19 | 2/3 | This paper |
| Yersina pestis KIM10+ | Gamma proteobacteria | 7 | 5 + 2 | 2 | 0.06 | 1 | This paper |
| Oceanobacillus iheyensis HTE831 | Firmicutes | 7 | 3 + 1 + 1 + 1 + 1 | 5 | 0.96/1.02 | 15/16 | This paper |
| Streptococcus agalactiae NEM316 | Firmicutes | 7 | 6 + 1 | 2 | 0.07 | 1 | This paper |
| Streptococcus agalactiae 2603 V/R | Firmicutes | 7 | 7 | 1 | 0 | 0 | This paper |
| Vibrio cholerae El Tor N16961 | Gamma proteobacteria | 8 | 0 | 8 | 0.91/1.04 | 12/14 | This paper |
| Shewanella oneidensis MR-1 | Gamma proteobacteria | 9 | 4 + 3 + 1 + 1 | 4 | 0.32 | 5 | This paper |
| Bacillus subtilis 168 | Firmicutes | 10 | 0 | 10 | 1.48/2.18 | 23/34 | This paper |
| Clostridium perfringens 13 | Clostridiales | 10 | 0 | 10 | 0.93/1.18 | 14/18 | 37; this paper |
| Vibrio parahaemolyticus (RIMB22106633) | Gamma proteobacteria | 11 | 2 + 2 + 2 + 1 + 1 + 1 + 1 + 1 | 8 | 0.61 | 9 | This paper |
| Clostridium acetobutylicum | Clostridiales | 11 | 7 + 1 + 1 + 1 +1 | 5 | 0.26/0.92 | 4/14 | This paper |
| Bacillus cereus ATCC 14579 | Firmicutes | 13 | 7 + 1 + 1 + 1 + 1 + 1 + 1 | 7 | 0.46 | 7 | This paper |
| Archaea (5 genomes) | |||||||
| Methanocaldococcus jannaschii | Methanococcales (Euryarchaeota) | 2 | 0 | 2 | 0.07/0.27 | 1/4 | rrndb; this paper |
| Methanothermobacter thermoauto- trophicus | Methanobacteriales (Euryarchaeota) | 2 | 0 | 2 | 0.14 | 2 | rrndb |
| Haloarcula marismortui | Halobacteriales (Euryarchaeota) | 2 | 0 | 2 | 5 | 74 | 28 |
| Methanosarcina acetivorans C2A | Methanosarcinales (Euryarchaeota) | 3 | 2 + 1 | 2 | 0.07 | 1 | This paper |
| Methanosarcina mazei Goe1 | Methanosarcinales (Euryarchaeota) | 3 | 3 | 1 | 0 | 0 | This paper |
Data were obtained from the NCBI (www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html) and TIGR (www.tigr.org/tdb/mdbcomplete.html) genome databases, the rrndb (http://rrndb.cme.msu.edu) (22), and multiple literature sources.
Different numbers indicate identical 16S rRNA gene sequences in genomes with multiple operons.
Ribotypes are defined as identical 16S rRNA sequences.
Divergence is represented by pairwise distances calculated from alignments of all 16S rRNA sequences for each genome. Single values indicate that there are no insertions or deletions (I/D). When two values are given, left values represent no I/D and right values are with I/D.
The numbers of polymorphisms represent the cumulative numbers of sequence positions in which the aligned sequences differed. Single values indicate that there are no insertions or deletions (I/D). When two values are given, left values represent no I/D and right values are with I/D.
ND, not determined. Analyses were performed on alignments generated for this paper unless other sources are listed.
Analysis of 16S rRNA interoperonic sequences.
16S rRNA sequences from all retrieved genomes were aligned and analyzed with the Sequencher 4.1 software package (Genes Codes, Ann Arbor, Mich.) or with CLUSTAL X (19). For each set of operons, the numbers of divergent and identical 16S rRNA genes were determined. For divergent genes, the percentage of nucleotide difference and the number of polymorphisms were calculated. In cases in which insertion-deletion (I/D) events of >1 bp were observed, two values expressing divergence between the genes were calculated, with the first excluding (−I/D) and the second including (+I/D) the inserted segments (see Table 3).
TABLE 3.
Range and averages of percentages of nucleotide divergence in bacterial 16S rRNA sequences within genomes with multiple rrn operonsa
| No. of rrn operons | No. of sequences | Range of divergence (%) (−I/D) | Average (%) (−I/D) | Range of divergence (%) (+I/D) | Average (%) (+I/D) |
|---|---|---|---|---|---|
| 2b | 15 | 0-1.2 | 0.09 | 0-1.2 | 0.1 |
| 3 | 7 | 0-0.13 | 0.018 | 0-0.13 | 0.018 |
| 4b | 7 | 0-0.07 | 0.03 | 0-0.07 | 0.03 |
| 5 | 12 | 0-1.23 | 0.34 | 0-1.36 | 0.363 |
| 6b | 10 | 0-0.33 | 0.162 | 0-0.39 | 0.176 |
| 7 | 13 | 0-1.23 | 0.396 | 0-1.36 | 0.487 |
| 8 | 2 | 0.91 | 0.91 | 1.04 | 1.04 |
| 9 | 1 | 0.32 | 0.32 | 0.32 | 0.32 |
| 10 | 3 | 0.97-1.48 | 1.15 | 0.97-2.18 | 1.46 |
| 11 | 2 | 0.26-0.61 | 0.435 | 0.61-0.92 | 0.765 |
| 12 | 1 | 0.33 | 0.33 | 0.33 | 0.33 |
| 13 | 2 | NA | NA | NA | NA |
| 15 | 1 | NA | NA | NA | NA |
Data were obtained from the NCBI (www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html) and TIGR (www.tigr.org/tdb/mdb/mdbcomplete.html) genome databases, the rrndb (http://rrndb.cme.msu.edu) (22), and multiple literature sources. NA, no rrn operon sequences were available.
Four cases of extreme divergence (sequences from microorganisms in which the percentage of nucleotide divergence was much higher than the average) were excluded from the calculation. These were Desulfotomaculum kuznestovii, with two rrn operons and an 8.3% difference, Thermobispora bispora R51 and Thermoanaerobacter tengcongensis, both with four rrn operons and 6.4 and 11.6% differences, respectively, and Thermonospora chromogena, with six rrn operons and 6% divergence.
RESULTS
Ribosomal operon copy number in bacterial and archaeal genomes.
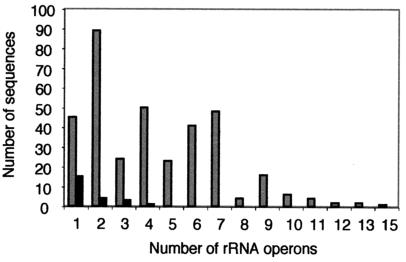
The information on rRNA operon copy numbers from the rrndb was amended with data obtained from the genomes retrieved so that a total of 355 bacterial strains were evaluated. This showed a range in rrn operon numbers from 1 to 15, with two rrn operon copies representing the most common class, with 25% of the total (Fig. 1). About 40% of the strains had either one or two operons. This was noted previously and demonstrates that despite a highly increased rate of genome sequencing and determination of operon copy numbers, this ratio has remained roughly constant over the last few years (6, 11). The next most abundant classes were four, seven, six, and three operons per genome, with frequencies of 14, 13.5, 11.5, and 6.7%, respectively (Fig. 1). Genomes with ≥10 operons were observed in only 4.3% of the cases. Although overall no clear correlation between rrn copy number and distantly related phylogenetic divisions was apparent from the data (21), a pattern of low operon copy number was observed for three domains. None of the 31 genomes belonging to the α-Proteobacteria had more than four operons. Furthermore, mycoplasma genomes displayed a maximum of three operons, and among the spirochetes, 20 of 24 genomes had a maximum of two operons.
FIG. 1.
Distribution of different rrn operon numbers among bacterial (gray bars) and archaeal (black bars) isolates. Data were obtained from complete genome sequences from the NCBI and TIGR genome databases, the rrndb database, and the literature.
The pattern of operon numbers among archaeal strains differed from that for Bacteria. Although information for only 23 strains could be retrieved from databases and the literature, a dominance of low rrn copy numbers was apparent (Fig. 1). The majority of genomes (65.2%) have a single operon (Fig. 1). Only one archaeon, Methanococcus vannielii, was found to have four operons.
rrn copy number variation between strains of the same bacterial species.
Although variations in operon numbers between different bacterial species have been well documented, variations between strains of the same species are considered less often. Sixteen examples of bacterial species with variable operon numbers in different strains were retrieved from databases and the literature (Table 2). Overall, the variation in operon numbers does not appear to be large, but the phenomenon is not restricted to a specific phylogenetic group since operon variation between strains of the same species was detected for diverse species (Table 2). For two species, Vibrio cholerae and Bacillus cereus, three different operon numbers have been reported, with B. cereus containing the highest variance in numbers. For all other bacteria, only two operon numbers which were different by a single operon were found (Table 2). Furthermore, for at least 42 species with multiple strain entries in the rrndb, only a single operon number was reported, indicating that operon copy variation in closely related bacteria may occur only in a minority of cases.
TABLE 2.
Strains of the same bacterial species with different numbers of rrn operonsa
| Microorganism name | Phylogenetic affiliation | No. of rrn operon copies |
|---|---|---|
| Borrelia burgdorferi | Spirochaetales | 1 and 2 |
| Chlamydia trachomatis | Chlamydiae | 1 and 2 |
| Rhodococcus fascians | Actinomycetales | 4 and 5 |
| Rhodopseudomonas palustris | Alpha proteobacteria | 1 and 2 |
| Bradyrhizobium japonicum | Alpha proteobacteria | 1 and 2 |
| Vibrio parahaemolyticus | Gamma proteobacteria | 9 and 11 |
| Vibrio cholerae | Gamma proteobacteria | 7, 8, and 9 |
| Pasteurella multocida | Gamma proteobacteria | 5 and 6 |
| Yersinia pestis | Gamma proteobacteria | 6 and 7 |
| Bacillus subtilis | Firmicutes | 9 and 10 |
| Bacillus anthracis | Firmicutes | 10 and 11 |
| Bacillus cereus | Firmicutes | 9, 12, and 13 |
| Staphylococcus aureus | Firmicutes | 5 and 6 |
| Enterococcus faecium | Firmicutes | 5 and 6 |
| Streptococcus agalactiae | Firmicutes | 6 and 7 |
| Streptococcus pyogenes | Firmicutes | 5 and 6 |
Data were obtained from the NCBI genome database (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html), the rrndb (http://rrndb.cme.msu.edu) (22), and the literature (18, 20).
Redundancy and divergence of 16S rRNA sequences within genomes.
For determination of the numbers of operons with identical and divergent 16S rRNA sequences, genomes with multiple operons were compiled primarily from the NCBI and TIGR databases, with some additional information from the rrndb and the literature (Table 1). The 76 bacterial genomes analyzed stem from 18 divergent phylogenetic groups; however, the Proteobacteria and Firmicutes divisions dominated the data set, with 33 and 21 genomes, respectively (Table 1). Within the Proteobacteria, the gamma and alpha subdivisions were overrepresented, with 20 and 7 genomes, respectively (Table 1). All other groups were represented by only one or two genomes, with the exception of the Actinomycetales, for which five genomes have been sequenced (Table 1). Among the Archaea, only five genomes were obtained from the databases, all of which belong to the Euryarchaeota and the four phylogenetic groups Methanococcales (1), Methanobacteriales (1), halophilic Archaea (1), and Methanosarcinales (2).
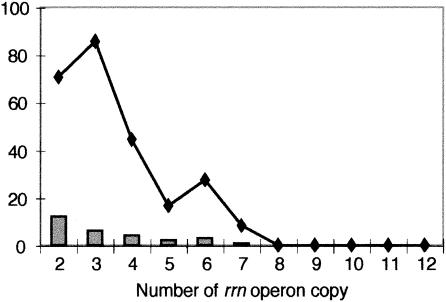
A total of 392 operons (380 from Bacteria and 12 from Archaea) with 230 (221 from Bacteria and 9 from Archaea) associated sequences were retrieved from the 81 (76 Bacteria and 5 Archaea) genomes. Among the Bacteria, 29 genomes (38% of the total) had completely invariant 16S rRNAs. In sum, over 43% of the sequences in bacterial genomes with multiple operons were identical, while for Archaea this number was 25%, or almost half that for Bacteria. A higher proportion of identical 16S rRNA sequences were found among bacterial genomes with fewer operons (Table 1; Fig. 2). Genomes with two and three operons showed identity in 70.6 and 85.7% of the cases (Fig. 2). Not surprisingly, this picture changed for genomes with higher numbers of operons; however, only relatively few complete genome sequences were available for each class (Fig. 2). Several genomes displayed high numbers of identical 16S rRNA sequences. Streptococcus agalactiae 2603 V/R has no differences in the 16S rRNAs of all seven of its rrn operons. Similarly, Clostridium acetobutylicum and B. cereus ATCC 14579 exhibited 7 identical operons of a total of 11 and 13 operon copies, respectively. However, examples of the other extreme were also found. For example, in Bacillus subtilis all 10 operons harbor different 16S rRNA sequences. Among the five archaeal genomes, only Methanosarcina mazei showed identity in all three rrn operons, despite the generally low number of operons per genome (Table 1).
FIG. 2.
Numbers (bars) and percentages of the total (line) of genomes with all identical 16S rRNA sequences among operons. Data were retrieved from the NCBI and TIGR genome databases and the rrndb database.
The averages and ranges of percent nucleotide divergence of multiple 16S rRNA genes within the genomes are shown in Table 3. Values were calculated with and without consideration of insertions and deletions, but the results differed only slightly. This indicates that divergence is largely caused by mutations and not by insertions or deletions (Table 3). For most classes of operon numbers, both the averages and ranges of nucleotide divergence remained under 1%, with lower operon number genomes displaying fewer differences in their 16S rRNA sequences. Cases for which the range of nucleotide divergence exceeded 1% were only found for genomes with 2, 5, 7, and 10 operons, but a comparison with the average values indicated that such a high level of divergence is rare. However, clear exceptions to the generally low level of divergence are four genomes which show extreme nucleotide differences among their 16S rRNA genes. These are the genomes of Desulfotomaculum kuznestovii (8.3% difference; two rrn operons), Thermobispora bispora R51 (6.4%; four rrn operons), Thermoanaerobacter tengcongensis (11.6%; four rrn operons), and Thermonospora chromogena (6%; six rrn operons). A 5% divergence between both rRNA operons was also reported for the archaeon Halobacterium marismortui (28). Since these four genomes fell clearly outside the range of values observed for all other genomes, they were excluded from Table 3.
Maximum interoperonic 16S rRNA divergence in bacterial genomes.
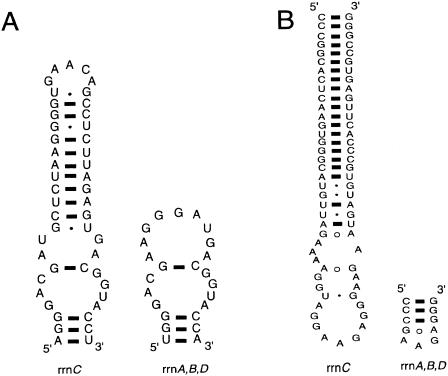
Our analysis revealed that, to date, the highest interoperonic divergence in 16S rRNA genes occurs in the genome of Thermoanaerobacter tengcongensis, with an 11.6% nucleotide difference. This extremely thermophilic bacterium was isolated from a Chinese hot spring (42), and its genome was completely sequenced by Bao et al. (3). It contains four rrn operon copies, and the 16S rRNA genes display a total of 188 polymorphisms (Table 1). The 16S rRNA genes clearly fall into two types, with the first representing three of the four operons (rrnA, rrnB, and rrnD). These contain only 1.1% divergent positions (17 polymorphisms), and operons rrnA and rrnD differ at only two nucleotides (0.13% divergence). In contrast, the second rrn type is represented by a single operon (rrnC) and contains 171 of the 188 polymorphic nucleotide sites, representing 90% of the total divergence. A large fraction of the variation is due to two significant length differences in variable stems, which give the molecule a total length of 1,620 nucleotides (Fig. 3).
FIG. 3.
Secondary structure model of the helices that differ in length between operons in Thermoanaerobacter tengcongensis. Nucleotide positions 444 to 490 (A) and 1,447 to 1,456 (B) are given according to Escherichia coli reference numbering.
We explored whether the highly divergent operon might have arisen via HGT or may represent a pseudogene, which accumulates mutations in the absence of functional constraints. A secondary structure analysis was carried out based on the rationale that mutations in a pseudogene should accumulate throughout the molecule and disrupt the secondary structure at multiple places. On the other hand, a functional or recently functional rRNA gene arisen via HGT would display nucleotide changes that are (i) concentrated in variable regions and (ii) compensated for if they are located in stem regions. This analysis detected only one nucleotide change of a total of 171 in an evolutionarily conserved position (base 94, C→A). In addition, 12 bp were identified as being compensated for in moderately variable stems. The remaining nucleotide substitutions were observed in variable regions and loops and did not disrupt the secondary structure of the 16S rRNA. About half of the extreme divergence of operon rrnC is associated with three inserted regions, of 24, 31, and 28 bases, distributed in different stem-loop regions. Even in the absence of these insertions, rrnC would still differ by 82 bases, or 6.5%, from the other operons. Overall, the secondary structure analysis provided no evidence that the molecule has lost its potential functionality, suggesting that rrnC does not represent a pseudogene.
DISCUSSION
Growing databases of completely sequenced genomes allow the exploration of patterns of interoperonic divergence among 16S rRNA sequences and provide critical information for the assessment of microbial diversity and evolution. Among Bacteria, classes of up to seven operons appear to be common, with no clear predominance of a single class of operon numbers (Fig. 1A). Nonetheless, as previously noted (11, 22), about 40% of bacteria have fewer than two operons. The picture is different for Archaea, among which the majority of strains have been shown to have a single operon and no genomes with more than four operons have been reported to date (Fig. 1). A detailed analysis of divergence among the 16S rRNA genes in completely sequenced bacterial genomes revealed that ∼40% of operons contain sequences identical to those of other operons (Table 1). This number appears to be much smaller for the Archaea; however, few completely sequenced genomes are available (Table 1). Overall, the large majority of 16S rRNA sequences from the same genome display very high similarities, with the ranges and averages remaining within a 1% nucleotide difference (Table 3). Only five genomes with extreme divergence among operons were detected, so overall, few incidences of HGT between divergent genomes are suggested.
Based on the level of divergence and redundancy among 16S rRNA sequences between operons of the same genome, more accurate bounds for diversity estimates of bacterial communities can be suggested. The analysis showed that 76 genomes with multiple operons contained 221 sequences (Table 1). Thus, if 16S rRNA gene diversity among these genomes were to be analyzed analogously to microbial communities by cloning and sequencing, a roughly threefold overestimation of diversity would result. However, this clearly represents an upper bound since a considerable fraction of genomes contain single operons. The magnitude of this fraction is difficult to estimate since genome sequences are currently derived from cultured strains. Among these, organisms with multiple operons are likely overrepresented since they appear to be more adaptable to changing environmental conditions and grow more readily on culture media (5, 21). This also makes it likely that environments which display more stable conditions overall harbor bacteria with fewer operons, leading to a less severe overestimation of diversity. However, there are currently 21 genomes with a single operon available. Adding these to the above estimate provides a lower bound of diversity overestimation of 2.5-fold. Thus, overall we suggest this value as a conservative bound for the correction of bacterial diversity estimates by cloning and sequencing.
Operon numbers appear conserved overall among closely related organisms, but even among strains of the same species small-scale variation is evident (Table 2). Among more distantly related organisms, no pattern of high or low operon numbers emerged from the analysis, so correction factors can only be applied to overall estimates of microbial diversity, not to individual phylogenetic groups. However, three notable exceptions were evident. Despite the considerable numbers of strains analyzed, the α-Proteobacteria, Spirochaetales, and mycoplasma strains appear to contain only low numbers of operons. For example, no α-Proteobacteria with more than four operons have been described to date, and the seven genomes available for α-Proteobacteria show high homogeneity, with only seven 16S rRNA sequences. This suggests that diversity estimates by clone libraries may be more accurate for this phylogenetic group than for others and that the α-Proteobacteria may overall be adapted to relatively stable environmental niches. In this context, it may be predicted that newly isolated representatives of the SAR11 clade, which dominates the open ocean environment (33), also contain few rRNA operons.
The operon comparison revealed the highest divergence to date among 16S rRNA genes within a single genome and showed that four of the five examples of highly divergent 16S rRNA sequences stem from thermophilic organisms. Thermoanaerobacter tengcongensis displayed 11.6% nucleotide divergence due to 188 polymorphic sites among its four 16S rRNA genes. A secondary structure analysis suggested that the rrnC operon arose via HGT since none of the divergent nucleotides appeared to disrupt the functional configuration of the molecule. Indeed, the three insertions in rrnC result in two longer but perfectly matched stems compared to the other operons (Fig. 3). Similar length differences have been detected in the thermophilic bacterium D. kuznetsovii (40). Additional evidence for HGT of the rrnC operon in Thermoanaerobacter tengcongensis is provided by its higher similarity (95%) to other Thermoanaerobacter species (T. subterraneus SL9 and T. keratinophilus 2KXI).
Whether there is an ecological significance to the occurrence of extreme divergence in thermophiles remains unknown, but for at least some strains it has been confirmed that the divergent rRNAs are transcribed and are thus likely functional (41). However, the pattern may suggest that genomes of thermophiles are prone to HGT. This is supported by the suggestion of HGT in other thermophiles (23, 25, 29, 38). For example, extensive studies of strains of a Thermotoga sp. showed that ∼25% of the genes are likely of archaeal origin (29), and a comparison of the genomes of “Pyrococcus abyssi,” Pyrococcus furiosus, and Pyrococcus horikoshii suggested the occurrence of extensive HGT (25).
Despite some extreme cases of 16S rRNA divergence among five genomes, overall a clear dominance of close relationships exists, with the vast majority of interoperonic sequence differences showing <1% divergence (Table 3). Thus, 16S rRNAs may primarily diverge due to mutation or HGT between closely related organisms only. This conforms to the complexity hypothesis, which states that successful HGT over large phylogenetic distances should be a rare occurrence for rRNA genes (1). Because the rRNAs are structural molecules, successful interactions with a large number of other gene products are dependent on the primary sequence of the rRNAs and should theoretically limit functionality in a highly heterologous genomic background. On the other hand, the rRNA genes, as members of a multigene family, are subjected to homogenization processes such as gene conversion (15, 26, 27). Were such processes to occur at high rates, they would relatively quickly erase traces of HGT even if it occurred between distantly related organisms. Nonetheless, genome sequences, taken as a snapshot of the incidence of HGT of 16S rRNA genes between phylogenetically distant organisms, currently confirm that the rRNAs provide a relatively solid framework for the estimation of phylogenetic relationships.
Acknowledgments
This work was partially supported by a grant from NSF-OCE to M.F.P. and a postdoctoral fellowship from the Spanish Ministry of Education (Ministerio de Educacion, Cultura y Deporte [MECD]) to S.G.A.
We are indebted to Francisco Rodríguez-Valera and Alex Mira for useful comments on the manuscript.
REFERENCES
- 1.Aguinaldo, A. M. A., and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human astric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Bao, Q., Y. Tian, W. Li, Z. Xu, Z. Xuan, S. Hu, W. Dong, J. Yang, Y. Chen, Y. Xu, X. Lai, L. Huang, X. Dong, Y. Ma, L. Ling, H. Tan, R. Chen, J. Wang, J. Yu, and H. Yang. 2002. A complete sequence of T. tengcongensis genome. Genome Res. 12:689-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casamayor, E. O., C. Pedrós-Alio, G. Muyzer, and R. Amann. 2002. Microheterogeneity in 16S ribosomal DNA-defined bacterial populations from a stratified planktonic environment is related to temporal changes and to ecological adaptations. Appl. Environ. Microbiol. 68:1706-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Codon, C., L. Dionysios, C. Squires, I. Schwartz, and C. L. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques 34:790-802. [DOI] [PubMed] [Google Scholar]
- 7.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daubin, V., N. A. Moran, and H. Ochman. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301:829-832. [DOI] [PubMed] [Google Scholar]
- 9.Dryden, S. C., and S. Kaplan. 1990. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 18:7267-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field, K. G., D. Gordon, T. Wright, M. Rappe, E. Urbach, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 65:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 12.Fox, G. E., J. D. Wisotzkey, and J. P. Jurtshuk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 13.García-Martinez, J., and F. Rodríguez-Valera. 2000. Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of group I. Mol. Ecol. 9:935-948. [DOI] [PubMed] [Google Scholar]
- 14.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 15.Hillis, D. M., C. Moritz, C. A. Porter, and R. J. Baker. 1990. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science 251:308-310. [DOI] [PubMed] [Google Scholar]
- 16.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. Dusko Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 19.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with ClustalX. Trends Biochem. Sci. 10:403-405. [DOI] [PubMed] [Google Scholar]
- 20.Johansen, T., C. R. Carlson, and A. B. Kolsto. 1996. Variable number of rRNA gene operons in Bacillus cereus strains. FEMS Microbiol. Lett. 136:325-328. [DOI] [PubMed] [Google Scholar]
- 21.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, et al. 1997. The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 24.Klepac-Ceraj, V., M. Bahr, B. C. Crump, A. P. Teske, J. E. Hobbie, and M. F. Polz. High overall diversity and dominance of microdiverse relationships in salt marsh sulfate-reducing bacteria. Environ. Microbiol., in press. [DOI] [PubMed]
- 25.Lecompte, O., R. Ripp, V. Puzos-Barbe, S. Duprat, R. Heiling, J. Dietrich, J. C. Thierry, and O. Poch. 2001. Genome evolution at the genus level: comparison of the three complete genomes of hyperthermophilic archaea. Genome Res. 11:981-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao, D. 1999. Concerted evolution: molecular mechanisms and biological implications. Am. J. Hum. Genet. 64:24-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao, D. 2000. Gene conversion drives within genomic sequences: concerted evolution of ribosomal RNA genes in Bacteria and Archaea. J. Mol. Evol. 51:305-317. [DOI] [PubMed] [Google Scholar]
- 28.Mylvaganam, S., and P. P. Dennis. 1992. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Halobacterium marismorturi. Genetics 130:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nesbo, C., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ninet, B., M. Monod, S. Embler, J. Pawlowski, C. Metral, P. Rohner, R. Auckenthaler, and B. Hirschel. 1996. Two different 16S rRNA genes in a mycobacterial strain. J. Clin. Microbiol. 34:2531-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Pósfal, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 32.Prüß, B. M., K. P. Francis, F. von Stetten, and S. Scherer. 1999. Correlation of 16S ribosomal DNA signature sequences with temperature-dependent growth rates of mesophilic and psychrotolerant strains of the Bacillus cereus group. J. Bacteriol. 181:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 34.Reischl, U., K. Feldman, L. Naumann, B. J. Gaugler, B. Ninet, and B. Hirschel. 1998. 16S rRNA sequence diversity in Mycobacterium celatum strains caused by presence of two different copies of the 16S rRNA gene. J. Clin. Microbiol. 36:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocap, G., F. W. Larimer, J. Lamerdin, M. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hausner, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424:1042-1047. [DOI] [PubMed] [Google Scholar]
- 36.Sass, H., E. Wieringa, H. Cypionka, H.-D. Babenzien, and J. Overmann. 1998. High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch. Microbiol. 170:243-251. [DOI] [PubMed] [Google Scholar]
- 37.Shimizu, T., S. Ohshima, K. Ohtani, K. Hoshino, K. Honjo, H. Hayashi, and T. Shimizu. 2001. Sequence heterogeneity of the ten rRNA operons in Clostridium perfringens. Syst. Appl. Microbiol. 24:149-156. [DOI] [PubMed] [Google Scholar]
- 38.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity: magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 40.Tuorova, T. P., B. B. Kuznetzov, E. V. Novikova, A. B. Poltaraus, and T. N. Nazina. 2001. Heterogeneity of the nucleotide sequence of the 16S rRNA genes of the type strain of Desulfotomaculum kuznetsovii. Microbiology 70:678-684. [PubMed] [Google Scholar]
- 41.Wang, Y., Z. S. Zhang, and N. Ramanan. 1997. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol. 179:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue, Y., Y. Xu, Y. Liu, Y. Ma, and P. Zhou. 2001. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 51:1335-1341. [DOI] [PubMed] [Google Scholar]
- 43.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal gene transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]